Abstract
Clonal haematopoiesis results from acquired mutations in haematopoietic stem and progenitor cells (HSPCs). These mutations can confer the HSPC with a competitive advantage, leading to their clonal expansion within the limiting bone marrow niche. This process is often insufficient to produce a haematologic malignancy; however, the expanding HSPC clones increasingly give rise to progeny leucocytes whose phenotypes can be altered by the somatic mutations that they harbour. Key findings from multiple human studies have shown that clonal haematopoiesis in the absence of overt haematologic alterations is common amongst the ageing population and associated with mortality and cardiovascular disease. Key findings from experimental studies have provided evidence for a causative role for clonal haematopoiesis in cardiovascular diseases, and aspects of these mechanisms have been elucidated. Whilst our understanding of the impact and biology of clonal haematopoiesis is in its infancy, analyses of some of the most commonly mutated driver genes suggest promising clinical scenarios involving the development of personalized therapies with immunomodulatory drugs that exploit the perturbation caused by the particular mutation. Herein, we review the accumulating epidemiological and experimental evidence, and summarize our current understanding of the importance of clonal haematopoiesis as a new causal risk factor for atherosclerotic cardiovascular disease and heart failure.
Keywords: ageing, cardiovascular disease, clonal haematopoiesis
Introduction:
Ageing, cardiovascular disease and clonal haematopoiesis
The major modifiable risk factors for cardiovascular disease include smoking, hypertension, hyperlipidaemia and diabetes mellitus [1]. However, advanced age is the major cardiovascular disease risk factor, and it is estimated that the prevalence of cardiovascular disease will markedly increase as the population ages. Since advanced age is viewed to be unmodifiable, there is a great need to better understand how the processes of biological ageing impact cardiovascular diseases at a mechanistic level. In this regard, it has long been appreciated that all cells in the body randomly accumulate mutations as the organism ages, and this leads to a condition referred to as somatic mosaicism [2, 3]. Recent advances in DNA sequencing technology show that somatic mosaicism is a widespread and an inevitable part of the ageing process. Moreover, it is common to find individuals with evidence of clonal amplification events in various tissues that arise from somatic mutations in ‘driver’ genes that confer a fitness advantage to the founder cell [4]. Whilst it is widely appreciated that the accumulation of multiple ‘driver’ gene mutations within the same clone can lead to the development of cancers, recent studies have shown that normal tissues often develops precancerous, clonal expansions of cells without changing the total mass of cells within a tissue [5]. The detection of these clonal expansions is dependent on the age of the individual and the proliferative capacity of the particular tissue, suggesting that mutations in driver genes accumulate with time and the number of cell divisions.
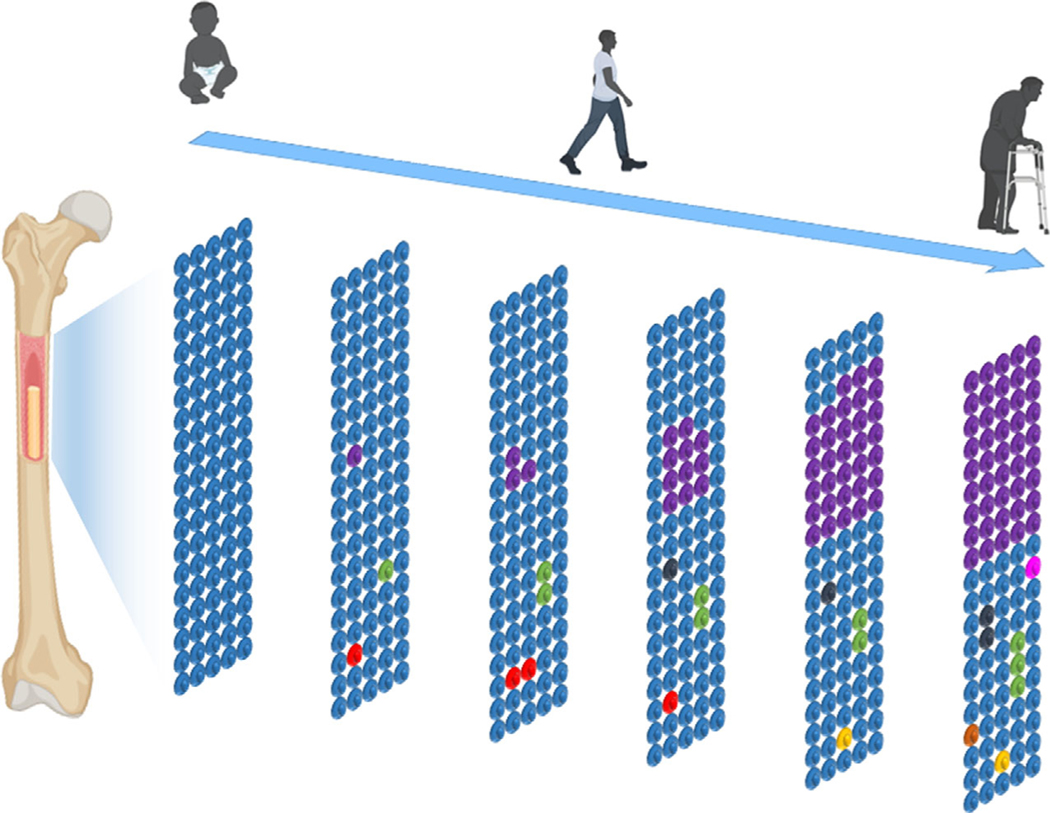
The haematopoietic system produces billions of blood cells per day, and thus, it is susceptible to the development of somatic mutations that can give rise to clonal events [6, 7]. This process is referred to as clonal haematopoiesis (Fig. 1). Clonal haematopoiesis was initially deduced decades ago by examining the skewing of X chromosome inactivation in the blood cells of women. Whilst X chromosome skewing is often associated with haematologic malignancies, it was noted that elderly women with no haematologic disorders also display this phenomenon at a high frequency [8]. A breakthrough was made when it was discovered that the asymptomatic women exhibiting X chromosome inactivation skewing in their blood were more likely to also possess a mutation in the ten eleven translocation 2 (TET2) driver gene [9], suggesting that genes recurrently mutated in haematologic malignancies could be responsible for the development of clonal haematopoiesis in otherwise healthy women. Subsequent analyses of chromosomal mosaicism also provided evidence for clonal expansion in the haematopoietic cells from elderly individuals [10–12].
Fig. 1.
Scheme of age-related clonal haematopoiesis. The accumulation of mutations in haematopoietic stem cell is an inevitable consequence of ageing. Small HSPC clones harbouring these mutations form by middle age. Some of these clones can be lost (red HSPC), whereas others are relatively stable (green, black and yellow HSPC). Some of these clones will enlarge in individuals (purple HSPC) such that a substantial fraction of progeny leucocytes can be derived from the mutant stem cells. This condition, referred to as clonal haematopoiesis, can ultimately affect leucocyte function and promote cardiovascular disease.
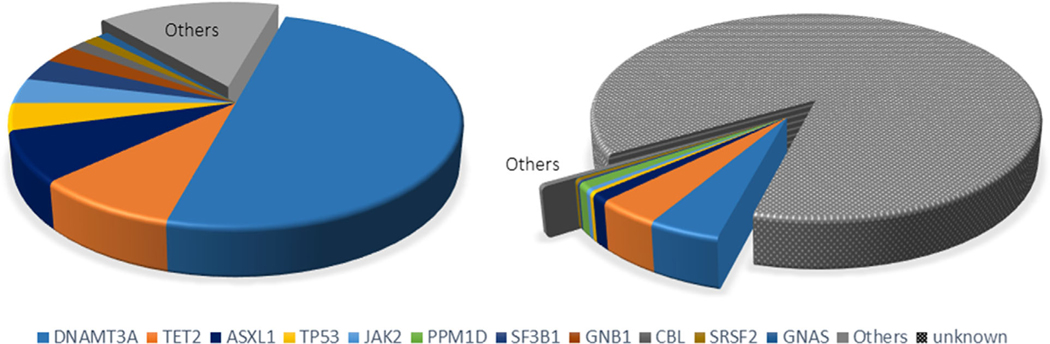
In 2014, three independent studies analysed the peripheral blood of individuals for clonal haematopoiesis by assessing mutations within multiple genes that are recurrently mutated in haematological malignancies [13–15]. Although many candidate ‘driver’ genes were found to be associated with these clonal events in an age-dependent manner, the majority of these mutations occurred in genes encoding the epigenetic regulators DNA methyltransferase 3 A (DNMT3A), TET2 and additional sex combs-like 1 (ASXL1) (Fig. 2). One of these studies determined that the frequency of driver gene-associated clonal events in the haematopoietic system was approximately 10% of individuals over the age of 70 [14]. However, this is likely to represent an underestimation of the actual frequency of clonal events due to limitations in DNA sequencing sensitivity and the focus on candidate driver genes that was employed by this study. Whilst it was found that individuals that harboured these driver gene mutations in their blood cells had an increased risk of developing a subsequent haematologic cancer [14], most will never develop this malignancy because this is a relatively rare condition compared to the observed frequencies of clonal haematopoiesis. Thus, this condition has been referred to as clonal haematopoiesis of indeterminate potential or CHIP. This definition has limitations, however, including its exclusive focus on known driver gene-associated haematologic cancer that may only account for a small fraction of the total clonal events [13, 16]. Thus, others have proposed the term age-related clonal haematopoiesis (ARCH) to describe this condition [17].
Fig. 2.
Comparison of frequencies of known and unknown driver genes assessed by exon sequencing of candidate driver genes versus whole genome analysis. (Left) The 2014 study of Jaiswal et al. [14] involved the sequencing of exonic regions of 160 candidate driver genes, that is genes known to be recurrently mutated in haematologic malignancies. Clonal events were associated with single nucleotide variants or small insertions and deletions in 68 distinct driver genes. The 10 most frequent driver genes are displayed individually in the pie diagram, and the 58 least prevalent driver genes (15.4% of total individuals) are grouped in ‘other’. (Right) The 2017 study of Zink et al. [16] employed a nonbiased, but less sensitive, whole genome analysis method to score clonal events. Based upon genome sequence analysis, it was deduced that approximately 14% of the clonal events could be associated with known haematologic cancer driver genes, amongst which 4% could be grouped in ‘other’. However, the majority of clonal events were of unknown origin.
Estimates of the prevalence of clonal haematopoiesis is dependent upon how one defines this condition and the methodology used to detect it.
In this regard, Zink et al. [16] employed a nonbiased whole genome sequencing approach on peripheral blood to analyse clonal haematopoiesis in 11,262 Icelanders. Although this method of detection is less sensitive than the targeted exon sequencing approaches employed previously, it was not limited to the detection of known driver genes. These investigators reported that the frequency of clonal events in the blood was more than 50% in individuals older than 85 years and that clonal haematopoiesis ‘trended toward inevitability’ [16]. In this analysis, mutations in known driver genes accounted for approximately 14% of the total number of clonal events (Fig. 2). What can account for the majority of clonal haematopoiesis events that are not associated with a known haematological malignancy driver mutation? Possibilities include the positive selection of unidentified driver genes or ‘genetic drift’ over time. A related issue is the sensitivity of the methods employed to detect clonal haematopoiesis. Using ultra-deep, error-corrected sequencing, which allows detection of clonal events as low as 0.03% variant allele fractions (VAF), it was found that mutations in a select group of candidate driver genes could be detected in the blood of 19 of 20 individuals examined between the ages 50–70 years [18]. Thus, it would seem that the development of multiple small clones is essentially ubiquitous by middle age, but that the expansion of one or a subset of these clones at later ages only occurs in a portion of individuals [19]. It is not known why large clonal expansions occur in some individuals but not others, but it is possible that environmental factors, such as smoking [20], participate in this process by altering the environment of the bone marrow niche such that it favours the expansion of the mutant HSPC clone [21].
Associations between clonal haematopoiesis, mortality and cardiovascular disease
Since 2014, a number of studies have associated clonal haematopoiesis with all-cause mortality [12–14, 16]. Importantly, this association can be made regardless of whether an individual’s clonal haematopoiesis can be attributed to a known haematological driver gene or not. Whilst these mutations increase the risk of blood cancer, the incidence of a haematologic malignancy is not sufficient to account for the relatively large increases in the observed mortality in individuals with clonal haematopoiesis [14]. An unplanned secondary analysis by Jaiswal et al. [14] suggested that the increased mortality in individuals with candidate driver gene-associated clonal haematopoiesis could be due to an increased incidence of coronary heart disease (hazard ratio, 2.0; 95% CI, 1.2 to 3.4) and ischaemic stroke (hazard ratio, 2.6; 95% CI, 1.4 to 4.8) after adjusting for age. This unexpected connection between clonal haematopoiesis and cardiovascular disease was confirmed in a subsequent study reported by Jaiswal et al. [22]. This study performed a meta-analysis encompassing 1010 individuals from two cohorts without cardiovascular disease at baseline and demonstrated that clonal haematopoiesis-positive individuals harbouring a mutation in the DNMT3A, TET2, ASXL1 or Janus kinase 2 (JAK2) valine to phenylalanine mutation at position 617 (JAK2V617F) had an increased risk for development of coronary heart disease after adjusting for the classical risk factors of age, sex, type 2 diabetes, total cholesterol, high-density lipoprotein, smoking and hypertension (hazard ratio of 1.9 (95% CI, 1.4–2.7)). Furthermore, in a meta-analysis of 7245 individuals below the age of 50, the prevalence of clonal haematopoiesis due to mutations in TET2, ASXL1 and JAK2 was 4 times higher than in younger individuals with early-onset myocardial infarction. Individuals with large clonal expansions (VAF > 10%) also displayed an association with coronary artery calcification score (odds ratio 12.0; 95% CI 2.4–64.0), that is indicative of vascular lesions, whereas individuals with smaller haematopoietic clones (VAF < 10%) did not (odds ratio 0.9; 95% CI 0.2–4.5), implying a dose-dependent effect of clone size. Overall, these population-based studies demonstrated an association between clonal haematopoiesis and coronary artery disease and stroke, presumably due to elevated levels of atherosclerosis.
Recent epidemiological studies also indicate an association between clonal haematopoiesis and heart failure. Dorsheimer et al. [23] performed deep, error-corrected DNA sequencing on 56 genes that are commonly mutated in myeloid malignancies in bone marrow-derived mononuclear cells from 200 patients who participated in one of four different clinical trials originally designed to examine the effect of intracoronary administration of autologous bone marrow cells on ischaemic heart failure after myocardial infarction. Using a VAF threshold of 2%, they found that 38 out of 200 patients were positive for clonal haematopoiesis where the clones most commonly exhibited mutations in DNMT3A and TET2. The patients with DNMT3A- or TET2-mediated clonal expansions showed significantly worse morbidity and mortality (death, HR 3.25; 95% CI 1.62–6.52, death combined with rehospitalization for heart failure, HR 3.25; 95% CI 1.71–6.19). Furthermore, the dose dependency of this effect was analysed by lowering the inclusion threshold of VAF from 2% to 0.5%, leading to the addition of 119 additional mutant clones of smaller size (66 DNMT3A and 53 TET2 mutations). This analysis found that, whilst VAF < 1% group showed almost comparable outcome with the no mutation cohort, both the VAF = 1 to 2% and VAF > 2% cohorts displayed worse outcomes, indicating a dose-dependent effect of clone size on mortality and morbidity in these heart failure patients. More recently, clonal haematopoiesis was shown to be predictive of worse prognosis in patients treated for valvular heart disease. During the study period, Mas-Peiro et al. [24] enrolled 279 consecutive individuals with severe aortic valve stenosis (AS) undergoing transfemoral aortic valve implantation (TAVI) and analysed their peripheral blood for clonal haematopoiesis by DNA sequence analysis of candidate driver gene mutations. This study focused on DNMT3A and TET2 mutations with a VAF greater than 2% and found that the prevalence of clones harbouring these mutations increased with age, from 25% in those between 55 and 69 years of age to 52.9% in those between the ages of 90–99 years. In comparison with previous reports, these levels of DNMT3A- and TET2-mediated clonal haematopoiesis are much more frequent than what is expected to be found in the general population, implying that clonal haematopoiesis is elevated in patients with severe AS. Furthermore, the analysis of medium-term outcome (to exclude procedure-related complications) revealed that clonal haematopoiesis is strongly predictive of death after adjustment for age, sex and NT-pro B-type natriuretic peptide (BNP) (HR 4.81, 95% CI 1.49–15.57). Further analyses revealed that patients with DNMT3A-mediated clonal haematopoiesis exhibited an elevated Th17/Treg ratio and those with TET2-mediated clonal haematopoiesis exhibited increased circulating CD14dimCD16+ nonclassical monocytes. These data, and similar analyses of TET2-mediated clonal haematopoiesis in heart failure patients [25], suggest that clonal haematopoiesis confers driver gene-specific effects on the immune system. Although the cause of death was not described in the TAVI study, noncardiac causes of mortality are prevalent in patients who undergo this procedure [26, 27], implying that clonal haematopoiesis could have a pleiotropic impact on this population.
Recently, Bick and Pirruccello et al. [28] analysed the prevalence of DNMT3A- and TET2-mediated clonal haematopoiesis in the UK Biobank and examined its association with incident cardiovascular disease including myocardial infarction, coronary revascularization stroke and death. It was found that clonal haematopoiesis associated with either of these two driver genes was predictive of a greater risk of developing cardiovascular disease, and this association was strongest in individuals with large clones (VAF > 10%) (HR = 1.59, 95% CI: 1.21–2.09, P < 0.001). Of interest, those who developed DNMT3A- or TET2-mediated clonal haematopoiesis with simultaneous genetic deficiency of IL-6 signalling, due to the presence of the heritable IL6R p.Asp358Ala allele, displayed a reduction in cardiovascular disease risk (HR = 0.46, 95% CI: 0.29–0.73, P < 0.001). This effect was corroborated in a second cohort. Notably, the IL6R p.Asp358Ala allele did not significantly impact cardiovascular disease risk in individuals who did not exhibit clonal haematopoiesis in the UK Biobank cohort (HR = 0.95, 95% CI: 0.89–1.06, P = 0.08). These compelling data suggest that inhibitors of IL-6 signalling may reduce cardiovascular disease risk in patients with and DNMT3A- or TET2-mediated clonal haematopoiesis.
Mechanistic links between clonal haematopoiesis and cardiovascular disease revealed by experimental studies
Epidemiological studies are inherently descriptive, and it is often difficult to discern causal relationships or the directionality between the associated events. Therefore, animal models can aid in developing evidence for causality and provide a molecular framework to define underlying mechanisms (Table 1). A study by Fuster et al. [29] provided the first evidence for a causal relationship between clonal haematopoiesis and cardiovascular disease. In this study, hyperlipidaemic atherosclerosisprone mice underwent bone marrow transplant (BMT) with 10% Tet2-deficient cells to recapitulate the clonal expansion of HSPC in humans. Consistent with the paradigm of clonal haematopoiesis caused by inactivating mutations in the human TET2 gene, the murine Tet2-deficient cells underwent expansion into multiple blood cell lineages with no alterations in total blood cell numbers or evidence of a systemic inflammatory response. However, atherosclerotic plaque size was increased in the aortae of mice that exhibited the expansion of Tet2-deficient haematopoietic cells, providing initial evidence that clonal haematopoiesis can contribute to atherosclerotic cardiovascular disease. This work was corroborated by an independent study that found an increase in vascular plaque size in following complete BMT of Tet2-deficient cells in hyperlipidaemic mice [22]; however, these experimental conditions also led to widespread tissue inflammation, a confounding condition that is typically not seen in individuals with clonal haematopoiesis. Both studies reported that myeloid-specific ablation of Tet2 was sufficient to confer the increased plaque size phenotype [22, 29]. Mechanistic analyses revealed that the increase in plaque size was associated with over-activation of the nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain-containing 3 (NLRP3) inflammasome-mediated signalling leading to elevated production of interleukin-1 beta (IL-1β) secretion by Tet2-deficient cells within the plaque [29]. Correspondingly, macrophages isolated from Tet2-deficient mice displayed an increase in the expression of IL-1β and the downstream cytokine IL-6. In the experimental model of atherosclerosis, the larger lesions caused by haematopoietic Tet2 deficiency could be ameliorated when mice were treated with the small-molecule NLRP3 inhibitor MCC950 [29]. In agreement with these experimental findings, it has been reported that individuals that harbour TET2-mediated clonal haematopoiesis exhibit significantly higher serum levels of IL-1β and IL-6 in the TOPMed cohort [30]. Finally, a recent study reported that rare germline variants in TET2 may contribute to pulmonary arterial hypertension via an IL-1β-dependent mechanism [31].
Table 1.
Summary of experimental studies that have examined clonal haematopoiesis and CVD end-points
| Driver gene | Clonal haematopoiesis model | CVD model | Disease phenotype | Suggested mechanisms | References |
|---|---|---|---|---|---|
| Tet2 | Myeloablative BMT KO donor |
Atherosclerosis | ↑Atherosclerosis | ↑Pro-inflammatory transcripts ↑Il1b, Il6, Nlrp3 in macrophages |
[22,29] |
| Tet2 | Myeloablative BMT KO donor |
MI (LAD ligation) PO (TAC) |
↑Cardiac remodelling | ↑Il1b, Il6 in macrophages | [32] |
| Tet2 | Myeloablative BMT ex vivo CRISPR | PO (AngII) | ↑Cardiac remodelling ↑Renal fibrosis |
↑Il1b, Il6 in RAW 264.7 cells | [33] |
| Tet2 | Adoptive BMT KO donor |
Ageing | ↑Cardiac remodelling | ↑Pro-inflammatory transcripts in cardiac macrophages | [52] |
| Dnmt3a | Myeloablative BMT ex vivo CRISPR | PO (AngII) | ↑Cardiac remodelling ↑Renal fibrosis |
↑Il6, Cxcl1/2, Ccl5 in RAW 264.7 cells | [33] |
| Jak2 | Myeloablative BMT Jak2V617F donor |
Venous thrombosis | ↑Thrombosis MP neoplasm | ↑NET formation | [37] |
| Jak2 | w/o BMT (Mx1-Cre) | Atherosclerosis | ↑Atherosclerosis MP neoplasm |
↓Efferocytosis ↑Il1b, Il6 in macrophages |
[39] |
| JAK2 | Myeloablative BMT ex vivo myeloid-specific promoter | MI (LAD ligation) PO (TAC) |
↑Cardiac remodelling | ↑Il1b, Il6, TNF in THP-1 cells | [45] |
MI, myocardial infarction; MP, myeloproliferative; PO, pressure overload; TAC, transverse aortic constriction.
Experimental studies have also explored the possibility that clonal haematopoiesis can contribute to heart failure phenotypes. Permanent ligation of the left anterior descending artery in mice that had received a partial BMT with Tet2-deficient cells resulted in greater myocardial infarct size, chamber remodelling and fibrosis, and a lower ejection fraction compared to control mice transplanted with wild-type bone marrow [32]. This same study corroborated and extended these findings in a second model of heart failure involving pressure-overload hypertrophy following transverse aortic constriction (TAC) surgery. As in the atherosclerosis study [29], myeloid-specific ablation of Tet2 was sufficient to confer more severe phenotypes in both heart failure models. In the heart failure models, the infiltration of Tet2-deficient inflammatory cells to the heart was accompanied by elevated expression levels of IL-1β. Accordingly, the exacerbated cardiac dysfunction caused by the clonal expansion of Tet2-deficient immune cells in either heart failure model was alleviated by treatment with a small-molecule NLRP3-inflammasome inhibitor, suggesting that Tet2-deficient immune cells exacerbate failing heart phenotypes through the over-activation of IL-1β signalling. In a separate study, Sano et al. also examined the role of Tet2-mediated clonal haematopoiesis in an angiotensin II infusion model of heart failure [33]. As in the pressure-overload hypertrophy model, a competitive BMT procedure was employed to mimic clonal expansion, and mice receiving Tet2-deficient bone marrow displayed increased cardiac hypertrophy, fibrosis and dysfunction [33].
A CRISPR/Cas9-mediated technique has been developed to more efficiently and economically manipulate clonal haematopoiesis driver genes in mouse models of disease [34]. In the initial study, this method was employed to introduce inactivating mutations in the Tet2 gene of HSPC prior to their implantation into irradiated mice [33]. As with the competitive BMT protocol, the acute ablation of the Tet2 by the CRISPR/Cas9 system led mutant cell expansion in the haematopoietic compartment, assessed with a lentivirus-encoded reporter gene, and resulted in greater cardiac remodelling when mice were subjected to the angiotensin II infusion model of heart failure. Sano et al. [33] then proceeded to employ the CRISPR/Cas9-mediated ablation technique to examine the impact of depleting haematopoietic cell Dnmt3a in the angiotensin II-induced heart failure model. Whilst the manipulation of Dnmt3a did not lead to a time-dependent increase in haematopoietic cell expansion, the relatively low level of Dnmt3a mutations that was achieved by the procedure was sufficient to confer greater cardiac hypertrophy and fibrosis, and a further reduction in cardiac function, in the angiotensin II-induced heart failure model. The enhanced heart failure phenotype was associated with greater macrophage infiltration in the heart and with an increase in markers of T-cell infiltration [33]. As noted previously, clinical evidence in support of these experimental findings was provided by the study of Dorsheimer et al. [23] who showed that chronic heart failure patients with TET2- or DNMT3A-mediated clonal haematopoiesis exhibit a worse prognosis.
Another clonal haematopoiesis driver gene is JAK2V617F. JAK2 is a nonreceptor tyrosine kinase that is ubiquitously expressed. The JAK2V617F gain-of-function mutation in haematopoietic cells is frequently associated with myeloproliferative neoplasms (MPN) that increase the risks of myocardial infarction, stroke and venous thrombosis due to increased blood viscosity, leucocytosis and tendency to form of neutrophil extracellular traps (NET) [35, 36]. Wolach et al. [37] have shown that neutrophils obtained from patients with MPN harbouring JAK2V617F mutation prone to form NET and Jak2V617F knock-in mice have an increased propensity for NET formation and thrombosis in addition to their well-described MPN phenotype. However, epidemiological studies have demonstrated that asymptomatic individuals can display an amplification of leucocytes harbouring the JAK2V617F allele (i.e. clonal haematopoiesis). For example, a recent study employing a sensitive droplet digital polymerase chain reaction assay found that the prevalence of the JAK2V617F mutation is 3.1% in a general population [38]. Whilst mutation-positive individuals can display a more hyperproliferative blood cell count profile compared with the nonmutated background, it is clear that many individuals that harbour this mutation have blood cell counts within the normal range and never go on to develop MPN. Yet, those individuals harbouring the JAK2V617F mutation within the confines of a clonal haematopoiesis diagnosis have an increased risk of coronary artery disease [21]. In light of these issues, it is difficult to model the effects of JAK2V617F-mediated clonal haematopoiesis on cardiovascular disease phenotypes because mouse models expressing the JAK2V617F allele in HSPC are confounded by strong myeloproliferative disease phenotypes [39–44]. Recently, Wang et al. [43] performed a complete BMT of Jak2V617F cells into an atherogenic mouse strain to assess vascular lesions and haematological phenotypes. Whilst these mice developed greater atherosclerotic lesion size, they also displayed MPN-like phenotype that confounds the interpretation of the role of the JAK2V617F-mediated clonal haematopoiesis-like state on cardiovascular disease. Similarly, JAK2V617F transgenic mice display cardiac hypertrophy [42], but it is unclear whether this is due to clonal haematopoiesis event per se or other features associated with the MPN phenotype. In an attempt to sort through these issues, Sano et al. established a murine model where HSPCs are transduced with a lentivirus vector that expresses the JAK2V617F allele from a synthetic myeloid-specific promoter [45]. The rationale for this study is based upon the supposition that the heterogeneity amongst the HSC subpopulations that acquire the JAK2V617F mutation may account for the discrepant phenotypes between JAK2V617F-mediated MPNs and clonal haematopoiesis [40], and the observation by Sano et al. [45] that JAK2V617F HSPC display a strong myeloid bias in competitive BMT studies in mice. Transplantation of HSPC expressing JAK2V617F in a myeloid-restricted manner led to greater cardiac dysfunction and inflammation in models of permanent LAD ligation or pressure-overload hypertrophy under conditions where mice did not exhibit overt haematologic abnormalities. Mechanistic analyses indicated that myeloid lineage-restricted expression of the JAK2V617F mutation can promote cardiac dysfunction through an increase in myocardial inflammation. Although highly speculative at this time, these data provide support for the notion that clonal haematopoiesis can arise from the acquisition of JAK2V617F mutations within a subpopulation of progenitor cells that preferentially give rise to circulating myeloid cells, and thereby contribute to myocardial disease independent of the thrombocytosis, erythrocytosis or leucocytosis phenotypes that are associated with JAK2V617F-mediated MPN.
To develop better models of clonal haematopoiesis, it is desirable to avoid the irradiation procedures that destroy the bone marrow niche and damage tissues throughout the body [46–49]. In avoiding myeloablative strategies, clonal haematopoiesis experiments can also be conducted under conditions that more faithfully mimic the regulatory mechanisms associated with native haematopoiesis [50, 51]. In view of these issues, Wang et al. [52] developed a new model of murine clonal haematopoiesis that employs the adoptive transfer of unfractionated bone marrow cells to mice that have not been preconditioned by a myeloablative strategy. In these experiments, the delivery of wild-type bone marrow cells led to a low but stable engraftment of donor-derived long-term haematopoietic stem cells (LT-HSC) and various HSPC fractions, and minimally contributed to circulating leucocyte populations and resident immune cell populations in the heart and other tissues. In contrast, the adoptive transfer of Tet2-deficient bone marrow cells led to progressive expansion of Tet2-deficient cells into various fractions of bone marrow HSPC, circulating leucocytes and tissue-resident immune cells. Consistent with the notion that Tet2 deficiency confers a competitive advantage to the HSPC, a higher expression levels of proliferation and fitness genes were observed in the LT-HSC fraction of bone marrow. Thus, the nonconditioned, adoptive transfer model can be useful for studies aimed at understanding the mechanisms by which the bone marrow microenvironment controls the process of clonal haematopoiesis, and how it may be influenced by ageing, exposure to genotoxic stressors or other environmental stimuli. Further, the benign nature of the adoptive transfer procedure could also enable the study of subtle disease processes. For example, using the adoptive transfer model, Wang et al. [52] found that Tet2-mediated clonal haematopoiesis is sufficient to accelerate age-related cardiac dysfunction in the absence of injurious stimuli in mice that were followed for as long as 18 months. Whilst this study only focused on cardiac dysfunction in ageing mice, it raises the possibility that clonal haematopoiesis can generally impact the processes that promote biological ageing and diminish health span. It has recently been reported that clonal haematopoiesis is associated with accelerated biological ageing based upon measurements of the ‘epigenetic clock’ that assesses the methylation patterns at specific CpG sites [53]. Although preliminary, the work of Wang et al. [52] provides evidence of a potential causal link between clonal haematopoiesis and the pace of biological ageing.
Clonal haematopoiesis and immunomodulatory therapies
Our increased understanding of the health effects of clonal haematopoiesis comes at a time when there is renewed interest in the application of anti-inflammatory drugs for the treatment of cardiovascular disease. Notably, the CANTOS (Canakinumab Anti-Inflammatory Thrombosis Outcome Study) trial enrolled patients with previous myocardial infarction and elevated levels of high-sensitivity C-reactive protein (hsCRP) to test whether a neutralizing antibody against IL-1β (i.e. canakinumab) could impact the primary composite outcome of myocardial infarction, stroke or cardiovascular death. Compared with placebo, the middle dose of canakinumab reduced the risk of major adverse events by 15% over the follow-up period. A subsequent analysis revealed that the patients could be subdivided into responders and nonresponders based upon an individual’s degree of hsCRP reduction following the administration of therapy [54]. For treated patients who exhibited a reduction in hsCRP to less than 2 mg L−1, there was also a 25%, 31% and 31% reductions in major adverse cardiovascular events, cardiovascular mortality and all-cause mortality, respectively, whereas there was no impact on the clinical end-points of the canakinumab-treated patients that did not show this reduction in hsCRP. This finding of responders vs. nonresponders indicated as-yet-unidentified differences in the immune systems of patients could dictate the efficacy of the anti-inflammatory therapy. Related to this issue, it was previously proposed from experimental studies that individuals with TET2-mediated clonal haematopoiesis would be superior responders to therapies that target the IL-1β/NLRP3 inflammasome pathway [29]. Support for this hypothesis was recently provided by an exploratory analysis of 3964 participants in the CANTOS trial that found individuals with TET2-mediated clonal haematopoiesis exhibit a 64% reduction in MACE in response to canakinumab therapy [55]. Notably, this association was only made with TET2-mediated clonal haematopoiesis, but not with clonal haematopoiesis derived from other known driver genes. This finding appears to dovetail with a recent study reporting that TET2-mediated clonal haematopoiesis, but not by mutations in other driver genes, leads to a detectable elevation in circulating IL-1β levels in the TOPMed cohort [30]. Thus, TET2 appears to uniquely impact the IL-1β signalling pathway. Although prospective randomized trials are warranted to confirm this result, it suggests that analysis of an individual’s clonal haematopoiesis status could provide a better understand how a particular patient might respond to specific immunomodulatory drugs. Finally, as mentioned above, it has been reported that individuals with clonal haematopoiesis associated with mutations in either DNMT3A or TET2 exhibit a greater risk of developing cardiovascular disease; however, individuals with a simultaneous genetic deficiency of IL-6 signalling display a reduction in cardiovascular disease risk [28]. Thus, IL-6 inhibitors may represent an alternative strategy for reducing cardiovascular disease risk in patients with and DNMT3A- or TET2-mediated clonal haematopoiesis.
Conclusion and future directions
Accumulating evidence shows that clonal haematopoiesis is a prevalent age-associated condition (Fig. 1). Numerous studies have shown that individuals with clonal haematopoiesis are at higher risk of mortality due in large part to the increased prevalence of cardiovascular disease. Experimental studies have shown that the driver gene mutations that lead to the clonal expansion of HSPC also corrupt the phenotypes of the progeny leucocytes. To the extent that it has been examined, leucocytes that harbour driver gene mutations exhibit exaggerated inflammatory responses to stimuli, and this contributes to cardiovascular pathologies including atherosclerosis and heart failure. In toto, these epidemiological and experimental studies suggest that clonal haematopoiesis is a new causal risk factor for cardiovascular disease that is prevalent in the elderly.
Research in the area of clonal haematopoiesis is in its infancy, and we have an incomplete understanding of the mechanisms that give rise to these phenomena and their ultimate health consequences. Numerous driver genes recurrently mutated in haematological malignancies have been associated with clonal haematopoiesis, and there is a growing understanding of their mechanistic similarities and differences (Table 1). In this regard, the known driver genes encode a diverse group of regulatory molecules, and it is likely that they confer divergent functions on haematopoietic cells and thereby have divergent roles in disease processes. Experimental and epidemiological studies are defining mechanistic differences and similarities between the TET2, DNMT3A and JAK2V617F driver genes that have received the greatest attention to date. In particular, multiple studies have shown that haematopoietic cell mutations in the TET2 driver gene will promote pro-inflammatory responses through activation of the NLRP3 inflammasome/IL-1β/IL-6 pathway, and it is conceivable that this information could be exploited to better treat patients with this form of clonal haematopoiesis once they develop a cardiovascular anomaly or other disease. On the other hand, we know very little about how the immune system and disease processes are impacted by clonal haematopoiesis caused by mutations in the dozens of less prevalent driver genes (Fig. 2). Some of these driver gene-mediated clonal events could be highly pathological, whereas others may be benign (or even cardioprotective). It is reasonable to speculate that driver genes will exhibit disease-specific actions and likely impact age-related diseases beyond the cardiovascular system. Given the large number of candidates, it will be a daunting task for epidemiological studies to assign disease associations to individual driver genes, particularly for the clonal events that result from the less prevalent mutant loci. With regard to mechanistic studies, most basic science laboratories incrementally build on well-established protocols and tend to operate on a single gene/single disease basis. However, a broader approach will be required to develop a comprehensive and discriminating view of clonal haematopoiesis that could allow for the development of therapies with greatest benefit to target individuals with specific somatic mutations. Finally, it appears that a relatively small fraction of the clonal haematopoiesis events can be explained by known driver genes that are recurrently mutated in haematological malignancies (Fig. 2), and we have little or no understanding of the mechanism that give rise to the more enigmatic clonal expansions. Clonal haematopoiesis with no known drivers has been associated with an increase in all-cause mortality, but the specific morbidities associated with this condition are unknown. Clearly, there is ample room for young, creative investigators to make significant progress in this new area of medical research.
Footnotes
Conflict of interest statement
The authors have no conflicts of interest to declare.
References
- 1.Virani SS, Alonso A, Benjamin EJ et al. Heart disease and stroke statistics-2020 update: a report from the American heart association. Circulation 2020; 141: e139–e596. [DOI] [PubMed] [Google Scholar]
- 2.Forsberg LA, Gisselsson D, Dumanski JP. Mosaicism in health and disease - clones picking up speed. Nat Rev Genet 2017; 18: 128–42. [DOI] [PubMed] [Google Scholar]
- 3.Vijg J. Somatic mutations, genome mosaicism, cancer and aging. Curr Opin Genet Dev 2014; 26: 141–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sano S, Wang Y, Walsh K. Somatic mosaicism: implications for the cardiovascular system. Eur Heart J 2019: ehz907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yizhak K, Aguet F, Kim J et al. RNA sequence analysis reveals macroscopic somatic clonal expansion across normal tissues. Science 2019; 364: eaaw0726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans MA, Sano S, Walsh K. Cardiovascular disease, aging, and clonal hematopoiesis. Annu Rev Pathol 2020; 15: 419–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuster JJ, Walsh K. Somatic mutations and clonal hematopoiesis: unexpected potential new drivers of age-related cardiovascular disease. Circ Res 2018; 122: 523–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busque L, Mio R, Mattioli J et al. Nonrandom X-inactivation patterns in normal females: lyonization ratios vary with age. Blood 1996; 88: 59–65. [PubMed] [Google Scholar]
- 9.Busque L, Patel JP, Figueroa ME et al. Recurrent somatic TET2 mutations in normal elderly individuals with clonal hematopoiesis. Nat Genet 2012; 44: 1179–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs KB, Yeager M, Zhou W et al. Detectable clonal mosaicism and its relationship to aging and cancer. Nat Genet 2012; 44: 651–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Laurie CC, Laurie CA, Rice K et al. Detectable clonal mosaicism from birth to old age and its relationship to cancer. Nat Genet 2012; 44: 642–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loh PR, Genovese G, Handsaker RE et al. Insights into clonal haematopoiesis from 8,342 mosaic chromosomal alterations. Nature 2018; 559: 350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genovese G, Kahler AK, Handsaker RE et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med 2014; 371: 2477–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jaiswal S, Fontanillas P, Flannick J et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med 2014; 371: 2488–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie M, Lu C, Wang J et al. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nat Med 2014; 20: 1472–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zink F, Stacey SN, Norddahl GL et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 2017; 130: 742–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shlush LI. Age-related clonal hematopoiesis. Blood 2018; 131: 496–504. [DOI] [PubMed] [Google Scholar]
- 18.Young AL, Challen GA, Birmann BM, Druley TE. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat Commun 2016; 7: 12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watson CJ, Papula AL, Poon GYP et al. The evolutionary dynamics and fitness landscape of clonal hematopoiesis. Science 2020; 367: 1449–54. [DOI] [PubMed] [Google Scholar]
- 20.Coombs CC, Zehir A, Devlin SM et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 2017; 21: 374–82.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henry CJ, Marusyk A, DeGregori J. Aging-associated changes in hematopoiesis and leukemogenesis: what’s the connection? Aging (Albany NY) 2011; 3: 643–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaiswal S, Natarajan P, Silver AJ et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017; 377: 111–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorsheimer L, Assmus B, Rasper T et al. Association of mutations contributing to clonal hematopoiesis with prognosis in chronic ischemic heart failure. JAMA Cardiol 2019; 4: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mas-Peiro S, Hoffmann J, Fichtlscherer S et al. Clonal haematopoiesis in patients with degenerative aortic valve stenosis undergoing transcatheter aortic valve implantation. Eur Heart J 2020; 41: 933–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dorsheimer L, Assmus B, Rasper T et al. Hematopoietic alterations in chronic heart failure patients by somatic mutations leading to clonal hematopoiesis. Haematologica 2019; 105: e328–e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bouleti C, Himbert D, Iung B et al. Long-term outcome after transcatheter aortic valve implantation. Heart 2015; 101: 936–42. [DOI] [PubMed] [Google Scholar]
- 27.Van Mieghem NM, van der Boon RM, Nuis RJ et al. Cause of death after transcatheter aortic valve implantation. Catheter Cardiovasc Interv 2014; 83: E277–82. [DOI] [PubMed] [Google Scholar]
- 28.Bick AG, Pirruccello JP, Griffin GK et al. Genetic interleukin 6 signaling deficiency attenuates cardiovascular risk in clonal hematopoiesis. Circulation 2020; 141: 124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fuster JJ, MacLauchlan S, Zuriaga MA et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 2017; 355: 842–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bick AG, Weinstock JS, Nandakumar SK et al. Inherited causes of clonal hematopoiesis of indeterminate potential in TOPMed whole genomes. bioRxiv (Bioarchive) 2019; 782748. [Google Scholar]
- 31.Potus F, Pauciulo MW, Cook EK et al. Novel mutations and decreased expression of the epigenetic regulator TET2 in pulmonary arterial hypertension. Circulation 2020;141:1986–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sano S, Oshima K, Wang Y et al. Tet2-mediated clonal hematopoiesis accelerates heart failure through a mechanism involving the IL-1beta/NLRP3 inflammasome. J Am Coll Cardiol 2018; 71: 875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sano S, Oshima K, Wang Y, Katanasaka Y, Sano M, Walsh K. CRISPR-mediated gene editing to assess the roles of Tet2 and Dnmt3a in clonal hematopoiesis and cardiovascular disease. Circ Res 2018; 123: 335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sano S, Wang Y, Evans MA et al. Lentiviral CRISPR/Cas9-mediated genome editing for the study of hematopoietic cells in disease models. J Vis Exp 2019; 152: e59977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spivak JL. Myeloproliferative neoplasms. N Engl J Med 2017; 376: 2168–81. [DOI] [PubMed] [Google Scholar]
- 36.Tefferi A, Pardanani A. Myeloproliferative neoplasms: a contemporary review. JAMA Oncol 2015; 1: 97–105. [DOI] [PubMed] [Google Scholar]
- 37.Wolach O, Sellar RS, Martinod K et al. Increased neutrophil extracellular trap formation promotes thrombosis in myeloproliferative neoplasms. Sci Transl Med 2018; 10: eaan8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cordua S, Kjaer L, Skov V, Pallisgaard N, Hasselbalch HC, Ellervik C. Prevalence and phenotypes of JAK2 V617F and calreticulin mutations in a Danish general population. Blood 2019; 134: 469–79. [DOI] [PubMed] [Google Scholar]
- 39.Akada H, Yan D, Zou H, Fiering S, Hutchison RE, Mohi MG. Conditional expression of heterozygous or homozygous Jak2V617F from its endogenous promoter induces a polycythemia vera-like disease. Blood 2010; 115: 3589–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mead AJ, Mullally A. Myeloproliferative neoplasm stem cells. Blood 2017; 129: 1607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullally A, Lane SW, Ball B et al. Physiological Jak2V617F expression causes a lethal myeloproliferative neoplasm with differential effects on hematopoietic stem and progenitor cells. Cancer Cell 2010; 17: 584–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi K, Zhao W, Chen Y, Ho WT, Yang P, Zhao ZJ. Cardiac hypertrophy associated with myeloproliferative neoplasms in JAK2V617F transgenic mice. J Hematol Oncol 2014; 7: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang W, Liu W, Fidler T et al. Macrophage inflammation, erythrophagocytosis, and accelerated atherosclerosis in Jak2 (V617F) mice. Circ Res 2018; 123: e35–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing S, Wanting TH, Zhao W et al. Transgenic expression of JAK2V617F causes myeloproliferative disorders in mice. Blood 2008; 111: 5109–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sano S, Wang Y, Yura Y et al. JAK2 (V617F) -mediated clonal hematopoiesis accelerates pathological remodeling in murine heart failure. JACC Basic Transl Sci 2019; 4: 684–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bajpai G, Bredemeyer A, Li W et al. Tissue resident CCR2− and CCR2+ cardiac macrophages differentially orchestrate monocyte recruitment and fate specification following myocardial injury. Circ Res 2019; 124: 263–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dick SA, Macklin JA, Nejat S et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol 2019; 20: 29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sasi SP, Yan X, Zuriaga-Herrero M et al. Different sequences of fractionated low-dose proton and single iron-radiation-induced divergent biological responses in the heart. Radiat Res 2017; 188: 191–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Q, Zhao K, Shen Q et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 2015; 525: 389–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez-Fraticelli AE, Wolock SL, Weinreb CS et al. Clonal analysis of lineage fate in native haematopoiesis. Nature 2018; 553: 212–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun J, Ramos A, Chapman B et al. Clonal dynamics of native haematopoiesis. Nature 2014; 514: 322–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Y, Sano S, Yura Y et al. Tet2-mediated clonal hematopoiesis in nonconditioned mice accelerates age-associated cardiac dysfunction. JCI Insight 2020; 5: e135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robertson NA, Hillary RF, McCartney DL et al. Age-related clonal haemopoiesis is associated with increased epigenetic age. Curr Biol 2019; 29: R786–R7. [DOI] [PubMed] [Google Scholar]
- 54.Ridker PM, MacFadyen JG, Everett BM et al. Relationship of C-reactive protein reduction to cardiovascular event reduction following treatment with canakinumab: a secondary analysis from the CANTOS randomised controlled trial. Lancet 2018; 391: 319–28. [DOI] [PubMed] [Google Scholar]
- 55.Svensson EC, Madar A, Campbell CD et al. TET2-driven clonal hematopoiesis predicts enhanced response to canakinumab in the CANTOS trial: an exploratory analysis. Circulation 2018; 138: A15111. [DOI] [PMC free article] [PubMed] [Google Scholar]