Abstract
Amyloid fibrils are ordered aggregates that may be formed from disordered, partially unfolded, and fragments of proteins and peptides. There are several diseases, which are due to the formation and deposition of insoluble β‐sheet protein aggregates in various tissue, collectively known as amyloidosis. Here, we have used bovine α‐lactalbumin as a model protein to understand the mechanism of amyloid fibril formation at pH 1.6 and 65°C under non‐reducing conditions. Amyloid fibril formation is confirmed by Thioflavin T fluorescence and atomic force microscopy (AFM). Our finding demonstrates that hydrolysis of peptide bonds occurs under these conditions, which results in nicking and fragmentation. The nicking and fragmentation have been confirmed on non‐reducing and reducing gel. We have identified the fragments by matrix‐assisted laser desorption ionization‐time of flight (MALDI‐TOF) mass spectrometry. The fragmentation may initiate nucleation as it coincides with AFM images. Conformational changes associated with monomer resulting in fibrillation are shown by circular dichroism and Raman spectroscopy. The current study highlights the importance of nicking and fragmentation in amyloid fibril formation, which may help understand the role of acidic pH and proteolysis under in vivo conditions in the initiation of amyloid fibril formation.
Keywords: aggregation, amyloid, fragmentation, nucleation, α‐lactalbumin
Abbreviations
- AFM
atomic force microscopy
- Asp
aspartic acid
- CD
circular dichroism
- HEWL
hen egg‐white lysozyme
- kDa
kilo Dalton
- MALDI‐TOF
matrix‐assisted laser desorption ionization‐time of flight
- SDS‐PAGE
sodium dodecyl sulphate‐poly acrylamide gel electrophoresis
- β‐ME
β‐mercaptoethanol
1. INTRODUCTION
Cellular protein folding is controlled and assisted by molecular chaperones, protein disulphide isomerases, and peptidyl‐prolyl isomerases. Sometimes, cellular machinery fails to control the folding process, which leads to the aggregation of proteins. These aggregates are associated with many neurodegenerative diseases like Alzheimer's, Parkinson's, Huntington's, prion‐related disease, and so forth. Apart from neurodegenerative diseases, non‐neuropathic systemic amyloidosis, for example, light chain amyloidosis, lysozyme amyloidosis, localize insulin amyloidosis are also caused by the deposition of protein aggregates in different tissues and organs of the body.1, 2 First two decades of the twenty‐first century leads to the explosion in our knowledge and understanding of amyloid fibril formation.3 Recent developments in amyloid fibril structures came from solution NMR, solid‐state NMR, and Cryo‐EM studies of different amyloid fibrils. At the molecular level, amyloid fibrils consist of the cross‐β sheet, in which polypeptide chains are arranged perpendicular to the fibril axis.4, 5 The orientation of monomers into amyloid fibril makes the amyloid fold more stable than their corresponding native state.6 Apart from disease‐causing, functional amyloids also exist in nature, for example, curli in bacteria, sup 35 in fungi, and Pmel17 in humans.7 Amyloids are also emerging as biomaterial due to their stability, mechanical strength, and adhesion properties.8
At the onset of aggregation, which may be due to mutations or environmental changes, protein undergoes conformational changes resulting into misfolding or proteolysis of the native state. Conformational changes in proteins initiate nucleation, which further recruit monomers or fragments to form oligomers. Elongation of the oligomers can further take place by addition of monomers or oligomers, which can lead to higher‐level well‐ordered amyloid fibrils.9, 10 The fragments, which are formed by proteolysis, have been involved in several amyloid‐related diseases. Alzheimer's β‐peptide (Aβ) is produced by proteolytic processing of the transmembrane protein called amyloid precursor protein.11, 12 Familial British dementia is due to the formation of the amyloid fibril of 34 residue amyloid‐Bri peptide fragment, which is derived by cleavage of C‐terminal mutated, extended transmembrane precursor protein (ABriPP) of 277 amino acid.13 The composition of aortic medial amyloid shows a 50 amino acid long peptide called medin, derived from a proteolytic fragment of lactadherin.14 β2‐microglobulin is the main component of amyloid fibrils deposited in patients with dialysis‐related amyloidosis, which undergoes proteolysis, covalent modifications, and conformational changes to form amyloid under physiological conditions.15 In an interesting study on β2‐microglobulin, the role of proline is demonstrated by mutating P32G leads to the identification of folding intermediate, which is the precursor of amyloid fibril formation at neutral pH.16 However, initially under in vitro conditions, this protein has been shown to form amyloid at acidic pH.17 The pH‐dependent cleavage of lithostathine is suggested to remove a charged polypeptide portion that usually contains a hydrophobic spot required for aggregation. In this case, the N‐terminally cleaved portion of lithostathine found in fibrillar aggregates of Alzheimer's disease and chronic calcifying pancreatitis.18 The Finnish type familial amyloidosis is caused by mutant gelsolin, where a 71‐residue fragment of 8 kDa arises by proteolytic cleavage in gelsolin mutants (D187N) that form amyloid fibrils. In vivo gelsolin fragmentation occurs in an acidic organelle, and in vitro, it has been shown that both the wild type and the mutant (D178N) gelsolin fragments form amyloid fibrils. This fragment adopts mostly unstructured conformation and does not form amyloid fibrils at pH 7.4, but surprisingly, incubation at low pH leads to the formation of well‐defined fibrils.19, 20
Bovine α‐lactalbumin is a small globular, acidic, and calcium‐binding protein having a molecular weight 14,175 Da and present in the mammalian milk whey. It acts as a component of lactose synthase and thus regulating lactose biosynthesis.21, 22 It is a member of the lysozyme family of protein.23 The atomic resolution structure of α‐lactalbumin shows that it consists of major α‐subdomain and minor β‐subdomain. The α‐subdomain has three main α‐helices consists of 5–11, 23–34, and 86–98 residues and two small 310‐helices of 18–20 and 115–118 residues. The minor β‐subdomain has three‐stranded antiparallel β‐pleated sheets of 41–44, 47–50, 55–56 residues and the short 310 helices of 77–80 residues. It has four disulphide bonds between Cys6‐Cys120, Cys28‐Cys111, Cys73‐Cys91, and Cys61‐Cys77 (Figure 1). Both subdomains are divided by a deep cleft and held together by two disulphide bonds (Cys73‐Cys91 and (Cys61‐Cys77).24, 25 It is one of the best‐studied model proteins from protein folding point of view. The concept of molten globule state in which secondary structure is compact and tertiary structure fluctuates is among the best described for α‐lactalbumin.26, 27
FIGURE 1.
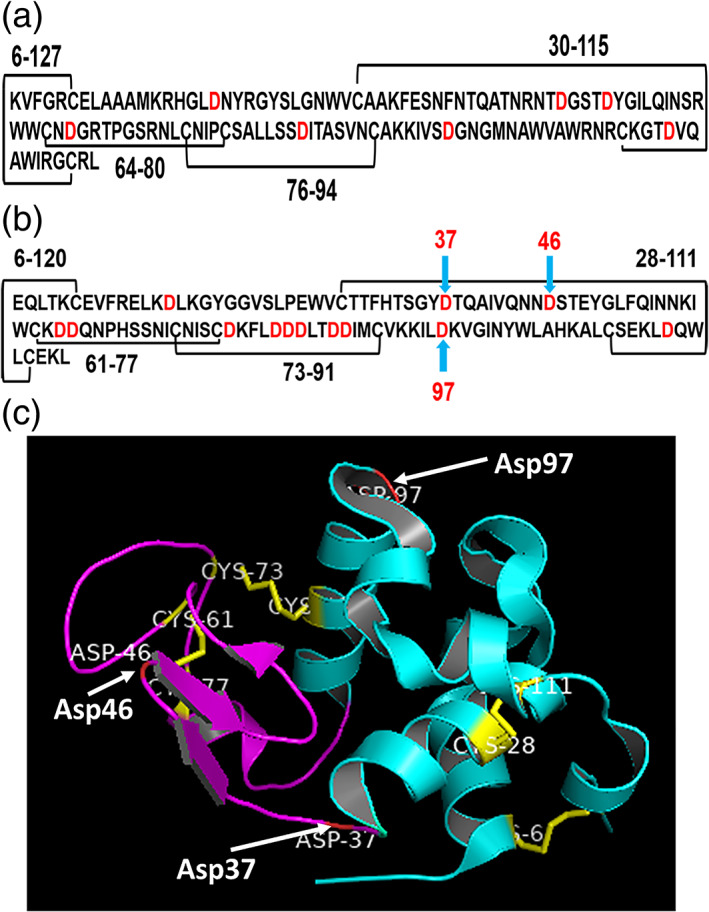
(a) Primary structure of hen egg‐white lysozyme. The disulphide bonds are indicated as black line and Asp indicated as red. (b) Primary structure of bovine α‐Lactalbumin.24 The disulphide bonds are indicated by the black line and hydrolysis prone sites are indicated by the light blue arrow. (c) The tertiary structure of bovine α‐Lactalbumin.25 The figure is made by using PDB file 1F6S with the help of software PyMOL version 1.8.4. The four disulphide bonds are shown by yellow stick and the hydrolysis sites are indicated by the white arrow. α Domain and 310‐helices are indicated in cyan and β‐domain in magenta color
The fragment of α‐lactalbumin involved in amyloid fibril formation was observed by treating with the serine protease from Bacillus licheniformis protease. Protein fragments of 8.8 and 9.8 kDa were observed in the presence of protease at pH 7.0 and 55°C. The molecular mechanism involved in the side‐by‐side assembly of dimer of these fragments into amyloid fibrils has been reported.28 To identify the region responsible for α‐lactalbumin amyloid fibril formation, it was found that asymmetrical peptide, identical to 35–51 amino acid residue of human α‐lactalbumin and homologous peptide to β‐domain of mammalian α‐lactalbumin can form amyloid fibrils. This leads to the conclusion that the amyloidogenic determinant is present in the β‐domain of α‐lactalbumin.29 The addition of small tetrapeptides (TDYG and TEYG) drastically increases the rate of fibrillogenesis corresponding to the 35–51 region of β‐subdomain of human α‐lactalbumin.30 In the acidic pH (2.0) at 37°C, α‐lactalbumin forms amyloid fibrils under partially reducing conditions.31 Partially reduced α‐lactalbumin (1SS‐α‐lactalbumin) is the state in which three out of four disulphide bonds are reduced, and free cysteines are carboxymethylated.32 By using proteases, such as proteinase K and chymotrypsin at neutral pH and pepsin at acidic pH, limited proteolysis was carried out for identification of the domain responsible for amyloid fibril formation.
Structure and dynamics of acid unfolded molten globule state upon proteolysis revealed that the region surrounding β‐sheet is flexible, which is removed first upon proteolysis.33 In the molten globule state, the α subdomain retains secondary structure while the β domain loses.34 Limited proteolysis in the partially folded state of α‐lactalbumin causes nicking at 40–41 and 52–53 positions, which shows that this region is flexible.35 The flexible conformation or partially unfolded structure could be the key to trigger amyloid fibril formation. Nicking at 40–41 position and deletion of 41–52 region accelerates the fibril formation.36 Nicking refers to the hydrolysis of peptide bond, which causes fragmentation if it occurs at least two points. However, if the nicked region is held by disulfide bonds, fragmentation does not occur. α‐lactalbumin aggregation has been prevented by using molecular chaperones like αB‐crystallin and β‐casein. The αB‐crystallin has been shown as an efficient chaperone under the conditions where amorphous aggregates are formed.37, 38 Membrane containing negatively charged phospholipid, phosphatidylserine enhances the amyloid fibril formation of holo α‐lactalbumin because it provides a physiological low‐pH environment on the cellular membrane, thus accelerates the fibril formation.39 The amyloid fibril formation of bovine apo α‐lactalbumin is inhibited by oleic acid at pH 4.5, where insoluble aggregates are formed. However, further lowering the pH between 2.0 and 3.0, the inhibitory effect of oleic acid is not effective in amyloid fibril formation.40 Slightly unfolded α‐lactalbumin forms a complex with fatty acid and kill cancerous cells.41, 42
Amino acid sequence, three‐dimensional structure, number, and location of disulfide bonds in α‐lactalbumin is similar to that of lysozyme, where out of 123 residues, 40 residues are identical, and 60 residues are similar.24, 43 The hen egg‐white lysozyme (HEWL) forms amyloid fibrils under the acidic pH and elevated temperature.44 It is observed that HEWL at acidic pH and elevated temperature undergoes stepwise hydrolysis, fragmentation, and assembly to form amyloid fibrils.45 Being similar to lysozyme, α‐lactalbumin is a suitable model protein to study the amyloid fibril formation at acidic pH, thus mimicking in vivo proteolysis conditions as shown in the human transthyretin amyloid and the AL‐amyloid formation.46, 47
In this study, we have focused on the amyloid‐forming property of α‐lactalbumin at acidic pH 1.6 and 65°C. We are showing for the first time that α‐lactalbumin undergoes nicking and fragmentation under these conditions. In the acidic pH and elevated temperature, more flexible conformation of α‐lactalbumin appears, which might be helping in the hydrolysis of Asp‐X peptide bond, resulting into nicking and fragmentation. These fragments may initiate nucleation, and the addition of nicked monomers and fragments lead to amyloid fibril formation.
2. RESULTS
2.1. Kinetics of amyloid fibril formation by ThT fluorescence
ThT fluorescence was carried out to confirm the amyloid fibril formation at acidic pH and elevated temperature. Due to the charged nature of ThT, it weakly interacts with amyloid fibrils in acidic condition.48 ThT fluorescence is more suited for fibril assay at neutral pH therefore, ThT assay was carried out at pH 7.0 (Figure 2). Our current understanding of the molecular mechanism of ThT is that in solution, both rings that are present in the ThT, freely rotate through their carbon–carbon single bond. When it is excited by a particular wavelength of light in the free state, it rapidly quenches the excited state due to rotation and resulting in low intensity of fluorescence emission.
FIGURE 2.
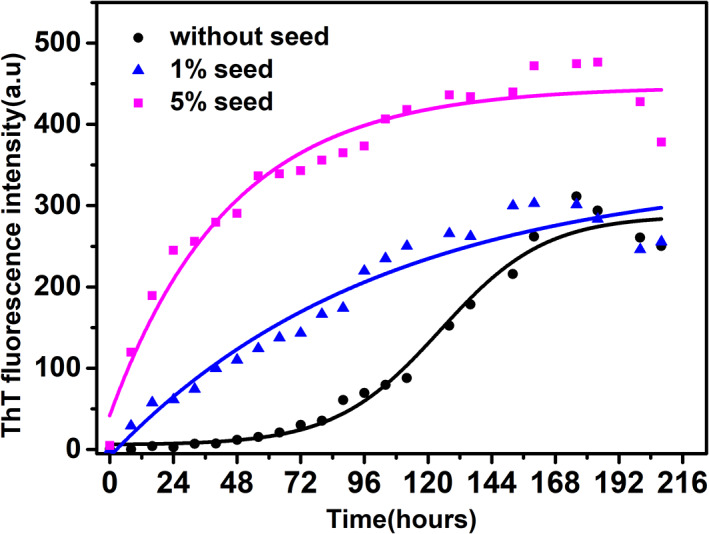
Kinetics of bovine α‐lactalbumin amyloid fibril formation by Thioflavin T. Amyloid fibril formation without seed (black), 1% seed (blue) and 5% seed (magenta)
On the other hand, in the bound state with amyloid fibrils, there is free rotation restriction about the single bond, resulting in increased fluorescence intensity upon excitation with a particular wavelength. Therefore, ThT intensity is increasing with time after the lag phase.49 The lag phase of the kinetics is 64 hr (Figure 2) which is calculated by fitting the kinetics data with non‐linear curve fitting and using the formula as described in Equation (1) where lag time was calculated as tlag= −2 . ThT fluorescence intensity increases over a period of time up to 175 hr and remains constant thereafter. Seeding with the preformed fibrils is known to accelerate the nucleation‐dependent elongation reaction and thus reducing the lag phase drastically.45 To see the effect of seeding during fibril formation, kinetics by ThT fluorescence was carried out. For seeding experiment, 232 hr mature fibrils were used as seed. Seeding of the fibrillation reaction with 1% seed completely abolishes the lag phase while 5% did the same with a higher slope (Figure 2). The effect of seeding in reducing the lag phase of fibrillation demonstrates the role of nucleation in the acceleration of amyloid fibril formation.
2.2. Morphology of oligomers and amyloid fibrils
To visualize and morphologically characterize oligomers and fibrils, AFM was carried out in the intermittent mode during amyloid fibril formation. The oligomer and the fibril heights were measured from the cross‐section perpendicular to the long axis of fibrils. Oligomerization started during the lag phase of fibrillation, which cannot be detected by ThT fluorescence. The oligomers of spherical shapes with the height of 2.5–3.0 nm were observed after 24 hr of fibrillation (Figure 3a). They were continued to be present during the entire lag phase at 48 (Figure 3b) and 64 hr (Figure 3c) along with the short‐length fibrils. Mature fibrils started dominating from 88 hr onward with the height of 3.7 nm; however, still few oligomers are visible along with the fibrils (Figure 3d). Fully mature and twisted fibrils having 3.0 nm height are present with long and unbranched structure at 196 hr (Figure 3e).
FIGURE 3.
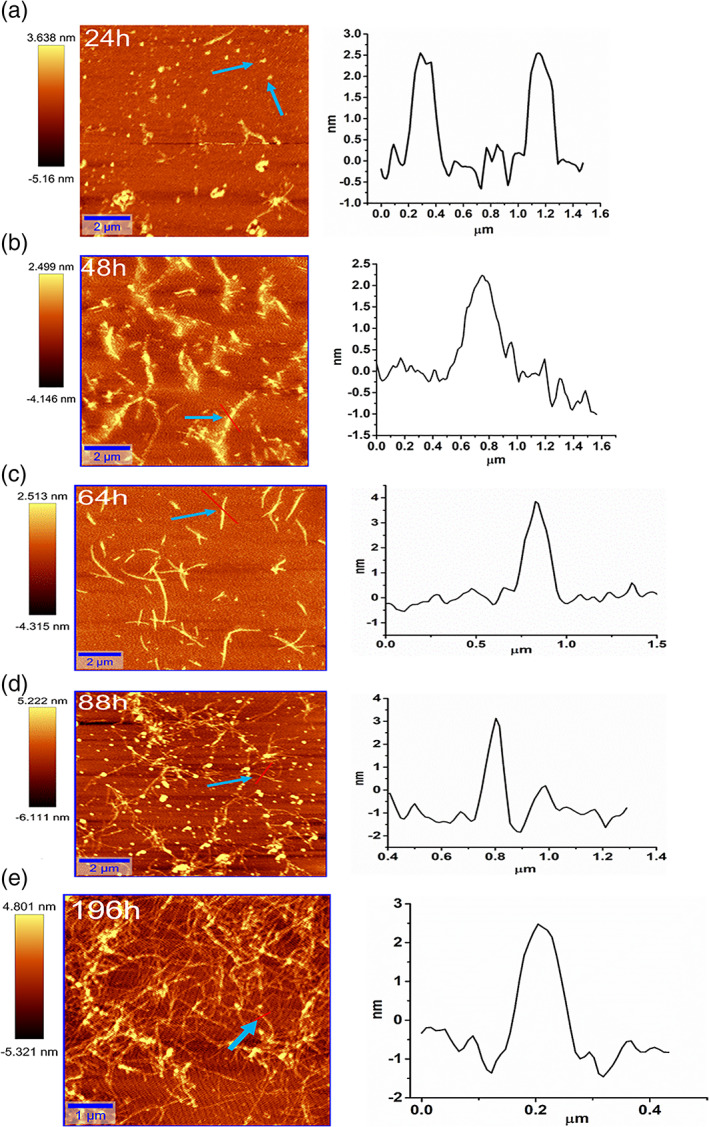
Atomic force microscopy (AFM) images of bovine α‐lactalbumin during amyloid formation at different time points. (a) 24 hr, (b) 48 hr, (c) 64 hr, (d) 88 hr, and (e) 196 hr. The height distribution of oligomers and fibrils is shown adjacent to each image wherein, the height and area scale bars are in nm and μm, respectively. The blue arrow indicates that the particular cross‐section of the image is selected for height distribution
2.3. Kinetics by sodium dodecyl sulphate‐poly acrylamide gel electrophoresis
To understand the amyloid fibril formation at the molecular level, the kinetics of fibril formation is followed by sodium dodecyl sulphate‐poly acrylamide gel electrophoresis (SDS‐PAGE) analysis. Under acidic condition and high temperature, peptide bond hydrolysis is the most susceptible at Asp residue. The heating of proteins under dilute acidic condition causes cleavage of peptide bonds at both ends and thus removal of Asp from proteins occur.50, 51 Bovine α‐lactalbumin has pI 4.5 and contains 13 Asp residues. In order to understand the role of nicking and fragmentation at acidic pH (1.6) and higher temperature (65°C) containing 100 mM NaCl, SDS‐PAGE was carried out. Acid hydrolysed samples were collected at different time points and analyzed on 16.5% Tricine gel. At acidic pH and 65°C, it is expected that bovine α‐lactalbumin may show fragmentation. Since α‐lactalbumin is an acidic protein, therefore 100 mM NaCl was added to the reaction mixture at acidic pH in order to accelerate the amyloid fibril formation.31 Kinetics of fibrillation was followed on Tricine SDS‐PAGE because of better separation of smaller molecular weight fragments. All the fibrillation kinetics was carried out under non‐reducing condition.
At the beginning of the fibrillation reaction at 0 and 12 hr, clearly, only the monomer band of 14.2 kDa is present. From 24 hr onward, apart from the monomer band, two fragments of less than 10 kDa have also started appearing. The monomer remains the dominant species up to 132 hr. However, out of the two fragments, the smaller fragment's intensity kept on increasing over a period of time till 132 hr (Figure 4a,b). Under acidic pH and elevated temperature, these fragments are formed due to hydrolysis of peptide bonds, causing nicking and fragmentation. Partial hydrolysis is leading to the formation of nicked α‐lactalbumin, which are held by four disulfide bonds and migrate as a full‐length 14.2 kDa protein.
FIGURE 4.
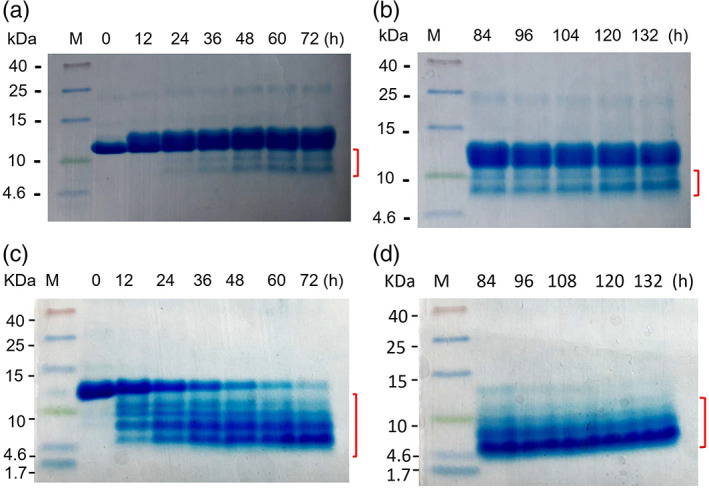
Hydrolysis kinetics of bovine α‐lactalbumin followed by Tricine sodium dodecyl sulphate‐poly acrylamide gel electrophoresis (SDS‐PAGE). (a) and (b) SDS‐PAGE run under non‐reducing conditions without β‐mercaptoethanol in protein sample buffer. (c) and (d) samples run under reducing condition in the presence of β‐mercaptoethanol in sample buffer. M denotes molecular weight marker in kDa and (h) denotes time in hour at which samples were collected during fibrillation. The red marks indicate the fragmentation in both non‐reducing (a and b) and reducing (c and d) gel
To confirm the nicking of partially hydrolysed monomer, which are held by disulfide bonds, samples were reduced by β‐mercaptoethanol (β‐ME). In the reducing gel, monomer intensity started rapidly decreasing from 12 hr onward, and almost no monomer left after 84 hr (Figure 4c,d). Instead of two fragments in the non‐reducing gel (Figure 4a,b), at least four fragments started appearing from 12 hr of the incubation in the reduced gel (Figure 4c,d). As the monomer intensity decreasing, fragments intensity increasing (Figure 4c,d). A completely different gel pattern on reducing gel compare to non‐reducing gel is due to the presence of four disulphide bonds in α‐lactalbumin. Four disulfide bonds hold the nicked region of α‐lactalbumin together in non‐reducing gel and, therefore, move as intact α‐lactalbumin and only two fragments (Figure 4a,b). While in the reducing gel, the disulphide bonds, which hold the different nicked region together, are broken and resulting in several smaller bands (Figure 4c,d).
2.4. Kinetics by matrix‐assisted laser desorption ionization‐time of flight
It is clearly visualized in the hydrolysis experiments (Figure 4) that the fragmentation occurs over the time period during which amyloid fibril formation is likely to occur. In order to identify the molecular species, present in fibrillating conditions, samples were collected at different time points and analyzed by matrix‐assisted laser desorption ionization‐time of flight (MALDI‐TOF) mass spectrometry. Samples were collected at different time points from the fibrillation reaction‐ under non‐reducing conditions (corresponding to non‐reducing gel, Figure 4) and mixed with the acidic matrix sinapinic acid and acetonitrile. MALDI‐TOF mass spectrometry is an excellent method to identify the molecular species, but it is not quantitative. In this method, small molecular fragments fly better than larger species. MALDI‐TOF is less precise than electrospray ionization‐mass spectrometry (ESI‐MS), and that is, why there is a slight deviation in measured mass than the theoretical mass. Due to low sample requirement and better accuracy than the SDS‐PAGE, we have decided to use this technique to identify the fragments during the fibrillating condition. In the control sample at the zero‐time point, full‐length α‐lactalbumin is present, displaying the single charged peak with 14,168 Da and also the double‐charged peak with the mass of 7,087 Da (Figure 5a) corresponding to the molecular mass of intact α‐lactalbumin. Apart from singly charged species, double‐charged peaks are also frequently observed in MALDI‐TOF spectra. At 24 hr, in addition to the single and double charge species, a tiny peak of 5,898 Da has started appearing in the mass spectrum (Figure 5b). At 48 hr of fibrillation, a very sharp peak of the monomer is present. In addition to the monomer peak, around 7,000 Da, couple of peaks are also present, including a prominent peak of 7,318 Da. Another group of peaks at 5,789/5,903 Da and a tiny peak of 8,285 Da are present at 48 hr sample. Several other smaller peaks of 2,615, 2,501, 3,134 Da are also present (Figure 5c), which may be in very small quantity but could not be detected by SDS‐PAGE. At 72 hr, similar pattern was observed where 7,021/7,301 Da and 5,800 Da peaks are present; however, the monomer peak's intensity has drastically decreased compared to fragments (Figure 5d). The same trend continues at 96 hr where further reduction in the intensity of monomer peak was observed, and strong peaks of 7,205/7,302 Da and 5,773 Da peaks are present. Apart from that, a small intensity peak of 8,285 Da is also present. During the later stage of fibrillation at 144 hr, the monomer peak is entirely missing, which is in contrast to the thick band present in SDS‐PAGE at 132 hr, which shows the limitation of MALDI‐TOF mass spectrometry (Figure 4b). However, 7,186/7,301 Da peaks are present at 144 hr.
FIGURE 5.
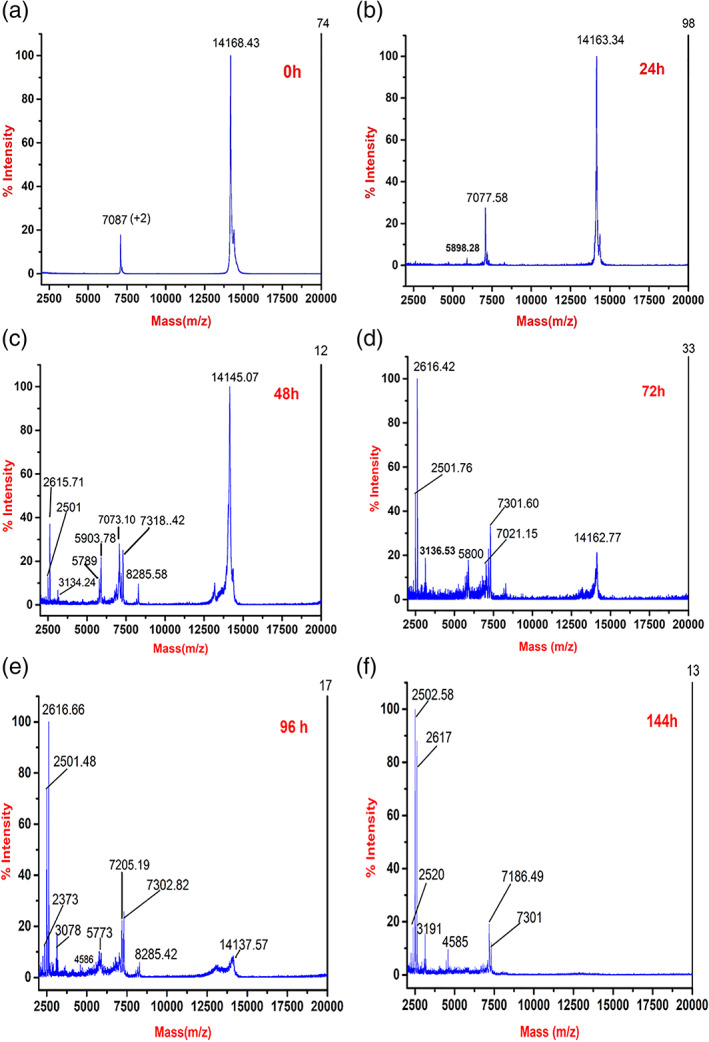
Kinetics of bovine α‐lactalbumin amyloid fibril formation by matrix‐assisted laser desorption ionization‐time of flight (MALDI‐TOF) mass spectrometry. Samples were collected at (a) 0 hr, (b) 24 hr, (c) 48 hr, (d) 72 hr, (e) 96 hr, and (f) 144 hr and diluted with acidic matrix
There are 13 Asp residues in bovine α‐lactalbumin, which provide the possible site of acid hydrolysis during fibrillating conditions. However, four disulfide bonds (6–120, 28–111, 61–77, and 73–91) are also present, which hold the molecule together.24 As described earlier,50, 51 peptide bonds involving Asp are most susceptible for acid hydrolysis. The N and the C terminus of bovine α‐lactalbumin are connected by disulfide bonds 6–120 and 28–111 (Figure 1). Taking together the presence of two fragments on SDS‐PAGE (Figure 4) and MALDI‐TOF mass spectra, it can be deduced that the monomer species are nicked at 37 and 97 positions thus give rise to the fragment. The smaller fragments may then be originating from 38–97 which contains 10 out of 13 Asp, corresponding to the peaks of 7,021 Da and 7,205/7,302/7,318 Da. Further nicking and fragmentation at Asp46 and Asp97 give rise to the fragments of 5,789/5,800/5,903 Da and 8,285 Da. The origin of slightly different molecular weight fragments may be also due to either‐N or‐C terminus peptide bond hydrolysis of Asp residues. The suggested fragments and their possible molecular weights have been given in Table 1. There are also small fragments that are present in MALDI‐TOF from 48 hr onward, varying between 2,500 and 3,000 Da, which cannot be detected on SDS PAGE due to very low concentration, which is beyond the detection limit of SDS PAGE. These small peaks may originate due to internal cleavage of Asp where the possibility of hydrolysis is maximum. They may also represent double‐charged peaks of fragments.
TABLE 1.
MALDI‐TOF suggested fragments and their theoretical molecular weight
| S. no | Fragment | Suggested molecular weight (Da) | Theoretical molecular weight (Da) |
|---|---|---|---|
| 1 | 38–97 | 7,021/7,073 | 6,895 |
| 2 | 1–37 and 98–123 | 7,302/7,318/7,205/7,186 | 7,309 |
| 3 | 47–97 | 5,773/5,898/5,903/5,789/5,800 | 5,911 |
| 4 | 1–46 and 98–123 | 8,285 | 8,293 |
Abbreviation: MALDI‐TOF, matrix‐assisted laser desorption ionization‐time of flight.
2.5. Secondary structure analysis by Raman spectroscopy
Raman spectroscopy was carried out to follow the secondary structure changes during α‐lactalbumin amyloid fibril formation. Raman spectroscopy measures vibrational transition in proteins, which mainly originates from the CONH group of the amide bond. The amide‐I band from 1,600 to 1,700 cm−1 is most commonly used to investigate the change in protein secondary structure. It originates from in‐plane vibration of C═O stretching and a very small contribution from out of phase C─N stretching in the peptide bond. The amide‐I band has sub‐band regions, which indicate the specific secondary structure of the protein like α‐helix, β‐sheet, and disordered region in Raman spectrum.52, 53
Raman spectra of monomer α‐lactalbumin at pH 1.6 (Figure 6a) and matured amyloid fibrils at pH 1.6 and 65°C (Figure 6b) showed apparent differences in the amide‐I and amide‐III region of the spectra. Amide‐I peaks do not overlap with other regions of the spectrum, thus give direct information about secondary structure. In the monomer, the amide‐I region has two prominent bands at 1,664 and 1,614 cm−1 (Figure 6a). Upon fibrillation, the band at 1,664 cm−1 shifted toward 1,671 cm−1, which is an indication of conformational changes at the secondary structure level and denotes the formation of the β sheet structure.54 However, the aromatic peak of monomer at 1,614 cm−1 has minor change and shifted to 1,615 cm−1 in the fibril, which again shows slight reorganization of aromatic residues in the amyloid fibrils. To identify the sub‐bands in the amide‐I region, deconvolution of monomer and fibril bands were carried out. Four sub‐bands were identified in the amide I region of the monomer and amyloid fibrils, as shown in Table 2. In the monomer, the major sub‐band in the amide‐I region is 1,665 cm−1 which shows the disordered region at acidic pH 1.6.55 The sub‐band at 1,680 cm−1 has been assigned as β‐turn and 1,647 cm−1 as α‐helix (Figure 6c). In the matured fibril, the 1,665 cm−1 band shifted to 1,673 cm−1 which indicates the presence of β‐sheet.56, 57 Sub‐band at 1654 cm−1 is the indication of the disordered region and 1,690 cm−1 as β‐turn in the amyloid fibrils (Figure 6d). 52, 58, 59 Effect of pH on α‐lactalbumin by Raman spectroscopy was reported that the major band in the amide‐I region is 1,660 cm−1 at pH 6.6. Upon decrease in pH to 2.1, this peak shifted to 1,664 cm−1, which indicates a disordered region.54, 60 The complex band of aromatic residues are lies in the range of 1,600–1,630 cm−1.61 The shift in the amide‐I region has been shown from 1,662 to 1,672 cm−1 when there is a conformational change from globular to amyloid fibrils in lysozyme.62 Band at 1,664 cm−1 represents predominantly disordered at acidic pH while 1,614 cm−1 band represents the aromatic region of the protein.55 Amide III band is also sensitive to conformational change in secondary structure and originates from in‐phase C─N stretching and in‐plane N─H bending. It is very complex to elucidate the amide III band because amide II and amide III bands are marginally superimposed. That is why generally amide I is used to analyze the change in the secondary structure of the protein. The amide III region is present in the range of 1,200–1,340 cm−1.52 There are signature peak range associated with secondary structure configuration like β‐sheet in the range of 1,230–1,240 cm−1, disorder 1,241–1,255 cm−1, and α‐helix 1,260–1,310 cm−1.63, 64, 65, 66 Our results show a prominent disorder region signature peak at 1243 cm−1 in the case of monomer and a 1,239 cm−1 signature peak corresponding to β‐sheet in the fibril, which indicates the rearrangement of peptide bonds to form β‐sheet in the amyloid fibrils (Figure 6a,b). Other peaks of this region, could not be assigned.
FIGURE 6.
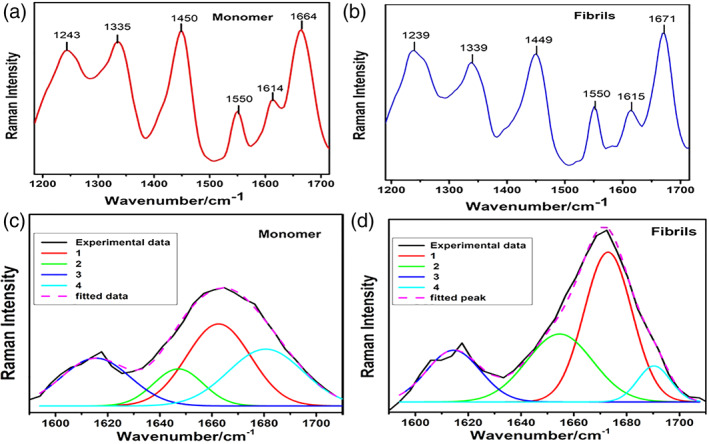
Raman spectra of (a) monomer and (b) fibrils acquired with excitation at 532 nm. Spectra were deconvoluted by fitting Gaussian function in the amide‐I region that is, 1,600–1,700 cm−1 of (c) monomer, and (d) fibrils
TABLE 2.
Deconvolution of amide‐I band of monomer α‐lactalbumin and mature fibrils showing amide‐I sub‐band positions
| Band number | Monomer | Fibrils | ||||
|---|---|---|---|---|---|---|
| Band position (cm−1) | Area | Width | Band position (cm−1) | Area | Width | |
| 1 | 1,665 disordered | 875 | 28 | 1,673 β sheet | 1,146 | 21 |
| 2 | 1,647 α helical | 304 | 22 | 1,654 disordered | 715 | 29 |
| 3 | 1,615 aromatic | 546 | 31 | 1,614 aromatic | 444 | 24 |
| 4 | 1,680 β turn | 708 | 31 | 1,690 β turn | 188 | 14 |
2.6. Kinetics of amyloid formation by CD spectroscopy
While Raman spectroscopy is an excellent method to follow the secondary structure for both soluble and insoluble proteins, circular dichroism (CD) is more suitable for soluble proteins. CD spectroscopy is a powerful technique to study the molten globule state of α‐lactalbumin.67 In this work, α‐lactalbumin fibrillation occurs at pH 1.6, where it is known to adopt the molten globule‐like structure in which secondary structure retains but tertiary structure lost.67 Far‐UV CD (200–250 nm) spectroscopy is used to follow the change in the secondary structure during α‐lactalbumin amyloid formation at pH 1.6 and 65°C. Samples were collected at different time intervals and stored at 4°C. Since the fibrillation was carried out at 400 μM, therefore samples were diluted to 15 μM, and spectra were collected at 25°C (Figure 7). CD spectrum of α‐lactalbumin at 0 hr shows a typical α‐helical signature having two prominent peaks at 208 and 222 nm. There was a gradual loss of secondary structure over a period from 8 to 72 hr where the negative peaks at 208 and 222 nm are decreasing. This time frame corresponds to the lag phase of fibrillation, as shown by ThT fluorescence (Figure 2). At 108 hr, 222 nm peak is completely lost while 208 nm peak shows decreased ellipticity and loss of about 40% secondary structure. The decrease of the CD bands at 208 and 222 nm points to a loss of soluble α‐helical protein content, thus indicating amyloid fibril formation, which is evident by the ThT fluorescence (Figure 2) and AFM data (Figure 3).
FIGURE 7.
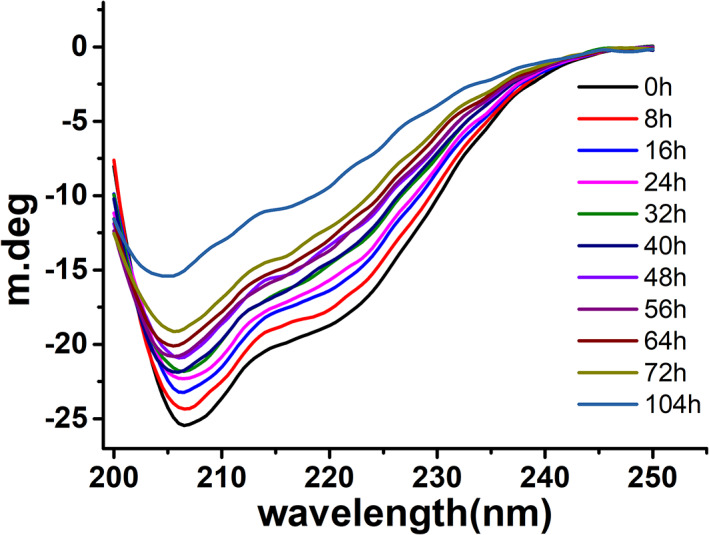
Kinetics of amyloid fibril formation of bovine α‐lactalbumin monitored by far‐UV circular dichroism (CD) (200–250 nm) spectroscopy. Samples at different time points were collected during fibrillation and measured at 25°C
2.7. Composition of amyloid fibrils
The amyloid fibrils were grown for 208 hr under the fibril forming conditions, that is, at pH 1.6 and 65°C. These fibrils were collected through centrifugation at 25,000 × g for 90 min at 4°C. The pellet was washed three times by dilute HCl (pH 1.6) containing 100 mM NaCl followed by centrifugation to remove the soluble fraction. The pellet containing fibrils were solubilized in 10 M urea for 3 days at room temperature. The composition of the fibril was analyzed by SDS‐PAGE under non‐reducing and reducing conditions. SDS‐PAGE (Figure 8) analysis shows that dissolved amyloid fibrils under non‐reducing condition are composed of the 14,175 Da monomer and two fragments of less than 10 kDa (Figure 8). The solubilized fibril fragments correspond to the approximately same molecular weight of the fragments observed during kinetics of amyloid fibril formation at 48 hr. To know the nature of monomeric species of solubilized fibrils, the sample was reduced by β‐mercaptoethanol. The reduced sample has almost no monomer left and several fragments appeared on the gel (Figure 8), which confirms nicking of the monomer. This suggests that the mature amyloid fibrils are composed of primarily the nicked monomer and two major fragments.
FIGURE 8.
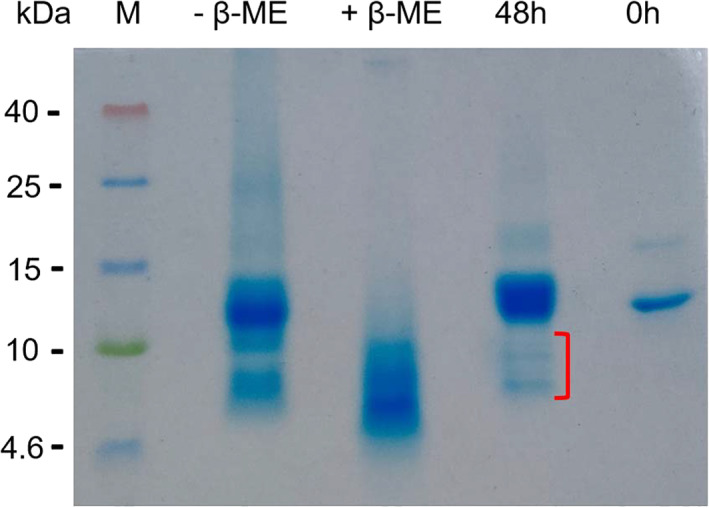
Composition of the mature amyloid fibrils by sodium dodecyl sulphate‐poly acrylamide gel electrophoresis (SDS‐PAGE). The mature fibrils were prepared in 100 mM NaCl pH 1.6 at 65°C for 208 hr. Fibrils were solubilized in 10 M urea before loading on the SDS‐PAGE gel under non‐reducing (−βME) and reducing (+βME) conditions. Samples collected after 48 hr of fibrillation and control of 0 hr sample are also shown in the gel. Red mark indicates the fragments
3. DISCUSSION
Amyloid fibrils can be formed from misfolded monomers, cleaved peptides, or protein fragments. In this study, model protein bovine α‐lactalbumin is used to understand the mechanism of α‐lactalbumin amyloid fibril formation at acidic pH and elevated temperature. At acidic pH, it adopts the molten globule like structure, where most of the secondary structure remains while the tertiary structure is lost. The thermal transition curve of α‐lactalbumin at acidic pH shows non‐cooperative behaviour.26 When it is incubated at pH 1.6 containing 100 mM NaCl and 65°C under static conditions, it forms amyloid fibrils.
At pH 1.6, bovine α‐lactalbumin becomes positively charged. Due to the presence of positive charge, monomers repel each other and do not associate thus prevent amyloid fibril formation. However, addition of 100 mM NaCl known to promote the amyloid fibril formation in α‐lactalbumin at low pH and elevated temperature.31 Addition of salt screens the positive charges and prevent the repulsive electrostatic inter‐molecular interaction and promotes favorable interactions such as hydrophobic interactions for the association to form amyloid fibrils in β2‐microglobulin. Therefore, a critical salt concentration is essential to balance the favorable interactions during amyloid fibril formation.68, 69 Another related protein, HEWL, under the same condition, forms amyloid fibrils without addition of NaCl and a shorter lag phase of 40 hr.45 This could be due to the differences in their amino acid sequence of the amyloidogenic region in α‐lactalbumin and HEWL.
Several studies have shown that the abundance of aromatic amino acids, β‐sheet forming residues, net charge, and hydrophobicity plays an important role in the amyloidogenicity of a protein.70, 71, 72 Loss of the secondary structure is gradual during amyloid fibril formation, as evident by CD spectroscopy (Figure 7). A significant amount of secondary structure is due to the soluble fraction present during amyloid fibrillation, as evident by SDS‐PAGE (Figure 4). Our CD data confirm the previous finding that the bovine α‐lactalbumin in acidic condition shows α‐ helical structure, which decreases during fibrillation.39
In fibrillating condition at acidic pH (1.6) and 65°C, side chains of Asp get protonated. The protonated side chains of Asp catalyze the hydrolysis of peptide bonds. During hydrolysis, an intermediate cyclic anhydride is formed between the β‐carboxylic group and the amide terminus, causing cleavage of Asp‐X peptide bond or X‐Asp bond.50, 51 This leads to nicking and fragmentation, as shown by SDS‐PAGE and MALDI‐TOF (Figures 4, 5). When nicking occurs, monomers are still held by four disulfide bonds present in α‐lactalbumin and acts as a ready substrate for the fibrillation (Figure 4). Under identical conditions, HEWL, which is similar to α‐lactalbumin in structure and having four disulfide bonds, also shows nicking and fragmentation due to hydrolysis of Asp‐X leading to the fibril formation.45
While following the kinetics on SDS‐PAGE, samples were reduced by adding β‐mercaptoethanol in the SDS‐PAGE sample buffer, which breaks the disulfide bonds and monomer no longer hold as intact molecule. In the reducing SDS‐PAGE, the monomer lasts only up to 84 hr, while in the non‐reduced gel, the monomer is dominant even at 132 hr (Figure 4). This confirms the nicking of α‐lactalbumin, which migrates as a monomer on SDS‐PAGE. Nicking is further confirmed on the gel as several bands appear on reducing gel while only two prominent bands are present in the non‐reduced gel.
In order to identify the region produced by nicking and fragmentation in bovine α‐lactalbumin, we mapped possible hydrolysis sites, analyzed the molecular mass, and compared with SDS‐PAGE and MALDI‐TOF data. It has a total of 13 Asp (Figure 1) which provides the site of hydrolysis as Asp‐X bond is most susceptible to acid hydrolysis. In principle, nicking can occur at any Asp‐X peptide bond, which may be held by disulfide bonds and still move as a monomer on non‐reducing SDS‐PAGE (Figure 4a,b). Apart from a monomer, two fragments on SDS‐PAGE also started appearing from 24 hr onward on the non‐reducing gel. The origin of these two bands can be traced by looking at the MALDI‐TOF mass spectra. Asp‐X hydrolysis yields two major fragments of approximately 6,900/7,300 and 5,800/5,900 and 8,285 Da, as shown by MALDI‐TOF (Figure 5). 6,900/7,300 Da fragment is most likely originating from hydrolysis at Asp37‐38 and Asp97‐98 sites containing 61–77 and 73–91 disulfide bonds. Out of 13 Asp, 11 Asp are present in the 37–97 region, making this region unstable and susceptible to further acid hydrolysis. The fragment 38–97 contains full β‐subdomain and almost complete α helix‐3 (86–98) of the α domain. Structural analysis revealed that the β‐subdomain is more flexible compare to the α‐subdomain. In the acidic molten globule, it is the β sub‐domain that becomes disordered first.33 Alternative hydrolysis at 46–47 and 97–98 positions, which contains 10 Asp, leads to 5,800/5,900 and 8,285 Da fragments as shown by MALDI‐TOF mass spectrometry (Figure 5).
The role of protein fragments in the α‐lactalbumin amyloid formation have been studied earlier also. Proteolytically cleaved, nicked α‐lactalbumin incorporates into amyloid fibrils.35 The symmetrical peptide, identical to 35–51 amino acid residue of human α‐lactalbumin and homologous peptide to β‐domain of mammalian α‐lactalbumin can form amyloid fibrils in 0.5 M glycine‐HCl, pH 2.4 containing 100 mM NaCl at 55°C showing the presence of amyloidogenic region in β‐domain.28 Evidence of seeding was observed by the addition of small tetrapeptide (TDYG and TEYG), which drastically increases the rate of fibrillogenesis of region corresponding to 35–51 of β‐domain of human α‐lactalbumin.29 In HEWL, it has been shown that the 49/53–101 fragment is amyloidogenic, which is rate‐determining for nucleation and amyloid fibril formation.44, 45
As shown in the SDS‐PAGE (Figure 4), fragmentation also coincides with AFM results where oligomerisation also starts at 24 hr (Figure 3a). Therefore, it may be concluded that the fragmentation of α‐lactalbumin initiates nucleation. However, at 24 hr, oligomers are not detected due to the limitation of ThT fluorescence as it only detects amyloid fibrils. CD spectroscopy confirms the secondary structure changes in the monomer leading to amyloid fibril formation. Raman spectroscopy data also confirm the change in secondary structure and presence of β‐sheet, which is observed in mature fibrils (Figure 6). The proposed mechanism of bovine α‐lactalbumin amyloid fibril formation has been described in Figure 9.
FIGURE 9.
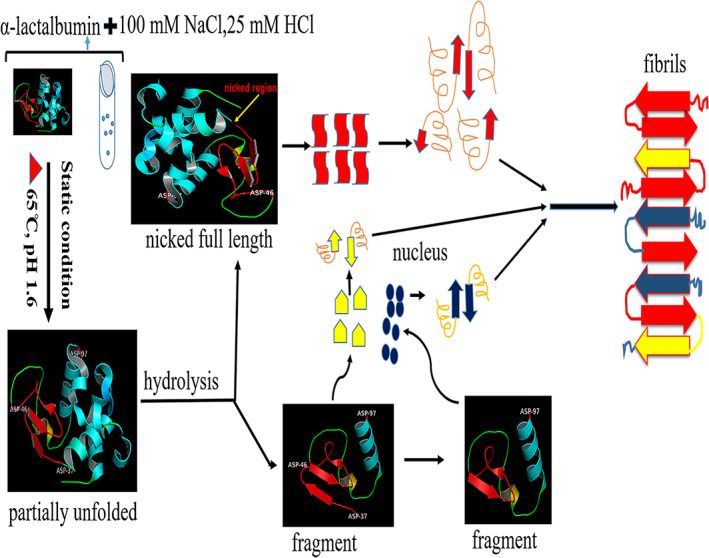
Proposed schematic representation of the mechanism of bovine α‐lactalbumin amyloid fibril formation at acidic (pH 1.6) and elevated temperature (65°C). The two fragments are shown as visible in sodium dodecyl sulphate‐poly acrylamide gel electrophoresis (SDS‐PAGE) gel. In this experimental condition, there are two possible pathways to form amyloid fibrils. The one pathway may be directly nicked α‐lactalbumin participates in fibril formation and the second pathway, after hydrolysis fragment may initiate nucleation which accelerate fibrillation of nicked as well as fragments
4. CONCLUSIONS
Given that bovine α‐lactalbumin is highly abundant in human food sources, it is of considerable interest to study amyloid fibril formation of α‐lactalbumin. While many reports exist in the literature our study emphasizes the role of specific hydrolytic fragmentation under acidic conditions and elevated temperatures. By considering amino acid sequence, location of disulfide bonds, non‐reduced and reduced SDS‐PAGE, MALDI‐TOF mass spectrometry, it can be concluded that at acidic pH and high‐temperature, hydrolysis of peptide bonds causes nicking and fragmentation of α‐lactalbumin which initiates nucleation and later both, nicked and fragmented α‐lactalbumin, participate in the amyloid fibril formation. Formation of amyloid fibril formation is confirmed by ThT fluorescence, AFM and secondary structure changes were observed by CD and Raman spectroscopy. This study may be helpful to understand the mechanism of amyloid fibril formation in the acidic condition of the cellular environment as well as protease‐induced fragmentation and amyloid fibril formation in a number of amyloidogenic proteins. These findings can also assist in identifying if current practices in milk processing is associated with α‐lactalbumin fibril formation.
5. MATERIALS AND METHODS
Bovine α‐lactalbumin (L6010), NaCl, Tris, glycerol, sodium phosphate dibasic and monobasic, SDS and Thioflavin‐T were purchased from Sigma–Aldrich (St. Louis, USA). Tricine, Coomassie brilliant blue G‐250, and R‐250, polyacrylamide, bisacrylamide, β‐mercaptoethanol were purchased from BIO‐RAD (California USA). Mica Sheet V1 grade (from TED PELLA Canada), Calcium Fluoride substrate Raman Grade obtained from Crystran Ltd, Poole, Dorset, UK. All other chemicals were of research‐grade.
5.1. α‐Lactalbumin preparation
Lyophilized powder of bovine α‐lactalbumin 10 mg/ml was dissolved in 25 mM HCl containing 100 mM NaCl. The dissolved protein solution was filtered through a 0.45 μm pore size filter (Millipore). The Protein concentration was measured by diluting the stock solution and taking the absorbance at 280 nm by using extinction coefficient ε = 28,500 M−1 cm−1.73 The stock solution was kept at 4°C for a maximum of 3 days and diluted further to make up 400 μM for fibrillation.
5.2. Amyloid fibril formation assay by Thioflavin‐T
To follow the kinetics of α‐lactalbumin amyloid fibril formation, 400 μM protein in 25 mM HCl (pH 1.6) containing 100 mM NaCl was incubated at 65°C under the static condition in a 1.5 ml microcentrifuge tube. The 20 μl sample was collected every 8/12 hr by vortexing the incubated sample for 7 s and stored at 4°C. The stock solution of Thioflavin‐T (ThT) was prepared in water by using a molar extinction coefficient 36,000 M−1 cm−1 at λ = 412 nm.74 To detect the amyloid fibril formation, 10 μM, equimolar ratio of both, ThT and α‐lactalbumin, were taken. ThT and α‐lactalbumin were mixed in the 20 mM sodium phosphate buffer (pH 7.0) to a final concentration of 10 μM monomer equivalent in 500 μl. The emission spectra were recorded from 450 to 600 nm by excitation at 440 nm using Cary Eclipse fluorescence spectrophotometer. The slits width for excitation and emission were 5 nm each. The seeding experiments were performed by using preformed matured fibril as a seed after the sonication.
The amyloid fibril formation is a nucleation‐dependent polymerization reaction in which monomer unfolds or forms fragments that initiate the nucleus formation, and finally, elongation occurs. The amyloid fibril formation kinetics follow a sigmoidal curve in which a lag phase, an elongation phase, and a stationary phase occur. The ThT intensity of fluorescence measurement was plotted as a function of time and fitted by sigmoidal curve by the following equation:75
| (1) |
There are following denotation explain are given below:
=fluorescence intensity as a function of time t, y i = initial intercept of baseline, the intercept of final baseline with the y‐axis, and are slope of the initial and final baselines, respectively, is the time required to attain halfway through the elongation phase and is the elongation time constant. The apparent rate constant, , for the growth of fibrils is given by 1/ , and the lag time generally described as t lag= −2 .
5.3. Atomic force microscopy
The protein samples collected at different time points were diluted 40 times in 25 mM HCl (pH 1.6). A 15 μl diluted sample was applied on freshly cleaved mica sheets of the highest grade V1 (TED PELLA Inc., USA). The samples were air‐dried for 45 min at room temperature and then gently washed with filtered 2 ml double distilled water and kept overnight for air‐drying. The air‐dried samples were scanned to capture the image under intermitted contact mode using Tips Nano NSG30 probe. Au reflective coated total tip shape is tetrahedral. The last 500 nm tip shape is cylindrical with height 14–16 μm and tip radius 6 nm with force constant 40 N/m. The resonate frequency of 320 kHz, driving amplitude (Vpp) 0.450, driving frequency (Hz) 285,973, point per line 256, line per image 256 are used. The initial scan size was 10 μm × 10 μm with 1,290 × 1,290 pixel were collected. The scan speed(s/line) were 0.500. WITec alpha 300 atomic force microscope (WITec GmbH, Germany) was used for imaging. Data were analyzed by Project 5 software, and image backgrounds were subtracted by line subtraction order of three. The height of the fibrils was measured after background subtractions applying the method image cross‐section.
5.4. Kinetics of α‐lactalbumin amyloid fibril characterized by SDS‐PAGE
Kinetics of α‐lactalbumin amyloid fibril formation was monitored by Tris‐Tricine SDS‐PAGE (16.5% Acrylamide wt/vol).76 The sample collected at the different time points for ThT kinetics were mixed with sample buffer to a final protein concentration of 18 μg/well. After the mixing, the sample was heated at 95°C for 5 min and loaded on the gel. Before staining the gel, it was fixed in 50% methanol and 10% glacial acetic acid for 1 hr, then stained with 0.025% Coomassie brilliant blue (G‐250). After staining, the gels were destained in the destaining solution (methanol: glacial acetic acid: water in the ratio of 50:40:10, respectively). To analyze the α‐lactalbumin sample on reducing gel, 1 μl of β‐mercaptoethanol was added in the protein sample and kept at 4°C for 1 hr, then heated to 95°C for 5 min and loaded on the gel.
5.5. MALDI‐TOF mass spectrometry
At different time points, samples were collected by vortexing the sample then centrifuging it at 25000xg. The matrix was prepared by mixing the 10 mg/ml sinapinic acid in 70% acetonitrile and 0.1% Trifluoroacetic acid (TFA) (vol/vol). The sample and the matrix were mixed in the ratio of 1:10 (vol/vol). After the drying of the matrix and sample, the measurements were run on a SCIEX TOF/TOF 5800 mass spectrometer equipped with a 349 nm of nitrogen laser. Spectra were recorded in the linear positive mode. Data were acquired by averaging 500 satisfactory shots.
5.6. Raman spectroscopy
The native protein solution and amyloid fibril sample of 30 μl each were poured on the Calcium fluoride substrate. The poured samples were left overnight for air drying. The system was calibrated using a silicon substrate in order to test the standard band position and the intensity. The spectra were collected on WITec alpha 300 R confocal Raman spectroscopy system (WITec GmbH Germany). A laser of 532 nm was excited on the sample with a power of 20.2 mW, 600 I/mm grating, and a 100 X plane Fluor objective with numerical aperture of 0.9 at a working distance of 4.7 mm, and data were collected over the range of 100–3,600 cm−1. Each spectrum consists of 20 accumulations with an integration time of 2 s. After collecting the spectra, all the data were analyzed in WITec project 5.0 software. All the original spectra were smoothened by Savitzky–Golay with order 4. Cosmic ray and background were removed by filter size 1, dynamic factor 15, and for background removal, shape size 300 and noise factor 100 were taken. After the smoothness, cosmic ray removal, and background subtraction, the spectra were deconvoluted by selecting the Gaussian peak type option. Here we have taken the amide‐I (1,600–1,700 cm−1) region for deconvolution to see the sub‐band with respect to their conformational change during amyloid fibril formation.
5.7. CD spectroscopy
CD measurement of α‐lactalbumin was done on a JASCO J‐815 CD Spectropolarimeter (Jasco, Japan) attached with a peltier. The aliquots were taken by vortexing the samples, which were incubated in amyloidogenic condition and diluted to 15 μM. Far‐UV CD (200–250 nm) spectra were recorded in a quartz cuvette of 1 mm path length. The data were collected with the step size of 0.1 nm, the bandwidth of 1 nm, and the scanning speed of 20 nm/min. Total seven accumulations were collected and averaged. All the data were subtracted with buffer as a reference.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Rahamtullah: Investigation; methodology; writing‐original draft; writing‐review & editing. Rajesh Mishra: Conceptualization; funding acquisition; investigation; supervision; writing‐original draft; writing‐review & editing.
ACKNOWLEDGMENTS
The authors acknowledge Mr Saroj K. Jha for technical help in AFM and Raman spectroscopy at the Advanced Instrumentation Research Facility (AIRF), Jawaharlal Nehru University. The authors also would like to acknowledge Dr Nipendra Sigh and Mr Subodh Jain for their technical assistance in MALDI‐TOF experiments at the advanced technology platform center, RCB, Faridabad, Haryana. We thank DBT‐BUILDER project (BT/PR/5006/INF/153/2012) and UPE‐II, JNU. Rahamtullah is a recipient of DBT fellowship.
Rahamtullah, Mishra R. Nicking and fragmentation are responsible for α‐lactalbumin amyloid fibril formation at acidic pH and elevated temperature. Protein Science. 2021;30:1919–1934. 10.1002/pro.4144
Funding information Department of Biotechnology, Ministry of Science and Technology, Grant/Award Number: BT/PR/5006/INF/153/2012
REFERENCES
- 1.Chiti F, Dobson CM. Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu Rev Biochem. 2017;86:27–68. [DOI] [PubMed] [Google Scholar]
- 2.Eisenberg D, Jucker M. The amyloid state of proteins in human diseases. Cell. 2012;148:1188–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gallardo R, Ranson NA, Radford SE. Amyloid structures: Much more than just a cross‐β fold. Curr Opin Struct Biol. 2020;60:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Iadanza MG, Jackson MP, Hewitt EW, Ranson NA, Radford SE. A new era for understanding amyloid structures and disease. Nat Rev Mol Cell Biol. 2018;19:755–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, Blake CCF. Common core structure of amyloid fibrils by synchrotron X‐ray diffraction. J Mol Biol. 1997;273:729–739. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin AJ, Knowles TPJ, Tartaglia GG, et al. Metastability of native proteins and the phenomenon of amyloid formation. J Am Chem Soc. 2011;133:14160–14163. [DOI] [PubMed] [Google Scholar]
- 7.Otzen D, Riek R. Functional amyloids. Cold Spring Harb Perspect Biol. 2019;11:a033860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thakur AK, Sinha N. ToxPoint: A need for regulatory thinking for amyloid‐based biomaterials. Toxicol Sci. 2021;179:1–2. [DOI] [PubMed] [Google Scholar]
- 9.Arosio P, Knowles TP, Linse S. On the lag phase in amyloid fibril formation. Phys Chem Chem Phys. 2015;17:7606–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cremades N, Cohen SIA, Deas E, et al. Direct observation of the interconversion of normal and toxic forms of α‐synuclein. Cell. 2012;149:1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Selkoe DJ, Hardy J. The amyloid hypothesis of Alzheimer's disease at 25 years. EMBO Mol Med. 2016;8:595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen GF, Xu TH, Yan Y, et al. Amyloid beta: Structure, biology and structure‐based therapeutic development. Acta Pharmacol Sin. 2017;38:1205–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vldal R, Franglone B, Rostagno A, et al. A stop‐codon mutation in the BRI gene associated with familial British dementia. Nature. 1999;399:776–781. [DOI] [PubMed] [Google Scholar]
- 14.Häggqvist BO, Näslund J, Sletten K, et al. Medin: An integral fragment of aortic smooth muscle cell‐produced lactadherin forms the most common human amyloid. Proc Natl Acad Sci U S A. 1999;96:8669–8674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichner T, Radford SE. Understanding the complex mechanisms of β2‐microglobulin amyloid assembly. FEBS J. 2011;278:3868–3883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jahn TR, Parker MJ, Homans SW, Radford SE. Amyloid formation under physiological conditions proceeds via a native‐like folding intermediate. Nat Struct Mol Biol. 2006;13:195–201. [DOI] [PubMed] [Google Scholar]
- 17.McParland VJ, Kad NM, Kalverda AP, et al. Partially unfolded states of β‐microglobulin and amyloid formation in vitro. Biochemistry. 2000;39:8735–8746. [DOI] [PubMed] [Google Scholar]
- 18.Cerini C, Peyrot V, Garnier C, et al. Biophysical characterization of lithostathine. Evidences for a polymeric structure at physiological pH and a proteolysis mechanism leading to the formation of fibrils. J Biol Chem. 1999;274:22266–22274. [DOI] [PubMed] [Google Scholar]
- 19.Solomon JP, Page LJ, Balch WE, Kelly JW. Gelsolin amyloidosis: Genetics, biochemistry, pathology and possible strategies for therapeutic intervention. Crit Rev Biochem Mol Biol. 2012;47:282–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ratnaswamy G, Koepf E, Bekele H, Yin H, Kelly JW. The amyloidogenicity of gelsolin is controlled by proteolysis and pH. Chem Biol. 1999;6:293–304. [DOI] [PubMed] [Google Scholar]
- 21.Hiraoka Y, Sewaga T, Kuwajima K, Sugai S, Murani N. α‐Lactalbumin: A calcium metalloprotein. Biochem Biophys Res Commun. 1980;95:1098–1104. [DOI] [PubMed] [Google Scholar]
- 22.Brodbeck U, Denton WL, Tanahashi N, Ebner KE. The isolation and identification of the B protein of lactose synthetase as alpha‐lactalbumin. J Biol Chem. 1967;242:1391–1397. [PubMed] [Google Scholar]
- 23.Sugai S, Ikeguchi M. Conformational comparison between α‐lactalbumin and lysozyme. Adv Biophys. 1994;30:37–84. [DOI] [PubMed] [Google Scholar]
- 24.Brew K, Castellino FJ, Vanaman TC, Hill RL. The complete amino acid sequence of bovine alpha‐lactalbumin. J Biol Chem. 1970;245:4570–4582. [PubMed] [Google Scholar]
- 25.Chrysina ED, Brew K, Acharya KR. Crystal structures of Apo‐ and holo‐bovine α‐lactalbumin at 2.2‐Å resolution reveal an effect of calcium on inter‐lobe interactions. J Biol Chem. 2000;275:37021–37029. [DOI] [PubMed] [Google Scholar]
- 26.Dolgikh DA, Gilmanshin RI, Brazhnikov EV, Bychkova VE, Semisotnov GV. α‐Lactalbumin: Compact state with fluctuating tertiary structure? FEBS Lett. 1981;136:311–315. [DOI] [PubMed] [Google Scholar]
- 27.Kuwajima K. The molten globule state of α‐actalbumin. FASEB J. 1996;10:102–109. [DOI] [PubMed] [Google Scholar]
- 28.Otte J, Ipsen R, Bauer R, Bjerrum MJ, Waninge R. Formation of amyloid‐like fibrils upon limited proteolysis of bovine α‐lactalbumin. Int Dairy J. 2005;15:219–229. [Google Scholar]
- 29.Egorov V, Solovyov K, Grudinina N, et al. Atomic force microscopy study of peptides homologous to beta‐domain of alpha‐lactalbumins. Protein Pept Lett. 2007;14:471–474. [DOI] [PubMed] [Google Scholar]
- 30.Egorov VV, Lebedev DV, Shaldzhyan AA, et al. A conservative mutant of a proteolytic fragment produced during fibril formation enhances fibrillogenesis. Prion. 2014;8:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goers J, Permyakov SE, Permyakov E a, Uversky VN, Fink AL. Conformational prerequisites for alpha‐lactalbumin fibrillation. Biochemistry. 2002;41:12546–12551. [DOI] [PubMed] [Google Scholar]
- 32.Bomhoff G, Sloan K, McLain C, Gogol EP, Fisher MT. The effects of the flavonoid baicalein and osmolytes on the Mg2+ accelerated aggregation/fibrillation of carboxymethylated bovine 1SS‐α‐lactalbumin. Arch Biochem Biophys. 2006;453:75–86. [DOI] [PubMed] [Google Scholar]
- 33.De Laureto PP, Scaramella E, Frigo M, et al. Limited proteolysis of bovine α‐lactalbumin: Isolation and characterization of protein domains. Protein Sci. 1999;8:2290–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Laureto PP, Vinante D, Scaramella E, Frare E, Fontana A. Stepwise proteolytic removal of the β subdomain in α‐lactalbumin: The protein remains folded and can form the molten globule in acid solution. Eur J Biochem. 2001;268:4324–4333. [DOI] [PubMed] [Google Scholar]
- 35.De Laureto PP, De Filippis V, Di Bello M, Zambonin M, Fontana A. Probing the molten globule state of a‐lactalbumin by limited proteolysis. Biochemistry. 1995;34:12596–12604. [DOI] [PubMed] [Google Scholar]
- 36.De Laureto PP, Frare E, Battaglia F, Mossuto MF, Uversky VN, Fontana A. Protein dissection enhances the amyloidogenic properties of α‐lactalbumin. FEBS J. 2005;272:2176–2188. [DOI] [PubMed] [Google Scholar]
- 37.Kulig M, Ecroyd H. The small heat‐shock protein αB‐crystallin uses different mechanisms of chaperone action to prevent the amorphous versus fibrillar aggregation of α‐lactalbumin. Biochem J. 2012;448:343–352. [DOI] [PubMed] [Google Scholar]
- 38.Sanders HM, Jovcevski B, Carver JA, Pukala TL. The molecular chaperone β‐casein prevents amorphous and fibrillar aggregation of α‐lactalbumin by stabilisation of dynamic disorder. Biochem J. 2020;477:629–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao H, Tuominen EKJ, Kinnunen PKJ. Formation of amyloid fibers triggered by phosphatidylserine‐containing membranes. Biochemistry. 2004;43:10302–10307. [DOI] [PubMed] [Google Scholar]
- 40.Yang F, Zhang M, Zhou BR, Chen J, Liang Y. Oleic acid inhibits amyloid formation of the intermediate of α‐lactalbumin at moderately acidic pH. J Mol Biol. 2006;362:821–834. [DOI] [PubMed] [Google Scholar]
- 41.Håkansson A, Zhivotovsky B, Orrenius S, Sabharwal H, Svanborg C. Apoptosis induced by a human milk protein. Proc Natl Acad Sci U S A. 1995;92:8064–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ho JCS, Nadeem A, Svanborg C. HAMLET – A protein‐lipid complex with broad tumoricidal activity. Biochem Biophys Res Commun. 2017;482:454–458. [DOI] [PubMed] [Google Scholar]
- 43.Vanaman TC, Brew K, Hill RL. The disulfide of bovine alpha‐lactalbumin. J Biol Chem. 1970;245:4583–4590. [PubMed] [Google Scholar]
- 44.Frare E, Polverino De Laureto P, Zurdo J, Dobson CM, Fontana A. A highly amyloidogenic region of hen lysozyme. J Mol Biol. 2004;340:1153–1165. [DOI] [PubMed] [Google Scholar]
- 45.Mishra R, Sörgjerd K, Nyström S, Nordigården A, Yu YC, Hammarström P. Lysozyme amyloidogenesis is accelerated by specific nicking and fragmentation but decelerated by intact protein binding and conversion. J Mol Biol. 2007;366:1029–1044. [DOI] [PubMed] [Google Scholar]
- 46.Bergström J, Gustavsson A, Hellman U, et al. Amyloid deposits in transthyretin‐derived amyloidosis: Cleaved transthyretin is associated with distinct amyloid morphology. J Pathol. 2005;206:224–232. [DOI] [PubMed] [Google Scholar]
- 47.Teng J, Turbat‐Herrera EA, Herrera GA. Extrusion of amyloid fibrils to the extracellular space in experimental mesangial AL‐amyloidosis: Transmission and scanning electron microscopy studies and correlation with renal biopsy observations. Ultrastruct Pathol. 2014;38:104–115. [DOI] [PubMed] [Google Scholar]
- 48.Lindgren M, Sörgjerd K, Hammarström P. Detection and characterization of aggregates, prefibrillar amyloidogenic oligomers, and protofibrils using fluorescence spectroscopy. Biophys J. 2005;88:4200–4212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Biancalana M, Koide S. Molecular mechanism of Thioflavin‐T binding to amyloid fibrils. Biochim Biophys Acta Proteins Proteom. 2010;1804:1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schultz J. Cleavage at aspartic acid. Methods Enzymol. 1967;11:255–263. [Google Scholar]
- 51.Inglis AS. Cleavage at aspartic acid. Methods Enzymol. 1983;91:324–332. [DOI] [PubMed] [Google Scholar]
- 52.Kurouski D, Van Duyne RP, Lednev IK. Exploring the structure and formation mechanism of amyloid fibrils by Raman spectroscopy: A review. Analyst. 2015;140:4967–4980. [DOI] [PubMed] [Google Scholar]
- 53.Rygula A, Majzner K, Marzec KM, Kaczor A, Pilarczyk M, Baranska M. Raman spectroscopy of proteins: A review. J Raman Spectrosc. 2013;44:1061–1076. [Google Scholar]
- 54.Ma CY, Rout MK, Chan WM, Phillips DL. Raman spectroscopic study of oat globulin conformation. J Agric Food Chem. 2000;48:1542–1547. [DOI] [PubMed] [Google Scholar]
- 55.Yu NT. Comparison of protein structure in crystals, in lyophilized state, and in solution by laser Raman scattering III α‐Lactalbumin. J Am Chem Soc. 1974;96:4664–4668. [DOI] [PubMed] [Google Scholar]
- 56.Shashilov VA, Sikirzhytski V, Popova LA, Lednev IK. Quantitative methods for structural characterization of proteins based on deep UV resonance Raman spectroscopy. Methods. 2010;52:23–37. [DOI] [PubMed] [Google Scholar]
- 57.Oshokoya OO, Roach CA, JiJi RD. Quantification of protein secondary structure content by multivariate analysis of deep‐ultraviolet resonance Raman and circular dichroism spectroscopies. Anal Methods. 2014;6:1691–1699. [Google Scholar]
- 58.Dolui S, Mondal A, Roy A, et al. Order, disorder, and reorder state of lysozyme: Aggregation mechanism by Raman spectroscopy. J Phys Chem B. 2020;124:50–60. [DOI] [PubMed] [Google Scholar]
- 59.Xing L, Fan W, Chen N, Li M, Zhou X, Liu S. Amyloid formation kinetics of hen egg white lysozyme under heat and acidic conditions revealed by Raman spectroscopy. J Raman Spectrosc. 2019;50:629–640. [Google Scholar]
- 60.Liang M, Chen VYT, Chen HL, Chen W. A simple and direct isolation of whey components from raw milk by gel filtration chromatography and structural characterization by Fourier transform Raman spectroscopy. Talanta. 2006;69:1269–1277. [DOI] [PubMed] [Google Scholar]
- 61.Mensch C, Johannessen C. The influence of the amino acid side chains on the Raman optical activity spectra of proteins. ChemPhysChem. 2019;20:5. [DOI] [PubMed] [Google Scholar]
- 62.Moretti M, Allione M, Marini M, et al. Raman study of lysozyme amyloid fibrils suspended on super‐hydrophobic surfaces by shear flow. Microelectron Eng. 2017;178:194–198. [Google Scholar]
- 63.Bhattacharya S, Ghosh S, Pandey NK, Chaudhury S, Dasgupta S, Roy A. Distribution of protein Ramachandran psi (ψ) angle using non‐resonance visible Raman scattering measurements. J Phys Chem B. 2013;117:13993–14000. [DOI] [PubMed] [Google Scholar]
- 64.Roach CA, Simpson JV, JiJi RD. Evolution of quantitative methods in protein secondary structure determination via deep‐ultraviolet resonance Raman spectroscopy. Analyst. 2012;137:555–562. [DOI] [PubMed] [Google Scholar]
- 65.JiJi RD, Balakrishnan G, Hu Y, Spiro TG. Intermediacy of poly(L‐proline) II and beta‐strand conformations in poly(L‐lysine) beta‐sheet formation probed by temperature‐jump/UV resonance Raman spectroscopy. Biochemistry. 2006;45:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Spiro TG, Gaber BP. Laser Raman scattering as a probe of protein structure. Annu Rev Biochem. 1977;46:553–572. [DOI] [PubMed] [Google Scholar]
- 67.Kuwajima K, Hiraoka Y, Ikeguchi M, Sugai S. Comparison of the transient folding intermediates in lysozyme and α‐lactalbumin. Biochemistry. 1985;24:874–881. [DOI] [PubMed] [Google Scholar]
- 68.Goto Y, Adachi M, Muta H, So M. Salt‐induced formations of partially folded intermediates and amyloid fibrils suggests a common underlying mechanism. Biophys Rev. 2018;10:493–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Raman B, Chatani E, Kihara M, et al. Critical balance of electrostatic and hydrophobic interactions is required for beta 2‐microglobulin amyloid fibril growth and stability. Biochemistry. 2005;44:1288–1299. [DOI] [PubMed] [Google Scholar]
- 70.DuBay KF, Pawar AP, Chiti F, Zurdo J, Dobson CM, Vendruscolo M. Prediction of the absolute aggregation rates of amyloidogenic polypeptide chains. J Mol Biol. 2004;341:1317–1326. [DOI] [PubMed] [Google Scholar]
- 71.Pawar AP, DuBay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. Prediction of “aggregation‐prone” and “aggregation‐ susceptible” regions in proteins associated with neurodegenerative diseases. J Mol Biol. 2005;350:379–392. [DOI] [PubMed] [Google Scholar]
- 72.Jones S, Manning J, Kad NM, Radford SE. Amyloid‐forming peptides from β2‐microglobulin ‐ insights into the mechanism of fibril formation in vitro. J Mol Biol. 2003;325:249–257. [DOI] [PubMed] [Google Scholar]
- 73.Mach H, Middaugh CR, Lewis RV. Statistical determination of the average values of the extinction coefficients of tryptophan and tyrosine in native proteins. Anal Biochem. 1992;200:74–80. [DOI] [PubMed] [Google Scholar]
- 74.Groenning M, Olsen L, van de Weert M, Flink JM, Frokjaer S, Jørgensen FS. Study on the binding of Thioflavin T to β‐sheet‐rich and non‐β‐sheet cavities. J Struct Biol. 2007;158:358–369. [DOI] [PubMed] [Google Scholar]
- 75.Gade Malmos K, Blancas‐Mejia LM, Weber B, et al. ThT 101: A primer on the use of thioflavin T to investigate amyloid formation. Amyloid. 2017;24:1–16. [DOI] [PubMed] [Google Scholar]
- 76.Schägger H. Tricine‐SDS‐PAGE. Nat Protoc. 2006;1:16–22. [DOI] [PubMed] [Google Scholar]


