Abstract
Background
Long-term outbreaks of multidrug-resistant Gram-negative bacilli related to hospital-building water systems have been described. However, successful mitigation strategies have rarely been reported. In particular, environmental disinfection or replacement of contaminated equipment usually failed to eradicate environmental sources of Pseudomonas aeruginosa.
Methods
We report the investigation and termination of an outbreak of P. aeruginosa producing VIM carbapenemase (PA-VIM) in the adult intensive care unit (ICU) of a Swiss tertiary care hospital with active case finding, environmental sampling and whole genome sequencing (WGS) of patient and environmental strains. We also describe the implemented control strategies and their effectiveness on eradication of the environmental reservoir.
Results
Between April 2018 and September 2020, 21 patients became either infected or colonized with a PA-VIM strain. For 16 of them, an acquisition in the ICU was suspected. Among 131 environmental samples collected in the ICU, 13 grew PA-VIM in sink traps and drains. WGS confirmed the epidemiological link between clinical and environmental strains and the monoclonal pattern of the outbreak. After removing sinks from patient rooms and implementation of waterless patient care, no new acquisition was detected in the ICU within 8 months after the intervention.
Discussion
Implementation of waterless patient care with removal of the sinks in patient rooms was successful for termination of a PA-VIM ICU outbreak linked to multiple environmental water sources. WGS provides highly discriminatory accuracy to investigate environment-related outbreaks.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13054-021-03726-y.
Keywords: Pseudomonas aeruginosa, VIM, Carbapememase, Sink, Waterless, Outbreak, Aquatic reservoir, cgMLST
Background
Pseudomonas aeruginosa is an important cause of hospital-acquired infections, affecting predominantly fragile or immunocompromised patients [1–3]. A major factor in its prominence as a pathogen is its intrinsic resistance to antibiotics and disinfectants [4]. Increasing incidence of multidrug resistant P. aeruginosa, in particular carbapenemase-producing strains, is a global concern [5]. The Verona Integron-encoded Metallo-beta-lactamase (VIM), the most widespread Metallo-beta-lactamase (MBL) produced by P. aeruginosa, hydrolyses all classes of beta-lactams except monobactams and cefiderocol [6], making therapeutic options in case of infection very limited [7]. Carbapenem-resistant P. aeruginosa has therefore been ranked as critical-priority bacteria in the WHO priority list of antibiotic resistant bacteria [8]. In this context, prevention of acquisition and spread of these strains is a priority.
Hospital water and water-related devices may serve as a reservoir of healthcare-associated pathogens [9]. Several clinically important Gram-negative bacterial species, including P. aeruginosa are well adapted to colonize the biofilms of water systems, and their presence has been associated with sporadic infections and outbreaks in hospitalized patients [10]. Eradication of water reservoirs is particularly challenging and mitigation strategies have been rarely reported to be successful in the long-term [11]. We report the investigation of an outbreak of VIM-53 positive P. aeruginosa (PA-VIM) in the intensive care unit (ICU) of a Swiss tertiary care hospital with the first case being detected in March 2018, the identification of an environmental aquatic source with molecular characterisation and genomic analysis confirming the monoclonal pattern of the outbreak and the effect of the mitigation strategy. In accordance with the ORION recommendations for outbreak reporting [12], we provide a detailed description of the outbreak and its source, the containment measures that were implemented, and the follow-up of colonized or infected patients.
Methods
Setting
Geneva University Hospitals (HUG) is a large tertiary care hospital located in Geneva, Switzerland, with approximately 1900 patient beds. The adult ICU is a mixed medical-surgical ICU, comprised of 2 subunits with a total of 30 beds and providing care for approximately 2500 patients annually with an average length of stay of 4 days. One ward contains only single patient rooms with a sink and sewage disposal in each patient room (Fig. 1). The other ward contains patient rooms with 3–4 beds and for each patient room, a medication preparatory area with a sink, and a separated soiled utility room with a sink and a sewage disposal. The incidence of carbapenemase-producing Enterobacteriacae or non-fermentative Gram-negative bacteria detected in our institution in 2018, when the outbreak started, was low with 1.3 cases per 1000 patient days. From March 9 to May 19, 2020, the ICU expanded its capacity to 110 beds by beds densification and surface extension to admit the 1st wave of severe COVID-19 patients; the single patient rooms were reserved to COVID negative and/or patients infected or colonised by multidrug resistance organisms (MDRO) [13].
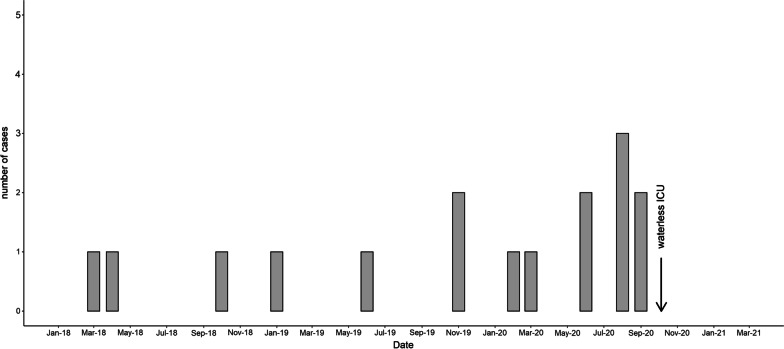
Fig. 1.
Epidemic curve of cases with infection and/or acquisition of Pseudomonas aeruginosa VIM in the intensive care unit (ICU). Between March 2018 and September 2020, 16 patients were newly colonized and/or infected with a P. aeruginosa producing VIM strain during their ICU stay. In October 2020, waterless patient care was implemented in the ICU with no new acquisition event in the ICU during the next 8 months
Infection control measures
Preventive measures were based on recent evidence and guidelines [14]. Weekly screening for intestinal carriage of MDRO was performed among all ICU patients. Targeted screening was performed at admission for patients presenting specific risk profiles [15]. In September 2020, due to the detection of further PA-VIM acquisition events, the ICU screening strategy was intensified with screening for MDRO upon admission, discharge and twice weekly for all patients present on the day of screening. In addition to rectal swabs, sampling of clinical sites (tracheal aspirations, urines, wounds) was encouraged. The enhanced screening was maintained for three months after the last case was detected. Detection of carbapenemase-producing (CP) P. aeruginosa prompted enhanced infection control procedures including contact precautions, single bed bedroom isolation and environmental chlorine cleaning. Patient status was marked with an alert in the electronic health record. Contact precautions and specific environmental cleaning procedures were maintained until 5 negative samples. As part of a ventilator-associated pneumonia (VAP) prevention bundle, patients likely to be ventilated more than 48 h receive three times per day selective oral-pharyngeal decontamination (SOD) with colistin, tobramycin and nystatin [16]. Daily SOD administration was maintained as long as the patient remained under mechanical ventilation. In case of newly identified colonisation by a MDR bacteria, the SOD regimen was interrupted. Due to a concomitant endemic problem with Serratia marcescens in the ICU detected in 2017 with a suspected water reservoir, several preventive interventions were already implemented in 2018 to mitigate contamination of sinks and reduce transmission of Gram-negative bacteria from potentially colonized sinks [17]. Educational rounds to reinforce compliance with hand hygiene, proper use of gloves and aseptic care procedures while using water were regularly performed. Mitigation strategies focused on re-enforced training of nursing staff on hand hygiene. Modification of behaviors to minimize drain colonisation were implemented, including limitation of the use of sinks for hand hygiene when specifically indicated only, procedures for patient bathing, separation of non-contaminated and contaminated areas and tasks, dedicated storage space > 1 m from sinks, and no use of sinks dedicated to direct care to the patient for hand washing. No disinfection of the sinks was performed.
Case definition
A case was defined as a patient with a clinical or screening sample positive for PA-VIM. A case was defined as definitely nosocomial if the patient was negative on a previous sample from the same body site during the same hospital stay. A case was defined as probably nosocomial when the patient had no previous negative sample during the same hospital stay and the first positive sample was retrieved more than 48 h after patient admission. A case was attributed to the ICU when the first positive sample was collected more than 48 h after the patient admission in the ICU, either during or after the stay in the ICU. All positive cases were followed until discharge or death.
Environmental sampling
Environmental sampling of water sources in the ICU was conducted between September and October 2020. Altogether, 131 environmental samples were collected. The samples were collected on sink drains (72), sink traps (36), sink elbows (3), washer decontaminators (8), water (6), water collected through bacterial filter (4), wipes (1) and ultrasound gel (1).
Bacterial identification, molecular characterization and genomic analysis
Clinical specimens, rectal swabs or stool samples and environmental specimens were inoculated on selective media (ESBL ChromID, McConkey and ChromID OXA-48). Specific search for P. aeruginosa was systematically performed. Bacterial isolates were identified by matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS, MALDI-Biotyper Bruker Daltonics). Antimicrobial susceptibility testing was performed in accordance with European Committee on Antimicrobial Susceptibility Testing guidelines by disk susceptibility testing. MICs for multidrug resistant isolates were determined by liquid susceptibility testing (Sensititre, ThemoFisher®). Selected isolates identified as carbapenem resistant by disk testing were additionally screened locally for carbapenemase production via polymerase chain reaction (PCR) (Amplex, eazyplex® SuperBug CRE for the detection of NDM, OXA-48, OXA-181 and VIM carbapenemase genes and home-made PCR for the detection of SME, IMP, GES, SPM, SIM, GIM). Strains were sequenced using the Illumina MiSeq platform. Sequence reads were analyzed using BioNumerics™ (version 7.6.3, created by bioMérieux, available at http://www.applied-maths.com). Core genome Multi Locus Sequence Typing (cgMLST) was performed as previously described [18].
Ethical considerations
This work was classified as service evaluation and outbreak investigation, and was therefore exempted from Ethics Committee Review.
Results
Outbreak description
Between March 2018 and September 2020, 21 patients newly colonised or infected with PA-VIM were identified at HUG. All PA-VIM strains were resistant to colistin. All cases were considered as definitely nosocomial or probably nosocomial, except one that was detected positive at admission screening but with several prior hospital stays; thus, prior hospital acquisition cannot be ruled out. Five patients never stayed in the ICU; however, a possible cross-transmission event outside the ICU with a positive case from the ICU was suspected for one of them; for the four other patients the source of acquisition could not be established. Acquisition was considered linked to the ICU for 16 patients out of 21 (76.2%) (Fig. 1). For 12 patients the first positive sample was collected during ICU stay. For 4 other patients, the detection of the first positive sample occurred after an ICU stay. The attack rate in the ICU for the entire period (from March 2018 to December 2020) was 2.3 /1000 admissions, with the highest rate occurring in August 2020 (18.4/1000 admissions). A cross-transmission event was suspected for one patient (case 11 in Table 1) because of the temporal and spatial link with another case (case 10) in the ICU.
Table 1.
Demographics and clinical characteristics of cases with an ICU-linked acquisition with date and site of isolation of VIM-PA (N = 16)
| Patient | Gender | Age (years) | Date of isolation of VIM-PA | Culture site | Initial site of infection or colonisation | Final outcome | Selective oral decontamination | Immunosupression/comorbidities | Site of acquisition |
|---|---|---|---|---|---|---|---|---|---|
| 1 | M | 61 | 2018-03-16 | Urine | Urine colonisation | Discharge | no | Chronic kidney disease | Possible ICU |
| 2 | F | 55 | 2018-04-09 | Urine | Urine colonisation | Discharge | no | Renal transplant | Possible ICU |
| 3 | M | 67 | 2018-10-15 | Perianal swab | Intestinal colonisation | Discharge | no | Severe aplastic anemia | ICU |
| 4 | F | 67 | 2019-01-21 | Wound | Wound colonisation | Discharge | no | Asthma, rheumatoid polyarthritis | Possible ICU |
| 5 | M | 33 | 2019-06-16 | Tracheal secretion, perianal swab | Respiratory tract colonisation and intestinal colonisation | Discharge | no | Tetraparesis | ICU |
| 6 | M | 61 | 2019-11-11 | Urine, wound, perianal swab | Urine and intestinal colonisation and sacral pressure ulcer | Discharge | no | HIV, Lymphoma B | ICU |
| 7 | M | 66 | 2019-12-31 | Urine | Urine colonisation | Discharge | no | Renal transplant | Possible ICU |
| 8 | M | 67 | 2020-02-03 | LBA, perianal swab | Peritonitis, intestinal colonisation | Discharge | yes | ICU | |
| 9 | F | 35 | 2020-03-21 | Blood | Bloodstream infection, septic choc | Death | yes | Renal transplant, hematologic malignancy | ICU |
| 10 | M | 56 | 2020-06-07 | Perianal swab, urine, sputum | Intestinal and urine colonisation | Discharge | no | Chronic cardiac disease | ICU |
| 11 | M | 75 | 2020-06-28 | Perianal swab | Intestinal colonisation | Death | no | ICU | |
| 12 | M | 68 | 2020-08-01 | Urine, blood | Urinary tract infection, bloodstream infection | Discharge | yes | Renal transplant | ICU |
| 13 | F | 16 | 2020-08-09 | Urine | Intestinal and urine colonisation | Discharge | yes | Renal and liver transplant | ICU |
| 14 | F | 43 | 2020-08-26 | Wound sacral | Intestinal colonisation and wound (sacral pressure ulcer) | Discharge | no | Denutrition | ICU |
| 15 | F | 75 | 2020-09-07 | Perianal swab | Intestinal colonisation | Discharge | yes | ICU | |
| 16 | M | 53 | 2020-09-14 | Perianal swab, urine | Intestinal and respiratory tract colonisation, orchi-epididymitis | Discharge | no | Stem cell transplant | ICU |
VIM-PA: Pseudomonas aeruginosa producing VIM carbapenemase
Demographics and clinical characteristics of the 16 patients with suspected or confirmed ICU acquisition are summarized in Table 1. Median age was 61 years (IQR, 50.5–67); 10 (62.5%) were male. Five of them (31.3%) were infected and 11 (68.7%) were colonized only. Sites of colonisation were intestinal colonisation for 10 (66.7%), urinary colonisation for 6 (37.5%), respiratory tract colonisation for 2 (12.5%) and wound colonisation for 3 (18.7%). Six patients had more than one site colonised. One patient presented a bloodstream infection and one died during the course of his infection. Another case died in the context of undocumented sepsis, with unclear attributability to PA-VIM. Five patients received selective oral decontamination (SOD) for VAP prevention.The median time lag between start of the SOD and first documentation of PA-VIM was 16 days (IQR 9–18). No decolonisation regimen was offered to patients, due to the lack of evidence of decolonisation strategies for carbapenemase producing bacteria [19, 20].
Environmental investigations
Among the 131 environmental samples collected, 16 were positive for PA-VIM (12.2%). Among the positive samples, 11 samples were taken from sink traps, 4 from sink strainers and 1 from a washer disinfector. Location of the positive and negative samples in the ICU are shown in Fig. 2.
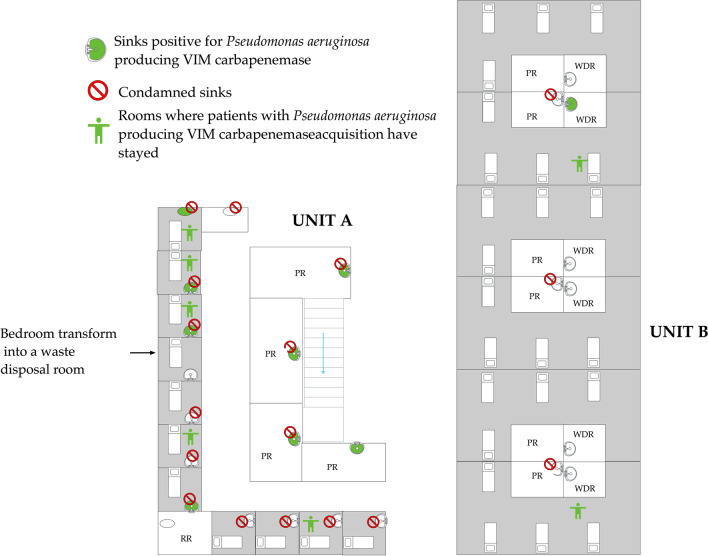
Fig. 2.
Map of the intensive care unit with the two subunits A and B. Sinks with samples positive for Pseudomonas aeruginosa VIM are filled in green. Sinks condemned are marked with a red forbidden sign. Bedrooms who hosted patients with newly P. aeruginosa VIM acquisition are marked with a green human symbol. PR: preparation room, WDR: waste disposal room, RR: rest room for healthcare workers
Sequencing results
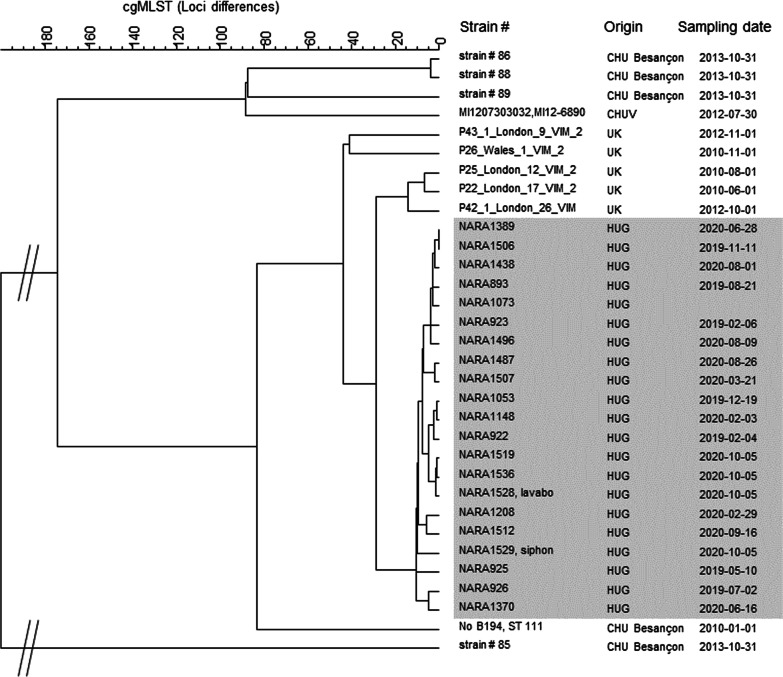
Clinical isolates from the 16 patients with suspected ICU acquisition and 2 isolates from environmental sampling were sequenced. All isolates belonged to the sequence type 111 (ST-111). Additionally, 3 other clinical isolates (one from a patient with suspected cross transmission outside ICU and 2 from patients with unknown source of acquisition) were sequenced and belonged to the same cluster. Isolates were compared with 6 ST-111 strains from a cluster in a UK hospital [21] and 5 ST-111 strains from a French hospital [22]. The analysis of these 32 genomes showed 1112/5932 variable loci. Clustering was performed using a categorical coefficient and an UPGMA similarity tree was constructed (Fig. 3). Differences of 0–15 loci differences were observed among the 21 HUG isolates, confirming that the HUG cluster was different from the 11 other strains.
Fig. 3.
UPGMA similarity tree based on cgMLST analysis of 32 ST111 strains of Pseudomonas aeruginosa. The tree includes 19 clinical strains and 2 environmental strains (sinks drains) from HUG, 5 strains from UK, 5 strains from Besançon (France) and 1 strain from Lausanne (Switzerland). The strain number, its origin and date of sampling are mentioned. The 21 strains from HUG belong to the same cluster. From the 19 clinical strains from HUG, 16 belong to patients with a suspected
source of acquisition in the ICU, 1 from a patient with suspected cross transmission outside ICU and 2 from patients with nosocomial but unknown source of acquisition
Mitigation strategies
In September 2020, due to the detection of three new acquisition events and in regard to the conclusions of the environmental and genomic investigations with a clear evidence for a water reservoir, we implemented “water-free” patient care with removal of all sinks from patient rooms and of most of the sinks in medication preparation areas. Sinks were kept only in rooms dedicated to waste disposal and two sinks were kept in preparation rooms. All patient care-related activities that take place in the patient room and that would normally involve the use of tap water were adapted to a ‘water-free’ alternative (table in Additional file 1: Appendix). A bathing protocol using disposable chlorhexidine-free washing gloves and shampoo cap (SINAQUA®, WELCARE INDUSTRIES S.p.A.) was implemented. In September 2020, we also encouraged intensivists to re-assess on a case-by-case basis the indication for SOD and limit its use.
Impact of the mitigation strategies and termination of the outbreak
After implementation of water-free patient care, no new acquisition event was detected in the ICU within the next 8 months (until May 2021). In January 2021, we discovered 4 new acquisition events of PA-VIM within HUG which occurred in a shared bedroom in an intermediate care unit hosting COVID-19 patients located in a different building. The first detected case had stayed in the ICU before but all screening samples collected during his stay in the ICU were negative. New environmental samples were collected in the sink located in the waste disposal room beside the ICU room the patient stayed and all were negatives.
Discussion
We report here the successful control of a sink-related PA-VIM outbreak and the eradication of the water reservoir by introducing waterless patient care in an ICU of a tertiary care hospital in Switzerland.
The role of the hospital water environment as a reservoir and transmission pathway for hospital-acquired infections outbreaks has received increasing attention over the last decade [9, 10, 23]. Several outbreaks linked to the different water reservoirs (faucets, sinks, potable water, sink surfaces and wastewater drainage system) have been reported [24, 25], most frequently involving multidrug resistant Gram-negative bacteria including Enterobacteriaceae and non-fermentative Gram-negative such as Pseudomonas sp, and Acinetobacter sp. and other Gram-negative bacilli. These bacteria are well adapted to survive for a long time in the water environment and create biofilms, making them resistant to usual disinfection procedures. Water-related outbreaks occurred frequently in ICU settings, but have also been reported in burn units or neonatal intensive care units [9]. The population affected are frequently immunocompromised patients (e.g. haematological malignancies, stem cell transplants), as observed in the current outbreak. Furthermore, exposure to multiple medical devices has clearly been associated with water-related HAI [26, 27]. Several mechanisms and infection control breaches can lead to patient contamination from the water sources. Among them, poor sink designs, use of sinks for contaminated clinical disposal, storage of clean patient materials around sinks, blocked sewage pipe and waste pipes leaks have been reported [10, 19]. Hota et al. demonstrated, with the use of fluorescein injection into sinks, splash-back up to 1 m from the sink when the water was running [24]. In this scenario, biofilms spread from the sink trap to the sink drain and organisms are dispersed when impacted by water from the skin faucet. Gram-negative organisms dispersed can directly contaminate the patient or its environment in case of close contact between the patient bed and the sink (as it was the case in some patient rooms of our ICU), or can transiently contaminate the hands of healthcare workers during handwashing who can then contaminate the patient or its environment [29].
Several mitigation strategies have been reported with more or less success in outbreak termination and eradication of environmental water reservoirs [11]. Some engineering modifications can minimize contamination of surrounding surfaces and objects. For example, better sink design to reduce splashing, appropriate use of splash guards or barriers or rearrangement of the surrounding space with dedicated storage space more than 1 m from sinks have been proposed [30–32]. Nevertheless, major structural modifications are not always feasible. In our case, initial mitigations strategies implemented in the context of an endemic S. marcescens situation and which aimed to reduce colonisation of surrounding objects from sinks trough dedicated training of healthcare workers and modification of behaviors did not prevent the PA-VIM outbreak. Sinks in patient rooms were located less than 1 m from patient beds which left little room for architectural reorganization. Other interventions such as disinfection of sink drains attempted to reduce biofilms formation in sinks once colonized with multidrug-resistant organisms, but they have usually failed in reported outbreaks [11]. Novel technologies such as installation of devices that heat and vibrate on the exterior of traps or ozone might be promising; however, efficacy and cost-effectiveness still need to be demonstrated [33]. Implementation of waterless care activities was a successful measure in the context of our ICU, and has already been reported in a similar outbreak [24]. Hopman et al. reported that removal of the sinks from all patient rooms and the introduction of ‘water-free’ patient care in 2014 was associated with a statistically significant lower number of ICU patients that become colonized with Gram negative bacteria, including MDR Gram negative bacteria [34]. Nevertheless, this strategy is usually not implementable outside the ICU context and alternative solutions are urgently needed.
Selection pressure from administration of broad-spectrum antibiotics is well described as being an important contributor for acquisition [26]. Five patients in this outbreak received SOD before acquisition of the PSA-VIM. Emergence of colistin resistance among ESBL-producing Klebsiella pneumoniae isolates has been reported after introduction of Selective Digestive Tract Decontamination in an ICU [35]. Even if the potential benefits of SOD on protection against VAP are well described in the literature, we recommend in the context of a MDR outbreak, to re-assess the balance between benefits and risks and consider, at least temporarily, to suspend its administration.
We suspected one cross-transmission event in summer 2020 between the 1st and 2nd COVID-19 waves. As reported in an MRSA outbreak in an ICU during the SARS-CoV-1 epidemic [36] changes in infection control measures and personal protective equipment with universal gloving policy, among other factors, may have favoured this transmission event. Interestingly only one PA-VIM environmental acquisition was documented during the high activity of the 1st COVID-19 wave before water free patient care implementation.
WGS performed on 19 patient strains and 2 environmental strains showed that they all belonged to the ST-111 and had only 1–15 loci differences with wgMLST. This strongly suggest they are epidemiologically linked and belonged to the same chain of transmission [18].
Inferring potential transmission events solely based on epidemiological data proved challenging due to overlap of patients in the hospital. Considering that phylogenetically closely related isolates have a common ancestor and thus derive from a chain of transmission, the high power of discrimination of WGS allows to unravel outbreaks and decipher transmission events [37, 38].
In contrast to current conventional wisdom, our active patient screening policy did not help to completely stop the outbreak—it helped to detect cases and guide us towards an environmental reservoir. Thus, additional environmental screening and interventions are crucial and should be emphasized, in addition to conventional IPC measures.
A limitation of this study is the relatively short time lag of 8 months to evaluate the sustainability of the measures in stopping the outbreak, considering also that seasonality may have play a role.
Conclusion
MDRO infections due to environmental sources is a major issue in ICU setting. We report an outbreak of PA-VIM in the ICU of a Swiss tertiary care hospital. Environmental and genomic investigations confirmed the aquatic source and the outbreak was controlled by implementation of waterless patient care, in addition to conventional infection control measures.
Supplementary Information
Additional file 1: Appendix. Waterless alternatives implemented in the ICU for patient care-related actions.
Acknowledgements
We would like to thank all the dedicated HCWs of the intensive care unit and members of the HUG SPCI team implicated in the outbreak (Americo Agostinho, Catherine Bandiera-Clerc, Véronique Camus, Claude Ginet, Dan Lebowitz, Delphine Pérréard, Marlène Fraccaro-Hamoneau, Carolina Frankhasuer-Rodriguez, Valérie Sauvan, Isabelle Soulake). We thank Daniel Teixera for his help with data extraction.
Authors' contributions
GC wrote the first draft of the manuscript. All the authors revised the manuscript and made significant contribution. All authors read and approved the final manuscript.
Funding
This work has not been funded by an external source.
Availability of supporting data
All supporting data will be made fully available.
Declarations
Ethical approval and consent to participate
This work was classified as service evaluation and outbreak investigation, and was therefore exempted from Ethics Committee Review.
Consent for publication
Not applicable.
Competing interests
All authors have no conflict of interest to declare.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Zawacki A, O’Rourke E, Potter-Bynoe G, Macone A, Harbarth S, Goldmann D. An outbreak of Pseudomonas aeruginosa pneumonia and bloodstream infection associated with intermittent otitis externa in a healthcare worker. Infect Control Hosp Epidemiol. 2004;25(12):1083–1089. doi: 10.1086/502348. [DOI] [PubMed] [Google Scholar]
- 2.Longtin Y, Troillet N, Touveneau S, Boillat N, Rimensberger P, Dharan S, et al. Pseudomonas aeruginosa outbreak in a pediatric intensive care unit linked to a humanitarian organization residential center. Pediatr Infect Dis J. 2010;29(3):233–237. doi: 10.1097/INF.0b013e3181bc24fb. [DOI] [PubMed] [Google Scholar]
- 3.Tissot F, Blanc DS, Basset P, Zanetti G, Berger MM, Que Y-A, et al. New genotyping method discovers sustained nosocomial Pseudomonas aeruginosa outbreak in an intensive care burn unit. J Hosp Infect. 2016;94(1):2–7. doi: 10.1016/j.jhin.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 4.Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, Hickey MJ, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406(6799):959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 5.Simner PJ, Opene BNA, Chambers KK, Naumann ME, Carroll KC, Tamma PD. Carbapenemase detection among carbapenem-resistant glucose-nonfermenting gram-negative bacilli. J Clin Microbiol. 2017;55(9):2858–2864. doi: 10.1128/JCM.00775-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jacoby GA, Munoz-Price LS. The New β-Lactamases. New England J Med. 2005;352(4):380–391. doi: 10.1056/NEJMra041359. [DOI] [PubMed] [Google Scholar]
- 7.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440–58. doi: 10.1128/CMR.00001-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tacconelli E, Carrara E, Savoldi A, Harbarth S, Mendelson M, Monnet DL, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 9.Kanamori H, Weber DJ, Rutala WA. Healthcare outbreaks associated with a water reservoir and infection prevention strategies. Clin Infect Dis. 2016;62(11):1423–1435. doi: 10.1093/cid/ciw122. [DOI] [PubMed] [Google Scholar]
- 10.Kizny Gordon AE, Mathers AJ, Cheong EYL, Gottlieb T, Kotay S, Walker AS, et al. The hospital water environment as a reservoir for carbapenem-resistant organisms causing hospital-acquired infections—a systematic review of the literature. Clin Infect Dis. 2017;64(10):1435–1444. doi: 10.1093/cid/cix132. [DOI] [PubMed] [Google Scholar]
- 11.Lewis SS, Smith BA, Sickbert-Bennett EE, Weber DJ. Water as a source for colonization and infection with multidrug-resistant pathogens: focus on sinks. Infect Control Hosp Epidemiol. 2018;39(12):1463–1466. doi: 10.1017/ice.2018.273. [DOI] [PubMed] [Google Scholar]
- 12.The ORION statement: guidelines for transparent reporting of Outbreak Reports and Intervention studies Of Nosocomial infection|The EQUATOR Network [Internet]. [cited 2021 Feb 16]. https://www.equator-network.org/reporting-guidelines/the-orion-statement-guidelines-for-transparent-reporting-of-outbreak-reports-and-intervention-studies-of-nosocomial-infection/.
- 13.Primmaz S, Le Terrier C, Suh N, Ventura F, Boroli F, Bendjelid K, et al. Preparedness and reorganization of care for coronavirus disease 2019 patients in a Swiss ICU: characteristics and outcomes of 129 patients. Crit Care Explor. 2020;2(8):e0173. doi: 10.1097/CCE.0000000000000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tomczyk S, Zanichelli V, Grayson ML, Twyman A, Abbas M, Pires D, et al. Control of carbapenem-resistant enterobacteriaceae, Acinetobacter baumannii, and Pseudomonas aeruginosa in healthcare facilities: a systematic review and reanalysis of quasi-experimental studies. Clin Infect Dis. 2019;68(5):873–884. doi: 10.1093/cid/ciy752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fankhauser C, Zingg W, Francois P, Dharan S, Schrenzel J, Pittet D, et al. Surveillance of extended-spectrum-beta-lactamase-producing Enterobacteriaceae in a Swiss Tertiary Care Hospital. Swiss Med Wkly. 2009;139(51–52):747–751. doi: 10.4414/smw.2009.12918. [DOI] [PubMed] [Google Scholar]
- 16.Landelle C, Nocquet Boyer V, Abbas M, Genevois E, Abidi N, Naimo S, et al. Impact of a multifaceted prevention program on ventilator-associated pneumonia including selective oropharyngeal decontamination. Intensive Care Med. 2018;44(11):1777–1786. doi: 10.1007/s00134-018-5227-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martischang R, Sauvan V, Leo S, Lazarevic V, Chraïti M-N, Martin Y, Soule H, Schrenzel J, Koyluk Tomsuk Z, Pugin J, Harbarth S. Abstracts from the 5th International Conference on Prevention & Infection Control (ICPIC 2019). P489 “behind the sinks”: endemic Serratia marcescens in a swiss intensive care unit (ICU). Antimicrob Resist Infect Control. 2019;8(1):148.
- 18.Blanc DS, Magalhães B, Koenig I, Senn L, Grandbastien B. Comparison of whole genome (wg-) and core genome (cg-) MLST (BioNumericsTM) versus SNP variant calling for epidemiological investigation of Pseudomonas aeruginosa. Front Microbiol. 2020;11:1729. doi: 10.3389/fmicb.2020.01729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catho G, Huttner BD. Strategies for the eradication of extended-spectrum beta-lactamase or carbapenemase-producing Enterobacteriaceae intestinal carriage. Expert Rev Anti Infect Ther. 2019;17(8):557–569. doi: 10.1080/14787210.2019.1645007. [DOI] [PubMed] [Google Scholar]
- 20.Huttner BD, de Lastours V, Wassenberg M, Maharshak N, Mauris A, Galperine T, et al. A five-day course of oral antibiotics followed by faecal transplantation to eradicate carriage of multidrug-resistant enterobacteriaceae: a randomized clinical trial. Clin Microbiol Infect. 2019;25:830–838. doi: 10.1016/j.cmi.2018.12.009. [DOI] [PubMed] [Google Scholar]
- 21.Turton JF, Wright L, Underwood A, Witney AA, Chan Y-T, Al-Shahib A, et al. High-resolution analysis by whole-genome sequencing of an international lineage (sequence type 111) of Pseudomonas aeruginosa associated with metallo-carbapenemases in the United Kingdom. J Clin Microbiol. 2015;53(8):2622–2631. doi: 10.1128/JCM.00505-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cholley P, Stojanov M, Hocquet D, Thouverez M, Bertrand X, Blanc DS. Comparison of double-locus sequence typing (DLST) and multilocus sequence typing (MLST) for the investigation of Pseudomonas aeruginosa populations. Diagn Microbiol Infect Dis. 2015;82(4):274–277. doi: 10.1016/j.diagmicrobio.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 23.Exner M, Kramer A, Lajoie L, Gebel J, Engelhart S, Hartemann P. Prevention and control of health care–associated waterborne infections in health care facilities. Am J Infect Control. 2005;33(5):S26–40. doi: 10.1016/j.ajic.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Hota S, Hirji Z, Stockton K, Lemieux C, Dedier H, Wolfaardt G, et al. Outbreak of multidrug-resistant Pseudomonas aeruginosa colonization and infection secondary to imperfect intensive care unit room design. Infect Control Hosp Epidemiol. 2009;30(1):25–33. doi: 10.1086/592700. [DOI] [PubMed] [Google Scholar]
- 25.Pournaras S, Maniati M, Petinaki E, Tzouvelekis LS, Tsakris A, Legakis NJ, et al. Hospital outbreak of multiple clones of Pseudomonas aeruginosa carrying the unrelated metallo-beta-lactamase gene variants blaVIM-2 and blaVIM-4. J Antimicrob Chemother. 2003;51(6):1409–1414. doi: 10.1093/jac/dkg239. [DOI] [PubMed] [Google Scholar]
- 26.Voor In ’t Holt AF, Severin JA, Lesaffre EMEH, Vos MC. A systematic review and meta-analyses show that carbapenem use and medical devices are the leading risk factors for carbapenem-resistant Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2014;58(5):2626–37. doi: 10.1128/AAC.01758-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perkins KM, Reddy SC, Fagan R, Arduino MJ, Perz JF. Investigation of healthcare infection risks from water-related organisms: summary of CDC consultations, 2014–2017. Infect Control Hosp Epidemiol. 2019;40(6):621–626. doi: 10.1017/ice.2019.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gestrich SA, Jencson AL, Cadnum JL, Livingston SH, Wilson BM, Donskey CJ. A multicenter investigation to characterize the risk for pathogen transmission from healthcare facility sinks. Infect Control Hosp Epidemiol. 2018;39(12):1467–1469. doi: 10.1017/ice.2018.191. [DOI] [PubMed] [Google Scholar]
- 29.Decker BK, Palmore TN. The role of water in healthcare-associated infections. Curr Opin Infect Dis. 2013;26(4):345–351. doi: 10.1097/QCO.0b013e3283630adf. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chia PY, Sengupta S, Kukreja A, Ponnampalavanar SS, Ng OT, Marimuthu K. The role of hospital environment in transmissions of multidrug-resistant gram-negative organisms. Antimicrob Resist Infect Control. 2020;9(1):29. doi: 10.1186/s13756-020-0685-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Department of Health and Social Care. Design for flooring, walls, ceilings, sanitary ware and windows (HBN 00-10) [Internet]. 2013 [cited 2021 April 13]. https://www.gov.uk/government/publications/guidance-on-flooring-walls-and-ceilings-and-sanitary-assemblies-in-healthcare-facilities.
- 32.Infection Control-International Health Facility Guidelines [Internet]. 2017 [cited 2021 April 13]. https://healthfacilityguidelines.com/GuidelineIndex/Index/Infection-Control.
- 33.Livingston S, Cadnum JL, Gestrich S, Jencson AL, Donskey CJ. Efficacy of automated disinfection with ozonated water in reducing sink drainage system colonization with Pseudomonas species and Candida auris. Infect Control Hosp Epidemiol. 2018;39(12):1497–1498. doi: 10.1017/ice.2018.176. [DOI] [PubMed] [Google Scholar]
- 34.Hopman J, Bos R, Voss A, Kolwijck E, Sturm P, Pickkers P, et al. Reduced rate of MDROs after introducing ‘water-free patient care’ on a large intensive care unit in the Netherlands. Antimicrob Resist Infect Control. 2015;4(Suppl 1):O40. doi: 10.1186/2047-2994-4-S1-O40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Halaby T, Al Naiemi N, Kluytmans J, van der Palen J, Vandenbroucke-Grauls CMJE. Emergence of colistin resistance in Enterobacteriaceae after the introduction of selective digestive tract decontamination in an intensive care unit. Antimicrob Agents Chemother. 2013;57(7):3224–3229. doi: 10.1128/AAC.02634-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yap FHY, Gomersall CD, Fung KSC, Ho P-L, Ho O-M, Lam PKN, et al. Increase in methicillin-resistant staphylococcus aureus acquisition rate and change in pathogen pattern associated with an outbreak of severe acute respiratory syndrome. Clin Infect Dis. 2004;39(4):511–516. doi: 10.1086/422641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Snyder LA, Loman NJ, Faraj LA, Levi K, Weinstock G, Boswell TC, et al. Epidemiological investigation of Pseudomonas aeruginosa isolates from a six-year-long hospital outbreak using high-throughput whole genome sequencing. Euro Surveill. 2013;18(42):20611. doi: 10.2807/1560-7917.ES2013.18.42.20611. [DOI] [PubMed] [Google Scholar]
- 38.Magalhães B, Valot B, Abdelbary MMH, Prod’hom G, Greub G, Senn L, et al. Combining standard molecular typing and whole genome sequencing to investigate Pseudomonas aeruginosa epidemiology in intensive care units. Front Public Health. 2020;8:3. doi: 10.3389/fpubh.2020.00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Appendix. Waterless alternatives implemented in the ICU for patient care-related actions.
Data Availability Statement
All supporting data will be made fully available.