Abstract
Natural products, like turmeric, are considered powerful antioxidants which exhibit tumor-inhibiting activity and chemoradioprotective properties. Nowadays, there is a great demand for developing novel, affordable, efficacious, and effective anticancer drugs from natural resources. In the present study, we have employed a stringent in silico methodology to mine and finally propose a number of natural products, retrieved from the biomedical literature. Our main target was the systematic search of anticancer products as anticancer agents compatible to the human organism for future use. In this case and due to the great plethora of such products, we have followed stringent bioinformatics methodologies. Our results taken together suggest that natural products of a great diverse may exert cytotoxic effects in a maximum of the studied cancer cell lines. These natural compounds and active ingredients could possibly be combined to exert potential chemopreventive effects. Furthermore, in order to substantiate our findings and their application potency at a systems biology level, we have developed a representative, user-friendly, publicly accessible biodatabase, NaturaProDB, containing the retrieved natural resources, their active ingredients/fractional mixtures, the types of cancers that they affect, and the corresponding experimentally verified target genes.
1. Introduction
Cancer represents one of the leading causes of death globally. Despite the availability of anticancer therapeutics, cancer incidence is increasing gradually. According to the World Cancer Report of the International Agency for Research on Cancer (IARC) and the World Health Organization (WHO), the global cancer burden is estimated to have risen to 18.1 million new cases and 9.6 million deaths in 2018 (GLOBOCAN 2018 database: https://gco.iarc.fr/). Nowadays, research efforts are directed towards the discovery of naturally derived chemical compounds with anticancer potential [1]. A great number of highly potent bioactive compounds, derived from plants, have been found to possess anticancer properties, and this number is increasing exponentially [2, 3]. For example, there are natural products with antioxidant and anti-inflammatory capacities [4, 5], thereby preventing oxidative stress and inflammation, which can cause damage to DNA, eventually leading to genomic instability and eventually to carcinogenesis [6, 7].)
Natural products derived from plants and natural product-based anticancer drugs have been associated with reduced cancer mortality and risk. Fruits and vegetables contain vitamins, minerals, folate, plant sterols, carotenoids, and various phytochemicals such as flavonoid and polyphenols, which are suggested to have cancer chemopreventive potential [8–11].
Another category of plants that possess antitumor, antioxidant, and anti-inflammatory properties is spices and herbs [12]. They contain tannins, alkaloids, phenolic diterpenes, vitamins, flavonoids, and polyphenols. Spices and herbs such as curcumin, clove, rosemary, sage, oregano, and cinnamon are excellent sources of antioxidants due to the high content of phenolic compounds [8, 13, 14]. The antioxidants in edible and medicinal plant extracts have been shown to counteract ROS-mediated damage in diverse human cancers [15].
Furthermore, nuts are enriched in nutrients of high biological value. Nuts contain high amounts of vegetable protein [16, 17] and fat, mostly unsaturated fatty acids [17, 18]. They are also rich in a variety of other nutrients, as well as dietary fiber [19], vitamins (e.g., folic acid, niacin, tocopherols, and vitamin B6), minerals (e.g., calcium, magnesium, and potassium), and many other bioactive constituents such as phytosterols [17, 20] and phenolic compounds [17, 21]. Among the dietary plants, nuts, like peanuts and walnuts, contain the highest total content of antioxidants [21]. Nuts are associated with cancer prevention and they exert their potential chemopreventive effect through documented anti-antioxidant, anti-inflammatory, proapoptotic, antiproliferation, and antimetastatic activities [22].
Notably, natural compounds with cancer chemopreventive potential have also been derived from nonplant resources. For example, many cyclic peptides and analogues derived from marine organisms have been shown to possess anticancer, antimicrobial, anti-inflammatory, antiproliferative, and antihypertensive properties [23–25]. In addition, lactoferrin, a multifunctional protein found in bovine and camel milk, possesses anticancer, antimicrobial, and immune system boosting effects [26–29].
In the present work, taking into account the numerous studies on the biological (chemopreventive) activities of natural products, we have employed an in silico methodology to determine an effective combination of such products, retrieved from the biomedical literature, affecting diverse types of cancers and cancer-related pathways. Furthermore, we have developed a representative database, NaturaProDB, containing the retrieved data, which is maintained by the National Technical University of Athens, Greece.
2. Materials and Methods
2.1. Data Acquisition and Compilation
To obtain a broad spectrum of naturally occurring products with demonstrated anticancer activity, the bibliographic database MEDLINE/PubMed (https://www.ncbi.nlm.nih.gov/pubmed) was searched manually for full-text articles (from January 2003 up to 20 October 2020) using relevant keywords, including “natural products” or “naturally occurring compounds” or “natural ingredients” or “natural agents” or “natural substances” or “natural extracts” or “superfoods” and “cancer” or “malignancy.” The criteria applied to assess the anticancer potential of the natural products/fractional extracts were based on their ability to (i) suppress the growth of cancer cell lines and (ii) alter the expression of target genes, either oncogenes or tumor suppressor genes. Genus and species names (binomial nomenclature) were assigned to the source organism of the natural products according to the NCBI Taxonomy database [30]. The different types of cancers were classified according to NCBI's MeSH [31]. The official HUGO Gene Nomenclature Committee (HGNC) [32] symbols were used for the human genes.
The adverse effects of the natural compounds/extracts on human health were also assessed via extensive literature mining using the keywords “adverse effect” or “toxic∗” or “side effect” and “natural product.” The articles retrieved from MEDLINE/PubMed were carefully examined for any association between the active compounds/extracts and toxicity in humans.
2.2. Pathway Enrichment Analysis
The retrieved target genes were provided as input to WebGestalt (WEB-based GEne SeT AnaLysis Toolkit) [33], an online tool for functional annotation enrichment analysis, to identify statistically significant overrepresented cancer-associated WikiPathways. The threshold for the FDR-adjusted p value was set at 10−3, and hypergeometric distribution was used.
2.3. Functional Association Network
The associations among the genes/proteins under study were investigated and visualized with the usage of STRING (Search Tool for Retrieval of Interacting Genes/Proteins) v11.0 [34, 35], a database of either known or predicted, direct or indirect, association data among genes or proteins. These data are derived from diverse resources, including text mining of the scientific literature, biological and biochemical pathways, gene coexpression, high-throughput experiments, and gene fusion. The confidence score for displaying interactions was set to 0.9.
The software platform Cytoscape (http://www.cytoscape.org/) [36] was used for network analysis, processing, and visualization.
2.4. Statistical Analyses
All statistical analyses were performed with the R package “Stats” and Microsoft Excel Macros.
To identify the minimum number of superfoods that target all cancer types, as well as the maximum number of target genes, we utilized the R package “RcppGreedySetCover” for resolving set cover problems.
2.5. Differential Gene Expression Analysis
RNA sequencing (RNA-seq) gene expression data for 27 tumor and corresponding normal tissue samples (Table S1), from the TCGA and GTEx databases, respectively, were downloaded from the GEPIA2 (Gene Expression Profiling Interactive Analysis) online web server [37] (http://gepia2.cancer-pku.cn/). The differentially expressed genes (DEGs) between tumor and normal samples were identified using one-way analysis of variance (ANOVA), by setting the cutoff value for absolute log fold change ∣log2FC | ≥2 and FDR-adjusted p value ≤ 0.05.
2.6. Database Design
2.6.1. Database Storage
“Cloud Firestore,” a NoSQL cloud database, was used to store and sync data for the client- and server-side web development. Cloud Firestore real-time data read-write feature was used for automatic data synchronization, thereby providing the user with the most updated data (https://firebase.google.com/docs/database).
2.6.2. Website Design
Google's “Firebase Hosting” was utilized to host NaturaProDB's static assets (HTML and JavaScript); the popular front-end framework VueJS was offered through officially maintained supporting packages for creating the website user interface. The generated networks were processed and analyzed by Cytoscape (http://www.cytoscape.org/); Cytoscape.js library (https://js.cytoscape.org/) was used for network implementation, visualization, and interaction in the designed user interface.
3. Results
3.1. Data Retrieval and Assembly
A total of 562 relevant articles were selected after thorough review; of those, 86 articles were included in our study according to the eligibility criteria. Data collected from those studies regarding the scientific name of the source organism or food, natural product or fractional extract mixture, target cancer type, cell lines used for assessing anticancer activity in in vitro experimental studies, and target gene symbol along with its expression status (up- or down-regulated) were merged and recorded in a table (Table S1). The natural products were divided into eight major groups: vegetables/fruits, herbs, spices, nuts, dairy products, cereals, marine organisms, and oil. The vast majority of natural products originated from plants; however, natural compounds were also extracted from fungi, bacteria, marine organisms, and dairy products. Collectively, 87 source organisms/foods, 19 types of cancers, 105 target genes, and 66 cell lines were retrieved from the eligible studies and the relevant data were recorded in an Excel worksheet. The distribution pattern of natural products or target genes with respect to types of cancers is shown in Figure 1. The greatest percentages of natural products and target genes are distributed within the solid tumor breast and colon cancer, as well as the blood cancer leukemia.
Figure 1.
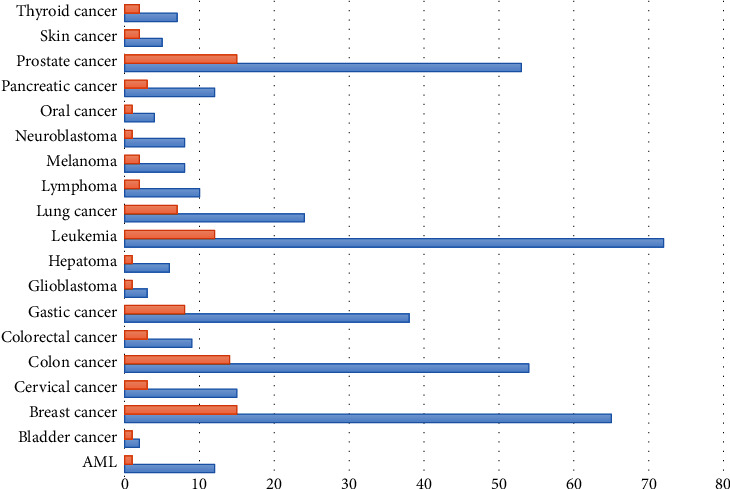
Bar graph depicting the distribution of natural products (orange) and genes (blue) with respect to cancer types.
A total of 81 pivotal target genes were found to participate in cancer-relevant, interdependent pathways. These 81 genes and their corresponding proteins appear to be highly interconnected within the functional network shown in Figure 2. The generated network is quite dense, with an average node degree of 12.4 (Figure 2), suggesting tight associations, either physical or functional, among molecules so as to exert their antineoplastic effect. The distribution of genes across pathways is depicted in Figure 3. Among the overrepresented pathways are those associated with genomic instability, prosurvival pathways, TP53-mediated signaling, apoptosis, and cell cycle.
Figure 2.
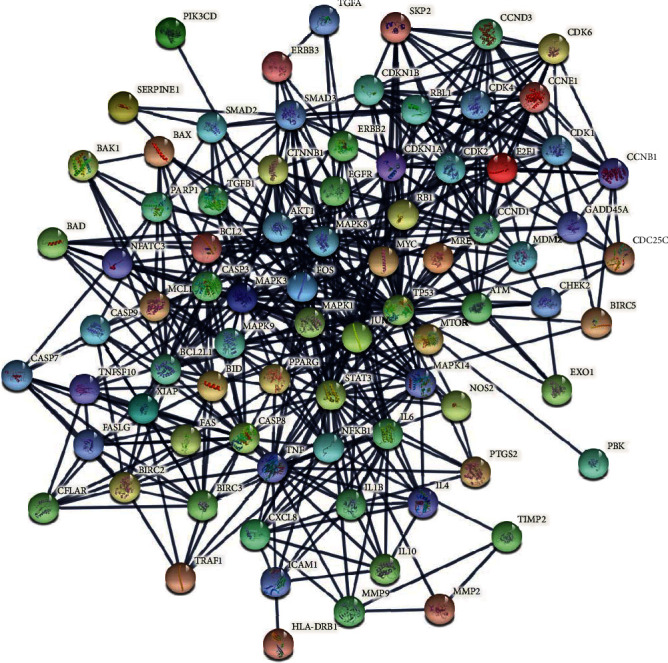
STRING output functional network of the 81 genes/gene products. The nodes represent molecules and the connecting lines (edges) indicate an interaction confidence score above 0.9.
Figure 3.
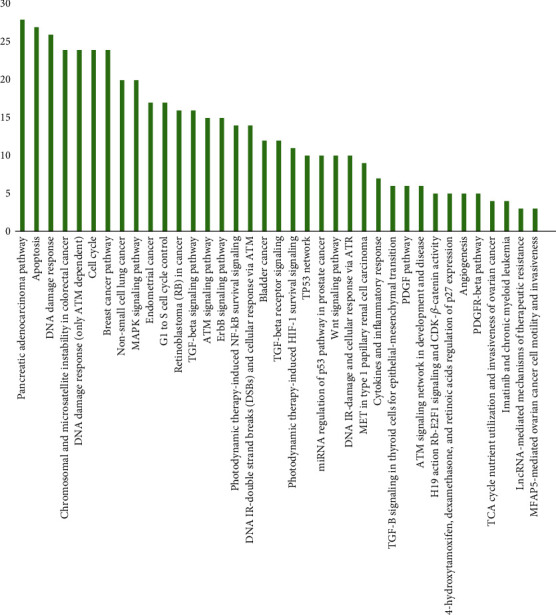
Bar plot depicting the overrepresented cancer-related pathways across target genes.
We found a total of 22 natural foods targeting 57 genes (Table 1), most of them plants, except the fungus hazel mushroom. The active substances/functional extracts of the corresponding natural foods, as well as the effective concentration needed for cell growth inhibition, are listed in the second and third columns of Table 1, respectively. Based on literature research, no adverse health effects of the natural compounds/functional extracts were reported for the same dose range as in the one in the third columns in Table 1. In addition, a bipartite network was generated (Figure 4) based on the identified DEGs in diverse cancer tissue samples (Table 2). Collectively, 45 out of the 57 target genes appear to be highly connected due to their differential coexpression in 27 types of cancers (Figure 4).
Table 1.
List of the selected organisms/foods, active compounds/extracts, effective inhibitory concentration, affected cancer type, and target genes.
| Source | Active substance/extract | Effective concentration | Cancer type | Target gene |
|---|---|---|---|---|
| Actinidia callosa var. callosa | Ethyl acetate fraction of Actinidia callosa var. callosa (EAAC) | IC50: 73.3 and 92.7 μg/ml for 24 h treatment | Hepatoma | AKT1, MAPK1, MMP2, MMP9, PIK3CD, TIMP2 |
| Aegle marmelos | Beta-caryophyllene and caryophyllene oxide fractions of Aegle marmelos extract | Maximum inhibition: 50 μg/ml for 24 h treatment | Lymphoma; neuroblastoma | ATM, BAK1, BAX, BCL2, CASP8, CASP9, MDM2, PTGS2 |
| Allium sativum (garlic) | Allicin | Maximum inhibition: 90 μg/ml for 24 h treatment | Glioblastoma | BAX, BCL2 |
| Allium sativum (garlic) | N-Benzyl-N-methyldecan-1-amine (NBNMA) | Maximum inhibition: 50 μg/ml for 24 h treatment | Leukemia | BAD, BAX, BCL2, BCL2L1, BIRC2, CASP3, CASP8, CASP9, CDK2, CDKN1A, XIAP |
| Aloe vera (Barbados aloe) | Aloin (AL) | IC50: 10 μmol/l for 72 h treatment | Colorectal cancer | BCL2L1, MYC, STAT3 |
| Anacardium occidentale (cashews) | Cardanol monoene (CM) extracted from cashew nut shell liquid (CNSL) | IC50: 23.15 ± 2.42 μM (24 h); 12.30 ± 1.67 μM (48 h) | Melanoma | BAX, BCL2, TP53, CASP3, PARP1 |
| Annona muricata | Ethyl acetate extract of Annona muricata leaves (EEAM) | IC50: 11.43 ± 1.87 μg/ml (HT-29); 8.98 ± 1.24 μg/ml (HCT-116) for 24 h treatment | Colon cancer | CASP3, CASP7, CASP8, CASP9, BAX, BCL2 |
| Arachis hypogaea (peanuts) | Resveratrol | IC50: 46.81 ± 1.26 μM (MCF-7); 53.02 ± 2.42 μM (HeLa) for 72 h treatment | Breast cancer; cervical cancer | CDK4, CDKN1A, MAPK3, TP53 |
| Arachis hypogaea (peanuts) | Peanut skin procyanidins (PSP) | IC50: 48.57 μg/ml for 24 h treatment | Prostate cancer | BAX, BCL2, CASP3, TP53 |
| Basella rubra (spinach) | Natural antioxidants (NAOs) from spinach extract | MIC: 3.2 mg/ml for 24 h treatment | Prostate cancer | CDK2, CDKN1A, E2F1, RB1, RBL1 |
| Black pepper | Piperine | IC50: 54 ± 5 μM to 126 ± 3 μM for 72 h treatment | Colon cancer | AKT1, BIRC5, CCND1, CCND3, CDKN1A, CDKN1B, MAPK8 |
| Black soybean | Black soybean extract | IC50: 3.69 mg/ml for 72 h treatment | Gastric cancer | BAX, BCL2, CASP3 |
| Brassica spp. vegetables (cabbage, cauliflower, and brussels sprouts) | Indole-3-carbinol (I3C) | Maximum inhibition: 50 μM for 24 h treatment | Acute myeloid leukemia (AML); leukemia | AKT1, BCL2, BIRC2, BIRC5, CCND1, MMP9, NFKB1, NOS2, PTGS2, TNF, TRAF1, XIAP |
| Castanea (chestnut) | Ethanol extracts of raw chestnut (RCE) | Maximum inhibition: 200 μg/ml for 24 h treatment | Gastric cancer | ASP3, CASP7, CASP8, FASLG, HLA-DRB1,PARP1, TNFSF10, XIAP |
| Citrus aurantium | Flavonoids | IC50: 99 μg/ml for 24 h treatment | Gastric cancer | CASP3, CCNB1, CDK1, PARP1 |
| Curcuma longa (turmeric) | Curcumin | Maximum inhibition: 25 μmol/l for 24 h treatment | Bladder cancer | NFKB1, PTGS2 |
| Curcuma longa (turmeric) | Curcumin | IC50: 11 to 46 μM (72 h) | Pancreatic cancer | CXCL8, NFKB1, PTGS2 |
| Curcuma longa (turmeric) | Curcumin | Maximum inhibition: 50 μM for 24 h treatment | Thyroid cancer | SMAD2, SMAD3, TGFB1 |
| Daphne genkwa | Yuanhuadine | Maximum inhibition: 32 nM for 72 h treatment | Lung cancer | AKT1, CDK2, CDK4, CDKN1A, MYC |
| Ipomoea batatas (sweet potato) | Sporamin | Maximum inhibition: 100 μM for 72 h treatment | Pancreatic cancer | BAX, BCL2, BCL2L1, NFKB1 |
| Juglans mandshruica (Manchurian walnut) | Juglanin | IC50: 20.07 to 29.13 μM for 24 h treatment | Breast cancer | BAD, BAX, BCL2, CASP3, CASP8, CASP9, CDC25C, CDK1, CDKN1B, CHEK2 |
| Juglans mandshruica (Manchurian walnut) | Juglone | IC50: 8 μM for 24 h treatment | Leukemia | AKT1, CASP3, MTOR, PIK3CD |
| Lebanese Daucus carota (wild carrot) | Daucus carota oil extract (DCOE) | IC50: 10.2 ± 0.90 to 19.1 ± 0.98 μM for 48 h treatment | Skin Cancer | AKT1, BCl2, BAX |
| Naematoloma sublateritium (hazel mushroom) | Hexane fraction of N. sublateritium extract (HFNS) | IC50: 200 μg/ml for 24 h treatment | Breast Cancer | JUN, MAPK14, MAPK8, MAPK9, MMP9, NFKB1, SERPINE1, TIMP2 |
| Pulsatilla koreana | Pulsatilla koreana extract (PKE) | IC50: 120 to 140 μg/ml for 24 h treatment | Thyroid cancer | BAX, BCL2, CASP3, PARP1 |
| Red sorghum bran | 3-Deoxyanthocyanidins | IC50: 300 μg/ml for 24 h treatment | Breast Cancer | BCL2, TP53 |
| Sanguinaria canadensis (bloodroot) | Sanguinarine | IC50: 1 μM; maximum inhibition: 2 μM for 24 h treatment | Oral cancer | AKT1, CASP3, CASP9, PIK3CD |
MIC: minimum inhibitory concentration. IC50: half-maximal effective inhibitory concentration.
Figure 4.
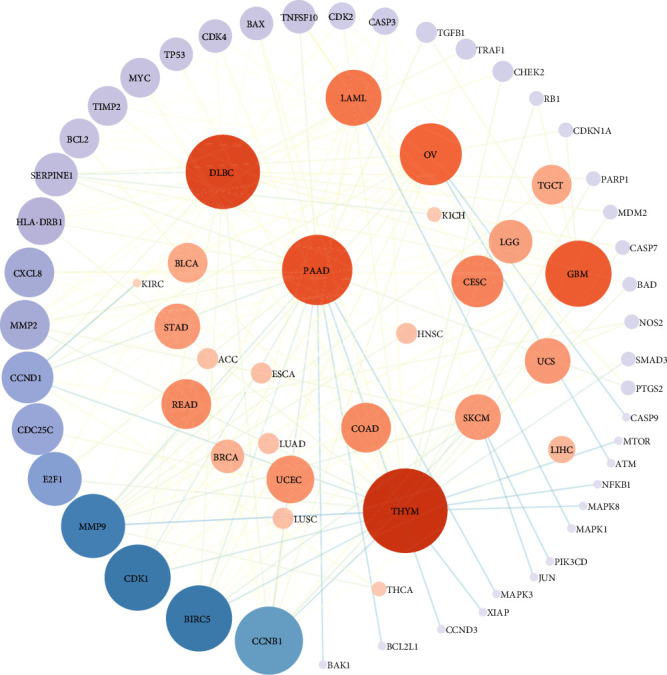
Bipartite network displaying TCGA-derived cancer-gene associations. The interactions are presented in a circular mode; the cancer types are shown at the center and the genes at the periphery. The size of the nodes is proportional to their connectivity degree.
Table 2.
Differentially expressed genes in diverse TCGA-derived cancers.
| Gene | TCGA cancer type | Status |
|---|---|---|
| ATM | OV | Down |
| BAD | DLBC,THYM | Up |
| BAK1 | PAAD | Up |
| BAX | DLBC,GBM,PAAD,TGCT,THYM | Up |
| BCL2 | CESC,OV,UCS | Down |
| BCL2 | DLBC,LAML,THYM | Up |
| BCL2L1 | PAAD | Up |
| BIRC5 | LAML | Down |
| BIRC5 | BLCA,BRCA,CESC,COAD,DLBC,GBM,LIHC,LUAD,LUSC,OV,PAAD,READ,SKCM,STAD,THYM,UCEC,UCS | Up |
| CASP3 | DLBC,GBM,PAAD,THYM | Up |
| CASP7 | DLBC,THYM | Up |
| CASP9 | OV | Down |
| CCNB1 | LAML | Down |
| CCNB1 | ACC,BLCA,BRCA,CESC,COAD,DLBC,GBM,LIHC,LUAD,LUSC,OV,PAAD,READ,SKCM,STAD,THYM,UCEC,UCS | Up |
| CCND1 | LAML | Down |
| CCND1 | COAD,DLBC,KIRC,LGG,OV,PAAD,READ,STAD,THCA,THYM | Up |
| CCND3 | PAAD | Up |
| CDC25C | LAML | Down |
| CDC25C | CESC,COAD,DLBC,GBM,OV,READ,STAD,THYM,UCEC,UCS | Up |
| CDK1 | LAML | Down |
| CDK1 | ACC,BLCA,BRCA,CESC,COAD,DLBC,GBM,LIHC,LUAD,LUSC,OV,PAAD,READ,STAD,THYM,UCEC,UCS | Up |
| CDK2 | DLBC,GBM,SKCM,THYM | Up |
| CDK4 | DLBC,GBM,LGG,SARC,TGCT,THYM | Up |
| CDKN1A | OV | Down |
| CDKN1A | GBM | Up |
| CHEK2 | DLBC,TGCT,THYM | Up |
| CXCL8 | CESC,COAD,ESCA,GBM,HNSC,PAAD,READ,SKCM,STAD,UCEC | Up |
| E2F1 | BLCA,BRCA,CESC,COAD,DLBC,LIHC,OV,PAAD,READ,THYM,UCEC,UCS | Up |
| HLA-DRB1 | DLBC,GBM,LAML,LGG,OV,PAAD,STAD,TGCT,THYM | Up |
| JUN | SKCM | Down |
| MAPK1 | LAML | Down |
| MAPK3 | PAAD | Up |
| MAPK8 | THYM | Up |
| MDM2 | DLBC,SARC,THYM | Up |
| MMP2 | ACC,CESC,OV,SKCM | Down |
| MMP2 | DLBC,GBM,LAML,LGG,PAAD,THYM | Up |
| MMP9 | THYM | Down |
| MMP9 | BLCA,BRCA,CESC,COAD,ESCA,GBM,HNSC,OV,PAAD,READ,SKCM,STAD,TGCT,THCA,UCEC,UCS | Up |
| MTOR | THYM | Up |
| MYC | COAD,DLBC,GBM,LGG,READ,THYM | Up |
| NFKB1 | THYM | Up |
| NOS2 | COAD,READ | Up |
| PARP1 | DLBC,THYM | Up |
| PIK3CD | SKCM | Up |
| PTGS2 | LAML,PAAD | Up |
| RB1 | GBM,THYM | Up |
| SERPINE1 | KICH,OV | Down |
| SERPINE1 | DLBC,ESCA,GBM,HNSC,PAAD | Up |
| SMAD3 | UCEC | Down |
| SMAD3 | THYM | Up |
| TGFB1 | GBM,LGG,PAAD | Up |
| TIMP2 | BLCA,CESC,OV,UCEC | Down |
| TIMP2 | LAML,PAAD | Up |
| TNFSF10 | KICH | Down |
| TNFSF10 | CESC,LAML,PAAD,TGCT | Up |
| TP53 | DLBC,GBM,LAML,LGG,THYM | Up |
| TRAF1 | OV,UCS | Down |
| TRAF1 | DLBC | Up |
| XIAP | THYM | Up |
Among the targeted genes are the key proapoptotic BAX (BCL2-associated X protein) [38], the caspases CASP3/8/9, and the cardinal player TP53 which are consistently upregulated in diverse cancer cell lines following treatment with natural compounds (Table 1). Furthermore, the antiapoptotic genes BCL2 and XIAP (X-linked inhibitor of apoptosis) [39, 40] are downregulated by natural products and upregulated in several cancer tissues (Tables 1 and 2). BCL2 can also suppress apoptosis by inhibiting the activity of caspases indispensable for apoptosis, such as CASP3 [41, 42]. The DNA damage response-associated gene PARP1 (poly(ADP-ribose) polymerase 1), pharmacological inhibitors of which are used in anticancer treatment [43], and CDKN1A (cyclin dependent kinase inhibitor 1A), a universal inhibitor of CDK/cyclin complexes [44, 45], are involved in DNA damage detection and DNA damage-induced cell cycle arrest [44, 46], respectively (Table 1). In addition, several natural products can potentially exert their antineoplastic effect on the oncogene AKT1 (AKT serine/threonine kinase 1) [47]. Among the genes that are targeted by the 22 superfoods is NFKB1, which plays a protagonistic role in inflammatory responses [48, 49] (Table 1). NFKB1 also plays a dual role in apoptosis, either as inducer or as inhibitor of apoptosis [50]; thus, NFKB1 was found to be both up- or downregulated by natural compounds in different cancers, as well as the same type of cancer (Table S1).
3.2. NaturaProDB
The data presented in Table S1 were deposited in a repository, referred to as the National Technical University of Athens Anticancer Products Database (NaturaProDB; https://naturaprodb.web.app/). This database has a user friendly interface and can be queried using different options, that is, by (a) source, (b) natural product, (c) target cancer, (d) target gene, and (e) expression status of target gene, as well as the combination of the above options (Figure 5(a)). The results appear in a new window, in a tabulated format. Each entry contains the (i) general class of natural sources, (ii) food or organism, (iii) constituent compound/functional extract, (iv) target cancer, (v) target gene, (vi) gene status (up- or downregulated), (vii) cell line tested, and (viii) a hyperlink to the corresponding PubMed webpage (Figure 5(b)). The search output is provided in a JSON or TSV format. By clicking on the “Networks” tab, two interactive networks are displayed: the “TCGA Network” shown in Figure 4 and the “Natural Products Network” which includes food-target cancer-target gene associations (listed in Table S1). The latter network is highly interconnected, suggesting that natural products target multiple and diverse cancer cells and corresponding genes (Figure 5(c)).
Figure 5.

NaturaProDB workflow showing the (a) example input query, (b) example results page, and (c) “Natural Products Network,” where the node size is proportional to their connectivity degree.
4. Discussion
There is an ongoing need for alternative, effective, economical resources for drug development. Natural compounds with antineoplastic potential, as compared to synthetic compounds, are considered to be more efficacious and bioavailable and cost effective, with less toxic adverse effects [1]. The constant demand for natural antioncogenic medicines is also reflected in the number of databases dedicated to natural products with anticancer activity, such as CancerHSP [51], CHMIS-C [52], InPACdb [53], NPACT [54], and NCARE [55].
Another parameter that must be taken into consideration is the cellular, genetic, and metabolic heterogeneity of cancers and the complex tumor microenvironment. Designing of broad spectrum therapeutic strategies represents an intriguing solution to this problem. The so-called “dirty” drugs have multiple, instead of single, molecular targets. Within this context, the anticancer activity of several natural compounds could be safely combined in such a way as to maximize their inherent additive and synergistic effects and avoid any side effects, towards the design of potent dirty drugs. In our study, we suggest a combination of 22 natural foods which, based on in vitro experimental studies, could potentially target all 19 types of cancers and 57 key cancer-relevant genes under investigation. These genes are implicated in interconnected pathways (Figure 2), including those related to genomic integrity and cell cycle control. In particular, chromosomal instability and DNA damage response and repair (DDR/R) pathways are known to contribute largely to carcinogenesis. One of the characteristics of cancer is the presence of genomic lesions, caused either directly or indirectly, through the generation of DNA-damaging intermediates, like reactive oxygen species (ROS) and in general free radicals (e.g., hydrogen ion and hydroxide) [56, 57]. These genomic lesions, if not properly processed, could lead to genomic instability and eventually to carcinogenesis [58]. ATM (Table 1) plays a protagonistic role in the initial stage of DDR/R, that is, DNA damage detection and stress-response signaling [59, 60]. ATM signaling is activated by a wide variety of DNA lesions and DNA replication stress [61, 62]. Cyclin D1 (CCND1) (Table 1) was demonstrated to induce post-DNA damage cell cycle arrest and apoptosis in different types of cancers [63, 64].
Moreover, several cell prosurvival pathways (Figure 2) were found to be enriched in the gene set. For example, the transforming growth factor beta (TGFB), MAPK, ERBB, HIF-1, WNT, PDGF/PDGFRB, and NFKB signaling pathways are implicated in several aspects of cancer initiation, promotion, and progression [65–71], by mediating survival of cancer cells. The apoptotic pathway is also overrepresented. Cancer cells have developed the ability to evade apoptosis, mainly due to deregulation of key proapoptotic molecules like TP53, BAX, BCL2, BIRC5, and XIAP (Table 1).
Several of the plants (e.g., garlic, peanuts, spinach, and black soybean) listed in Table 1 contain more than one active compound or phytochemicals with anticancer activity (Table S1), suggesting that they exert their anticancer effect in an additive or synergistic manner. One of the garlic clove ingredients, the phenylamine NBNMA (Table 1), was shown to induce cell cycle arrest, via downregulation of the cell cycle M-phase inducer cyclin dependent kinase 2 (CDK2) and overexpression of CDKN1A, as well as apoptosis, by activation of the proapoptotic factors CASP3, -8, -9, BAX, and BAD and, conversely, inactivation of the antiapoptotic BCL2, BCL2L1, BIRC2, and XIAP in leukemic cells [72]. As it is shown in Table 2, BCL2 is overexpressed in acute myeloid leukemia. Another garlic compound, allicin was shown to sensitize hepatocellular carcinoma cells to anticancer agents via a ROS-dependent signaling pathway [73]. Allicin, also, increases the sensitivity of colorectal cancer cells to radiation through the inhibition of a NFKB1-mediated pathway [74]. Furthermore, allicin potentiates apoptosis in human glioblastoma cells, by elevating the expression of BAX and downregulating BCL2 (Table 1) [75]. Peanuts, which contain procyanidins in their skin, can inhibit the proliferation of prostate cancer cells and promote apoptotic cell death by downregulating BCL2 and upregulating the proapoptotic factors BAX, CASP3, and the TP53 [76]. The phenolic compounds, which comprise the largest group of phytochemicals, are known to exert their antineoplastic effect by contributing to cell proliferation, apoptosis, angiogenesis, metastasis, and inflammation under oxidative stress [77–81]. The phenolic antioxidant resveratrol in peanuts (Table 1) was shown to exhibit antiproliferative activity in cervical and breast cancer cells, by decreasing the expression of the DNA damage-induced prosurvival protein kinases MAPK3 and CDK4 and, conversely, elevating TP53 and CDKN1A [82].
Juglone, a phenolic compound in the Manchurian walnut, was shown to inhibit the proliferation of human leukemia cells and enhance apoptosis [83]. In particular, Juglone (Table 1) markedly inhibited the phosphorylation of PI3K/AKT/mTOR, a major antiapoptotic and prosurvival signaling pathway which is overactivated in multiple cancers [84], and induced the cleavage and activation of the proapoptotic procaspase-3. Moreover, juglanin was shown to inhibit the proliferation of breast cancer cell proliferation through the differential regulation of cell cycle-associated proteins (i.e., CDC25C, CDK1, CDKN1B, and CHEK2), the activation of the proapoptotic factors BAD, BAX, CASP3, -8 and -9, and, conversely, the suppression of the antiapoptotic protein BCL2 [85]. Of note, CDK1 (cyclin-dependent kinase 1), which is downregulated by the flavonoid compound juglanin (Table 1), was found to be overexpressed in invasive breast cancer (Table 2); Izadi et al. suggest that CDK1 is the best CDK target for breast cancer therapy [86].
The cancer-preventive properties of the cruciferous vegetables (belonging to the family Brassicaceae) have been acknowledged for a long time. The active ingredient indole-3-carbinol (I3C) has been documented to play a role in the prevention of several cancers [87]. For example, I3C was found to inhibit the growth of prostate cancer cells and induce G1 cell cycle arrest and apoptotic cell death [88]. Furthermore, it was demonstrated by Takada and colleagues [89] that I3C (Table 1) blocks the expression of NFKB1-regulated prometastatic, proproliferation, and antiapoptotic gene products (i.e., AKT1, BCL2, BIRC2, BIRC5, CCND1, MMP9, PTGS2, TNF, and XIAP) in myeloid and leukemia cells. Of those genes, BCL2 and PTGS2 were shown to be upregulated in acute myeloid leukemia (Table 2). In the same study, the gene encoding NOS2 (nitric oxide synthase 2), which catalyzes the production of the reactive free radical nitric oxide, was downregulated, leading to the suggestion that I3C might exert antioxidant activity.
Flavonoids present in citrus fruits have been documented to exhibit cancer-preventive potential by participating in cell cycle inhibition, suppression of metastasis, and angiogenesis, as well as anti-inflammatory signal transduction pathways [90]. In particular, flavonoids isolated from Citrus aurantium were shown to inhibit the growth of human gastric cancer cells, by suppressing the proteins CCNB1 (cyclin B1) and CDK1 (Table 1) which control cell cycle progression; the corresponding genes CCNB1 and CDK1 were found to be upregulated in stomach cancer (Table 2). In addition, activation of CASP3 together with the inactivation of PARP1, which is involved in DNA damage repair, potentiated apoptosis [91].
The spice Curcuma longa, commonly known as turmeric or “Indian saffron,” has been used in the folk medicine of India for thousands of years. Curcumin, the major active ingredient of turmeric, is a powerful antioxidant, with well-documented anti-inflammatory and anticancer potential [92]. Curcumin was shown to suppress growth of bladder and pancreatic cancer cell lines through the inhibition of NFKB1-regulated proinflammatory and proproliferative gene products PTGS2 [93] and CXCL8 [94] (Table 1); both CXCL8 and PTGS2 were also found to be downregulated in pancreatic cancer (Table 2). Moreover, curcumin was demonstrated to inhibit metastasis in human papillary thyroid carcinoma cells by downregulating components of the prometastatic signaling pathway TGFB1/SMAD2/SMAD3 [95] (Table 1).
Other phytocompounds, like sporamin in sweet potato [96–98], piperine in black pepper [99], sanguinarine in bloodroot [100–102], 3-deoxyanthocyanins in red sorghum bran [103], and aloin in Aloe vera [104], have been documented to exhibit anticancer effects in several cancers. Of importance, these agents possess antioxidant properties [105–108]. Aloin [104] and piperine [109] can inhibit proliferation of (colo)rectal cancer cells; the genes MYC and BIRC5, the expression of which is decreased by the two compounds (Table 1), are otherwise overexpressed in colon adenocarcinoma (Table 2). Similarly, the antiapoptotic BCL2L1 gene, which was found downregulated in sporaminin-induced apoptotic pancreatic cancer cells [97] (Table 1), is overexpressed in pancreatic adenocarcinoma (Table 2). Furthermore, β-caryophyllene and β-caryophyllene oxide extracted from Aegle marmelos are suggested to possess anti-inflammatory potential and were demonstrated to induce apoptosis in cancer cells of diverse tissue origin, that is, lymphoma (i.e., haematological cancer) and neuroblastoma (i.e., nerve tissue neoplasm), through overexpression of the proapoptotic (ATM, BAK1, BAX, and CASP8/9) and underexpression of the antiapoptotic (BCL2, MDM2 proto-oncogene, and PTGS2) genes [110] (Table 1). The genes BCL2 and MDM2 are also upregulated in lymphoid neoplasm diffuse large B-cell lymphoma (Table 2).
5. Conclusions
Nowadays, there is a great need for drugs derived from nature that can be 100% compatible if possible to the human organism, with a potential applicability in the fight against human diseases. In this study, we have targeted human cancer. Based on systematic searches and use of current bioinformatics methodologies, we have designed a biodatabase exceeding the existing standards of natural product databases, i.e., at a systems biology level. We have found that diverse natural products, with no observed adverse health effects, can target dissimilar cancer types through the significant alteration of the expression of multiple, common genes, which are involved in shared interconnected cancer-relevant pathways. Therefore, the efficient and safe combination of bioactive compounds derived from natural resources can be potentially applied to exert cytotoxic effects on diverse types of cancer cells and regulate the expression of numerous target genes which play a central role in cancer pathways. Further studies in animal models should be directed towards the investigation of the chemopreventive actions and safety of those compounds. The aforementioned findings should be taken into consideration in the rational design of drugs with broad spectrum activity, such as “dirty” drugs. Lastly, NaturaProDB was developed to facilitate the retrieval of relevant information. In conclusion, in this study, we provide a novel and effective systems biology approach to investigate the potential value of the combined activity of natural products in cancer chemoprevention, which can be exploited in the development of anticancer multitargeted therapies.
Acknowledgments
A.P. would like to extend her thanks to Izmir Biomedicine and Genome Center (IBG), Izmir, Turkey, for its support in carrying out the project. I.T. acknowledges the YÖK (Yükseköğrenim Kurulu) scholarship.
Abbreviations
- ACC:
Adrenocortical carcinoma
- BLCA:
Bladder urothelial carcinoma
- BRCA:
Breast-invasive carcinoma
- CESC:
Cervical squamous cell carcinoma and endocervical adenocarcinoma
- COAD:
Colon adenocarcinoma
- DLBC:
Lymphoid neoplasm diffuse large B-cell lymphoma
- ESCA:
Esophageal carcinoma
- GBM:
Glioblastoma multiforme
- HNSC:
Head and neck squamous cell carcinoma
- KICH:
Kidney chromophobe
- KIRC:
Kidney renal clear cell carcinoma
- LAML:
Acute myeloid leukemia
- LGG:
Brain lower-grade glioma
- LIHC:
Liver hepatocellular carcinoma
- LUAD:
Lung adenocarcinoma
- LUSC:
Lung squamous cell carcinoma
- OV:
Ovarian serous cystadenocarcinoma
- PAAD:
Pancreatic adenocarcinoma
- READ:
Rectum adenocarcinoma
- SARC:
Sarcoma
- SKCM:
Skin cutaneous melanoma
- STAD:
Stomach adenocarcinoma
- TGCT:
Testicular germ cell tumors
- THCA:
Thyroid carcinoma
- THYM:
Thymoma
- UCEC:
Uterine corpus endometrial carcinoma
- UCS:
Uterine carcinosarcoma.
Contributor Information
Athanasia Pavlopoulou, Email: athanasia.pavlopoulou@ibg.edu.tr.
Alexandros G. Georgakilas, Email: alexg@mail.ntua.gr.
Data Availability
The data used to support the findings of this study are available from the corresponding authors upon logical request.
Conflicts of Interest
The authors declare no conflicts of interest.
Authors' Contributions
Conceptualization was done by A.G.G., D.T., and A.P.; methodology was done by D.T., I.T., G.K., and A.P.; software was acquired by D.T., I.T., G.M.B., and V.Z.; validation was done by D.T., I.T., V.Z., and A.P.; formal analysis was done by D.T., I.T., G.K., and A.P.; investigation was done D.T.; data curation was done by D.T., I.T., G.M.B., and A.P.; writing and original draft preparation were done by A.G.G., D.T., I.T., and A.P.; writing and review and editing were done by all authors.; visualization was done by I.T., G.M.B., G.K., and A.P.; supervision was done by A.G.G. and A.P.; project administration was done by A.G.G. and A.P.; all authors have read and agreed to the current version of the manuscript. Dionysia Theofylaktou and Işıl Takan contributed equally to this work.
Supplementary Materials
Table S1: the class, natural source, active ingredient, the affected cancer type(s), target gene(s), gene expression status, cell line used in the experimental study, and corresponding PubMed link are listed.
References
- 1.Block K. I., Gyllenhaal C., Lowe L., et al. Designing a broad-spectrum integrative approach for cancer prevention and treatment. Seminars in cancer biology. 2015;35(Suppl):S276–S304. doi: 10.1016/j.semcancer.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cragg G. M., Newman D. J. Plants as a source of anti-cancer agents. Journal of Ethnopharmacology. 2005;100(1-2):72–79. doi: 10.1016/j.jep.2005.05.011. [DOI] [PubMed] [Google Scholar]
- 3.da Rocha A. B., Lopes R. M., Schwartsmann G. Natural products in anticancer therapy. Current Opinion in Pharmacology. 2001;1(4):364–369. doi: 10.1016/S1471-4892(01)00063-7. [DOI] [PubMed] [Google Scholar]
- 4.Bacchetti T., Turco I., Urbano A., Morresi C., Ferretti G. Relationship of fruit and vegetable intake to dietary antioxidant capacity and markers of oxidative stress: a sex-related study. Nutrition. 2019;61:164–172. doi: 10.1016/j.nut.2018.10.034. [DOI] [PubMed] [Google Scholar]
- 5.Jurenka J. S. Anti-inflammatory properties of curcumin, a major constituent of Curcuma longa: a review of preclinical and clinical research. Alternative Medicine Review : A Journal of Clinical Therapeutic. 2009;14(2):141–153. [PubMed] [Google Scholar]
- 6.Coussens L. M., Werb Z. Inflammation and cancer. Nature. 2002;420(6917):860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kryston T. B., Georgiev A. B., Pissis P., Georgakilas A. G. Role of oxidative stress and DNA damage in human carcinogenesis. Mutation Rsesearch. 2011;711(1-2):193–201. doi: 10.1016/j.mrfmmm.2010.12.016. [DOI] [PubMed] [Google Scholar]
- 8.Jiang T. A. Health benefits of culinary herbs and spices. Journal of AOAC International. 2019;102(2):395–411. doi: 10.5740/jaoacint.18-0418. [DOI] [PubMed] [Google Scholar]
- 9.Key T. J. Fruit and vegetables and cancer risk. British Journal of Cancer. 2011;104(1):6–11. doi: 10.1038/sj.bjc.6606032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee J., Shin A., Oh J. H., Kim J. Colors of vegetables and fruits and the risks of colorectal cancer. World Journal of Gastroenterology. 2017;23(14):2527–2538. doi: 10.3748/wjg.v23.i14.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raffa D., Maggio B., Raimondi M. V., Plescia F., Daidone G. Recent discoveries of anticancer flavonoids. European Journal of Medicinal Chemistry. 2017;142:213–228. doi: 10.1016/j.ejmech.2017.07.034. [DOI] [PubMed] [Google Scholar]
- 12.Steinmetz K. A., Potter J. D. Vegetables, fruit, and cancer prevention: a review. Journal of the American Dietetic Association. 1996;96(10):1027–1039. doi: 10.1016/S0002-8223(96)00273-8. [DOI] [PubMed] [Google Scholar]
- 13.Neveu V., Perez-Jiménez J., Vos F., et al. Phenol-Explorer: an online comprehensive database on polyphenol contents in foods. Database : the journal of biological databases and curation. 2010;2010, article bap024 doi: 10.1093/database/bap024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin K. Y. The spice for joint inflammation: anti-inflammatory role of curcumin in treating osteoarthritis. Drug Design, Development and Therapy. 2016;Volume 10:3029–3042. doi: 10.2147/DDDT.S117432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sammar M., Abu-Farich B., Rayan I., Falah M., Rayan A. Correlation between cytotoxicity in cancer cells and free radical-scavenging activity: <i>In vitro</i> evaluation of 57 medicinal and edible plant extracts. Oncology Letters. 2019;18(6):6563–6571. doi: 10.3892/ol.2019.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brufau G., Boatella J., Rafecas M. Nuts: source of energy and macronutrients. The British Journal of Nutrition. 2006;96(Suppl 2):S24–S28. doi: 10.1017/BJN20061860. [DOI] [PubMed] [Google Scholar]
- 17.Ros E. Health benefits of nut consumption. Nutrients. 2010;2(7):652–682. doi: 10.3390/nu2070652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ros E., Mataix J. Fatty acid composition of nuts--implications for cardiovascular health. The British Journal of Nutrition. 2006;96(Suppl 2):S29–S35. doi: 10.1017/BJN20061861. [DOI] [PubMed] [Google Scholar]
- 19.Salas-Salvado J., Bullo M., Perez-Heras A., Ros E. Dietary fibre, nuts and cardiovascular diseases. The British Journal of Nutrition. 2006;96(Suppl 2):S46–S51. doi: 10.1017/bjn20061863. [DOI] [PubMed] [Google Scholar]
- 20.Segura R., Javierre C., Lizarraga M. A., Ros E. Other relevant components of nuts: phytosterols, folate and minerals. The British Journal of Nutrition. 2006;96(Suppl 2):S36–S44. doi: 10.1017/BJN20061862. [DOI] [PubMed] [Google Scholar]
- 21.Blomhoff R., Carlsen M. H., Andersen L. F., Jacobs D. R., Jr. Health benefits of nuts: potential role of antioxidants. The British Journal of Nutrition. 2006;96(Suppl 2):S52–S60. doi: 10.1017/BJN20061864. [DOI] [PubMed] [Google Scholar]
- 22.Falasca M., Casari I. Cancer chemoprevention by nuts: evidence and promises. Frontiers in Bioscience. 2012;4:109–120. doi: 10.2741/254. [DOI] [PubMed] [Google Scholar]
- 23.Aneiros A., Garateix A. Bioactive peptides from marine sources: pharmacological properties and isolation procedures. Journal of Chromatography B, Analytical Technologies in the Biomedical and Life Sciences. 2004;803(1):41–53. doi: 10.1016/j.jchromb.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Braña A. F., Fiedler H. P., Nava H., et al. Two Streptomyces species producing antibiotic, antitumor, and anti-inflammatory compounds are widespread among intertidal macroalgae and deep-sea coral reef invertebrates from the central Cantabrian Sea. Microbial Ecology. 2015;69(3):512–524. doi: 10.1007/s00248-014-0508-0. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y., Phat C., Hong S. C. Structural diversity of marine cyclic peptides and their molecular mechanisms for anticancer, antibacterial, antifungal, and other clinical applications. Peptides. 2017;95:94–105. doi: 10.1016/j.peptides.2017.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Shyu P. T., Oyong G. G., Cabrera E. C. Cytotoxicity of probiotics from Philippine commercial dairy products on cancer cells and the effect on expression of cfos and cjun early apoptotic-promoting genes and interleukin-1 beta and tumor necrosis factor-alpha proinflammatory cytokine genes. BioMed Research International. 2014;2014:9. doi: 10.1155/2014/491740.491740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hasson S. S., Al-Busaidi J. Z., Al-Qarni Z. A., et al. In vitro apoptosis triggering in the BT-474 human breast cancer cell line by lyophilised camel’s milk. Asian Pacific Journal of Cancer Prevention: APJCP. 2015;16(15):6651–6661. doi: 10.7314/APJCP.2015.16.15.6651. [DOI] [PubMed] [Google Scholar]
- 28.de Mejia E. G., Dia V. P. The role of nutraceutical proteins and peptides in apoptosis, angiogenesis, and metastasis of cancer cells. Cancer Metastasis Reviews. 2010;29(3):511–528. doi: 10.1007/s10555-010-9241-4. [DOI] [PubMed] [Google Scholar]
- 29.Tung Y. T., Chen H. L., Yen C. C., et al. Bovine lactoferrin inhibits lung cancer growth through suppression of both inflammation and expression of vascular endothelial growth factor. Journal of Dairy Science. 2013;96(4):2095–2106. doi: 10.3168/jds.2012-6153. [DOI] [PubMed] [Google Scholar]
- 30.Federhen S. The NCBI taxonomy database. Nucleic Acids Research. 2012;40(D1):D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayers E. W., Agarwala R., Bolton E. E., et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Research. 2019;47(D1):D23–D28. doi: 10.1093/nar/gky1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Braschi B., Denny P., Gray K., et al. Genenames. org: the HGNC and VGNC resources in 2019. Nucleic Acids Research. 2019;47(D1):D786–D792. doi: 10.1093/nar/gky930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y., Wang J., Jaehnig E. J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Research. 2019;47(W1):W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Szklarczyk D., Gable A. L., Lyon D., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szklarczyk D., Gable A. L., Nastou K. C., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Research. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon P., Markiel A., Ozier O., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang Z., Kang B., Li C., Chen T., Zhang Z. GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Research. 2019;47(W1):W556–W560. doi: 10.1093/nar/gkz430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pawlowski J., Kraft A. S. Bax-induced apoptotic cell death. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(2):529–531. doi: 10.1073/pnas.97.2.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deveraux Q. L., Reed J. C. IAP family proteins--suppressors of apoptosis. Genes & Development. 1999;13(3):239–252. doi: 10.1101/gad.13.3.239. [DOI] [PubMed] [Google Scholar]
- 40.Duckett C. S., Nava V. E., Gedrich R. W., et al. A conserved family of cellular genes related to the baculovirus iap gene and encoding apoptosis inhibitors. The EMBO Journal. 1996;15(11):2685–2694. doi: 10.1002/j.1460-2075.1996.tb00629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Porter A. G., Janicke R. U. Emerging roles of caspase-3 in apoptosis. Cell Death and Differentiation. 1999;6(2):99–104. doi: 10.1038/sj.cdd.4400476. [DOI] [PubMed] [Google Scholar]
- 42.Swanton E., Savory P., Cosulich S., Clarke P., Woodman P. Bcl-2 regulates a caspase-3/caspase-2 apoptotic cascade in cytosolic extracts. Oncogene. 1999;18(10):1781–1787. doi: 10.1038/sj.onc.1202490. [DOI] [PubMed] [Google Scholar]
- 43.Malyuchenko N. V., Kotova E. Y., Kulaeva O. I., Kirpichnikov M. P., Studitskiy V. M. PARP1 inhibitors: antitumor drug design. Acta Naturae. 2015;7(3):27–37. doi: 10.32607/20758251-2015-7-3-27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Waldman T., Kinzler K. W., Vogelstein B. p21 is necessary for the p53-mediated G1 arrest in human cancer cells. Cancer Research. 1995;55(22):5187–5190. [PubMed] [Google Scholar]
- 45.Xiong Y., Hannon G. J., Zhang H., Casso D., Kobayashi R., Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366(6456):701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
- 46.Bunz F., Dutriaux A., Lengauer C., et al. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282(5393):1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 47.Staal S. P., Hartley J. W., Rowe W. P. Isolation of transforming murine leukemia viruses from mice with a high incidence of spontaneous lymphoma. Proceedings of the National Academy of Sciences of the United States of America. 1977;74(7):3065–3067. doi: 10.1073/pnas.74.7.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lawrence T. The nuclear factor NF-kappaB pathway in inflammation. Cold Spring Harbor Perspectives in Biology. 2009;1:p. a001651. doi: 10.1101/cshperspect.a001651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu T., Zhang L., Joo D., Sun S. C. NF-κB signaling in inflammation. Signal Transduction and Targeted Therapy. 2017;2(1) doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Concetti J., Wilson C. L. NFKB1 and Cancer: friend or foe? Cell. 2018;7 doi: 10.3390/cells7090133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tao W., Li B., Gao S., et al. CancerHSP: anticancer herbs database of systems pharmacology. Scientific Reports. 2015;5(1):p. 11481. doi: 10.1038/srep11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang X., Shao L., Zhang H., Wang S. CHMIS-C: a comprehensive herbal medicine information system for cancer. Journal of Medicinal Chemistry. 2005;48(5):1481–1488. doi: 10.1021/jm049838d. [DOI] [PubMed] [Google Scholar]
- 53.Vetrivel U., Subramanian N., Pilla K. InPACdb--Indian plant anticancer compounds database. Bioinformation. 2009;4(2):71–74. doi: 10.6026/97320630004071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mangal M., Sagar P., Singh H., Raghava G. P., Agarwal S. M. NPACT: naturally occurring plant-based anti-cancer compound-activity-target database. Nucleic Acids Research. 2013;41(D1):D1124–D1129. doi: 10.1093/nar/gks1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi H., Cho S. Y., Pak H. J., et al. NPCARE: database of natural products and fractional extracts for cancer regulation. Journal of Cheminformatics. 2017;9(1):p. 2. doi: 10.1186/s13321-016-0188-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mikkelsen R. B., Wardman P. Biological chemistry of reactive oxygen and nitrogen and radiation-induced signal transduction mechanisms. Oncogene. 2003;22(37):5734–5754. doi: 10.1038/sj.onc.1206663. [DOI] [PubMed] [Google Scholar]
- 57.Yamamori T., Yasui H., Yamazumi M., et al. Ionizing radiation induces mitochondrial reactive oxygen species production accompanied by upregulation of mitochondrial electron transport chain function and mitochondrial content under control of the cell cycle checkpoint. Free Radical Biology & Medicine. 2012;53(2):260–270. doi: 10.1016/j.freeradbiomed.2012.04.033. [DOI] [PubMed] [Google Scholar]
- 58.Abdel-Rahman W. M. Genomic instability and carcinogenesis: an update. Current Genomics. 2008;9(8):535–541. doi: 10.2174/138920208786847926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Iliakis G., Wang Y., Guan J., Wang H. DNA damage checkpoint control in cells exposed to ionizing radiation. Oncogene. 2003;22(37):5834–5847. doi: 10.1038/sj.onc.1206682. [DOI] [PubMed] [Google Scholar]
- 60.Yang J., Xu Z. P., Huang Y., Hamrick H. E., Duerksen-Hughes P. J., Yu Y. N. ATM and ATR: sensing DNA damage. World Journal of Gastroenterology. 2004;10(2):155–160. doi: 10.3748/wjg.v10.i2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Burger K., Ketley R. F., Gullerova M. Beyond the trinity of ATM, ATR, and DNA-PK: multiple kinases shape the DNA damage response in concert with RNA metabolism. Frontiers in Molecular Biosciences. 2019;6:p. 61. doi: 10.3389/fmolb.2019.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Marechal A., Zou L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harbor Perspectives in Biology. 2013;5(9) doi: 10.1101/cshperspect.a012716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cai C. K., Zhao G. Y., Tian L. Y., et al. miR-15a and miR-16-1 downregulate CCND1 and induce apoptosis and cell cycle arrest in osteosarcoma. Oncology Reports. 2012;28(5):1764–1770. doi: 10.3892/or.2012.1995. [DOI] [PubMed] [Google Scholar]
- 64.Smith D., Mann D., Yong K. Cyclin D type does not influence cell cycle response to DNA damage caused by ionizing radiation in multiple myeloma tumours. British Journal of Haematology. 2016;173(5):693–704. doi: 10.1111/bjh.13982. [DOI] [PubMed] [Google Scholar]
- 65.Syed V. TGF‐β signaling in cancer. Journal of Cellular Biochemistry. 2016;117(6):1279–1287. doi: 10.1002/jcb.25496. [DOI] [PubMed] [Google Scholar]
- 66.Arteaga C. L., Engelman J. A. ERBB receptors: from oncogene discovery to basic science to mechanism-based cancer therapeutics. Cancer Cell. 2014;25(3):282–303. doi: 10.1016/j.ccr.2014.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen S., Sang N. Hypoxia-inducible factor-1: a critical player in the survival strategy of stressed cells. Journal of Cellular Biochemistry. 2016;117(2):267–278. doi: 10.1002/jcb.25283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lu Z., Xu S. ERK1/2 MAP kinases in cell survival and apoptosis. IUBMB Life. 2006;58(11):621–631. doi: 10.1080/15216540600957438. [DOI] [PubMed] [Google Scholar]
- 69.El-Sahli S., Xie Y., Wang L., Liu S. Wnt signaling in cancer metabolism and immunity. Cancers. 2019;11(7):p. 904. doi: 10.3390/cancers11070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Farooqi A. A., Siddik Z. H. Platelet-derived growth factor (PDGF) signalling in cancer: rapidly emerging signalling landscape. Cell Biochemistry and Function. 2015;33(5):257–265. doi: 10.1002/cbf.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Piva R., Belardo G., Santoro M. G. NF-kappaB: a stress-regulated switch for cell survival. Antioxidants & Redox Signaling. 2006;8(3-4):478–486. doi: 10.1089/ars.2006.8.478. [DOI] [PubMed] [Google Scholar]
- 72.Jeong J. W., Park S., Park C., et al. N-benzyl-N-methyldecan-1-amine, a phenylamine derivative isolated from garlic cloves, induces G2/M phase arrest and apoptosis in U937 human leukemia cells. Oncology Reports. 2014;32(1):373–381. doi: 10.3892/or.2014.3215. [DOI] [PubMed] [Google Scholar]
- 73.Zou X., Liang J., Sun J., et al. Allicin sensitizes hepatocellular cancer cells to anti-tumor activity of 5-fluorouracil through ROS-mediated mitochondrial pathway. Journal of Pharmacological Sciences. 2016;131(4):233–240. doi: 10.1016/j.jphs.2016.04.017. [DOI] [PubMed] [Google Scholar]
- 74.Huang W. L., Wu S. F., Xu S. T., et al. Allicin enhances the radiosensitivity of colorectal cancer cells via inhibition of NF‐κB signaling pathway. Journal of Food Science. 2020;85(6):1924–1931. doi: 10.1111/1750-3841.15156. [DOI] [PubMed] [Google Scholar]
- 75.Cha J. H., Choi Y. J., Cha S. H., Choi C. H., Cho W. H. Allicin inhibits cell growth and induces apoptosis in U87MG human glioblastoma cells through an ERK-dependent pathway. Oncology Reports. 2012;28(1):41–48. doi: 10.3892/or.2012.1772. [DOI] [PubMed] [Google Scholar]
- 76.Chen L., Yan F., Chen W., et al. Procyanidin from peanut skin induces antiproliferative effect in human prostate carcinoma cells DU145. Chemico-Biological Interactions. 2018;288:12–23. doi: 10.1016/j.cbi.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 77.Cardeno A., Sanchez-Hidalgo M., Alarcon-de-la-Lastra C. An up-date of olive oil phenols in inflammation and cancer: molecular mechanisms and clinical implications. Current Medicinal Chemistry. 2013;20(37):4758–4776. doi: 10.2174/09298673113209990159. [DOI] [PubMed] [Google Scholar]
- 78.Duluc L., Jacques C., Soleti R., Iacobazzi F., Simard G., Andriantsitohaina R. Modulation of mitochondrial capacity and angiogenesis by red wine polyphenols _via_ estrogen receptor, NADPH oxidase and nitric oxide synthase pathways. The International Journal of Biochemistry & Cell Biology. 2013;45(4):783–791. doi: 10.1016/j.biocel.2013.01.007. [DOI] [PubMed] [Google Scholar]
- 79.Losada-Echeberria M., Herranz-Lopez M., Micol V., Barrajon-Catalan E. Polyphenols as promising drugs against main breast cancer signatures. Antioxidants. 2017;6(4):p. 88. doi: 10.3390/antiox6040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mileo A. M., Miccadei S. Polyphenols as modulator of oxidative stress in cancer disease: new therapeutic strategies. Oxidative Medicine and Cellular Longevity. 2016;2016:17. doi: 10.1155/2016/6475624.6475624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanagornmeatar K., Chaotham C., Sritularak B., Likhitwitayawuid K., Chanvorachote P. Cytotoxic and anti-metastatic activities of phenolic compounds from Dendrobium ellipsophyllum. Anticancer Research. 2014;34(11):6573–6579. [PubMed] [Google Scholar]
- 82.Saenglee S., Jogloy S., Patanothai A., Leid M., Senawong T. Cytotoxic effects of peanut phenolics possessing histone deacetylase inhibitory activity in breast and cervical cancer cell lines. Pharmacological Reports: PR. 2016;68(6):1102–1110. doi: 10.1016/j.pharep.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 83.Xu H. L., Yu X. F., Qu S. C., Qu X. R., Jiang Y. F., Sui da Y. Juglone, from _Juglans mandshruica Maxim_ , inhibits growth and induces apoptosis in human leukemia cell HL-60 through a reactive oxygen species- dependent mechanism. Food and Chemical Toxicology. 2012;50(3-4):590–596. doi: 10.1016/j.fct.2012.01.002. [DOI] [PubMed] [Google Scholar]
- 84.Zou Z., Tao T., Li H., Zhu X. mTOR signaling pathway and mTOR inhibitors in cancer: progress and challenges. Cell & Bioscience. 2020;10(1):p. 31. doi: 10.1186/s13578-020-00396-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sun Z. L., Dong J. L., Wu J. Juglanin induces apoptosis and autophagy in human breast cancer progression via ROS/JNK promotion. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie. 2017;85:303–312. doi: 10.1016/j.biopha.2016.11.030. [DOI] [PubMed] [Google Scholar]
- 86.Izadi S., Nikkhoo A., Hojjat-Farsangi M., et al. CDK1 in breast cancer: implications for theranostic potential. Anti-Cancer Agents in Medicinal Chemistry. 2020;20(7):758–767. doi: 10.2174/1871520620666200203125712. [DOI] [PubMed] [Google Scholar]
- 87.Dashwood R. H. Indole-3-carbinol: anticarcinogen or tumor promoter in brassica vegetables? Chemico-Biological Interactions. 1998;110(1-2):1–5. doi: 10.1016/S0009-2797(97)00115-4. [DOI] [PubMed] [Google Scholar]
- 88.Chinni S. R., Li Y., Upadhyay S., Koppolu P. K., Sarkar F. H. Indole-3-carbinol (I3C) induced cell growth inhibition, G1 cell cycle arrest and apoptosis in prostate cancer cells. Oncogene. 2001;20(23):2927–2936. doi: 10.1038/sj.onc.1204365. [DOI] [PubMed] [Google Scholar]
- 89.Takada Y., Andreeff M., Aggarwal B. B. Indole-3-carbinol suppresses NF-kappaB and IkappaBalpha kinase activation, causing inhibition of expression of NF-kappaB-regulated antiapoptotic and metastatic gene products and enhancement of apoptosis in myeloid and leukemia cells. Blood. 2005;106(2):641–649. doi: 10.1182/blood-2004-12-4589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koolaji N., Shammugasamy B., Schindeler A., Dong Q., Dehghani F., Valtchev P. Citrus peel flavonoids as potential cancer prevention agents. Current developments in nutrition. 2020;4(5, article nzaa025) doi: 10.1093/cdn/nzaa025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee D. H., Park K. I., Park H. S., et al. Flavonoids isolated from Korea Citrus aurantium L. induce G2/M phase arrest and apoptosis in human gastric cancer AGS cells. Evidence-based complementary and alternative medicine: eCAM. 2012;2012, article 515901:1–11. doi: 10.1155/2012/515901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giordano A., Tommonaro G. Curcumin and Cancer. Nutrients. 2019;11(10):p. 2376. doi: 10.3390/nu11102376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kamat A. M., Sethi G., Aggarwal B. B. Curcumin potentiates the apoptotic effects of chemotherapeutic agents and cytokines through down-regulation of nuclear factor-kappaB and nuclear factor-kappaB-regulated gene products in IFN-alpha-sensitive and IFN-alpha-resistant human bladder cancer cells. Molecular Cancer Therapeutics. 2007;6(3):1022–1030. doi: 10.1158/1535-7163.MCT-06-0545. [DOI] [PubMed] [Google Scholar]
- 94.Li L., Aggarwal B. B., Shishodia S., Abbruzzese J., Kurzrock R. Nuclear factor-kappaB and IkappaB kinase are constitutively active in human pancreatic cells, and their down-regulation by curcumin (diferuloylmethane) is associated with the suppression of proliferation and the induction of apoptosis. Cancer. 2004;101(10):2351–2362. doi: 10.1002/cncr.20605. [DOI] [PubMed] [Google Scholar]
- 95.Zhang L., Cheng X., Gao Y., et al. Curcumin inhibits metastasis in human papillary thyroid carcinoma BCPAP cells _via_ down-regulation of the TGF- β/Smad2/3 signaling pathway. Experimental Cell Research. 2016;341(2):157–165. doi: 10.1016/j.yexcr.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 96.Li P. G., Mu T. H., Deng L. Anticancer effects of sweet potato protein on human colorectal cancer cells. World Journal of Gastroenterology. 2013;19(21):3300–3308. doi: 10.3748/wjg.v19.i21.3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Qian C., Chen X., Qi Y., et al. Sporamin induces apoptosis and inhibits NF-kappaB activation in human pancreatic cancer cells. Tumour Biology. 2017;39:p. 1010428317706917. doi: 10.1177/1010428317706917. [DOI] [PubMed] [Google Scholar]
- 98.Yao J., Qian C. Sporamin induce apoptosis in human tongue carcinoma cells by down-regulating Akt/GSK-3 signaling. Fundamental & Clinical Pharmacology. 2011;25(2):229–236. doi: 10.1111/j.1472-8206.2010.00830.x. [DOI] [PubMed] [Google Scholar]
- 99.Manayi A., Nabavi S. M., Setzer W. N., Jafari S. Piperine as a potential anti-cancer agent: a review on preclinical studies. Current Medicinal Chemistry. 2018;25(37):4918–4928. doi: 10.2174/0929867324666170523120656. [DOI] [PubMed] [Google Scholar]
- 100.Gong X., Chen Z., Han Q., et al. Sanguinarine triggers intrinsic apoptosis to suppress colorectal cancer growth through disassociation between STRAP and MELK. BMC Cancer. 2018;18(1):p. 578. doi: 10.1186/s12885-018-4463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sun M., Lou W., Chun J. Y., et al. Sanguinarine suppresses prostate tumor growth and inhibits survivin expression. Genes & Cancer. 2010;1(3):283–292. doi: 10.1177/1947601910368849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xu J. Y., Meng Q. H., Chong Y., et al. Sanguinarine inhibits growth of human cervical cancer cells through the induction of apoptosis. Oncology Reports. 2012;28(6):2264–2270. doi: 10.3892/or.2012.2024. [DOI] [PubMed] [Google Scholar]
- 103.Suganyadevi P., Saravanakumar K. M., Mohandas S. The antiproliferative activity of 3-deoxyanthocyanins extracted from red sorghum (Sorghum bicolor) bran through P53-dependent and Bcl-2 gene expression in breast cancer cell line. Life Sciences. 2013;92(6-7):379–382. doi: 10.1016/j.lfs.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 104.Pan Q., Pan H., Lou H., Xu Y., Tian L. Inhibition of the angiogenesis and growth of Aloin in human colorectal cancer in vitro and in vivo. Cancer Cell International. 2013;13(1):p. 69. doi: 10.1186/1475-2867-13-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Awika J. M., Rooney L. W., Waniska R. D. Properties of 3-deoxyanthocyanins from sorghum. Journal of Agricultural and Food Chemistry. 2004;52(14):4388–4394. doi: 10.1021/jf049653f. [DOI] [PubMed] [Google Scholar]
- 106.Bavarsadi M., Mahdavi A. H., Ansari-Mahyari S., Jahanian E. Effects of different levels of sanguinarine on antioxidant indices, immunological responses, ileal microbial counts and jejunal morphology of laying hens fed diets with different levels of crude protein. Journal of Animal Physiology and Animal Nutrition. 2017;101(5):936–948. doi: 10.1111/jpn.12528. [DOI] [PubMed] [Google Scholar]
- 107.Mittal R., Gupta R. L. In vitro antioxidant activity of piperine. Methods and Findings in Experimental and Clinical Pharmacology. 2000;22(5):271–274. doi: 10.1358/mf.2000.22.5.796644. [DOI] [PubMed] [Google Scholar]
- 108.Teka T., Kassahun H. <p>Characterization and evaluation of antioxidant activity of <em>Aloe schelpei</em> Reynolds</p>. Drug Design, Development and Therapy. 2020;Volume 14:1003–1008. doi: 10.2147/DDDT.S241412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Yaffe P. B., Power Coombs M. R., Doucette C. D., Walsh M., Hoskin D. W. Piperine, an alkaloid from black pepper, inhibits growth of human colon cancer cells via G1 arrest and apoptosis triggered by endoplasmic reticulum stress. Molecular Carcinogenesis. 2015;54(10):1070–1085. doi: 10.1002/mc.22176. [DOI] [PubMed] [Google Scholar]
- 110.Sain S., Naoghare K. P., Saravana Devi S., et al. Beta caryophyllene and caryophyllene oxide, isolated from Aegle marmelos, as the potent anti-inflammatory agents against lymphoma and neuroblastoma cells. Anti-Inflammatory & Anti-Allergy Agents in Medicinal Chemistry. 2014;13(1):45–55. doi: 10.2174/18715230113129990016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: the class, natural source, active ingredient, the affected cancer type(s), target gene(s), gene expression status, cell line used in the experimental study, and corresponding PubMed link are listed.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding authors upon logical request.


