Abstract
DNA methylation on cytosines of CpG dinucleotides is well established as a basis of epigenetic regulation in mammalian cells. Since aberrant regulation of DNA methylation in promoters of tumor suppressor genes or proto-oncogenes may contribute to the initiation and progression of various types of human cancer, sequence-specific methylation and demethylation technologies could have great clinical benefit. The CRISPR-Cas9 protein with a guide RNA can target DNA sequences regardless of the methylation status of the target site, making this system superb for precise methylation editing and gene regulation. Targeted methylation-editing technologies employing the dCas9 fusion proteins have been shown to be highly effective in gene regulation without altering the DNA sequence. In this review, we discuss epigenetic alterations in tumorigenesis as well as various dCas9 fusion technologies and their usages in site-specific methylation editing and gene regulation.
Methylation of DNA on cytosines in CpG dinucleotides is well established as a basis of epigenetic regulation in development and human cancers (Jones and Baylin 2007; Greenberg and Bourc’his 2019; Park and Han 2019). Dysregulation of DNA methylation has been shown to contribute to the initiation and progression of various cancers, and thus sequence-specific methylation editing technologies could have broad and great clinical impact. The clustered regulatory interspaced short palindromic repeat (CRISPR)-Cas9 system has been used in many applications, including genome editing, gene regulation and genetic screens (Jinek et al. 2012; Cho et al. 2013; Cong et al. 2013; Mali et al. 2013; Kweon and Kim 2018; Lee et al. 2018; Sato et al. 2018). In this system, a guide RNA (gRNA) binds to the target site and recruits the Cas9 nuclease protein for gene editing or deletion. The nuclease-inactivated Cas9 protein (dCas9) is a useful methylation editing tool to selectively target DNA sites with high specificity and binding efficiency (Amabile et al. 2016; Liu et al. 2016; McDonald et al. 2016; Vojta et al. 2016; Huang et al. 2017; Stepper et al. 2017; Xiong et al. 2017; Pflueger et al. 2018). The dCas9 protein with a gRNA can target DNA sequences regardless of the methylation status of the target site, making this system superb for precise methylation editing and gene regulation (Hsu et al. 2013). In this review, we discuss epigenetic alterations in carcinogenesis as well as CRISPR-based methylation editing technologies and their regulatory mechanisms. The in vivo applications and off-target effects of the epigenetic tools are also discussed.
Dysregulation of methylation in carcinogenesis
Aberrant regulation of DNA methylation has been observed in various human carcinomas, including breast cancer (Fackler et al. 2004), colorectal cancer (Cui et al. 2002; Cui et al. 2003), lung cancer (Belinsky et al. 2002a; Jarmalaite et al. 2003), liver cancer (Shen et al. 1998; Tao et al. 2000), ovarian cancer (Sung et al. 2013), and glioblastoma (Weller et al. 2010). Abnormal hypermethylation of CpG-rich regions (CpG islands) in the promoters of tumor suppressor genes and hypomethylation at highly or moderately repeated heterochromatin DNA sequences in oncogenes (Ehrlich 2002; Ehrlich et al. 2002) are associated with increased malignancy in ovarian cancer (Sung et al. 2013), breast cancer (Widschwendter and Jones 2002; Fackler et al. 2004), lung cancer (Belinsky et al. 2002a; Jarmalaite et al. 2003), and glioblastoma (Weller et al. 2010). Basically, DNA methylation is an epigenetic regulatory mechanism for gene silencing through transcriptional repression, which occurs at the DNA base cytosine mainly within CpG-rich regions, producing 5’-methylcytosine (5-mC) catalyzed by DNA methyltransferases (DNMTs). DNA demethylation is catalyzed by ten-eleven translocation methylcytosine dioxygenase (TET) for the conversion from 5-methylcytosine (5-mC) to 5-hydroxymethylcytosine (5-hmC). These represent essential epigenetic physiological processes that ensure both cellular and tissue homeostasis (Jones and Baylin 2007; Greenberg and Bourc’his 2019; Park and Han 2019). DNA methylation status can be characterized by the balance between methylation and demethylation status at the locus for the biological effects, but it has been widely established that promoter methylation status is correlated with the levels of gene expression.
The abnormal DNA methylation appears to be an early event in carcinogenesis, and normal methylation is disrupted during carcinogenesis. Generally, promoter hypermethylation of tumor suppressor genes results in silencing of tumor suppressor genes, while hypomethylation of oncogenes leads to activating oncogenes, which are common events in carcinogenesis (Jones 2002; Choi et al. 2017). Consequently, tumor cells acquire advantages for selective growth through the genetic instability of the tumor (Jones 2002). Many cancer studies have demonstrated that the tumor suppressor genes that are methylated are frequently involved in cell cycle arrest (e.g. TP53 (Chuikov et al. 2004), SALL2 (Sung et al. 2013), p16 INK4A, p14 ARF (Belinsky et al. 2002b; Jarmalaite et al. 2003)), DNA repair (MGMT (Weller et al. 2010), hMLH1 (Capel et al. 2007)), and so on. Hypomethylation of promoter regions of proto-oncogenic genes such as c-Myc, N-Ras, and c-Jun increased expression of the corresponding genes at both RNA and protein levels in carcinoma (Shen et al. 1998; Tao et al. 2000). Cancer-associated promoter hypomethylation is often related with decreases of overall genomic or satellite DNA methylation. In many studies, both cancer-associated DNA hypomethylation and hypermethylation are altered in the genome of tumors. However, hypomethylation and hypermethylation are generally independent events in cancer. High frequencies of these alterations in cancer increase malignancy and may eventually lead to tumor cell heterogeneity.
Epigenetic alterations in advanced cancer and chemotherapy
Differences in epigenetic expression in primary and metastatic tumors have been suggested. Hypermethylation of tumor suppressor genes is more often observed in metastatic cancer than primary cancer. The frequency of hypermethylation of cyclin D2, RAR-beta, Twist, RASSF1A, and HIN-1 promoters was examined in primary breast cancer and metastatic sites (Mehrotra et al. 2004). All five genes had higher frequencies of hypermethylation in metastatic bone, brain, and lung compared with the primary breast carcinoma. Absence of gene expressions correlated to hypermethylation of their promoters in metastatic carcinoma cells microdissected from lymph nodes (Mehrotra et al. 2004). In addition, epigenomic reprogramming during pancreatic cancer metastasis demonstrated that specified malignant epigenetic alterations were targeted to thousands of chromatin domains across the genome (McDonald et al. 2017).
Chemoresistant cancer is also regulated by an epigenetic network. The global DNA methylation patterns in adriamycin-resistant human breast cancer and paclitaxel-resistant breast cancer cells are similar. However, these patterns are significantly different from the parental breast cancer, indicating that DNA methylation is changed in chemoresistant cancer cells (Gu et al. 2016; He et al. 2016). Genome-wide profiling of methylation and gene expression in chemoresistant breast cancer revealed that methylation plays a role in gene silencing during the acquisition of chemoresistance, because gene expression in chemoresistant cancer cells is negatively correlated with the promoter and 5’UTR methylation compared with parent cells (He et al. 2016). Methylation of Notch3, a tumor suppressor and inhibitor of MDR1, is inactivated by DNA hypermethylation in adriamycin-resistant human breast cancer cells, and is related with the expression of the multidrug-resistant gene, MDR1 (Gu et al. 2016). In ovarian cancer, dysregulation of DNA hypermethylation is observed in platinum drug-resistant cells. In an analysis of large-scale transcriptome changes in cisplatin-resistant ovarian cancer, resistance was related with loss of hypermethylation at several CpG sites primarily localized in the intergenic regions of the genome (Lund et al. 2017). Changes in KLF4 and IL6 from aberrant methylation in platinum-resistant ovarian cancer cells were observed as potential key drivers of drug resistance.
DNA methylation can also change depending on the chemotherapy. Among anticancer drugs, 5-fluorouracil (5-FU) changes DNA methylation in gastric cancer patients (Mitsuno et al. 2007). In a study of 56 gastric cancer patients, methylation of p16INK4a displayed a significant correlation with longer survival in the 38 patients in the 5-FU chemotherapy group, but not in the 18 patients of the non-treated group, suggesting that p16INK4a methylation is induced by 5-FU-based chemotherapy (Mitsuno et al. 2007). In ovarian cancer, platinum-based chemotherapy leads to different methylation status, which is associated with patient overall survival (Flanagan et al. 2017). Cell based-experiments revealed that functional DNA mismatch repair increases the frequency of platinum-induced DNA methylation alterations. Therefore, these results suggest that detection of DNA methylation in blood following chemotherapy could be useful as a noninvasive method of monitoring patients’ epigenetic responses after chemotherapy. Together these studies indicate that epigenetic regulation plays an important role in cancer progression and chemoresistance, indicating its potential application in cancer diagnosis, prognosis, and chemotherapy.
CRISPR-mediated promoter methylation and demethylation technologies
Since abnormal DNA methylation is linked to the initiation and progression of various human cancers, site-specific methylation editing tools could have great clinical benefit. Many groups have used DNA-binding proteins, including zinc finger protein (ZNF) and transcription activator-like effectors (TALEs), fused to a methyltransferase or demethylase enzyme for targeted DNA methylation editing (Li et al. 2007; Rivenbark et al. 2012; Maeder et al. 2013; Siddique et al. 2013; Nunna et al. 2014; Bernstein et al. 2015). While these approaches with engineered nucleotide-binding proteins efficiently edited methylation states at the target sites, these methods also displayed limitations, showing non-specific binding to the genome with high off-target effects and requiring labor-intensive design of each DNA-binding motif. Liu et al. directly compared TALE-based approaches with CRISPR-based methods and evaluated their methylation specificity and gene regulation efficiency in human cells (Liu et al. 2016). The authors constructed TALE- and CRISPR-demethylase fusion vectors that target the same promoter region (RHOXF2) and determined the methylation rates of the RHOXF2 promoter by conducting a bisulfite sequencing assay. The results revealed that the demethylation activity of the CRISPR fusion at the target sequence was two-fold higher than that of the TALE vector, suggesting that the CRISPR vector system may have better methylation editing efficiency. In addition, the authors performed a chromatin immunoprecipitation sequencing assay with an anti-Cas9 antibody and found that the CRISPR system achieved high target specificity. Furthermore, among various gene editing technologies, the CRISPR-Cas9 system with a gRNA was a superior tool for DNA methylation editing since it can target DNA sequences regardless of the methylation status of the target site (Hsu et al. 2013). Many research groups have used a nuclease-inactivated Cas9 protein (dCas9) fused to methyltransferase or demethylase for selective DNA methylation or demethylation and regulation of target gene expression.
Targeted promoter methylation
Development and maintenance of many human cancers are due, in part, to overexpression of proto-oncogenes. Therefore, targeting the promoters of proto-oncogenes for DNA methylation is an attractive therapeutic strategy to block their transcription and target cancer cell populations. DNA methylation is facilitated by the DNA methyltransferase enzymes DNMT3A and DNMT3B, and methylation is maintained by the enzyme DNMT1 (Smith and Meissner 2013; Greenberg and Bourc’his 2019). Fusion proteins of dCas9 with a methyltransferase enzyme, such as dCas9-DNMT3A and -DNMT3B fusion clones, have been used for site-specific promoter methylation to suppress downstream genes (Table 1) (Amabile et al. 2016; Liu et al. 2016; McDonald et al. 2016; Vojta et al. 2016; Huang et al. 2017; Stepper et al. 2017; Xiong et al. 2017; Pflueger et al. 2018). In this gene regulation approach, the fusion proteins are recruited to the target site by a gRNA for sequence-specific DNA methylation (Fig.. 1A).
Table 1.
The dCas9 vectors for site-specific methylation and gene silencing.
| dCas9 fusion vectors | Description | Targeted gene/promoter | References |
|---|---|---|---|
| dCas9-DNMT3A | DNMT3A fused to dCas9 | CDKN2A, Cdkn1a, ARF, uPA, TGFBR3 | (Amabile et al. 2016; Liu et al. 2016; McDonald et al. 2016; Vojta et al. 2016) |
| dCas9-DNMT3B | DNMT3B fused to dCas9 | uPA, TGFBR3 | (Lin et al. 2018) |
| dCas9-DNMT3A R887E-DNMT3L | DNMT3A mutant for lower off-target effects | VEGFA | (Hofacker et al. 2020) |
| dCas9-DNMT3A-DNMT3L | DNMT3A-3L fusion for higher methylation rates | EpCAM, CXCR4, TFRC | (Stepper et al. 2017) |
| dCas9-DNMT3A-DNMT3L, dCas9-Ezh2, dCas9-KRAB | Combination of histone and DNA methyltransferases for gene suppression | HER2, SNURF | (O’Geen et al. 2019) |
| dCas9-SunTag-DNMT3A | dCas9 fused to SunTag epitopes for recruiting multiple copies of antibody-fused DNMT3A | HOXA5 | (Huang et al. 2017) |
| dCas9-Split M.SssI | Methyltransferase is separated for lower off-target effects | SALL2 | (Xiong et al. 2017) |
| dCas9-MQ1 | Bacterial DNA methylase MQ1 fused to dCas9 | HOXA5 | (Lei et al. 2017) |
| dCas9-MQ1 Q147E | MQ1 mutant for higher methylation efficiencies and lower off-target effects | H0XA5, H0XA4, EYA4, RUNX1, Igf2/H19 | (Lei et al. 2017) |
dCas9, nuclease-inactivated Cas9; CD, catalytic domain
Figure 1. dCas9-mediated targeted DNA methylation technologies for gene silencing.
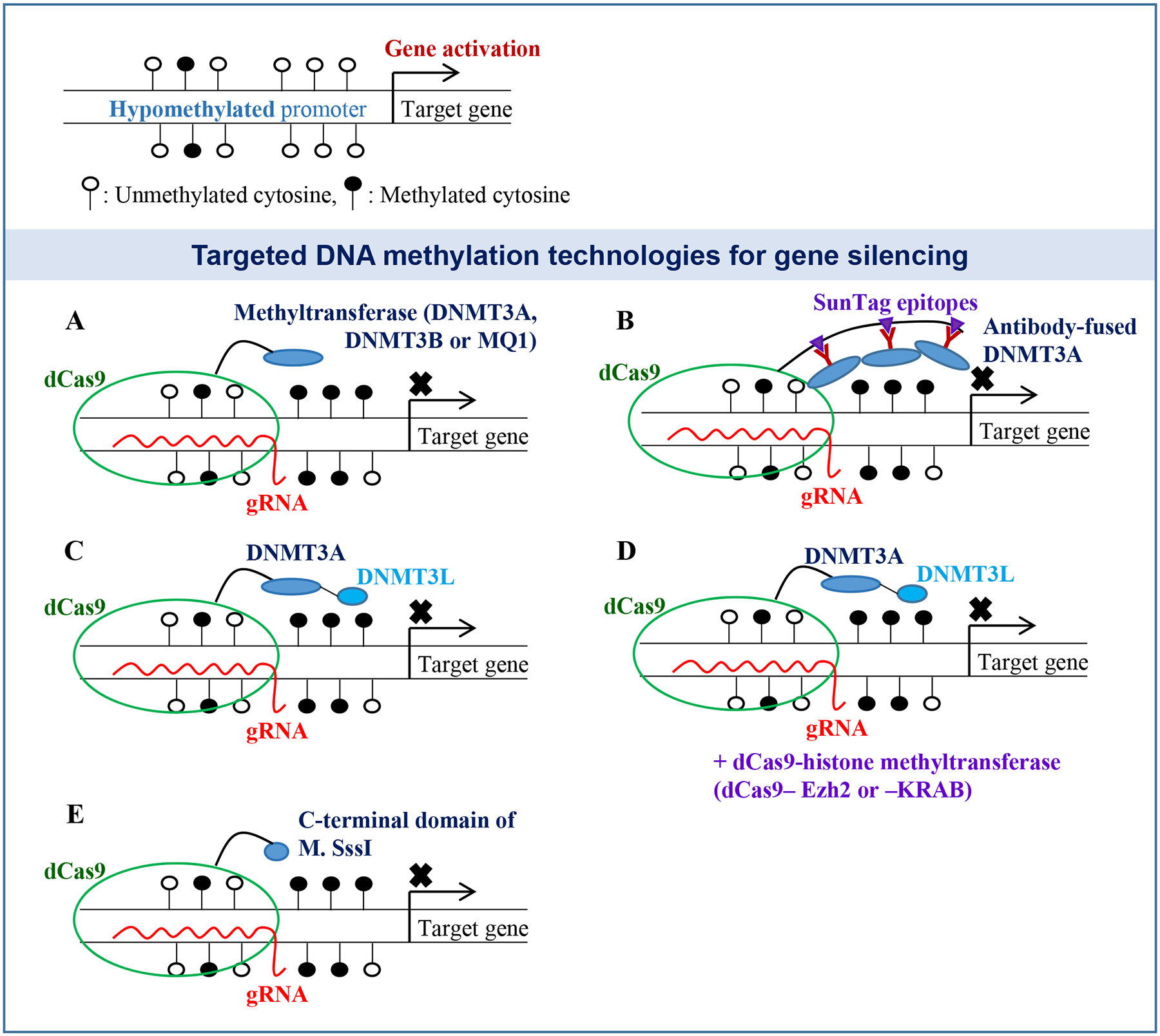
(A) The dCas9 fusion protein is recruited to the target site by a gRNA, allowing the fused enzyme to methylate the promoter region and block gene expression. (B) The dCas9 protein fused to Sun-tag epitopes has been used to recruit multiple copies of antibody-fused DNMT3A to increase the DNA methylation rates at the target site. (C) The dCas9-DNMT3A-DNMT3L proteins have been also successfully used for targeted DNA methylation. (D) An additional histone methyltransferase vector (dCas9-Ezh2 or -KRAB) has been used for effective gene suppression. (E) C-terminal domain of M. SssI (CpG methyltransferase) fused to dCas9 has been also used for promoter methylation. Open-circles, unmethylated cytosines; Closed-circles, methylated cytosines.
Some modifications in the system appeared to be helpful to boost the methylation efficiency and accuracy. Huang et al. constructed the dCas9-SunTag-DNMT3A vector by fusing repetitive peptide epitopes (SunTag) with the dCas9 protein for recruiting multiple copies of the antibody-fused DNMT3A protein to the target site (Fig. 1B). Together with a gRNA targeting HOXA5, this SunTag epigenetic tool displayed superior DNA methylation and gene suppression without significant off-target effects (Huang et al. 2017). Lei et al. adapted a bacterial (Mollicutes spiroplasma) DNA methyltransferase, MQ1, for site-directed promoter methylation in human cells and mouse embryos (Lei et al. 2017). The authors generated dCas9-MQ1 fusions and targeted HOXA4, HOXA5, and RUNX1 for promoter methylation. This epigenetic tool with a bacterial enzyme allowed significant DNA methylation within 24 h post-introduction (Fig. 1A).
Stepper et al. constructed a vector expressing dCas9 fused to a single-chain DNA methyltransferase DNMT3A-3L (Fig. 1C). The authors targeted the promoter regions of human EpCAM, CXCR4 and TFRC genes for DNA methylation and found that dCas9-DNMT3A3L led to promoter methylation with high and broad efficiencies (Stepper et al. 2017). O’Geen et al. also used the single-chain DNA methyltransferase DNMT3A-3L but combined it with a histone methyltransferase Ezh2 or KRAB to increase the methylation activities at the target sites (O’Geen et al. 2019). Both dCas9-Ezh2 and -KRAB fusion clones together with dCas9-DNMT3A-3L led to short-term repression of HER2 in human cells. Long-term suppression of HER2, however, was only shown with dCas9-Ezh2, but not dCas9-KRAB, indicating that selecting optimum combinations of histone and DNA methyltransferases is necessary to achieve maximal methylation and gene suppression rates (Fig. 1D).
Targeted promoter demethylation
Silencing of tumor suppressor genes due to promoter hypermethylation has been observed in various human cancers. These observations have indicated the potential for developing targeted demethylation technologies to reactivate tumor suppressor genes and inhibit cancer cells. The TET enzymes play a key role in DNA demethylation, facilitating the initial process of DNA demethylation. Choudhury et al., Okada et al. and Halmai et al. used the TET1 catalytic domain (TET1CD) fused to the dCas9 protein for targeted promoter demethylation and gene activation in mammalian cells (Fig. 2A and Table 2) (Choudhury et al. 2016; Okada et al. 2017; Halmai et al. 2020).
Figure 2. dCas9-mediated targeted DNA demethylation technologies for gene activation.
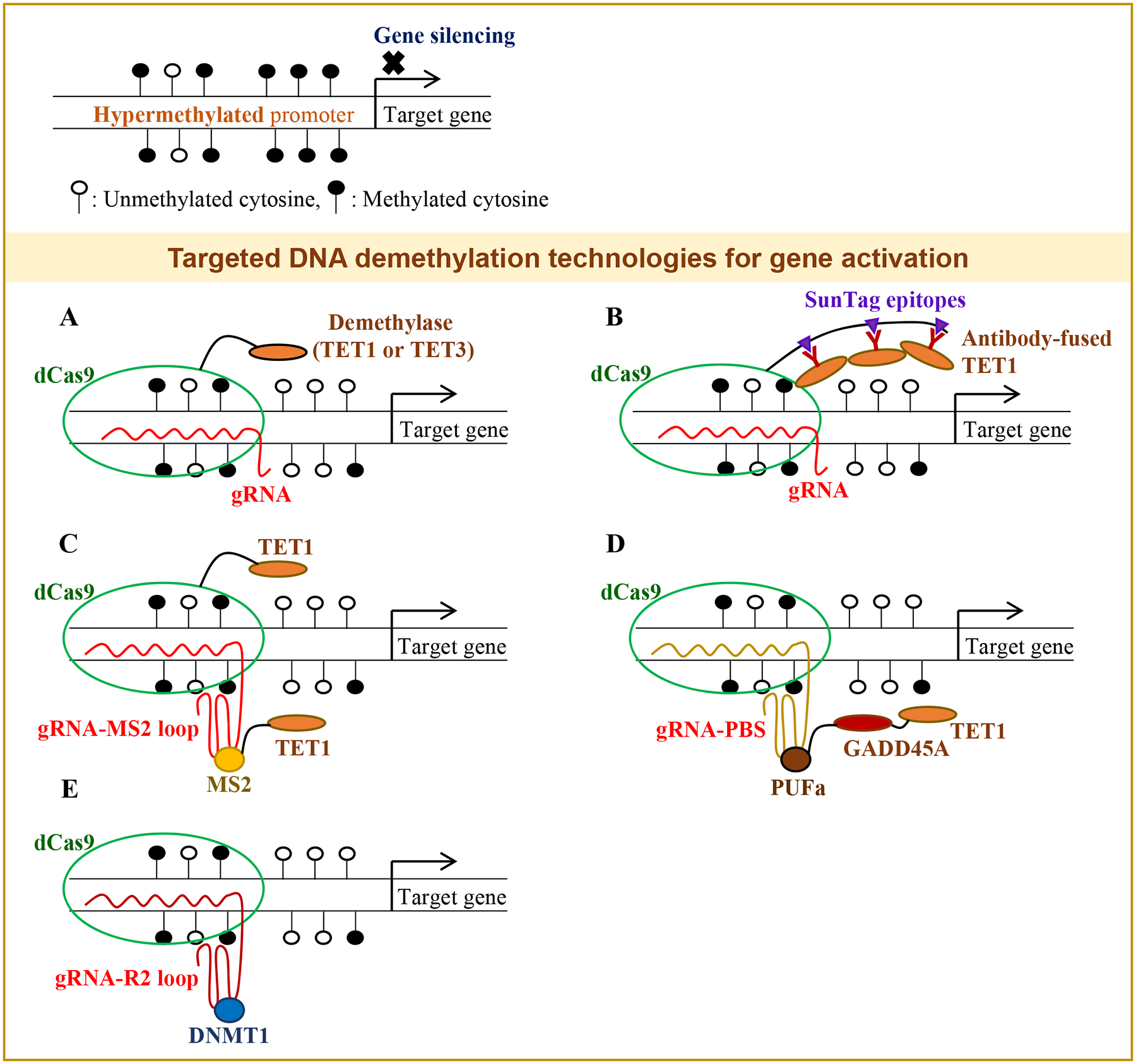
(A) The dCas9-demethylase fusion proteins with a gRNA have been used for site-specific promoter demethylation and gene activation. (B) The dCas9 protein with the Sun-tag epitopes has been used to recruit multiple copies of antibody-fused TET1. (C) A gRNA having MS2 RNA loops, which have a high binding affinity to the MS2 coat protein, has been used to recruit an additional TET1 enzyme to enhance the demethylation rates at the target site. (D) A gRNA containing PUF-binding sites (PBS) that recruit the PUFa-GADD45A-TET1 fusion proteins has been also used for promoter demethylation. (E) A gRNA having R2 loops has been employed to sequester DNMT1 at the target site to inhibit the DNA methylation process. Open-circles, unmethylated cytosines; Closed-circles, methylated cytosines.
Table 2.
The dCas9 vectors for site-specific demethylation and gene activation.
| dCas9 fusion vectors | Description | Targeted gene/promoter | References |
|---|---|---|---|
| dCas9-TET1 | TET1CD fused to dCas9 | CDKL5, BRCA, Foxp3 | (Choudhury et al. 2016; Okada et al. 2017; Halmai et al. 2020) |
| dCas9-TET3 | TET3CD fused to dCas9 | KLOTHO | (Xu et al. 2018) |
| dCas9-TET1 & MS2-TET1 | dCas9-TET1 and MS2-TET1 recruited to the target site for better methylation efficiencies | RANKL, MAGEB2, MMP2 | (Xu et al. 2016) |
| dCas9, gRNA-PBS, PUFa-GADD45A-TET1 | PUFa-GADD45A-TET1 recruited to the target site with a gRNA-PBS for synergistic gene activation effects | MLH1 | (Taghbalout et al. 2019) |
| dCas9-R2 | Lower DNA methylation via mactivation of DNMT1 at the target site | RANKL | (Lu et al. 2019) |
| dCas9-SunTag-scFv-sfGFP-TET1CD | dCas9 fused to SunTag epitopes for recruiting multiple copies of antibody-fused TET1 | Fgf21 | (Hanzawa et al. 2020) |
dCas9, nuclease-inactivated Cas9; CD, catalytic domain
Hanzawa et al. used the dCas9-SunTag-scFv-sfGFP-TET1CD fusion vector for demethylation of the Fgf21 promoter (Table 2) (Hanzawa et al. 2020). In this system, dCas9 was fused to SunTag epitopes for recruiting multiple copies of antibody-fused TET1 for enhanced demethylation activities at the target site (Fig. 2B). Xu et al. used dCas9-TET3CD and targeted the hypermethylated promoter regions of RASAL1, EYA1, and LRFN2, leading to promoter demethylation and gene expression (Fig. 2A) (Xu et al. 2018).
Xu et al. also used dCas9-TET1, but they employed an additional fusion vector (MS2-TET1) for a higher DNA demethylation efficiency (Xu et al. 2016). Two copies of bacteriophage MS2 RNA elements were fused with the gRNA sequence. Since the MS2 coat protein has a high binding affinity to the MS2 RNA elements, once gRNA binds to the target site, it recruits the MS2-TET1 fusion for stronger DNA demethylation efficiency (Fig. 2C). With this dual vector system, the authors targeted the promoter regions of RANKL, MAGEB2, and MMP2 and reported significant promoter demethylation and gene activation (Xu et al. 2016). Another system using a unique RNA sequence and RNA-binding protein was developed for targeted promoter demethylation and gene induction (Taghbalout et al. 2019). This system used dCas9, the Pumilio/FBF (PUF) domain fused with TET1-GADD45A, and a gRNA containing PUF-binding sites (PBS). The authors successfully targeted the MLH1 promoter for demethylation with the PUF-TET1-GADD45A fusion protein and a gRNA containing the PBS sequence (Fig. 2D). Since GADD45A enhances the activity of TET1, this system with dual effectors led to significantly higher gene reactivation rates.
Lu et al. developed an approach different than TET enzyme-mediated demethylation strategies. DNMT1 is the most abundant methyltransferase that is required for the maintenance of DNA methylation. The authors developed a dCas9-R2 system that harbors the R2 stem-loop structure for inhibiting the enzymatic activity of DNMT1, thus lowering the methylation rates at the target site with high accuracy (Fig. 2E) (Lu et al. 2019).
Off-target effects of CRISPR technologies
The dCas9-methyltrasferease vectors have been shown to be effective in targeted promoter methylation and gene silencing (Liu et al. 2016; McDonald et al. 2016; Vojta et al. 2016; Lei et al. 2017). However, concerns have been raised that the dCas9-methyltransferase system could cause off-target methylation (Galonska et al. 2018; Lin et al. 2018; Pflueger et al. 2018). Galonska et al. used dCas9-methyltransferases in pluripotent cells to measure global off-target effects of the dCas9 fusion protein. Their whole genome studies showed that widespread off-target activities of the dCas9-methyltransferases in tested cells. This potential off-target problem could be addressed by mutant forms of DNMT3A and MQ1, which have significantly low off-target effects while maintaining the same levels of methylation activities (Table 1) (Lei et al. 2017; Hofacker et al. 2020). Lei et al. generated a single amino acid mutant of MQ1 fused to dCas9, and this dCas9-MQ1 Q147L clone led to significant CpG methylation in 24 h without off-target effects in human cells and mouse embryos (Lei et al. 2017). Hofacker et al. constructed mutant forms of the DNMT3A protein and evaluated if they showed lower off-target effects while retaining the same level of DNA methylation activity (Hofacker et al. 2020). A single amino acid mutant dCas9-DNMT3A R887E showed significantly low off-target effects while its methylation activity remained unaltered (Table 1). Xiong et al. divided the M.SssI CpG methyltransferase enzyme into two domains (between residues 272 and 273) and fused the C-terminal domain with dCas9 for lower off-target effects (Fig. 1E) (Xiong et al. 2017). The authors targeted the SALL2 promoter region, which is hypomethylated in HEK293T cells, and the engineered vectors led to methylation of the SALL2 promoter within 48 h.
In vivo DNA methylation and demethylation studies
The dCas9 system has been shown to be effective for methylation editing in animal studies as well. The dCas9-MQ1 Q147L plasmid and gRNAs targeting the Igf2/H19 locus were introduced into mouse zygotes followed by embryo transfer to female mice, birth of the engineered mice, and epigenome typing analyses (Lei et al. 2017). The dCas9-MQ1 Q147L fusion led to significant increases of methylation at the target sites in newborn mice. This in vivo study demonstrated that the CRISPR-based approach is applicable to DNA methylation of endogenous gene loci in mice (Lei et al. 2017).
Liu et al. introduced dCas9-Tet1CD into engineered mice with a methylation-sensitive GFP reporter and tested if the CRISPR vector could lead to demethylation of the target site, thus activating the reporter gene (Fig. 2A). The results demonstrated that the dCas9-demethylase fusions with gRNAs can be employed to edit methylation status in vivo (Liu et al. 2016). The dCas9-SunTag-TET1 vector with a single-chain variable fragment (scFv) was used to target the Fgf21 promoter for demethylation in the adult mouse liver (Fig. 2B) (Hanzawa et al. 2020). This study also showed that regulation of gene expression is achieved by a site-specific epigenetic tool without altering the DNA sequence in vivo.
Conclusion and future direction
Various dCas9-methyltransferase and -demethylase fusion proteins have been used to modify promoter methylation and subsequent gene expression. Since many human diseases are caused by alteration of the methylation status of key genes, these site-specific epigenetic tools employing dCas9 and gRNA could have great clinical impact. Future investigations may focus on the development of effective delivery systems that allow the CRISPR vector to reach the targeted cellular site for tissue-specific methylation editing. Furthermore, editing tools for conditional regulation of gene expression will be highly useful, especially for genes with contrasting functional roles depending on the cellular context. For example, KLF4 and SALL2 act as tumor suppressor genes in one context and oncogenes in another context (Rowland and Peeper 2006; Sung and Yim 2015; Sung and Yim 2017). Since these genes have a CpG island in their promoter regions that are involved in methylation-mediated gene regulation (Sung et al. 2013; Yang and Zheng 2014), cell-specific promoter-driven CRISPR-demethylase and -methyltransferase fusions will enable targeting of their promoter regions to correctly edit the methylation states and eliminate the pathological cells.
Another aspect of the future investigations would be development of the effective dCas9-gRNA delivery systems that can be safely used in vivo for therapeutic applications. Although various viral and non-viral delivery vehicles and technologies have been developed and successfully used for CRISPR-mediated gene editing, limitations have been identified, including strong immune responses, packaging limit of viral vectors, cell damage caused by microinjection, degradation of vehicles and inability to reach cells’ nuclei (Follenzi et al. 2007; Wu et al. 2010; Ahi et al. 2011; Horii et al. 2014; Wang et al. 2016; Liu et al. 2019). Therefore, theses technical challenges should be addressed for safe applications of the dCas9-methylation editing tools in therapeutic approaches for human cancer patients.
Acknowledgement
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (to CKS; SC2GM122686) and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (to HY; NRF-2020R1A2C2008672).
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- Ahi YS, Bangari DS, and Mittal SK (2011) Adenoviral vector immunity: its implications and circumvention strategies. Curr Gene Ther 11: 307–20. doi: 10.2174/156652311796150372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amabile A, Migliara A, Capasso P, Biffi M, Cittaro D, Naldini L, and Lombardo A (2016) Inheritable Silencing of Endogenous Genes by Hit-and-Run Targeted Epigenetic Editing. Cell 167: 219–232 e14. doi: 10.1016/j.cell.2016.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinsky SA, Palmisano WA, Gilliland FD, Crooks LA, Divine KK, Winters SA, Grimes MJ, Harms HJ, Tellez CS, Smith TM, Moots PP, Lechner JF, Stidley CA, and Crowell RE (2002a) Aberrant promoter methylation in bronchial epithelium and sputum from current and former smokers. Cancer Res 62: 2370–7. doi: https://www.ncbi.nlm.nih.gov/pubmed/11956099 [PubMed] [Google Scholar]
- Belinsky SA, Snow SS, Nikula KJ, Finch GL, Tellez CS, and Palmisano WA (2002b) Aberrant CpG island methylation of the p16(INK4a) and estrogen receptor genes in rat lung tumors induced by particulate carcinogens. Carcinogenesis 23: 335–9. doi: 10.1093/carcin/23.2.335 [DOI] [PubMed] [Google Scholar]
- Bernstein DL, Le Lay JE, Ruano EG, and Kaestner KH (2015) TALE-mediated epigenetic suppression of CDKN2A increases replication in human fibroblasts. J Clin Invest 125: 1998–2006. doi: 10.1172/JCI77321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel E, Flejou JF, and Hamelin R (2007) Assessment of MLH1 promoter methylation in relation to gene expression requires specific analysis. Oncogene 26: 7596–600. doi: 10.1038/sj.onc.1210581 [DOI] [PubMed] [Google Scholar]
- Cho SW, Kim S, Kim JM, and Kim JS (2013) Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology 31: 230–2. doi: 10.1038/nbt.2507 [DOI] [PubMed] [Google Scholar]
- Choi SJ, Shin YS, Kang BW, Kim JG, Won KJ, Lieberman PM, Cho H, and Kang H (2017) DNA hypermethylation induced by Epstein-Barr virus in the development of Epstein-Barr virus-associated gastric carcinoma. Arch Pharm Res 40: 894–905. doi: 10.1007/s12272-017-0939-5 [DOI] [PubMed] [Google Scholar]
- Choudhury SR, Cui Y, Lubecka K, Stefanska B, and Irudayaraj J (2016) CRISPR-dCas9 mediated TET1 targeting for selective DNA demethylation at BRCA1 promoter. Oncotarget 7: 46545–46556. doi: 10.18632/oncotarget.10234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuikov S, Kurash JK, Wilson JR, Xiao B, Justin N, Ivanov GS, Mckinney K, Tempst P, Prives C, Gamblin SJ, Barlev NA, and Reinberg D (2004) Regulation of p53 activity through lysine methylation. Nature 432: 353–60. doi: 10.1038/nature03117 [DOI] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, and Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339: 819–23. doi: 10.1126/science.1231143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, and Feinberg AP (2003) Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science 299: 1753–5. doi: 10.1126/science.1080902 [DOI] [PubMed] [Google Scholar]
- Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, and Feinberg AP (2002) Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res 62: 6442–6. doi: https://www.ncbi.nlm.nih.gov/pubmed/12438232 [PubMed] [Google Scholar]
- Ehrlich M (2002) DNA hypomethylation, cancer, the immunodeficiency, centromeric region instability, facial anomalies syndrome and chromosomal rearrangements. J Nutr 132: 2424S–2429S. doi: 10.1093/jn/132.8.2424S [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Jiang G, Fiala E, Dome JS, Yu MC, Long TI, Youn B, Sohn OS, Widschwendter M, Tomlinson GE, Chintagumpala M, Champagne M, Parham D, Liang G, Malik K, and Laird PW (2002) Hypomethylation and hypermethylation of DNA in Wilms tumors. Oncogene 21: 6694–702. doi: 10.1038/sj.onc.1205890 [DOI] [PubMed] [Google Scholar]
- Fackler MJ, Mcveigh M, Mehrotra J, Blum MA, Lange J, Lapides A, Garrett E, Argani P, and Sukumar S (2004) Quantitative multiplex methylation-specific PCR assay for the detection of promoter hypermethylation in multiple genes in breast cancer. Cancer Res 64: 4442–52. doi: 10.1158/0008-5472.CAN-03-3341 [DOI] [PubMed] [Google Scholar]
- Flanagan JM, Wilson A, Koo C, Masrour N, Gallon J, Loomis E, Flower K, Wilhelm-Benartzi C, Hergovich A, Cunnea P, Gabra H, Braicu EI, Sehouli J, Darb-Esfahani S, Vanderstichele A, Vergote I, Kreuzinger C, Castillo-Tong DC, Wisman GBA, Berns EM, Siddiqui N, Paul J, and Brown R (2017) Platinum-Based Chemotherapy Induces Methylation Changes in Blood DNA Associated with Overall Survival in Patients with Ovarian Cancer. Clin Cancer Res 23: 2213–2222. doi: 10.1158/1078-0432.CCR-16-1754 [DOI] [PubMed] [Google Scholar]
- Follenzi A, Santambrogio L, and Annoni A (2007) Immune responses to lentiviral vectors. Curr Gene Ther 7: 306–15. doi: 10.2174/156652307782151515 [DOI] [PubMed] [Google Scholar]
- Galonska C, Charlton J, Mattei AL, Donaghey J, Clement K, Gu H, Mohammad AW, Stamenova EK, Cacchiarelli D, Klages S, Timmermann B, Cantz T, Scholer HR, Gnirke A, Ziller MJ, and Meissner A (2018) Genome-wide tracking of dCas9-methyltransferase footprints. Nat Commun 9: 597. doi: 10.1038/s41467-017-02708-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MVC, and Bourc’his D (2019) The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol 20: 590–607. doi: 10.1038/s41580-019-0159-6 [DOI] [PubMed] [Google Scholar]
- Gu X, Lu Y, He D, Lu C, Jin J, Lu X, and Ma X (2016) Methylation of Notch3 modulates chemoresistance via P-glycoprotein. Eur J Pharmacol 792: 7–14. doi: 10.1016/j.ejphar.2016.10.024 [DOI] [PubMed] [Google Scholar]
- Halmai J, Deng P, Gonzalez CE, Coggins NB, Cameron D, Carter JL, Buchanan FKB, Waldo JJ, Lock SR, Anderson JD, O’geen H, Segal DJ, Nolta J, and Fink KD (2020) Artificial escape from XCI by DNA methylation editing of the CDKL5 gene. Nucleic acids research 48: 2372–2387. doi: 10.1093/nar/gkz1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzawa N, Hashimoto K, Yuan X, Kawahori K, Tsujimoto K, Hamaguchi M, Tanaka T, Nagaoka Y, Nishina H, Morita S, Hatada I, Yamada T, and Ogawa Y (2020) Targeted DNA demethylation of the Fgf21 promoter by CRISPR/dCas9-mediated epigenome editing. Sci Rep 10: 5181. doi: 10.1038/s41598-020-62035-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He DX, Gu F, Gao F, Hao JJ, Gong D, Gu XT, Mao AQ, Jin J, Fu L, and Ma X (2016) Genome-wide profiles of methylation, microRNAs, and gene expression in chemoresistant breast cancer. Sci Rep 6: 24706. doi: 10.1038/srep24706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker D, Broche J, Laistner L, Adam S, Bashtrykov P, and Jeltsch A (2020) Engineering of Effector Domains for Targeted DNA Methylation with Reduced Off-Target Effects. Int J Mol Sci 21. doi: 10.3390/ijms21020502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T, Arai Y, Yamazaki M, Morita S, Kimura M, Itoh M, Abe Y, and Hatada I (2014) Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci Rep 4: 4513. doi: 10.1038/srep04513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, Cradick TJ, Marraffini LA, Bao G, and Zhang F (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nature biotechnology 31: 827–32. doi: 10.1038/nbt.2647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YH, Su J, Lei Y, Brunetti L, Gundry MC, Zhang X, Jeong M, Li W, and Goodell MA (2017) DNA epigenome editing using CRISPR-Cas SunTag-directed DNMT3A. Genome Biol 18: 176. doi: 10.1186/s13059-017-1306-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarmalaite S, Kannio A, Anttila S, Lazutka JR, and Husgafvel-Pursiainen K (2003) Aberrant p16 promoter methylation in smokers and former smokers with nonsmall cell lung cancer. Int J Cancer 106: 913–8. doi: 10.1002/ijc.11322 [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816–21. doi: 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA (2002) DNA methylation and cancer. Oncogene 21: 5358–60. doi: 10.1038/sj.onc.1205597 [DOI] [PubMed] [Google Scholar]
- Jones PA, and Baylin SB (2007) The epigenomics of cancer. Cell 128: 683–92. doi: 10.1016/j.cell.2007.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kweon J, and Kim Y (2018) High-throughput genetic screens using CRISPR-Cas9 system. Arch Pharm Res 41: 875–884. doi: 10.1007/s12272-018-1029-z [DOI] [PubMed] [Google Scholar]
- Lee JG, Sung YH, and Baek IJ (2018) Generation of genetically-engineered animals using engineered endonucleases. Arch Pharm Res 41: 885–897. doi: 10.1007/s12272-018-1037-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Zhang X, Su J, Jeong M, Gundry MC, Huang YH, Zhou Y, Li W, and Goodell MA (2017) Targeted DNA methylation in vivo using an engineered dCas9-MQ1 fusion protein. Nat Commun 8: 16026. doi: 10.1038/ncomms16026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Papworth M, Minczuk M, Rohde C, Zhang Y, Ragozin S, and Jeltsch A (2007) Chimeric DNA methyltransferases target DNA methylation to specific DNA sequences and repress expression of target genes. Nucleic acids research 35: 100–12. doi: 10.1093/nar/gkl1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Liu Y, Xu F, Huang J, Daugaard TF, Petersen TS, Hansen B, Ye L, Zhou Q, Fang F, Yang L, Li S, Floe L, Jensen KT, Shrock E, Chen F, Yang H, Wang J, Liu X, Xu X, Bolund L, Nielsen AL, and Luo Y (2018) Genome-wide determination of on-target and off-target characteristics for RNA-guided DNA methylation by dCas9 methyltransferases. Gigascience 7: 1–19. doi: 10.1093/gigascience/giy011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Chang J, Jiang Y, Meng X, Sun T, Mao L, Xu Q, and Wang M (2019) Fast and Efficient CRISPR/Cas9 Genome Editing In Vivo Enabled by Bioreducible Lipid and Messenger RNA Nanoparticles. Adv Mater 31: e1902575. doi: 10.1002/adma.201902575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, Shu J, Dadon D, Young RA, and Jaenisch R (2016) Editing DNA Methylation in the Mammalian Genome. Cell 167: 233–247 e17. doi: 10.1016/j.cell.2016.08.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu A, Wang J, Sun W, Huang W, Cai Z, Zhao G, and Wang J (2019) Reprogrammable CRISPR/dCas9-based recruitment of DNMT1 for site-specific DNA demethylation and gene regulation. Cell Discov 5: 22. doi: 10.1038/s41421-019-0090-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund RJ, Huhtinen K, Salmi J, Rantala J, Nguyen EV, Moulder R, Goodlett DR, Lahesmaa R, and Carpen O (2017) DNA methylation and Transcriptome Changes Associated with Cisplatin Resistance in Ovarian Cancer. Sci Rep 7: 1469. doi: 10.1038/s41598-017-01624-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, Costello JF, Wilkinson MF, and Joung JK (2013) Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nature biotechnology 31: 1137–42. doi: 10.1038/nbt.2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, and Church GM (2013) CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nature biotechnology 31: 833–8. doi: 10.1038/nbt.2675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdonald JI, Celik H, Rois LE, Fishberger G, Fowler T, Rees R, Kramer A, Martens A, Edwards JR, and Challen GA (2016) Reprogrammable CRISPR/Cas9-based system for inducing site-specific DNA methylation. Biol Open 5: 866–74. doi: 10.1242/bio.019067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mcdonald OG, Li X, Saunders T, Tryggvadottir R, Mentch SJ, Warmoes MO, Word AE, Carrer A, Salz TH, Natsume S, Stauffer KM, Makohon-Moore A, Zhong Y, Wu H, Wellen KE, Locasale JW, Iacobuzio-Donahue CA, and Feinberg AP (2017) Epigenomic reprogramming during pancreatic cancer progression links anabolic glucose metabolism to distant metastasis. Nat Genet 49: 367–376. doi: 10.1038/ng.3753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra J, Vali M, Mcveigh M, Kominsky SL, Fackler MJ, Lahti-Domenici J, Polyak K, Sacchi N, Garrett-Mayer E, Argani P, and Sukumar S (2004) Very high frequency of hypermethylated genes in breast cancer metastasis to the bone, brain, and lung. Clin Cancer Res 10: 3104–9. doi: 10.1158/1078-0432.ccr-03-0118 [DOI] [PubMed] [Google Scholar]
- Mitsuno M, Kitajima Y, Ide T, Ohtaka K, Tanaka M, Satoh S, and Miyazaki K (2007) Aberrant methylation of p16 predicts candidates for 5-fluorouracil-based adjuvant therapy in gastric cancer patients. J Gastroenterol 42: 866–73. doi: 10.1007/s00535-007-2113-1 [DOI] [PubMed] [Google Scholar]
- Nunna S, Reinhardt R, Ragozin S, and Jeltsch A (2014) Targeted methylation of the epithelial cell adhesion molecule (EpCAM) promoter to silence its expression in ovarian cancer cells. PloS one 9: e87703. doi: 10.1371/journal.pone.0087703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’geen H, Bates SL, Carter SS, Nisson KA, Halmai J, Fink KD, Rhie SK, Farnham PJ, and Segal DJ (2019) Ezh2-dCas9 and KRAB-dCas9 enable engineering of epigenetic memory in a context-dependent manner. Epigenetics Chromatin 12: 26. doi: 10.1186/s13072-019-0275-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Kanamori M, Someya K, Nakatsukasa H, and Yoshimura A (2017) Stabilization of Foxp3 expression by CRISPR-dCas9-based epigenome editing in mouse primary T cells. Epigenetics Chromatin 10: 24. doi: 10.1186/s13072-017-0129-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JW, and Han JW (2019) Targeting epigenetics for cancer therapy. Arch Pharm Res 42: 159–170. doi: 10.1007/s12272-019-01126-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflueger C, Tan D, Swain T, Nguyen T, Pflueger J, Nefzger C, Polo JM, Ford E, and Lister R (2018) A modular dCas9-SunTag DNMT3A epigenome editing system overcomes pervasive off-target activity of direct fusion dCas9-DNMT3A constructs. Genome Res 28: 1193–1206. doi: 10.1101/gr.233049.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivenbark AG, Stolzenburg S, Beltran AS, Yuan X, Rots MG, Strahl BD, and Blancafort P (2012) Epigenetic reprogramming of cancer cells via targeted DNA methylation. Epigenetics 7: 350–60. doi: 10.4161/epi.19507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland BD, and Peeper DS (2006) KLF4, p21 and context-dependent opposing forces in cancer. Nature reviews. Cancer 6: 11–23. doi: 10.1038/nrc1780 [DOI] [PubMed] [Google Scholar]
- Sato M, Ohtsuka M, Nakamura S, Sakurai T, Watanabe S, and Gurumurthy CB (2018) In vivo genome editing targeted towards the female reproductive system. Arch Pharm Res 41: 898–910. doi: 10.1007/s12272-018-1053-z [DOI] [PubMed] [Google Scholar]
- Shen L, Fang J, Qiu D, Zhang T, Yang J, Chen S, and Xiao S (1998) Correlation between DNA methylation and pathological changes in human hepatocellular carcinoma. Hepatogastroenterology 45: 1753–9. doi: https://www.ncbi.nlm.nih.gov/pubmed/9840141 [PubMed] [Google Scholar]
- Siddique AN, Nunna S, Rajavelu A, Zhang Y, Jurkowska RZ, Reinhardt R, Rots MG, Ragozin S, Jurkowski TP, and Jeltsch A (2013) Targeted methylation and gene silencing of VEGF-A in human cells by using a designed Dnmt3a-Dnmt3L single-chain fusion protein with increased DNA methylation activity. J Mol Biol 425: 479–91. doi: 10.1016/j.jmb.2012.11.038 [DOI] [PubMed] [Google Scholar]
- Smith ZD, and Meissner A (2013) DNA methylation: roles in mammalian development. Nat Rev Genet 14: 204–20. doi: 10.1038/nrg3354 [DOI] [PubMed] [Google Scholar]
- Stepper P, Kungulovski G, Jurkowska RZ, Chandra T, Krueger F, Reinhardt R, Reik W, Jeltsch A, and Jurkowski TP (2017) Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic acids research 45: 1703–1713. doi: 10.1093/nar/gkw1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CK, Li D, Andrews E, Drapkin R, and Benjamin T (2013) Promoter methylation of the SALL2 tumor suppressor gene in ovarian cancers. Mol Oncol 7: 419–27. doi: 10.1016/j.molonc.2012.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung CK, and Yim H (2015) The tumor suppressor protein p150(Sal2) in carcinogenesis. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine 36: 489–94. doi: 10.1007/s13277-014-3019-1 [DOI] [PubMed] [Google Scholar]
- Sung CK, and Yim H (2017) Roles of SALL2 in tumorigenesis. Arch Pharm Res 40: 146–151. doi: 10.1007/s12272-016-0874-x [DOI] [PubMed] [Google Scholar]
- Taghbalout A, Du M, Jillette N, Rosikiewicz W, Rath A, Heinen CD, Li S, and Cheng AW (2019) Enhanced CRISPR-based DNA demethylation by Casilio-ME-mediated RNA-guided coupling of methylcytosine oxidation and DNA repair pathways. Nat Commun 10: 4296. doi: 10.1038/s41467-019-12339-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao L, Yang S, Xie M, Kramer PM, and Pereira MA (2000) Effect of trichloroethylene and its metabolites, dichloroacetic acid and trichloroacetic acid, on the methylation and expression of c-Jun and c-Myc protooncogenes in mouse liver: prevention by methionine. Toxicol Sci 54: 399–407. doi: 10.1093/toxsci/54.2.399 [DOI] [PubMed] [Google Scholar]
- Vojta A, Dobrinic P, Tadic V, Bockor L, Korac P, Julg B, Klasic M, and Zoldos V (2016) Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic acids research 44: 5615–28. doi: 10.1093/nar/gkw159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Zuris JA, Meng F, Rees H, Sun S, Deng P, Han Y, Gao X, Pouli D, Wu Q, Georgakoudi I, Liu DR, and Xu Q (2016) Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proceedings of the National Academy of Sciences of the United States of America 113: 2868–73. doi: 10.1073/pnas.1520244113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M, Stupp R, Reifenberger G, Brandes AA, Van Den Bent MJ, Wick W, and Hegi ME (2010) MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol 6: 39–51. doi: 10.1038/nrneurol.2009.197 [DOI] [PubMed] [Google Scholar]
- Widschwendter M, and Jones PA (2002) DNA methylation and breast carcinogenesis. Oncogene 21: 5462–82. doi: 10.1038/sj.onc.1205606 [DOI] [PubMed] [Google Scholar]
- Wu Z, Yang H, and Colosi P (2010) Effect of genome size on AAV vector packaging. Mol Ther 18: 80–6. doi: 10.1038/mt.2009.255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong T, Meister GE, Workman RE, Kato NC, Spellberg MJ, Turker F, Timp W, Ostermeier M, and Novina CD (2017) Targeted DNA methylation in human cells using engineered dCas9-methyltransferases. Sci Rep 7: 6732. doi: 10.1038/s41598-017-06757-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Tan X, Tampe B, Wilhelmi T, Hulshoff MS, Saito S, Moser T, Kalluri R, Hasenfuss G, Zeisberg EM, and Zeisberg M (2018) High-fidelity CRISPR/Cas9- based gene-specific hydroxymethylation rescues gene expression and attenuates renal fibrosis. Nat Commun 9: 3509. doi: 10.1038/s41467-018-05766-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Tao Y, Gao X, Zhang L, Li X, Zou W, Ruan K, Wang F, Xu GL, and Hu R (2016) A CRISPR-based approach for targeted DNA demethylation. Cell Discov 2: 16009. doi: 10.1038/celldisc.2016.9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WT, and Zheng PS (2014) Promoter hypermethylation of KLF4 inactivates its tumor suppressor function in cervical carcinogenesis. PloS one 9: e88827. doi: 10.1371/journal.pone.0088827 [DOI] [PMC free article] [PubMed] [Google Scholar]


