Abstract
Hepatocellular carcinoma (HCC) is one of the tumors with a higher mortality rate globally, which significantly threatens people's health. Hepatitis C virus (HCV) infection is a major driving factor of HCC. This study aims to determine the key microRNA (miRNA), hub genes, and related pathways, construct potential miRNA–mRNA regulatory networks, and clarify the new molecular mechanism of HCV-related HCC. In this study, 16 differentially expressed miRNAs (DE miRNAs) were identified. The prediction of potential transcription factors and target genes not only found that SP1 and ERG1 may potentially regulate most of the screened DE miRNAs, but it also obtained 2923 and 1782 predicted target genes for the up-regulation and down-regulation of DE miRNAs, respectively. Subsequently, the introduction of differentially expressed genes dataset GSE62232 for target gene verification yielded 98 and 147 potential up-regulation and down-regulation target genes. The gene ontology (GO) and Kyoto encyclopedia of genes and genomes pathway enrichment analysis showed that they were mainly enriched in the cell cycle process, that is, subsequently, 20 hub genes were screened out through the protein–protein interaction network, and related genes were further evaluated using the GEPIA database. Based on the above analysis, the miRNA-hub gene regulatory network was constructed. In short, this research's hub genes and miRNAs closely related to HCV-related HCC were screened and identified through bioinformatics analysis and then built their connection. These results are expected to find potential therapeutic targets for HCV-related HCC.
Keywords: bioinformatic analysis, hepatitis C virus, hepatocellular carcinoma, microRNA–mRNA
1. Introduction
Hepatocellular carcinoma (HCC) is the most common form of primary liver cancer and ranks second in mortality from cancer.[1] Infection with hepatitis C virus (HCV) is currently the dominant risk factor for HCC.[2] Thus, increasing efficacious hepatitis C treatments may contribute to improving the global epidemiology status quo of HCC. In recent years, despite much research work that has been done, there is still largely unknown on the underlying mechanisms of progression in HCV-related HCC. Therefore, it is crucial to explore and develop effective diagnostic and therapeutic methods for HCV-related HCC.
Microribonucleic acid (miRNA) is a small noncoding RNA of about 21 nucleotides, a key posttranscriptional regulator of gene expression.[3] Previous studies have found that most protein-coding genes’ activity may be regulated by miRNA.[4] It has been reported that some miRNAs activate mRNA translation[5,6]; it can be seen that the interaction between miRNA and mRNA is very close. Interestingly, multiple genes exert important functions in the HCC process.[7] Many studies have also proved that there is a complex relationship between miRNA–mRNA with cancer.[8,9]
With the development of microarray and bioinformatics analysis in genetic screening, mining big data to identify new biomarkers and therapeutic ideas is the latest strategy.[10] In this study, the original microarray data set GSE40744 was selected from the Gene Expression Omnibus (GEO) database.[11] These data sets were further applied to screen out differentially expressed genes (DEGs) in HCV-related HCC and normal liver tissue. Next, we predicted the potential of DE miRNA upstream transcription factors and target genes, using the GSE62232 data set from the GEO database to further verify the target genes.[12] After enrichment analysis, protein–protein interaction (PPI) network analysis, and expression level evaluation, the potential miRNA–mRNA was successfully established in the HCV-related HCC regulatory network.
2. Materials and methods
2.1. Preprocessing of microarray dataset
The clinical samples of HCV-related HCC were retrieved from the GEO database, a common tool for storing, screening, and finding high-throughput microarray gene data sets.[13] One dataset (GSE40744) depended on the GPL14613 platform and met the inclusion criteria. Sixteen blood samples were selected from a total of 76 blood samples of GSE40744, including 7 normal liver samples and 9 HCV-related HCC samples. All sample data in this study were obtained from a public database, which allowed researchers to download datasets for relevant research and analysis, ethical review, and approval were thus not required.
2.2. Screening and visualization of differentially expressed microribonucleic acids
Screening DE miRNAs in HCV-related HCC and normal liver samples were carried out by GEO2R (http://www.ncbi.nlm.nih.gov/geo/geo2r).[13] The t test method was used to calculate the P-value of genes and corrected the false-positive results of the P value of genes using Benjamini and Hochberg methods. The following selection criteria screened out the DE miRNAs: adjusted P value < .05, | log2 FC | ≥2 were defined as the screening thresholds.
2.3. Predicting potential transcription factors and target genes of differentially expressed microribonucleic acids
We employed FunRich software to predicted the upstream transcription factors of screened up-regulated and down-regulated DE miRNAs, respectively (P < .05).[14] We present the results of the top 10 upstream transcription factors, according to the P value. Then, the miRNet database (http://www.mirnet.ca), a tool for functional analysis and visual exploration of miRNA,[15] was used to predict its downstream target genes by inputting the screened DE miRNAs related data.
2.4. Identification of differentially expressed genes
To further construct the regulatory network with our target genes of screened DE miRNAs, we employed the GEO database to download the HCV-related HCC mRNA expression datasets GSE62232. The R software “LIMMA” package was utilized to identify DEGs between HCV-related HCC and normal liver samples, and an adjusted P value < .05, | log2 FC | ≥1 were set as the thresholds. Due to the negative feedback relationship between miRNA and mRNA, we employed Venny 2.1.0 (https://bioinfogp.cnb.csic.es/tools/venny/) to intersect between down-regulated (up-regulated) DEGs with target genes of screened up-regulated (down-regulated) DE miRNAs to obtain the final target genes.[16]
2.5. Functional enrichment analysis of differentially expressed genes
GO enrichment analysis, including molecular function (MF), biological process (BP), and cell composition (CC), can conduct functional mining and exploration of gene data to promote a deeper understanding of the underlying disease mechanisms of DEGs.[17] Kyoto encyclopedia of genes and genomes (KEGG) is a database resource utilized to understand DEGs’ functions at the molecular level.[18] GO and KEGG enrichment pathways of the DEGs in HCV-related HCC were analyzed by the DAVID database.[19] And then, we employed the ggplot2 package in the R software to visualize these results above. P < .05 were considered statistically significant.
2.6. Protein–protein interaction network construction and hub gene analyses
The STRING database (http://www.string-db.org/) can perform interaction analysis to acquire the PPI network for the DEGs.[20] Then, visualize the network using Cytoscape software, a convenient and utility tool for the visualization of various interacting proteins or genes.[21] The top 10 hub genes in the PPI network with MCC methods of cytoHubba plug-in were selected for further analysis and screened hub genes serve as the miRNA final candidate target genes.
2.7. Validating the hub gene expression levels
The expression levels of hub genes in HCV-related HCC and normal liver samples were verified by the GEPIA database, a newly developed interactive web server based on TCGA and genotype-tissue expression data.[22] The down-regulated and the up-regulated hub genes were all imported into GEPIA, respectively. P < .01 was considered statistically significant.
2.8. Identification of potential microRNA-mRNA regulatory network
To explore the possible molecular mechanisms of HCV. Therefore, the miRNA and miRNA-target with negatively correlated regulatory networks were established and visualized using Cytoscape software.
3. Results
3.1. Screening of differentially expressed microribonucleic acids
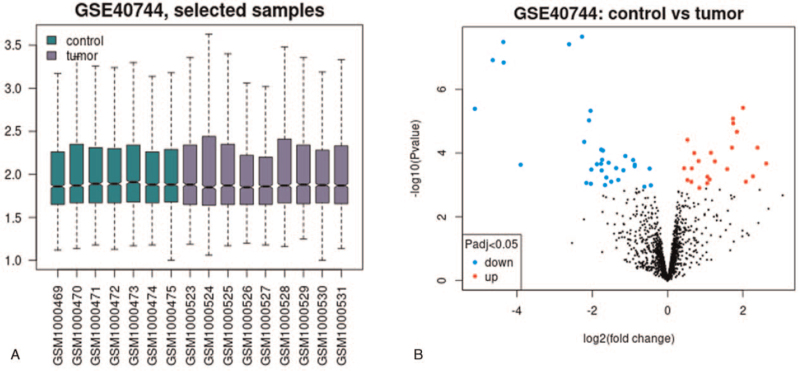
To investigate various approaches to further understand HCV development's possible mechanism to the HCC stage, we selected GSE40744 to screen DE miRNAs. The GSE40744 dataset included 7 normal liver tissues and 9 HCV-related HCC tissue samples, and the sample list of these 16 samples after correction and standardization were shown in Figure 1A. According to the set DE miRNAs thresholds, the GEO2R was applied to investigate the DE miRNAs. Finally, 11 up-regulated and 5 down-regulated DE miRNAs were identified, and their differential genes were visualized by a volcano map (Fig. 1B). The specific information of DE miRNAs was listed in Table 1.
Figure 1.
Identification of DE miRNAs: (A) sample list Boxplot about HCV related HCC DE miRNAs of the GSE40744 database. (B) Volcano map about HCV related HCC DE miRNAs of the GSE40744 database. Adjusted P value < .05, | log2 FC | ≥1, were set as the thresholds for identifying DE miRNAs. DE miRNAs = differentially expressed miRNAs, HCC = hepatocellular carcinoma, HCV = hepatitis C virus.
Table 1.
List of consistent DE miRNAs.
| DE miRNAs | miRNA name |
| Up-regulated (n = 11) | hsa-mir-10b, hsa-mir-1269, hsa-mir-146b, hsa-mir-182, hsa-mir-21, hsa-mir-221, hsa-mir-224, hsa-mir-3175, hsa-mir-3195, hsa-mir-34a, hsa-mir-452. |
| Down-regulated (n = 5) | hsa-mir-122, hsa-mir-139, hsa-mir-378, hsa-mir-422a, hsa-mir-424 |
DE-miRNAs = differentially expressed miRNAs, miRNA = microRNA/microribonucleic acid.
3.2. Predict the potential transcription factors of differentially expressed microribonucleic acids
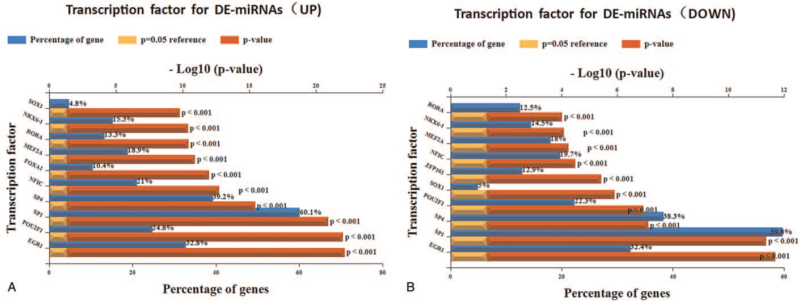
To find more HCV-related HCC molecules that have mutual regulatory effects, we predicted the upstream transcription factors that regulate DE miRNAs by FunRich software. The top 10 transcription factors involved in up-regulating and down-regulating these DE miRNAs are shown in Figure 2A and B.
Figure 2.
Predicted transcription factors of DE miRNAs: (A) top 10 upstream transcription factors of up-regulated DE miRNAs. (B) Top 10 upstream transcription factors of down-regulated DE miRNAs. DE miRNAs = differentially expressed miRNAs.
3.3. Predict the target genes of differentially expressed microribonucleic acids and differentially expressed genes identification
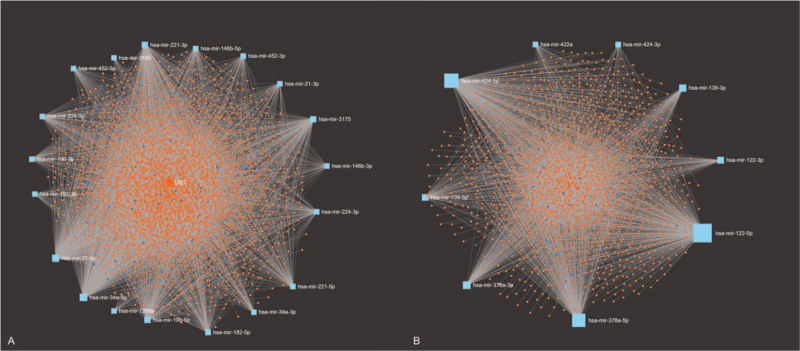
Generally, miRNAs perform posttranscriptional functions by base-pairing to the mRNA 3’untranslated regions.[23] Therefore, the miRNet database was applied to predict the downstream target genes of up-regulated and down-regulated DE miRNAs, respectively. Finally, as shown in Figure 3A and B, 2923 and 1782 target genes of DE miRNAs were acquired.
Figure 3.
Potential target genes of DE miRNAs predicted by miRNet database: (A) up-regulated miRNA-target gene network constructed. (B) Down-regulated miRNA-target gene network constructed. DE miRNAs = differentially expressed miRNAs, miRNA = microRNA/microribonucleic acid.
3.4. Identification of differentially expressed genes and validation of the target genes
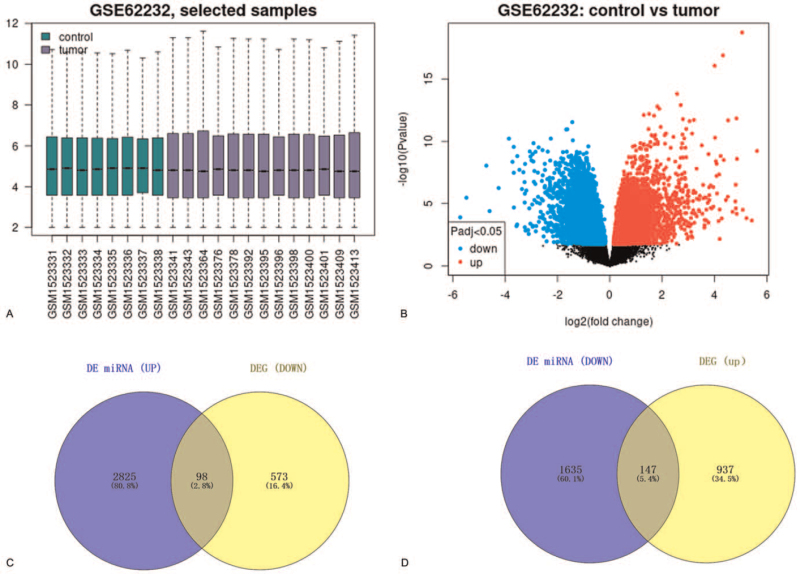
Subsequently, we intended to identify DEGs between HCV-related HCC and normal liver samples using GSE62232 downloaded from the GEO database. The visualization of data normalization and DEGs’ information was depicted in Figure 4A and B. Previous studies have shown a negative feedback relationship between miRNA and mRNA. Thus, Venny 2.1.0 was utilized to take the intersection between down-regulated (up-regulated) DEGs with target genes of up-regulated (down-regulated) DE miRNAs; 98 and 147 candidate target genes were eventually acquired, respectively (Fig. 4C, D).
Figure 4.
DEGs identification and the target genes validation: (A) sample list of GSE62232. (B) Volcano map to visualize DEGs in HCV-related HCC in the GSE62232 database. (C) The intersection of up-regulated miRNA target genes and down-regulated genes. (D) The intersection of down-regulated miRNA target genes and up-regulated genes. DEGs = differentially expressed genes, HCC = hepatocellular carcinoma, HCV = hepatitis C virus, miRNA = microRNA/microribonucleic acid.
3.5. The function and pathway enrichment analysis of differentially expressed genes
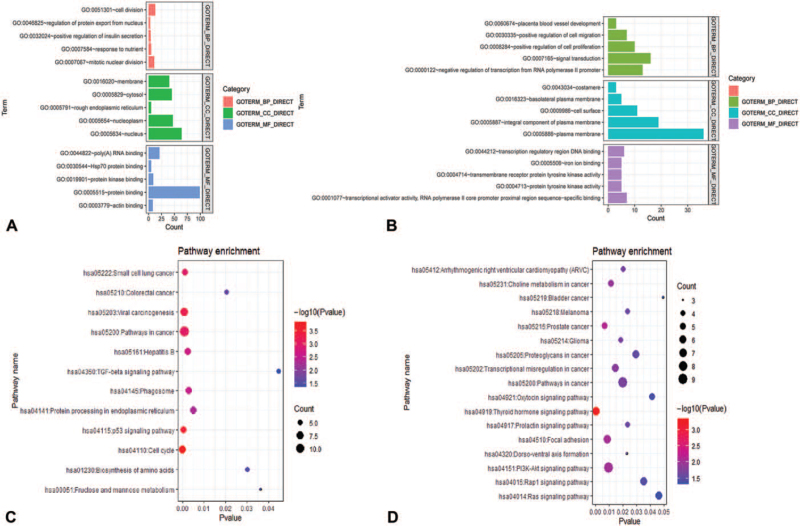
The GO enrichment results show that the first 5 functional enrichment results were selected. The BP of up-regulated DEGs were mainly enriched in cell division, mitotic nuclear division, and the CC was enriched in the nucleoplasm and membrane; MF was mainly involved in protein binding (Fig. 5A). The BP of down-regulated DEGs were mainly enriched in positive regulation of cell migration; regarding the CC, it was mainly enriched in the integral component of the plasma membrane and cell surface; the MF were mainly involved in transmembrane receptor protein tyrosine kinase activity and transcriptional activator activity (Fig. 5B).
Figure 5.
GO and KEGG enrichment analysis of candidate genes. (A) The top 10 enriched BP, CC, MF of up-regulated candidate genes. (B) The top 10 enriched BP, CC, MF of down-regulated candidate genes. (C) KEGG results of up-regulated candidate genes. (D) KEGG results of up-regulated candidate genes. P < .05 was considered statistically significant. BP = biological process, CC = cell composition, GO = gene ontology, KEGG = Kyoto encyclopedia of genes and genomes, MF = molecular function.
The KEGG pathway enrichment analysis results showed that the difference was statistically significant when P < .05. It was found that the up-regulated DEGs were mainly enriched in the cell cycle, p53 signaling pathway, viral carcinogenesis, pathways in cancer, and small cell lung cancer (Fig. 5C); the down-regulated DEGs were mainly enriched in thyroid hormone signaling pathway and PI3K-Akt signaling pathway (Fig. 5D).
3.6. Protein–protein interaction network construction and hub gene analyses
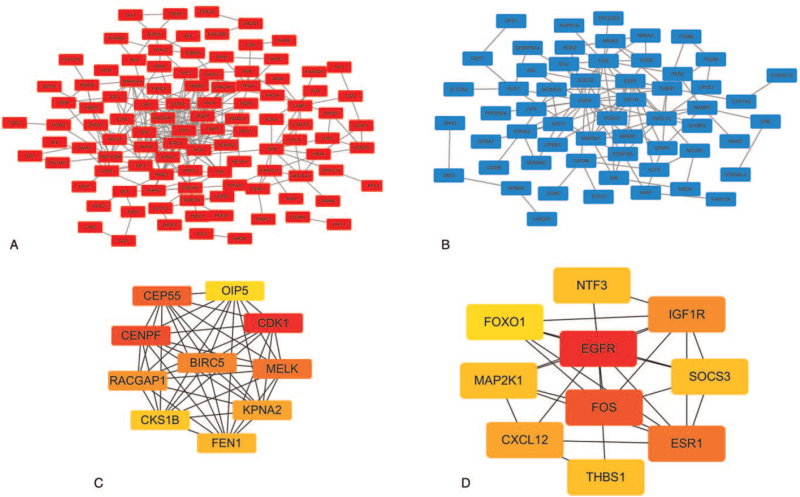
To further investigate the interaction among the 245 DEGs (147 up-regulated DEGs and 98 down-regulated DEGs), and to construct and visualize their PPI network, we will take these 245 DEGs and input them into a STRING database, and uploaded the obtained protein interaction data file to the Cytoscape database to complete. The PPI network composed of up-regulated DEGs contains 9 points and 2 edges. Next, we knew that the PPI network of up-regulated DEGs containing 106 nodes and 279 edges (Fig. 6A); and the PPI network composed of down-regulated DEGs contains 58 points and 112 edges (Fig. 6B), and then we employed the cytoHubba plug-in to identify the hub genes from it. For the target genes of up-regulated DE miRNAs, the results are as follows: CDK1, CENPF, CEP55, MELK, BIRC5, RACGAP1, KPNA2, FEN1, CKS1B, and OIP5 (Fig. 6C). For the target genes of down-regulated DE miRNAs, the results are as follows: EGFR, FOS, ESR1, IGF1R, CXCL12, NTF3, MAP2K1, SOCS3, THBS1, and FOXO1 (Fig. 6D).
Figure 6.
PPI network construction and hub gene screening. (A) PPI network of up-regulated DEGs. (B) PPI network of down-regulated DEGs, the red nodes indicated up-regulated genes, and the blue nodes indicated downregulated genes. (C) Top 10 hub genes with a higher degree of connectivity screened from the PPI network of up-regulated DEGs. (D) Top 10 hub genes with a higher degree of connectivity screened from the PPI network of down-regulated DEGs. DEGs = differentially expressed genes, PPI = protein–protein interaction.
3.7. Comparison of hub gene expression level
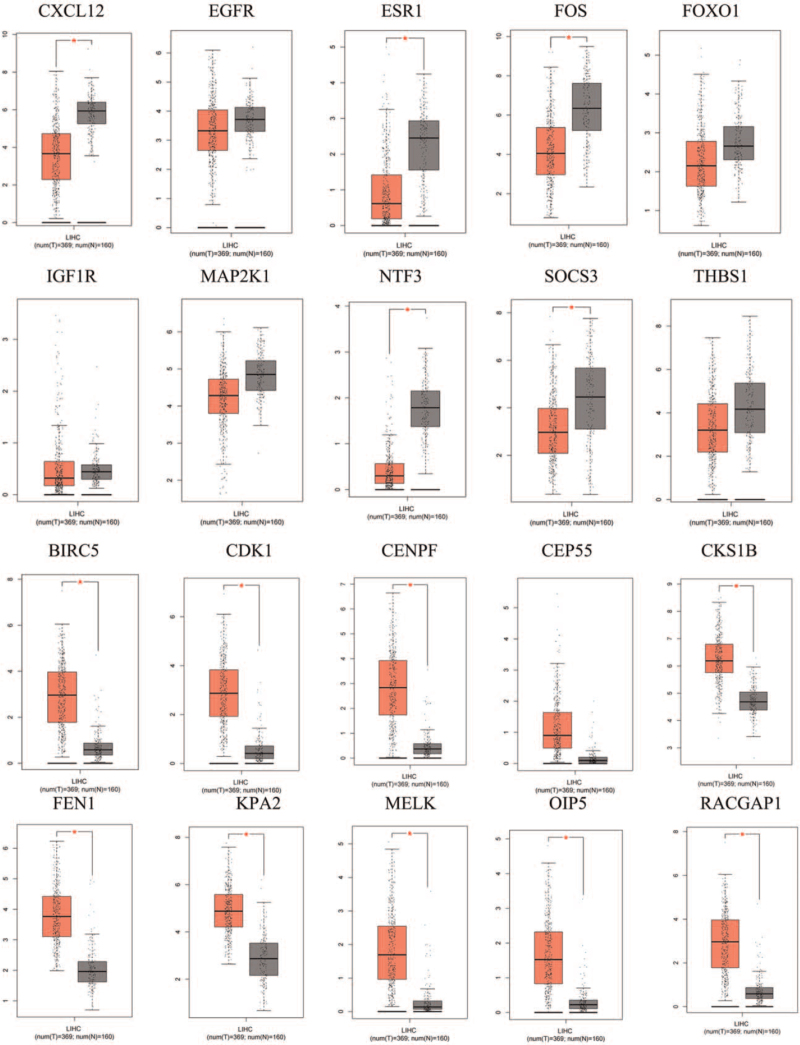
To further examine the role of DE miRNAs and these target DEGs in HCV-related HCC, we performed expression levels detection of the 20 hub genes through the GEPIA database. As shown in Figure 7, compared with normal tissues, the expression levels of 5 of 10 hub genes of down-regulated DE miRNA were obviously up-regulated in HCV-related HCC tissues. Nine of 10 hub genes of up-regulated DE miRNAs were evidently lower in HCV-related HCC tissues than in normal liver tissues. However, there are 6 hub gene expression levels with no significant difference, including EGFR, FOXO1, IGF1R, MAP2K1, THBS, and CEP55.
Figure 7.
The expression levels of hub genes from the GEPIA database: the first 10 images are the expression levels of down-regulated hub genes, and the last 10 images are the expression levels of up-regulated hub genes. ∗P < .05 indicates a significant difference.
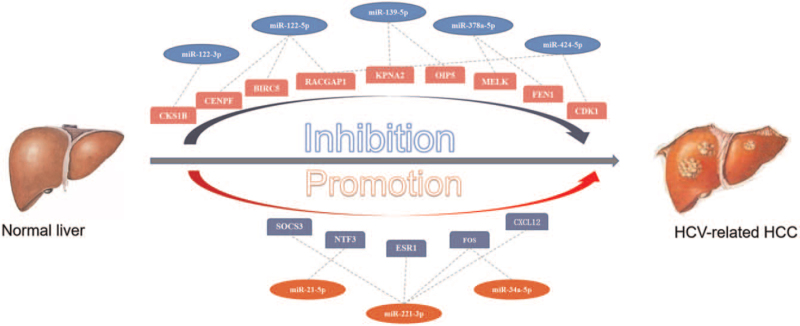
3.8. Identification of potential microRNA-mRNA regulatory network
We combined it with the prediction analysis of DE miRNAs target genes. In the pair degree's central gene expression analysis of the pair degree, the candidate miRNA-hub gene regulatory network was identified. On the basis of these findings, it was finally successfully constructed a potential miRNA–mRNA regulatory network associated with the development of HCV-related HCC, as presented in Figure 8.
Figure 8.
The candidate miRNA-hub gene regulatory network in HCV related HCC. The upward adjustment is represented by red, and the downward adjustment is represented by blue. HCC = hepatocellular carcinoma, HCV = hepatitis C virus, miRNA = microRNA/microribonucleic acid.
4. Discussion
Hepatitis C infection is a thorny public health problem and one of the leading causes of liver disease and liver cancer.[24] The second generation of direct-acting antivirals first came out in 2013,[25] generally became the main clinical therapy with lower side effects and a high response rate. They fundamentally change the natural course of HCV-related chronic hepatitis/cirrhosis and reduce the risk of HCC development.[26–28] Previous studies have shown that procession of HCV infection to HCC could be promoted by a direct or an indirect way, such as chronic inflammation, oxidative stress, and genetic mutations, as well as the interplay of host, virus, and environmental factors.[29,30] All in all, the occurrence and development of HCV-related HCC is usually a complex process involving multiple genes and multiple factors. Thus, the combined analysis of multiple genes in the era of big data is crucial. In this study, based on bioinformatics analysis methods, a total of 16 miRNAs in HCV-related HCC were screened in HCV-related HCC that differed from normal liver tissue expression. Predicting these 16 DE miRNAs’ upstream transcription factors found that EGR1 and SP1 have the most obvious regulation of up-regulation and down-regulation of DE miRNAs. Studies have shown that growth factors can induce EGR1 to transduce proliferation signals. Its role in proliferation and apoptosis has been proven.[31] The interaction between EGR1 and miRNA-181a-5p can affect the normal cell cycle of the HCC process.[32] Besides, SP1 is a zinc finger protein that can activate the transcription of many genes rich in CG-rich Sp binding sites.[33] Sp1 can bind to the promoter region of DHCR24 and inhibit hepatocyte apoptosis by destroying p53, thereby increasing the association with HCV the incidence of HCC.[34] The above research reports support our prediction results, which indicate that EGR1 and SP1 may regulate these DE miRNAs and affect the occurrence and development of HCV-related HCC.
Next, the differential genes from the GSE62232 data of the GEO data improve further improve the reliability of predicting target genes. After cross-validation, 98 down-regulated candidate target genes and 147 down-regulated candidate target genes were obtained. The GO and KEGG enrichment analysis showed that the GO mainly enriched cell division, nucleoplasm, and protein binding in up-regulated DEGs. This indicates that some of these DEGs may exist in the nucleus and participate in cell division to promote liver cells’ proliferation by binding to proteins. For example, TFAP4 can promote the carcinogenic ability of HCC.[35] On the contrary, the down-regulated DEGs mainly focus on the positive regulation of cell migration, the components of the plasma membrane, and the activity of transmembrane receptor protein tyrosine kinases. This indicates that some DEGs may be an organic part of the plasma membrane and have tyrosine kinase activity to promote cell signal transduction, phosphorylation that is, thereby promoting HCC occurrence and development. For example, tyrosine kinases play a vital role in liver cancer occurrence and metastasis.[36] The tyrosine kinase inhibitor sorafenib is used clinically to treat patients with advanced HCC.[37] Therefore, many genes in up-regulation and down-regulation are expected to become therapeutic targets. KEGG's results show that the up-regulated DEGs were mainly enriched in the cell cycle, p53 signal transduction pathway, and viral carcinogenesis. The down-regulated DEGs were mainly enriched in thyroid hormone signaling pathway, focal adhesion, PI3K-Akt signaling pathway. As we all know, the cell cycle plays an important role in maintaining normal life or tumors, and maintaining balance is particularly important for normal physiological activities. p53 is a tumor suppressor gene, and it needs to be balanced to play an active role. However, when certain key genes that are overexpressed or down-regulated appear, they may disrupt the balance and transform into tumors. For example, the cell cycle process is related to the overexpression of cyclophilin A in liver cancer cells.[38] The low expression of RDM1 can promote the occurrence of HCC through p53 and other pathways.[39] Additionally, some genes are enriched in the carcinogenic effects of viruses, the PI3K-Akt signaling pathway, that is, may play a key role in the occurrence of HCC after HCV infection. Similar to our predicted results, it has been reported that the PI3K-Akt signaling pathway can be activated by HCV and further promote HCV replication.[40] In short, GO, and KEGG's results reveal the specific molecular mechanism of HCC related to HCV to a certain extent.
Subsequently, using string and Cytoscape software, the PPI interaction network and hub genes of the up-regulated and down-regulated target genes were obtained. The expression levels of these central genes were further evaluated. Encouragingly, we found that most of the central genes have expression differences between HCV-related HCC patients and normal samples. Compared with normal tissues, 9 of the 10 central genes up-regulated by DE miRNA were significantly down-regulated in HCC tissues. Five of the 10 central genes of the down-regulated DE miRNA were significantly up-regulated in HCV-related HCC tissues. The finally up-regulated candidate hub genes include CDK1, CENPF, MELK, BIRC5, RACGAP1, KPNA2, FEN1, CKS1B, and OIP5. The down-regulated candidate hub genes are FOS, ESR1, CXCL12, NTF3, and SOCS3. These hub genes significantly influence the occurrence and development of HCV-related HCC from different perspectives. For example, CDK1 is cyclin-dependent kinase 1, and its overexpression can induce cell cycle progression and promote the proliferation of HCC cells.[41] The high expression of BIRC5 promotes the occurrence and development of HCC,[42] while RACGAP1 can affect HCV replication, and the up-regulation of its expression is significantly related to human HCC.[43]
In recent years, a large amount of evidence has shown the important role of miRNA-miRNA in HCV-related HCC. For example, miRNA-152 is produced due to its interaction with DNMT-1. It has a reverse correlation effect and can be used as a prognostic biomarker for HCV-related HCC.[44] The expression level of miR-138 can regulate TERT protein expression, thereby inhibiting the replication and senescence of HCV-related HCC cells.[45] Therefore, constructing a potential miRNA–mRNA regulatory network is essential for the early treatment of HCV-related HCC. Based on the above analysis, we constructed a potential miRNA–mRNA network. The network regulation diagram analysis showed that the up-regulated DE miRNA that regulates the number of mRNAs was hsa-mir-221-3p, which constitutes hsa-mir-221-3p/FOS, ESR1, CXCL12, SOCS3. The most down-regulated DE miRNAs that regulate the number of mRNAs were hsa-mir-122-5p and hsa-mir-424-5p, which constitute hsa-mir-122-5p/CENPF, BIRC5, and RACGAP1. Some related studies have shown that similar to our analysis results, miR-221 was one of the most significantly up-regulated miRNAs in HCC, and ESR1, FOS, and CXCL12 were its target genes.[46] It was found that miR-122 was regulated by DCAF1, participated in HCV replication, and induced the occurrence of HCC.[47,48] It was reported that the down-regulated miR-122 and miR-424 could be used as early biomarkers of liver cancer.[49] Besides, miR-21 is significantly elevated in the serum of patients with HCC and can inhibit the activation of the NOTCH1 pathway, thereby inhibiting sorafenib's antitumor effect.[50] Therefore, the above reports support the regulatory role of these regulatory axes in HCV-related HCC. Although results in the study with integrated bioinformatics analysis are of significance for understanding the interaction of HCV-related HCC, certain limitations still exist. The results from the sample enrolled need to be testified to apply to more related clinical samples. And the miRNA-hub gene interactions need to be confirmed through in vivo or in vitro experiments to make the HCV-related HCC-miRNA–mRNA network more comprehensive. Therefore, further studies are highly needed.
In summary, we have verified the respective expression and potential MFs of miRNA–mRNA pairs by integrating mining data and using comprehensive biometric analysis methods and constructed a potential regulatory network to help determine early biomarkers and therapeutic targets for HCV-related HCC. The choice also provides a theoretical basis and new strategies for future research on HCV-related HCC.
5. Conclusion
The identified key DEGs and their path analysis can further understand the specific molecular mechanism through comprehensive bioinformatic analysis. Besides, according to the regulation of DE miRNA, and we have gradually identified 10 key down-regulated and up-regulated target genes. Based on their miRNA–mRNA pairs, we successfully constructed a potential HCV-related HCC regulatory network. It provides a comprehensive reference and theoretical basis for finding HCV-related drug treatment targets and developing HCC.
Author contributions
Conceptualization: Zeng Tu.
Data curation: Rui Hao, Qianqian Liu, Lu Wang, Yang Wang.
Formal analysis: He Lu, Yanan Guo, Ailong Huang.
Investigation: Qianqian Liu, Lu Wang, Yang Wang.
Methodology: Rui Hao.
Project administration: Zeng Tu.
Resources: Zeng Tu.
Software: Rui Hao.
Supervision: He Lu, Yanan Guo, Ailong Huang, Zeng Tu.
Validation: Rui Hao.
Visualization: Rui Hao.
Writing – original draft: Rui Hao.
Writing – review & editing: Zeng Tu.
Footnotes
Abbreviations: BP = biological process, CC = cell composition, DE miRNAs = differentially expressed miRNAs, DEGs = differentially expressed genes, GEO = gene expression omnibus, GO = gene ontology, HCC = hepatocellular carcinoma, HCV = hepatitis C virus, KEGG = Kyoto encyclopedia of genes and genomes, MF = molecular function, miRNA = microRNA/microribonucleic acid, PPI = protein–protein interaction.
How to cite this article: Hao R, Lu H, Guo Y, Liu Q, Wang L, Wang Y, Huang A, Tu Z. Bioinformatics analysis of constructing a HCV-related hepatocellular carcinoma miRNA–mRNA regulation network. Medicine. 2021;100:33(e26964).
The authors have no conflicts of interests to disclose.
The datasets generated during and/or analyzed during the current study are publicly available.
References
- [1].Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer 2015;136:E359–86. [DOI] [PubMed] [Google Scholar]
- [2].Taura N, Yatsuhashi H, Nakao K, Ichikawa T, Ishibashi H. Long-term trends of the incidence of hepatocellular carcinoma in the Nagasaki prefecture, Japan. Oncol Rep 2009;21:223–7. [PubMed] [Google Scholar]
- [3].Ambros V. The functions of animal microRNAs. Nature 2004;431:350–5. [DOI] [PubMed] [Google Scholar]
- [4].Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle 2008;7:1545–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Niepmann M. Activation of hepatitis C virus translation by a liver-specific microRNA. Cell Cycle 2009;8:1473–7. [DOI] [PubMed] [Google Scholar]
- [7].Ozen C, Yildiz G, Dagcan AT, et al. Genetics and epigenetics of liver cancer. N Biotechnol 2013;30:381–4. [DOI] [PubMed] [Google Scholar]
- [8].O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature 2005;435:839–43. [DOI] [PubMed] [Google Scholar]
- [9].Ota A, Tagawa H, Karnan S, Tsuzuki S, et al. Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res 2004;64:3087–95. [DOI] [PubMed] [Google Scholar]
- [10].Marquardt JU, Galle PR, Teufel A. Molecular diagnosis and therapy of hepatocellular carcinoma (HCC): an emerging field for advanced technologies. J Hepatol 2012;56:267–75. [DOI] [PubMed] [Google Scholar]
- [11].Diaz G, Melis M, Tice A, et al. Identification of microRNAs specifically expressed in hepatitis C virus-associated hepatocellular carcinoma. Int J Cancer 2013;133:816–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Schulze K, Imbeaud S, Letouzé E, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet 2015;47:505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Barrett T, Wilhite SE, Ledoux P, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res 2013;41:D991–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pathan M, Keerthikumar S, Ang CS, et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015;15:2597–601. [DOI] [PubMed] [Google Scholar]
- [15].Fan Y, Xia J. miRNet-functional analysis and visual exploration of miRNA-Target interactions in a network context. Methods Mol Biol 2018;1819:215–33. [DOI] [PubMed] [Google Scholar]
- [16].Oliveros JC. Venny. An interactive tool for comparing lists with Venn's diagrams. (2007–2015). [Google Scholar]
- [17].Zhu J, Zhao Q, Katsevich E, Sabatti C. Exploratory gene ontology analysis with interactive visualization. Sci Rep 2019;9:7793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 2012;40:D109–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57. [DOI] [PubMed] [Google Scholar]
- [20].Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47:D607–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Su G, Morris JH, Demchak B, Bader GD. Biological network exploration with Cytoscape 3. Curr Protoc Bioinformatics 2014;47:01–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45:W98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell 2009;136:215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Gower E, Estes C, Blach S, Razavi-Shearer K, Razavi H. Global epidemiology and genotype distribution of the hepatitis C virus infection. J Hepatol 2014;61:S45–57. [DOI] [PubMed] [Google Scholar]
- [25].Liang TJ, Ghany MG. Current and future therapies for hepatitis C virus infection. N Engl J Med 2013;368:1907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cabibbo G, Celsa C, Calvaruso V, et al. Direct-acting antivirals after successful treatment of early hepatocellular carcinoma improve survival in HCV-cirrhotic patients. J Hepatol 2019;71:265–73. [DOI] [PubMed] [Google Scholar]
- [27].Calvaruso V, Cabibbo G, Cacciola I, et al. Incidence of hepatocellular carcinoma in patients with HCV-associated cirrhosis treated with direct-acting antiviral agents. Gastroenterology 2018;155:411.e4–21.e4. [DOI] [PubMed] [Google Scholar]
- [28].Cabibbo G, Petta S, Calvaruso V, et al. Is early recurrence of hepatocellular carcinoma in HCV cirrhotic patients affected by treatment with direct-acting antivirals? A prospective multicentre study. Aliment Pharmacol Ther 2017;46:688–95. [DOI] [PubMed] [Google Scholar]
- [29].Irshad M, Gupta P, Irshad K. Molecular basis of hepatocellular carcinoma induced by hepatitis C virus infection. World J Hepatol 2017;9:1305–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bertino G, Malaguarnera G, Frazzetto E, et al. Responsibility of hepatitis C virus in the development of hepatocellular carcinoma: from molecular alterations to possible solutions. World J Hepatol 2018;10:448–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Adamson ED, Mercola D. Egr1 transcription factor: multiple roles in prostate tumor cell growth and survival. Tumour Biol 2002;23:93–102. [DOI] [PubMed] [Google Scholar]
- [32].Bi JG, Zheng JF, Li Q, et al. MicroRNA-181a-5p suppresses cell proliferation by targeting Egr1 and inhibiting Egr1/TGF-β/Smad pathway in hepatocellular carcinoma. Int J Biochem Cell Biol 2019;106:107–16. [DOI] [PubMed] [Google Scholar]
- [33].Vizcaíno C, Mansilla S, Portugal J. Sp1 transcription factor: a long-standing target in cancer chemotherapy. Pharmacol Ther 2015;152:111–24. [DOI] [PubMed] [Google Scholar]
- [34].Tsukiyama-Kohara K. Role of oxidative stress in hepatocarcinogenesis induced by hepatitis C virus. Int J Mol Sci 2012;13:15271–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Song J, Xie C, Jiang L, et al. Transcription factor AP-4 promotes tumorigenic capability and activates the Wnt/β-catenin pathway in hepatocellular carcinoma. Theranostics 2018;8:3571–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Huang X, Gan G, Wang X, Xu T, Xie W. The HGF-MET axis coordinates liver cancer metabolism and autophagy for chemotherapeutic resistance. Autophagy 2019;15:1258–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].de Rosamel L, Blanc JF. Emerging tyrosine kinase inhibitors for the treatment of hepatocellular carcinoma. Expert Opin Emerg Drugs 2017;22:175–90. [DOI] [PubMed] [Google Scholar]
- [38].Gong Z, Chi C, Huang X, et al. Cyclophilin A is overexpressed in hepatocellular carcinoma and is associated with the cell Cycle. Anticancer Res 2017;37:4443–7. [DOI] [PubMed] [Google Scholar]
- [39].Chen SL, Liu LL, Wang CH, et al. Loss of RDM1 enhances hepatocellular carcinoma progression via p53 and Ras/Raf/ERK pathways. Mol Oncol 2020;14:373–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Shi Q, Hoffman B, Liu Q. PI3K-Akt signaling pathway upregulates hepatitis C virus RNA translation through the activation of SREBPs. Virology 2016;490:99–108. [DOI] [PubMed] [Google Scholar]
- [41].Yang WX, Pan YY, You CG. CDK1, CCNB1, CDC20, BUB1, MAD2L1, MCM3, BUB1B, MCM2, and RFC4 may be potential therapeutic targets for hepatocellular carcinoma using integrated bioinformatic analysis. Biomed Res Int 2019;2019:1245072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wu MJ, Ke PY, Horng JT. RacGTPase-activating protein 1 interacts with hepatitis C virus polymerase NS5B to regulate viral replication. Biochem Biophys Res Commun 2014;454:19–24. [DOI] [PubMed] [Google Scholar]
- [43].Bai G, Zheng W, Ma W. Identification and functional analysis of a core gene module associated with hepatitis C virus-induced human hepatocellular carcinoma progression. Oncol Lett 2018;15:6815–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].El-Araby RE, Khalifa MA, Zoheiry MM, et al. The interaction between microRNA-152 and DNA methyltransferase-1 as an epigenetic prognostic biomarker in HCV-induced liver cirrhosis and HCC patients. Cancer Gene Ther 2020;27:486–97. [DOI] [PubMed] [Google Scholar]
- [45].Shiu TY, Shih YL, Feng AC, et al. HCV core inhibits hepatocellular carcinoma cell replicative senescence through downregulating microRNA-138 expression. J Mol Med (Berl) 2017;95:629–39. [DOI] [PubMed] [Google Scholar]
- [46].Mou T, Zhu D, Wei X, et al. Identification and interaction analysis of key genes and microRNAs in hepatocellular carcinoma by bioinformatics analysis. World J Surg Oncol 2017;15:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Yan Y, Li C, Sun B, Yang R. DCAF1 is involved in HCV replication through regulation of miR-122. Arch Virol 2018;163:977–85. [DOI] [PubMed] [Google Scholar]
- [48].El-Ahwany EGE, Mourad L, Zoheiry MMK, et al. MicroRNA-122a as a non-invasive biomarker for HCV genotype 4-related hepatocellular carcinoma in Egyptian patients. Arch Med Sci 2019;15:1454–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Amr KS, Elmawgoud Atia HA, Elazeem Elbnhawy RA, Ezzat WM. Early diagnostic evaluation of miR-122 and miR-224 as biomarkers for hepatocellular carcinoma. Genes Dis 2017;4:215–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang S, Cai L, Zhang F, Shang X, Xiao R, Zhou H. Inhibition of EZH2 attenuates sorafenib resistance by targeting NOTCH1 activation-dependent liver cancer stem cells via NOTCH1-related microRNAs in hepatocellular carcinoma. Transl Oncol 2020;13:100741. [DOI] [PMC free article] [PubMed] [Google Scholar]