Abstract
Background:
Sublingual buprenorphine-naloxone (BUP-NX), an FDA-approved treatment for opioid use disorder (OUD) combines buprenorphine, a partial mu/kappa agonist with naloxone, a mu/kappa antagonist. Extended-release injection naltrexone (XR-NTX), also an FDA-approved treatment for OUD, is an antagonist at the mu receptor and partial agonist at the kappa receptor. However, only a fraction of patients responds well, while some drop out of treatment and relapse.
Objectives:
Determine whether gene variants in the opioid gene system are associated with better or worse treatment response.
Methods:
In a 24-week, multisite, randomized, comparative effectiveness trial of daily, sublingual self-administration of BUP-NX versus monthly injection of XR-NTX conducted in the National Drug Abuse Clinical Trials Network, DNA was collected and four opioid gene variants were evaluated: (1) mu opioid receptor 118A>G; (2) 68-bp repeat in prodynorphin; (3) prodynorphin SNP rs910080; and (4) kappa opioid receptor SNP rs6473797. In non-Hispanic Caucasians (N = 334), separate logistic regressions were used to model two outcomes: (1) receiving the first dose of medication, and among those (2) receiving the last dose of medication (or completion of treatment) as a function of treatment (XR-NTX vs BUP-NX), each gene variant, and their interaction.
Results:
There were no significant gene variant by treatment interactions, nor were there significant main effects on receiving the first dose, nor completion of treatment.
Conclusions:
The outcome of treatment of OUD with medications is likely a complex function of multiple factors, environmental, psychosocial, and possibly genetic, such that major effects of genetic variants may be unlikely.
Introduction:
The opioid crisis with high rates of death from opioid overdose has focused attention on improving the effectiveness of medication treatments for opioid use disorder (OUD). Maintenance treatments with methadone, buprenorphine, or extended-release injection naltrexone are all effective treatments in some persons. However, the rate of dropout from these medication treatments is high, particularly for buprenorphine and injection naltrexone [1–4], and stopping medication is associated with relapse [5–7]. Thus, research is needed to reduce dropout and improve the effectiveness of each of these treatments. One approach would be patient-treatment matching - i.e., determining what types of patients would respond best to each of these medications. However, to date, little is known about patient characteristics that would predict differential response.
Gene variants, particularly those affecting pharmacodynamics and/or pharmacokinetics, can influence pharmacotherapy outcomes [8–12]. Both buprenorphine and naltrexone are partial kappa agonists and both act at the mu opioid receptor, buprenorphine as a partial agonist and naltrexone as an antagonist. Thus, one might expect gene variants in the mu opioid receptor to be associated with better response to one than the other - i.e., gene variant by treatment interaction. Both medications also act as kappa opioid receptor antagonists, and thus one might expect variants in the kappa system to predict outcome similarly for both medications, or perhaps to predict differently (i.e., gene variant by treatment interaction) if, for example, kappa antagonism has different effects in the setting of mu partial agonism versus antagonism.
A recently completed 24-week, randomized, comparative effectiveness trial of daily sublingual self-administration of buprenorphine-naloxone (BUP-NX) versus monthly injection of extended-release naltrexone (XR-NTX) offers the first opportunity to examine effects of gene variants, or gene variant by treatment interaction, on response to these medications. The primary outcome analysis found that XR-NTX was more difficult to initiate, due to the need for patients to tolerate detoxification prior to starting naltrexone, but once initiated both medications showed similar 24-week relapse rates [13]. An analysis examining baseline demographic and clinical characteristics as moderators of differential treatment effects, found only a few moderators with modest effects (manuscript in preparation). Here, we examine four gene variants as predictors or moderators of the effects of BUP-NX and XR-NTX on the outcomes receiving the first dose of study medication and completion of treatment, with the following hypotheses:
Individuals with the functional mu opioid receptor gene (OPRM1) variant 118A>G (rs1799971) will have an increased risk of relapse on XR-NTX, based on the greater opioid antagonist driven hypothalamic-adrenal axis activation, a hallmark of opioid withdrawal, previously shown in carriers of this variant [14–16].
Variants of the prodynorphin gene (PDYN) associated with brain expression of dynorphin have been associated with depressive symptoms [17,18]. Previously, our group along with others found that the number of PDYN 68-bp repeats may alter dynorphin gene expression [19]. The short form of the PDYN 68-bp repeat was associated with greater expression of dynorphin, possibly leading to dysphoria and more depressive symptoms, which may contribute to relapse. Our hypothesis is that PDYN variants 68 bp repeat (SS/SL vs LL), and rs910080 (AA/AG vs GG) which yield an increase in dynorphin peptides, will be associated with worse treatment outcome.
The kappa opioid receptor gene (OPRK1) rs6473797 variant, associated with various drug addictions [20], may affect treatment response, as both BUP-NX and XR-NTX are partial agonists of the kappa opioid receptor.
Methods:
Clinical Trial Methodology:
Detailed methods of the comparative effectiveness trial (NCT02032433), upon which this genetic analysis is based, and its primary outcome findings have been previously published [13]. Briefly, adults with DSM-5 opioid use disorder (OUD) were recruited from eight US detoxification/short-term residential treatment settings. Those meeting eligibility criteria were randomized 1:1 to treatment with XR-NTX or BUP-NX, the randomization stratified by site and by OUD severity (intravenous use of >6 bags/day vs all other use). Following randomization, assigned treatment was initiated as soon as possible, i.e., for BUP-NX usually as soon as mild to moderate withdrawal symptoms were present, for XR-NTX only after detoxification was completed, a urine sample negative for opioids was collected, and a negative naloxone challenge was completed. Treatment was continued in an outpatient setting for 24 weeks, with daily self- administered sublingual BUP-NX, flexible dosing with providers encouraged to increase the dose as rapidly as possible, or with monthly 380 mg XR-NTX injections. The primary outcome was Relapse at any time across the 24-week trial defined as 4 consecutive weeks of opioid use by self-report or urine toxicology, or 7 consecutive days of self-reported use.
A blood sample for genetic analysis was collected, in almost all cases during baseline procedures. All sites obtained local IRB approvals and consent for genetic testing was included as part of the main trial consent. Altogether, 772 individuals were consented and screened; 570 were randomized, 283 to XR-NTX (of whom 204 received the first dose) and 287 to BUP-NX (of whom 270 received a first dose).
The genetic analyses on induction success were limited to the subsample of non-Hispanic, Caucasians to limit heterogeneity of ancestry (n=334; 164 in XR-NTX, 170 in BUP-NX). Further, the genetic analyses on completion of treatment were limited to non-Hispanic, Caucasians who received a first dose of treatment (i.e., successfully inducted onto treatment: 132 XR-NTX, 158 BUP-NX).
DNA preparation:
Coded blood samples were sent to the NIDA genetic repository where genomic DNA was isolated by standard methods. Purified genomic DNA (30 ug per subject) from 636 consented individuals was received from the repository. DNA quality was assessed by gel electrophoresis and DNA quantity was determined using a NanoDrop® 1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA).
Genotyping of the single nucleotide polymorphisms (SNPs):
SNPs rs1799971 (A118G) in OPRM1, rs910080 in PDYN and rs6473797 in OPRK1 were genotyped using pre-designed TaqMan® SNP Genotyping Assays and Universal PCR master mix (all from Applied Biosystems, Foster City, CA) in a 384-well plate. Polymerase chain reaction (PCR) cycling was done on an ABI Prism® 9700 Thermocycler (Applied Biosystems, Foster City, CA) and the endpoint analysis was performed using an ABI Prism® 7900HT Sequence Detection System using Sequence Detection Software v.2.3 (Applied Biosystems, Foster City, CA).
Microsatellite analysis:
Genotyping of the PDYN 68-bp repeat region was performed as described previously [21]. Briefly, DNA was PCR amplified in two replicates: first, using oligonucleotide primers
5’-CTGTGTATGGAGAGGCTGAGT-3’ labeled with 5’-FAM and 5’-AGGCGGTTAGGTAGAGTTGTC-3’, yielding a PCR product with the fluorescently labeled forward strand, and second, using 5’-CTGTGTATGGAGAGGCTGAGT-3’ and 5’-AGGCGGTTAGGTAGAGTTGTC-3’ labeled with 5’-FAM, yielding a PCR product with fluorescently labeled reverse strand. Fragments were analyzed by capillary electrophoresis using a 3730xl DNA Analyzer and GeneMapper® v.4.0 software (both from Applied Biosystems, Foster City, CA) as previously described [22]. The “alleles” for the 68-base pair repeat were determined by the number of repeat fragments, where 1 or 2 copies of the repeat were designated as “Short” (S) allele, and 3 or 4 copies of the repeat were designated as the “Long” (L) allele. Genotypes were grouped as short/short (SS) (1,1; 1,2; or 2,2 copies), short/long (SL) (1,3; 1,4; 2,3; of 2,4 copies) and long/long (LL) (3,3; 3,4; or 4,4 copies).
Outcome Measures:
Received first dose:
We examine receiving first dose (binary: yes/no) of randomized medication (vs not receiving first dose) as an outcome. Some randomized patients never initiated randomized medication, particularly naltrexone, which has a well know induction hurdle. Patients need to be fully detoxified prior to initiating naltrexone and some patients do not tolerate the detoxification or period of waiting for opioids to clear their system.
Received last dose:
We also examine a binary indicator of good clinical response, defined as receiving last dose of medication (binary: yes/no) based on the administration of a final naltrexone injection or a final refill of buprenorphine prescription. This reflects retention in treatment, which is of paramount importance in the medical treatment of opioid use disorder. Additionally, we sought to examine the primary outcome measure for the trial, defined as ‘relapse’ at any point over the 24-week trial (binary: yes/no), indicating either return to regular opioid use or dropout from treatment.
Patients were seen weekly during the trial during which self-reported substance use was collected with the ‘Time-Line Follow Back’ method, and urine was collected for testing for opioids (buprenorphine, methadone, morphine [heroin, codeine, morphine], oxycodone) and other drugs. Relapse was operationalized as 1) Four or more consecutive weeks of any non-study opioid use (by urine toxicology, or self-report, or failure to provide a urine sample); or 2) Seven or more consecutive days of self-reported non-study opioid use. In the primary outcome analysis [13] ‘time to relapse’ was analyzed with survival analysis. For the present analysis, ‘relapse’ is a binary outcome— ‘relapse’ vs ‘no relapse’ across the 24-week trial—with absence of ‘relapse’ across 24-weeks conceptualized as indicative of good clinical response. As it turned out, the outcomes ‘received last dose’ and ‘relapse’ were highly overlapping. There was only one instance where a participant did not receive the last dose but did not meet ‘relapse’ criteria; conversely, all of those who ‘relapsed’ did not receive the last dose. Thus, results are only presented for ‘received last dose’.
Data Analysis:
Power analyses were conducted prior to sample collection for the primary analyses for this trial, details are provided in full in Lee et al, 2017 [13]. Baseline differences between treatment groups in the non-Hispanic, Caucasian sample were assessed using chi-square tests for categorical measures and t-test for continuous measures. Chi-square tests were also used to assess whether each genetic variant did not violate Hardy-Weinberg Equilibrium among the non-Hispanic, Caucasian subsample. This was done by assessing if there were significant differences in the observed versus expected frequencies of each genetic variant, based on the allele proportions of the sample.
Among all randomized participants in the non-Hispanic, Caucasian subsample (N = 334), separate logistic regression models were fitted to assess whether either of the four gene variants moderated the treatment effect on failure to receive first dose onto treatment drug. These regression models were estimating the odds of failing to receive first dose. Each unadjusted model included the effect of treatment (XR-NTX vs BUP-NX), gene variant, and their 2-way interaction with a separate model fit for each gene variant. If the 2-way interaction was significant then the treatment effect was computed for each level of the gene variant, and if the 2-way interaction was not significant, only the main effects of treatment and gene variant were assessed. Each model was additionally fit adjusting for the covariates age, sex, IV use, and other drug use (either stimulant, sedative, heavy alcohol use, or cannabis).
Further, among randomized Caucasians who successfully received their first dose of treatment medication (N = 290), logistic regression models were then fitted to assess whether either of the four gene variants moderated the treatment effect on the main outcome of interest: failure to receive last dose (yes/no). The regression models were estimating the odds of failure to receive last dose. Unadjusted models were run similarly to above with the effect of treatment (XR-NTX vs BUP-NX), gene variant, and their 2-way interaction fitted separately for each gene variant. Only the main effects of treatment and gene variant were assessed if the 2-way interaction was not significant. Adjusted models were additionally fit controlling for the covariates age, sex, IV use, and other drug use. We additionally fitted similar models instead using the primary outcome in the parent clinical trial, 24-week relapse (yes/no), but due to there being only one discrepant case between the ‘received last dose’ and ‘relapse’ outcomes, results for ‘relapse’ are not reported. .
All hypothesis tests were two-sided at a significance level of 5% without correction for multiple tests due to the exploratory nature of this analysis. All analyses were done in SAS version 9.4.
Results
Sample Characteristics:
Of the 570 subjects randomized in the parent trial, 334 were non-Hispanic, Caucasian (164 randomized to XR-NTX, 170 randomized to BUP-NX) based on self-report (Figure 1). Of those non-Hispanic, Caucasians randomized to XR-NTX, 81% (132 out of 164) received their first study medication dose, and of the 170 non-Hispanic, Caucasians that were randomized to BUP-NX, 158 (93%) received their first dose of study medication. Baseline demographic and clinical characteristics for the non-Hispanic, Caucasian subsample are presented in Table 1. There were no significant differences between treatment groups on any baseline or clinical characteristics among the subsample. Overall, the patients were on average 33 years old, mostly male (67%), and the majority had never been married (67%) and were unemployed (64%). The majority were IV users (66%) whose primary opioid was heroin (79%). About half currently used cannabis or stimulants, and about a quarter were current sedative users or heavy drinkers. Additionally, there were no violations of Hardy-Weinberg Equilibrium for the genetic variants in this overall sample (all p > 0.05). Genotype frequencies for each genetic variant by treatment assignment and outcome are presented in Table 2.
Figure 1.
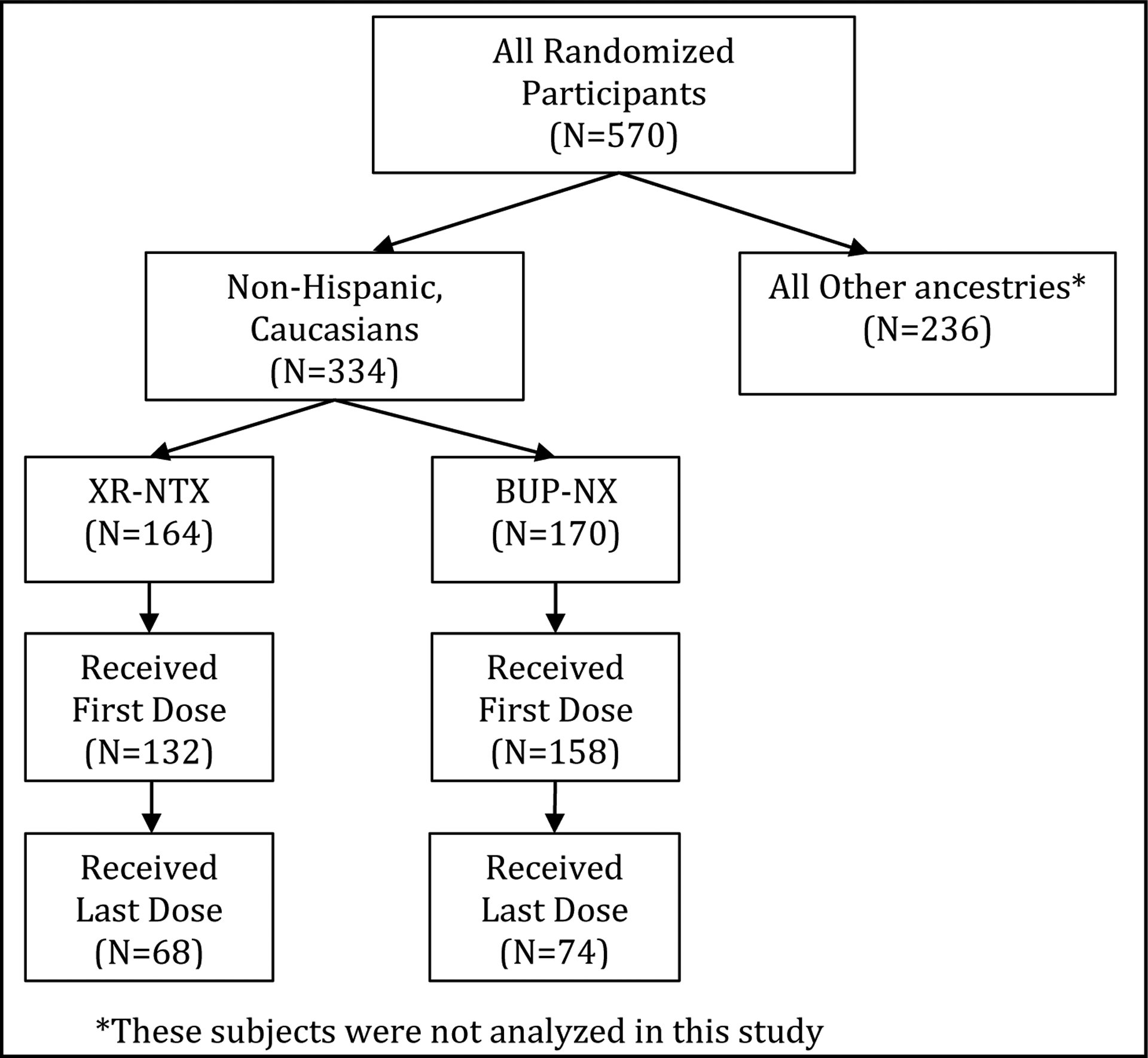
Subject numbers
Table 1.
Baseline demographic and clinical characteristics by treatment group (N=334).
| XR-NTX (N=164) | BUP-NX (N=170) | |||
|---|---|---|---|---|
| Measure | N | % or M (SD) | N | % or M (SD) |
| Received first dose (% yes) | 132 | 80.5% | 158 | 92.9% |
| Demographic Characteristics | ||||
| Gender (% male) | 109 | 66.5% | 115 | 67.6% |
| Age | 164 | 32.8 (8.5) | 170 | 33.0 (9.0) |
| Marital status (% never married) | 109 | 66.5% | 114 | 67.1% |
| Employment (% unemployed) | 105 | 64.0% | 110 | 64.7% |
| Clinical Characteristics | ||||
| IV Use (% yes) | 111 | 67.7% | 111 | 65.3% |
| Primary Opioid | ||||
| Buprenorphine | 4 | 2.5% | 1 | 0.6% |
| Opioid analgesics | 26 | 16.0% | 32 | 18.9% |
| Methadone | 3 | 1.8% | 4 | 2.4% |
| Heroin | 130 | 79.8% | 132 | 78.1% |
| Primary opioid cost ($/day) | 163 | 92.7 (70.3) | 169 | 104.7 (84.0) |
| Age at onset of opioid use | 164 | 20.6 (6.5) | 170 | 21.2 (7.1) |
| Duration of opioid use (years) | 164 | 12.2 (8.0) | 170 | 11.7 (8.1) |
| First treatment episode (% yes) | 59 | 36.0% | 70 | 41.2% |
| Stimulant use (% yes)a | 80 | 48.8% | 93 | 54.7% |
| Sedative use (% yes)a | 41 | 25.0% | 53 | 31.2% |
| Heavy alcohol use (% yes)a | 38 | 23.2% | 42 | 24.7% |
| Cannabis use (% yes)a | 73 | 44.5% | 78 | 45.9% |
| Clinical | ||||
| HAM-Db Score (Range: 0–52) | 164 | 8.1 (6.1) | 170 | 8.8 (6.5) |
| Any psych disorder (% yes) | 113 | 68.9% | 116 | 68.2% |
| SOWSc (Range: 0–64) | 164 | 16.0 (12.9) | 170 | 15.7 (13.2) |
30-days prior to admission for detox
Hamilton Depression Rating Scale
Subjective Opiate Withdrawal Scal
Table 2.
Genotype frequencies by treatment and outcome (N=334).
| XR-NTX (N=164) | BUP-NX (N=170) | |||||||
|---|---|---|---|---|---|---|---|---|
| Randomized | Failed to receive first dose | Failed to receive last dose | Randomized | Failed to receive first dose | Failed to receive last dose | |||
| Variant | Genotype | N (frequency) | N (frequency) | N (frequency | N (frequency | N (frequency | N (frequency | |
| OPRM1 rs1799971 | AA | 126 (0.78) | 26 (0.84) | 47 (0.76) | 132 (0.78) | 11 (0.92) | 69 (0.82) | |
| AG | 31 (0.19) | 4 (0.13) | 13 (0.21) | 36 (0.21) | 1 (0.08) | 15 (0.18) | ||
| GG | 4 (0.02) | 1 (0.03) | 2 (0.03) | 2 (0.01) | 0 (0) | 0 (0) | ||
| minor allele, G | 39 (0.12) | 6 (0.1) | 17 (0.14) | 40 (0.12) | 1 (0.04) | 15 (0.09) | ||
| PDYN 68-bp repeat | SS | 13 (0.08) | 2 (0.06) | 6 (0.1) | 14 (0.08) | 2 (0.17) | 11 (0.13) | |
| SL | 62 (0.38) | 14 (0.44) | 29 (0.46) | 82 (0.49) | 3 (0.25) | 38 (0.46) | ||
| LL | 87 (0.54) | 16 (0.5) | 28 (0.44) | 73 (0.43) | 7 (0.58) | 34 (0.41) | ||
| minor allele, S | 88 (0.27) | 18 (0.28) | 41 (0.33) | 110 (0.33) | 7 (0.29) | 60 (0.32) | ||
| PDYN rs910080 | TT | 91 (0.56) | 13 (0.41) | 37 (0.58) | 93 (0.55) | 8 (0.67) | 43 (0.52) | |
| TC | 62 (0.38) | 18 (0.56) | 23 (0.36) | 67 (0.4) | 4 (0.33) | 34 (0.41) | ||
| CC | 10 (0.06) | 1 (0.03) | 4 (0.06) | 8 (0.05) | 0 (0) | 6 (0.07) | ||
| minor allele, C | 82 (0.25) | 20 (0.31) | 31 (0.24) | 83 (0.25) | 4 (0.17) | 46 (0.28) | ||
| OPRK1 rs6473797 | AA | 90 (0.55) | 18 (0.56) | 36 (0.56) | 89 (0.52) | 83 (0.53) | 41 (0.49) | |
| AG | 63 (0.39) | 13 (0.41) | 24 (0.38) | 66 (0.39) | 62 (0.39) | 34 (0.4) | ||
| GG | 10 (0.06) | 1 (0.03) | 4 (0.06) | 15 (0.09) | 13 (0.08) | 9 (0.11) | ||
| minor allele, G | 83 (0.25) | 15 (0.23) | 32 (0.25) | 96 (0.28) | 88 (0.28) | 52 (0.31) | ||
Effects of Treatment by Genotype on Receiving First Dose of Medication:
Model results for unadjusted logistic regression models on failure to receive first dose for each genetic variant are presented in Table 3. The 2-way interactions between treatment and genotype on induction were not significant for any of the gene variants suggesting that there are no differential effects of treatment between genotypes for these gene variants. When assessing the main effects on the odds of failing to receive first dose, there were no significant differences by genotypes (all p > 0.05), but there were significant differences between treatments, as expected with higher odds of failing to receive the first dose when randomized to XR-NTX, compared to those randomized to BUP-NX (see Table 3, all p < 0.05), consistent with the findings of the primary outcome analysis [13]. Analyses adjusted by age, sex, IV use, and other drug use showed similar results to unadjusted analyses.
Table 3.
Results of logistic regression models predicting failure to receive first dose of treatment drug (left panel) and failure to receive last dose (right panel) for each genetic variant.
| Outcome: Failed to receive first dose (Non-Hispanic, Caucasians, n=334) |
Outcome: Failed to receive last dose (Non-Hispanic, Caucasians who received first dose, n=290) |
||||||
|---|---|---|---|---|---|---|---|
| Gene Variant | Treatment by Gene variant Interaction χ2(df), p-value |
Main effect of Treatmenta OR (95% CI) |
Main effect of Gene Variant OR (95% CI) |
Treatment by Gene variant Interaction χ2(df), p-value |
Main effect of Treatmenta OR (95% CI) |
Main effect of Gene Variant OR (95% CI) |
|
|
OPRM1 A118G (GG/AG vs AA) |
χ2(1) = 0.42, p = 0.517 |
3.15b
(1.55, 6.39) |
0.53 (0.21, 1.33) |
χ2(1) = 1.93, p = 0.164 |
0.80 (0.50, 1.28) |
0.73 (0.42, 1.27) |
|
|
PDYN 68 bp repeat (SS/SL vs LL) |
χ2(1) = 1.35, p = 0.245 |
3.20b (1.58, 6.48) |
0.93 (0.49, 1.79) |
χ2(1) = 2.17, p = 0.141 |
0.88 (0.55, 1.41) |
1.52 (0.95, 2.43) |
|
|
PDYN rs910080 (CC/TC vs TT) |
χ2(1) = 2.91, p = 0.088 |
3.20b
(1.58, 6.46) |
1.47 (0.77, 2.80) |
χ2(1) = 0.04, p = 0.850 |
0.85 (0.53, 1.35) |
1.21 (0.76, 1.93) |
|
|
OPRK1 rs6473797 (GG/AG vs AA) |
χ2(1) = 0.05, p = 0.831 |
3.22b (1.59, 6.50) |
1.00 (0.52, 1.90) |
χ2(1) = 0.78, p = 0.378 |
0.84 (0.53, 1.34) |
1.14 (0.72, 1.81) |
|
Odds-ratio is XR-NTX vs BUP-NX
Significant at p-value < 0.05
χ2 = chi-square, df = degree of freedom, OR = odds ratio, CI = confidence interval
Effects of Treatment by Genotype on Receiving Last Dose of Medication:
Results for unadjusted logistic regression models on failure to receive last dose for each genetic variant are presented in Table 3. For all four variants, the 2-way interactions between treatment and genotype were not significant when estimating failure to receive last dose. This suggests that there are no differential effects of treatment between genotypes for any of these gene variants. When assessing the main effects of treatment and genotype, there were no significant differences in the odds of failure to receive last dose between treatment or between genotypes for any of the variants. Models additionally controlling for age, sex, IV use and other drug use showed similar results to unadjusted analyses.
Discussion:
We examined four common important gene variants of the opioid system, OPRM1 variant 118A>G, PDYN 68-bp repeat, PDYN rs910080, and OPRK1 rs6473797, as moderators or predictors of treatment outcome among a non-Hispanic Caucasian subsample in a comparative effectiveness trial of sublingual buprenorphine-naloxone (BUP-NX) vs extended-release injection naltrexone (XR-NTX). None of these variants was found to be a significant moderator of differential response to BUP-NX vs XR-NTX, i.e., no genotype by treatment interactions were detected. None of the variants examined had a significant main effect on any of the outcomes as well.
From a personalized- or precision- medicine vantage point, the absence of gene-variant-based moderator effects (prediction of a better response to one medication versus the other, buprenorphine vs naltrexone), or response to either buprenorphine or naltrexone (predictor effects, aside from the PDYN 68 bp repeat finding discussed above) is a disappointment. However, the dearth of effects observed here is consistent with the generally small and often elusive effects that have characterized psychiatric genetics. A relevant example is the association of the OPRM1 variant 118A>G (rs1799971, Asn40Asp) with response to naltrexone in treatment of alcohol use disorder, which appeared promising in some datasets, yet failed to replicate in others. Larger samples, additional variants, and combinations of genetic and clinical predictors may be needed to arrive at a clinically meaningful prediction of treatment effects.
A limitation of the current study is the relatively small sample size. Analysis was restricted to non-Hispanic Caucasian patients and there were too few African Americans or other ethnic cultural groups for a meaningful analysis. The sample analyzed for the outcome of ‘received last dose’ excludes those patients who were randomized but failed to initiate medication. Since genotype effects explored here were hypothesized to depend on physiological interactions between the medications and the opioid system, it made sense to include only those patients exposed to the medication. However, this is not an intent to treat sample of all randomized patients and thus could be biased.
Disclosure/Funding:
Supported by the National Institute on Drug Abuse (NIDA) through a collaborative clinical trial mechanism, PAR-07-232 (R01DA024554, to Dr. Nunes; and R01DA024555, to Dr. Lee), and additional support (K24DA022412, to Dr. Nunes). Trial medication was provided in kind from an investigator-initiated grant from Alkermes. Dr. Lee reports receiving grant support and study medication from Alkermes and study medication from Indivior (formerly Reckitt Benckiser). Dr. Nunes reports serving on an advisory board for Alkermes, receiving study medication from Reckitt Benckiser and Duramed Pharmaceuticals, being lead investigator for a NIDA-funded study of a computer-delivered behavioral intervention supplied by HealthSim, and being site principal investigator for a study funded by, and receiving travel support from, Brainsway. Dr. Rotrosen reports study support in his role as PI or Investigator in the form of funds or donated medication from Alkermes (manufacturer of Vivitrol) and from Indivior (manufacturer of Suboxone) and serves as a non-paid member of an Alkermes study steering committee. Indivior donated Suboxone for the present study. No paid consulting, speaker or other activities, no equity. No other potential conflict of interest relevant to this article was reported.
References
- 1.Hser YI, Saxon AJ, Huang D, et al. Treatment retention among patients randomized to buprenorphine/naloxone compared to methadone in a multi-site trial. Addiction. 2014January;109(1):79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krupitsky E, Nunes EV, Ling W, et al. Injectable extended-release naltrexone for opioid dependence: a double-blind, placebo-controlled, multicentre randomised trial. Lancet. 2011April30;377(9776):1506–13. [DOI] [PubMed] [Google Scholar]
- 3.Lee JD, Friedmann PD, Kinlock TW, et al. Extended-Release Naltrexone to Prevent Opioid Relapse in Criminal Justice Offenders. The New England journal of medicine. 2016March31;374(13):1232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattick RP, Breen C, Kimber J, et al. Buprenorphine maintenance versus placebo or methadone maintenance for opioid dependence. Cochrane database of systematic reviews (Online). 2014February6(2):CD002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hser YI, Evans E, Huang D, et al. Long-term outcomes after randomization to buprenorphine/naloxone versus methadone in a multi-site trial. Addiction. 2016April;111(4):695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nunes EV, Gordon M, Friedmann PD, et al. Relapse to opioid use disorder after inpatient treatment: Protective effect of injection naltrexone. Journal of substance abuse treatment. 2018February;85:49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weiss RD, Potter JS, Griffin ML, et al. Long-term outcomes from the National Drug Abuse Treatment Clinical Trials Network Prescription Opioid Addiction Treatment Study. Drug and alcohol dependence. 2015May1;150:112–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoffmeyer S, Burk O, von Richter O, et al. Functional polymorphisms of the human multidrug-resistance gene: Multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. PNAS. 1999;97(7):3473–34787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim RB, Leake BF, Choo EF, et al. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clinical pharmacology and therapeutics. 2001August;70(2):189–99. [DOI] [PubMed] [Google Scholar]
- 10.Levran O, O’Hara K, Peles E, et al. ABCB1 (MDR1) genetic variants are associated with methadone doses required for effective treatment of heroin dependence. Human molecular genetics. 2008July15;17(14):2219–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levran O, Peles E, Hamon S, et al. CYP2B6 SNPs are associated with methadone dose required for effective treatment of opioid addiction. Addict Biol. 2013July;18(4):709–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levran O, Peles E, Hamon S, et al. Nerve growth factor beta polypeptide (NGFB) genetic variability: association with the methadone dose required for effective maintenance treatment. The pharmacogenomics journal. 2012August;12(4):319–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee JD, Nunes EV, Novo P, et al. Comparative effectiveness of extended-release naltrexone versus buprenorphine-naloxone for opioid relapse prevention (X:BOT): a multicentre, open-label, randomised controlled trial. The Lancet. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bond C, LaForge KS, Tian M, et al. Single-nucleotide polymorphism in the human mu opioid receptor gene alters beta-endorphin binding and activity: possible implications for opiate addiction. Proceedings of the National Academy of Sciences of the United States of America. 1998August4;95(16):9608–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kranzler H, Gelernter J, O’Malley S, et al. Association of Alcohol or Other Drug Dependence with Alleles of the mu Opioid Receptor Gene (OPRMl). Alcoholism: Clinical and Experimental Research. 1998;22:1359–1362. [PubMed] [Google Scholar]
- 16.Wand GS, McCaul M, Yang X, et al. The Mu-Opioid Receptor Gene Polymorphism (A118G) Alters HPA Axis Activation Induced by Opioid Receptor Blockade. Neuropsychopharmacology. 20022002/January/01;26(1):106–114. [DOI] [PubMed] [Google Scholar]
- 17.Karpyak VM, Winham SJ, Preuss UW, et al. Association of the PDYN gene with alcohol dependence and the propensity to drink in negative emotional states. The international journal of neuropsychopharmacology / official scientific journal of the Collegium Internationale Neuropsychopharmacologicum. 2013June;16(5):975–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Preuss UW, Winham SJ, Biernacka JM, et al. PDYN rs2281285 Variant Association with Drinking to Avoid Emotional or Somatic Discomfort. PloS one. 2013;8(11):e78688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rouault M, Nielsen DA, Ho A, et al. Cell-specific effects of variants of the 68-base pair tandem repeat on prodynorphin gene promoter activity. Addict Biol. 2011April;16(2):334–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Levran O, Londono D, O’hara K, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes, Brain and Behavior. 2008;7(7):720–729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Williams TJ, LaForge KS, Gordon D, et al. Prodynorphin gene promoter repeat associated with cocaine/alcohol codependence. Addict Biol. 2007September;12(3–4):496–502. [DOI] [PubMed] [Google Scholar]
- 22.Proudnikov D, Kroslak T, Sipe JC, et al. Association of polymorphisms of the cannabinoid receptor (CNR1) and fatty acid amide hydrolase (FAAH) genes with heroin addiction: impact of long repeats of CNR1. The pharmacogenomics journal. 2010June;10(3):232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]


