Abstract
The single‐stranded DNA binding protein (SSB) is essential to all aspects of DNA metabolism in bacteria. This protein performs two distinct, but closely intertwined and indispensable functions in the cell. SSB binds to single‐stranded DNA (ssDNA) and at least 20 partner proteins resulting in their regulation. These partners comprise a family of genome guardians known as the SSB interactome. Essential to interactome regulation is the linker/OB‐fold network of interactions. This network of interactions forms when one or more PXXP motifs in the linker of SSB bind to an OB‐fold in a partner, with interactome members involved in competitive binding between the linker and ssDNA to their OB‐fold. Consequently, when linker‐binding occurs to an OB‐fold in an interactome partner, proteins are loaded onto the DNA. When linker/OB‐fold interactions occur between SSB tetramers, cooperative ssDNA‐binding results, producing a multi‐tetrameric complex that rapidly protects the ssDNA. Within this SSB‐ssDNA complex, there is an extensive and dynamic network of linker/OB‐fold interactions that involves multiple tetramers bound contiguously along the ssDNA lattice. The dynamic behavior of these tetramers which includes binding mode changes, sliding as well as DNA wrapping/unwrapping events, are likely coupled to the formation and disruption of linker/OB‐fold interactions. This behavior is essential to facilitating downstream DNA processing events. As OB‐folds are critical to the essence of the linker/OB‐fold network of interactions, and they are found in multiple interactome partners, the SSB interactome is classified as the first family of prokaryotic, oligosaccharide/oligonucleotide binding fold (OB‐fold) genome guardians.
Keywords: genome guardian, OB‐fold, PXXP, SH3 domain, SSB
1. INTRODUCTION
The single‐stranded DNA binding protein (SSB) interactome is essential to maintaining genome integrity in bacteria.1, 2 The interactome consists of at least 20 DNA‐binding proteins that includes exonucleases, DNA helicases, DNA polymerases, DNA primases, recombination mediators, DNA repair enzymes, and topoisomerases (Figure 1, Table 1). The central player regulating interactome function is also an interactome member and is the product of the essential ssb gene, the highly stable, tetrameric, SSB.1, 3, 4, 5, 6, 7 The link between SSB function and interactome regulation was revealed in the sequence and structure of the protein as explained below.
FIGURE 1.
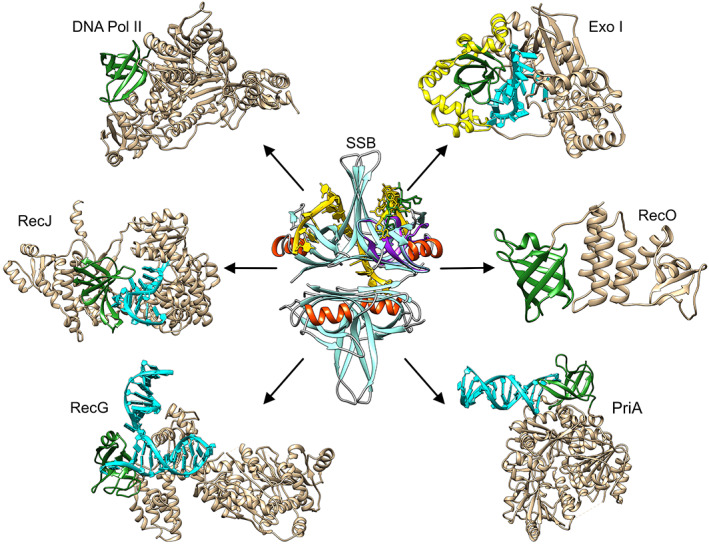
The single‐strand binding protein is the central player of the single‐stranded DNA binding protein (SSB) interactome. Each protein is presented as a ribbon diagram generated in Chimera.135 For SSB (PDB file 1EYG; colored cyan and red) the DNA in one‐half of the tetramer is colored yellow. In the monomer on the top right, an SH3 domain (PDB file: 2KXC; purple) was structurally aligned using TM‐align to show both the alignment and the possible location of a linker position (PXXP ligand; green).9, 136, 137 For the interactome partners, OB‐folds are colored green, DNA where present, is shown in cyan. The following PDB files were used to generate this image: Exonuclease I (PDB file: 4JS5),95 the extended SH3 domain is colored yellow according to Reference 138; RecO (3Q8D)27; PriA (6DGD)96; RecG (1GM5)71; Pol II (PDB file 3K5O)139; and RecJ (5F55)84
TABLE 1.
The majority of interactome partners have OB‐folds
| Protein | Has an OB‐folda | Aligns with SH3 domainb | Binds SSBc |
|---|---|---|---|
| SSB | YesC,T | Yes | YesL |
| AlkB | YesT | Yes | YesL |
| DinG | NDd | No | YesL |
| DnaG | YesC,T | Yes | YesA |
|
Exonuclease I |
YesC,T |
Yes |
YesA |
|
Exonuclease IX |
No |
No |
YesU |
| Pol II | YesC,T | Yes | YesU |
| Pol III (α) | YesT | Yes | Yes |
| Pol III (χ) | YesT | Weake | YesA |
| Pol III (ψ) | YesT | Weake | YesA |
| Pol IV | PossibleT | Weake | YesU |
| PriA | YesC,T | Yes | YesL,A |
| PriB | YesC,T | Yes | Yes |
| PriC | Unknown | NPf | Yes |
| RadD | YesT | Yes | Yes |
| RecG | YesC,T | Yes | YesL |
| RecJ | YesC,T | Yes | YesA |
| RecO | YesC,T | Yes | YesL,A |
| RecQ | ND | No | YesA |
| RNaseHI | ND | No | YesA |
| Topo III | YesT | Weake | YesU |
| Uracil glycosylase | PossibleT | Weake | YesL or A |
The presence of an OB‐fold was either readily observable in the crystal structures (C) or detected by TM‐align (T). OB‐folds were identified in in 6 partners by Inoue et. al. (144).
Two SH3 domains were used separately in TM‐align to determine of OB‐folds were present. These are Abl kinase and the insulin receptor tyrosine kinase substrate (IRTKS) SH3 domain.119, 123 These proteins bind ligands with either one or two PXXP motifs, respectively.
Binding between linker domain (L) or acidic tip (A) or unknown (U).
ND, no OB‐fold detected either by examination of the crystal structure or using TM‐align.117
TM‐alignment is weak and detects a possible SH3‐fold.
NP, not possible.
SSB monomers are divided into three regions each with a unique structure and function critical to the roles the protein plays in the cell (Figure 2a).8 Region one, known as the core domain of the protein, contains one oligonucleotide‐oligosaccharide binding fold (OB‐fold; four per tetramer) that is responsible for binding to single‐strand DNA (ssDNA), the linker of a nearby tetramer or the acidic phospholipids of the inner membrane (Figure 1, center).9, 10, 11, 12, 13, 14, 15, 16 Region 2, an intrinsically disordered linker or linker, contains three, conserved PXXP motifs whose role in protein function is to mediate protein–protein interactions.14 Region 3, known as the acidic tip (or tip), is positioned at the C‐terminus of the protein and functions as a regulator of the C‐terminal tail and as a secondary binding site.
FIGURE 2.
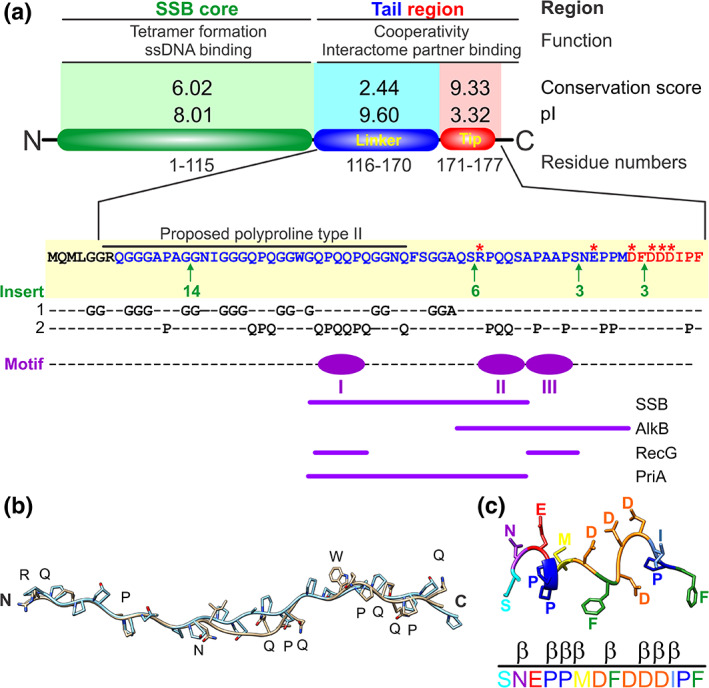
The three regions of an single‐stranded DNA binding protein (SSB) monomer each have critical roles in protein function. (a) A single monomer is divided by trypsin cleavage into the SSB core and tail. The tail is further divided into the intrinsically disordered linker (linker; blue) and acidic tip (tip; red). The conservation scores were previously published and generated using Praline and the pI of each region was calculated using the ProtParam tool of Expasy.15, 55, 140 The sequence of the tail of SSB is shown in blue and red, corresponding to the linker and tip, respectively. The stars indicate charged residues with the position and sites and sizes of the inserts indicated in green (for details see Reference 15). Over‐represented residues are highlighted on lines 1 and 2.15 The positions of the PXXP motifs are shown in purple. The roles of each motif in partner binding are indicated.14, 37, 65 (b) The N‐terminal half of the linker can be modeled on collagen.15 Collagen is colored in cyan, and the N‐terminal half of the linker is presented in neutral coloring. (c) A possible structure of the SSB acidic tip. The C‐terminal 14 residues were used as a tag for purification of myosin II.64 The tip was retained during structure determination and only the C‐terminal residues of PDB file 2JHR are shown with residues colored to enable visualization
It has long been thought that the eight‐residue acidic tip or C‐terminal peptide, mediated all SSB‐protein interactions as its removal eliminates partner binding and, as a peptide, it can bind target proteins.17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 However, recent evidence suggests that this is no longer an accurate depiction of SSB function. Hence, the linker/OB‐fold model was proposed to explain how both SSB and the interactome, function.29
This model provides a convincing rationale for mechanism of action of SSB and its cross‐talk with the interactome members containing OB‐folds, using a mechanism similar to eukaryotic Src homology 3 (SH3) domains binding PXXP motifs to mediate target protein function.30, 31 This makes sense because SH3 domains are structurally almost identical to OB‐folds and, there are three PXXP motifs in each of the linker regions of SSB.15, 32 Consequently, when a linker from one of the tetramers binds to the OB‐fold in another SSB, cooperative ssDNA‐binding results, rapidly producing a stable complex that protects the ssDNA from damage or nuclease digestion.14 In contrast, when an SSB linker binds to an OB‐fold in an interactome partner, proteins are loaded onto DNA, their functions are regulated, and, in some cases, this is accompanied by SSB dissociation.14, 29, 33, 34, 35, 36, 37
Molecular modeling, recently validated by experiments, has further revealed that ssDNA and the PXXP‐ligand can compete for binding to the OB‐fold so that when SSB coats ssDNA, some OB‐folds bind DNA, and some, linker PXXP‐motifs (Figure 1, center).14, 15 Thus, during cooperative ssDNA binding, the association of the first tetramer with DNA exposes its C‐termini.20, 38 Exposed linkers bind to OB‐folds in an adjacent tetramer, and this is repeated multiple times, creating an extensive network of linker/OB‐fold interactions that protect the ssDNA and require elevated concentrations of salt or powerful DNA motor proteins to disrupt them.15, 29, 39, 40, 41 Concurrently, the remaining exposed linkers within the SSB‐ssDNA complex are also available to bind interactome partner OB‐folds.33, 34
As linkers can bind to the SSB OB‐folds, the primary role of the acidic tip is to regulate the structure of the C‐terminal regions of the protein to prevent OB‐fold binding from happening until required.42 This is critical as if linkers bind stably to SSB OB‐folds in the absence of ssDNA, the protein may be inactivated and eliminate interactome function resulting in cell death. Thus, once linker/OB‐fold mediated, complex formation between SSB and a partner has occurred, the secondary function of the acidic tip, that is, binding to partners at sites distal to the OB‐fold, is utilized.
In both SSB‐SSB and SSB‐partner binding, OB‐folds may function as scaffolds for the PXXP‐containing linker, likely contributing to complex stability, as observed for c‐Src.43 The presence of multiple OB‐folds in SSB and its interactome partners has led to the proposal that this family of proteins is the first family of OB‐fold genome guardians discovered in prokaryotes.14, 29, 44, 45, 46 The SSB protein and the mechanism of interactome regulation are the topics of this review.
1.1. Organization of an SSB monomer
SSB exists as a 75.4 kDa homo‐tetramer that is the active form of the protein.47, 48 Each 18,844 Da monomer is comprised of three distinct regions with region one being easily separated from regions two and three by proteolytic cleavage (Figure 2a).4, 8, 15, 38, 49 Region one, known as the core domain of the protein, is comprised of the N‐terminal 115 amino acids and contains the information required for tetramer formation and, the OB‐folds which bind in competitive fashion to ssDNA, the linker region of the other SSB tetramers or, the acidic phospholipids of the inner membrane (Figure 1, center, 2A).14, 15, 38, 49, 50, 51, 52, 53, 54 Not surprisingly, sequence analyses reveal that the first 115 residues are well conserved, with a conservation score of 6 out of 10 and the pI of this domain is 8.01.15, 16 The OB‐fold is structurally almost identical to eukaryotic SH3 domains.32 The importance of this point in SSB function is elaborated upon in the section “The mechanism of linker/OB‐fold binding”.
Region 2 of the SSB monomer is comprised of the intrinsically disordered linker.56 It is part of the C‐terminal one‐third of the protein that was initially proposed to be non‐essential, functioning as a spacer.8 However, it is now clear that this region of SSB is essential for cooperative ssDNA binding mediated via protein–protein interactions.14, 29, 56, 57, 58 Sequence analysis revealed that the overall sequence of the linker is poorly conserved with a conservation score of 2.44 out of 10 and a pI of 9.6.15 The overall poor sequence conservation of the linker region is the result of 4, variably sized insertions (Figure 2a, green arrows). When inserts are accounted for in alignments, the presence of 3, well‐conserved PXXP motifs is revealed.15 These motifs are well‐known for their ability to bind SH3 domains in eukaryotes.59, 60 Further, PXXP motifs have a high propensity for adopting a left‐handed PPII helix, suggesting that this region plays a role in mediating protein–protein interactions.61 Consistent, the PXXP motifs bind to OB‐folds in either another SSB tetramer or interactome partner.14 This is explained in more detail in the section “The mechanism of linker/OB‐fold binding.”
In addition to the critical PXXP motifs, the linker sequence also has a high over‐representation for Gly (27.8%), Gln (20.4%), Pro (16.7%) and, Ser (7.4%) (Figure 2a).15 The presence and spacing of these over‐represented residues in the N‐terminal half of this region ending at amino acid 148, is consistent with the formation of a polyproline II helix (PPII).62 Modeling shows that this part of the linker can adopt a PPII helix that superimposes well with a collagen peptide, with an RMSD = 0.8 Å for the backbone atoms (Figure 2b).15 The similarity to collagen may provide a mechanism for the flexibility associated with the linker. Also, the over‐represented residues are arranged in repeats similar to those found in spider silk, the X‐type HMW subunit of wheat gluten and, the ω‐protein.63 These have been proposed to impart similar elastomeric properties to the linker of SSB.15 Collectively, this flexibility may be necessary to bind partners of different sizes, present in different conditions and, to enable the linker to readily dock onto the partner OB‐folds.
Although region 3 or the acidic tip of each SSB monomer is only 8 residues in length, it is the most well‐conserved part of the protein.1, 15 In an alignment of the C‐terminal tails of 251 Proteobacterial SSB proteins, the conservation score for the tip was 9.33 out of 10 and this region has a pI of 3.32 (Figure 2a).15 The low pI is consistent with this region containing 67% of the charged residues in the C‐terminal one‐third of the protein and this plays a role in SSB regulation.42 One possible structure of the C‐terminal 14 residues of Escherichia coli SSB was revealed in the structure of myosin II where it was used a purification tag.64 In this structure, the tip does not contact the N‐terminal SH3 domain of myosin II and is predominantly β‐turn with three of the four Asp residues on one side of the structure and the terminal Phe positioned in a planar configuration at the C‐terminus (Figure 2c). It had long been thought that the tip mediated all SSB‐protein interactions as its removal eliminates partner binding and, as a peptide, it can bind target proteins sometimes in multiple locations.17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28 However, this is no longer an accurate depiction of SSB protein function. Instead, this region of the protein functions as a regulator of SSB and as a secondary binding site as explained in subsequent sections.
1.2. The linker is the primary protein–protein interaction domain of SSB
In 2017, the Bianco group showed using linker‐swapping and deletion mutants, that the intrinsically disordered linker of SSB was required for binding to RecG and RecO, and that the tip was likely not involved.29 This follows because each mutant SSB used in this study retained a functional acidic tip but was defective for partner binding. It was concluded that the acidic tip cannot be responsible for mediating protein–protein interactions.
Consequently, the linker/OB‐fold model of interactions was proposed as being responsible for mediating SSB‐partner binding, thereby linking the ssDNA and partner binding activities of the protein together for the first time.29 This model proposed that for SSB‐SSB interactions, binding of the linker of one tetramer to the OB‐fold in another tetramer, results in cooperative ssDNA‐binding, producing a stable complex that rapidly protects the DNA from damage or nuclease digestion.14 It also proposed that, when SSB linker‐binding to an OB‐fold in interactome partners occurs, proteins are loaded onto DNA, their functions regulated, and, in some cases, binding results in SSB dissociation.14, 29, 33, 34, 35, 36, 37
The linker/OB‐fold model was corroborated by the Huang and Varshney groups working with PriA and uracil DNA glycosylase, respectively.29, 65, 66 Again, linker‐swapping studies were done, and importantly, a species‐specific acidic tip was present in each chimeric SSB protein tested. Also, Huang found that linker length may also play a role in the ability of SSB to bind partner proteins. Recently, the Anindya group demonstrated that the acidic tip is dispensable for binding to the DNA alkylation repair protein AlkB and instead, binding required residues 152–169 of the linker.37 Collectively, these studies show that the linker is responsible for mediating protein–protein interactions.
1.3. The mechanism of linker/OB‐fold binding
Analysis of the linker sequence revealed the presence of the three conserved PXXP motifs thereby providing the first insight into the mechanism of action of linker/OB‐fold binding (Figure 2a).15 PXXP motifs are well known in eukaryotic systems as they are the ligand for SH3 domains.59, 60, 67 The importance of this result for interactome function became clear when it was realized that SH3 domains are structurally almost identical to OB‐folds.32 When these domains are superimposed, they differ by less than 2 Å for the β‐strands (Figure 3a‐c).29, 32
FIGURE 3.
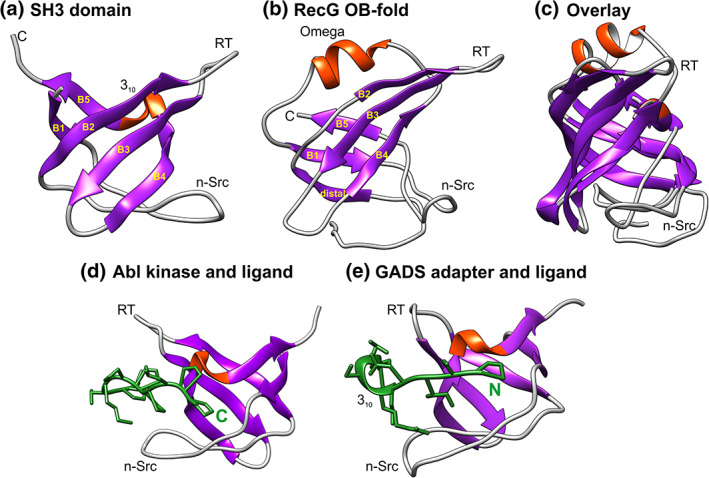
SH3 domains and OB‐folds are structurally almost identical. Images were generated using Chimera with helices colored red and β‐sheets in purple. The labeling of strands, helices, and loops in panels A and B is taken from Reference 32. Loop nomenclature is from the Src protein (RT‐Src and nSrc, respectively)/68 The RT‐loop connects β2 and 3 while the nSrc loop connects β‐strands 1 and 2. (a) The SH3 domain shown is from the insulin receptor tyrosine kinase substrate (IRTKS) SH3 domain (PDB file:2KXC).137 (b) The OB‐fold is from the T. maritima RecG (PDB file:1GM5).71 (c) Structural alignment of the SH3 domain and with the RecG OB‐fold. The alignment was done using TM‐align.136 (d) and (e) SH3 domains can bind PXXP ligands in opposite orientations. The structures are from PDB files 1ABO and 2DON, respectively70, 141
Furthermore, SH3 domains bind PXXP‐containing ligands in a pocket sandwiched between the RT‐Src (RT) and nSrc loops (Figure 3d,e).68 Binding can occur in one of the two orientations with either the C‐terminus proximal to the 310‐helix as seen for Abl kinase or, the N‐terminus of the ligand proximal to this helix as observed in the GADS adapter protein.67, 69, 70 For Abl kinase, the peptide ligand binds over its entire length and interacts with three major sites on the SH3 domain using both hydrogen‐bonding and van der Waals contacts (Figure 3d). Residues 4–10 of the peptide used to determine the co‐crystal structure, adopt the conformation of a left‐handed polyproline helix type II. In contrast, in addition to being bound in the opposite orientation to Abl kinase, ligand binding by the GADS adapter protein is unique among SH3 domains as the peptide has a 310‐helix in its approximate center (Figure 3e). This helix positions the two proline residues towards the face of the SH3 domain to facilitate binding.
The binding of PXXP ligands by SH3 domains and the structural similarity of these domains to OB‐folds suggested a mechanism for SSB‐partner interactions.15, 29 First, the partner should contain an OB‐fold, and second, one or more PXXP motifs in the linker of SSB should mediate binding to that OB‐fold. The importance of the partner OB‐fold in SSB binding was demonstrated in vivo. First, deletion of OB‐folds in PriA, RecG, and RecO eliminated SSB binding.14 Second, mutation of single residues in the RecG OB‐fold predicted to be involved in forming the linker/OB‐fold interface, reduced SSB binding 10‐ to 20‐fold (Figure 4a,b; residues F75, M80, R95 and, F97). These residues are also part of the binding site of the helicase for the leading strand arm of the fork.71 Notably, F97 mutants are defective for both SSB and DNA binding, consistent with the model that that these binding sites overlap and that DNA and SSB binding is competitive.33, 72, 73
FIGURE 4.
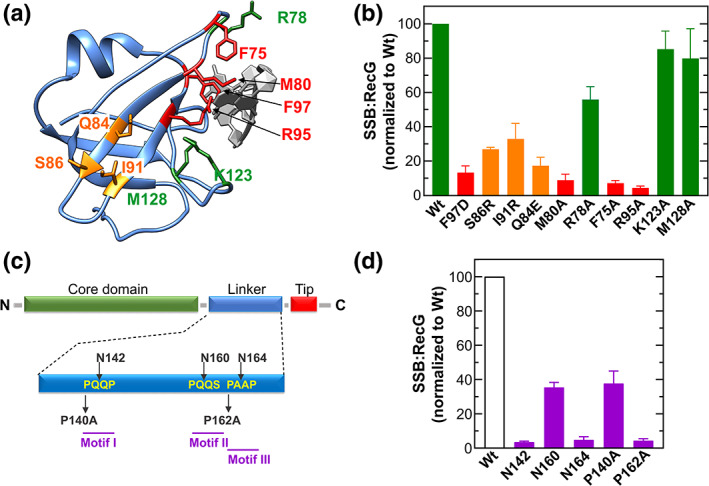
The single‐stranded DNA binding protein (SSB)‐partner interface is comprised of the partner OB‐fold on one side and the PXXP motifs of the linker on the other. (a) and (b) Residues of the RecG OB‐fold comprise one side of the SSB‐RecG interface. (a) Location of residues in the RecG OB‐fold that were predicted to be involved in SSB binding. The DNA is colored in light grey. (b) Analysis of SDS‐PAGE gels from in vivo binding studies.14 Residues are colored according to their importance in SSB binding: green (unimportant); red (critical) and orange (intermediate). (c) and (d) The PXXP motifs comprise one side of the SSB‐RecG interface. (c) Location of mutations in the PXXP motifs. (d) Analysis of SDS‐PAGE gels of in vivo binding studies14
Next, it was shown that SSB uses PXXP motifs I and III to bind RecG, as when these are mutated either by disrupting PXXP spacing or by altering the structure when Pro is replaced by Ala, binding is reduced 20‐fold (Figure 4c,d). Linker swapping experiments show that either PXXP motif I, II, or both, are required for stimulation of the ATPase activity of Klebsiella pneumoniae PriA by its cognate SSB.65 Finally, the binding site of E. coli AlkB was mapped to a region spanning motifs II and III of the E. coli SSB linker.37 Finally, cooperative ssDNA binding by SSB is eliminated when the proline spacing of PXXP motif I is interrupted.14 This result makes sense as SSB‐SSB interactions, mediated by linker/OB‐fold binding, are required for cooperative ssDNA binding.
Eukaryotic PXXP motifs have been classified into different classes.74 Similarly, the SSB PXXP motifs may be classified into two of these classes. Motif I represents a unique class (WGQPxxPQG) whereas motifs II and III more closely resemble Class I (+xxPXXP; for SSB ‐ QxRPxxP and QxxPxxP, respectively).14, 15 Based on their sequence contexts, which together with the unique sequences of each partner OB‐fold, enables one SSB linker to bind to multiple partners. The utilization of different motifs for binding to different partners results in a unique protein–protein interface in each case.
1.4. The acidic tip can bind to interactome partners
Shortly after its discovery, SSB was shown to interact with DNA polymerase II.75 It was later shown to bind to Exonuclease I and protein n (PriB).76, 77 Using SSB‐affinity chromatography, three additional, but unidentified partners of MWr = 25, 32 and, 36 kDa were shown to bind.78 Several years later, the McHenry and O'Donnell groups showed that SSB bound to the χψ‐heterodimer and, separately to the ψ‐subunit of DNA polymerase III and, that a peptide comprising the C‐terminal 15 residues was sufficient for binding to psi.21, 79 Also, binding was impaired when a mutant in the acidic tip, SSB113 (P176S), was used. Later, the Sandigursky and Genshcel groups confirmed SSB‐ExoI binding in vivo and in vitro.80, 81 Further, Genshcel demonstrated that binding to Exonuclease I in the absence of DNA was eliminated when either the SSB113 (P176S), SSB F177C, or SSBΔC10 (lacks the C‐terminal 10 residues) mutant proteins were used. In contrast, SSB G15D (an OB‐fold mutant) retained binding ability. Finally, the binding of Exo I to SSB was inhibited by a 9‐residue peptide corresponding to the acidic tip. Next binding of SSB to the χ‐subunit only of DNA polymerase III was shown and, when a mutant lacking the C‐terminal 26 residues of SSB (SSBΔC26) was used to replace wild type, binding was lost.22 Subsequent studies re‐confirmed binding to Exonuclease I and demonstrated binding to the RecQ DNA helicase.26, 82 Binding was studied using full‐length SSB and RecQ and, C‐terminal peptides and full‐length partners. It was also shown that binding was eliminated when a mutant SSB protein lacking only the acidic tip was used (SSBΔC8). Additional studies have adopted the peptide‐partner approach to demonstrate binding to RecO, PriA and, the χ‐subunit of DNA polymerase III.17, 20, 27 Separate studies used SSBΔC8 to show the importance of the tip in partner binding in vitro and in vivo.23, 24, 83 However, a recent study shows that the use of SSB tip mutant proteins is problematic as changing a single residue in the tip can have a significant impact on the structure of the C‐terminal region of SSB, as shown for SSB 113 (P176S).14 One explanation of the effects of tip mutations on SSB function is presented in the section “The acidic tip is not the primary protein‐protein interaction domain of SSB.”
Further evidence showing that the tip can form a complex with a partner came from several structural studies using tip peptides and a full‐length partner (Figure 5).19, 25, 26, 27, 84, 85 Surprisingly, and even though the sequence of the acidic tip is almost invariant, the primary amino acid sequence of the binding pockets is not well conserved. Instead, the structural features of the binding pocket are conserved so that the terminal F177 sits in the base of a hydrophobic pocket and is flanked by basic residues that are proposed to contact the acidic residues of the tip. However, in all structures to date, only the last 3 or 4 residues are visible (either DIPF or IPF; Figure 5, insets).84 For Exonuclease I, the structure showed two binding sites for the acidic tip, with one of these serving as the biologically relevant site.26
FIGURE 5.
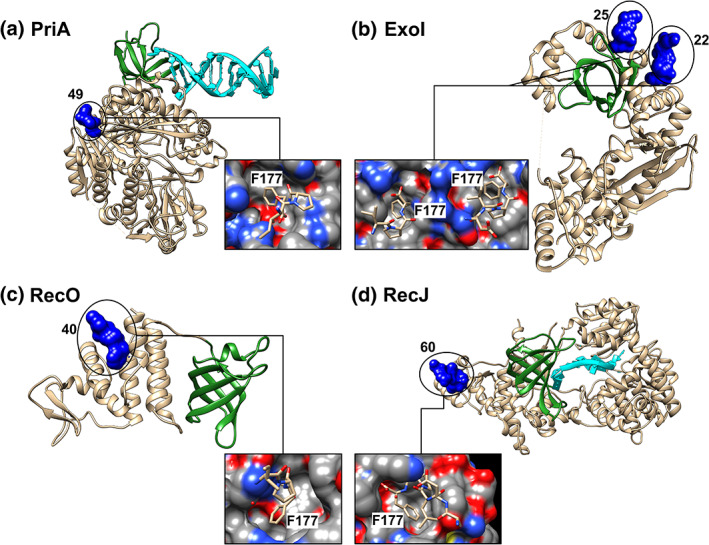
The locations of single‐stranded DNA binding protein (SSB) acidic tip binding pockets on partners are distant from the OB‐folds. The structures of four tip‐partner complexes are shown with the OB‐folds colored in green and DNA in cyan. The tip peptides are presented as surfaces and are colored blue. The numbers associated with each panel are the distances in Angstroms from F177 of the peptide in its binding pocket to the RT‐loop of the OB‐fold in each protein. Loop nomenclature is from the Src protein (RT, RT‐Src and nSrc, respectively).68 Insets: the tip binding pockets in each of the partner proteins with surfaces colored according to charge. PDB files used to create this image were (a) PriA (6DGD and 4NL8)25, 96; (b) Exo I (3C94)26; (c) RecO (3Q8D)27 and (d, RecJ (5F56)84
It is important to note that the binding sites on partners for the acidic tip are positioned 22–60 Å from the partner OB‐fold where DNA binding occurs (Figure 5).68 Consequently, the current peptide‐partner structures do not provide a satisfactory explanation for the mechanism of the functional interaction of SSB with a partner. Instead, it is the binding of the linker of SSB to the partner OB‐fold that likely results in the functional interaction. For the DNA helicases PriA and RecG, this has been called remodeling and takes place during the loading of these enzymes onto DNA and includes enhancing their ATPase and helicase activities.33, 34 For Exo I, Pol II, and RecJ this results in regulation of nuclease activity.23, 65, 76, 83, 86 For RecO, likely the binding of its OB‐fold with the linker of ssDNA‐bound SSB, results in tetramer dissociation.35, 36
1.5. The acidic tip is a regulator of SSB function
Multiple studies have shown that the acidic tip can bind to partner proteins (see the preceding). These studies were done using isolated tip or C‐terminal region peptides and an intact partner. However, in the absence of four core domains, these isolated peptides may function differently, as explained below. The structural studies show that the peptide binds in a pocket on the partner with the terminal F177 positioned in the base of the pocket (Figure 5, insets).19, 25, 26, 27, 84, 85 It was proposed that the F177‐pocket interaction is essential and supported by the association of the charged residues in the tip with appropriately positioned residues in the partner pocket.
This model was tested using full‐length, mutant SSB proteins and full‐length RecG as the partner. First, an extra residue was added after the terminal Phe to create SSB S178 and separately, SSB A178. Second, SSBΔC8 was used and third, the aspartic acid residues were changed to alanine to create SSB D4A4. These four mutants do not bind RecG in the absence of DNA whereas wild type can do so (Figure 6a).14 The simple, but the incorrect interpretation of these results, is that the acidic tip is the primary binding site and, when it is mutated, binding is eliminated.
FIGURE 6.
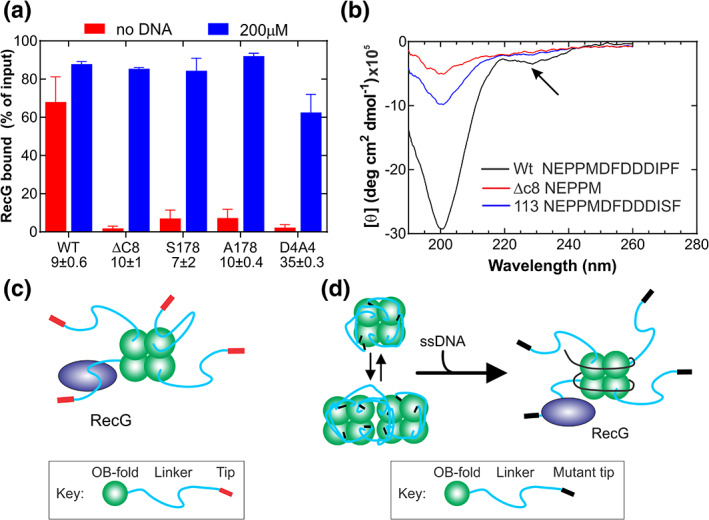
The acidic tip is not required for partner binding but instead regulates SSB tail structure. (a) The results from bead binding assays are shown.14 In these assays, his‐SSB proteins were bound to magnetic beads and then either RecG added (red bars) or, in separate experiments poly d(T) followed by RecG (blue bars). The values below each column title are the intrinsic site sizes for each protein determined in intrinsic fluorescence quenching experiments. (b) Mutations in the acidic tip affect the structure of the tail of SSB. Circular dichroism of peptides corresponding to the C‐terminal 30 peptides of SSB was done.14 Only the last 13 (Wt and SSB113) or 5 residues (SSBΔC8) are shown in the graph. Arrow, peak corresponding to α‐helical content. (c) Schematic showing RecG binding to SSB in the absence of ssDNA. Here binding to exposed linkers occurs. When ssDNA is added, additional RecG can bind, possibly because a linker previously and transiently bound to an OB‐fold, becomes exposed (subunit in the top right of the SSB tetramer). This may contribute to the 20% increase in RecG binding observed for WtSSB in panel B. (d) When acidic tips are mutated, regulation of the tail of SSB is lost, enabling linkers to bind to SSB OB‐folds. These linkers cannot bind RecG but when ssDNA is added, it binds to SSB OB‐folds with high affinity, outcompeting the linkers. Once free of the OB‐folds, the linkers bind RecG
However, when stoichiometric ssDNA was bound to each SSB first, the level of RecG binding increased 1,417‐fold for SSBΔC8; 400‐ to 460‐fold for SSB S‐ and A‐178, respectively and 900‐fold for SSB D4A4. The increase in binding is not due to RecG binding to ssDNA as assays were done in 600 mM NaCl, a condition in which RecG‐DNA binding is inhibited.23, 87 For the first three mutants, which have a site size comparable to wild type, stoichiometric ssDNA restored SSB‐RecG binding to wild type levels (Figure 6a).14 In contrast, for the quadruple aspartic acid mutant, binding was restored to only 72% of wild type, as the amount of DNA added was sub‐stoichiometric. This follows because SSB D4A4 has a four‐fold larger site size, up from 9 ± 0.6 for wild type to 35 ± 0.3 nt/monomer.14 The restoration of RecG binding in the presence of ssDNA for these mutants demonstrates that the acidic tip is not essential for partner binding when this sequence is present in the context of the full‐length SSB protein. Identical results were observed with the recombination mediator RecO. Here, SSBΔC8 bound to immobilized DNA on beads, bound to RecO in the absence and presence of 200mM NaCl, with an efficiency comparable to wild type.28 If the acidic tip is the primary binding site as has long been thought, then these mutants should not bind RecG or RecO, regardless of the presence of DNA. Instead, ssDNA binding to SSB OB‐folds exposed the linkers of these mutant proteins, and the linkers bound to the RecG and separately, RecO OB‐folds using their PXXP motifs. Therefore, as SSBΔC8 binds to RecO and separately RecG in the presence of ssDNA, the acidic tip is not required for partner binding in the context of the SSB protein.14,28
Consequently, the primary role of the acidic tip is as a regulator of protein function as first proposed by the Dixon group.42 In this role, the tip uses long‐range electrostatic effects provided by the four aspartic acid residues to regulate the activity of SSB. One aspect of this regulation is to prevent the linker from stably binding to the OB‐fold of SSB in the absence of ssDNA. This is a critical function as if linker/OB‐fold binding were to take place in vivo, it would likely be a lethal event as SSB would be inactivated and be defective for partner binding. The ability of the acidic tip to prevent the linker from binding the OB‐fold of SSB was shown recently using GFP‐SSB tail fusions.14 Here, wild type and separately, SSBΔC8 C‐terminal tails were fused to GFP, which is a β‐barrel protein and does not contain an OB‐fold.88 Binding to the core region of SSB was only observed for the mutant tail (ΔC8) GFP‐fusion as the wild‐type acidic tip sequence prevented OB‐fold binding.
The second aspect of tip regulation is to influence the overall structure of the C‐terminal tail so that SSB, OB‐fold binding is minimized. Consistent, this region of SSB is very sensitive to even single amino acid mutations as shown by circular dichroism (CD).89 For the wild‐type peptide comprised of the C‐terminal 30 residues of SSB, the CD spectrum exhibits negative peaks at 200 and 230 nm, consistent with the presence of either a partially unfolded poly‐l‐proline type II (PPII) helix or random coil and, α‐helical structure, respectively (Figure 6b).90 The mutation of P176 to S in SSB113 eliminated the α‐helical content and caused a significant reduction in the negative peak at 200 nm. The deletion of the acidic tip eliminated virtually all secondary structures in the peptide.90 Structural changes in CD spectra were also obtained when comparing the WT and SSBΔC8 proteins. Collectively these results show the acidic tip regulates the structure of the SSB tail consistent with Dixon's model.42
These results also provide insight into the partner binding defects observed for SSB tail mutants, either substitutions (SSB P176S; F177C), additions (S/A178), or deletions (SSB ΔC8, 10 or 26). These effects of these mutations are usually, but incorrectly, interpreted as the acidic tip being the primary binding site so that when mutated, partner binding is lost. However, an alternative model which provides a more convincing rationale, supported by the data discussed above, is the following. Mutations in the acidic tip of the full‐length SSB protein result in dramatic changes in the overall structure of the C‐terminal tail resulting in the linker being sequestered by the SSB OB‐fold so that in the absence of ssDNA, partner binding is prevented (Figure 6c,d).14 This is consistent with a loss in the regulation of SSB function.42 When ssDNA is added, it binds to the SSB OB‐folds, displacing the linkers so that they can bind to partner proteins. This is observed for both the DNA helicase RecG and the recombination mediator RecO.
The impact of the elimination of the long‐range electrostatic effects in the acidic tip on the tail and thus control of SSB function was revealed in studies with SSB D4A4.14 In the context of the full‐length SSB protein, the absence of the electrostatic charges in the C‐terminal tail of this mutant results in an increase in pI of this region from 3.9 ± 0.5 to 6.2 ± 0.5 and for the entire protein, 5.41 ± 0.09 to 8.95 ± 0.10.91 The effect of the loss of 67% of the electrostatic charges in the tail region is the binding of the linker to the SSB OB‐fold, thus explaining the four‐fold increase in the intrinsic site size of the protein. Because the linker is sequestered by the OB‐fold, the protein cannot bind RecG in the absence of DNA. In contrast, when ssDNA is added, it binds to the OB‐folds likely with greater affinity than the linkers (Kd,ssDNA = 1.45 × 10−7–4.04 × 10−9 M).92, 93 This displaces the linkers which then bind to the RecG OB‐fold. This makes sense as the binding of ssDNA to SSB is known to induce a conformational change in the protein that exposes the tail regions of each monomer. This change is manifested as an increase in proteolytic cleavage of the tails as well as an increase in the affinity of SSB for partner proteins.20, 38, 94 For wild type SSB, linker/OB‐fold binding does occur albeit transiently and this may explain the 20% increase in RecG binding when ssDNA is added to binding assays (Figure 6a,c).
The loss of the acidic tip reveals insight into the third aspect of the regulation of SSB function by this region of the protein. That is, it controls the affinity of SSB for ssDNA. The absence of the acidic tip results in a protein (SSBΔC8) that binds ssDNA with higher affinity than wild type.57 Consequently, it is more difficult for the RecG DNA helicase to displace SSBΔC8, requiring 25% longer than wild type.94 This aspect of regulation is important for SSB function as while it must bind to ssDNA with high affinity, it must also at the same time, be able to vacate the DNA so that downstream DNA processing can occur to restore the structure of duplex DNA. This regulation of the C‐terminal tail region is provided by the acidic tip using a mechanism that is yet to be revealed. The linker is involved as when it is absent, the SSB core with only an acidic tip (SSB125) binds with even greater affinity to ssDNA so that RecA and separately, RecG have great difficulty in displacing this mutant.94 For RecG, it takes 14‐fold longer to displace SSB125 than wild type (32 ± 7 vs. 2.3 ± 0.6 sec, respectively).
1.6. OB‐folds are present in many SSB interactome members
For the linker/OB‐fold model to serve as the primary mechanism of interactome regulation by SSB, interactome partners should contain an OB‐fold to bind to the PXXP motifs of the linker. For the six partners shown in Figure 1, the OB‐folds are readily visible (highlighted in dark green). For Exonuclease I, it is in the approximate center of the extended SH3 domain; for PriA, RecG, and RecO the OB‐fold is positioned the N‐terminus; for RecJ it is in the center of the structure and for Pol II, it is the N‐terminal domain and is comprised on non‐contiguous residues (2–38 and 122–147, respectively). Analysis of the remaining structures of interactome partners, reveals that as many as 9 additional partners likely contain an OB‐fold (Table 1). Thus 15 of 20 interactome members contain an OB‐fold. Furthermore, for Exonuclease I, PriA, RecG, and RecJ, the OB‐folds are involved in DNA binding (Figure 1).71, 84, 95, 96 This suggests that linkers and ssDNA may compete for binding to OB‐folds.
For SSB, the competition for OB‐fold binding between ssDNA and the linker was first predicted using modeling.15, 72 Here structural alignments of SH3 domains bound to peptide ligands with an OB‐fold of SSB show that the binding sites of a PXXP ligand and ssDNA overlap (Figure 1, SSB structure in the center; ssDNA in yellow and ligand in green; SH3 domain in purple). The competition between the linker and ssDNA binding to the OB‐folds of SSB was demonstrated experimentally using the tip mutant, SSB D4A4.14 For this mutant, long‐range electrostatic interactions are absent which then allows the SSB OB‐folds to sequester the linker. The effect of linker sequestration by the OB‐fold is observed as a four‐fold increase in site‐size. The consequence of this effect is a loss of RecG binding in the absence of ssDNA (Figure 6a,d). In contrast, when ssDNA is present to compete with linkers for OB‐fold binding, the DNA binds with higher affinity to the OB‐fold, resulting in linker displacement followed by binding to the OB‐fold in RecG.
Linker‐ssDNA competition was also shown experimentally for RecG during helicase loading onto forks by SSB, and later for PriA. Here the binding of the linker to the helicase OB‐fold results in remodeling of each enzyme.33, 34 For RecG, the SSB‐remodeled enzyme binds only to the duplex DNA of the parental arm of the fork. The only way for RecG to bind to the DNA is by using its helicase domains.71, 97 This follows because the wedge domain, which is required for fork binding, is bound to the linker instead during remodeling. For PriA, linker/OB‐fold binding enables the helicase to bind dsDNA directly. This is a significant change induced in the helicase as in the absence of SSB, PriA does not bind duplex DNA.34, 98, 99
It is conceivable that when the PXXP motifs mediate linker binding to an OB‐fold in the interactome partner, this could position the acidic tip of that linker into its binding pocket positioned nearby. However, analysis of the available crystal structures reveals that tip binding pockets are positioned 22–60 Å away from the RT‐loop of each partner OB‐fold (Figure 5).68 This indicates that for the linker bound to the OB‐fold, its acidic tip is not involved in partner binding. However, as the tip binds partner proteins and, as the binding pockets are distal to the OB‐fold where ssDNA binding takes occurs, it is conceivable that the tip from a second monomer within the tetramer also functions as a secondary binding site. Here the initial binding requires interactions between the linker of one subunit of the tetramer and the OB‐fold of the partner. The tip from the tail of a second SSB monomer binds to its pocket position distal to the OB‐fold, possibly stabilizing the complex. This would result in a stoichiometry of 2 SSB monomers per partner as shown for ExoI, Pol II, and RecG.23, 76
1.7. Linker OB‐fold interactions explain cooperative ssDNA binding by SSB
SSB was originally identified as a DNA unwinding protein because of its ability to passively unwind or destabilize the DNA double helix.100 When the gene was discovered, the term SSB was proposed to indicate that the protein could bind both ssDNA and RNA.101, 102, 103 Once it was known that SSB could bind ssDNA, studies were targeted at understanding the mechanism of binding.75 Very early on it was shown that binding to ssDNA was cooperative and that binding resulted in shortening of the DNA length.104 Solution binding studies showed that SSB could wrap ssDNA around itself and this was later visualized by electron microscopy thereby explaining the decrease in ssDNA length associated with protein binding.105, 106
Early studies to identify the DNA binding domain of SSB used a combination of mutagenesis, protein‐DNA crosslinking, and proteolysis to show that the N‐terminal 115 residues contained the DNA binding site (reviewed in Reference 4). Several years later the analysis of the crystal structure of a tetramer comprised of N‐terminal core monomers revealed the presence of four OB‐folds (one per subunit in the tetramer). These OB‐folds were intimately associated with the ssDNA which wrapped around the tetramer using an extensive network of electrostatic and base‐stacking interactions with the phosphodiester backbone and nucleotide bases, respectively (Figure 1; SSB center).9, 53 This structure explained how the binding of SSB to ssDNA resulted in shortening of the overall length of the DNA.104, 105, 106
However, SSB tetramers bind to ssDNA cooperatively.12, 104, 106 In electron microscopy images, SSB tetramers were observed to be unevenly distributed on the ssDNA, with sections of the nucleic acid‐containing clusters of protein, bound in juxtaposition and others lacking SSB altogether. This facet of SSB protein behavior can be expressed by ϖ, the cooperativity parameter, which ranges from 50 to 105.39, 104, 107
Implicit in cooperative binding, is the interaction between tetramers. The mechanism for these interactions initially remained elusive, although several models were proposed. The first model invoked a tetramer‐tetramer interface while the second proposed that the linkers wrap around each other to enable the acidic tips to join tetramers together by binding to SSB core regions.9, 56 The tetramer‐tetramer interface model has not been confirmed, there are no data available to support the linker‐wrapping model and, a biologically relevant binding site for the acidic tip on SSB has not been identified.
However, at a non‐physiological pH of 3.4, where SSB dissociates into monomers, the acidic tip can make intra‐molecular contacts with Valines 29 and 58 of the OB‐fold.108 The biological relevance of these interactions is unclear as SSB was monomeric and thus inactive and, the optimized structural model has the C‐terminus of the tip pointing away from the OB‐fold in an orientation opposite to that observed in tip‐partner crystal structures. Furthermore, in a subsequent study done at pH 7.6 where SSB is a tetramer, it was found that the affinity of tip peptides for SSB ranged between 13 ± 3 and 19 ± 5 mM.42 As the binding is so weak, the majority of acidic tips are unbound even in the absence of ssDNA, leading the authors to propose that the tip does not bind the SSB OB‐fold but instead, uses long‐range electrostatic effects to regulate SSB activity. Thus, the acidic tip is likely not involved in mediating SSB‐SSB interactions required for cooperative ssDNA binding.
The Lohman group found the first evidence for the role of the linker in cooperative ssDNA by showing that linker deletions impaired cooperativity.56 They also demonstrated that SSB‐SSB interactions can be mediated by linker regions.109 The Bianco group extended these studies and revealed that the linker/OB‐fold model provides a convincing explanation for the mechanism of cooperative ssDNA binding by SSB that has subsequently been validated experimentally.14, 15 In this model, tetramer‐tetramer interactions are provided by PXXP motifs in the linker(s) from one SSB tetramer binding to the OB‐fold of an adjacent tetramer (Figure 7a; SSB35 mode).29 This is repeated multiple times, creating an extensive network of linker/OB‐fold interactions.14, 94 Not surprisingly, insertion of an asparagine residue in the center of PXXP motif I, which increases the spacing between the prolines, eliminates cooperative binding to ssDNA.14 These effects were observed in 10 mM NaCl where the SSB35 binding mode predominates.110, 111 The binding of linkers to OB‐folds would position adjacent tetramers closer together so that bridge‐interface residues, Y22 and K73, can be brought into proximity.112 This could further enhance cooperative binding mediated by the initial linker/OB‐fold interactions.
FIGURE 7.
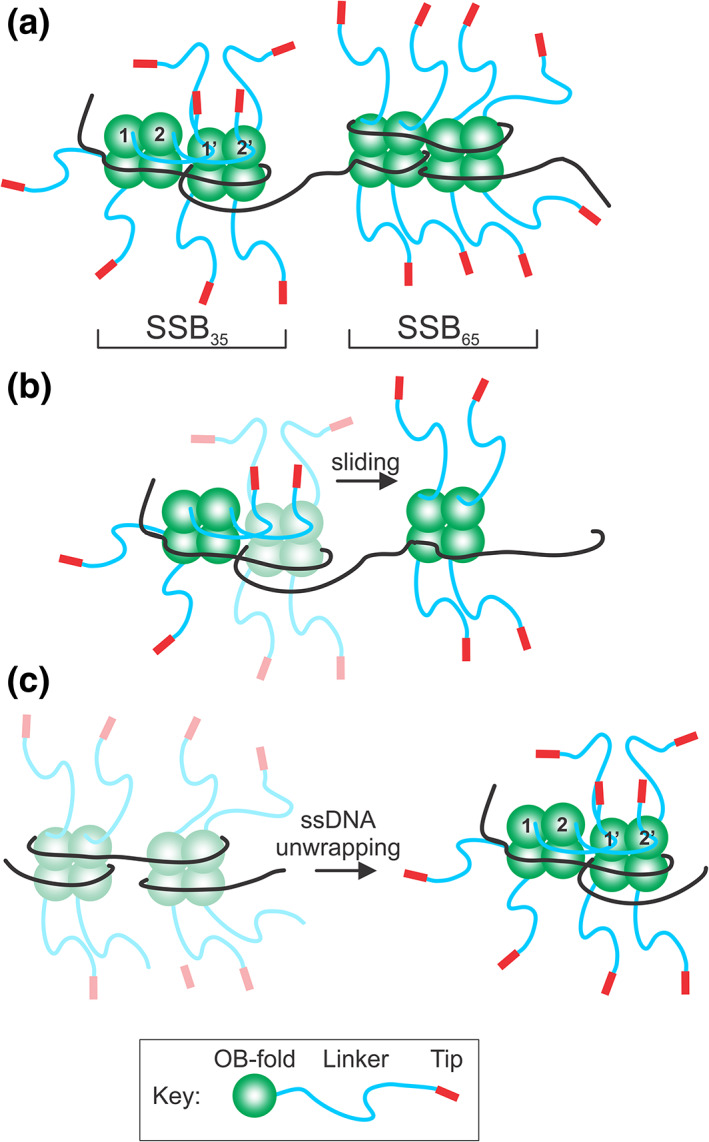
The linker/OB‐fold model can explain the dynamic behavior of SSB. (a) Single‐stranded DNA binding protein (SSB) can interconvert between ssDNA binding modes, defined by the number of nucleotides of ssDNA occluded by a tetramer. Here only two modes are shown: SSB35, where cooperative binding occurs involving the linkers of SSB 1 and 2, binding to the OB‐folds of SSB 1′ and 2′, respectively. For the SSB65 mode, cooperative ssDNA binding does not occur, and linkers do not bind nearby OB‐folds. (b) SSB protomers can slide on ssDNA. In the initial complex shown in SSB35 mode, linkers of the left SSB bind to the OB‐folds of the central SSB tetramer. For sliding to occur, these linker/OB‐fold interactions must disengage so that this tetramer can slide to the position on the right. The sliding tetramer is colored as a faded complex to show the transition from the initial to the final position (bright colors). (c) SSB complexes undergo unwrapping and wrapping transitions during complex assembly. Tetramers are colored as faded complexes (wrapped) to show the transition from wrapped to unwrapped complexes (SSB35 mode; bright colors). In the ssDNA‐unwrapped state, linkers can bind to exposed OB‐folds as protein density on the ssDNA increases. This figure was adapted from Reference 142
In addition to stimulating ssDNA binding, linker/OB‐fold interactions are also required for efficient displacement of SSB from DNA. Deletion of a region of the linker comprising PXXP motifs II and III increases the lag time for displacement by RecG 1.4‐fold, whereas deletion of a region comprising all three motifs increases the lag time 14‐fold.94 Similar results were observed for displacement by RecA during in vitro DNA strand exchange.94 These results point to a role for the linker in SSB dissociation but the PXXP motifs are yet to be directly implicated in this process.
1.8. A role for linker/OB‐fold interactions in the dynamic behavior of SSB
SSB exists as a tetramer and binds tightly and cooperatively to ssDNA (Kd = 1.45x10−7 to 4.04 x 10−9 M), requiring as high as 2 M salt for dissociation.38, 47, 92, 93, 100, 113 Depending on solution conditions such as varying [NaCl], temperature, pH, and applied force, each tetramer can occlude 8–65 nt of ssDNA, and the protein can also form octamers via long‐range intramolecular interactions between non‐nearest neighbor SSB tetramers.11, 35, 39, 106, 114, 115, 116 Importantly, SSB does not form a static complex on DNA.
Instead, single‐molecule studies have shown that SSB‐ssDNA complexes are dynamic with binding modes interconverting in a salt‐dependent manner; that the protein can slide on DNA and is undergoes intersegment transfer (Figure 7b).117, 118, 119, 120 Recently, it was found that ssDNA‐binding by SSB is biphasic, with the initial ssDNA wrapping events being followed by unwrapping events as protein density on the DNA increases (Figure 7c). The unwrapping energy cost increases as more ssDNA is successively unraveled.11, 116 This dynamic behavior is intrinsic to SSB function as the protein must bind cooperatively to ssDNA to rapidly protect it.121 Equally important, SSB must provide access to other proteins, either by disengaging from the DNA or, by rearranging on the nucleic acid lattice first before finally disengaging.
The mechanism of the dynamic behavior of SSB can be explained by the linker/OB‐fold model. For example, in the SSB8 to SSB35 modes, the commonly held view is that not all OB‐folds are bound to ssDNA (Figure 7a). Cooperative ssDNA binding is observed when multiple linker/OB‐fold interactions form in the SSB35 mode.111 Consistent, mutations in PXXP motif I eliminate cooperative ssDNA binding.14 As the [NaCl] increases, SSB converts from the SSB35 to the SSB65 mode, where it is thought that all OB‐folds are bound to ssDNA and cooperative ssDNA binding is reduced.39, 111, 122 Under these conditions, linker/OB‐fold interactions may either not occur or are reduced in number. When potassium glutamate replaced NaCl, cooperativity is still observed at elevated salt where the SSB65 mode formed and required functional linkers.109 To explain their data, Kozlov et al. proposed a model whereby linker‐linker interactions position acidic tips to bind OB‐folds. However, there is no evidence of linker‐linker interactions and PXXP motifs are not known to bind to each other. Instead, the preferred target for a PXXP motif is either an OB‐fold or an SH3 domain.14, 59 Thus, under these conditions, it is more likely that linker/OB‐fold interactions mediate cooperative binding.
Linker/OB‐fold binding likely facilitates the recruitment of additional tetramers to ssDNA and once bound, the juxtaposed SSB molecules could form a compact complex on the ssDNA. For SSB to slide away from this complex, linkers would have to disengage from the OB‐folds (Figure 7b). In similar fashion, the sliding of a tetramer towards this complex could be facilitated by linkers reaching out and contacting the lone SSB, resulting in reeling in of this tetramer into the complex.
The wrapping/unwrapping events that occur as the protein density on ssDNA increases may also be coupled to linker/OB‐fold interactions (Figure 7c).11, 116 At low density, the chance of finding a partner is low and ssDNA wrapping may occur. Once protein density increases, DNA unwrapping coupled to linker/OB‐fold binding may then take place. Finally, in addition to mediating interactions between juxtaposed tetramers, linker/OB‐fold binding may also be responsible for mediating long‐range intramolecular interactions between non‐nearest neighbor SSB tetramers that result in compacted SSB‐ssDNA structures.35
1.9. The SSB OB‐folds play additional roles in interactome regulation
In exponentially growing cells, there are 2,058 SSB tetramers per cell.121 There are also on average 2–4 DNA replication forks per cell, with anywhere from 500 to 1,000 nucleotides of ssDNA exposed at each fork.123 Using a site size of 40 nucleotides occluded per tetramer, there would be up to 25 SSB proteins bound per fork or 100 per cell, leaving as many as 1,958 free tetramers. If these free tetramers are not sequestered away from the genome, they could cause excessive strand separation and/or spurious melting of duplex DNA that otherwise might be lethal to the cell as suggested previously.124, 125, 126
Consequently, in the absence of exogenous DNA damage, the storage site of a large fraction of this free SSB is at the inner membrane where the protein binds to the three major, inner membrane phospholipids: phosphatidylethanolamine, phosphatidylglycerol, and cardiolipin with Kd values below 1 μM.16 As SH3 domains also bind phospholipids it was proposed that membrane binding by SSB is mediated by the OB‐folds.16, 127, 128 This makes sense as within 5 min of the genome being damaged, virtually all SSB transfers from the inner membrane to the genome to facilitate DNA repair processes. The transfer is likely driven by the 117 to 260‐fold higher affinity of SSB for exposed ssDNA relative to phospholipids.92, 93, 129 Finally, once the genome is repaired, SSB localizes to the inner membrane. As SSB binds to interactome partners, these also colocalize to the membrane as shown for the repair DNA helicases RecG and PriA, which are present at 7–15 and 2–4 molecules per cell, respectively.24, 121 The SSB‐RecG complex can transfer directly to forks, whereas it is more likely that PriA is transferred from the complex with SSB to tetramers bound to forks, but this remains an open question.24, 130
In addition to being sequestered at the inner membrane in the absence of DNA damage, some fraction of SSB undergoes liquid–liquid phase‐separation (LLPS) into condensates containing protein, ssDNA, and some interactome partners.131 In vitro, LLPS was not observed when SSBΔC8 or only the core region of SSB was used, and the authors concluded that the acidic tip was essential for condensate formation. However, this interpretation is likely incorrect as it is now known that linker/OB‐fold binding is required for SSB‐SSB interactions.14 Thus, the linker/OB‐fold model also applies to the sequestration of SSB into condensates. It follows then, in these condensates, some OB‐folds will be occupied by linkers while others are bound to sub‐stoichiometric ssDNA. At present, it is unknown whether exogenous DNA damage results in dissociation of condensates resulting in the transfer of SSB to the genome to facilitate DNA repair processes.
1.10. Summary and future perspectives
The presence of at least one OB‐fold in multiple interactome members has led to the proposal that the SSB interactome is the first OB‐fold family of genome guardians identified in prokaryotes.14 This is analogous to the OB‐fold family of genome guardians in eukaryotes.44, 45 In eukaryotic cells, many of the proteins functioning in DNA replication, telomere homeostasis, activation of the DNA‐damage checkpoint, and DNA repair contain OB‐folds.132, 133, 134 Not surprisingly, these OB‐folds play critical roles in DNA binding, protein complex assembly, and are involved in regulating complex protein–protein interactions.45 For the SSB interactome, maintenance of genomic stability relies on the coordinated actions of the OB‐fold family with SSB as the central player. Essential to this coordination is the network of linker/OB‐fold interactions. These interactions ensure that interactome partners are loaded onto the DNA at the right time to guarantee cell viability.
Crucial components of the linker/OB‐fold mechanism are the inherent flexibility imparted by the repetitive elements in the SSB linker and the polyproline type II helix; the specificity imparted by linker PXXP motifs binding to unique OB‐folds in partners, and C‐terminal tail regulation by the acidic tip. Flexibility is needed for cooperative ssDNA binding and dissociation, as well as enabling SSB binding to target proteins of various sizes and, possibly occurring in different structural configurations, that is, bound to ssDNA, in solution or, within multi‐subunit complexes such as the replisome. Further, the sequence context of each PXXP motif enables one SSB protein to bind to multiple partners at different times, and the sequences of the structurally conserved OB‐folds enable the formation of unique protein–protein interfaces that facilitates distinctive functional outcomes. Inherent to the linker/OB‐fold model is the competition between ssDNA and linkers, and separately, phospholipids and ssDNA for the OB‐folds, which adds an additional level of regulation to the SSB interactome.
While the linker/OB‐fold model provides a satisfactory explanation for many facets of SSB and interactome function, there is still much work to be done. First, there is a large apparent discrepancy between results obtained with C‐terminal peptides of different lengths and the full length, SSB protein. This disagreement must be resolved using carefully characterized, full‐length mutant SSB proteins only, as peptides likely behave differently in isolation than within the context of the SSB tetramer. Second, studies have demonstrated that SSB‐partner interactions involve the PXXP motifs of the linker binding to a nearby OB‐fold. What remains to be ascertained is the orientation of binding and whether it is the same for all SSB‐partner interactions. Third, the roles of the PXXP motifs in SSB dynamics remain to be established and if they are involved, do different motifs have distinct or overlapping functions? Fourth, do the linkers and acidic tips coordinate their actions both within one subunit and between subunits of the same tetramer to ensure interactome regulation? Finally, if partners do not contain canonical OB‐folds, what is their mechanism of SSB binding? Is it exclusively acidic tip‐mediated or is the linker involved as well? The answers to these and other questions, likely to be revealed in the answering process, will provide a complete picture of the dynamic SSB interactome.
AUTHOR CONTRIBUTIONS
Piero Bianco: Conceptualization; writing‐review & editing.
ACKNOWLEDGMENTS
Work in the Bianco Laboratory is supported by the National Institutes of Health Grant; Grant number: GM100156.
Bianco PR. The mechanism of action of the SSB interactome reveals it is the first OB‐fold family of genome guardians in prokaryotes. Protein Science. 2021;30:1757–1775. 10.1002/pro.4140
Funding information National Institute of General Medical Sciences, Grant/Award Number: GM10056
REFERENCES
- 1.Shereda RD, Kozlov AG, Lohman TM, Cox MM, Keck JL. SSB as an organizer/mobilizer of genome maintenance complexes. Crit Rev Biochem Mol Biol. 2008;43:289–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lecointe F, Serena C, Velten M, et al. Anticipating chromosomal replication fork arrest: SSB targets repair DNA helicases to active forks. EMBO J. 2007;26:4239–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chase JW, Williams KR. Single‐stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. [DOI] [PubMed] [Google Scholar]
- 4.Meyer RR, Laine PS. The single‐stranded DNA‐binding protein of Escherichia coli . Microbiol Rev. 1990;54:342–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli . Microbiol Rev. 1994;58:401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carlini LE, Porter RD. Analysis of SSB mutations in vivo implicates SSB protein in two distinct pathways of SOS induction and in recombinational DNA repair. Mol Microbiol. 1997;24:129–139. [DOI] [PubMed] [Google Scholar]
- 7.Porter RD, Black S, Pannuri S, Carlson A. Use of the escherichia coli ssb gene to prevent bioreactor takeover by plasmidless cells. Nature Biotechnology, 1990;8(1):47–51. 10.1038/nbt0190-47. [DOI] [PubMed] [Google Scholar]
- 8.Curth U, Genschel J, Urbanke C, Greipel J.In vitro and in vivo function of the C‐terminus of escherichia coli single‐stranded DNA binding protein. Nucleic Acids Research, 1996;24:2706–2711. 10.1093/nar/24.14.2706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raghunathan S, Kozlov A, Lohman T, Waksman G. Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nat Struct Biol. 2000;7:648–652. [DOI] [PubMed] [Google Scholar]
- 10.Kuznetsov S, Kozlov A, Lohman T, Ansari A. Microsecond dynamics of protein‐DNA interactions: Direct observation of the wrapping/unwrapping kinetics of single‐stranded DNA around the E. coli SSB tetramer. J Mol Biol. 2006;359:55–65. [DOI] [PubMed] [Google Scholar]
- 11.Naufer MN, Morse M, Möller GB, et al. Multiprotein E. coli SSB–ssDNA complex shows both stable binding and rapid dissociation due to interprotein interactions. Nucleic Acids Research, 2021;49:1532–1549. 10.1093/nar/gkaa1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lohman TM, Bujalowski W. Escherichia coli single strand binding protein: Multiple single‐stranded DNA binding modes and cooperativities. In: Revzin A, editor. The biology of nonspecific DNA‐protein interactions. Boca Raton: CRC Press, Inc., 1990; p. 131–170. [Google Scholar]
- 13.Lohman TM, Overman LB. Two binding modes in Escherichia coli single strand binding protein‐single stranded DNA complexes. Modulation by NaCl concentration. J Biol Chem. 1985;260:3594–3603. [PubMed] [Google Scholar]
- 14.Ding W, Tan HY, Zhang JX, et al. The mechanism of Single strand binding protein–RecG binding: Implications for SSB interactome function. Protein Science, 2020;29:1211–1227. 10.1002/pro.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bianco PR. The tale of SSB. Prog Biophys Mol Biol. 2017;127:111–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao T, Liu Y, Wang Z, et al. Super‐resolution imaging reveals changes inEscherichia coliSSB localization in response to DNA damage. Genes to Cells, 2019;24:814–826. 10.1111/gtc.12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shinn MK, Kozlov AG, Nguyen B, Bujalowski WM, Lohman TM. Are the intrinsically disordered linkers involved in SSB binding to accessory proteins?. Nucleic Acids Research, 2019. 10.1093/nar/gkz606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu D, Windsor MA, Gellman SH, Keck JL. Peptide inhibitors identify roles for SSB C‐terminal residues in SSB/exonuclease I complex formation. Biochemistry, 2009;48;6764–6771. 10.1021/bi900361r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shereda RD, Reiter NJ, Butcher SE, Keck JL. Identification of the SSB binding site on E. coli RecQ reveals a conserved surface for binding SSB's C terminus. J Mol Biol. 2009;386:612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozlov AG, Jezewska MJ, Bujalowski W, Lohman TM. Binding specificity of escherichia coli single‐stranded DNA Binding protein for the χ subunit of DNA pol III holoenzyme and PriA helicase. Biochemistry, 2010;49:3555–3566. 10.1021/bi100069s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelman Z, Yuzhakov A, Andjelkovic J, O'Donnell M. Devoted to the lagging strand‐the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J. 1998;17:2436–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Witte G, Urbanke C, Curth U. DNA polymerase III chi subunit ties single‐stranded DNA binding protein to the bacterial replication machinery. Nucleic Acids Res. 2003;31:4434–4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buss J, Kimura Y, Bianco P. RecG interacts directly with SSB: Implications for stalled replication fork regression. Nucleic Acids Res. 2008;36:7029–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu C, Tan HY, Choi M, et al. SSB binds to the RecG and PriA helicases in vivo in the absence of DNA. Genes Cells. 2016;21:163–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhattacharyya B, George NP, Thurmes TM, et al. Structural mechanisms of PriA‐mediated DNA replication restart. Proc Natl Acad Sci U S A. 2014;111:1373–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu D, Keck JL. Structural basis of Escherichia coli single‐stranded DNA‐binding protein stimulation of exonuclease I. Proc Natl Acad Sci U S A. 2008;105:9169–9174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ryzhikov M, Koroleva O, Postnov D, Tran A, Korolev S. Mechanism of RecO recruitment to DNA by single‐stranded DNA binding protein. Nucleic Acids Res. 2011;39:6305–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chilingaryan Z, Headey SJ, Lo ATY, et al. Fragment‐based discovery of inhibitors of the bacterial DnaG‐SSB interaction. Antibiotics. 2018;7:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bianco PR, Pottinger S, Tan HY, Nguyenduc T, Rex K, Varshney U. The IDL of E. coli SSB links ssDNA and protein binding by mediating protein‐protein interactions. Protein Sci. 2017;26:227–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu H, Chen JK, Feng S, Dalgarno DC, Brauer AW, Schreiber SL. Structural basis for the binding of proline‐rich peptides to SH3 domains. Cell. 1994;76:933–945. [DOI] [PubMed] [Google Scholar]
- 31.Saksela K, Permi P. SH3 domain ligand binding: What's the consensus and where's the specificity? FEBS Lett. 2012;586:2609–2614. [DOI] [PubMed] [Google Scholar]
- 32.Agrawal V, Kishan RK. Functional evolution of two subtly different (similar) folds. BMC Struct Biol. 2001;1:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun Z, Tan HY, Bianco PR, Lyubchenko YL. Remodeling of RecG helicase at the DNA replication fork by SSB protein. Sci Rep. 2015;5:9625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, Sun Z, Bianco PR, Lyubchenko YL. Atomic force microscopy‐based characterization of the interaction of PriA helicase with stalled DNA replication forks. J Biol Chem. 2020;295:6043–6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bell JC, Liu B, Kowalczykowski SC. Imaging and energetics of single SSB‐ssDNA molecules reveal intramolecular condensation and insight into RecOR function. Elife. 2015;4:e08646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang J, Kim JY, Kim C, et al. Single‐molecule observation of ATP‐independent SSB displacement by RecO in Deinococcus radiodurans. Elife. 2020;9:e50945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nigam R, Mohan M, Shivange G, Dewangan PK, Anindya R. Escherichia coli AlkB interacts with single‐stranded DNA binding protein SSB by an intrinsically disordered region of SSB. Mol Biol Rep. 2018;45:865–870. [DOI] [PubMed] [Google Scholar]
- 38.Williams KR, Spicer EK, LoPresti MB, Guggenheimer RA, Chase JW. Limited proteolysis studies on the Escherichia coli single‐stranded DNA binding protein. Evidence for a functionally homologous domain in both the Escherichia coli and T4 DNA binding proteins. J Biol Chem. 1983;258:3346–3355. [PubMed] [Google Scholar]
- 39.Lohman T, Ferrari M. Escherichia coli single‐stranded DNA‐binding protein: Multiple DNA‐binding modes and cooperativities. Annu Rev Biochem. 1994;63:527–570. [DOI] [PubMed] [Google Scholar]
- 40.Green M, Hatter L, Brookes E, Soultanas P, Scott DJ. Defining the intrinsically disordered C‐terminal domain of SSB reveals DNA‐mediated compaction. J Mol Biol. 2016;428:357–364. [DOI] [PubMed] [Google Scholar]
- 41.Manosas M, Perumal SK, Bianco PR, Ritort F, Benkovic SJ, Croquette V. RecG and UvsW catalyse robust DNA rewinding critical for stalled DNA replication fork rescue. Nat Commun. 2013;4:2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su XC, Wang Y, Yagi H, et al. Bound or free: Interaction of the C‐terminal domain of Escherichia coli single‐stranded DNA‐binding protein (SSB) with the tetrameric core of SSB. Biochemistry. 2014;53:1925–1934. [DOI] [PubMed] [Google Scholar]
- 43.Maffei M, Arbesu M, Le Roux AL, Amata I, Roche S, Pons M. The SH3 domain acts as a scaffold for the N‐terminal intrinsically disordered regions of c‐Src. Structure. 2015;23:893–902. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen DD, Kim EY, Sang PB, Chai W. Roles of OB‐fold proteins in replication stress. Front Cell Dev Biol. 2020;8:574466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Flynn RL, Zou L. Oligonucleotide/oligosaccharide‐binding fold proteins: A growing family of genome guardians. Crit Rev Biochem Mol Biol. 2010;45:266–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bianco PR. DNA helicase‐SSB interactions critical to the regression and restart of stalled DNA replication forks in Escherichia coli . Genes. 2020;11:471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molineux IJ, Friedman S, Gefter ML. Purification and properties of the Escherichia coli deoxyribonucleic acid‐unwinding protein. Effects on deoxyribonucleic acid synthesis in vitro. J Biol Chem. 1974;249:6090–6098. [PubMed] [Google Scholar]
- 48.Weiner JH, Bertsch LL, Kornberg A. The deoxyribonucleic acid unwinding protein of Escherichia coli. Properties and function in replication. J Biol Chem. 1975;250:1972–1980. [PubMed] [Google Scholar]
- 49.Sancar A, Williams KR, Chase JW, Rupp WD. Sequences of the ssb gene and protein. Proc Natl Acad Sci U S A. 1981;78:4274–4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Casas‐Finet JR, Khamis MI, Maki AH, Chase JW. Tryptophan 54 and phenylalanine 60 are involved synergistically in the binding of E. coli SSB protein to single‐stranded polynucleotides. FEBS Lett. 1987;220:347–352. [DOI] [PubMed] [Google Scholar]
- 51.Merrill BM, Williams KR, Chase JW, Konigsberg WH. Photochemical cross‐linking of the Escherichia coli single‐stranded DNA‐binding protein to oligodeoxynucleotides. Identification of phenylalanine 60 as the site of cross‐linking. J Biol Chem. 1984;259:10850–10856. [PubMed] [Google Scholar]
- 52.Carlini L, Curth U, Kindler B, Urbanke C, Porter RD. Identification of amino acids stabilizing the tetramerization of the single stranded DNA binding protein from Escherichia coli . FEBS Lett. 1998;430:197–200. [DOI] [PubMed] [Google Scholar]
- 53.Raghunathan S, Ricard C, Lohman T, Waksman G. Crystal structure of the homo‐tetrameric DNA binding domain of Escherichia coli single‐stranded DNA‐binding protein determined by multiwavelength x‐ray diffraction on the selenomethionyl protein at 2.9‐A resolution. Proc Natl Acad Sci USA. 1997;94:6652–6657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Savvides SN, Raghunathan S, Futterer K, Kozlov AG, Lohman TM, Waksman G. The C‐terminal domain of full‐length E. coli SSB is disordered even when bound to DNA. Protein Sci. 2004;13:1942–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilkins MR, Gasteiger E, Bairoch A, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–552. [DOI] [PubMed] [Google Scholar]
- 56.Kozlov AG, Weiland E, Mittal A, et al. Intrinsically disordered C‐terminal tails of E. coli single‐stranded DNA binding protein regulate cooperative binding to single‐stranded DNA. J Mol Biol. 2015;427:763–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kozlov AG, Cox MM, Lohman TM. Regulation of single‐stranded DNA binding by the C termini of Escherichia coli single‐stranded DNA‐binding (SSB) protein. J Biol Chem. 2010;285:17246–17252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Antony E, Weiland E, Yuan Q, et al. Multiple C‐terminal tails within a single E. coli SSB homotetramer coordinate DNA replication and repair. J Mol Biol. 2013;425:4802–4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kaneko T, Li L, Li SS. The SH3 domain—a family of versatile peptide‐ and protein‐recognition module. Front Biosci. 2008;13:4938–4952. [DOI] [PubMed] [Google Scholar]
- 60.Kurochkina N, Guha U. SH3 domains: Modules of protein‐protein interactions. Biophys Rev. 2013;5:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Meirson T, Bomze D, Kahlon L, Gil‐Henn H, Samson AO. A helical lock and key model of polyproline II conformation with SH3. Bioinformatics. 2020;36:154–159. [DOI] [PubMed] [Google Scholar]
- 62.Adzhubei AA, Sternberg MJ, Makarov AA. Polyproline‐II helix in proteins: Structure and function. J Mol Biol. 2013;425:2100–2132. [DOI] [PubMed] [Google Scholar]
- 63.Matsushima N, Yoshida H, Kumaki Y, et al. Flexible structures and ligand interactions of tandem repeats consisting of proline, glycine, asparagine, serine, and/or threonine rich oligopeptides in proteins. Curr Protein Pept Sci. 2008;9:591–610. [DOI] [PubMed] [Google Scholar]
- 64.Fedorov R, Bohl M, Tsiavaliaris G, et al. The mechanism of pentabromopseudilin inhibition of myosin motor activity. Nat Struct Mol Biol. 2009;16:80–88. [DOI] [PubMed] [Google Scholar]
- 65.Huang Y, Huang C. The glycine‐rich flexible region in SSB is crucial for PriA stimulation. RSC Adv. 2018;8:35280–35288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Handa P, Acharya N, Varshney U. Chimeras between single‐stranded DNA‐binding proteins from Escherichia coli and Mycobacterium tuberculosis reveal that their C‐terminal domains interact with uracil DNA glycosylases. J Biol Chem. 2001;276:16992–16997. [DOI] [PubMed] [Google Scholar]
- 67.Musacchio A, Noble M, Pauptit R, Wierenga R, Saraste M. Crystal structure of a Src‐homology 3 (SH3) domain. Nature. 1992;359:851–855. [DOI] [PubMed] [Google Scholar]
- 68.Yu H, Rosen MK, Shin TB, Seidel‐Dugan C, Brugge JS, Schreiber SL. Solution structure of the SH3 domain of Src and identification of its ligand‐binding site. Science. 1992;258:1665–1668. [DOI] [PubMed] [Google Scholar]
- 69.Feng S, Chen JK, Yu H, Simon JA, Schreiber SL. Two binding orientations for peptides to the Src SH3 domain: Development of a general model for SH3‐ligand interactions. Science. 1994;266:1241–1247. [DOI] [PubMed] [Google Scholar]
- 70.Dimasi N. Crystal structure of the C‐terminal SH3 domain of the adaptor protein GADS in complex with SLP‐76 motif peptide reveals a unique SH3‐SH3 interaction. Int J Biochem Cell Biol. 2007;39:109–123. [DOI] [PubMed] [Google Scholar]
- 71.Singleton MR, Scaife S, Wigley DB. Structural analysis of DNA replication fork reversal by RecG. Cell. 2001;107:79–89. [DOI] [PubMed] [Google Scholar]
- 72.Bianco PR, Lyubchenko YL. SSB and the RecG DNA helicase: An intimate association to rescue a stalled replication fork. Protein Sci. 2017;26:638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Briggs GS, Mahdi AA, Wen Q, Lloyd RG. DNA binding by the substrate specificity (wedge) domain of RecG helicase suggests a role in processivity. J Biol Chem. 2005;280:13921–13927. [DOI] [PubMed] [Google Scholar]
- 74.Teyra J, Huang H, Jain S, et al. Comprehensive analysis of the human SH3 domain family reveals a wide variety of non‐canonical specificities. Structure. 2017;25:1598–1610. [DOI] [PubMed] [Google Scholar]
- 75.Molineux IJ, Gefter ML. Properties of the Escherichia coli in DNA binding (unwinding) protein: Interaction with DNA polymerase and DNA. Proc Natl Acad Sci U S A. 1974;71:3858–3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Molineux IJ, Gefter ML. Properties of the Escherichia coli DNA‐binding (unwinding) protein interaction with nucleolytic enzymes and DNA. J Mol Biol. 1975;98:811–825. [DOI] [PubMed] [Google Scholar]
- 77.Low RL, Shlomai J, Kornberg A. Protein n, a primosomal DNA replication protein of Escherichia coli. Purification and characterization. J Biol Chem. 1982;257:6242–6250. [PubMed] [Google Scholar]
- 78.Perrino FW, Meyer RR, Bobst AM, Rein DC. Interaction of a folded chromosome‐associated protein with single‐stranded DNA‐binding protein of Escherichia coli, identified by affinity chromatography. J Biol Chem. 1988;263:11833–11839. [PubMed] [Google Scholar]
- 79.Glover B, McHenry C. The chi psi subunits of DNA polymerase III holoenzyme bind to single‐stranded DNA‐binding protein (SSB) and facilitate replication of an SSB‐coated template. J Biol Chem. 1998;273:23476–23484. [DOI] [PubMed] [Google Scholar]
- 80.Sandigursky M, Mendez F, Bases RE, Matsumoto T, Franklin WA. Protein‐protein interactions between the Escherichia coli single‐stranded DNA‐binding protein and exonuclease I. Radiat Res. 1996;145:619–623. [PubMed] [Google Scholar]
- 81.Genschel J, Curth U, Urbanke C. Interaction of E. coli single‐stranded DNA binding protein (SSB) with exonuclease I. the carboxy‐terminus of SSB is the recognition site for the nuclease. Biol Chem. 2000;381:183–192. [DOI] [PubMed] [Google Scholar]
- 82.Shereda RD, Bernstein DA, Keck JL. A central role for SSB in Escherichia coli RecQ DNA helicase function. J Biol Chem. 2007;282:19247–19258. [DOI] [PubMed] [Google Scholar]
- 83.Cadman CJ, McGlynn P. PriA helicase and SSB interact physically and functionally. Nucleic Acids Res. 2004;32:6378–6387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Cheng K, Xu H, Chen X, et al. Structural basis for DNA 5 ‐end resection by RecJ. Elife. 2016;5:e14294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Petzold C, Marceau AH, Miller KH, Marqusee S, Keck JL. Interaction with single‐stranded DNA‐binding protein stimulates Escherichia coli ribonuclease HI enzymatic activity. J Biol Chem. 2015;290:14626–14636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Han ES, Cooper DL, Persky NS, et al. RecJ exonuclease: Substrates, products and interaction with SSB. Nucleic Acids Res. 2006;34:1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Slocum SL, Buss JA, Kimura Y, Bianco PR. Characterization of the ATPase activity of the Escherichia coli RecG protein reveals that the preferred cofactor is negatively supercoiled DNA. J Mol Biol. 2007;367:647–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang F, Moss LG, Phillips GN Jr. The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. [DOI] [PubMed] [Google Scholar]
- 89.Greenfield NJ. Using circular dichroism spectra to estimate protein secondary structure. Nat Protoc. 2006;1:2876–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Greenfield NJ. Circular dichroism (CD) analyses of protein‐protein interactions. Methods Mol Biol. 2015;1278:239–265. [DOI] [PubMed] [Google Scholar]
- 91.Kozlowski LP. IPC 2.0: Prediction of isoelectric point and pKa dissociation constants. Nucleic Acids Res. 2021;gkab29. 10.1093/nar/gkab295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ehn M, Nilsson P, Uhlen M, Hober S. Overexpression, rapid isolation, and biochemical characterization of Escherichia coli single‐stranded DNA‐binding protein. Protein Exp Purif. 2001;22:120–127. [DOI] [PubMed] [Google Scholar]
- 93.Wu HY, Lu CH, Li HW. RecA‐SSB interaction modulates RecA nucleoprotein filament formation on SSB‐wrapped DNA. Sci Rep. 2017;7:11876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tan HY, Wilczek LA, Pottinger S, et al. The intrinsically disordered linker of E. coli SSB is critical for the release from single‐stranded DNA. Protein Sci. 2017;26:700–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Korada SK, Johns TD, Smith CE, Jones ND, McCabe KA, Bell CE. Crystal structures of Escherichia coli exonuclease I in complex with single‐stranded DNA provide insights into the mechanism of processive digestion. Nucleic Acids Res. 2013;41:5887–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Windgassen TA, Leroux M, Satyshur KA, Sandler SJ, Keck JL. Structure‐specific DNA replication‐fork recognition directs helicase and replication restart activities of the PriA helicase. Proc Natl Acad Sci U S A. 2018;115:E9075–E9084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mahdi AA, McGlynn P, Levett SD, Lloyd RG. DNA binding and helicase domains of the Escherichia coli recombination protein RecG. Nucleic Acids Res. 1997;25:3875–3880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tanaka T, Taniyama C, Arai K, Masai H. ATPase/helicase motif mutants of Escherichia coli PriA protein essential for recombination‐dependent DNA replication. Genes Cells. 2003;8:251–261. [DOI] [PubMed] [Google Scholar]
- 99.Nurse P, Liu J, Marians KJ. Two modes of PriA binding to DNA. J Biol Chem. 1999;274:25026–25032. [DOI] [PubMed] [Google Scholar]
- 100.Sigal N, Delius H, Kornberg T, Gefter ML, Alberts B. A DNA‐unwinding protein isolated from Escherichia coli: Its interaction with DNA and with DNA polymerases. Proc Natl Acad Sci U S A. 1972;69:3537–3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meyer RR, Glassberg J, Kornberg A. An Escherichia coli mutant defective in single‐strand binding protein is defective in DNA replication. Proc Natl Acad Sci U S A. 1979;76:1702–1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Molineux IJ, Pauli A, Gefter ML. Physical studies of the interaction between the Escherichia coli DNA binding protein and nucleic acids. Nucleic Acids Res. 1975;2:1821–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ruyechan WT, Wetmur JG. Studies on the noncooperative binding of the Escherichia coli DNA unwinding protein to single‐stranded nucleic acids. Biochemistry. 1976;15:5057–5064. [DOI] [PubMed] [Google Scholar]
- 104.Ruyechan WT, Wetmur JG. Studies on the cooperative binding of the Escherichia coli DNA unwinding protein to single‐stranded DNA. Biochemistry. 1975;14:5529–5534. [DOI] [PubMed] [Google Scholar]
- 105.Krauss G, Sindermann H, Schomburg U, Maass G. Escherichia coli single‐strand deoxyribonucleic acid binding protein: Stability, specificity, and kinetics of complexes with oligonucleotides and deoxyribonucleic acid. Biochemistry. 1981;20:5346–5352. [DOI] [PubMed] [Google Scholar]
- 106.Chrysogelos S, Griffith J. Escherichia coli single‐strand binding protein organizes single‐stranded DNA in nucleosome‐like units. Proc Natl Acad Sci U S A. 1982;79:5803–5807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Greipel J, Maass G, Mayer F. Complexes of the single‐stranded DNA‐binding protein from Escherichia coli (eco SSB) with poly(dT). An investigation of their structure and internal dynamics by means of electron microscopy and NMR. Biophys Chem. 1987;26:149–161. [DOI] [PubMed] [Google Scholar]
- 108.Shishmarev D, Wang Y, Mason CE, et al. Intramolecular binding mode of the C‐terminus of Escherichia coli single‐stranded DNA binding protein determined by nuclear magnetic resonance spectroscopy. Nucleic Acids Res. 2014;42:2750–2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kozlov AG, Shinn MK, Weiland EA, Lohman TM. Glutamate promotes SSB protein‐protein interactions via intrinsically disordered regions. J Mol Biol. 2017;429:2790–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bujalowski W, Lohman TM. Escherichia coli single‐strand binding protein forms multiple, distinct complexes with single‐stranded DNA. Biochemistry. 1986;25:7799–7802. [DOI] [PubMed] [Google Scholar]
- 111.Lohman TM, Overman LB, Datta S. Salt‐dependent changes in the DNA binding co‐operativity of Escherichia coli single strand binding protein. J Mol Biol. 1986;187:603–615. [DOI] [PubMed] [Google Scholar]
- 112.Dubiel K, Myers AR, Kozlov AG, et al. Structural mechanisms of cooperative DNA binding by bacterial single‐stranded DNA‐binding proteins. J Mol Biol. 2019;431:178–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liu J, Choi M, Stanenas AG, et al. Novel, fluorescent, SSB protein chimeras with broad utility. Protein Sci. 2011;20:1005–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lohman T. Kinetics of protein‐nucleic acid interactions: Use of salt effects to probe mechanisms of interaction. CRC Crit Rev Biochem. 1986;19:191–245. [DOI] [PubMed] [Google Scholar]
- 115.Lohman TM, Bujalowski W, Overman LB, Wei TF. Interactions of the E. coli single strand binding (SSB) protein with ss nucleic acids. Binding mode transitions and equilibrium binding studies. Biochem Pharmacol. 1988;37:1781–1782. [DOI] [PubMed] [Google Scholar]
- 116.Suksombat S, Khafizov R, Kozlov AG, Lohman TM, Chemla YR. Structural dynamics of E. coli single‐stranded DNA binding protein reveal DNA wrapping and unwrapping pathways. Elife. 2015;4:e08193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Roy R, Kozlov AG, Lohman TM, Ha T. Dynamic structural rearrangements between DNA binding modes of E. coli SSB protein. J Mol Biol. 2007;369:1244–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhou R, Kozlov AG, Roy R, et al. SSB functions as a sliding platform that migrates on DNA via reptation. Cell. 2011;146:222–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Roy R, Kozlov AG, Lohman TM, Ha T. SSB protein diffusion on single‐stranded DNA stimulates RecA filament formation. Nature. 2009;461:1092–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lee KS, Marciel AB, Kozlov AG, Schroeder CM, Lohman TM, Ha T. Ultrafast redistribution of E. coli SSB along long single‐stranded DNA via intersegment transfer. J Mol Biol. 2014;426:2413–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Schmidt A, Kochanowski K, Vedelaar S, et al. The quantitative and condition‐dependent Escherichia coli proteome. Nat Biotechnol. 2016;34:104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bujalowski W, Lohman TM. Limited co‐operativity in protein‐nucleic acid interactions. A thermodynamic model for the interactions of Escherichia coli single strand binding protein with single‐stranded nucleic acids in the "beaded", (SSB)65 mode. J Mol Biol. 1987;195:897–907. [DOI] [PubMed] [Google Scholar]
- 123.Lewis JS, Jergic S, Dixon NE. The E. coli DNA replication fork. Enzyme. 2016;39:31–88. [DOI] [PubMed] [Google Scholar]
- 124.Pant K, Karpel RL, Rouzina I, Williams MC. Mechanical measurement of single‐molecule binding rates: Kinetics of DNA helix‐destabilization by T4 gene 32 protein. J Mol Biol. 2004;336:851–870. [DOI] [PubMed] [Google Scholar]
- 125.Shokri L, Marintcheva B, Richardson CC, Rouzina I, Williams MC. Single molecule force spectroscopy of salt‐dependent bacteriophage T7 gene 2.5 protein binding to single‐stranded DNA. J Biol Chem. 2006;281:38689–38696. [DOI] [PubMed] [Google Scholar]
- 126.von Hippel P, Delagoutte E. A general model for nucleic acid helicases and their "coupling" within macromolecular machines. Cell. 2001;104:177–190. [DOI] [PubMed] [Google Scholar]
- 127.Heuer K, Arbuzova A, Strauss H, Kofler M, Freund C. The helically extended SH3 domain of the T cell adaptor protein ADAP is a novel lipid interaction domain. J Mol Biol. 2005;348:1025–1035. [DOI] [PubMed] [Google Scholar]
- 128.Perez Y, Maffei M, Igea A, et al. Lipid binding by the unique and SH3 domains of c‐Src suggests a new regulatory mechanism. Sci Rep. 2013;3:1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Kunzelmann S, Morris C, Chavda AP, Eccleston JF, Webb MR. Mechanism of interaction between single‐stranded DNA binding protein and DNA. Biochemistry. 2010;49:843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tan HY, Bianco PR. SSB facilitates fork substrate discrimination by PriA. ACS Omega. 2021. 10.1021/acsomega.1c00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Harami GM, Kovacs ZJ, Pancsa R, et al. Phase separation by ssDNA binding protein controlled via protein‐protein and protein‐DNA interactions. Proc Natl Acad Sci U S A. 2020;117:26206–26217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. [DOI] [PubMed] [Google Scholar]
- 133.Bonetti D, Colombo CV, Clerici M, Longhese MP. Processing of DNA ends in the maintenance of genome stability. Front Genet. 2018;9:390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bochkarev A, Bochkareva E. From RPA to BRCA2: Lessons from single‐stranded DNA binding by the OB‐fold. Curr Opin Struct Biol. 2004;14:36–42. [DOI] [PubMed] [Google Scholar]
- 135.Pettersen EF, Goddard TD, Huang CC, et al. UCSF chimera—a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. [DOI] [PubMed] [Google Scholar]
- 136.Zhang Y, Skolnick J. TM‐align: A protein structure alignment algorithm based on the TM‐score. Nucleic Acids Res. 2005;33:2302–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Aitio O, Hellman M, Kazlauskas A, et al. Recognition of tandem PxxP motifs as a unique Src homology 3‐binding mode triggers pathogen‐driven Actin assembly. Proc Natl Acad Sci U S A. 2010;107:21743–21748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Breyer WA, Matthews BW. Structure of Escherichia coli exonuclease I suggests how processivity is achieved. Nat Struct Biol. 2000;7:1125–1128. [DOI] [PubMed] [Google Scholar]
- 139.Wang F, Yang W. Structural insight into translesion synthesis by DNA pol II. Cell. 2009;139:1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Simossis VA, Heringa J. PRALINE: A multiple sequence alignment toolbox that integrates homology‐extended and secondary structure information. Nucleic Acids Res. 2005;33:W289–W294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Musacchio A, Saraste M, Wilmanns M. High‐resolution crystal structures of tyrosine kinase SH3 domains complexed with proline‐rich peptides. Nat Struct Biol. 1994;1:546–551. [DOI] [PubMed] [Google Scholar]
- 142.Bianco PR, Lu Y. Single molecule studies of stalled DNA replication fork rescue in E. coli . Nucleic Acids Res. 2021;49:4220–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Inoue J, Nagae T, Mishima M, Ito Y, Shibata T, Mikawa T. A mechanism for single‐stranded DNA‐binding protein (SSB) displacement from single‐stranded DNA upon SSB‐RecO interaction. Journal of Biological Chemistry, 2011;286:6720–6732. 10.1074/jbc.m110.164210. [DOI] [PMC free article] [PubMed] [Google Scholar]


