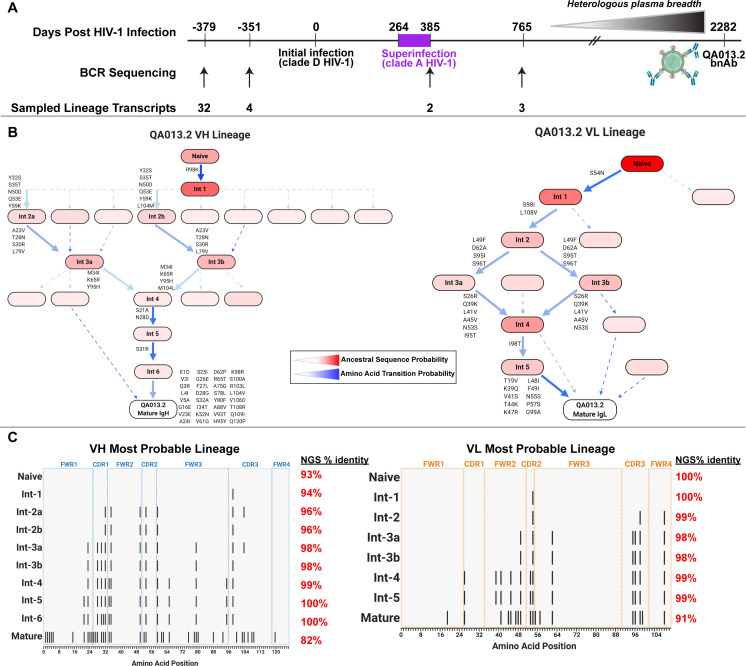
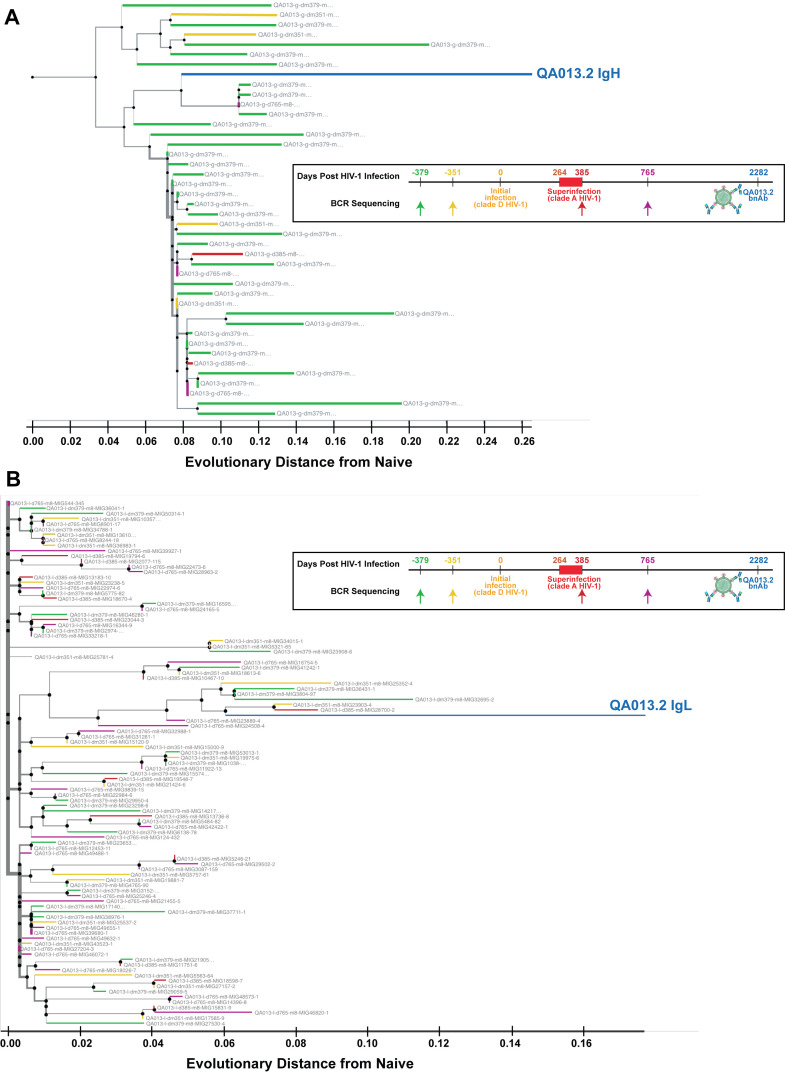
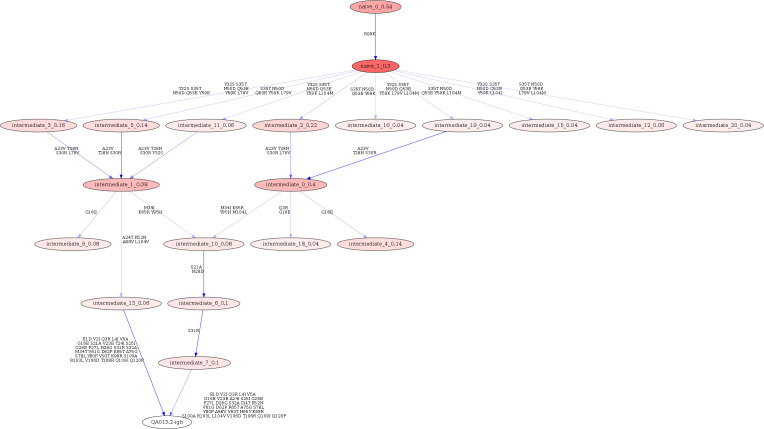
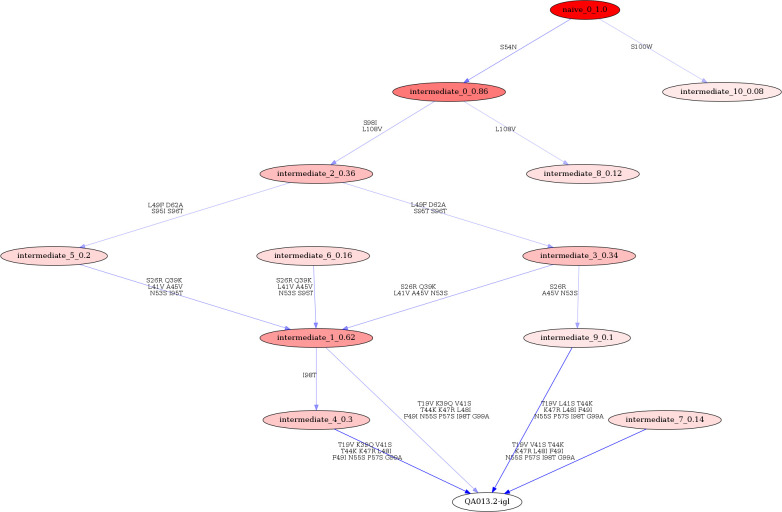
Figure 1. QA013.2 inferred clonal lineage development.
(A) Timeline of HIV infection, isolation of QA013.2 bnAb, and longitudinal PBMC samples used for antibody variable region sequencing. Days post initial HIV infection are listed above the timeline and black arrows below the timeline correspond to the specific PBMC samples that were used for deep sequencing. Approximate heterologous plasma breadth as determined previously (Cortez et al., 2012) is depicted above the timeline, which is not to scale. Estimated timing of superinfection as previously described (Chohan et al., 2005) is shown in purple on the timeline, with the timing of superinfection estimated as the midpoint, 324.5 days post initial infection. The QA013.2 bnAb was not isolated until 2282 days, or about 6.2 years post initial infection, as indicated. (B) Lineage graphics representing probable developmental paths for heavy and light chain lineages between inferred naïve BCR and mature QA013.2 sequences. Nodes represent inferred ancestral sequences, while arrows in between each node represent amino acid transitions. The red shading of nodes is proportional to the posterior probability that this ancestral sequence was present in the true lineage. For each given node, the blue shading of arrows arising from that node is proportional to the corresponding transition probability. Low probability nodes were filtered out, resulting in some incomplete pathways within the graphics. Dashed arrows indicate possible developmental paths that were not chosen based on low probability. See Figure 1—figure supplements 1–4. (C) Probable lineage sequences for the developing QA013.2 heavy (blue) and light chains (orange) are displayed in their inferred chronological order. With respect to the inferred naive sequence at the top of each lineage, variable region amino acid substitutions are indicated by black lines. Dashed lines demarcate the CDRs that are flanked by FWRs. Red percentages to the right of each sequence represent the nucleotide identity of each computationally inferred lineage member to sampled NGS sequences present in the longitudinal B cell repertoire of QA013, rounded to the nearest percent. See also Supplementary files 1 and 2 for fasta files of computationally inferred lineage members and sampled NGS sequences with high nucleotide identity to inferred lineage members.