Abstract
Background:
Radiomic descriptors from MRI are promising for disease diagnosis and characterization but may be sensitive to differences in imaging parameters.
Objective:
To evaluate the repeatability and robustness of radiomic descriptors within healthy brain tissue regions on prospectively acquired MRI scans; in a test-retest setting, under controlled systematic variations of MRI acquisition parameters, and after post-processing.
Study Type:
Prospective
Subjects:
15 healthy participants
Field Strength/Sequence:
3.0 T, axial T2-weighted 2D turbo spin-echo pulse sequence, 181 scans acquired (two test/retest reference scans and twelve with systematic variations in contrast weighting, resolution, and acceleration per participant; removing scans with artifacts).
Assessment:
146 radiomic descriptors were extracted from a contiguous 2D region of white matter in each scan, before and after post-processing.
Statistical Tests:
Repeatability was assessed in a test/retest setting and between manual and automated annotations for the reference scan. Robustness was evaluated between the reference scan and each group of variant scans (contrast weighting, resolution, and acceleration). Both repeatability and robustness were quantified as the proportion of radiomic descriptors that fell into distinct ranges of the concordance correlation coefficient (CCC): excellent (CCC > 0.85), good (0.7 ≤ CCC ≤ 0.85), moderate (0.5 ≤ CCC < 0.7), and poor (CCC < 0.5); for unprocessed and post-processed scans separately.
Results:
Good to excellent repeatability was observed for 52% of radiomic descriptors between test/retest scans, and 48% of descriptors between automated vs manual annotations, respectively. Contrast weighting (TR/TE) changes were associated with the largest proportion of highly robust radiomic descriptors (21%, after processing). Image resolution changes resulted in the largest proportion of poorly robust radiomic descriptors (97%, before post-processing). Post-processing of images with only resolution/acceleration differences resulted in 73% of radiomic descriptors showing poor robustness.
Data Conclusions:
Many radiomic descriptors appear to be non-robust across variations in MR contrast weighting, resolution, and acceleration, as well in test-retest settings, depending on feature formulation and post-processing.
Keywords: radiomics, MRI, robustness, repeatability, reproducibility
Introduction
Radiomics, or the computerized extraction of image intensity, shape, appearance, and texture descriptors from routine MR or CT imaging(1), has recently demonstrated great success for building analytic models for characterizing disease presence or predicting response to therapy across multiple organs(2, 3). However, wider adoption of radiomic descriptors as in vivo imaging markers of disease requires a comprehensive evaluation of their repeatability and robustness under different imaging conditions(4). Thus far, such studies(5) have primarily been conducted using CT imaging of diseased patients or phantoms where radiomic descriptors have been evaluated under variations of well-understood acquisition parameters(6, 7) (such as reconstruction settings, tube currents, radiation doses, or slice thicknesses) in images that have been acquired in a prospectively controlled fashion. Given the even greater number of acquisition parameters associated with an MRI scan, there is a need to similarly interrogate the impact of differences in these parameters on radiomic descriptors.
For instance, T2-weighted (T2w) MRI scans are often widely available in retrospectively pooled cohorts, although with significant variations in acquisition or reconstruction parameters(8). As there is no routinely used reference standard T2w MR imaging protocol, a pooled cohort of T2w MR images is likely to have significant heterogeneity in terms of acquisition parameters such as contrast weighting (repetition time (TR) and echo time (TE)), spatial resolution, and reconstruction approaches (parallel imaging). Critically, while radiologists may be able to adapt the resulting minor image differences due to prior experience and training, the sensitivity of radiomic descriptors to such MR acquisition differences has not been deeply explored.
Systematically evaluating the impact of individual MR acquisition parameters on radiomic descriptors requires a controlled approach, where in vivo MRI scans are prospectively acquired such that only one acquisition parameter is changed at a time (e.g. acquire MRI scans where only TR values are changed while holding TE, resolution, and all other parameters constant). To minimize the impact of disease heterogeneity in such a controlled study and for generalizable results, the performance of radiomic descriptors need to be examined using healthy tissue within a fixed body region (e.g. white or grey matter in the brain), rather than using phantoms or simulation data(9, 10). Thus far, robustness of radiomic descriptors has primarily been examined in the context of how they vary within the same subject between test/re-test brain MRI scans(11) (where the acquisition parameters are the same in both scans, also termed repeatability) or across retrospectively curated multi-site or multi-scanner cohorts(12) (where acquisition parameters may not be controlled). Radiomic analysis also typically includes several post-processing operations(12, 13) (such as bias correction(14), intensity standardization(15), and resolution resampling) which are applied to MR images prior to extracting a series of different types (or “families”) of radiomic descriptors. A detailed study of how post-processing steps impact the robustness of radiomic descriptors from different acquisition variants in a controlled setting would be beneficial(16).
Therefore, the aim of this study was to assess the repeatability and robustness of widely used radiomic descriptors within well-defined healthy brain tissue regions; both in a test-retest setting as well as under controlled, systematic variations of acquisition parameters using prospectively acquired T2w MRI scans. A secondary aim was to investigate the impact of post-processing steps on the robustness of radiomics descriptors. he overall goal was to determine which radiomic descriptors were robust across imaging variants, which descriptors benefit from post-processing, and which imaging variants could potentially be pooled for wider radiomic analyses.
Materials and Methods
Data acquisition
Institutional review board approval and informed consent were obtained. Fifteen healthy volunteers (6 females, 9 males, age 29.4±14 years) were recruited prospectively for MR imaging between September 2018 and November 2018. All MR imaging data were acquired in a single session for each participant, on the same 3T imaging unit (MAGNETOM Skyra; Siemens Healthcare, Erlangen, Germany) and by the same operator. Up to 15 different MRI scans were acquired for each participant and exported as DICOM images for further analysis. These T2-weighted (T2w) acquisitions were based on a standard or reference scan, specifically an axial 2D turbo spin-echo pulse sequence with the following parameters: TR=5740ms, TE=94ms, 4mm slice thickness, 0.7mm in-plane resolution, 31 slices (image sections). These parameters were selected based on the default protocol used clinically at our institution. The total scan time for the reference T2w acquisition was 63 seconds. The reference scan was repeated once for each participant following which an additional 12 variant scans were also acquired by altering parameters individually with respect to the reference scan: TR (3000ms, 4000ms, 5000ms, 7000ms, 8000ms), TE (84ms, 103ms, 112ms), high in-plane resolution (HR; 0.35mm, 0.5mm), low in-plane resolution (LR; 0.9mm), and R=2 parallel imaging acceleration (GRAPPA(17)). These variations in image acquisition parameters were chosen based on the range of parameters observed in brain tumor scans available(8) in The Cancer Imaging Archive (TCIA). The total scan time was 22 minutes and 10 seconds per participant. After data collection, imaging volumes with obvious motion artifacts were excluded, resulting in 11-15 usable images per variant scan (see Table E1 in the Appendix for details of the total of 181 usable scans). The first scan acquired for each participant was considered the reference scan, with respect to which the retest reference scan as well as all variant scans were to be evaluated for repeatability and robustness. Figure 1 provides an overview of the study workflow and experimental design.
Figure 1:
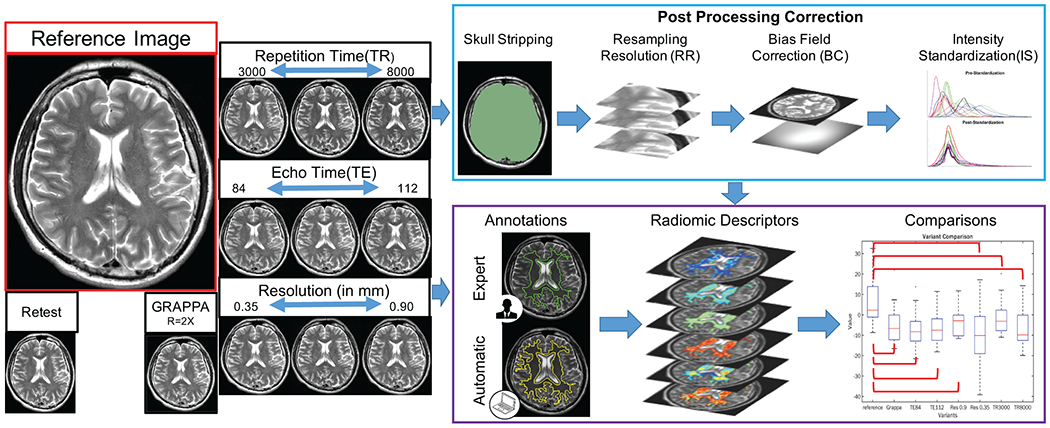
Study workflow showing (left) prospective MRI data acquisition with controlled variations of sequence parameters, and (right) processing steps. Reference images were acquired using a T2w turbo spin echo sequence using our institution’s default clinical protocol, which was repeated once for test/retest evaluation. Variant images were acquired by altering a single acquisition parameter individually, yielding a total of 181 brain MRI scans from 15 healthy participants. Primary experimental workflow (purple box) involved annotation of white matter on each MRI scan followed by radiomic feature extraction within this region. For robustness analysis, feature values from each variant scan were compared to those from the reference scan to quantify the impact of acquisition variations. For repeatability analysis, descriptor performance was compared between manually annotated (green outline) as well as automatically delineated (yellow outline) white matter regions, in a test/retest evaluation of the reference scan, as well as before and after coregistration of contrast variants and the reference scan. Descriptor repeatability and robustness was assessed on unprocessed images as well as after all images had undergone post-processing (blue box).
Annotation of white matter regions on MRI scans
The reference MRI scan for each participant was annotated for white matter (WM) extent by a radiologist (KB) with 5 years of experience using 3D Slicer(18) (v4.5, www.slicer.org). WM was annotated on a single 2D image section (on each reference scan for each participant) approximately 8 mm below the top of the ventricles. This section contained a large region of white matter and was easily identifiable across all participant MRI volumes. Manual WM segmentations were morphologically eroded by a disk element (3-pixel radius) to reduce the impact of very small contour variations and underwent connected component analysis to ensure only large contiguous regions were considered (average size 7362±1234 pixels). Pruned WM annotations were mapped onto all variant and repeat scans for each participant. To ensure that only WM regions were included for further radiomic analysis, the mapped regions were manually inspected and corrected as needed. An automated annotation was also performed, using the automated segmentation module(19) within 3D Slicer to delineate the WM region on the reference scan for each participant (Section E2 in the Appendix summarizes implementation details).
Post-processing of MRI scans
Prior to radiomics feature extraction, MRI scans are typically subjected to a series of post-processing steps to overcome image appearance differences. In this work, the set of operations applied to the images differed slightly between acquisition variant groups. All scans first underwent skull stripping(20) to ensure the bright skull did not affect further corrections. The remaining operations included: (a) bias correction(14) to remove smooth variations in MR intensities across the image (typically introduced by the receiver coils); (b) linearly resampling the DICOM images to ensure that the nominal resolution matched that of the reference scan (0.7 mm in-plane); and (c) intensity standardization(15) to ensure that MR intensities in all the volumes had consistent WM- and gray matter-specific ranges. Section E4 in the Appendix provides further details on the implementation of all post-processing operations. The order of these post-processing operations for all acquisition variants is summarized in Table 1, based on previous studies in the literature(13, 21, 22).
Table 1: Summary of experiments conducted in this work.
Radiomic descriptor repeatability was evaluated between: (a) expert and automated annotations across variant and reference scans, (b) test/retest scans based on reference parameters, (c) contrast variants and reference before and after coregistration. For robustness analysis, imaging acquisition parameters were grouped, and the bolded parameter set corresponds to the variant scans evaluated with respect to the reference scan (TR=5740ms, TE=94ms, Size=0.7mm, R=1). LR = lower resolution, HR = higher resolution, BC = bias correction, IS = intensity standardization, RR = resolution resampling.
| Experiment | Parameter variants considered | Number of comparisons | Post-processing steps applied |
|---|---|---|---|
| Repeatability | Reference Repeated reference |
146 | BC+IS |
| Expert annotation Automated Annotation |
146 | BC+IS | |
| Unregistered contrast variants Coregistered contrast variants |
146*8=1168 | Registration | |
| Image contrast parameters (TR/TE) |
TE= [84,103,112] TR= 5740 Size=0.7 R=1 |
146*3 = 438 | BC+IS |
| TE=94 TR= [3000,4000,5000,7000,8000] Size=0.7 R=1 |
146 *5 = 730 | ||
| Voxel resolution parameters | TE=94 TR=5740 Size= [0.9], termed LR R=1 |
146 | RR+BC+IS |
| TE=94 TR=5740 Size= [0.35,0.5], termed HR R=1 |
146*2=292 | ||
| Acceleration parameters (GRAPPA) | TE=94 TR=5740 Size=0.7 R=[2] |
146 | BC+IS |
Radiomic Feature Extraction
A total of 146 pixel-wise radiomic descriptors from six different families (including variations in 2D window sizes (WS) between 3-7 pixels) were extracted from the manually annotated WM region on each MR image (both before and after post-processing) using in-house MATLAB (The MathWorks, Inc., Natick, MA) implementations. Table E3 in the Appendix gives a description of each radiomic feature family and their associated parameters based on the Image Biomarker Standardization Initiative guidelines(23). These features can be broadly grouped into six families: histogram, gradient(24) and Laws(25) (edge-based), Gabor(26) (wavelet-based), and Haralick(27) and COLLAGE(28) (co-occurrence based) descriptors. Pixel-wise feature maps were computed for all 181 datasets included in this study,
Statistical Analysis
To quantify the repeatability and robustness of the different feature families, the concordance correlation coefficient(29) (CCC) was computed between each pair of reference and variant images, and for each feature separately. If σr and σv are the variances and μr and μv are the means for the radiomic descriptor in the reference and variant images, respectively, and ρr,v is the covariance between them, the CCC is computed as:
As CCC ranges between 0 and 1, it was subdivided into four robustness ranges for easier interpretability(30): excellent (CCC > 0.85), good (0.7 ≤ CCC ≤ 0.85), moderate (0.5 ≤ CCC < 0.7), and poor (CCC < 0.5).
Agreement between manual and automated annotations was assessed via the Dice coefficient:
where WMa corresponds to the automated WM annotations, WMm to the manual WM annotations, and |.| is the cardinality operator.
Radiomic descriptor repeatability was evaluated in a test/retest setting for reference parameters as well as between annotation sources (on the reference image). First, CCC was computed for each of the 146 radiomic descriptors based on comparing the reference image and the repeated reference image using manual WM annotations. Next, CCC was calculated for each of the 146 radiomic descriptors between manual and automated WM annotations on the first reference image. Repeatability was evaluated for unprocessed and post-processed images separately and visualized via a thermometer plot for the different CCC ranges. Each thermometer was shaded in based on the proportion of descriptors from that family that fell within a specific CCC range.
Additionally, the impact of minor differences between image contrast (TR/TE) variants and the reference image was evaluated by comparing the repeatability of radiomic descriptors before and after coregistration. CCC was calculated for each of the 146 descriptors by comparing the reference image to each of the eight unregistered TR/TE variants using manual annotations. Next, each TR and TE volume was affinely coregistered to the corresponding reference volume, for each participant separately (via 3D Slicer). CCC was then again calculated for all 146 descriptors between the reference image and eight coregistered TR/TE variant images using manual annotations. No additional post-processing was applied in this experiment to specifically evaluate the impact of coregistration alone. Thermometer plots were used to visualize the proportion of radiomic descriptors that fell into each range of CCC values per feature family, for unregistered and coregistered scans separately.
Radiomic descriptor robustness was similarly evaluated for each acquisition variant with respect to the reference image; for unprocessed and post-processed images separately. Variant images were grouped by parameter (TR, TE, LR, HR, GRAPPA; see Table 1) and the number of radiomic descriptors per feature family that fell into each CCC range were counted and normalized by the total number of comparisons conducted. The proportion of radiomic descriptors that fell into each range of CCC values were visualized via thermometer plots per feature family and for each acquisition variant group separately.
Results
Repeatability of radiomic descriptors in test/retest evaluation, between annotation sources, and before/after coregistration
Figure 2(A) depicts a thermometer plot summarizing the results of test/retest evaluation of the reference acquisition parameters, using unprocessed and post-processed images. Overall, 77-78 descriptors out of a total set of 146 (53%) showed good to excellent repeatability (regardless of post-processing) while 28/146 descriptors (19%) on unprocessed and 22/146 descriptors (15%) on post-processed images showed poor repeatability. When examined by feature family, 75% of the Gabor descriptors showed excellent test/retest repeatability, followed by histogram (54%), and COLLAGE (35%) on unprocessed images. However, the proportions of repeatable descriptors in each of these feature families were markedly reduced in post-processed images to 33% (Gabor), 0% (histogram), and 15% (COLLAGE). Gradient descriptors were consistently poorly repeatable in test/retest evaluation, on both unprocessed (100% poor) and post-processed images (90% poor). Similarly, test/retest repeatability measurements in Laws descriptors remained largely unchanged between unprocessed (38%, good to excellent) and post-processed (32%, good to excellent) images. While 38% of Haralick descriptors showed good to excellent test/retest repeatability on unprocessed images, this proportion increased to 79% on post-processed images.
Figure 2:

Thermometer plot for repeatability experiments showing results of (A) test/retest evaluation (between reference and the repeated reference images) within manual WM annotations, (B) manual and automated WM annotations on the first reference image, and (C) before/after coregistration of contrast variants with the first reference image. Plots are shaded based on proportion of radiomic descriptors from different families (in different colors) that fall within different CCC-based robustness ranges, with exact numerical percentages included. Note that CCC ranges were defined as follows: excellent (CCC > 0.85), good (0.7 ≤ CCC ≤ 0.85), moderate (0.5 ≤ CCC < 0.7), and poor (CCC < 0.5).
Figure 2(B) shows a thermometer plot summarizing the results of comparing radiomic descriptors between manual and automated WM annotations on the reference image. Manual and automated WM annotations showed reasonable overlap with a Dice coefficient of 0.77±0.05 across all participants. Overall, while 47/146 descriptors (32%) showed excellent repeatability and 57/146 descriptors (57%) showed poor repeatability between annotation sources on unprocessed images, these proportions worsened after post-processing (17% or 25 descriptors excellent, 58% or 84 descriptors poor). Good to excellent maual/automated repeatability on unprocessed images was primarily observed for Gabor (100%), histogram (54%), and Haralick (62%) descriptors. Post-processing resulted in fewer good to excellent Gabor (33%) and histogram (15%) descriptors, although the number of Haralick descriptors (62%) with good to excellent manual/automated repeatability remained unchanged. Similarly, the proportion of COLLAGE descriptors within different repeatability ranges also remained relatively unchanged between unprocessed (50%, good to excellent) and post-processed images (43%, good to excellent). Finally, edge-based descriptors showed poor repeatability between annotation sources, on both unprocessed (gradient: 90%, Laws: 94%) and post-processed images (gradient: 90%, Laws: 100%).
Figure 2(C) shows a thermometer plot summarizing the impact of image coregistration on the repeatability of radiomic features between TR/TE variants and the reference scan. Across all feature families, only 15% of descriptors showed good-excellent repeatability prior to registration which was markedly reduced on coregistered scans (9% with good-excellent repeatability). Among feature families, while 51% of COLLAGE descriptors showed good-excellent repeatability on unregistered scans (and comprised the largest proportion of such features), no COLLAGE descriptors (0%) were repeatable after coregistration. Coregistration also worsened the performance of histogram (83% before, 92% after), Gabor (84% before, 90% after), and Haralick (75% before, 95% after) descriptors; all of which showed markedly poorer repeatability on coregistered scans. Only the edge-based feature families appeared to benefit from coregistration and showed improved repeatability, seen in the performance of gradient descriptors (100% poor, 0% excellent before to 90% poor, 4% excellent after) as well as Laws descriptors (84% poor, 15% good-moderate before to 40% poor, 58% good-moderate after).
Radiomic descriptor robustness between different MR acquisition variant groups
Thermometer plots showing the proportion of radiomic descriptors within each robustness range for each group of acquisition variants (TR, TE, LR, HR, GRAPPA) are depicted in Figure 3, with different colors corresponding to different feature families. These are examined in more detail in the context of each acquisition variant group, as follows.
Figure 3:
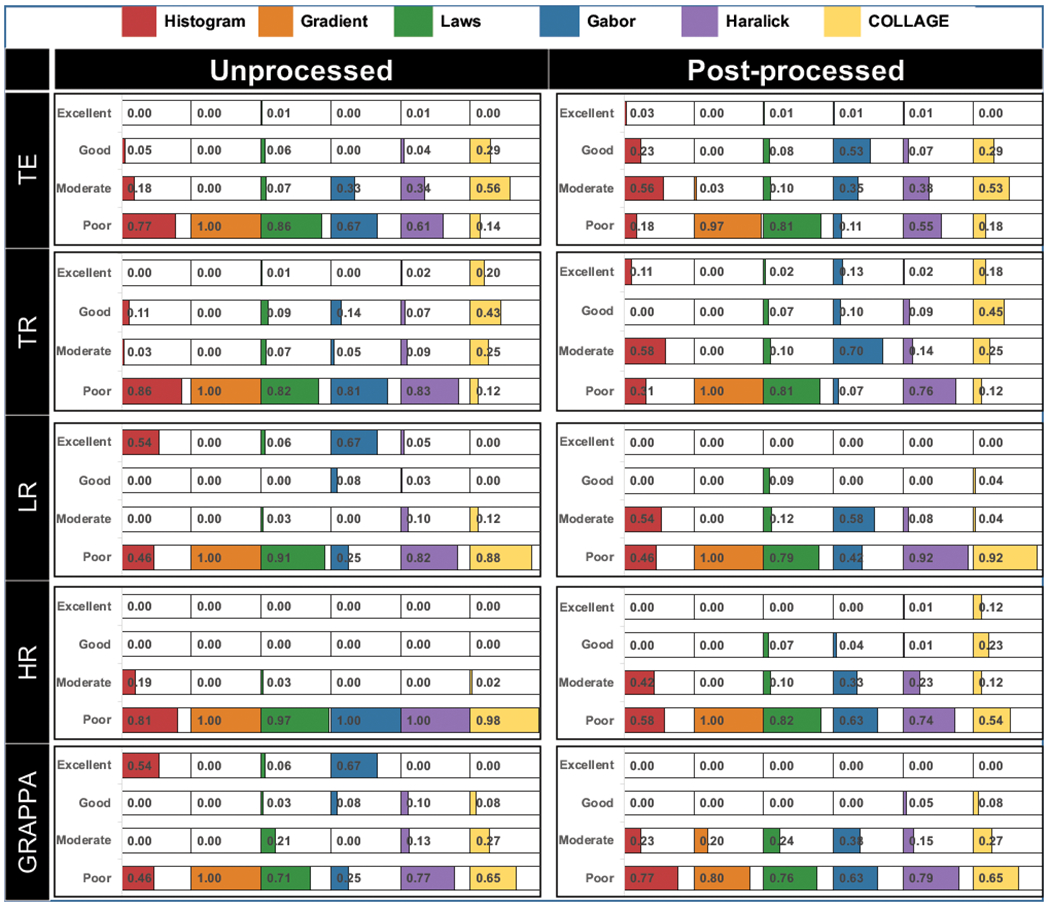
Thermometer plots for robustness experiments depicting proportions of radiomic descriptors from different families (shaded in different colors) falling within different CCC-based robustness ranges, with exact numerical percentages also indicated. All descriptors were compared between reference and variant images within expert WM annotations, with plots grouped by acquisition variant as summarized in Table 1. Note CCC ranges were defined as follows: excellent (CCC > 0.85), good (0.7 ≤ CCC ≤ 0.85), moderate (0.5 ≤ CCC < 0.7), and poor (CCC < 0.5).
Robustness of radiomic feature families with respect to variations in MR image contrast acquisition parameters
The only descriptor family with moderate to excellent performance under TR/TE contrast variations was COLLAGE (TE: 85%; TR: 68%). The proportion of COLLAGE descriptors within each robustness range also remained largely unchanged between unprocessed and post-processed images. Additionally, a larger number of COLLAGE descriptors exhibited higher robustness across changes in TR (18-20% excellent, 43-45% good) as compared to changes in TE (0% excellent, 29% good). Figure 4(A) shows an expression heatmap for a representative COLLAGE descriptor (entropy WS=5) with good robustness across changes in TE and excellent robustness across changes in TR.
Figure 4:
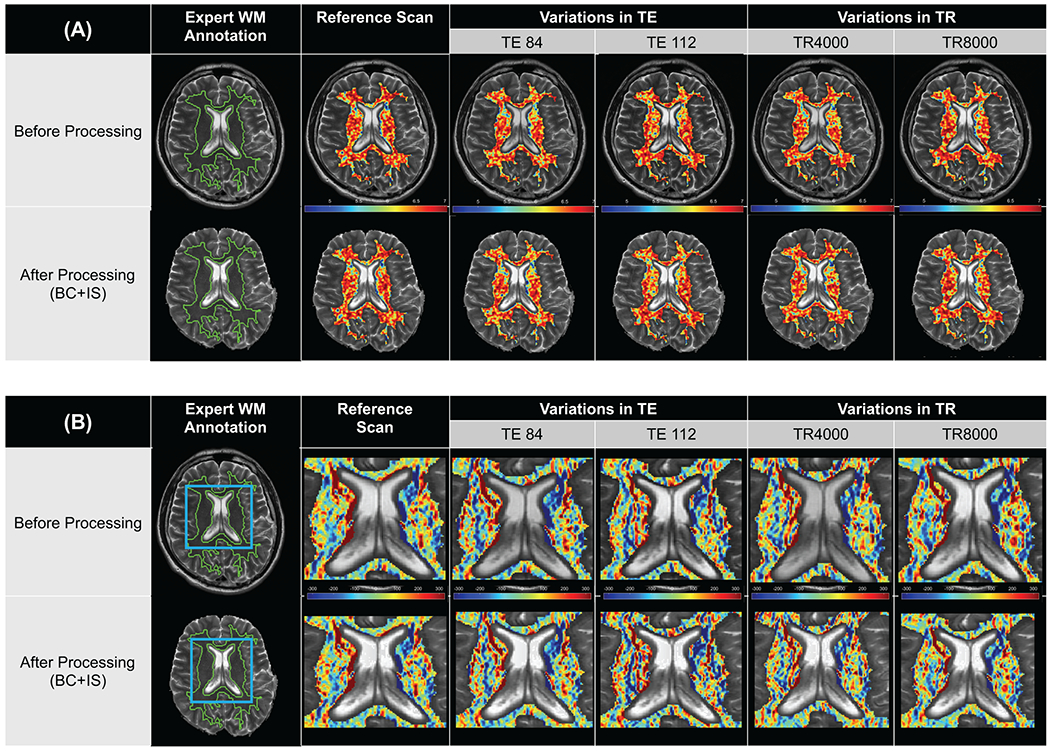
Fig. 4. Representative radiomic heatmaps for (A) a COLLAGE descriptor (entropy WS=5, co-occurrence family) exhibiting good to excellent robustness across variations in TR and TE, and (B) representative radiomic heatmaps for a Laws descriptor (L5E5, edge-based family) exhibiting poor robustness across variations in TR and TE. Post-processing does not appear to markedly affect the appearance of feature heatmaps when compared between top (unprocessed) and bottom (post-processed), across all columns.
While also in the co-occurrence family, a majority of Haralick descriptors showed poor robustness across changes in TE and TR, both before (61% and 83%, respectively) and after (55% and 76%, respectively) processing. The gradient and Laws operator families (edge-based) were poorly robust across all image contrast variations, whether on unprocessed (82-100% poor) or post-processed images (81%-100% poor). Figure 4(B) shows a representative edge-based descriptor (Laws L5E5) as an expression heatmap, illustrating poor robustness of feature expression across TR and TE changes, both for the unprocessed images as well as after post-processing.
The histogram and wavelet feature families largely exhibited poor robustness across changes in TR (81% and 86%, respectively) as well as TE (67% and 77%, respectively), on unprocessed images. Post-processing slightly improved the robustness in both feature families across TE variations, with 79% of histogram descriptors and 88% of Gabor descriptors showing good to moderate robustness. A larger number of descriptors in these families were robust after post-processing the images with TR variations, seen by the increased proportions in excellent (histogram: 11%, Gabor: 13%) as well as good to moderate (histogram: 58%, Gabor: 80%) CCC ranges.
Robustness between radiomic feature families with respect to differences in nominal MR image resolutions
Histogram and Gabor feature families comprised the largest proportion of descriptors with excellent robustness across lower resolution variants (54% and 67%, respectively) on unprocessed images. Post-processing severely impacted both feature families, resulting in 54% of histogram descriptors and 58% of Gabor descriptors demonstrating moderate robustness (no descriptors showed excellent robustness in either family). When considering higher resolution imaging variants, 81% of histogram descriptors and 100% of Gabor descriptors showed poor robustness on unprocessed images. These proportions were slightly improved after post-processing, with only 58% of histogram descriptors and 63% of Gabor descriptors showing poor robustness, and the rest showing moderate robustness. Figure 5 shows a representative wavelet descriptor (Gabor WS=3, Orientation=0°) illustrating the change in robustness for higher- and lower-resolution variants compared to the reference, both before and after post-processing.
Figure 5:
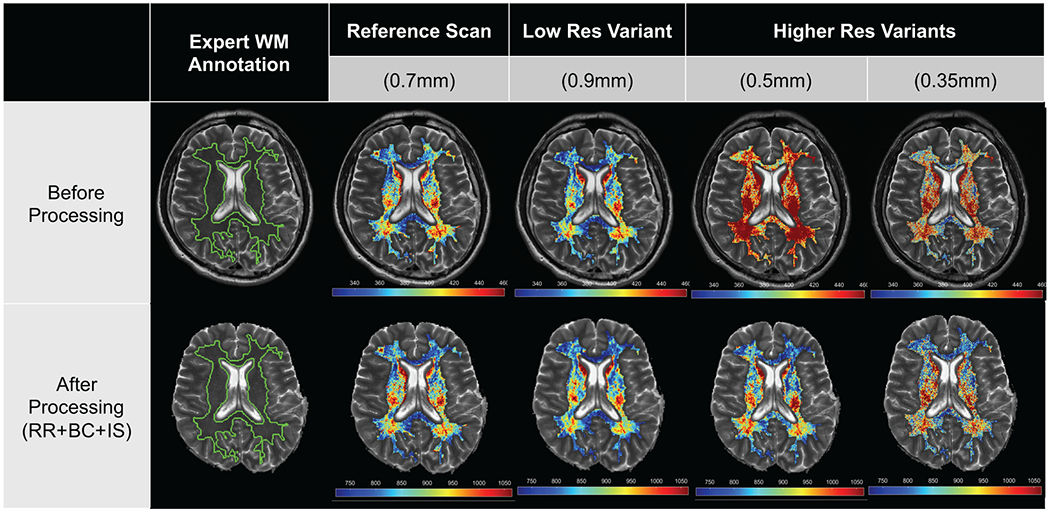
Representative radiomic heatmaps for a Gabor descriptor (WS=3, Orientation=0°, wavelet family) on unprocessed (top row) and post-processed images (bottom row), within the expert WM annotation. Feature heatmaps on unprocessed LR variant images are more consistent with the reference (excellent robustness) while unprocessed HR variant images are relatively inconsistent (poor robustness). Post-processing causes the feature heatmap to appear similar across LR and HR variant images with respect to the reference (moderate robustness in both cases).
The Haralick and COLLAGE families had marginally fewer descriptors with poor robustness on unprocessed LR variants (Haralick: 82%, COLLAGE: 88%) compared to unprocessed HR variants (Haralick: 100%, COLLAGE:98%). However, while the proportion of poorly robust Haralick (92%) and COLLAGE (92%) descriptors remained relatively unchanged on post-processed LR variants, post-processed HR variants exhibited a marked reduction in the number of poorly robust co-occurrence descriptors (Haralick: 74%, COLLAGE: 54%). The proportion of COLLAGE descriptors with good to excellent robustness increased after post-processing across both resolution variants (LR: 4% good, HR: 35% good to excellent), compared to Haralick descriptors (LR: 0%, HR: 2%; good to excellent). Gradient descriptors were 100% poorly robust across all resolution variants, whether on unprocessed or post-processed images. By comparison, while 91% to 97% of Laws descriptors were poorly robust on unprocessed LR and HR variant images respectively, only 79% (LR) and 82% (HR) of these descriptors were poorly robust after post-processing.
Robustness between radiomic feature families with respect to changes in parallel imaging reconstruction
Most descriptors in the Haralick and COLLAGE families were poorly robust between accelerated variants and the non-accelerated reference, both on unprocessed(Haralick: 77%, COLLAGE: 65%) and post-processed(Haralick: 79%, COLLAGE: 65%). The proportion of descriptors in different robustness ranges did not markedly change between unprocessed and post-processed images, though COLLAGE (35% for both unprocessed images) had a marginally higher number of descriptors with good to moderate robustness compared to Haralick (23% for unprocessed images, 20% for post-processed images). Figure 6 shows a representative Haralick descriptor (Information Measure 2) with poor robustness between the GRAPPA variant and the non-accelerated reference image.
Figure 6:
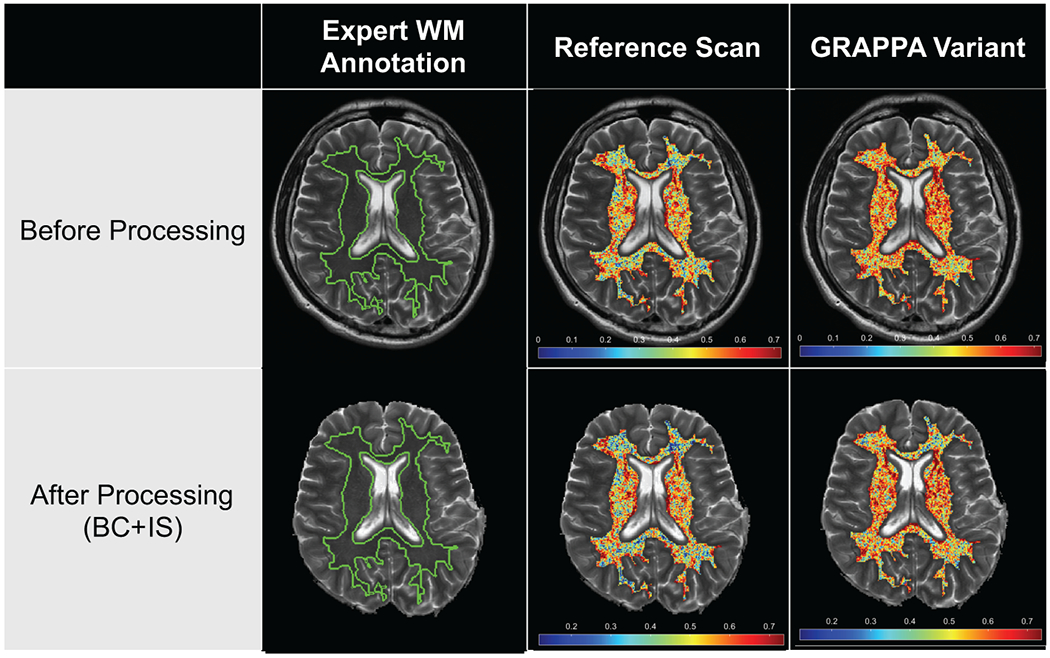
Representative radiomic heatmaps for Haralick descriptor (Information Measure 2, co-occurrence family) showing poor robustness between variant (GRAPPA reconstruction, R=2) and reference (non-accelerated, R=1) images, on unprocessed (top row) and post-processed images (bottom row). Note marked variations in the feature heatmaps within expert WM annotations on variant images compared to the corresponding reference image, which remains inconsistent despite post-processing.
The edge-based feature family were similarly poorly robust on unprocessed (gradient: 100%, Laws: 71%) and post-processed images (gradient: 80%, Laws: 76%). However, while 6% of Laws descriptors showed excellent robustness on unprocessed GRAPPA variants, no Laws descriptors showed excellent robustness after post-processing. Conversely, 20% of gradient descriptors appeared moderately robust on post-processed GRAPPA variants compared to 0% on unprocessed images. Histogram (54%) and Gabor (67%) descriptors were the only feature families with excellent robustness on unprocessed GRAPPA variants, compared to the reference. After post-processing, none of these descriptors showed excellent robustness, although 23% of histogram and 33% of Gabor descriptors show moderate robustness.
Discussion
Wider clinical use of radiomic descriptors for characterizing tissue and disease on imaging is contingent on understanding their repeatability in test-retest settings and robustness across variations in image acquisition parameters(5, 16). In this study, an in vivo MR imaging cohort was prospectively accrued to study (1) which radiomic descriptors were repeatable in a test-retest setting and between different annotation sources, (2) how robust different families of radiomic descriptors were across controlled, systematic variations in individual MRI acquisition parameters and whether post-processing steps improved their robustness, and (3) which imaging variants could potentially be pooled for wider radiomic analyses. To minimize the impact of disease heterogeneity and to have generalizable results in such a controlled study, performance of radiomic descriptors was studied within well-defined white matter brain tissue regions on MRI scans from healthy volunteers.
Repeatability analysis involved two distinct comparisons. First, when comparing radiomic descriptors between the reference acquisition and a second repetition of the reference, approximately half of descriptors showed good to excellent repeatability while nearly 20% showed poor repeatability.. Second, when comparing radiomic descriptors between manually and automatically generated white matter annotations on the same reference images, approximately 40% of descriptors showed poor repeatability and nearly half showed good to excellent repeatability. Radiomic descriptors thus exhibited poorer repeatability between manual and automated annotations than between test/retest scans, as has been observed previously (31, 32) and potentially due to the moderate overlap between the two sets of annotations. Among feature families, co-occurrence based Haralick and COLLAGE descriptors consistently showed good to excellent repeatability performance which was only marginally changed after post-processing; in-line with previous findings across a number of different organs(9, 10, 32, 33). By contrast, gradient and Laws descriptors were poorly repeatable in both comparisons (both before and after post-processing) which resonates with studies suggesting their sensitivity to even marginal imaging or annotation differences (34, 35). The difference in repeatability performance between edge-based and co-occurrence descriptors further suggests that first-order derivatives (used in Laws and gradient operators) may be more sensitive than higher-order derivatives (used in Haralick and COLLAGE). Interestingly, coregistration of contrast variants to the reference resulted in edge-based descriptors (gradient, Laws), in turn showing a marked increase in repeatability with respect to the reference. The sensitivity of these descriptors may thus be a function of subtle shifts and artifacts which may be introduced within a scan or between scans. Histogram and Gabor descriptors demonstrated excellent repeatability on unprocessed images in both the test/retest and expert/automated evaluation, but post-processing reduced their level of repeatability. Since no contrast differences are expected in repeatability analysis, the different post-processing steps may have introduced subtle variations in the intensity distributions(21) which are known to directly impact the repeatability of histogram and Gabor descriptors (9).
In this study, radiomic descriptor robustness was evaluated by comparing the reference scan with scans having systematic changes in each of TR, TE, HR, LR, GRAPPA, using the original unprocessed images as well as after post-processing. For unprocessed image contrast variants, only ~1% of the descriptors showed excellent robustness across changes in TE while 4% showed excellent robustness when considering changes in TR. Radiomic descriptors have previously demonstrated poor robustness to changes in TE(30) and our study suggests that they may be sensitive to changes in TR as well, especially when the resulting contrast differences are not accounted for via post-processing. After applying post-processing, there was a marked increase in the proportions of radiomic descriptors with good to excellent robustness across varying TR and TE values and a reduced proportion of descriptors with poor robustness. The large differences in signal intensities in image contrast variants (compared to the reference) appear to thus only be partially accounted for via the post-processing steps of bias correction and intensity standardization (12).
When considering unprocessed resolution variants, ~20% of descriptors showed excellent robustness between scans at a lower voxel resolution and the reference while no descriptors showed good or excellent robustness between unprocessed variants with a higher voxel resolution and the reference. The impact of nominal voxel resolution on radiomic descriptors has been noted previously(36, 37), and is likely due to corresponding variations in the number of voxels and the concomitant differences in spatial extent when computing voxel-wise feature responses. Post-processing LR variants resulted in no descriptors showing excellent robustness and two feature families (histogram and Gabor) exhibiting worsened robustness compared to unprocessed images. By contrast, post-processing HR variants modestly improved robustness where ~10% of descriptors exhibited good to excellent robustness. In other words, linearly up-sampling a lower-resolution image to match the higher-resolution reference had an overall negative impact while downsampling a higher-resolution image to match a lower-resolution reference had only a marginally positive impact. Post-processing interpolation does not fully account for differences between scans acquired at different image resolutions, where interpolated voxels via up-sampling appear to worsen the robustness of radiomic descriptors (also noted previously(38)) while downsampling higher-resolution images appears to marginally reduce resolution-related differences.
Finally, when comparing accelerated variants (GRAPPA) to the reference, good to excellent robustness was exhibited by ~25% of descriptors. Parallel imaging reconstruction is intended to result in almost identical average signal intensity values compared to the reference(10), which appears to result in robust descriptors in families that are most dependent on the underlying intensity profiles (histogram and Gabor, similar to findings from our repeatability analysis). Post-processing of the reference and GRAPPA-accelerated images resulted in an increased proportion of poorly robust descriptors, due to histogram and Gabor descriptors exhibiting worsened robustness. Similar to findings from repeatability analysis, applying bias correction and intensity standardization appeared to worsen the performance of radiomics descriptors when no contrast differences are present between the reference and the variant scan; likely indicating that additional variations were introduced by post-processing operations.
In this study, we further evaluated the robustness of individual radiomic feature families with respect to the different acquisition variants to understand their robustness to changes in the imaging protocol. Histogram and Gabor descriptors were the only families to show excellent robustness across multiple unprocessed imaging variants. Post-processing modestly improved the robustness of both feature families across contrast differences, but also reduced robustness in the lower resolution and GRAPPA-accelerated variants. These two feature families may thus be most robust on MRI scans with minimal image contrast differences (i.e. unprocessed test-retest images, different annotation sources, images with parallel imaging or of a lower resolution). Haralick descriptors (intensity co-occurrences) showed poor robustness across almost all acquisition variants, unlike their good to excellent repeatability performance. COLLAGE descriptors (gradient orientation co-occurrences) were split between good, moderate, and poor robustness for almost all imaging variants, similar to their repeatability performance. The differing performance between the two types of co-occurrence descriptors may be because intensity co-occurrences (used in Haralick) are more dependent on absolute image intensity values than gradient co-occurrences (used in COLLAGE, based on relative differences between adjacent pixels), lessening the impact of small image contrast changes on the latter. Overall, good to moderate robustness was achieved by a majority of histogram, Gabor, COLLAGE, and Haralick (only TE changes) descriptors when images of different contrasts (TR and TE) had been post-processed. Gradient features were universally poorly robust across all variants, with a small fraction (~20%) becoming slightly more robust to changes in parallel imaging acceleration with the application of post-processing. Similarly, Laws features were also poorly robust across all imaging variants; however, this did not change as a result of post-processing. Similar to their repeatability performance, first-order derivatives in these feature families appeared to be highly sensitive to both image contrast and resolution differences between MR scans.
Limitations
Diseased individuals were not included in our cohort in order to carefully study radiomic descriptor robustness in as controlled a fashion as possible within healthy brain tissue regions. We evaluated a single tissue type (white matter) in our experiments as this typically comprises a large contiguous region on a brain MRI section and was easily identifiable. The repeatability and robustness of radiomic features identified in our study thus need to be confirmed for other tissue regions (grey matter, cerebrospinal fluid), for other acquisition sequences(39) (e.g. T1-weighted, diffusion-weighted), as well as in diseased individuals in the future. In addition, in this study, we opted to evaluate the robustness of radiomic descriptors based on defining ranges for the CCC measure alone, as has been commonly reported in the literature(30, 40). Our study was also limited to a subset of possible variations in the T2-weighted brain MR imaging acquisition, this subset being based on the range of values found in brain MRI scans in TCIA(8). Other common variations in the MR acquisition such as the number of averages, sampling bandwidth, or motion could be studied in future work. While only a single reader’s manual annotations were used in this study, these were compared against an automated annotation approach (in terms of overlap as well as descriptor repeatability). An expansion on this study may include additional readers to more fully assess the impact of interobserver variation in this context. This study also used DICOMs for analysis and not images directly reconstructed from the raw data. As different vendors use different algorithms and filters to generate DICOM images from k-space data, this may be an additional source of variation that requires a more detailed interrogation in the future. Additional factors which could be explored include the software package used, additional feature families, parameters such as bin size or neighborhood window, as well as comparing voxel- and region-wise descriptors. Finally, the sequence of post-processing operations used in our experiments was determined based on the literature(13, 21, 22). These could be further permuted to identify a post-processing sequence to optimally account for imaging differences due to variations in acquisition parameters, potentially further improving the robustness of radiomic descriptors.
Conclusions
In conclusion, acquisition parameter changes in T2-weighted MR images can have a significant impact on the repeatability and robustness of derived radiomic descriptors. Only certain subsets of imaging variants should be safely considered for pooled analysis, but only for a subset of radiomic descriptors and potentially with better post-processing. Improved quality control of acquisition parameters and incorporation of descriptor robustness are hence critical to ensure clinically relevant and generalizable radiomic analysis and machine learning performance via MRI.
Supplementary Material
Grant Support:
Research reported in this publication was supported by the National Cancer Institute under 1U24CA199374-01, R01CA202752-01A1, R01CA208236-01A1, R01CA216579-01A1, R01CA220581-01A1, 1U01CA239055-01, 1U01CA248226-01, 1U54CA254566-01, the National Heart, Lung, and Blood Institute under 2R01HL094557-06, 1R01HL151277-01A1, the National Science Foundation (CBET) under 1553441, the National Institute for Biomedical Imaging and Bioengineering (NIBIB) CWRU Interdisciplinary Biomedical Imaging Training Program under award number 5T32EB00750912, the NIH Training Program in Musculoskeletal Research Grant at CWRU 5T32AR007505-32, the NIBIB under 1R43EB028736-01, the National Center for Research Resources under award number 1C06RR12463-01, the VA Merit Review Award IBX004121A from the United States Department of Veterans Affairs Biomedical Laboratory Research and Development Service, the Office of the Assistant Secretary of Defense for Health Affairs, through the Breast Cancer Research Program (W81XWH-19-1-0668), the Prostate Cancer Research Program (W81XWH-15-1-0558, W81XWH-20-1-0851), the Lung Cancer Research Program (W81XWH-18-1-0440, W81XWH-20-1-0595), the DOD/CDMRP Peer Reviewed Cancer Research Program (W81XWH-18-1-0404, W81XWH-16-1-0329), the Kidney Precision Medicine Project (KPMP) Glue Grant, the Ohio Third Frontier Technology Validation Fund, the Dana Foundation David Mahoney Neuroimaging Program, Johnson & Johnson WiSTEM2D Award , the V Foundation Translational Research Award, the Wallace H. Coulter Foundation Program in the Department of Biomedical Engineering at Case Western Reserve University, and the Clinical and Translational Science Collaborative of Cleveland (UL1TR0002548) from the National Center for Advancing Translational Sciences component of the National Institutes of Health and NIH roadmap for Medical Research. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, the Department of Defense, or the United States Government.
Conflicts of Interest:
Dr. Madabhushi is an equity holder in Elucid Bioimaging and in Inspirata Inc. In addition he has served as a scientific advisory board member for Inspirata Inc, Astrazeneca, Bristol Meyers-Squibb and Merck. Currently he serves on the advisory board of Aiforia Inc. He also has sponsored research agreements with Philips, AstraZeneca, Boehringer-Ingelheim and Bristol Meyers-Squibb. His technology has been licensed to Elucid Bioimaging. He is also involved in a NIH U24 grant with PathCore Inc, and 3 different R01 grants with Inspirata Inc. Dr. Viswanath and Dr. Madabhushi have had technology licensed to Elucid Bioimaging. Dr. Seiberlich has received research support from Siemens Healthineers. Dr. Tiwari has received a research award from Johnson & Johnson.
References
- 1.Lambin P, Rios-Velazquez E, Leijenaar R, et al. : Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer Oxf Engl 1990 2012; 48:441–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lambin P, Leijenaar RTH, Deist TM, et al. : Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 2017; 14:749–762. [DOI] [PubMed] [Google Scholar]
- 3.Limkin EJ, Sun R, Dercle L, et al. : Promises and challenges for the implementation of computational medical imaging (radiomics) in oncology. Ann Oncol Off J Eur Soc Med Oncol 2017; 28:1191–1206. [DOI] [PubMed] [Google Scholar]
- 4.O’Connor JPB, Aboagye EO, Adams JE, et al. : Imaging biomarker roadmap for cancer studies. Nat Rev Clin Oncol 2017; 14:169–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Traverso A, Wee L, Dekker A, Gillies R: Repeatability and Reproducibility of Radiomic Features: A Systematic Review. Int J Radiat Oncol Biol Phys 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berenguer R, Pastor-Juan MDR, Canales-Vázquez J, et al. : Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters. Radiology 2018; 288:407–415. [DOI] [PubMed] [Google Scholar]
- 7.Meyer M, Ronald J, Vernuccio F, et al. : Reproducibility of CT Radiomic Features within the Same Patient: Influence of Radiation Dose and CT Reconstruction Settings. Radiology 2019; 293:583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scarpace L, Mikkelsen T, Cha S, et al. : Radiology Data from The Cancer Genome Atlas Glioblastoma Multiforme [TCGA-GBM] collection. 2016. [Google Scholar]
- 9.Ford J, Dogan N, Young L, Yang F: Quantitative Radiomics: Impact of Pulse Sequence Parameter Selection on MRI-Based Textural Features of the Brain. Contrast Media Mol Imaging 2018; 2018:1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang F, Dogan N, Stoyanova R, Ford JC: Evaluation of radiomic texture feature error due to MRI acquisition and reconstruction: A simulation study utilizing ground truth. Phys Medica PM Int J Devoted Appl Phys Med Biol Off J Ital Assoc Biomed Phys AIFB 2018; 50:26–36. [DOI] [PubMed] [Google Scholar]
- 11.Li Z, Duan H, Zhao K, Ding Y: Stability of MRI Radiomics Features of Hippocampus: An Integrated Analysis of Test-Retest and Inter-Observer Variability. IEEE Access 2019; 7:97106–97116. [Google Scholar]
- 12.Um H, Tixier F, Bermudez D, Deasy JO, Young RJ, Veeraraghavan H: Impact of image preprocessing on the scanner dependence of multi-parametric MRI radiomic features and covariate shift in multi-institutional glioblastoma datasets. Phys Med Biol 2019; 64:165011. [DOI] [PubMed] [Google Scholar]
- 13.Moradmand H, Aghamiri SMR, Ghaderi R: Impact of image preprocessing methods on reproducibility of radiomic features in multimodal magnetic resonance imaging in glioblastoma. J Appl Clin Med Phys 2020; 21:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tustison NJ, Avants BB, Cook PA, et al. : N4ITK: improved N3 bias correction. IEEE Trans Med Imaging 2010; 29:1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nyúl LG, Udupa JK, Zhang X: New variants of a method of MRI scale standardization. IEEE Trans Med Imaging 2000; 19:143–150. [DOI] [PubMed] [Google Scholar]
- 16.Cattell R, Chen S, Huang C: Robustness of radiomic features in magnetic resonance imaging: review and a phantom study. Vis Comput Ind Biomed Art 2019; 2:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griswold MA, Jakob PM, Heidemann RM, et al. : Generalized autocalibrating partially parallel acquisitions (GRAPPA). Magn Reson Med 2002; 47:1202–1210. [DOI] [PubMed] [Google Scholar]
- 18.Fedorov A, Beichel R, Kalpathy-Cramer J, et al. : 3D Slicer as an image computing platform for the Quantitative Imaging Network. Magn Reson Imaging 2012; 30:1323–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pohl KM, Bouix S, Nakamura M, et al. : A hierarchical algorithm for MR brain image parcellation. IEEE Trans Med Imaging 2007; 26:1201–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bauer S, Fejes T, Reyes M: A Skull-Stripping Filter for ITK. In Insight J; 2012. [Google Scholar]
- 21.Madabhushi A, Udupa JK: Interplay between intensity standardization and inhomogeneity correction in MR image processing. IEEE Trans Med Imaging 2005; 24:561–576. [DOI] [PubMed] [Google Scholar]
- 22.Schwier M, van Griethuysen J, Vangel MG, et al. : Repeatability of Multiparametric Prostate MRI Radiomics Features. Sci Rep 2019; 9:9441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zwanenburg A, Vallières M, Abdalah MA, et al. : The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020:191145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duda R, Hart P: Pattern Classification and Scene Analysis. 1st edition. New York Wiley; . [Google Scholar]
- 25.Laws KI: Textured Image Segmentation. IPI Report. University of Southern California; 1980:186 p. [Google Scholar]
- 26.Jain AK, Farrokhnia F: Unsupervised texture segmentation using Gabor filters. Pattern Recognit 1991; 24:1167–1186. [Google Scholar]
- 27.Haralick RM, Shanmugam K, Dinstein I: Textural Features for Image Classification. IEEE Trans Syst Man Cybern 1973; SMC-3:610–621. [Google Scholar]
- 28.Prasanna P, Tiwari P, Madabhushi A: Co-occurrence of Local Anisotropic Gradient Orientations (CoLlAGe): A new radiomics descriptor. Sci Rep 2016; 6:37241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin LI: A concordance correlation coefficient to evaluate reproducibility. Biometrics 1989; 45:255–268. [PubMed] [Google Scholar]
- 30.Bianchini L, Santinha J, Loução N, et al. : A multicenter study on radiomic features from T2-weighted images of a customized MR pelvic phantom setting the basis for robust radiomic models in clinics. Magn Reson Med 2020. [DOI] [PubMed] [Google Scholar]
- 31.Kocak B, Durmaz ES, Kaya OK, Ates E, Kilickesmez O: Reliability of Single-Slice-Based 2D CT Texture Analysis of Renal Masses: Influence of Intra- and Interobserver Manual Segmentation Variability on Radiomic Feature Reproducibility. AJR Am J Roentgenol 2019; 213:377–383. [DOI] [PubMed] [Google Scholar]
- 32.Pati S, Verma R, Akbari H, et al. : Reproducibility analysis of multi-institutional paired expert annotations and radiomic features of the Ivy Glioblastoma Atlas Project (Ivy GAP) dataset. Med Phys 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gourtsoyianni S, Doumou G, Prezzi D, et al. : Primary Rectal Cancer: Repeatability of Global and Local-Regional MR Imaging Texture Features. Radiology 2017; 284:552–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tunali I, Hall LO, Napel S, et al. : Stability and reproducibility of computed tomography radiomic features extracted from peritumoral regions of lung cancer lesions. Med Phys 2019; 46:5075–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu H, Parra NA, Qi J, et al. : Repeatability of Quantitative Imaging Features in Prostate Magnetic Resonance Imaging. Front Oncol 2020; 10:551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Molina D, Pérez-Beteta J, Martínez-González A, et al. : Lack of robustness of textural measures obtained from 3D brain tumor MRIs impose a need for standardization. PloS One 2017; 12:e0178843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Molina D, Pérez-Beteta J, Martínez-González A, et al. : Influence of gray level and space discretization on brain tumor heterogeneity measures obtained from magnetic resonance images. Comput Biol Med 2016; 78:49–57. [DOI] [PubMed] [Google Scholar]
- 38.Chirra P, Leo P, Yim M, et al. : Multisite evaluation of radiomic feature reproducibility and discriminability for identifying peripheral zone prostate tumors on MRI. J Med Imaging Bellingham Wash 2019; 6:024502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baeßler B, Weiss K, Pinto Dos Santos D: Robustness and Reproducibility of Radiomics in Magnetic Resonance Imaging: A Phantom Study. Invest Radiol 2019; 54:221–228. [DOI] [PubMed] [Google Scholar]
- 40.Hu P, Wang J, Zhong H, et al. : Reproducibility with repeat CT in radiomics study for rectal cancer. Oncotarget 2016; 7:71440–71446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


