Abstract
Salmonella enterica serovar Gallinarum biovar Pullorum (bvP) and biovar Gallinarum (bvG) are the etiological agents of pullorum disease (PD) and fowl typhoid (FT) respectively, which cause huge economic losses to poultry industry especially in developing countries including India. Vaccination and biosecurity measures are currently being employed to control and reduce the S. Gallinarum infections. High endemicity, poor implementation of hygiene and lack of effective vaccines pose challenges in prevention and control of disease in intensively maintained poultry flocks. Comparative genome analysis unravels similarities and dissimilarities thus facilitating identification of genomic features that aids in pathogenesis, niche adaptation and in tracing of evolutionary history. The present investigation was carried out to assess the genotypic differences amongst S.enterica serovar Gallinarum strains including Indian strain S. Gallinarum Sal40 VTCCBAA614. The comparative genome analysis revealed an open pan-genome consisting of 5091 coding sequence (CDS) with 3270 CDS belonging to core-genome, 1254 CDS to dispensable genome and strain specific genes i.e. singletons ranging from 3 to 102 amongst the analyzed strains. Moreover, the investigated strains exhibited diversity in genomic features such as virulence factors, genomic islands, prophage regions, toxin-antitoxin cassettes, and acquired antimicrobial resistance genes. Core genome identified in the study can give important leads in the direction of design of rapid and reliable diagnostics, and vaccine design for effective infection control as well as eradication. Additionally, the identified genetic differences among the S. enterica serovar Gallinarum strains could be used for bacterial typing, structure based inhibitor development by future experimental investigations on the data generated.
1. Introduction
Pullorum disease (PD) and fowl typhoid (FT) are two distinct septicaemic diseases caused by non-motile Salmonella enterica subsp. enterica serovar Gallinarum (S. Gallinarum) biovar Pullorum (bvP) and biovar Gallinarum (bvG), respectively, which exhibit host-specificity towards poultry and aquatic birds [1–3]. The diseases caused by these invasive avian pathogens cause high morbidity and acute mortality in poultry in India and various countries of Asia, Africa and South America. Pullorum disease (PD) occurs in young birds and persists for long periods in spleen and reproductive tract which leads to high mortality [4]. The disease is characterised by white diarrhoea chaperoned with increased antimicrobial resistance (AMR) and high infection rates [5, 6]. On the other hand, FT can affect birds of all ages but primarily occurs in adult birds and results in variable morbidity depending on the age, species, and breed of the bird [7]. Salmonella Enteritidis, a major food-borne pathogen causes infection which leads to enteritis in multiple hosts in addition to poultry [8]. Although S. Gallinarum has negligible importance in humans, S. Enteritidis is an important zoonoses [8, 9]. The S. Gallinarum, in addition has been found to be a recently evolved ancestors of S. Enteritidis [10].
The virulence of Salmonella spp. is mediated by an arsenal of genes which are capable of invasion, replication, and colonization inside the host cells [11, 12]. The genetic factors involved in pathogenesis of FT and PD at molecular and cellular mechanisms are still under elucidation [2, 13]. The genomic sources of virulence systems in Salmonella enterica serovars are mainly divided into two elements, one of which is horizontally acquired chromosomally located mobile genetic elements known as Salmonella pathogenicity Islands (SPIs), and prophage elements, whereas other is plasmids associated with salmonellae [14]. These genetic elements have played a seminal role in various ecological niche adaptations in different host and pathogenicity life-style in Salmonella by encoding for proteins carrying out cell-adherence, cellular invasion at host level, induction of innate immune and/or pathophysiological responses to infection [15, 16]. Two pathogenicity islands, Salmonella pathogenicity island 1 (SPI-1) and SPI-2 have been described, which play roles in mediating disease by Salmonella enterica through their respective type III secretion systems (TTSS). SPI which encode T6SSs have been detected in SPI6, SPI19, 20, 21 in S. Gallinarum and S. Enteritidis [17].
Additionally, prophage lysogeny is a rich contributor of Salmonella genome diversity, and their acquisition leads to enhanced virulence and pathogenicity [18, 19]. The prophage elements are being increasingly utilised as molecular markers for strain discrimination [20, 21]. Another crucial genetic element that is involved in persistence, virulence and AMR includes toxin-antitoxin system (TA system) [22–24]. Furthermore, the Salmonella serovars including the Gallinarum and Enteritidis are increasingly reporting incidence of antimicrobial resistance (AMR) [25, 26]. The comparative genomic analysis of pathogens helps in elucidating the diversity and depth of functional set of genes in form of pan-genome and decoding of evolutionary history [27, 28]. Moreover, it aids in functional annotation, as well as decodes molecular mechanisms underlying pathogenesis and niche adaptation by precise measurement of genetic variation within and between pathogenic groups [29–31].
Recently we determined the first draft genome sequence of an Indian strain of S. Gallinarum VTCCBAA614 isolated from diseased poultry [32]. The present investigation dealt with whole genome characterization and assessment of genotypic differences amongst bvG and bvP strains including in house S. Gallinarum Sal40 VTCCBAA614 strain (Table 1). The study unravelled pan-genome, core-genome, dispensable genome, and strain specific genes of the analyzed S. Gallinarum strains. Moreover, the investigation decoded diversity in virulence factors, genomic islands, prophage regions, TA systems, and acquired AMR genes in the investigated strains via employment of various computational tools. The genotypic differences revealed by the study will serve as the genomic resource for the identification and discrimination of biovars, and could form the basis of structure based inhibitors design and that would be beneficial to poultry industry. Although comparative genomics analysis of host-adapted salmonellae genomes have elucidated various evolutionary and virulence themes [5, 8], this is the first report for the assessment of genotypic differences, which includes an Indian strain among the investigated strains from America and China.
Table 1. Genome statistical information of nine investigated Salmonella strains as obtained by Prokka pipeline and ANI calculator.
| #Organism name | Geographic location | Accession number | Assembly status | Total length | Size | GC | CDS | tRNA | tmRNA | rRNA | rr | SNP detected |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| (MB) | Content % | |||||||||||
| S. enterica Enteritidis | United Kingdom | NC_011294.1 | Complete | 4,685,848 | 4.68 | 52.17 | 4352 | 85 | 1 | 22 | 2 | - |
| str. P125109 | ||||||||||||
| S. enterica Gallinarum | Brazil | NC_011274.1 | Complete | 4,658,697 | 4.65 | 52.20 | 4452 | 77 | 1 | 22 | 2 | 7832 |
| str. 287/91 | ||||||||||||
| S. enterica Gallinarum | Not available | NZ_CP019035.1 | Complete | 4,609,911 | 4.60 | 52.20 | 4399 | 78 | 1 | 22 | 2 | 7505 |
| str. 9184 | ||||||||||||
| S.enterica Gallinarum | India | JSWQ00000000.1 | Contig | 4,598,206 | 4.59 | 52.22 | 4455 | 68 | 1 | 4 | 2 | 7946 |
| str. Sal40 | ||||||||||||
| S.enterica Pullorum | USA | NZ_CP012347.1 | Complete | 4,694,842 | 4.69 | 52.19 | 4474 | 81 | 1 | 23 | 2 | 7424 |
| str. ATCC 9120 | ||||||||||||
| S.enterica Pullorum | China | NC_021984.1 | Complete | 4,682,599 | 4.68 | 52.14 | 4635 | 80 | 1 | 22 | - | 12726 |
| str. S06004 | ||||||||||||
| S.enterica Pullorum | China | NZ_CP022963.1 | Complete | 4,728,875 | 4.72 | 52.17 | 4548 | 75 | 1 | 22 | 2 | 7543 |
| str. QJ-2D-Sal | ||||||||||||
| S.enterica Gallinarum/Pullorum | China | NC_022221.1 | Complete | 4,623,089 | 4.62 | 52.23 | 4397 | 78 | 1 | 22 | 2 | 7786 |
| str. CDC1983-67 | ||||||||||||
| S.enterica Gallinarum/pullorum | Not available | NC_016831.1 | Complete | 4,637,962 | 4.63 | 52.21 | 4451 | 75 | 1 | 22 | 2 | 8038 |
| str. RKS5078 |
2. Material and methods
2.1 Data collection of Salmonella genomes
The complete genome sequences of eight S. Gallinarum strains i.e., S. Gallinarum str. 287/91 (NC_011274.1), S. Gallinarum str. 9184 (NZ_CP019035.1), S. Pullorum str. ATCC 9120 (NZ_CP012347.1), S. Pullorum str. S06004 (NC_021984.1), S. Pullorum QJ-2D-Sal (NZ_CP022963.1), S. Gallinarum/pullorum str. CDC1983-67 (NC_022221.1), and S. Gallinarum/pullorum str. RKS5078 (NC_016831.1) including draft genome sequence of S. Gallinarum Strain VTCCBAA614 (isolated from chicken in India from FT inflicted broiler flock) were mined from NCBI database to decode genotypic differences among the S. enterica serovar Gallinarum strains. In addition the genome sequence of S. Enteritidis str. P125109 (NC_011294.1) [10] a well studied pathogenic Salmonella strain was also retrieved from NCBI for employment as reference for comparative genome analysis (Table 1). Sequence quality of genome assemblies is dependent on the sequencing technology used, genome coverage and aim of the sequencing. Information regarding the investigated genomes such as sequencing technology used, genome coverage, as well as assembly method was mined from literature as well as NCBI database and is listed in S2 Table. In addition, measures of genome quality such as completeness, contamination, coarse consistency, and fine consistency as identified by EvalG and EvalCon tools of PATRIC database are also listed (S2 Table)
The Indian strain S. Gallinarum VTCCBAA614 [32] is available from our culture collection at NCVTC, NRCE Hisar. All the sequences were extracted in fasta and GenBank (gbk) format.
2.2 Genome statistics and visualization
Genomic features such as number of CDS, tRNA, tmRNA, rRNA and repeat region (rr) were determined by running Prokka pipeline at UseGalaxy webinterface (Table 1) [33]. The GC% of the genomes was determined by ANI Calculator which uses OrthoANIu algorithm [34]. Genomes were aligned with S. Enteritidis P125109 genome taken as reference genome. Alignment and single nucleotide polymorphism (SNP) detection was performed by using nucmer and show-snps (with Clr parameter) components of MUMmer package V 3.1 [35]. In addition, circular plot visualization of CDS, GC content, and GC skew with S. Enteritidis str. P125109 as reference genome against other Salmonella genomes was generated by EDGAR [36].
2.3 Pan- and core-genome calculations
Pan-genome is the complete set of orthologous genes (OGGs) harboured within a collection of investigated genomes, whereas, the core-genome refers to the set of genes present in all the genomes of a collection. On the other hand, dispensable genome refers to the set of genes harboured by one or a subset of investigated genomes. Singleton genes are the unique genes that do not have any homologs in the investigated genomes [37, 38]. EDGAR served as the resource for pan-genome, core-genome, and singleton genes calculations for the selected Salmonella strains [36]. A customised project was set up by EDGAR to conduct all the calculations for pan-genome, core-genome, dispensable genome, and singletons with selection of S. Enteritidis str. P125109 as reference genome.
2.4 Functional annotation of pan- and core-genome
The functional annotation of core-genome and complete set of strain-specific genes via orthology assignment was carried by eggNOG-mapper v2 online portal (http://eggnog-mapper.embl.de) [39, 40]. The tool employs precomputed clusters and phylogenies from its in house database and provides orthology assignment to a large set of sequences via fast orthology mapping. Out of the total 29,430 CDS that comprised core-genome of nine investigated Salmonella genomes, 29,125 (98.96%) CDS were queried by eggNOG-mapper v2 for orthology mapping. On the other side, 153 (48.11%) out of the total 318 strain specific CDS (singletons) were queried for orthology mapping.
2.5 Phylogenetic analysis of the strains
The identified core-genome of the investigated Salmonella strains was employed by EDGAR to decipher phylogenetic relationship among the strains. Alignment of each CDS set was obtained by MUSCLE [41], and then further joined to form one huge alignment. The FastTree software was used to construct the tree. Program employs Shimodaira-Hasegawa (SH) branch support values to verify the tree topology [42]. Conservation of gene order and genome rearrangements among Salmonella genomes were also explored by using EDGAR wherein S. Enteritidis P125109 was chosen as a reference to create synteny plots. The Indian strain, bvG VTCCBAA614, was not included, as the use of draft genomes is not recommended in synteny plot representation.
2.6 Detection of virulence factors
VFanalyzer tool available at VFDB (virulence factor database) was utilized to detect putative virulence factors in the investigated Salmonella genomes [43]. The first step in the detection of virulence factors by the tool involves construction of orthologous groups within the query genome sequence as well as in the pre-investigated Salmonella reference genomes archived in the database to avoid detection of paralogs. Next, it carries iterative and exhaustive sequence similarity search against the VFDB database for accurate detection of virulence factors. Finally, a context-based data refinement process is performed for virulence factors encoded by gene clusters [43]. We also carried out additional NCBI BLAST searches for the virulence genes that showed differential distribution after manual curation of VFanalyzer tool results.
2.7 Detection of Salmonella pathogenicity islands (SPI)
Two approaches were employed to detect SPI’s -the major determinant of S. enterica virulence via usage of web tool SPIFinder 1.0 available at (https://cge.cbs.dtu.dk/services/SPIFinder) [44] with default parameters, and NCBI BLAST search of known SPIs extracted from PAIDB v2.0 (http://www.paidb.re.kr/) against investigated genomes [45, 46].
2.8 Identification and in silico characterization of prophage sequences
Potential prophage sequences within the Salmonella genomes were detected and annotated by employing PHASTER (Phage Search Tool Enhanced Release). PHASTER, an improved version of PHAST phage search tools detects candidate prophage regions in the bacterial genomes and then classifies the identified regions into three classes i.e. intact, incomplete and questionable on the basis score obtained [47, 48].
2.9 Detection and analysis of Type II Toxin-Antitoxin (TA) gene cassettes
Type II TA loci were predicted in the Salmonella genomes by employing TAfinder [49]. The parameters used for the detection were BLAST e value: 0.01; HMMer E-value: 1; Maximum length for candidate toxin/antitoxin: 300; maximum overlap between candidate toxin and antitoxin: (-20–150).
2.10 Screening of acquired antimicrobial resistance (AMR) genes
Resfinder available at (https://cge.cbs.dtu.dk/services/ResFinder/) was employed to investigate the presence of acquired antimicrobial resistance (AMR) genes in the selected Salmonella strains [50]. Resfinder identifies acquired antibiotic resistance genes foraminoglycoside, β-lactam, colistin, fluoroquinolone, fosfomycin, fusidic acid, glycopeptide, MLS-macrolide, lincosamide and streptogramin B nitroimidazole, oxazolidinone, phenicol, rifampicin, sulphonamide, tetracycline and trimethoprim. Moreover, both known and unknown chromosomal point mutations were also detected in AMR resistance genes such as gyrA, parE, pmrA, pmrB, parC and 16S_rrsD.
3. Results
3.1 Comparative genome statistics
Comparative genome analysis of selected nine strains of Salmonella species (Table 1) was carried out by employing various bioinformatics tools. The average genome size of the investigated Salmonella strains was 4,657,781 bp, ranging from 4,598,206 bp (S. Gallinarum Sal40 strain VTCCBAA614) to 4,728,875 bp (S. Pullorum QJ-2D-Sal). The average GC content of all the analyzed genomes was 52.19%, ranging from 52.14% (S. Pullorum str. S06004) to 52.23% (S. Gallinarum/pullorum str. CDC1983-67) (Table 1). Genomic features of the investigated Salmonella strains were determined by running Prokka pipeline at Use Galaxy webinterface. The average number of CDS was observed to be 4463 with highest number detected in S. Pullorum str. S06004 (4635) and the lowest in S. Enteritidis str.
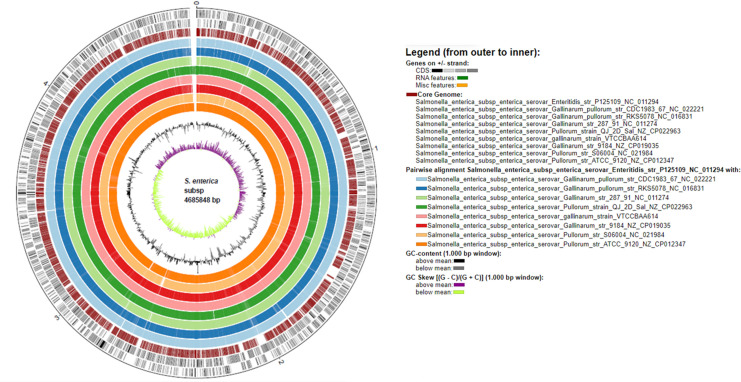
P125109 (4352) (Table 1). The tRNA sequences identified by Prokka pipeline in the S. Gallinarum genomes fell in the range of 75–81 i.e. 75 in S. Gallinarum/pullorum str. RKS5078 and 81 in S. Pullorum ATCC 9120. It was 85 in S. Enteritidis str. P125109 (Table 1). In addition, all the investigated Salmonella genomes were detected to possess only one tmRNA. Notably, only four rRNA sequence were detected in S. Gallinarum Sal40 strain VTCCBAA614 in comparison to possession of 23 rRNA sequences in S. Pullorum str. ATCC 9120 and presence of 22 rRNA sequences in rest of the genomes (Table 1). The circular plot visualization of investigated genomes with reference to S. Enteritidis str. P125109 depicts varied GC content and GC skew along with predominant similarity of core-regions of the investigated genomes (Fig 1).
Fig 1.
Circular plot genome representation of nine investigated Salmonella strains i.e. a) S. Gallinarum str. 287/91 b) S. Gallinarum str. 9184 b) S. Gallinarum Sal40 strain d) S. Pullorum str. ATCC 9120 e) S. Pullorum str. S06004 f) S. Pullorum QJ-2D-Sal g) S. Gallinarum/Pullorum str. CDC1983-67 and h) S. Gallinarum/Pullorum str. RKS5078 with demonstration of GC content, GC skew and CDS in reference to S. Enteritidis str. P125109.
3.2 Pan and core-genome analysis unravels strain specific genes
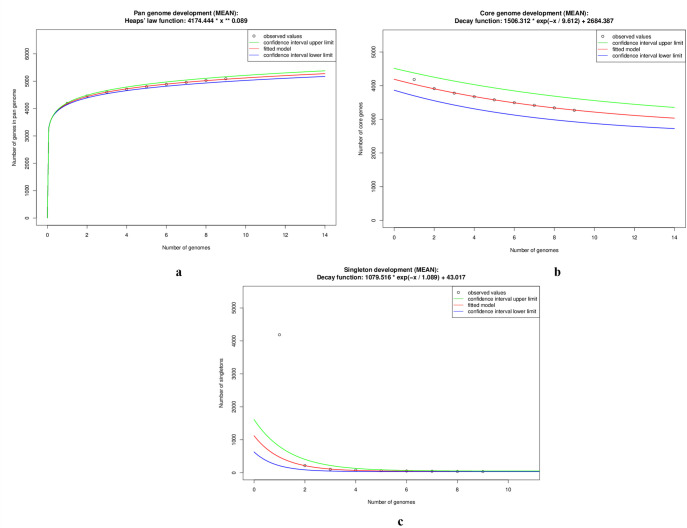
The pan-genome calculation by customised set up in EDGAR led us to have discrete idea of the total genetic repository of the Salmonella genomes under study. The pan-genome of S. Gallinarum was found to be in open state with growth exponent value of 0.089 (95% confidence interval 0.084 to 0.093) (S1 Table). The pan-genome development plot showed steady growth with addition of each new genome and reached 5091 on addition of ninth genome which is nearly 1.1 times the average number of genes of nine strains (Fig 2A, S1 Table). On the other hand, core-genome development plot became limited to 3270 genes, with shared genes decreasing with new genome addition. The extrapolated core-genome size was 2684 (95% confidence interval 2507.66 to 2861.11) (Fig 2B, S1 Table). In addition, singleton development plot also indicated S. enterica serovar Gallinarum to be in an open pan-genome state as 43 new genes were predicted to be found at every genome addition (Fig 2C, S1 Table).
Fig 2. Core genome development plot for nine Salmonella genomes.
The red curve shows the fitted model. Green represents confidence interval upper limit and blue demonstrates confidence interval lower limit.
The pan-genome of investigated Salmonella strains with the reference to S. Enteritidis str. P125109 strain is composed of 5,091 coding sequences (CDS) which include a core-genome of 3,270 (64.2%) CDS, a dispensable genome of 1254 (24.6%) CDS, and 567 (11.1%) singletons. The complete list of CDS detected as part of pan-genome, core-genome, and singletons with their function are listed in S3, S4 and S5A–S5I Tables. Notably, a total of 318 CDS in the range of 3–102 (Table 2, S5 Table) were found as strain specific genes aka singletons in the investigated genomes. Significantly, S. Gallinarum Sal40 strain VTCCBAA614 harboured the highest number (102) of singletons among the investigated Salmonella strains (Table 2, S5 Table). The singletons detected were identified to be as hypothetical proteins (61), transposase (13), membrane proteins (8), ATP binding proteins (3) among others. Interestingly, genomes of S. Enteritidis str. P125109 and S. Pullorum QJ-2D-Sal possessed 96 and 85 singletons each, respectively.
Table 2. Number of CDS identified in pan genome, core genome and singletons of the investigated Salmonella genomes.
| Organism and strain | Accession number | Pan genome | Core genome | No of singletons |
|---|---|---|---|---|
| S.enterica Enteritidis str. P125109 | NC_011294.1 | 5091 | 3270 | 96 |
| S. enterica Gallinarum str. 287/91 | NC_011274.1 | 3 | ||
| S.enterica Gallinarum str. 9184 | NZ_CP019035.1 | 4 | ||
| S. enterica Gallinarum str. Sal40 strain | JSWQ00000000.1 | 102 | ||
| S.enterica Pullorum str. ATCC 9120 | NZ_CP012347.1 | 10 | ||
| S. enterica Pullorum str. S06004 | NC_021984.1 | 10 | ||
| S. enterica Pullorum str. QJ-2D-Sal | NZ_CP022963.1 | 85 | ||
| S. enterica Gallinarum/pullorum str. CDC1983-67 | NC_022221.1 | 3 | ||
| S. enterica Gallinarum/pullorum str. RKS5078 | NC_016831.1 | 5 |
On the other hand, S. Pullorum str. ATCC 9120 and S. Pullorum str. S06004 genomes harboured ten singletons each, whereas, the rest of the genomes of possessed ≤5 singletons each (Table 2, S5A–S5I Table).
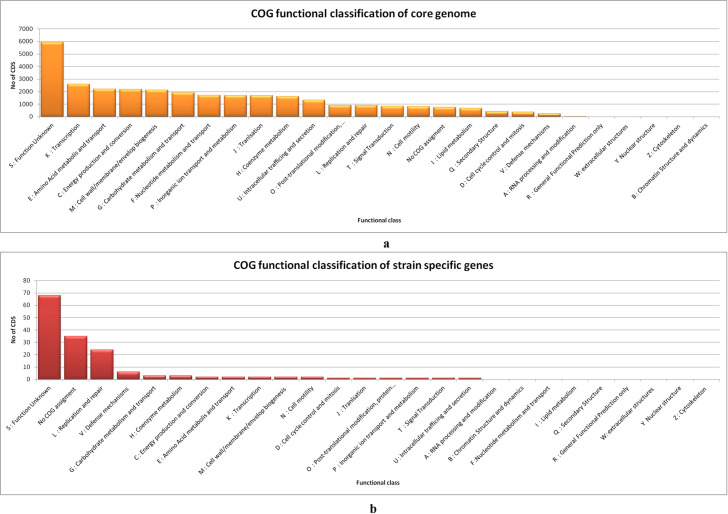
Functional annotation of core-genome and strain specific CDS by assignment of orthology (COG) was performed by eggNOG-mapper v2. The annotation by eggNOG-mapper provided detailed description of GO term, EC number, annotation level, COG category and description (S6 Table). 29,125 CDS (98.96%) out of the total 29,430 (core-genome sequences) were queried by eggNOG-mapper v2 for orthology mapping. The highest number among them belonged to the functional classes of function unknown (5923) transcription (2592), amino acid metabolism and transport (2196), energy production and conversion (2178), and cell wall/membrane/envelop biogenesis (2133) (Fig 3A, S6A Table). On the other hand, 153 out of the 318 CDS (complete set of identified singleton sequences) were queried by eggNOG-mapper v2. The highest number amongst them belonged to the classes of function unknown (68), replication and repair (24) and defence mechanism (6) among others. Thirty five singletons were not provided any COG category by eggNOG-mapper (Fig 3B, S6B Table)).
Fig 3.
a) Distribution of COG functional classes in core genome of investigated Salmonella strains by eggnog mapper b) Distribution of COG functional categories in strain specific genes by eggnog mapper.
3.3 Phylogenetic and synteny plot analysis
A phylogenetic tree was constructed on the basis of core-genome comprising of 9,83,299 aa-residues/bp per genome and 88,49,691 in total of nine Salmonella genomes by EDGAR (Fig 4) using S. enterica serotype Enteritidis strain P125109 as a reference sequence to demonstrate the evolutionary relationship among the S. Gallinarum strains used in the study.SH support values for our tree were very good in general, with a minimum value of 0.772 and only two values below the maximum of 1.00. The bvG and bvP formed two distinct and strongly supported clades in the phylogeny (Fig 4). In concordance with their taxonomic classification bvG strains clustered together i.e. S. Gallinarum str. 287/91, S. Gallinarum str. 9184 and Gallinarum Sal40 strain VTCCBAA614) and formed one clade. Whereas, the rest of the bvP strains bifurcated into 2 clades (Fig 4).
Fig 4. Unrooted phylogenetic tree showing taxonomic positions of the investigated Salmonella strains constructed by EDGAR on the basis of core genome set.
Branch support values that were lower than the maximum values of 1.0 are shown at the respective branches.
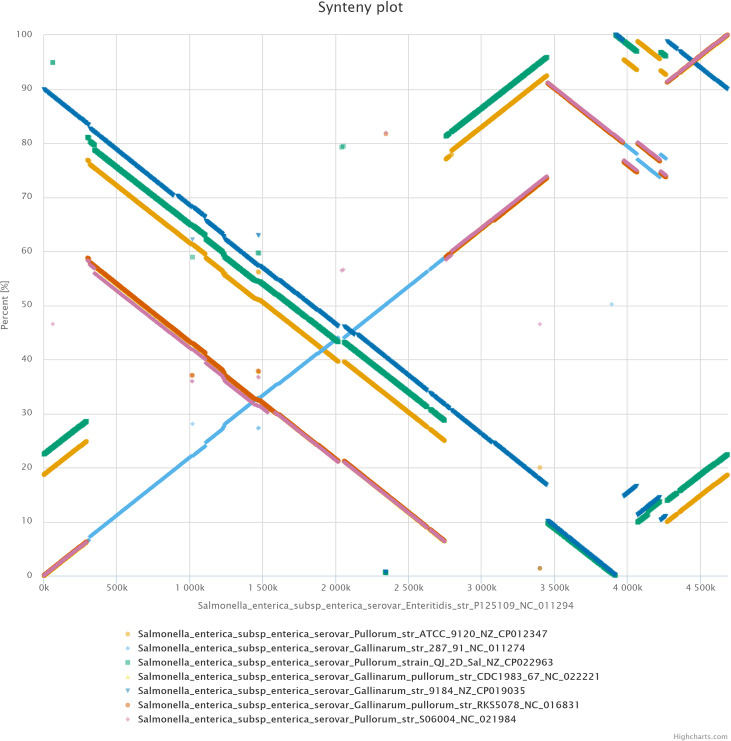
The synteny plot analysis performed by EDGAR of the investigated Salmonella genomes in reference to S. Enteritidis str. P125109 depicted large scale genomic rearrangements which included relocations, inversions, duplications and deletions (Fig 5). It was observed that all bvP strains genomes showed high degree of conservation of gene order among genomes. The highly similar genomic rearrangement within bvP comprised of inversion and duplication. However, the bvG strain 9184 was characterized by large region of inversion, which was not observed in bvG 287/91 in reference to S. Enteritidis str. P125109 strain. Synteny plot of individual investigated strains against the reference genome of S. Enteritidis str. P125109 is shown in S1 Fig.
Fig 5.
Synteny plot constructed by EDGAR of eight S. Gallinarum strains i.e. a) S. Pullorum str. ATCC 9120 (b) S. Gallinarum str. 287/91 c) S. Pullorum QJ-2D-Sal (d) S. Gallinarum/Pullorum str. CDC1983-67 (e) S. Gallinarum str. 9184, (f) S. Gallinarum/Pullorum str. RKS5078 (g) S. Pullorum str. S06004 against the reference of S. Enteritidis str. P125109.
3.4 Virulence factor detection
Putative virulence factors were detected by VFanalyzer tool in investigated Salmonella genomes. The tool identified candidate virulence factors related to capsule, fimbrial adherence, macrophage inducible genes, magnesium uptake, non-fimbrial adherence, regulation, secretion system, stress adaptation, autotransporter and invasion as listed (S7A–S7C Table). Fimbrial adherence genes which includes agf/csg, bcf, fim, lpf, peg, pef, saf, sef, sta, stb, stc, std, ste, stf, stg, sth, sti, stj, stk and tcf were searched for in the investigated genomes. Notably, pef, sta, stc, stg, stj, stk and tcf operons were not observed in any of the analyzed genomes. Manual curation of the results obtained from the tool alongwith BLASTN searches revealed that among the fimbrial adherence determinants, chaperone-usher fimbrial gene operons of bcf, fim, lpf, peg, saf, stb, std, ste, stf, sth and sti were present in the S. Enteritidis str. P125109, however genes in the fimbrial operon sef were predicted to be missing in this strain (S7C Table).
On the other hand, among the investigated S. Gallinarum genomes, chaperone-usher fimbrial gene operons, bcf, fim, lpf, peg, saf, stb, ste, stf, sth and sti were present. Fimbrial operon sef and std operon were additionally predicted to be disrupted in the S. Gallinarum genomes, with stdA and stdB showing no significant similarity and stdC homolog showing divergent sequence (69.11% similarity) on BLASTN analysis (S7B Table). The operon of adhesin called thin curled fimbriae (curli), which is encoded by the agf /csg fimbrial gene cluster was detected in all the 9 genomes analysed (S7C Table).
The std operon was detected in 3 out of 5 bvP genomes (S. Pullorum str. ATCC 9120, S. Pullorum str. S06004 and S. Pullorum str. QJ-2D-Sal) as well as S. Enteritidis str. P125109 and was absent in genomes of S. Gallinarum str. 287/91, S. Gallinarum str. 9184, S. Gallinarum Sal40 str. VTCCBAA614, S. Gallinarum/pullorum str. CDC1983-67 and S. Gallinarum/pullorum str. RKS5078 (S7 Table). The sef operon was disrupted showing complete absence of sefA and sefD genes in all the genomes. Sef operon was not detected in S. Gallinarum Sal40 str. VTCCBAA614 strain. The disruption in fim operon was only observed in S. Pullorum str. S06004 (S7C Table).
3.5 Detection of Salmonella Pathogenic Islands (SPIs)
A total of 113 homologs of SPIs with an average of 14.77 SPIs were detected in the investigated Salmonella genomes by the combined usage of web tool SPIFinder 1.0and NCBI BLAST search. The integrated approach revealed presence of high sequence identity SP-1, SP-2, SP-3, SP-4, SPI-5, SPI-12, SPI-13, SP-14, C63PI and SPCS54 islands in their genomes (S8A and S8B Table). The detected SPIs varied in the range of 0.3 to 41.8 kb in terms of size. Interestingly, 3 SPI-13 (AY956834, AY956833, AY956832) and 2 SPI-14 (AY956835, AY956836) were observed in each of the investigated genome with the exception of S. Gallinarum str. 9184 genome, wherein SPI-14 (AY956836) (0.4 kb) was not detected. Moreover, SPI-12 was also not observed in S. Gallinarum Sal40 strain VTCCBAA614. Notably, various other SPIs and resistance islands reported in S. enterica serovar such as SESS LEE, SGI-1, SPI-10, SPI-11, SPI-2, SPI-6, SPI-7, and HP1 whose sequence were extracted from PAIDB database were not found to be present by both the approaches. The features of detected SPI homologs in their respective genomes such as starting position, end position, size, and external annotation if any are detailed in (S8A and S8B Table).
3.6 Identification and analysis of prophage and prophage remnant regions
Each of the analyzed Salmonella genome was detected to possess at least one candidate prophage region by PHASTER (S2 Fig, S9 Table). In total, 23 prophage regions were identified in which fifteen were classified as putative intact regions and eight as incomplete prophage regions by PHASTER (S2 Fig, S9A Table). The detailed information of the identified prophage regions i.e., region name, region length, completeness, score, total protein, region position and GC percentage in the respective Salmonella genomes as determined by the web server are listed in S9A Table. The highest number of prophage region were detected in S. Pullorum QJ-2D-Sal and S. Pullorum str. S06004 with each possessing three complete prophages and one incomplete prophage region. In total, there were 5 different propahge elements with Gifsy_2 being the most common, which was detected in all genomes analysed. On the other hand, all 3 bvG possessed only a single (Gifsy_2) intact prophage element. Whereas bvP harboured a variety of phage elements including Gifsy_2. The average size of identified prophage regions was 36.31 kb in which largest (64.3 kb) and smallest (8.5 kb) candidate prophage regions were observed in S. Enteritidis str. P125109. On the other hand, the average size of identified complete prophage sequences in the analyzed Salmonella genomes was found to be 44.98 kb with lowest size of 29.2 kb belonging to S. Pullorum str. ATCC 9120. (S9A and S9B Table). Whereas, largest intact prophage region of 64.3 kb was observed in S. Enteritidis str. P125109 genome. The GC percentage in identified prophage region varied from 45.59% to 53.21% (S9A Table). On the other hand GC% of complete prophage regions varied from 47.23% to 53.21% (S9B Table).
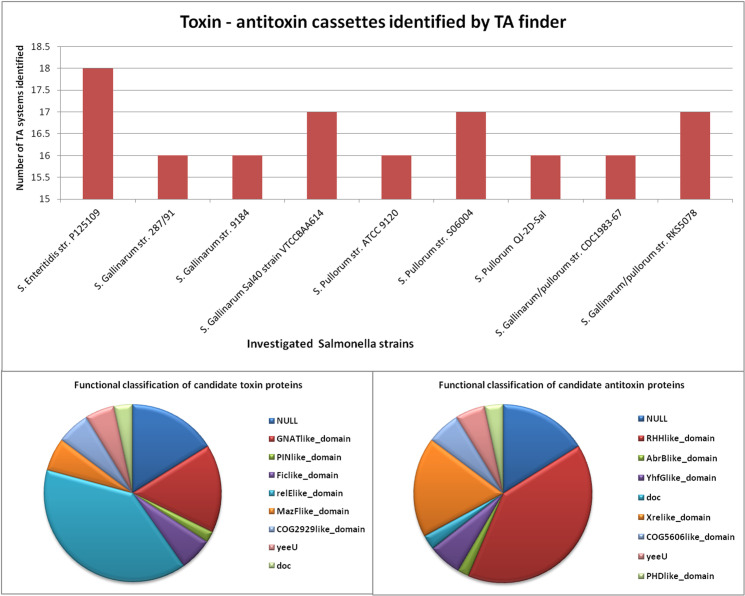
3.7 Presence of toxin antitoxin cassettes
TA loci were identified in all the investigated Salmonella genomes. A total of 149 Type II TA loci were identified, ranging from 16 to 18 in each of the genome analysed (Fig 6, S10 Table). The length of the identified toxin and antitoxin proteins varied from 73–475 and 55–319 aa, respectively each. Majority of the identified candidate toxin proteins (58) harboured relElike_domain in their structure. While, other candidate toxin proteins showed the presence of GNATlike_domain (24), MazFlike_domain (9), COG2929like_domain (9), yeeU (8), doc (5), PINlike_domain (3), and NULL (24) in their structures respectively (Fig 6, S10 Table). On the other hand, identified candidate antitoxin proteins majorly possessed RHHlike_domain (60). Whilst, other domains that were observed included Xrelike_domain(27), COG5606like_domain(9), YhfGlike_domain (9), yeeU (8), PHDlike_domain (5), doc (4), AbrBlike_domain (3) and NULL (24) in rest of the investigated candidate antitoxins (Fig 6, S10 Table). The detailed description of identified TA loci in Salmonella genomes which includes location in the genome, length, strand, family and domain are summarised in S10 Table.
Fig 6. Toxin-antitoxin cassettes identified by TA finder in investigated Salmonella strains with functional classification of candidate toxin and antitoxin proteins.
3.8 Detection of acquired antibiotic resistance genes/chromosomal point mutations
Interestingly, all the analyzed Salmonella genomes harboured acquired aminoglycoside resistance gene (aac(6’)-Iaa) (S3 Fig, S11 Table) as determined by Resfinder. The information related to identity, contig and position of the identified aminoglycoside gene in their respective genomes is listed in S11 Table. On the other hand, no acquired resistance gene for beta-lactam, colistin, fluoroquinolone, fosfomycin, fusidic acid, glycopeptide, MLS-macrolide, lincosamide and streptogramin B nitroimidazole, oxazolidinone, phenicol, rifampicin, sulphonamide, tetracycline, and trimethoprim was detected in the analyzed Salmonella genomes by Resfinder. Strikingly, known chromosomal point mutation in gyrA was detected in 2 bvG and 2 bvP genomes out of the nine analysed i.e., S. Gallinarum str. 287/91, S. Gallinarum Sal40 strain VTCCBAA614, S. Pullorum str. S06004 and S. Pullorum QJ-2D-Sal (S11 Table). In addition, unknown mutations were detected in 16S_rrsD gene of all the analysed genomes (S11 Table). Moreover, unknown mutations in parE were detected by Resfinder in S. Gallinarum Sal40 strain VTCCBAA614 and S. Pullorum str. S06004. A total of six antimicrobial resistance genes (acquired, known and unknown chromosomal point mutations) were identified among the nine investigated strains.
4. Discussion
Salmonella enterica subsp. enterica comprises of both host-adapted and host-promiscuous pathotypes that causes a spectrum of diseases depending on the serovar or host [51, 52]. Pullorum Disease (PD) and Fowl Typhoid (FT) caused by S. Gallinarum biovars Pullorum (bvP) and Gallinarum (bvG) respectively are endemic in countries of Asia and South America leading to huge economic losses to poultry industry [1, 5, 53]. It is interesting to note that as compared to FT, the PD reports have been low in India [54]. On the other hand, PD is frequent in China, being prevalent in every province [5]. Biosecurity and vaccinations applied in combination are important strategies to prevent and control these diseases in commercial and breeding flocks. Most commercially available vaccines are killed vaccines, although vaccination strategies for effective Salmonella control mainly include use of live attenuated strains capable of inducing a cellular immune response [55] still concerns about poor protection, lack of understanding of genetic basis of attenuation and residual pathogenicity remain [56–58].A progress can be made by a thorough understanding of the genetic virulence makeup of prevalent strains. Recently, we had obtained the first draft genome sequence of an Indian strain of S. Gallinarum VTCCBAA614 isolated from diseased poultry [32]. In the present investigation, we performed comparative genome analysis of eight strains of S. Gallinarum bvG and bvP originating from different countries (Brazil, USA, China and India) including in house strain with host-promiscuous S. Enteritidis str. P125109 taken as reference genome to characterize their genomes and gain insights into pan genome, pathogenesis, mobiliome, resistome, and taxonomy.
For a comprehensive analysis in an open pan-genome, determination of minimum number of necessary strains is difficult [27]. The present analysis entails sufficient genotypic variability, as our selected eight genomes represent a single serovar Gallinarum of Salmonella enterica ssp. enterica, and the total SNP count in each genome varied from 7424 to 12,726 (Table 1). The pan-genome of investigated strains of S. Gallinarum comprises of 5091 CDS with a core-genome of 3270 CDS and an indispensible genome of 1254 CDS (Table 2). With a Rcp of 64%, the genomes depict a fairly high degree of genomic diversity and asymmetry [59].
The pan-genome and core-genome development plot analysis in the present investigation revealed S. Gallinarum to possess an open pan-genome state (Fig 2A and 2B). A steady growth with addition of new genomes is depicted in the pan-genome development plot, which indicates the capacity of this sympatric species to rapidly acquire exogenous DNA [60]. The core development plot becomes limited to about 2600 genes (Fig 2B). The acquisition of exogenous DNA poses obvious challenge for control strategies against FT/PD, however core-genome based antigen selection also offers insight for vaccine candidates. It is important to examine these core genes for reverse vaccinology application as their presence in all strains and high degree of conservation makes them effective source of potentially universal antigens [61]. Although the dispensable genes are strain-restricted, they can also be explored and exploited for immune-antigens [61] (Fig 3, S6B Table). Interestingly, strain specific genes were also detected in all the investigated genomes in the range of 3–102 (S5 Table). Furthermore, singleton development plot demonstrated the possibility of finding 43 new genes with each newly sequenced genome (Fig 2C). Indian strain S. Gallinarum Sal40 strain VTCCBAA614 harboured the highest number of singletons i.e. 102 whilst S. Enteritidis str. P125109 possessed 96 singletons (S5 Table). Whether the presence of large number of singletons in S. Gallinarum Sal40 strain VTCCBAA614 and S. Pullorum QJ-2D-Sal, in comparison to other investigated strains is related to higher acquisition of foreign genes requires to be investigated.
Lower core-genome to pan-genome ratio and presence of large number of singletons in S. Enteritidis str. P125109 and other investigated strains suggests the dynamic state of S. Gallinarum genome. Salmonella Gallinarum has undergone extensive degradation and also acquired specific genes related to virulence, non-virulence, metabolism, information storage by horizontal gene transfer which may contribute to its avian host specificity [10, 27, 62].
Studies on very large number of Salmonella enterica genomes such as one employing 4893 genomes detected a pan-genome of 25.3 Mbp, a strict core of 1.5 Mbp present in all genomes, and a conserved core of 3.2 Mbp found in at least 96% of these genomes [63]. This makes for a strong case for creating a database of serovar specific genome components such as for S. Gallinarum, a host specific pathogen.
Following core-genome identification, its functional analysis by employment of eggNOG-mapper v2revealed that large number of the CDS (20%) belonged to function unknown category (5923), which indicates that our understanding of genetic repertoire of Salmonella genome is limited. It is probable that a sizeable number of such CDS may be mutant phenotypes for protein-coding genes [64]. Otherwise the maximum genes were related to transcription, cell-wall and membrane biogenesis, energy metabolism, amino acid metabolism and transportation among others (Fig 3, S6A Table). The identified core-genome is also dominated by ATP-binding cassette (ABC) exporters which could be explored as novel drug targets [65]. On the other hand, the functional annotation of complete set of strain specific CDS of investigated strains fell in the categories of function unknown, replication and repair, and defence mechanism among others (Fig 3, S6B Table).
The core-genome based phylogeny (Fig 4) of S. Gallinarum biovars. from different geographical origins has clearly divided strains into divergent clades, and Indian strain was grouped externally in the S. Gallinarum clade, as in previous study [1]. Similarly, the phylogenetic relationship between S. Gallinarum biovars and S. Enteritidis has been genomically analysed, which highlighted the shared ancestry of bvP and bvG with S. Enteritidis descendent originating in a ‘second’ clade [8]. In a recent study, the core-genome SNP phylogeny of only bvP strains from China has divided it into four lineages [5], which suggests that the study of within biovar genomic epidemiology of Indian bvG strains will similarly need inclusion of more number of local strains. The genomic diversity within the investigated S. Gallinarum strains is supported by large scale genomic rearrangement events including inversions, deletions, relocations and duplications detected in synteny analysis (Fig 5, S1 Fig).
The evolution of host-adapted serovars of salmonellae has traversed a path of periodic gene acquisition and gene disruption as a mechanism of niche adaptation away from enteric location to systemic one [8]. These genetic elements contribute to the bacterial virulence, pathogenesis, fitness in specific niche, and evolution [8, 12]. Putative virulence factors in the investigated Salmonella genomes related to fimbrial adherence and secretion system among others were searched and analysed (S7A–S7C Table).
The fimbrial antigens play an important role in adherence of bacteria to the cell surface, which is essential to the pathogenesis of the disease, leading to bacterial invasion [66]. Although a number of fimbriae have been described in Salmonella, the chaperone-usher pathway represent the major fimbriae encoded by Salmonella [67–72].
The degradation in fimbrial operon in S. Gallinarum has been associated with departure from intestinal colonization [5]. In our investigation, the fimbrial operons Agf/Csg, bcf, lpf, peg, saf, stb, ste, stf, sth and sti were observed to be present in all of the S. Gallinarum genomes. Suez et al, (2013) [73] performed Comparative Genomic Hybridization (CGH) on 5656 Salmonella ORF representing 12 invasive Salmonella serovars including serovar Dublin and Cholerasuis, 2 host-associated serovars, and detected five fimbrial clusters (bcf, csg, stb, sth and sti) to be part of core genome for invasion and systemic disease in humans. In our study, among the S. Gallinarum genomes, chaperone-usher fimbrial gene operons, bcf, lpf, peg, saf, stb, ste, stf, sth and sti were detected to be part of the core genome. Notably, in the S. Gallinarum genomes studied, operon fim, sef, and std were predicted to be disrupted (S7C Table).
Hu et al, (2019) [5], reported std along with saf and csg to be intact in almost all the S. Pullorum strains in their study. On the other hand, we detected differential distribution of std operon in our study. The std operon, which was detected in S. Enteritidis str. P125109 was not detected in any of the 3 bvG, and 2 of the 5 bvP genomes studied (i.e., S. Gallinarum/pullorum str. CDC1983-67 and S. Gallinarum/pullorum str. RKS5078). In addition to std operon, the sef operon, sefA and D genes were not detected in S. Enteritidis str. P125109 and all other investigated genomes.SEF14 plays a role in the colonization of Peyer’s patches and in the adhesion and invasion of intestine epithelial cells [74–76].
The lpf, fim, and agf /csg operons, which were detected in all of our investigated genomes, encode fimbrial proteins which mediate attachment of S. enterica serotype Typhimurium in epithelial cell lines in vitro for long-term intestinal persistence [77]. The gene csgD of agf /csg operon activates the production of curli fimbriae by transcriptional activation of the csgBAC operon encoding the structural genes of curli fimbriae [78]. The csgA gene of agf /csgoperon has also been evidenced to be involved in biofilm formation in S. Pullorum, which may contribute to the virulence [79].
The type 1 fimbriae (T1F) are important virulence factors in Enterobactericeae including Salmonella spp., and is commonly expressed in virulent strains of Salmonella spp. [80, 81]. In our study, the fim operon was detected in all genomes except absence of fimA and I genes in S. Pullorum str. S06004 genome. The genes responsible for synthesis of T1F are fimI, fimC, fimD, fimH, and fimF, and fimA promoter region making a single operon [82].
In all the genomes studied, pegABCD operon was detected to be the part of core genome. The peg fimbrial gene cluster originally discovered in S. enterica subsp. enterica serovars Paratyphi A, Enteritidis, and Gallinarum [10], is considered to influence caecal colonisation of chickens by S. Enteritidis [83]. However, Hu et al, 2019 [5], found the peg operon, disrupted in all strains of S. Pullorum, suggesting their redundancy.
The Salmonella pathogenicity island 1 (SPI-1), encoding type III secretion system (T3SS) secreted effector genes, consisting of sopA, sopB, sopD, sopD2, sopE and sopE2 are involved in the inflammation and diahhorea [84]. sopA, sopB, sopD, sopE and sopE2 were detected in all genomes except in S. Gallinarum strain 9184. Among the Salmonella pathogenicity island II (SPI-II) encoding T3SS effectors (pipB2, sseG, sseI, ssek2, and ssaS), the sseI and ssek2 were detected in S. Enteritidis str. P125109, however, sseI was not detected in any bvG and bvP strains, and sseK2 was absent in 3 bvG (str. 287/91, str.9184, str. VTCCBAA614) and 1 bvP strain (str. CDC1983-67). The SPI-2 effectors (sseI, sifA, sseF, pipB2, spiC, sspH2, and slrP) have been shown to play a role in inflammation, particularly in inhibition of dendritic cell migration from intestinal environment [85]. Recently, it has been demonstrated that pseudogenization of sseI in sequence type 313 (ST313) Salmonella Typhimurium has rendered it invasive in sub-Saharan African region human subjects [86]. Our BLAST results clearly show the genes to be truncated in all bvG/bvP strains studied (S7B Table). sseI is a also a pseudogene in S. Enteritidis isolates which cause systemic infection within immunocompromised hosts [87].
We next focussed our investigation on identification and analysis of candidate virulence factors that were previously not identified in the genomes of the analyzed strains which included genomic islands, prophages, TA cassettes and AMR genes.
The acquisition of virulence capability enhancement en masse by acquisition of SPI has been hall mark of S. Gallinarum [17, 88]. A total of 113 SPI homologs with nearly 15 SPIs per genome were detected by combinatorial usage of SPIfinder and BLAST searching (S8A and S8B Table), which uncovered the presence of homologs of SP-1, SP-2, SP-3, SP-4, SPI-5, SPI-12, SPI-13, SP-14, C63PI and SPCS54 island in the investigated genomes (S8A and S8B Table). The presence of particular SPIs in each of the genomes indicates their specificity towards S. Gallinarum and S. Enteritidis and thus corroborates with previously published reports of SPIs showing serovar specificity [44, 89, 90].Notably, the role of SPI’s other than SP1 and SP2 in virulence and pathology of S. Gallinarum infections has not been analyzed in detail. The elucidation of functional role of other SPIs thus requires further experimental investigations with functional knockout mutations or via targeting and deletion of SPIs by utilization of CRISPR-Cas systems [91–94]. The limitation of manually screening mutant phenotypes in functional knockout mutation studies has led to use of high-thoroughput sequencing technologies in conjunction with transposon mutagenesis i.e., transposon sequencing (Tn-seq) [95, 96]. Transposon-directed insertion site sequencing (TraDIS) was used to study the mechanisms used by Salmonella to enter and persist within the bovine lymphatic system in comparison to intestinal colonization [97]. Several variations of Tn-seq have been devised which mainly differ in transposon junction sequence amplification techniques, which can be applied to obtain global gene functional information in Salmonella.
Notably, SPI-12 (NC_006905_P4, 11.1 kb), and SPI-14 (AY956836, 0.4kb) were not detected in S. Gallinarum Sal40 strain VTCCBAA614 and S. Gallinarum str. 9184, respectively. SPI-12 has been shown to be crucial for bacterial survival in the host [98]. SPI-14 has been recently identified as being involved in Salmonella intestinal survival and invasion via activation of SPI-1 genes [99].
Prophages contribute to pathogenicity, bacterial fitness, diversity, resistance to phages, antimicrobial resistance, evolution as well as aid in increased environmental tolerance [20, 100, 101]. In our study, the intact prophages were detected in the range of 1–3 with the highest number in bvP QJ-2D-Sal and bvP str. S06004. As in our study, a recent study also detected only a single phage in all of their bvG genomes, which is less than 5±3 prophage regions per genome detected in 1,760 S. enterica genomes, using Phaster [102]. However the observed prophage diversity in our study is in contrast with lower diversity reported by [103], using 2 strains of S. Gallinarum. The gifsy_2 prophage element which was detected in all genomes analysed in this study was earlier detected predominantly in S. Typhimurium genome [102]. Significantly prophage element ST104 detected in bvP QJ-2D-Sal has been previously reported in MDR S. Typhimurium DT104 as phage ST104. Notably, prophage and plasmids acquisition has been demonstrated in S. Pullorum leading to emergence of multi-drug resistant (MDR) strains [5, 104]. Our study also highlights variable number of propahges detected in bvP as compared to single prophage element detected in bvG. Hu et al, 2019 [5] also detected multiple prophage elements in their bvP genomes, which leads to high genetic diversity besides AMR. The differential presence of prophages in the current study indicates the ability of S. Gallinarum genomes to acquire new gene content, and introduce diversity in them which is in accordance with the singleton development plot that depicted the finding of 43 new genes with each newly sequenced genome. This is further refurbished by detection of holin, phage-like protein, phage membrane protein, side tail fibre protein genes among 104 singletons detected in S. Gallinarum Sal40 strain by EDGAR (S4I Table)). The differential distribution of prophages in the current study can be utilized as markers for strain characterization, diversity assessment, tracking of strains, determination of transmission history of S. enterica Gallinarum strains as phage typing has been widely employed by various investigators for these goals in various bacterial species [21, 105–109]. Moreover, cryptic prophages identified in the study can be utilized as drug target after determination of their role and essentiality status in the genome as they are permanent resident of microbial genomes [110].
In order to elucidate the role of prophages of S. Gallinarum, prophage induction may be achieved using the SOS response activated by chemicals such as mitomycin C, hydrogen peroxide and UV radiation, which cause DNA damage, apart from instances of spontaneous induction, and inflammation [111–113]. Induced prophages can be further studied by genomic sequencing and bioinformatic methods [112]. Further, specific prophage methylation pattern and repertoire of methyltransferase motif estimation can give useful insights on prophage gene expression [114]. Nothwithstanding the widespread repression of prophage genes, transcriptomics data analysis of propahge regions can be an important way forward [112, 115].
Toxin-antitoxin operons which are implicated in pathogenicity, virulence, persistence, stress endurance, and antibiotic resistance in bacteria and archae were searched for in the investigated Salmonella genomes [116–118]. The computational search revealed a total of 149 TA cassettes in the range of 16–18 in the investigated S. Gallinarum genomes (S10 Table). Significantly, a majority of the toxins of identified TA systems belonged to relElike family and highest number of identified antitoxins possessed RHH like domain (S10 Table, Fig 6). The presence of 149 Toxin antitoxin operons in the range of 16–18 in each of the analyzed S. Gallinarum genomes indicates their possible role in growth regulation, niche adaptation, encountering of various stress responses inside macrophage as well as in persistence, as has been observed in various bacteria and archaea including S. Typhimurium LT2, [24, 117, 119]. Moreover, the biological significance of presence of large number of TA’s may be due to their potential involvement in number of underlying regulatory pathways, as can be inferred by identification of different domains such as GNATlike_domain (24 nos.), MazFlike_domain (9 nos.), COG2929 like_domain (9 nos.), yeeU (8 nos.), doc (5 nos.), and PINlike_domain (3 nos.) in the structure of the toxins detected in the study. These TA domains have been previously reported to be associated with distinct features and different functional roles [117, 120]. As the acquisition of TA operons can be either through horizontal gene transfer or gene amplification [121], therefore the diversity of TA operons observed in Salmonella genomes reemphasizes the flexible nature of S. Gallinarum genome. Elucidation of their role requires further experimental analysis to be carried out.
The in silico identified TA systems also requires to be experimentally characterized before considering them as bonafide TA operons by designing assays to test whether toxicity of over-expressed toxin protein of TA pair is abrogated by over-expression of putative antitoxin protein in heterologous systems [122–124]. Moreover, experimental investigation is required to decode their role in pathogenesis. In addition, to knock out studies a number of powerful genetic tools can be employed to elucidate the function of detected virulence factors. Ectopic expression of TA systems can be studied to decipher their role in growth regulation [125, 126]. In addition, site directed mutagenesis can be utilized for detection of residues crucial for toxin activity for functional characterization of TA systems [127, 128]. Moreover, expression profile of identified TA modules in response to various stresses at multiple time points can be detected by using whole genome microarray platform comprising of TA loci probes and that could lead to co-expressed genes/pathways [129]. The Salmonella TA systems can be considered as potent targets for structure based inhibitor design as has been previously employed against various other bacteria [130, 131].
Although plasmids are important carriers of AMR genes in Salmonella serovars, [132], we detected the presence of acquired aminoglycoside resistance gene (aac(6’)-Iaa) in all the investigated Salmonella genomes (S3 Fig, S11A Table). Neuert et al., 2018 [133] found in their study on Non-typhoidal Salmonella (NTS), that nearly all but eight of the total 3,491 isolates carried an aminoglycoside acetyltransferase aac(6′)-type gene. However, only eleven showed phenotypic resistance to an aminoglycoside antimicrobial. In another study, the genes associated with ant and aph were found in all aminoglycoside-resistant Salmonella isolates, but aac genes were only found in Salmonella ser. Typhimurium and Salmonella ser. Heidelberg from chicken-sourced isolates [134]. The observation is in concordance with previously published reports stating that aminoglycosides exhibit weak bactericidal activity against intracellular S. enterica serovars [135]. Strikingly, known mutation in gyrA were also observed in four genomes out of the nine investigated i.e., S. Gallinarum str. 287/91, S. Gallinarum Sal40 strain VTCCBAA614, S. Pullorum str. S06004 and S. Pullorum QJ-2D-Sal (S11B Table). Koerich et al (2018) [58] also reported high resistance to drugs from macrolide and quinolone groups in Salmonella Gallinarum field isolates. Literature mining has revealed that mutations in gyrA are associated with resistance to fluoroquinolones in eight species of Enterobacteriaceae [136]. Chromosonal point mutations in rrsD gene in the helix 34 region, which results in changes to the drug-binding site in the ribosomal 16S rRNA has been reported to confer resistance against aminoglycoside spectinomycin of Salmonella enterica serovar Typhimurium gene [137, 138]. Workers have also reported mutations in rrsD genes in 2 S. enterica isolates from cattle and chicken in South Africa [139].
Antimicrobial resistance in Salmonella serovars is a serious poultry husbandry and public health problem all over the world including Asian, and South American region [58, 140, 141]. The observation of variable Antimicrobial resistant genes in the study can be utilized as AMR markers. The presence of additional AMR genes in S. enterica Gallinarum str. Sal40 and S. enterica Pullorum str. S06004 indicates the ability of acquiring exogenous DNA for adaptation in response to environmental requirements. The differential presence underlines the need of NGS based mapping of AMR genes in S. Gallinarum serovars to better manage their control measures [142].
A number of investigators have previously carried out comparative genome analysis of S. Gallinarum with several other serovars which includes S. Enteritidis, S. Typhimurium to decode the genomic differences amongst them. The comparative genome analysis of S. Enteritidis PT4, S. Typhimurium LT2 and S. Gallinarum 287/91 was carried by Thomson et al., (2008) [10] wherein they reported predominant similarity and synteny in their core genomes and overrepresentation of putative pseudogenes in S. Gallinarum 287/91 in comparison to S. Enteritidis str. P125109. Similarly, a genomic comparison between S. Gallinarum and S. Pullorum was carried out by Feng and associates [143] and they included S. Gallinarum str. CDC1983-67, S. Gallinarum str. RKS5078, S. Gallinarum str. 287/91, and S. Enteritidis str. P125109 and reported a high number and differential distribution of pseudogenes in bvG and bvP strains in reference to S. Enteritidis strain. On the other hand, genome comparison between closely related S. enterica serovars Enteritidis, Dublin, and Gallinarum revealed differential distribution of prophages and pseudogenes in their genomes along besides SNPs, insertions, and deletions amongst the investigated strains [103]. In a similar manner, to decode different host-specificities, genes related to SPI of S. Enteritidis PT4 (NCTC 13349) and S. Gallinarum 287/91 NCTC 13346 were compared by Eswarappa et al. 2009 [144] and detected 24 positively selected genes that included SPI-2 TTSS and effector proteins of SPI-1 TTSS.
The present study suggests that the host restricted S. Gallinarum strains harbour strain specific genes and they exhibit differential distribution of putative virulence factors such as genomic islands, prophage regions, TA cassettes, and acquired AMR genes in their genomes. This study also highlights the need to analyse more number of S. Gallinarum genomes to understand the phylogeny and better capture of its pan-genome biodiversity. This comparative genomic analysis of S. Gallinarum serovar has provided a valuable insight and laid foundation for future experimental studies to be carried out to decipher the underlying mechanisms driving the pathogenesis and virulence of this avian restricted pathogen.
5. Conclusion
Salmonella enterica serovar Gallinarum biovar Pullorum (bvP) and biovar Gallinarum (bvG), are the causative agents of disease (PD) and fowl typhoid (FT) respectively, which causes considerable economic losses to poultry industry worldwide especially in developing countries including India. Comprehensive comparative genome analysis of eight S. enterica serovar Gallinarum strains originating from different geographical regions including Indian strain S. Gallinarum Sal40 VTCCBAA614 was carried out to decode the genotypic differences amongst them with a focus on detection and analysis of candidate virulence factors. The investigation revealed an open pan-genome for S. Gallinarum encompassing 5091 coding sequence (CDS) with 3270 CDS belonging to core-genome, 1254 CDS to dispensable genome and strain specific genes amongst the analyzed strains. Furthermore, analysis of distribution of candidate virulence factors in the investigated strains revealed diversity and differential distribution of genomic features such as genomic islands, prophage regions, toxin-antitoxin operons, and acquired antimicrobial resistance genes. The fimbrial operons Agf/Csg, bcf, lpf, peg, saf, stb, ste, stf, sth and sti were observed to be present in all of the S. Gallinarum genomes, whereas operon sef, and std showed differential distribution. The computational search unravelled the existence of high sequence identity genomic islands SP-1, SP-2, SP-3, SP-4, SPI-5, SPI-12, SPI-13, SP-14, C63PI and SPCS54 in their genomes. Additionally, 23 prophage regions and 149 Type II TA loci were also identified and characterized in the investigated genomes. The genomic variability detected among the S. enterica serovar Gallinarum strains will form the basis for future experimental investigations and could be used for bacterial typing. The core genome assignment can be utilised towards design of diagnostics, as well as drug and vaccine target prediction for effective surveillance, prevention, and control.
Supporting information
(a-h) Synteny plot analysis of investigated Salmonella strains with reference to S. Enteritidis str. P125109. Depicts synteny plot analysis of investigated Salmonella strains.
(TIFF)
Details of prophages detected.
(TIF)
Detects acquired antimicrobial resistance genes.
(TIF)
Pan-genome and core genome development projections.
(DOCX)
Quality features of analyzed genomes.
(XLSX)
Pan genome CDS list of genomes.
(XLSX)
Core genome CDS list of genomes.
(XLSX)
(a to i) Description of singletons detected in investigated Salmonella Gallinarum (a) S. Enteritidis str. P125109 (b) S. Gallinarum/pullorum str. CDC1983-67 (c) S. Gallinarum/pullorum str. RKS5078 (d) S. Gallinarum str. 287/91 (e) S. Gallinarum str. 9184 (f) S. Pullorum str. ATCC 9120 (g) S. Pullorum str. S06004 (h) S. Pullorum QJ-2D-Sal (i) S. Gallinarum Sal40 strains by EDGAR with their name and function with reference to S. Enteritidis str. P125109. Detected Singletons listing of all genomes.
(XLSX)
(a-b) The functional annotation of (a) Suppl Table 6 (a): This data sheet consists of functional annotation of core genome of analyzed Salmonella strains performed by eggNOG-mapper v2 and lists query, seed ortholog, e-value, score, best taxonomy level, preferred name, GO terms, EC number, annotation level, COG category and description. (b) Suppl Table 6 (b): This data sheet consists of functional annotation of strain specific coding sequences of analyzed Salmonella strains performed by eggNOG-mapper v2. Functional annotation of CDS of analysed genomes.
(XLSX)
a. The table list virulence factor (VF) identified by VFDB database in the investigated Salmonella genomes. b. Details of virulence factor (VF) identified by BLASTN searches showing differential distribution in VFDB results in the investigated Salmonella genomes. c. Consolidated details of virulence factor (VF) identified by VF analyzer tool and BLASTN searches in the investigated Salmonella genomes. Virulence genes.
(XLSX)
(a-b) The datasheet lists homologs of Salmonella pathogenic islands (SPIs) detected by (a) SPIFinder 1.0 and (b) BLAST search, in the investigated Salmonella genomes. Listing of Salmonella pathogenicity island detected by SPI Finder and by BLAST search.
(XLSX)
(a-b) The identified (a) complete list of prophage regions and (b) intact prophages, in the genomes of nine Salmonella strains that were investigated in the study with description of a) region number b) region length c) completeness d) score e) total proteins f) region position g) most common phage and h) GC%. The colour coding of the rows represents the classification of identified prophage regions by PHASTER, green- intact prophage region, red- incomplete prophage region and blue- questionable prophage region. Description of prophages detected in all genomes with details.
(XLSX)
TA gene listing detected in all genomes.
(XLSX)
(a-c) This datasheet details (a) acquired antimicrobial resistance genes, (b) known chromosomal mutations, and (c) unknown chromosomal mutations detected in investigated Salmonella strains by Resfinder. Details of AMR genes detected in genomes.
(XLSX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
This work was partly supported by Department of Biotechnology, Govt. of India, New Delhi (Garnt No. BT/BI/25/07/2012-BIF) (URL: http://dbtindia.gov.in/) award to RKV and Director, National Research Centre on Equines, National Centre for Veterinary Type Cultures, Hisar, Haryana, India. ZT acknowledges DBT “Bioinformatics infrastructure facility (BIF)” for providing Research Associateship. Funding source(s) had no involvement if any, in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the article preparation or decision to submit the article for publication.
References
- 1.De Carli S, Gräf T, Kipper D, Lehmann FK, Zanetti N, Siqueira FM, et al. Molecular and phylogenetic analyses of Salmonella Gallinarum trace the origin and diversification of recent outbreaks of fowl typhoid in poultry farms. Veterinary microbiology. 2017Dec1;212:80–6. doi: 10.1016/j.vetmic.2017.11.001 [DOI] [PubMed] [Google Scholar]
- 2.Alves Batista DF, de Freitas Neto OC, Maria de Almeida A, Maboni G, de Carvalho TF, de Carvalho TP, et al. Evaluation of pathogenicity of Salmonella Gallinarum strains harbouring deletions in genes whose orthologues are conserved pseudogenes in S. Pullorum. PloS one. 2018Jul20;13(7):e0200585. doi: 10.1371/journal.pone.0200585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wilson RL, Elthon J, Clegg S, Jones BD. Salmonella enterica serovars Gallinarum and Pullorum expressing Salmonella enterica serovar Typhimurium type 1 fimbriae exhibit increased invasiveness for mammalian cells. Infection and immunity. 2000Aug1;68(8):4782–5. doi: 10.1128/IAI.68.8.4782-4785.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wigley P, Berchieri A Jr, Page KL, Smith AL, Barrow PA. Salmonella enterica serovar Pullorum persists in splenic macrophages and in the reproductive tract during persistent, disease-free carriage in chickens. Infection and immunity. 2001Dec1;69(12):7873–9. doi: 10.1128/IAI.69.12.7873-7879.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu Y, Wang Z, Qiang B, Xu Y, Chen X, Li Q, et al. Loss and gain in the evolution of the Salmonella enterica serovar Gallinarum biovar Pullorum genome. Msphere. 2019Apr3;4(2):e00627–18. doi: 10.1128/mSphere.00627-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo R, Li Z, Zhou X, Huang C, Hu Y, Geng S, et al. Induction of arthritis in chickens by infection with novel virulent Salmonella Pullorum strains. Veterinary microbiology. 2019Jan1;228:165–72. doi: 10.1016/j.vetmic.2018.11.032 [DOI] [PubMed] [Google Scholar]
- 7.Barrow PA, Neto OF. Pullorum disease and fowl typhoid—new thoughts on old diseases: a review. Avian pathology. 2011Feb1;40(1):1–3. doi: 10.1080/03079457.2010.542575 [DOI] [PubMed] [Google Scholar]
- 8.Langridge GC, Fookes M, Connor TR, Feltwell T, Feasey N, Parsons BN, et al. Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proceedings of the National Academy of Sciences. 2015Jan20;112(3):863–8. doi: 10.1073/pnas.1416707112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moutaouakkil Y, Frikh M, Maleb A, Lemnouer A, Elouennass M. Infection caused by Salmonella Gallinarum emergent bacteria. Saudi J Health Sci 2017;6:68–70 [Google Scholar]
- 10.Thomson NR, Clayton DJ, Windhorst D, Vernikos G, Davidson S, Churcher C, et al. Comparative genome analysis of Salmonella Enteritidis PT4 and Salmonella Gallinarum 287/91 provides insights into evolutionary and host adaptation pathways. Genome research. 2008Oct1;18(10):1624–37. doi: 10.1101/gr.077404.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uzzau S, Leori GS, Petruzzi V, Watson PR, Schianchi G, Bacciu D, et al. Salmonella enterica serovar-host specificity does not correlate with the magnitude of intestinal invasion in sheep. Infection and immunity. 2001May1;69(5):3092–9. doi: 10.1128/IAI.69.5.3092-3099.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ilyas B, Tsai CN, Coombes BK. Evolution of Salmonella-host cell interactions through a dynamic bacterial genome. Frontiers in cellular and infection microbiology. 2017Sep29;7:428. doi: 10.3389/fcimb.2017.00428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z, Qin Y, Wang Y, Li X, Cao H, Zheng SJ. A Critical Role of Bacterioferritin in Salmonella pullorum-Induced IFN-β Expression in DF-1 Cells. Frontiers in microbiology. 2016Feb3;7:20. doi: 10.3389/fmicb.2016.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moreno Switt AI, den Bakker HC, Cummings CA, Rodriguez-Rivera LD, Govoni G, Raneiri ML, et al. Identification and characterization of novel Salmonella mobile elements involved in the dissemination of genes linked to virulence and transmission. PloS one. 2012Jul20;7(7):e41247. doi: 10.1371/journal.pone.0041247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haraga A, Ohlson MB, Miller SI. Salmonellae interplay with host cells. Nature Reviews Microbiology. 2008Jan;6(1):53–66. doi: 10.1038/nrmicro1788 [DOI] [PubMed] [Google Scholar]
- 16.Jantsch J, Chikkaballi D, Hensel M. Cellular aspects of immunity to intracellular Salmonella enterica. Immunological reviews. 2011Mar;240(1):185–95. doi: 10.1111/j.1600-065X.2010.00981.x [DOI] [PubMed] [Google Scholar]
- 17.Blondel CJ, Jiménez JC, Contreras I, Santiviago CA. Comparative genomic analysis uncovers 3 novel loci encoding type six secretion systems differentially distributed in Salmonella serotypes. BMC genomics. 2009Dec;10(1):1–7. doi: 10.1186/1471-2164-10-354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Figueroa‐Bossi N, Uzzau S, Maloriol D, Bossi L. Variable assortment of prophages provides a transferable repertoire of pathogenic determinants in Salmonella. Molecular microbiology. 2001Jan;39(2):260–72. doi: 10.1046/j.1365-2958.2001.02234.x [DOI] [PubMed] [Google Scholar]
- 19.Thomson N, Baker S, Pickard D, Fookes M, Anjum M, Hamlin N, et al. The role of prophage-like elements in the diversity of Salmonella enterica serovars. Journal of molecular biology. 2004May28;339(2):279–300. doi: 10.1016/j.jmb.2004.03.058 [DOI] [PubMed] [Google Scholar]
- 20.Bruüssow H, Canchaya C, Hardt WD. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiology and molecular biology reviews. 2004Sep1;68(3):560–602. doi: 10.1128/MMBR.68.3.560-602.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colavecchio A, D’Souza Y, Tompkins E, Jeukens J, Freschi L, Emond-Rheault JG, et al. Prophage integrase typing is a useful indicator of genomic diversity in Salmonella enterica. Frontiers in microbiology. 2017Jul10;8:1283. doi: 10.3389/fmicb.2017.01283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huguet KT, Gonnet M, Doublet B, Cloeckaert A. A toxin antitoxin system promotes the maintenance of the IncA/C-mobilizable Salmonella Genomic Island 1. Scientific reports. 2016Aug31;6(1):1–0. doi: 10.1038/s41598-016-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Cesare A, Losasso C, Barco L, Eckert EM, Conficoni D, Sarasini G, et al. Diverse distribution of Toxin-Antitoxin II systems in Salmonella enterica serovars. Scientific reports. 2016Jun30;6(1):1–9. doi: 10.1038/s41598-016-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.VanDrisse CM, Parks AR, Escalante-Semerena JC. A toxin involved in Salmonella persistence regulates its activity by acetylating its cognate antitoxin, a modification reversed by CobB sirtuin deacetylase. MBio. 2017May30;8(3):e00708–17. doi: 10.1128/mBio.00708-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan Z, Wang X, Zhang X, Geng S, Chen X, Pan W, et al. Changes in antimicrobial resistance among Salmonella enterica subspecies enterica serovar Pullorum isolates in China from 1962 to 2007. Veterinary microbiology. 2009May12;136(3–4):387–92. doi: 10.1016/j.vetmic.2008.11.015 [DOI] [PubMed] [Google Scholar]
- 26.Jain P, Chowdhury G, Samajpati S, Basak S, Ganai A, Samanta S, et al. Characterization of non-typhoidal Salmonella isolates from children with acute gastroenteritis, Kolkata, India, during 2000–2016. Brazilian Journal of Microbiology. 2020Jun;51(2):613–27. doi: 10.1007/s42770-019-00213-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rouli L, Merhej V, Fournier PE, Raoult D. The bacterial pangenome as a new tool for analysing pathogenic bacteria. New Microbes New Infect 20157: 72–85. doi: 10.1016/j.nmni.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sahl JW, Gillece JD, Schupp JM, Waddell VG, Driebe EM, Engelthaler DM, et al. Evolution of a pathogen: a comparative genomics analysis identifies a genetic pathway to pathogenesis in Acinetobacter. PloS one. 2013Jan24;8(1):e54287. doi: 10.1371/journal.pone.0054287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu QF, Cao DM, Su LL, Li SB, Ye GB, Zhu XY, et al. Genus-wide comparative genomics analysis of Neisseria to identify new genes associated with pathogenicity and niche adaptation of Neisseria pathogens. International journal of genomics. 2019Jan15;2019. doi: 10.1155/2019/6015730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rasko DA, Rosovitz MJ. Myers GSA, Mongodin EF, Fricke WF, Gajer P, et al. The Pangenome Structure of Escherichia coli: Comparative Genomic Analysis of E. coli Commensal and Pathogenic Isolates. J Bacteriol. 2008;190(20):6881. doi: 10.1128/JB.00619-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Armstrong J, Fiddes IT, Diekhans M, Paten B. Whole-genome alignment and comparative annotation. Annual review of animal biosciences. 2019Feb15;7:41–64. doi: 10.1146/annurev-animal-020518-115005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaid RK, Jindal N, Anand T, Bera BC, Riyesh T, Virmani N, et al. First draft genome sequence of Salmonella enterica serovar Gallinarum strain VTCCBAA614, isolated from chicken in India. Genome announcements. 2015Oct22;3(5):e01221–15. doi: 10.1128/genomeA.01221-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014Jul15;30(14):2068–9. doi: 10.1093/bioinformatics/btu153 [DOI] [PubMed] [Google Scholar]
- 34.Yoon SH, Ha SM, Lim J, Kwon S, Chun J. A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek. 2017Oct;110(10):1281–6. doi: 10.1007/s10482-017-0844-4 [DOI] [PubMed] [Google Scholar]
- 35.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome biology. 2004Jan;5(2):1–9. doi: 10.1186/gb-2004-5-2-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blom J, Kreis J, Spänig S, Juhre T, Bertelli C, Ernst C, et al. EDGAR 2.0: an enhanced software platform for comparative gene content analyses. Nucleic acids research. 2016Jul8;44(W1):W22–8. doi: 10.1093/nar/gkw255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proceedings of the National Academy of Sciences. 2005Sep27;102(39):13950–5. doi: 10.1073/pnas.0506758102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlos Guimaraes L, Benevides de Jesus L, Vinicius Canario Viana M, Silva A, Thiago Juca Ramos R, de Castro Soares S, et al. Inside the pan-genome-methods and software overview. Current genomics. 2015Aug1;16(4):245–52. doi: 10.2174/1389202916666150423002311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, Von Mering C, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Molecular biology and evolution. 2017Aug1;34(8):2115–22. doi: 10.1093/molbev/msx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic acids research. 2019Jan8;47(D1):D309–14. doi: 10.1093/nar/gky1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC bioinformatics. 2004Dec;5(1):1–9. doi: 10.1186/1471-2105-5-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Systematic biology. 2002May1;51(3):492–508. doi: 10.1080/10635150290069913 [DOI] [PubMed] [Google Scholar]
- 43.Liu B, Zheng D, Jin Q, Chen L, Yang J. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic acids research. 2019Jan8;47(D1):D687–92. doi: 10.1093/nar/gky1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roer L, Hendriksen RS, Leekitcharoenphon P, Lukjancenko O, Kaas RS, Hasman H, et al. Is the evolution of Salmonella enterica subsp. enterica linked to restriction-modification systems?. Msystems. 2016May1;1(3):e00009–16. doi: 10.1128/mSystems.00009-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoon SH, Hur CG, Kang HY, Kim YH, Oh TK, Kim JF. A computational approach for identifying pathogenicity islands in prokaryotic genomes. Bmc Bioinformatics. 2005Dec;6(1):1–1. doi: 10.1186/1471-2105-6-184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon SH, Park YK, Lee S, Choi D, Oh TK, Hur CG, et al. Towards pathogenomics: a web-based resource for pathogenicity islands. Nucleic acids research. 2007Jan1;35(suppl_1):D395–400. doi: 10.1093/nar/gkl790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, et al. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic acids research. 2016May3;44(W1):W16–21. doi: 10.1093/nar/gkw387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou Y, Liang Y, Lynch KH, Dennis JJ, Wishart DS. PHAST: a fast phage search tool. Nucleic acids research. 2011Jun14;39(suppl_2):W347–52. doi: 10.1093/nar/gkr485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makarova KS, Wolf YI, Koonin EV. Comprehensive comparative-genomic analysis of type 2 toxin-antitoxin systems and related mobile stress response systems in prokaryotes. Biology direct. 2009Dec;4(1):1–38. doi: 10.1186/1745-6150-4-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, et al. Identification of acquired antimicrobial resistance genes. Journal of antimicrobial chemotherapy. 2012Nov1;67(11):2640–4. doi: 10.1093/jac/dks261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jajere SM. A review of Salmonella enterica with particular focus on the pathogenicity and virulence factors, host specificity and antimicrobial resistance including multidrug resistance. Veterinary world. 2019;12(4):504. doi: 10.14202/vetworld.2019.504-521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng RA, Eade CR, Wiedmann M. Embracing diversity: differences in virulence mechanisms, disease severity, and host adaptations contribute to the success of nontyphoidal Salmonella as a foodborne pathogen. Frontiers in microbiology. 2019Jun26;10:1368. doi: 10.3389/fmicb.2019.01368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shivaprasad HL. Fowl typhoid and pullorum disease. Revue scientifique et technique (International Office of Epizootics). 2000Aug1;19(2):405–24. doi: 10.20506/rst.19.2.1222 [DOI] [PubMed] [Google Scholar]
- 54.Rajagopal R, Mini M. Outbreaks of salmonellosis in three different poultry farms of Kerala, India. Asian Pacific journal of tropical biomedicine. 2013Jun1;3(6):496–500. doi: 10.1016/S2221-1691(13)60103-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mastroeni P, Chabalgoity JA, Dunstan SJ, Maskell DJ, Dougan G. Salmonella: immune responses and vaccines. The Veterinary Journal. 2001Mar1;161(2):132–64. doi: 10.1053/tvjl.2000.0502 [DOI] [PubMed] [Google Scholar]
- 56.Silva EN, Snoeyenbos GH, Weinack OM, Smyser CF. Studies on the use of 9R strain of Salmonella Gallinarum as a vaccine in chickens. Avian diseases. 1981Jan1:38–52. [PubMed] [Google Scholar]
- 57.Kwon HJ, Cho SH. Pathogenicity of SG 9R, a rough vaccine strain against fowl typhoid. Vaccine. 2011Feb1;29(6):1311–8. doi: 10.1016/j.vaccine.2010.11.067 [DOI] [PubMed] [Google Scholar]
- 58.Koerich PK, Fonseca BB, Balestrin E, Tagliari V, Hoepers PG, Ueira-Vieira C, et al. Salmonella Gallinarum field isolates and its relationship to vaccine strain SG9R. British poultry science. 2018Mar4;59(2):154–9. doi: 10.1080/00071668.2017.1406062 [DOI] [PubMed] [Google Scholar]
- 59.Ghatak S, Blom J, Das S, Sanjukta R, Puro K, Mawlong M, et al. Pan-genome analysis of Aeromonas hydrophila, Aeromonas veronii and Aeromonas caviae indicates phylogenomic diversity and greater pathogenic potential for Aeromonas hydrophila. Antonie Van Leeuwenhoek. 2016Jul;109(7):945–56. doi: 10.1007/s10482-016-0693-6 [DOI] [PubMed] [Google Scholar]
- 60.Medini D, Donati C, Tettelin H, Masignani V, Rappuoli R. The microbial pan-genome. Current opinion in genetics & development. 2005Dec1;15(6):589–94. [DOI] [PubMed] [Google Scholar]
- 61.Rinaudo CD, Telford JL, Rappuoli R, Seib KL. Vaccinology in the genome era. The Journal of clinical investigation. 2009Sep1;119(9):2515–25. doi: 10.1172/JCI38330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bacciu D, Falchi G, Spazziani A, Bossi L, Marogna G, Leori GS, et al. Transposition of the heat-stable toxin astA gene into a gifsy-2-related prophage of Salmonella enterica serovar Abortusovis. Journal of bacteriology. 2004Jul15;186(14):4568–74. doi: 10.1128/JB.186.14.4568-4574.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Laing CR, Whiteside MD, Gannon VP. Pan-genome analyses of the species Salmonella enterica, and identification of genomic markers predictive for species, subspecies, and serovar. Frontiers in microbiology. 2017Jul31;8:1345. doi: 10.3389/fmicb.2017.01345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Price MN, Wetmore KM, Waters RJ, Callaghan M, Ray J, Liu H, et al. Mutant phenotypes for thousands of bacterial genes of unknown function. Nature. 2018May;557(7706):503–9. doi: 10.1038/s41586-018-0124-0 [DOI] [PubMed] [Google Scholar]
- 65.Rempel S, Gati C, Nijland M, Thangaratnarajah C, Karyolaimos A, de Gier JW, et al. A mycobacterial ABC transporter mediates the uptake of hydrophilic compounds. Nature. 2020Apr;580(7803):409–12. doi: 10.1038/s41586-020-2072-8 [DOI] [PubMed] [Google Scholar]
- 66.Finlay BB, Falkow S. Common themes in microbial pathogenicity. Microbiological reviews. 1989Jun;53(2):210–30. doi: 10.1128/mr.53.2.210-230.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nuccio SP, Baumler AJ. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiology and Molecular Biology Reviews. 2007Dec;71(4):551–75. doi: 10.1128/MMBR.00014-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yue M, Rankin SC, Blanchet RT, Nulton JD, Edwards RA, Schifferli DM. Diversification of the Salmonella fimbriae: a model of macro-and microevolution. PloS one. 2012Jun12;7(6):e38596. doi: 10.1371/journal.pone.0038596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Desai PT, Porwollik S, Long F, Cheng P, Wollam A, Clifton SW, et al. Evolutionary genomics of Salmonella enterica subspecies. MBio. 2013Mar5;4(2):e00579–12. doi: 10.1128/mBio.00579-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aviv G, Elpers L, Mikhlin S, Cohen H, Vitman Zilber S, Grassl GA, et al. The plasmid-encoded Ipf and Klf fimbriae display different expression and varying roles in the virulence of Salmonella enterica serovar Infantis in mouse vs. avian hosts. PLoS pathogens. 2017Aug17;13(8):e1006559. doi: 10.1371/journal.ppat.1006559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng RA, Wiedmann M. Recent Advances in Our Understanding of the Diversity and Roles of Chaperone-Usher Fimbriae in Facilitating Salmonella Host and Tissue Tropism. Frontiers in Cellular and Infection Microbiology. 2021Feb3;10:920. doi: 10.3389/fcimb.2020.628043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rehman T, Yin L, Latif MB, Chen J, Wang K, Geng Y, et al. Adhesive mechanism of different Salmonella fimbrial adhesins. Microbial pathogenesis. 2019Dec1;137:103748. doi: 10.1016/j.micpath.2019.103748 [DOI] [PubMed] [Google Scholar]
- 73.Suez J, Porwollik S, Dagan A, Marzel A, Schorr YI, Desai PT, et al. Virulence gene profiling and pathogenicity characterization of non-typhoidal Salmonella accounted for invasive disease in humans. PloS one. 2013Mar7;8(3):e58449. doi: 10.1371/journal.pone.0058449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Feutrier JO, Kay WW, Trust TJ. Cloning and expression of a Salmonella enteritidis fimbrin gene in Escherichia coli. Journal of bacteriology. 1988Sep;170(9):4216–22. doi: 10.1128/jb.170.9.4216-4222.1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thiagarajan D, Thacker HL, Saeed AM. Experimental infection of laying hens with Salmonella enteritidis strains that express different types of fimbriae. Poultry science. 1996Nov1;75(11):1365–72. doi: 10.3382/ps.0751365 [DOI] [PubMed] [Google Scholar]
- 76.Ogunniyi AD, Kotlarski I, Morona R, Manning PA. Role of SefA subunit protein of SEF14 fimbriae in the pathogenesis of Salmonella enterica serovar Enteritidis. Infection and immunity. 1997Feb;65(2):708–17. doi: 10.1128/iai.65.2.708-717.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weening EH, Barker JD, Laarakker MC, Humphries AD, Tsolis RM, Bäumler AJ. The Salmonella enterica serotype Typhimurium lpf, bcf, stb, stc, std, and sth fimbrial operons are required for intestinal persistence in mice. Infection and immunity. 2005Jun;73(6):3358–66 doi: 10.1128/IAI.73.6.3358-3366.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Römling U, Bian Z, Hammar M, Sierralta WD, Normark S. Curli fibers are highly conserved between Salmonella Typhimurium and Escherichia coli with respect to operon structure and regulation. Journal of bacteriology. 1998Feb1;180(3):722–31. doi: 10.1128/JB.180.3.722-731.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.El Hag M, Feng Z, Su Y, Wang X, Yassin A, Chen S, et al. Contribution of the csgA and bcsA genes to Salmonella enterica serovar Pullorum biofilm formation and virulence. Avian pathology. 2017Sep3;46(5):541–7. doi: 10.1080/03079457.2017.1324198 [DOI] [PubMed] [Google Scholar]
- 80.Duguid JP, Anderson ES, Campbell I. Fimbriae and adhesive properties in Salmonellae. The Journal of pathology and bacteriology. 1966Jul;92(1):107–37. doi: 10.1002/path.1700920113 [DOI] [PubMed] [Google Scholar]
- 81.Werneburg GT, Thanassi DG. Pili assembled by the chaperone/usher pathway in Escherichia coli and Salmonella. EcoSal Plus. 2018Mar13;8(1). doi: 10.1128/ecosalplus.ESP-0007-2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hahn E, Wild P, Hermanns U, Sebbel P, Glockshuber R, Häner M, et al. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. Journal of molecular biology. 2002Nov8;323(5):845–57. doi: 10.1016/s0022-2836(02)01005-7 [DOI] [PubMed] [Google Scholar]
- 83.Clayton DJ, Bowen AJ, Hulme SD, Buckley AM, Deacon VL, Thomson NR, et al. Analysis of the role of 13 major fimbrial subunits in colonisation of the chicken intestines by Salmonella enterica serovar Enteritidis reveals a role for a novel locus. BMC microbiology. 2008Dec;8(1):1–5. doi: 10.1186/1471-2180-8-228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, Higashide W, Dai S, Sherman DM, Zhou D. Recognition and ubiquitination of Salmonella type III effector SopA by a ubiquitin E3 ligase, HsRMA1. Journal of Biological Chemistry. 2005Nov18;280(46):38682–8. doi: 10.1074/jbc.M506309200 [DOI] [PubMed] [Google Scholar]
- 85.McLaughlin LM, Xu H, Carden SE, Fisher S, Reyes M, Heilshorn SC, et al. A microfluidic-based genetic screen to identify microbial virulence factors that inhibit dendritic cell migration. Integrative Biology. 2014Apr1;6(4):438–49. doi: 10.1039/c3ib40177d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carden SE, Walker GT, Honeycutt J, Lugo K, Pham T, Jacobson A, et al. Pseudogenization of the secreted effector gene sseI confers rapid systemic dissemination of S. Typhimurium ST313 within migratory dendritic cells. Cell host & microbe. 2017Feb8;21(2):182–94. doi: 10.1016/j.chom.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Klemm EJ, Gkrania-Klotsas E, Hadfield J, Forbester JL, Harris SR, Hale C, et al. , Goulding D. Emergence of host-adapted Salmonella Enteritidis through rapid evolution in an immunocompromised host. Nature microbiology. 2016Jan25;1(3):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Groisman EA, Ochman H. Pathogenicity islands: bacterial evolution in quantum leaps. Cell. 1996Nov29;87(5):791–4. doi: 10.1016/s0092-8674(00)81985-6 [DOI] [PubMed] [Google Scholar]
- 89.Hensel M. Evolution of pathogenicity islands of Salmonella enterica. International Journal of Medical Microbiology. 2004Sep24;294(2–3):95–102. doi: 10.1016/j.ijmm.2004.06.025 [DOI] [PubMed] [Google Scholar]
- 90.Gerlach RG, Hensel M. Salmonella pathogenicity islands in host specificity, host pathogen-interactions and antibiotics resistance of Salmonella enterica. Berliner und Munchener tierarztliche Wochenschrift. 2007Jul1;120(7/8):317. [PubMed] [Google Scholar]
- 91.Jones MA, Wigley P, Page KL, Hulme SD, Barrow PA. Salmonella enterica serovar Gallinarum requires the Salmonella pathogenicity island 2 type III secretion system but not the Salmonella pathogenicity island 1 type III secretion system for virulence in chickens. Infection and immunity. 2001Sep1;69(9):5471–6. doi: 10.1128/IAI.69.9.5471-5476.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wigley P, Jones MA, Barrow PA. Salmonella enterica serovar Pullorum requires the Salmonella pathogenicity island 2 type III secretion system for virulence and carriage in the chicken. Avian Pathology. 2002Oct1;31(5):501–6. doi: 10.1080/0307945021000005879 [DOI] [PubMed] [Google Scholar]
- 93.Selle K, Barrangou R. Harnessing CRISPR–Cas systems for bacterial genome editing. Trends in microbiology. 2015Apr1;23(4):225–32. doi: 10.1016/j.tim.2015.01.008 [DOI] [PubMed] [Google Scholar]
- 94.Standage-Beier K, Zhang Q, Wang X. Targeted large-scale deletion of bacterial genomes using CRISPR-nickases. ACS synthetic biology. 2015Nov20;4(11):1217–25. doi: 10.1021/acssynbio.5b00132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barquist L, Langridge GC, Turner DJ, Phan MD, Turner AK, Bateman A, et al. A comparison of dense transposon insertion libraries in the Salmonella serovars Typhi and Typhimurium. Nucleic acids research. 2013Apr1;41(8):4549–64. doi: 10.1093/nar/gkt148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jayeola V, McClelland M, Porwollik S, Chu W, Farber J, Kathariou S. Identification of novel genes mediating survival of Salmonella on low-moisture foods via transposon sequencing analysis. Frontiers in microbiology. 2020May15;11:726. doi: 10.3389/fmicb.2020.00726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Vohra P, Chaudhuri RR, Mayho M, Vrettou C, Chintoan-Uta C, Thomson NR, et al. Retrospective application of transposon-directed insertion-site sequencing to investigate niche-specific virulence of Salmonella Typhimurium in cattle. BMC genomics. 2019Dec;20(1):1–2. doi: 10.1186/s12864-018-5379-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tomljenovic-Berube AM, Henriksbo B, Porwollik S, Cooper CA, Tuinema BR, McClelland M, et al. Mapping and regulation of genes within Salmonella pathogenicity island 12 that contribute to in vivo fitness of Salmonella enterica Serovar Typhimurium. Infection and immunity. 2013Jul;81(7):2394–404. doi: 10.1128/IAI.00067-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jiang L, Feng L, Yang B, Zhang W, Wang P, Jiang X, et al. Signal transduction pathway mediated by the novel regulator LoiA for low oxygen tension induced Salmonella Typhimurium invasion. PLoS pathogens. 2017Jun 2;13(6):e1006429. doi: 10.1371/journal.ppat.1006429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fu Y, Wu Y, Yuan Y, Gao M. Prevalence and diversity analysis of candidate prophages to provide an understanding on their roles in Bacillus thuringiensis. Viruses. 2019Apr;11(4):388. doi: 10.3390/v11040388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Colavecchio A, Cadieux B, Lo A, Goodridge LD. Bacteriophages contribute to the spread of antibiotic resistance genes among foodborne pathogens of the Enterobacteriaceae family–a review. Frontiers in Microbiology. 2017Jun20;8:1108. doi: 10.3389/fmicb.2017.01108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mottawea W, Duceppe MO, Dupras AA, Usongo V, Jeukens J, Freschi L, et al. Salmonella enterica prophage sequence profiles reflect genome diversity and can be used for high discrimination subtyping. Frontiers in microbiology. 2018May4;9:836. doi: 10.3389/fmicb.2018.00836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Matthews TD, Schmieder R, Silva GG, Busch J, Cassman N, Dutilh BE, et al. Genomic comparison of the closely-related Salmonella enterica serovars Enteritidis, Dublin and Gallinarum. PloS one. 2015Jun3;10(6):e0126883. doi: 10.1371/journal.pone.0126883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tanaka K, Nishimori K, Makino SI, Nishimori T, Kanno T, Ishihara R, et al. Molecular characterization of a prophage of Salmonella enterica serotype Typhimurium DT104. Journal of clinical microbiology. 2004Apr;42(4):1807–12. doi: 10.1128/JCM.42.4.1807-1812.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shan J, Patel KV, Hickenbotham PT, Nale JY, Hargreaves KR, et al. Prophage carriage and diversity within clinically relevant strains of Clostridium difficile. Applied and environmental microbiology. 2012Sep1;78(17):6027–34. doi: 10.1128/AEM.01311-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dai Z, Wu F, Zheng Z, Yokomi R, Kumagai L, Cai W, et al. Prophage diversity of ‘Candidatus Liberibacter asiaticus’ strains in California. Phytopathology. 2019Apr20;109(4):551–9. doi: 10.1094/PHYTO-06-18-0185-R [DOI] [PubMed] [Google Scholar]
- 107.Dini M, Shokoohizadeh L, Jalilian FA, Moradi A, Arabestani MR. Genotyping and characterization of prophage patterns in clinical isolates of Staphylococcus aureus. BMC research notes. 2019Dec;12(1):1–6. doi: 10.1186/s13104-018-4038-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Crestani C, Forde TL, Zadoks RN. Development and application of a prophage integrase typing scheme for group B Streptococcus. Frontiers in microbiology. 2020Aug31;11:1993. doi: 10.3389/fmicb.2020.01993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Uelze L, Grützke J, Borowiak M, Hammerl JA, Juraschek K, Deneke C, et al. Typing methods based on whole genome sequencing data. One Health Outlook. 2020Dec;2(1):1–9. doi: 10.1186/s42522-020-0010-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang X, Wood TK. Cryptic prophages as targets for drug development. Drug Resistance Updates. 2016Jul1;27:30–8. doi: 10.1016/j.drup.2016.06.001 [DOI] [PubMed] [Google Scholar]
- 111.Miura S, Tamamura Y, Takayasu M, Sasaki M, Nishimura N, Tokugawa K, et al. Influence of SOS-inducing agents on the expression of ArtAB toxin gene in Salmonella enterica and Salmonella bongori. Microbiology. 2020Aug1;166(8):785–93. doi: 10.1099/mic.0.000939 [DOI] [PubMed] [Google Scholar]
- 112.Owen SV, Wenner N, Canals R, Makumi A, Hammarlöf DL, Gordon MA, et al. Characterization of the prophage repertoire of African Salmonella Typhimurium ST313 reveals high levels of spontaneous induction of novel phage BTP1. Frontiers in microbiology. 2017Feb23;8:235. doi: 10.3389/fmicb.2017.00235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Diard M, Bakkeren E, Cornuault JK, Moor K, Hausmann A, Sellin ME, et al. Inflammation boosts bacteriophage transfer between Salmonella spp. Science. 2017Mar17;355(6330):1211–5. doi: 10.1126/science.aaf8451 [DOI] [PubMed] [Google Scholar]
- 114.Pirone-Davies C, Hoffmann M, Roberts RJ, Muruvanda T, Timme RE, Strain E, et al. Genome-wide methylation patterns in Salmonella enterica subsp. enterica serovars. PloS one. 2015Apr10;10(4):e0123639. doi: 10.1371/journal.pone.0123639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Colgan AM, Kröger C, Diard M, Hardt WD, Puente JL, Sivasankaran SK, et al. The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS genetics. 2016Aug26;12(8):e1006258. doi: 10.1371/journal.pgen.1006258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chimal-Cázares F, Hernández-Martínez G, Pacheco S, Ares MA, Soria-Bustos J, Sánchez-Gutiérrez M, et al. Molecular characterization of SehB, a type II antitoxin of Salmonella enterica serotype Typhimurium: amino acid residues involved in DNA-binding, homodimerization, toxin interaction, and virulence. Frontiers in microbiology. 2020Apr9;11:614. doi: 10.3389/fmicb.2020.00614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thakur Z, Saini V, Arya P, Kumar A, Mehta PK. Computational insights into promoter architecture of toxin-antitoxin systems of Mycobacterium tuberculosis. Gene. 2018Jan30;641:161–71. doi: 10.1016/j.gene.2017.10.054 [DOI] [PubMed] [Google Scholar]
- 118.Yamaguchi Y, Park JH, Inouye M. Toxin-antitoxin systems in bacteria and archaea. Annual review of genetics. 2011Dec15;45:61–79. doi: 10.1146/annurev-genet-110410-132412 [DOI] [PubMed] [Google Scholar]
- 119.Fraikin N, Goormaghtigh F, Van Melderen L. Type II toxin-antitoxin systems: evolution and revolutions. Journal of bacteriology. 2020Mar11;202(7):e00763–19. doi: 10.1128/JB.00763-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sala A, Bordes P, Genevaux P. Multiple toxin-antitoxin systems in Mycobacterium tuberculosis. Toxins. 2014Mar;6(3):1002–20. doi: 10.3390/toxins6031002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tandon H, Sharma A, Wadhwa S, Varadarajan R, Singh R, Srinivasan N, et al. Bioinformatic and mutational studies of related toxin–antitoxin pairs in Mycobacterium tuberculosis predict and identify key functional residues. Journal of Biological Chemistry. 2019Jun7;294(23):9048–63. doi: 10.1074/jbc.RA118.006814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ramage HR, Connolly LE, Cox JS. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS genetics. 2009Dec11;5(12):e1000767. doi: 10.1371/journal.pgen.1000767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lobato-Márquez D, Moreno-Córdoba I, Figueroa V, Díaz-Orejas R, García-del Portillo F. Distinct type I and type II toxin-antitoxin modules control Salmonella lifestyle inside eukaryotic cells. Scientific reports. 2015Mar20;5(1):1–0. doi: 10.1038/srep09374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chan WT, Yeo CC, Sadowy E, Espinosa M. Functional validation of putative toxin-antitoxin genes from the Gram-positive pathogen Streptococcus pneumoniae: phd-doc is the fourth bona-fide operon. Frontiers in microbiology. 2014Dec5;5:677. doi: 10.3389/fmicb.2014.00677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Daimon Y, Narita SI, Akiyama Y. Activation of toxin-antitoxin system toxins suppresses lethality caused by the loss of σE in Escherichia coli. Journal of bacteriology. 2015Jul15;197(14):2316–24. doi: 10.1128/JB.00079-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Zhang SP, Wang Q, Quan SW, Yu XQ, Wang Y, Guo DD, et al. Type II toxin–antitoxin system in bacteria: Activation, function, and mode of action. Biophysics Reports. 2020Jun;6(2):68–79. [Google Scholar]
- 127.Armalyte J, Jurenaite M, Beinoraviciute G, Teišerskas J, Sužiedėlienė E. Characterization of Escherichia coli dinJ-yafQ toxin-antitoxin system using insights from mutagenesis data. Journal of bacteriology. 2012Mar15;194(6):1523–32. doi: 10.1128/JB.06104-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yu X, Gao X, Zhu K, Yin H, Mao X, Wojdyla JA, et al. Characterization of a toxin-antitoxin system in Mycobacterium tuberculosis suggests neutralization by phosphorylation as the antitoxicity mechanism. Communications biology. 2020May7;3(1):1–5. doi: 10.1038/s42003-019-0734-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Gupta A, Venkataraman B, Vasudevan M, Bankar KG. Co-expression network analysis of toxin-antitoxin loci in Mycobacterium tuberculosis reveals key modulators of cellular stress. Scientific reports. 2017Jul19;7(1):1–4. doi: 10.1038/s41598-016-0028-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chan WT, Balsa D, Espinosa M. One cannot rule them all: Are bacterial toxins-antitoxins druggable?. FEMS microbiology reviews. 2015Jul1;39(4):522–40. doi: 10.1093/femsre/fuv002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Równicki M, Pieńko T, Czarnecki J, Kolanowska M, Bartosik D, Trylska J. Artificial activation of Escherichia coli mazEF and hipBA toxin–antitoxin systems by antisense peptide nucleic acids as an antibacterial strategy. Frontiers in microbiology. 2018Nov26;9:2870. doi: 10.3389/fmicb.2018.02870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Feng Y, Liu J, Li YG, Cao FL, Johnston RN, Zhou J, et al. Inheritance of the Salmonella virulence plasmids: mostly vertical and rarely horizontal. Infection, Genetics and Evolution. 2012Jul1;12(5):1058–63. doi: 10.1016/j.meegid.2012.03.004 [DOI] [PubMed] [Google Scholar]
- 133.Neuert S, Nair S, Day MR, Doumith M, Ashton PM, Mellor KC, et al. Prediction of phenotypic antimicrobial resistance profiles from whole genome sequences of non-typhoidal Salmonella enterica. Frontiers in microbiology. 2018Mar27;9:592. doi: 10.3389/fmicb.2018.00592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hu L, Cao G, Brown EW, Allard MW, Ma LM, Khan AA, et al. Antimicrobial resistance and related gene analysis of Salmonella from egg and chicken sources by whole-genome sequencing. Poultry Science. 2020Dec1;99(12):7076–83. doi: 10.1016/j.psj.2020.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Menashe O, Kaganskaya E, Baasov T, Yaron S. Aminoglycosides affect intracellular Salmonella enterica serovars typhimurium and virchow. Antimicrobial agents and chemotherapy. 2008Mar;52(3):920–6. doi: 10.1128/AAC.00382-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Weigel LM, Steward CD, Tenover FC. gyrA mutations associated with fluoroquinolone resistance in eight species of Enterobacteriaceae. Antimicrobial Agents and Chemotherapy. 1998Oct1;42(10):2661–7. doi: 10.1128/AAC.42.10.2661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Anderson P. Jr Sensitivity and resistance to spectinomycin in Escherichia coli. Journal of bacteriology. 1969Nov;100(2):939–47. doi: 10.1128/jb.100.2.939-947.1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.O’connor M, Dahlberg AE. Isolation of spectinomycin resistance mutations in the 16S rRNA of Salmonella enterica serovar Typhimurium and expression in Escherichia coli and Salmonella. Current microbiology. 2002Dec;45(6):0429–33. doi: 10.1007/s00284-002-3684-y [DOI] [PubMed] [Google Scholar]
- 139.El Zowalaty ME, Hickman RA, Mthembu TP, Zishiri OT, El Zowalaty AE, Järhult JD. Genome sequences of two Salmonella enterica strains (MEZSAL74 and MEZSAL81) harbouring multiple antimicrobial resistance genes isolated from livestock in South Africa. Journal of Global Antimicrobial Resistance. 2020Jun1;21:396–8. doi: 10.1016/j.jgar.2020.04.001 [DOI] [PubMed] [Google Scholar]
- 140.Taddele MH, Rathore R, Dhama K. Antibiogram assay of Salmonella Gallinarum and other Salmonella enterica serovars of poultry origin in India. Asian Journal of Animal and Veterinary Advances. 2012;7(4):309–17. [Google Scholar]
- 141.Parveen S, Taabodi M, Schwarz JG, Oscar TP, Harter-Dennis J, White DG. Prevalence and antimicrobial resistance of Salmonella recovered from processed poultry. Journal of food protection. 2007Nov;70(11):2466–72. doi: 10.4315/0362-028x-70.11.2466 [DOI] [PubMed] [Google Scholar]
- 142.Hendriksen RS, Bortolaia V, Tate H, Tyson GH, Aarestrup FM, McDermott PF. Using Genomics to Track Global Antimicrobial Resistance. Front Public Health. 2019;7:242. Published 2019 Sep 4. doi: 10.3389/fpubh.2019.00242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Feng Y, Johnston RN, Liu GR, Liu SL. Genomic comparison between Salmonella Gallinarum and Pullorum: differential pseudogene formation under common host restriction. PLoS One. 2013Mar15;8(3):e59427. doi: 10.1371/journal.pone.0059427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Eswarappa SM, Janice J, Balasundaram SV, Dixit NM, Chakravortty D. Host-specificity of Salmonella enterica serovar Gallinarum: insights from comparative genomics. Infection, genetics and evolution. 2009Jul1;9(4):468–73. doi: 10.1016/j.meegid.2009.01.004 [DOI] [PubMed] [Google Scholar]