Abstract
Cross seeding between amyloidogenic proteins in the gut is receiving increasing attention as a possible mechanism for initiation or acceleration of amyloid formation by aggregation‐prone proteins such as αSN, which is central in the development of Parkinson's disease (PD). This is particularly pertinent in view of the growing number of functional (i.e., benign and useful) amyloid proteins discovered in bacteria. Here we identify two amyloidogenic proteins, Pr12 and Pr17, in fecal matter from PD transgenic rats and their wild type counterparts, based on their stability against dissolution by formic acid (FA). Both proteins show robust aggregation into ThT‐positive aggregates that contain higher‐order β‐sheets and have a fibrillar morphology, indicative of amyloid proteins. In addition, Pr17 aggregates formed in vitro showed significant resistance against FA, suggesting an ability to form highly stable amyloid. Treatment with proteinase K revealed a protected core of approx. 9 kDa. Neither Pr12 nor Pr17, however, affected αSN aggregation in vitro. Thus, amyloidogenicity does not per se lead to an ability to cross‐seed fibrillation of αSN. Our results support the use of proteomics and FA to identify amyloidogenic protein in complex mixtures and suggests that there may be numerous functional amyloid proteins in microbiomes.
Keywords: fibrillation, functional amyloid, microbiome, Parkinson's disease, proteomics
Abbreviations
- aa
amino acids
- aSN
a‐synuclein
- CBB
Coomassie Brilliant Blue
- CFU
colony forming unit
- CNS
central nervous system
- E. coli
Escherichia coli
- EECs
enteroendocrine cells
- ENS
enteric nervous system
- FA
formic acid
- FTIR
Fourier transform infrared
- FuBAs
functional bacterial amyloids
- GI
gastrointestinal
- LFQ
label‐free quantification
- MAMPs
microbe‐associated molecular patterns
- MQ
milli‐Q water
- PD
Parkinson's disease
- ProtK
proteinase K
- RT
room temperature
- SDS‐PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SEC
size‐exclusion chromatography
- SP
signal peptide
- TEM
transmission electron microscopy
- Tg
transgenic
- ThT
thioflavin T
- TLRs
Toll‐like receptors
- UPLC
ultra high‐performance liquid chromatography
- WT
wild type
1. INTRODUCTION
Parkinson's disease (PD) is a neurodegenerative movement disorder that affects 1–2% of all individuals above the age of 60.1 PD is characterized by the loss of dopaminergic neurons in the substantia nigra pars compacta (SNpc) of the midbrain and the accumulation of intracytoplasmic protein deposits referred to as Lewy bodies (LBs) or Lewy neurites (LNs).2 The LBs and LNs have been shown to be composed primarily of β‐sheet‐rich amyloid deposits of the intrinsically disordered protein α‐synuclein (αSN)3 which is believed to be a key player in the development of PD.
Within the cell, αSN is found in high concentrations at presynaptic structures.4, 5 It has been suggested that αSN can spread via neuron‐to‐neuron propagation6 and progression of sporadic PD has been proposed to happen as a caudo‐rostral process (moving from the lower brain stem via the basal midbrain and forebrain to the cerebral cortex).2 Outside the central nervous system (CNS), LBs and LNs have also been observed in the enteric nervous system (ENS) in early stages of PD.7 Recently it has even been proposed that there are two subtypes of PD, namely brain‐first and body‐first, depending on their place of origin.8 Spreading from the ENS to the CNS has been suggested to happen through the vagus nerve as vagotomised individuals have a decreased risk for subsequently developing PD,9 and this has been confirmed in mouse models.10 Further, enteric neurons secrete αSN under neuronal control.11 Studies in rats have also shown that human αSN, injected into the intestinal wall, can be transported to the dorsal motor nucleus of the vagus via the vagal nerve.12 Similarly, injection of recombinant αSN pre‐formed fibrils into the gastric wall of mice was shown to result in LB pathology in the brainstem, and this was dependent on retrograde transport through the vagus nerve.13
The great majority of neurons in the ENS is located in the myenteric and submucosal plexuses in the wall of the gastrointestinal (GI) tract.14 Enteric nerve fibers extend through the different layers of the GI tract and have been shown to connect directly with enteroendocrine cells (EECs).15 Both EECs and intestinal epithelial cells express Toll‐like receptors (TLRs) 1, 2 and 416 and are therefore able to recognize different structures, often referred to as microbe‐associated molecular patterns (MAMPs), expressed by, for example, gut bacteria. One example is the extracellular curli fibrils, which are aggregates of the CsgA protein produced by Escherichia coli (E. coli) and a wide range of other bacteria within the Enterobacteriaceae.17 These functional amyloid fibrils are recognized by TLR1 and TLR2 and mediate interleukin 1β production.18 This suggests that MAMPs produced by bacteria in our microbiome could interact with our CNS through the ENS, raising the possibility that αSN aggregation could be initiated through contacts with, for example, microbial proteins. In support of the pro‐aggregatory role of CsgA, the amount of αSN deposits in aged rats and C. elegans PD models was increased after being orally fed with curli‐producing E. coli compared to animals that had been fed with a curli‐deficient E. coli strain.19 Similar experiments have been performed in an transgenic (Tg) mouse model overexpressing αSN and again showed that exposure to curli correlated with increased αSN pathology in the gut and brain together with GI and motor deficits.20 Introducing curli‐producing E. coli in wild‐type (WT) mice had no effect on motor functions, suggesting that curli works in concert with other predisposing factors (like αSN overexpression) to cause disease.20
Functional bacterial amyloid (FuBA) such as CsgA and its counterpart FapC in Pseudomonas species serve multiple roles in bacteria. They are unusually stable and often resist high concentrations of surfactant, denaturant or organic acids as well as being resistant to proteolysis. These robust properties may explain their ability to strengthen bacterial biofilms mechanically21, 22 and increase their resistance to, for example, antibiotics.23 Whether bacterial biofilms are actually formed in the (healthy) human GI tract is unclear. Conditions like the high flow rate through parts of the GI tract (2–4 hr through the small intestine), gastric acid and the production of mucus by goblet cells are believed to make it difficult for bacteria to attach to at least the upper part of the digestive tract. However, the transit time is markedly longer in the colon (~60 hr),24 and some bacteria produce hydrolytic enzymes that can break down the glycoproteins in the mucus and use the mucus layer as an energy and carbon source.25 In addition, many bacteria within the GI tract can form biofilm on medical devices like feeding tubes26 and food particles27, and E. coli strains isolated from the GI tract can produce both extracellular cellulose and curli.28
Here we build on the observed correlation between FuBA and αSN aggregation. We analyze fecal samples from three transgenic PD rats and three WT rats for the presence of FuBA and their possible link to αSN aggregation. Due to the high stability of the FuBA structure, the amyloid state is insoluble under most denaturing conditions and only dissociate into monomers at high concentrations (80–100%) of formic acid (FA).29, 30 Thus, by treating complex samples with increasing concentrations of FA and identifying soluble proteins by trypsin digestion and LC–MS/MS analysis, potential FuBA can be identified by their markedly increased abundance at high (80–100%) FA concentrations.31 In this way, we identify 365 candidates, which were further analyzed bioinformatically to narrow the list to the two most promising candidates (here called Pr12 and Pr17). These were expressed recombinantly and examined biophysically, demonstrating significant tendencies to form amyloid. However, Pr12/Pr17 seeds did not promote αSN fibrillation. Thus, amyloidogenicity per se does not imply an ability to cross‐seed fibrillation of αSN.
2. RESULTS
2.1. Identification of potential amyloid protein candidates in the rat microbiome
Several studies have suggested that the human microbiome is changed in PD patients compared to healthy controls.32, 33, 34, 35, 36, 37, 38, 39, 40, 41, 42 The object of this study was not to attempt to elucidate statistically significant differences between healthy and PD‐afflicted rats, but rather to carry out a more general assessment of the occurrence of amyloidogenic proteins (and thus potentially functional amyloid) in the microbiome. Nevertheless, given the potential involvement of the microbiome in the development of PD, we decided to compare healthy rats with a rat model of PD that overexpresses full‐length human αSN and develops synucleinopathies both in the CNS and in the PNS.43 Microbial community profiling using amplicon primers targeting the V1–V3 variable region of the 16S rRNA gene revealed an increased abundance of Lachnospiraceae in the PD rats, whereas Lactobacillaceae dominated the WT rats (Figure 1a). However, the relative abundance of most other taxa varied greatly between replicates. Principal coordinate and ANOSIM analyses were carried out on the exact amplicon sequence variants (ASVs) to investigate if there was a statistical difference between the WT and PD rats (Figure S1). A separation along the first principal coordinate (PCo1) was observed between the WT and PD samples. However, the separation was not significant according to the permutations tests on the ordinated data (R = 0.7037; p‐value = .1; 719 permutations), reflecting the small sample size. Nevertheless, the samples provided an opportunity to search for possible FuBA through the differential solubilization of proteins at higher concentrations of FA, as described in Section 4.
FIGURE 1.
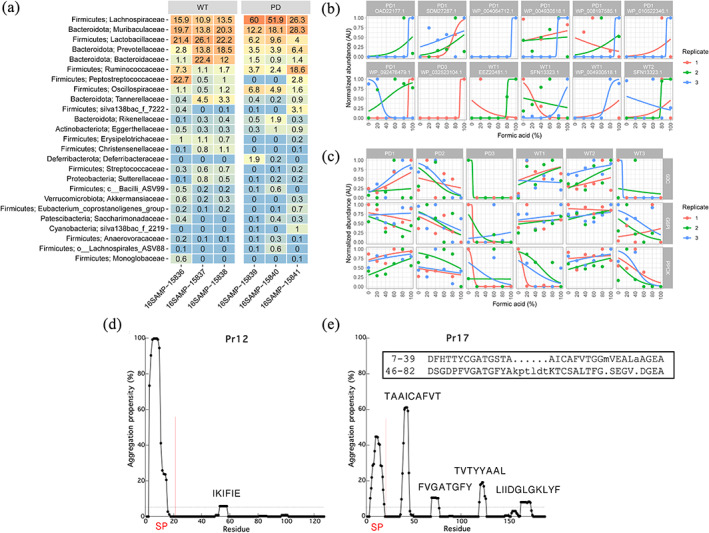
Identification of potential amyloid protein candidates. (a) Percent relative abundance of the top 25 bacterial families in wild type (WT) and Parkinson's disease (PD) rats. The data is based on V1–V3 ASVs classified based on the AutoTax processed SILVA 138 SSURef Nr99 database (77). Phylum names are provided together with the family names. (b) The 11 hit proteins proposed to contain a secretion signal peptide (there are 12 panels because SFN13323.1 is found in both WP1 and WT2). Data points can be hidden behind each other in some of the graphs. Not all proteins were identified in all three replicates. (c) Examples of negative controls including the household proteins glycose‐6‐phosphate isomerase (G6PI), 60 kDa chaperonin (60C) and phosphoenolpyruvate carboxykinase (PPCK). (d) TANGO analysis of Pr12 and (e) Pr17. The vertical red lines show the location of the signal peptide (SP). The horizontal dashed line represents an aggregation propensity value of 5%. In addition, both protein sequences were analyzed with the RADAR tool (49) but imperfect repeats could only be identified for the Pr17 sequence (insert in [e])
Protein identification with MaxQuant resulted in the identification of between 447 (sample WT3) and 1,314 (sample PD2) proteins in each of the six samples (Table S1). The proteins were quantified using the LQF algorithm, and their stability toward FA was determined as described.31 Comparing all identified proteins from the WT samples with those identified in the PD samples showed no obvious distinction between the two phenotypes. Moreover, the LQF results were characterized by relatively low reproducibility, as a large fraction of proteins were not identified in all replicates, probably due to low abundance (Figure S2). As the protocol for identification of putative functional amyloids requires a large number of replicates with a sigmoidal signature,31 comparison between WT and PD samples was inconclusive. Therefore, we decided to manually inspect the data and hand‐pick the most probable amyloid candidates based on their stability toward FA and ignore the number of replicates by which they were identified. We identified between 22 (sample PD3) and 110 (sample PD1) amyloid candidates based on their characteristic sigmoidal signature in plots of normalized protein abundance versus FA concentration (Figure 1b, Table S1). Of these 365 proteins, 27 had at least two replicates showing a sigmoidal signature (Table S2). One particularly interesting protein is elongation factor Tu (EF‐Tu) which appears three times in the PD1 sample. In fact, EF‐Tu appears as a hit 46 times (although in most of the cases only a single replicate shows the sigmoidal signature) across all samples and equally divided between PD (22 times) and WT samples (24 times) (data not shown). EF‐Tu is interesting because the Gallibacterium anatis EF‐Tu (ID: KGQ60852.1) was recently identified as a potential functional amyloid‐forming protein,44 indicating that this FA‐approach is able to identify amyloid‐relevant proteins. Household proteins glucose‐6‐phosphate isomerase (G6PI), 60 kDa chaperonin (60C) and phosphoenolpyruvate carboxykinase (PPCK)—which were identified 71, 38 and 143 times, respectively, across all six samples—generally do not show any specific signature across the FA concentration range (Figure 1c) (though a few peptides of PPCK were identified to follow a sigmoidal behavior and consequently led to the protein's inclusion in Table S1).
Because many functional amyloid systems are dependent on secretion through the Sec translocon, all protein hits were analyzed with the three different signal peptide prediction tools SignalP 4.1,45 DeepSig46 and SignalP 547 and ranked according to the probability of containing a signal peptide. From this analysis, 11 proteins were identified (Table 1). Out of these 11 proteins, only three proteins were predicted to contain a signal peptide with all three tools. We decided to select two proteins for recombinant expression to investigate their amyloidogenicity experimentally. We chose to limit ourselves to proteins predicted to contain a signal peptide by all three signal peptide predictors and to exclude large (> ca. 450 residues) proteins to avoid low recombinant expression levels. This left proteins WP_032523104.1 (12 kDa without the 22 aa signal peptide) and OAD22177.1 (17 kDa without the 18 aa signal peptide) which we will from now on refer to as Pr12 and Pr17, respectively (sequences provided in Figure S3a). Pr12 was identified in sample PD3 while Pr17 was found in sample PD1. According to the Uniprot database Pr12 is a DUF1499 domain‐containing protein produced by the cyanobacterium Prochlorococcus marinus while Pr17 is a secreted protein produced by the vacuolate sulfur bacteria Candidatus Thiomargarita nelsonii. Both these organisms are aquatic, making it unlikely that these particular organisms were found in the fecal matter. Furthermore, Pr12 and Pr17 were both identified based on a single peptide hit. Thus it is most likely that the identified proteins are homologues to the two proteins identified in P. marinus and C. Thiomargarita nelsonii and derive from organisms whose genomes are not yet available. To obtain residue‐specific information on the potential aggregation of Pr12 and Pr17, we used the TANGO web server software48 to predict aggregation‐prone regions. For both proteins it was seen that the signal peptide (SP) gave a high aggregation propensity (Figure 1d,e), but interestingly, Pr17 also showed four separated stretches of 8–12 residues with high aggregation propensity (aggregation propensity value >5%, dashed line) throughout the proteins sequence (Figure 1e). Both proteins were also investigated for the presence of imperfect repeats using the RADAR tool.49 Whereas no repeats could be found for Pr12, two repeats were identified in the Pr17 sequence (Figure 1e, insert). The repeats showed low conservation but, interestingly, two of the highly amyloidogenic stretches (TAAICAFVT and FVGATGFY) were covered by the repeat sequences. Note that neither Pr12 nor Pr17 showed any significant sequence similarity with either CsgA or FapC according to the Clustal Omega web server.
TABLE 1.
Eleven proteins from the PD microbiome containing at least two replicates with sigmoidal solubility curves and predicted to have a signal peptide
| Protein ID | Software with a positive signal peptide prediction | Mature protein length (aa) |
|---|---|---|
| OAD22177.1 (Pr17) | DeepSig, SignalP 4.1, SignalP 5 | 170 |
| SDM27287.1 | DeepSig, SignalP 4.1, SignalP 5 | 480 |
| WP_032523104.1 (Pr12) | DeepSig, SignalP 4.1, SignalP 5 | 105 |
| WP_004064712.1 | SignalP 4.1, SignalP 5 | 1,475 |
| WP_010522346.1 | SignalP 4.1, SignalP 5 | 808 |
| EEZ20481.1 | SignalP 5 | 380 |
| SFN13323.1 | SignalP 4.1 | 411 |
| SFR13334.1 | SignalP 5 | 361 |
| WP_004930518.1 | SignalP 4.1 | 172 |
| WP_008197585.1 | SignalP 5 | 121 |
| WP_092476479.1 | SignalP 4.1 | 537 |
2.2. Pr12 and Pr17 both form ThT‐positive aggregates with amyloid characteristics
It was possible to recombinantly produce both proteins in E. coli at yields of 3.5–4.5 mg/L culture. The proteins were then purified to satisfying purity, as visualized by SDS‐PAGE, by Ni‐NTA chromatography via an N‐terminal His6 tail (Figure S3b). To analyze the two protein's propensity to form amyloid, we started by analyzing their behavior in a standard ThT fibrillation assay. ThT is a dye commonly used to study amyloid fibrillation as it shows a bright fluorescence when bound the amyloid fibrils and therefore is a suitable reporter for fibril formation. After desalting into PBS, the concentration of the two proteins was measured and aggregation of the two proteins was immediately investigated with ThT. Note that when measuring protein concentration directly after desalting, the protein solution showed light scattering (tailing of the protein peak at wavelengths >320 nm), indicating protein aggregation (data not shown). An instant increase in ThT signal could be observed for both proteins (Figure 2a,b, left panels) but where Pr17 reaches a stable end level, the signal for Pr12 starts decreasing after reaching a maximum intensity after ~6 hr. No lag phase (which is otherwise usually seen during protein fibrillation) was observed for either protein. We note a short dip in the fluorescence signal around t = 0 (Figure S4) which we ascribe to fluorescence quenching as the temperature increases to 37°C. Far‐UV circular dichroism spectra recorded for these samples did not reveal any pronounced β‐sheet peak around 215 nm, probably because only a small fraction of the protein aggregated (see below). Instead we turned to FTIR which is more suitable for analysis of insoluble material. When the samples were spun down and the pellets resuspended, FTIR analysis showed a pronounced peak around 1,622 cm−1 for Pr12 and between 1,624–1,627 cm−1 for Pr17 (Figure 2a,b, right panels). Amyloid fibrils normally absorb strongly in the 1,615–1,630 cm−1 region50 while native β‐sheets absorb at wavenumbers in the 1,630–1,643 cm−1 range.51 This indicates that Pr12 and Pr17 aggregates are amyloid‐like.
FIGURE 2.
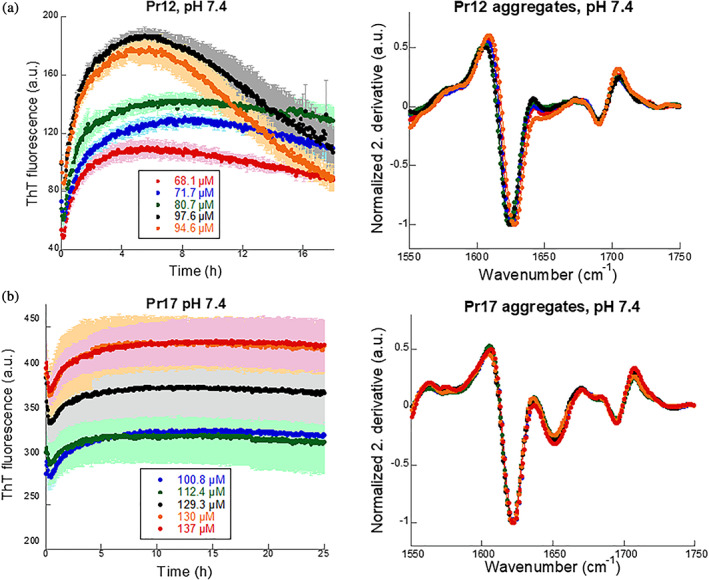
Pr12 and Pr17 readily form ThT‐positive aggregates. (a) Pr12 and (b) Pr17 were subjected to a ThT fibrillation assay (left panel) at varying concentrations immediately after desalting. FTIR (right panel) was performed on the pellet after fibrillation and showed cross‐β characteristic peaks between 1,620 and 1,630 cm−1
2.3. Fibrillation of Pr12 is sensitive to urea
To avoid premature aggregation of the proteins, the denaturant urea was added to the desalting buffer. The two proteins differed somewhat in their urea sensitivity. While ≥1.5 M urea completely inhibited aggregation of Pr12 (Figure 3a), 0.5 mg/ml Pr17 maintained aggregation up to 2.5 M urea (Figure 3c). Following fibrillation, the samples were spun down and the supernatants were analyzed with SDS‐PAGE. For Pr12, a good correlation was seen between the samples showing aggregation (0.2 M, 0.5 M, and 1 M urea) and the loss of protein from the supernatant (Figure 3b, Figure S5). However, for Pr17 all protein stayed in solution after centrifugation (data not shown) despite the increase in ThT fluorescence seen for the 0.4–2.5 M urea samples. To find out whether the difference in ThT fluorescence was caused by the formation of different species of Pr17, we analyzed these samples with SEC. The results confirmed that this was the case: Pr17 samples giving high ThT fluorescence signals (0.4–2.5 M urea) contained higher order oligomers that eluted from the column very close to the void volume, while samples with low/no ThT fluorescence signal (4 and 6 M urea) only show the Pr17 monomer peak eluting around 17 ml (Figure 3d). We cannot directly determine the size of the oligomers as they elute so close to the void volume (simple extrapolation based on calibrated standards suggest a molecular weight around 105 kDa). Given that there are no insoluble species of Pr17 under these conditions, we conclude that ThT must be binding to these oligomeric species to cause an increase in fluorescence. We ascribe the shift in monomer elution peak position with rising [urea] to either increased expansion of the protein or reduced interactions with the column. Based on calibrated standards, the apparent molecular weight of the monomer is around 60 kDa at low urea concentrations and around 15–18 kDa at higher urea concentrations, suggesting that Pr17 binds to the column unless urea is added. Earlier elution is even more pronounced for free ThT which elutes around 37 ml in 0.4 M urea but around 24 ml in 6 M urea (Figure S6a). Interestingly, ThT elution volume and end point ThT levels of Pr17 decline in a similar fashion as a function of [urea] (Figure 3e, red curve). Plotting the monomer/oligomer intensity as a function of urea concentration shows a cross‐over (i.e., roughly equal amounts of both species) at 2.25 M urea (Figure 3f).
FIGURE 3.
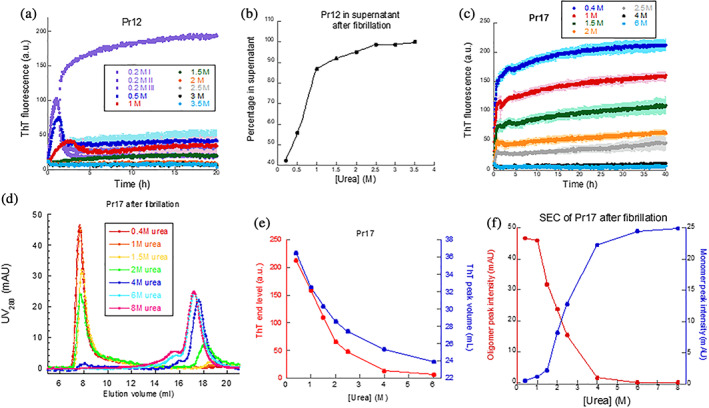
Fibrillation of Pr12 and Pr17 in the presence of urea. (a) Investigation of Pr12 aggregation in the presence of increasing concentrations of urea. (b) Amount of soluble protein left after aggregation determined by SDS‐PAGE of the supernatants after centrifugation of samples in (a). (c) Investigation of Pr17 aggregation in the presence of increasing concentrations of urea followed by (d) SEC analysis of the same samples including a control of Pr17 in 8 M urea (to keep it monomeric). Note that ThT elution peaks after 22 ml are omitted for clarity. (e) Correlation between the ThT end levels from (c) and the ThT elution volumes obtained from SEC profile in (d). (f) Correlation between the decrease in Pr17 oligomer and the increase in monomeric Pr17 with increasing urea concentrations
Pr17 contains four Cys residues (Position 32, 42, 87, and 103). SDS‐PAGE of the SEC samples under both non‐reducing (Figure S6b) and reducing (Figure S6c) conditions revealed a disulphide‐bonded dimer band under non‐reducing conditions (absent under reducing conditions), which is also seen in SEC as a shoulder to the monomer peak (Figure 3d). Oligomers form a smear in the top part of the non‐reducing gel. Note that the amount of monomer is essentially constant over 0.4–8 M urea on the reducing gel. This indicates that oligomers and dimers dissociate to monomers when exposed to SDS and reducing conditions. Under non‐reducing conditions, essentially all Pr17 is oligomeric at low [urea] (cfr. Figure 3d) but most of the oligomers dissociate on SDS‐PAGE gels, indicating that they are largely held together by non‐covalent interactions (rather than intermolecular disulphide bonds) which are disrupted by SDS.
2.4. pH dependence of Pr12 and Pr17 fibril formation
We also investigated the pH sensitivity of the two proteins' aggregation in view of the variable pH environment encountered in the GI tract. Pr12 showed a rapid increase in fluorescence within a few hours, particularly between pH 4 and 7.4, but again no lag phase could be observed (Figure 4a). Fibrillation was essentially abolished at pH 3 and pH 8. CD spectra (Figure S7a) confirm a classical β‐sheet spectrum formed by Pr12 at pH 4 after 24 hr while the spectrum at pH 6 is dominated by random coil but with features of β‐sheet structure; random coil predominates at pH 2 and above pH 6. At pH 7.4, the fibrillation curve showed an instant increase followed by a decrease after 5 hr. We tested if this was an effect of the buffer by carrying out fibrillation in 0–3.5 M urea with 60 mM Tris buffer instead of PBS at pH 7.4 but still observed the same “overshoot” behavior (Figure S8a) though with significant variation between the triplicates (Figure S8b). The increased solubilization of Pr12 at higher [urea] in Tris buffer (Figure S8c) was essentially identical to that in PBS buffer (Figure 3b). A sevenfold replication of aggregation of 0.5 mg/ml Pr12 in 60 mM Tris, pH 7.4 showed variation largely in their ThT endpoint (Figure S8d) although FTIR analysis showed identical secondary structure (Figure S8e). We therefore conclude that the ‘overshoot’ behavior reflects a stochastic aggregation behavior at this pH to form different levels of higher‐order aggregates (with consequent different levels of precipitation and light‐scattering). This indicates that Pr12 has a different aggregation mechanism at neutral pH compared to both more basic and more acidic conditions where the aggregation curves are more reproducible. Expasy predicts Pr12 to have a theoretical pI of 4.98 (https://web.expasy.org/protparam/) which cannot account for the big difference seen between pH 6 and pH 7.4.
FIGURE 4.
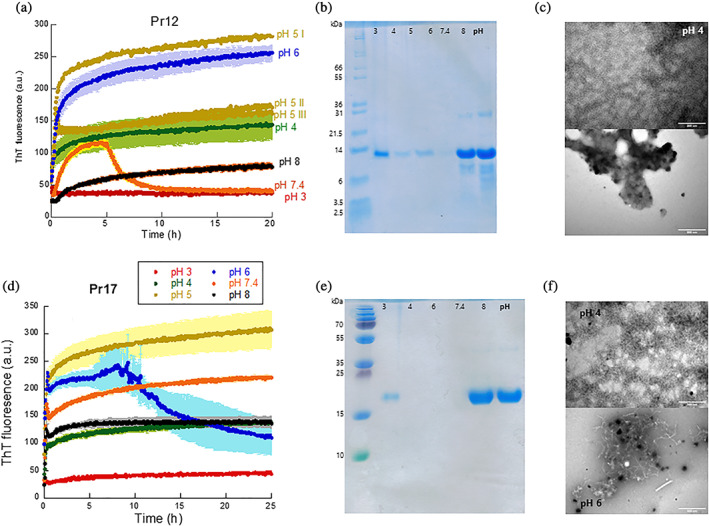
pH dependence of Pr12 and Pr17 aggregation. Pr12 (a) and Pr17 (d) were aggregated in the presence of ThT at different pH values and the samples were afterwards separated into soluble and insoluble fractions by centrifugation. The soluble part (the supernatant) was analyzed with SDS‐PAGE for Pr12 (b) and Pr17 (e) (the omitted Pr17 sample at pH 5 showed no soluble material). TEM images of Pr12 (c) and Pr17 (f) aggregates formed at either pH 4 (top) or pH 6 (bottom)
Pr17 aggregation is relatively robust to pH changes and shows an increase in ThT fluorescence at all pH values tested, although only to a very small extent at pH 3 (Figure 4d). The theoretical pI of Pr17 is 4.27. At pH 6, the aggregation curves always showed great variation between different experiments and sometimes gave an overshoot (Figure 4d) but with great variation between individual runs (Figure S9). CD spectra indicated the most pronounced β‐sheet signal at pH 4, a smaller signal at pH 6 and more random‐coil like structures above pH 6 (Figure S7b).
The extent of aggregation was measured by SDS‐PAGE of soluble (supernatant) and insoluble (pellet) protein after the ThT assay both for Pr12 (Figure 4b) and Pr17 (Figure 4e). Pr12 showed very low levels of soluble protein between pH 4 and 6, corresponding nicely with the high ThT end levels. At pH 7.4 and 8 the protein stays soluble despite the initial increase in ThT seen at pH 7.4. Similarly, for Pr17 after 25 hr fibrillation, we saw very low solubility at pH 3, 4 and 6 and much higher solubility at pH 7.4 and pH 8 despite the fact that the end‐point ThT levels are identical for pH 4 and pH 8. TEM analysis (Figure 4c,f) also showed pH‐dependent differences in aggregate structure: at pH 4, both proteins formed a mesh of very thin, worm‐like structures while at pH 6, Pr12 showed more amorphous aggregates with no apparent fibrils being present and Pr17 showed a mix of curly fibrils or worm‐like structures together with amorphous structures.
2.5. Pr17 aggregates are more stable than Pr12 toward urea and FA but not proteinase K
The high stability of the FuBA structure ensures that functional amyloids stay insoluble under denaturing conditions and only dissociate into monomers at high concentrations (80–100%) of FA.22, 29, 30 We, therefore, decided to investigate the stability of Pr12 and Pr17 aggregates toward FA. Aggregates were mixed with increasing concentrations of either urea (only Pr17, since Pr12 did not fibrillate >1 M urea) or FA (both Pr12 and Pr17) and the supernatants were analyzed with SDS‐PAGE (Figure S10, 5a,b). By normalizing to the 100% FA band (where we assume 100% of the fibrils dissolve) we can determine the denaturant concentration required to dissolve half of the fibrils, [den]50%. For Pr17 we determined [urea]50% to be ~4.4 M (as determined by visual inspection) and [FA]50% to be 32.7 ± 3.7% after fitting the data points to a sigmoidal curve. Pr12, in contrast, was highly sensitive toward FA with a [FA]50% around 0.1% (Figure 5b). The [FA]50% value of ~30% for Pr17 is only slightly lower than the stabilities of [FA]50% of around 50% we normally see for functional amyloids.52 In contrast, the [FA]50% value of 0.1% for Pr12 is similar the [FA]50% value determined for αSN fibrils.52 Thus in vitro aggregates of Pr17 show aspects of a classical functional amyloid protein while Pr12 aggregates appear much less stable.
FIGURE 5.
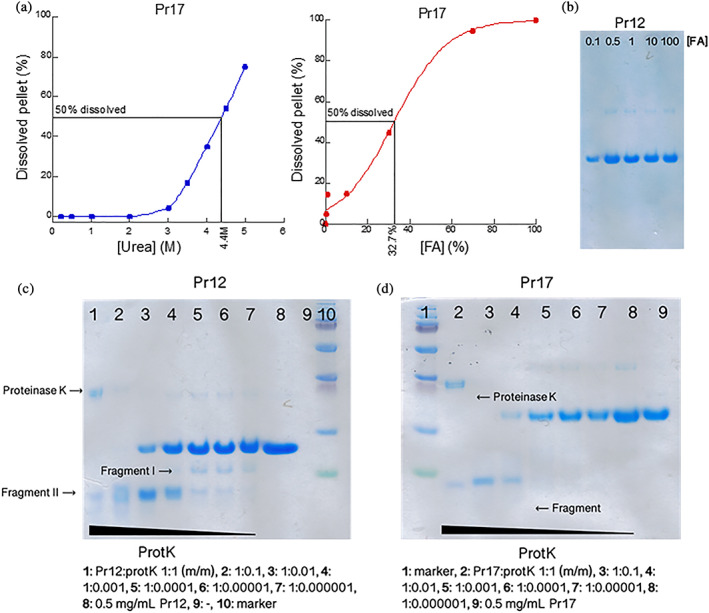
Stability investigations of Pr12 and Pr17 aggregates. (a) Aggregates formed by Pr17 were dissolved in increasing concentrations of urea (left) or formic acid (right) and the amount of dissolved protein was visualized using SDS‐PAGE after lyophilization. Quantification was performed on these gels using ImageJ. (b) Aggregates formed by Pr12 were dissolved in increasing concentrations of formic acid and dissolved protein was visualized with SDS‐PAGE after lyophilization. The Pr12 aggregates dissolved at very low formic acid concentrations. Aggregates of Pr12 (c) and Pr17 (d) were subjected to digestion with increasing ratios of protein:proteinase K up to 1:1 (m/m) followed by SDS‐PAGE analysis
We also treated protein aggregates with increasing concentrations of proteinase K (ProtK) to elucidate if the aggregates formed by the proteins—especially the more stable Pr17 aggregates—contained a protected core. In this case, we would expect to see a clear band on SDS‐PAGE after ProtK treatment. High concentrations of proteinase K (Pr12/Pr17:protK ratios of 1:1 (m/m) and 1:0.1 (m/m)) resulted in a break‐down to smaller fragment of ~7 kDa for Pr12 (Figure 5c, fragment II) and ~ 9 kDa for Pr17 (Figure 5d) at sample/ProtK mass ratios down to 1:0.01 (lane 4). This indicates a structured core present in both protein aggregates. At sample:protK (m/m) ratios of 1:0.001, appr. 40% of Pr12 (Figure 5c, lane 4) and 15% of Pr17 (Figure 5d, lane 5) remained full‐length after treatment. Interestingly, for Pr12 a slightly bigger fragment of ~11 kDa appeared when treated with lower proteinase K concentrations (Figure 5c, lane 4–7). However, the full‐length protein still dominated. This 11 kDa fragment is the first to be cleaved off (but only to a low degree as no apparent change can be seen in the intensity of the full‐length protein band) by ProtK and is fully degraded at higher ProK concentrations. The 9 kDa fragment of Pr17 appears to be relatively protected from the proteolytic attack when in the aggregated structure. However, the intensity of the band is lower at the 1:1 ratio (lane 2) than the 1:0.1 ratio (lane 3), so it is not completely resistant toward proteolytic degradation.
2.6. Co‐incubation of Pr12 and Pr17 with αSN
To study the effects of Pr12 and Pr17 on fibrillation of αSN, we compared the fibrillation of monomeric αSN in the absence and presence of these two proteins. We incubated monomeric αSN with seeds made from aggregated Pr12 or Pr17. As seen in Figure S11, neither Pr17 seeds (left) nor Pr12 seeds (right) had any significant effect on αSN fibrillation which in all cases fibrillated with a lag phase of ~5 hr.
3. DISCUSSION
3.1. Bacterial biofilm in the GI as a source of functional amyloid
Functional amyloids such as curli fibrils are often part of bacterial biofilms. Up to 40% of all bacteria form biofilms53 and it therefore comes as no surprise that biofilms and bacterial microcolonies have been observed in the human GI tract54, 55, 56 where up to 1011 colony forming units (CFU) are found per ml gut content in the colon.57 In addition, bacterial biofilms have been found associated to food particles in fecal samples from healthy individuals27, 58 and have also been visualized directly in mice.59 To date, however, studies of mucosal biofilms in the GI tract are largely limited to diseases like ulcerative colitis,60, 61 peptic ulcer disease,62 Crohn's disease and cases of diarrhea.63
Here we sought to identify amyloidogenic proteins in the microbiome of PD Tg rats and their healthy WT counterparts. One aspiration was to establish if the PD microbiome was enriched for amyloidogenic proteins, which might in turn indicate that microbial products produced in the GI tract could play a role in neurodegenerative diseases like Alzheimer's64 and PD12, 65 as suggested. However, we observed strikingly similar numbers of hit proteins identified from either of the two groups (181 hits for the three PD samples vs. 186 hits for the WT) and therefore cannot make any definite conclusions in this regard. Two proteins, here referred to as Pr12 and Pr17, were identified based on a combination of experimental (FA solubility/MS), bioinformatics analysis (the presence of a signal peptide and high amyloidogenicity across the amino acid sequence) and feasibility (a modest size compatible with recombinant expression). Both proteins were originally identified from the PD samples (Pr12 from the PD3 sample and Pr17 from the PD1 sample). The two proteins have not been biophysically characterized before but we show that both proteins readily form ThT‐positive aggregates to some extent at all pH values investigated (pH 3–8). Pr17 even aggregates in the presence of high concentrations of urea. The aggregation process, however, did not follow classical sigmoidal kinetics which involve an initial lag phase where nuclei are formed and grow.66 The absence of a lag phase is indicative of amorphous aggregation, even though fast amyloid formation has also been reported.67, 68 The aggregates formed at pH 7.4 for both proteins show a major band between 1,621 and 1,627 cm−1 when analyzed with FTIR, indicating extended β‐sheet structures like those present in amyloid fibrils. TEM images also revealed worm‐like fibril structures for both proteins. However, when the stability of the aggregates toward FA was investigated, Pr12 showed extreme sensitivity and dissolved at very low FA concentrations ([FA]50% ~ 0.1%) while Pr17 aggregates were more robust and required 32.7% FA for half of the aggregated material to dissolve. This value is lower than the 80–100% required to dissolve functional amyloid fibrils such as CsgA and FapC into protein monomers22, 29 (and also lower than the 50% value used as a criterion when searching the MS data for possible hits, cf. Section 4 and 31). However, other functional amyloids, such as the TasA fibrils produced by Bacillus subtilis, have also been found to dissolve at lower (~20%) FA concentrations69 and we therefore cannot exclude that a given protein is an amyloid based on the FA stability alone. The presence of several amyloidogenic hotspots and two imperfect repeats in the Pr17 sequence further support its robust fibrillation behavior.
3.2. pH sensitivity of fibrillation and the GI tract
It is noteworthy that Pr12 and Pr17 behave differently in terms of forming soluble versus insoluble aggregates between pH 3 and 8 in view of the great variation of intraluminal pH throughout the GI tract.70 In the stomach, the pH can be as low as 1.5 in the fasting state (though food consumption increases this within minutes to around pH 5)71 but rapidly changes to pH 6 in the duodenum and increases further to around neutral at the end of the small intestine. This is followed by a second drop to pH 5.7 in the cecum and a final increase to 6.7 when reaching the rectum.70 Since the bacterial count in the stomach is very low (< 103 CFU/ml72), we believe that the conditions in the small and large intestines (pH values ranging from 5.7 to 7.4) are particularly relevant.
3.3. Perspectives on the role of functional amyloid in PD
Based on the results presented here, we conclude that Pr17 holds great promise as a novel functional amyloid protein while Pr12 instead appears to form less structured β‐aggregates. The presence of a relatively protease‐resistant aggregation core indicates however that both proteins are able to form regular intermolecular contacts. We speculate that Pr12 might form even more stable amyloid fibrils in the presence of co‐factors that could be available in vivo. Neither of the two proteins had any seeding effect on αSN fibrillation. This, however, does not necessarily mean that the microbiome is not involved in the development of PD. First of all, the rats investigated were relatively young (3 months) and the signs of disease at this age is still very modest, as evaluated by subtle locomotor deficits, reduced ability to discriminate smells and more dot‐like appearance of αSN in the striatum nerve end terminals.43 At this age, no deficits in locomotor activity could be detected and the amount of insoluble (urea‐treated) αSN in most brain regions investigated (olfactory bulb, striatum, substantia nigra and hippocampus) was unchanged.43
4. MATERIALS AND METHODS
4.1. Materials
Fecal samples from three WT rats (WT1, WT2, and WT3) and three BAC‐SNCA Tg PD rats overexpressing human αSN (PD1, PD2, and PD3)43 were kindly provided by Olaf Riess's lab in Tübingen, Germany. All rats were female, 3 months old and kept in regular 12 hr light/dark cycles. All animals had free access to both water and food.
4.2. DNA extraction from WT and PD microbiome samples
Three WT samples and three PD samples were analyzed. DNA was extracted from the microbiome samples with a FastDNA spin kit (MP Biomedicals), following the manufacturers' instructions. Briefly, 50 mg of fecal pellet was subjected to bead beating (4 × 40 s at 6 m/s with 5 min incubations in between) in a Lysing Matrix E tube (MP Biomedicals) using a FastPrep‐24 instrument. Protein was precipitated, after which DNA was bound to a binding matrix suspension and eluted. DNA concentrations were determined with a Qubit dsDNA BR kit and the quality confirmed using gel electroporation on a 2200 TapeStation with D1000 Screentapes (Agilent Genomics).
4.3. V1–V3 16S rRNA amplicon sequencing
All DNA samples were diluted to 5 ng/μl and mixed with barcode adaptors and a master mix containing dNTPs, MgSO4 and Platinum® Taq DNA polymerase high fidelity (Thermo Fischer Scientific) as previously described.73 The following PCR conditions were used: 2 min incubation at 95°C followed by 30 cycles of [20 s at 95°C, 30 s at 56°C, 60 s at 72°C] and 5 min at 72°C. PCR products, now referred to as libraries, were purified using an Agencourt AMPure XP bead solution (Beckman Coulter) and a magnetic rack. Concentrations were determined with a Qubit dsDNA HS kit and amplification was confirmed on a 2,200 TapeStation (Agilent Genomics). Of note, 30 ng of each library were pooled and submitted for MiSeq sequencing.
Forward reads were processed using usearch v.11.0.667. Raw fastq files were filtered for phiX sequences using ‐filter_phix, trimmed to 200 bp using ‐fastx_truncate ‐trunclen 200, and quality filtered using ‐fastq_filter with ‐fastq_maxee 1.0. The sequences were dereplicated using ‐fastx_uniques with ‐sizeout ‐relabel Uniq. Exact ASVs were generated using ‐unoise3.74 ASV‐tables were created by mapping the raw reads to the ASVs using ‐otutab with the ‐zotus and ‐strand both options. Taxonomy was assigned to ASVs using ‐sintax with ‐strand both and ‐sintax_cutoff 0.875 and the Autotax processed SILVA 138 SSURef Nr99 database.76 The raw sequencing data is available at the SRA with the accession IDs ERR3477180‐ERR3477185.
4.4. Direct identification of amyloidogenic proteins in fecal samples by label‐free quantitative MS
Amyloidogenic proteins were identified as previously described.31 In short, 200 mg of each fecal pellet also used for DNA extraction were dissolved in 1 ml buffer (10 mM Tris–HCl, pH 8.0) containing 1X Halt™ protease inhibitor cocktail (Thermo Scientific). Cell lysis was achieved by bead beating in a Lysing Matrix E tube (MP Biomedicals) for 4 × 20 s at 6 m/s with 5 min breaks on ice in between beatings. Aliquots of 25 μl were transferred to 18 eppendorf tubes per sample (six different FA concentrations in three technical triplicates) before 1 hr of lyophilization. Samples were then mixed with FA at concentrations of 0% (ultraclean water), 20, 40, 60, 80, and 100% and lyophilized overnight. Lyophilized material was resuspended in a special reducing sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS‐PAGE) loading buffer77 and run on AnyKD gels (Biorad) for 10 min at 120 V, just allowing the samples to enter the gel. The gels were stained with Coomassie Brilliant Blue (CBB) G250. Gel bands were then cut into smaller pieces and reduced in a 10 mM DTT/0.1 M NH4HCO3 solution for 45 min at 56°C after several washing steps. In each washing step the gel pieces were rehydrated with 0.1 M NH4HCO3 for 5 min before addition of concentrated acetonitrile (1:1 vol/vol). Proteins were then alkylated for 30 min at room temperature (RT) in a solution of 55 mM iodoacetamide/0.1 M NH4HCO3. Finally, the gel pieces were washed thoroughly with 0.1 M NH4HCO3 and 1:1 vol/vol 0.1 M NH4HCO3/acetonitrile before drying in a vacuum centrifuge. In‐gel digestion was performed overnight at 37°C by rehydrating the gel particles in 12.5 ng/μl trypsin in a 0.1 M NH4HCO3 buffer. Peptides were recovered mainly from the overnight buffer but also extracted from the gel particles by incubation in 5% FA followed by addition of acetonitrile. This extraction was performed twice and all supernatants were pooled and dried by vacuum centrifugation overnight. Peptides were reconstituted in 0.1% trifluoroacetic acid and 2% acetonitrile and subjected to ultra‐performance liquid chromatography (UPLC) tandem mass spectrometry analysis by injection and concentration on a trapping column before separation on a separation column (Pepmap™ C18, Thermo Scientific) with a gradient of buffer B (100% acetonitrile) of 2–8% during the first minute and then from 8 to 30% in the following 39 min. Buffer B was then increased from 30% to 90% within 5 min. The UPLC system was coupled online to a Q Exactive Plus mass spectrometer (Thermo Scientific). The mass spectrometry data have been deposited to the ProteomeXchange Consortium (http://www.proteomexchange.org) via the PRIDE partner repository78 with the dataset identifier PXD014649.
4.5. Data analysis of LC–MS/MS data
Protein identification and quantification were performed as previously described31 using the MaxQuant v1.5.8.3 software79 and the label‐free quantification (LFQ) algorithm.80 The search was performed against the NCBI non‐redundant protein sequence database. To prevent systematic errors, LFQ values were normalized between individual measurements and each protein is identified based on at least two detectable peptides. This allows for relative abundances to be compared across samples. Amyloid proteins are expected to give a sigmoidal appearance when their relative abundances are plotted against FA concentration (with higher FA concentrations, more protein monomer will be released from the fibrils and enter the SDS‐PAGE gel). With an automated R‐markdown script the data can be separated for each protein and the highest concentration will be given a value of 1. Hit proteins should fulfill the following requirements: 60 < f50 < 100, f′(f50) > 0.025 where f50 is the FA concentration required to depolymerize half of the amyloid fibrils and f'(f50) is the slope of the fit at this concentration.31 The fit used was:
| (1) |
4.6. Bioinformatic analysis
Data analysis of the entire MS dataset were done using R.81 All proteins were fitted as described.31 We consider a protein to be a hit if at least one of the three triplicates shows a sigmoidal increase in abundance with FA concentration. Because well‐characterized FuBA systems like Fap in Pseudomonas and curli in E. coli all require Sec‐dependent secretion of the amyloid proteins, the hits were all analyzed by the SignalP 4.1, SignalP 5 and DeepSig algorithms45 to restrict the list to candidates containing a signal peptide. RADAR,49 AmylPred282 and Clustal2.1 were used to identify imperfect repeats, amyloidogenic amino acid sequences and sequence alignments, respectively.
4.7. Expression and purification of candidate amyloid proteins
pET30a vectors encoding proteins WP_032523104.1 and OAD22177.1 (referred to as Pr12 and Pr17, respectively) were prepared by Genscript. Both proteins were produced without signal peptides (residues 1–22 for Pr12 and residues 1–18 for Pr17) but with an N‐terminal His6 tag. E. coli BL21(DE3) bacteria were transformed with the expression vectors, spread on LB agar plate with 50 μg/ml kanamycin, grown up 24 hr and then transferred to LB medium containing 0.1% glycerol, 50 μg/ml kanamycin and 4 mM MgSO4. Cells were grown up in an incubator at 37°C and 125 rpm and induced with 1 mM IPTG at OD600 of 0.6–0.8. After 3 hr, cells were harvested by centrifugation at 4000 g for 20 min. Pellets were suspended in 50 ml of denaturation buffer (8 M GdmCl, 50 mM Tris–HCl, pH 8.0) with one protease inhibitor tablet (Roche) per L medium, lysed by slow stirring of the GdmCl‐cell pellet suspension overnight at 4°C and spun down at ~12,500 g for 30 min at 15°C. The supernatant was incubated with nickel‐nitrilotriacetic acid beads (typically 45 ml supernatant to 5 ml beads) for 1 hr at 4°C on rolling table with 60 rpm, after which the beads were packed in an empty PD10 column and washed with washing buffer (8 M GdmCl, 50 mM Tris–HCl, 30 mM imidazole, pH 8.0) before elution of the protein into elution buffer (8 M GdmCl, 50 mM Tris–HCl, 300 mM imidazole, pH 8.0). Protein elution fractions of ~2 ml were immediately frozen in liquid nitrogen and stored at −80°C.
4.8. Purification and preparation of αSN
αSN was prepared as described83 and protein concentration was determined at 280 nm using an extinction coefficient of 5,960 M−1 cm−1 and the molecular weight of 14,460 Da after the lyophilized αSN had been dissolved in 60 mM Tris buffer, pH 7.4, and filtered through a 0.2 μm filter.
4.9. Thioflavin T (ThT) assays of Pr12 and Pr17
The two proteins were first desalted into 1X PBS, pH 7.4, and protein concentration was determined at 280 nm using extinction coefficients of 14,100 and 14,690 L mol−1 cm−1 for Pr12 and Pr17, respectively, based on their amino acid composition. The proteins were then immediately transferred to a 96‐well Nunc optical bottom plate (Corning) containing 40 μM ThT. Fibrillation was followed using 445 nm excitation and 485 nm emission at 37°C on an Infinite Pro 23 (Tecan, Männersdorf) plate reader with 120 rpm shaking. Due to precipitation issues in 1X PBS, we started desalting the proteins from the elution buffer (8 M GdmCl, 50 mM Tris–HCl, 300 mM imidazole, pH 8.0) into 4 M urea in 1X PBS or 60 mM Tris, pH 7.4 (Pr12) or 8 M urea in 1X PBS, pH 7.4 (Pr17) to keep the proteins monomeric. Using Amicon Ultra spin filters (Merck) with a 3 K (Pr12) or 10 K cutoff (Pr17) both proteins were concentrated to either 7.5 or 10 mg/ml. The aggregation behavior of the two proteins (final concentration of 0.5 mg/ml) were then investigated in the presence of different urea concentrations or at different pH values. Because the proteins had been concentrated in urea, 0.2 M urea (Pr12) or 0.4 M urea (Pr17) were also present in all ThT assays. The buffers used to investigate pH effects were 80 mM citric acid (pH 3), 60 mM citric acid (pH 4), 60 mM MES (pH 6), 1X PBS (pH 7.4), and 60 mM Tris (pH 8).
4.10. Size‐exclusion chromatography (SEC) analysis of urea‐treated Pr17
After fibrillation of Pr17 in the presence of increasing [urea], the samples were subjected to SEC using a Superose 6 10/300 GL column (GE Healthcare). For each sample, the column was equilibrated with a buffer containing the same urea concentration in 1X PBS as the sample to be investigated. Then, 500 μl sample was injected and run with a flow rate of 0.5 ml/min on an ÄKTA Pure protein purification system (GE Healthcare, Chicago, IL). Control samples containing ThT alone at the lowest (0.4 M urea) and the highest (6 M urea) were also included.
4.11. Stability studies of Pr12 and Pr17 aggregates
Pr12 and Pr17 fibrils were produced by desalting the protein directly into 60 mM MES, pH 6, and incubating the samples overnight at 37°C at 300 rpm. Aggregates were pelleted by centrifugation (13,500 rpm, 15 min) in a MicroStar12 table centrifuge (VWR) and washed twice in milli‐Q water (MQ) before equal volumes of aggregates were aliquoted into new tubes, Pellets were then treated with equal amounts of solutions containing increasing [urea] or [FA] and incubated for 10 min at RT before pelleted again. Of note, 20 μl of the supernatant was then transferred to new tubes and either mixed with R loading buffer (the urea‐treated samples) or subjected to lyophilization (the FA‐treated samples). After lyophilization the samples were mixed with 20 μl of a special loading buffer containing 8 M urea. Both the urea‐ and FA‐treated samples were analyzed with SDS‐PAGE and quantified with ImageJ (https://imagej.net/). The percentage of solubilized Pr12/Pr17 aggregates was normalized to the 100% FA band and fitted to the following equation:
| (2) |
where [FA]50% is the FA concentration where half of the aggregates are dissolved, that is, the two states (aggregated state and monomeric state) are equally stable. The m‐value, as in conventional protein unfolding, is a measure of the potency of the denaturant, here FA.52 For treatment with proteinase K (ProtK), the protein mass in each pellet was first determined by treating one protein pellet with 100% FA and running this sample on SDS‐PAGE together with samples of known concentrations of 0.1, 0.2, 0.5, and 0.8 mg/ml. Pellets were then treated with different concentrations of ProtK in a buffer containing 60 mM Tris and 5 mM CaCl2 (pH 8) and incubated at 37°C for 30 min before the reactions were stopped by adding concentrated FA (giving a final [FA] of 50%, equal to 13.25 M). Samples were lyophilized in a Scanvac Coolsafe freeze dryer (Labogene) and subsequently analyzed on SDS‐PAGE.
4.12. Cross‐interactions between microbiome proteins and αSN
αSN of 1 mg/ml was fibrillated in the presence of Pr12 or Pr17 seeds using 40 μM ThT in a 96‐well plate. A 3 mm glass bead was added to each well, and the program used allowed 300 rpm orbital shaking with readings every 1,070 s and 600 s shaking between readings. Pr12 and Pr17 seeds were prepared by incubation overnight at 37°C with slow shaking (300 rpm). Aggregates were then spun down (13,500 rpm, 15 min) and washed twice with MQ before aliquoted into new tubes. Aggregates were pelleted again, and all supernatant was removed. Concentration in the pellet was determined in the same was as described above. To prepare seeds, aggregates of Pr12 and Pr17 were sonicated for 3*10 s with 10 s on ice in between using a QSonica Sonicator and subsequently added at 10, 5, or 1% (mass/mass) to 1 mg/ml monomeric αSN.
4.13. Transmission electron microscopy (TEM) analysis of protein fibrils
The same Pr12 and Pr17 aggregates used to study aggregate stability were also imaged with TEM. Pellets were resuspended in MQ and images were recorded as described.84
4.14. Fourier transform infrared (FTIR) spectroscopy of protein fibrils
FTIR spectra were recorded on a Tensor 27 instrument (Bruker Optics). Sample of 2 μl was deposited on the surface of an attenuated total reflection crystal and dried with nitrogen gas. The system was continuously purged with nitrogen gas. Background and water vapor subtractions were performed to obtain a straight baseline. The samples were analyzed with OPUS 5.5. Only the amide I region (1600–1700 cm−1) was used for analysis.
4.15. Circular dichroism (CD) spectroscopy of protein fibrils
All samples were sonicated before CD measurements. Far‐UV CD spectra were recorded on a Jasco J‐810 spectrophotometer (Jasco Spectroscopic Co. Ltd., Japan) in a 0.1 cm cuvette at a scan speed of 10 nm/min from 260 to 195 nm at 37°C with 0.1 mg/ml protein. Three spectra were accumulated and averaged. Buffer spectra were subtracted.
AUTHOR CONTRIBUTIONS
Line Christensen: Investigation; writing‐original draft. Saeid Alijanvand: Investigation. Michal Burdukiewicz: Formal analysis; methodology. Florian‐Alexander Herbst: Methodology. Henrik Kjeldal: Methodology. Morten Dueholm: Formal analysis; methodology; writing‐review & editing. Daniel E. Otzen: Conceptualization, formal analysis, funding acquisition, methodology, project administration, supervision, writing‐original draft, writing‐review& editing.
Supporting information
Appendix S1. Supporting Information includes two tables on the number and identity of proteins and putative amyloids identified by MS as well as 11 figures describing PCA analysis of bacterial composition in microbiome samples, occurrence and abundance of identified proteins, sequences of Pr12 and Pr17, SDS‐PAGE analysis of protein purification, close‐up views of protein aggregation at early time points, solubility of Pr12 in urea measured by SDS‐PAGE, effect of urea on protein elution, far‐UV CD curves of Pr12 and PR17 after 24 hr incubation at pH 4–10, ThT time curves of protein aggregation at different pH and urea concentrations, dissolution of Pr17 by urea and formic acid and ThT curves demonstrating lack of cross‐interaction between αSN and Pr12 or Pr17.
ACKNOWLEDGMENTS
D.E.O. acknowledges support from Innovation Foundation Denmark (Grant 5188‐00003B) through the Joint Programme on Neurodegenerative Diseases (aSynProtec), the Lundbeck Foundation (Grant R276‐2018‐671) and the Independent Research Council Denmark, Natural Sciences (Grant 8021‐00208B). M.B. acknowledges support from the National Science Centre (Grant 2019/35/B/NZ2/03997).
Christensen LFB, Alijanvand SH, Burdukiewicz M, et al. Identification of amyloidogenic proteins in the microbiomes of a rat Parkinson's disease model and wild‐type rats. Protein Science. 2021;30:1854–1870. 10.1002/pro.4137
Funding information Innovationsfonden, Grant/Award Number: 5188‐00003B; Lundbeckfonden, Grant/Award Number: R276‐2018‐671; Natur og Univers, Det Frie Forskningsråd, Grant/Award Number: 8021‐00208B; National Science Centre, Grant/Award Number: 2019/35/B/NZ2/03997
REFERENCES
- 1.Pringsheim T, Jette N, Frolkis A, Steeves TD. The prevalence of Parkinson's disease: A systematic review and meta‐analysis. Mov Disord. 2014;29:1583–1590. [DOI] [PubMed] [Google Scholar]
- 2.Braak H, Del Tredici K, Rub U, de Vos RA, Jansen Steur EN, Braak E. Staging of brain pathology related to sporadic Parkinson's disease. Neurobiol Aging. 2003;24:197–211. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini MG, Schmidt ML, Lee VM, Trojanowski JQ, Jakes R, Goedert M. Alpha‐synuclein in Lewy bodies. Nature. 1997;388:839–840. [DOI] [PubMed] [Google Scholar]
- 4.Braak H, Del Tredici K, Gai WP, Braak E. Alpha‐synuclein is not a requisite component of synaptic boutons in the adult human central nervous system. J Chem Neuroanat. 2000;20:245–252. [DOI] [PubMed] [Google Scholar]
- 5.Taguchi K, Watanabe Y, Tsujimura A, Tanaka M. Brain region‐dependent differential expression of alpha‐synuclein. J Comp Neurol. 2016;524:1236–1258. [DOI] [PubMed] [Google Scholar]
- 6.Masuda‐Suzukake M, Nonaka T, Hosokawa M, et al. Prion‐like spreading of pathological alpha‐synuclein in brain. Brain. 2013;136:1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Braak H, de Vos RA, Bohl J, Del Tredici K. Gastric alpha‐synuclein immunoreactive inclusions in Meissner's and Auerbach's plexuses in cases staged for Parkinson's disease‐related brain pathology. Neurosci Lett. 2006;396:67–72. [DOI] [PubMed] [Google Scholar]
- 8.Horsager J, Andersen KB, Knudsen K, et al. Brain‐first versus body‐first Parkinson's disease: A multimodal imaging case‐control study. Brain. 2020;143:3077–3088. [DOI] [PubMed] [Google Scholar]
- 9.Liu B, Fang F, Pedersen NL, et al. Vagotomy and Parkinson disease: A Swedish register‐based matched‐cohort study. Neurology. 2017;88:1996–2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan‐Montojo F, Schwarz M, Winkler C, et al. Environmental toxins trigger PD‐like progression via increased alpha‐synuclein release from enteric neurons in mice. Sci Rep. 2012;2:898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Paillusson S, Clairembault T, Biraud M, Neunlist M, Derkinderen P. Activity‐dependent secretion of alpha‐synuclein by enteric neurons. J Neurochem. 2013;125:512–517. [DOI] [PubMed] [Google Scholar]
- 12.Holmqvist S, Chutna O, Bousset L, et al. Direct evidence of Parkinson pathology spread from the gastrointestinal tract to the brain in rats. Acta Neuropathol. 2014;128:805–820. [DOI] [PubMed] [Google Scholar]
- 13.Uemura N, Yagi H, Uemura MT, Hatanaka Y, Yamakado H, Takahashi R. Inoculation of alpha‐synuclein preformed fibrils into the mouse gastrointestinal tract induces Lewy body‐like aggregates in the brainstem via the vagus nerve. Mol Neurodegen. 2018;13:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Furness JB, Callaghan BP, Rivera LR, Cho HJ. The enteric nervous system and gastrointestinal innervation: Integrated local and central control. Adv Exp Med Biol. 2014;817:39–71. [DOI] [PubMed] [Google Scholar]
- 15.Bohorquez DV, Shahid RA, Erdmann A, et al. Neuroepithelial circuit formed by innervation of sensory enteroendocrine cells. J Clin Invest. 2015;125:782–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogunovic M, Dave SH, Tilstra JS, et al. Enteroendocrine cells express functional toll‐like receptors. Am J Physiol Gastrointest Liver Physiol. 2007;292:G1770–G1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dueholm MS, Albertsen M, Otzen D, Nielsen PH. Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One. 2012;7:e51274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rapsinski GJ, Wynosky‐Dolfi MA, Oppong GO, et al. Toll‐like receptor 2 and NLRP3 cooperate to recognize a functional bacterial amyloid, curli. Infect Immun. 2015;83:693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen SG, Stribinskis V, Rane MJ, et al. Exposure to the functional bacterial amyloid protein curli enhances alpha‐synuclein aggregation in aged Fischer 344 rats and Caenorhabditis elegans . Sci Rep. 2016;6:34477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sampson TR, Challis C, Jain N, et al. A gut bacterial amyloid promotes alpha‐synuclein aggregation and motor impairment in mice. Elife. 2020;9:e53111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng G, Vad BS, Dueholm MS, et al. Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front Microbiol. 2015;6:1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collinson S, Emödy L, Müller K, Kay W. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis . J Bacteriol. 1991;173:4773–4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stenvang M, Dueholm MS, Vad BS, et al. Epigallocatechin gallate remodels Pseudomonas aeruginosa functional amyloids and increases biofilm susceptibility to antibiotic treatment. J Biol Chem. 2016;291:26540–26553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cummings JH, Bingham SA, Heaton KW, Eastwood MA. Fecal weight, colon cancer risk, and dietary intake of nonstarch polysaccharides (dietary fiber). Gastroenterology. 1992;103:1783–1789. [DOI] [PubMed] [Google Scholar]
- 25.Macfarlane GT, Hay S, Macfarlane S, Gibson GR. Effect of different carbohydrates on growth, polysaccharidase and glycosidase production by Bacteroides ovatus, in batch and continuous culture. J Appl Bacteriol. 1990;68:179–187. [DOI] [PubMed] [Google Scholar]
- 26.Hurrell E, Kucerova E, Loughlin M, et al. Neonatal enteral feeding tubes as loci for colonisation by members of the Enterobacteriaceae. BMC Infect Dis. 2009;9:146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Macfarlane S, McBain AJ, Macfarlane GT. Consequences of biofilm and sessile growth in the large intestine. Adv Dent Res. 1997;11:59–68. [DOI] [PubMed] [Google Scholar]
- 28.Bokranz W, Wang X, Tschape H, Romling U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J Med Microbiol. 2005;54:1171–1182. [DOI] [PubMed] [Google Scholar]
- 29.Dueholm MS, Petersen SV, Sonderkaer M, et al. Functional amyloid in pseudomonas. Mol Microbiol. 2010;77:1009–1020. [DOI] [PubMed] [Google Scholar]
- 30.Jordal PB, Dueholm MS, Larsen P, et al. Widespread abundance of functional bacterial amyloid in mycolata and other gram‐positive bacteria. Appl Environ Microbiol. 2009;75:4101–4110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Danielsen HN, Hansen SH, Herbst FA, et al. Direct identification of functional amyloid proteins by label‐free quantitative mass spectrometry. Biomolecules. 2017;7:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheperjans F, Aho V, Pereira PA, et al. Gut microbiota are related to Parkinson's disease and clinical phenotype. Mov Disord. 2015;30:350–358. [DOI] [PubMed] [Google Scholar]
- 33.Hasegawa S, Goto S, Tsuji H, et al. Intestinal dysbiosis and lowered serum lipopolysaccharide‐binding protein in Parkinson's disease. PLoS One. 2015;10:e0142164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keshavarzian A, Green SJ, Engen PA, et al. Colonic bacterial composition in Parkinson's disease. Mov Disord. 2015;30:1351–1360. [DOI] [PubMed] [Google Scholar]
- 35.Gorecki AM, Preskey L, Bakeberg MC, et al. Altered gut microbiome in Parkinson's disease and the influence of lipopolysaccharide in a human alpha‐synuclein over‐expressing mouse model. Front Neurosci. 2019;13:839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin A, Zheng W, He Y, et al. Gut microbiota in patients with Parkinson's disease in southern China. Parkinsonism Relat Disord. 2018;53:82–88. [DOI] [PubMed] [Google Scholar]
- 37.Pietrucci D, Cerroni R, Unida V, et al. Dysbiosis of gut microbiota in a selected population of Parkinson's patients. Parkinsonism Relat Disord. 2019;65:124–130. [DOI] [PubMed] [Google Scholar]
- 38.Hopfner F, Künstner A, Müller SH, et al. Gut microbiota in Parkinson disease in a northern German cohort. Brain Res. 2017;1667:41–45. [DOI] [PubMed] [Google Scholar]
- 39.Petrov VA, Saltykova IV, Zhukova IA, et al. Analysis of gut microbiota in patients with Parkinson's disease. Bull Exp Biol Med. 2017;162:734–737. [DOI] [PubMed] [Google Scholar]
- 40.Bedarf JR, Hildebrand F, Coelho LP, et al. Functional implications of microbial and viral gut metagenome changes in early stage L‐DOPA‐naive Parkinson's disease patients. Genome Med. 2017;9:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qian Y, Yang X, Xu S, et al. Alteration of the fecal microbiota in Chinese patients with Parkinson's disease. Brain Behav Immun. 2018;70:194–202. [DOI] [PubMed] [Google Scholar]
- 42.Barichella M, Severgnini M, Cilia R, et al. Unraveling gut microbiota in Parkinson's disease and atypical parkinsonism. Mov Disord. 2019;34:396–405. [DOI] [PubMed] [Google Scholar]
- 43.Nuber S, Harmuth F, Kohl Z, et al. A progressive dopaminergic phenotype associated with neurotoxic conversion of alpha‐synuclein in BAC‐transgenic rats. Brain. 2013;136:412–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez‐Ochoa J, Montes‐Garcia JF, Vazquez C, et al. Gallibacterium elongation factor‐Tu possesses amyloid‐like protein characteristics, participates in cell adhesion, and is present in biofilms. J Microbiol. 2017;55:745–752. [DOI] [PubMed] [Google Scholar]
- 45.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. [DOI] [PubMed] [Google Scholar]
- 46.Savojardo C, Martelli PL, Fariselli P, Casadio R. DeepSig: Deep learning improves signal peptide detection in proteins. Bioinformatics. 2018;34:1690–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Almagro Armenteros JJ, Tsirigos KD, Sonderby CK, et al. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol. 2019;37:420–423. [DOI] [PubMed] [Google Scholar]
- 48.Fernandez‐Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence‐dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. [DOI] [PubMed] [Google Scholar]
- 49.Heger A, Holm L. Rapid automatic detection and alignment of repeats in protein sequences. Proteins. 2000;41:224–237. [DOI] [PubMed] [Google Scholar]
- 50.Moran SD, Zanni MT. How to get insight into amyloid structure and formation from infrared spectroscopy. J Phys Chem Lett. 2014;5:1984–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zandomeneghi G, Krebs MR, McCammon MG, Fandrich M. FTIR reveals structural differences between native beta‐sheet proteins and amyloid fibrils. Protein Sci. 2004;13:3314–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Christensen LFB, Nowak JS, Sonderby TV, Frank SA, Otzen DE. Quantitating denaturation by formic acid: Imperfect repeats are essential to the stability of the functional amyloid protein FapC. J Biol Chem. 2020;295:13031–13046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Larsen P, Nielsen JL, Dueholm MS, Wetzel R, Otzen D, Nielsen PH. Amyloid adhesins are abundant in natural biofilms. Environ Microbiol. 2007;9:3077–3090. [DOI] [PubMed] [Google Scholar]
- 54.Bollinger RR, Barbas AS, Bush EL, Lin SS, Parker W. Biofilms in the normal human large bowel: Fact rather than fiction. Gut. 2007;56:1481–1482. [PMC free article] [PubMed] [Google Scholar]
- 55.Macfarlane S, Furrie E, Cummings JH, Macfarlane GT. Chemotaxonomic analysis of bacterial populations colonizing the rectal mucosa in patients with ulcerative colitis. Clin Infect Dis. 2004;38:1690–1699. [DOI] [PubMed] [Google Scholar]
- 56.Hartley CL, Neumann CS, Richmond MH. Adhesion of commensal bacteria to the large intestine wall in humans. Infect Immun. 1979;23:128–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. [DOI] [PubMed] [Google Scholar]
- 58.Walker AW, Duncan SH, Harmsen HJ, Holtrop G, Welling GW, Flint HJ. The species composition of the human intestinal microbiota differs between particle‐associated and liquid phase communities. Environ Microbiol. 2008;10:3275–3283. [DOI] [PubMed] [Google Scholar]
- 59.Sonnenburg JL, Angenent LT, Gordon JI. Getting a grip on things: How do communities of bacterial symbionts become established in our intestine? Nat Immunol. 2004;5:569–573. [DOI] [PubMed] [Google Scholar]
- 60.Johansson ME, Gustafsson JK, Holmen‐Larsson J, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Swidsinski A, Weber J, Loening‐Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clyne M, Drumm B. Adherence of Helicobacter pylori to primary human gastrointestinal cells. Infect Immun. 1993;61:4051–4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Knutton S, Lloyd DR, McNeish AS. Adhesion of enteropathogenic Escherichia coli to human intestinal enterocytes and cultured human intestinal mucosa. Infect Immun. 1987;55:69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Minter MR, Zhang C, Leone V, et al. Antibiotic‐induced perturbations in gut microbial diversity influences neuro‐inflammation and amyloidosis in a murine model of Alzheimer's disease. Sci Rep. 2016;6:30028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sampson TR, Debelius JW, Thron T, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson's disease. Cell. 2016;167:1469–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arosio P, Knowles TP, Linse S. On the lag phase in amyloid fibril formation. Phys Chem Chem Phys. 2015;17:7606–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fowler DM, Koulov AV, Alory‐Jost C, Marks MS, Balch WE, Kelly JW. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dueholm MS, Larsen P, Finster K, et al. The tubular sheaths encasing Methanosaeta thermophila filaments are functional amyloids. J Biol Chem. 2015;290:20590–20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc Natl Acad Sci U S A. 2010;107:2230–2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fallingborg J. Intraluminal pH of the human gastrointestinal tract. Dan Med Bull. 1999;46:183–196. [PubMed] [Google Scholar]
- 71.Dressman JB, Berardi RR, Dermentzoglou LC, et al. Upper gastrointestinal (GI) pH in young, healthy men and women. Pharm Res. 1990;7:756–761. [DOI] [PubMed] [Google Scholar]
- 72.Simon GL, Gorbach SL. Intestinal flora in health and disease. Gastroenterology. 1984;86:174–193. [PubMed] [Google Scholar]
- 73.Kirkegaard RH, McIlroy SJ, Kristensen JM, et al. The impact of immigration on microbial community composition in full‐scale anaerobic digesters. Sci Rep. 2017;7:9343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Edgar RC. UNOISE2: improved error‐correction for Illumina 16S and ITS amplicon sequencing. bioRxiv:081257. 2016.
- 75.SINTAX: a simple non‐Bayesian taxonomy classifier for 16S and ITS sequences. bioRxiv:074161. 2016.
- 76.Dueholm MS, Andersen KS, McIlroy SJ, et al. Generation of comprehensive ecosystems‐specific reference databases with species‐level resolution by high‐throughput full‐length 16S rRNA gene sequencing and automated taxonomy assignment (AutoTax). bioRxiv:672873. 2020. [DOI] [PMC free article] [PubMed]
- 77.Dueholm M, Søndergaard MT, Nilsson M, et al. Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiol Open. 2013;2:365–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vizcaino JA, Csordas A, Del‐Toro N, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44:11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry‐based shotgun proteomics. Nat Protoc. 2016;11:2301–2319. [DOI] [PubMed] [Google Scholar]
- 80.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome‐wide label‐free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13:2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Team RC . R: A language and environment for statistical computing. (2018). R Foundation for Statistical Computing, Vienna, Austria. Available from: http://www.R-project.ord/. [Google Scholar]
- 82.Tsolis AC, Papandreou NC, Iconomidou VA, Hamodrakas SJ. A consensus method for the prediction of 'aggregation‐prone' peptides in globular proteins. PLoS One. 2013;8:e54175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lorenzen N, Nielsen SB, Buell AK, et al. The role of stable α‐synuclein oligomers in the molecular events underlying amyloid formation. J Am Chem Soc. 2014;136:3859–3868. [DOI] [PubMed] [Google Scholar]
- 84.Mohammad‐Beigi H, Morshedi D, Shojaosadati SA, et al. Gallic acid loaded onto polyethylenimine‐coated human serum albumin nanoparticles (PEI‐HSA‐GA NPs) stabilizes α‐synuclein in the unfolded conformation and inhibits aggregation. RSC Adv. 2016;6:85312–85323. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Supporting Information includes two tables on the number and identity of proteins and putative amyloids identified by MS as well as 11 figures describing PCA analysis of bacterial composition in microbiome samples, occurrence and abundance of identified proteins, sequences of Pr12 and Pr17, SDS‐PAGE analysis of protein purification, close‐up views of protein aggregation at early time points, solubility of Pr12 in urea measured by SDS‐PAGE, effect of urea on protein elution, far‐UV CD curves of Pr12 and PR17 after 24 hr incubation at pH 4–10, ThT time curves of protein aggregation at different pH and urea concentrations, dissolution of Pr17 by urea and formic acid and ThT curves demonstrating lack of cross‐interaction between αSN and Pr12 or Pr17.


