Abstract
Ribosome-inactivating proteins (RIPs) are enzymes that irreversibly inactivate ribosomes as a consequence of their N-glycosylase (EC 3.2.2.22) activity. The enzyme cleaves the N-glycosidic bond between the adenine No. 4324 from the 28S rRNA and its ribose in rat ribosomes (or the equivalent adenine in sensitive ribosomes from other organisms). This adenine is located in the α-sarcin-ricin loop (SRL) that is crucial for anchoring the elongation factor (EF) G and EF2 on the ribosome during mRNA-tRNA translocation in prokaryotes and eukaryotes, respectively. RIPs have been isolated mainly from plants and examples of these proteins are ricin or Pokeweed Antiviral Protein (PAP). These proteins, either alone or as a part of immunotoxins, are useful tools for cancer therapy. The following protocol describes a method to detect the RNA fragment released when the RIP-treated apurinic RNA from rabbit reticulocyte lysate is incubated in the presence of acid aniline by electrophoresis on polyacrylamide gels. The fragment released (Endo’s fragment) is diagnostic of the action of RIPs.
Keywords: Ribosome-inactivating protein (RIP), rRNA N-glycosylase, Protein synthesis (inhibition), Sarcin-ricin loop, Polynucleotide:adenosine glycosylase, Ricin, Pokeweed Antiviral Protein (PAP), Beetin 27
Background
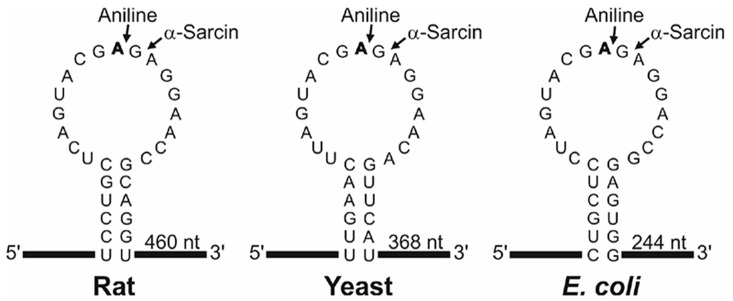
N-glycosylase activity of RIPs on the eukaryotic 28S rRNA was first described by Endo and Tsurugi for ricin (Endo and Tsurugi, 1988) in rat ribosomes; subsequently it was shown that some RIPs can also depurinate ribosomes from plants, bacteria and fungi. The result of this effect, upon treatment with aniline, is the release of an RNA fragment of between 240 and 500 nucleotides (depending on species) from the rRNA of the large subunit (Figure 1). Most RIPs depurinate ribosomes at one site (the adenine 4,324), whereas other RIPs such as saporins, PAP-R and trichokirin depurinate the rRNA at multiple sites. The protocol described here uses rabbit reticulocyte lysate, a eukaryotic cell-free model system very sensitive to the action of RIPs, which shows high rates of depurination for most RIPs. This allows obtaining a large amount of fragment which facilitates its detection by electrophoresis. In this procedure we use denaturing polyacrylamide minigels which require small quantities of sample and have higher resolution than the agarose gels when staining with fluorescent dyes.
Figure 1. Sarcin Ricin Loop of the large rRNA from rat, yeast and Escherichia coli.
The sequences (accession numbers NR_046246, J01355 and AB035926) were downloaded from the NCBI sequence database (http://www.ncbi.nlm.nih.gov/nucleotide/). The adenine released by the RIP action (boldfaced), the site of splitting by either the aniline or α-sarcin (arrows) and the size of the generated fragment are also indicated. Partial sequence of rabbit 28S rRNA (AF460236) indicates that rat and rabbit share the same SRL sequence.
Materials and Reagents
Note: All the reagents used in preparing buffers should be of molecular biology grade purity (RNase, DNase-free). Water and solutions should be autoclaved at 120 °C for 15 min (except of aniline and ethanol).
Disposable gloves
Pipettes and tips (either RNase, DNase-free or autoclaved)
Eppendorf tubes (1.5 ml polypropylene microcentrifuge tubes, either RNase, DNase-free or autoclaved)
Paper towels
Corning 50 ml PP centrifuge tubes (Corning, catalog number: 430291)
Pasteur pipettes
Cotton swabs
Rabbit reticulocyte lysate, untreated with micrococcal nuclease, either obtained as indicated by Pelham and Jackson (1976) or purchased from a biochemical supplier (for example: Promega, catalog number: L4151)
RIP either obtained as indicated by Barbieri et al. (2001) or purchased from a biochemical supplier (for example Ricin A chain: Sigma-Aldrich, catalog number: L9514)
Crushed ice
Phenol/TRIS saturated sol., for molecular biology, stabilized, DNAse, RNase and Protease free (ACROS Organics, catalog number: 327125000)
Ethanol absolute (EMD Millipore, catalog number: 100983)
-
Deionized, RNase and DNase-free water
Note: For example, Millipore Elix 5 (UV) water autoclaved 120 °C 15 min.
Aniline (Sigma-Aldrich, catalog number: 242284)
Urea (Thermo Fisher Scientific, Affymetrix, catalog number: 75826)
Ammonium persulfate (Sigma-Aldrich, catalog number: A3678)
N,N,N’,N’-tetramethylethylenediamine (TEMED) (Sigma-Aldrich, catalog number: T9281)
Acrylamide bis-acrylamide 19:1, 40% (w/v) solution (Thermo Fisher Scientific, Affymetrix, catalog number: 75848)
Either ethidium bromide (Sigma-Aldrich, catalog number: E7637) or GelRed (Biotium, catalog number: 41003)
EDTA.2H2O
NaOH pellets
Hydrochloric acid (HCl) (EMD Millipore, catalog number: 100317)
Sodium dodecyl sulfate (SDS) (Thermo Fisher Scientific, Affymetrix, catalog number: 75819)
Sodium acetate trihydrate (EMD Millipore, catalog number: 106267)
Glacial acetic acid (EMD Millipore, catalog number: 100063)
Diethyl pyrocarbonate (Sigma-Aldrich, catalog number: D5758)
Diethyl ether (EMD Millipore, catalog number: 100921)
Trimethylol aminomethane (Tris base) (Fisher Scientific, catalog number: BP154-1)
Boric acid (Sigma-Aldrich, catalog number: B6768)
Sucrose (Sigma-Aldrich, catalog number: 84097)
-
RNA markers (Roche Diagnostics, catalog number: 1062 638)
Note: This product has been discontinued. Can be replaced by Low Range ssRNA Ladder (New England Biolabs, catalog number: N0364S)
Bromophenol blue (Bio-Rad Laboratories, catalog number: 161-0404)
0.5 M EDTA (pH 8.0) (see Recipes)
50 mM Tris/0.5% SDS (pH 7.8) (see Recipes)
3 M sodium acetate (pH 5.2) (see Recipes)
70% ethanol (see Recipes)
2 M aniline (pH 4.5) (see Recipes)
Water saturated ether (see Recipes)
10x TBE buffer (see Recipes)
2x gel loading buffer (see Recipes)
Equipment
Deep freezer (-80 °C freezer) (Thermo Fisher Scientific, Thermo ScientificTM, model: TSE Series, catalog number: TSE400SSV)
Fume hood (BURDINOLA, model: V21-Space BAJA ST 1500)
Two microcentrifuges (DJB Labcare, model: Heraeus Biofuge Pico, catalog number: 75003235), one of them kept at 4 °C
30 °C water bath (JP SELECTA, catalog number: 6000140)
Vortex mixer for test tubes (IKA, catalog number: 0003617000)
BECKMAN DU-640 spectrophotometer (Beckman Coulter, model: DU-640)
Quartz cuvette (Hellma Analytics, model: 104-QS, catalog number: 104-10-40)
Polyacrylamide gel electrophoresis system (GE Healthcare, catalog number: 80-6418-77)
Power supply (GE Healthcare, catalog number: 18-1130-01)
Molecular Imager® Gel DocTM XR+ System with Image LabTM Software (Bio-Rad Laboratories, catalog number: 1708195)
Gel staining tray
Magnetic stirrer
Autoclave (JP Selecta, catalog number: 4002516)
Procedure
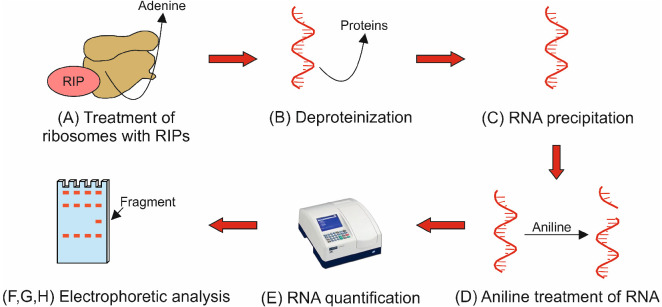
The general procedure to assay N-glycosylase activity is illustrated in a flowchart in Figure 2.
Figure 2. Flowchart illustrating protocol Procedure parts A-H.
The flowchart shows the key steps describing the eight main parts of the protocol procedure. Capital letters (A-H) refer to the subsections in the Procedure section.
-
Treatment of ribosomes with RIPs
Thaw frozen aliquots of both rabbit reticulocyte lysate (stored at -80 °C) and RIP (stored at -20 °C) at room temperature and place them on crushed ice.
-
Reactions are prepared on crushed ice into four Eppendorf tubes as indicated in the following:
Tube 1 (control) 2 (control) 3 (+RIP) 4 (+RIP) lysate 40 µl 40 µl 40 µl 40 µl RIP (1 mg/ml) - - 1 µl 1 µl Pipette 40 µl of rabbit reticulocyte lysate into the four tubes.
Add 1 µl of 1 mg/ml RIP into tubes 3 and 4.
Mix by gently vortexing.
Incubate at 30 °C for 1 h in a water bath.
Stop the reaction by adding 2 µl of 0.5 M EDTA (pH 8.0).
Mix by gently vortexing.
Add 500 µl of 50 mM Tris/0.5% SDS.
Vortex hard for 30 sec (with the purpose of denaturing proteins with the aid of SDS).
-
Deproteinization
Note: This process should be carried out under a fume hood.
Add 500 µl of phenol.
Vortex hard for 30 sec.
Centrifuge at 16,060 × g (13,000 rpm) for 5 min at room temperature.
Recover the aqueous (upper) phase (400 µl) to a new Eppendorf tube without disturbing the protein interphase.
-
RNA precipitation
-
To carry out RNA precipitation with 3 volumes of ethanol in 1.5 ml tubes, the aqueous phase is split into two Eppendorf tubes (A and B), with 200 µl in each. Perform ethanol precipitation by mixing the following:
Tube 1A (control) 1B (control) 2A (control) 2B (control) Aqueous phase 200 µl 200 µl 200 µl 200 µl 3 M sodium acetate, pH 5.2 20 µl 20 µl 20 µl 20 µl Absolute ethanol 600 µl 600 µl 600 µl 600 µl Tube 3A (+RIP) 3B (+RIP) 4A (+RIP) 4B (+RIP) Aqueous phase 200 µl 200 µl 200 µl 200 µl 3 M sodium acetate, pH 5.2 20 µl 20 µl 20 µl 20 µl Absolute ethanol 600 µl 600 µl 600 µl 600 µl Mix by vortexing.
Keep overnight at -80 °C (or at least 3 h).
Centrifuge at 16,060 × g (13,000 rpm) for 15 min at 4 °C. Orient each tube in the rotor with the cap hinge pointing outward, this will indicate the position of the pelleted RNA since pellets will be scarcely visible.
Take the tubes out. Carefully decant the supernatant liquid and discard it into paper towel without losing sight of the off-white pellet avoiding its displacement.
Add 250 µl of 70% ethanol.
Centrifuge at 16,060 × g (13,000 rpm) for 15 min at 4 °C. Orient each tube in the rotor with the cap hinge pointing outward.
Carefully decant the supernatant liquid and discard it into paper towel avoiding pellet displacement. Remove any droplet on the tube wall with the aid of a cotton swab.
Place the tubes up-side-down onto a clean paper towel and allow the pellet to air-dry for 15 min.
Resuspend the pellet in 10 µl of RNase-free deionized water. Mix the contents of tubes 1A and 1B, and 3A and 3B, obtaining now the tubes 1 and 3 with 20 µl each. Store these samples at -80 °C. Tubes 2A, 2B, 4A and 4B still remain separated.
-
-
Aniline treatment of RNA
-
Add one volume (10 µl) of 2 M aniline (pH 4.5) to tubes 2A, 2B, 4A and 4B.
Note: Reaction is carried out on crushed ice.
Mix by vortexing.
Incubate for 10 min on ice.
Stop the reaction by adding 200 µl of RNase-free deionized water and mix by vortexing.
Add 200 µl of water saturated ether.
Vortex hard for 20 sec.
Let the sample stand for 20 sec to allow phase separation.
Discard the ether (upper) phase.
Repeat steps D5 to D8.
Add 20 µl of 3 M sodium acetate (pH 5.2) and 600 µl of absolute ethanol.
Mix by vortexing.
Keep overnight at -80 °C (or at least 3 h).
Centrifuge at 16,060 × g (13,000 rpm) for 15 min at 4 °C. Orient each tube in the rotor with the cap hinge pointing outward.
Take the tubes out. Carefully decant the supernatant liquid and discard it into paper towel without losing sight of the off-white pellet avoiding its displacement.
Add 250 µl of 70% ethanol.
Centrifuge at 16,060 × g (13,000 rpm) for 15 min at 4 °C. Orient each tube in the rotor with the cap hinge pointing outward.
Carefully decant the supernatant liquid and discard it into paper towel avoiding pellet displacement. Remove any droplet on the tube wall with the aid of a cotton swab.
Place the tubes up-side-down onto a clean paper towel and allow the pellet to air-dry for 15 min.
Resuspend the pellet in 10 µl of RNase-free, deionized water. Mix the contents of tubes 2A and 2B, and 4A and 4B, obtaining now the tubes 2 and 4 with 20 µl each. Store these samples at -80 °C.
-
-
Spectrophotometric quantification of RNA
Thaw frozen samples 1-4 with 20 µl each and place them on ice.
Determine the RNA concentration of each sample in the spectrophotometer at 260 nm (in quartz cuvette of 10 mm path length, using water as blank). If A260 = 1, then the RNA concentration is 40 µg RNA/ml. Usually the yield of the process is 45-50 µg RNA for samples treated with aniline and 50-60 µg RNA for untreated samples.
-
Preparation of 7 M urea/5% (w/v) polyacrylamide gel
Prepare 25 ml of mix for two gels (glass plates of 10 x 10.5 cm, spacers of 0.1 cm) in a RNase-free Corning tube: 2.5 ml 10x TBE; 3.1 ml acrylamide/bisacrylamide (40%); 10.5 g urea; 12.3 ml RNase-free, deionized water; 0.2 ml 10% ammonium persulfate; 0.05 ml TEMED.
-
Preparation of samples
Samples are prepared into Eppendorf tubes placed on crushed ice.
Add 3 µg of RNA.
Complete the volume to 5 µl with water.
Add 5 µl of 2x gel loading buffer.
Mix by vortexing.
Boil 30 sec (in a 100 °C water bath) and place the tubes on crushed ice immediately.
-
Electrophoresis
Attach the gel plates to the gel apparatus.
Add TBE electrophoresis buffer (900 ml water and 100 ml 10x TBE) to the top and bottom chambers.
Remove the comb and rinse the wells with buffer to remove residual urea (using a Pasteur pipette).
Using a micropipettor, load the samples.
Run the gel at 15 mA for 1 h 50 min.
Disassemble the gel.
Stain the gel with either 0.5 µg/ml ethidium bromide or GelRed solution at room temperature for 40 min with gentle shaking (destaining is not needed after staining).
-
Place the gel on an ultraviolet transilluminator. Visualize and photograph RNA bands.
Note: Be aware that RNA will diffuse within the gel over time, therefore examination and photography should take place shortly after the end of electrophoresis.
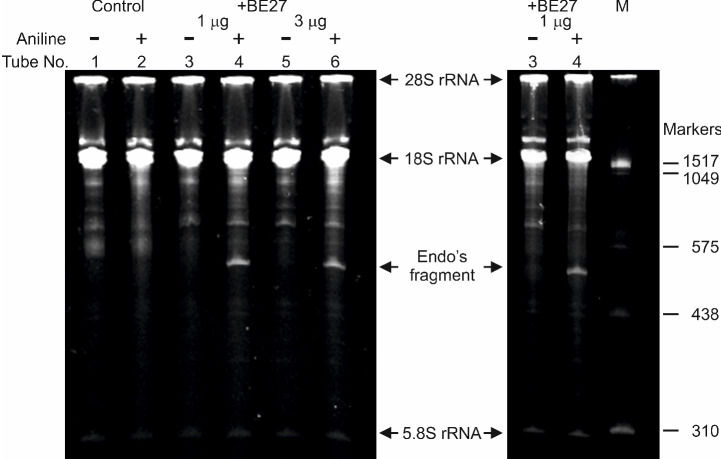
A representative data is shown in Figure 3.
Figure 3. rRNA N-glycosylase activity of BE27 in rabbit reticulocyte lysate.
Each lane contained 3 µg of RNA isolated from either untreated (control) or RIP (+BE27) treated ribosomes from rabbit. The arrows indicate the 28S, 18S and 5.8S rRNAs, and the RNA fragment released as a result of RIP action after aniline treatment (+). The number of the corresponding tube in section A is indicated. Samples from lanes 5 and 6 were processed as samples from lanes 3 and 4, respectively, but the ribosomes were treated with 3 µg instead of 1 µg of BE27. The size of RNA markers in nucleotides are also indicated (M). The gel to the left was stained with ethidium bromide and the gel to the right with GelRed.
Data analysis
This procedure is very specific for identifying rRNA N-glycosylase activity and the results are very reliable. Controls are included to rule out nonspecific RNA breaks produced at random by aniline treatment. On the other hand, a comparative analysis of RIP treated-rRNA in the absence and presence of aniline is carried out in each experiment to exclude unspecific effects due to possible contaminants that could fragment or degrade the RNA. Besides, the result can be confirmed by treating the RNA with different RIP concentrations. RNA molecular weight markers can be used to determine the size of the RNA fragment released (which would depend on the type of ribosomes used in the assay). It is also recommended to include in the experiment a previously published RIP (e.g., ricin) to unequivocally localize the fragment.
Notes
Due to its sensitivity and simplicity, rabbit reticulocyte lysate is the best system to check rRNA N-glycosylase activity. This method can also be used to test specific RNase activity (alpha sarcin-like activity) but without aniline treatment. Other sensitive ribosomes can also be used with minor modifications but because many RNA bands can be seen after gel electrophoresis, optimization is required before the Endo’s fragment can be detected. Such modified protocols have been used successfully for yeast, Penicillium digitatum, Escherichia coli, Agrobacterium tumefaciens, Streptomyces lividans, Brevibacterium lactofermentum, Vicia sativa, and Colo 320 and HeLa cell cultures.
Recipes
-
0.5 M EDTA (pH 8.0)
Add 186.1 g of disodium EDTA.2H2O to 800 ml of ddH2O
Stir vigorously on a magnetic stirrer
Adjust the pH to 8.0 with NaOH (~20 g of NaOH pellets)
Dispense into aliquots and sterilize by autoclaving
Note: The disodium salt of EDTA will not go into solution until the pH of the solution is adjusted to ~8.0 by the addition of NaOH. Autoclave at 120 °C for 15 min.
-
50 mM Tris/0.5% SDS
Mix 45 ml of ddH2O, 2.5 ml of 1 M Tris-HCl (adjusted to pH 7.8 with HCl) and 2.5 ml of 10% SDS. Autoclave 120 °C 15 min
-
3 M sodium acetate (pH 5.2)
20.412 g sodium acetate trihydrate
Add approximately 6 ml glacial acetic acid to pH 5.2 and up to 50 ml with ddH2O
Autoclave at 120 °C for 15 min
-
70% ethanol
70% ethanol absolute and 30% deionized, RNase and DNase-free water
-
2 M aniline (pH 4.5)
Add 18.25 ml of aniline to RNase and DNase-free water
Adjust to pH 4.5 with glacial acetic acid using an RNase-free electrode washed with either RNase and DNase-free water or with 1% diethyl pyrocarbonate in water
Add double distilled water up to 100 ml
Aliquot in Eppendorf tubes and store at -20 °C
-
Water saturated ether
Add one volume of deionized, RNase and DNase-free water to one volume of diethyl ether and mix. The upper phase is the water saturated ether
Note: It is stored in small quantities (10 ml) at room temperature under a fume hood.
-
10x TBE buffer
Dissolve (use magnetic stirrer) 108 g Tris base and 55 g boric acid in 800 ml deionized, RNase and DNase-free water
Add 40 ml 0.5 M EDTA (pH 8.0)
Adjust volume to 1 L
Autoclave at 120 °C for 15 min
Store at room temperature
-
2x gel loading buffer
TBE buffer
100 mg/ml sucrose
7 M urea
0.4 µg/ml bromophenol blue
Store at 4 °C until use
Acknowledgments
This protocol was modified from previous publications (Endo and Tsurugi, 1988; Sallustio and Stanley, 1990). This work was supported by grants BIO39/VA39/14 and BIO/VA17/15 to L.C.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Barbieri L., Bolognesi A. and Stirpe F.(2001). Purification and conjugation of type 1 ribosome-inactivating proteins. In: Hall, W. A.(Ed.). Immunotoxin Methods and Protocols-Series Methods in Molecular Biology. Humana Press, pp: 71-85. [DOI] [PubMed] [Google Scholar]
- 2.Citores L., Iglesias R., Gay C. and Ferreras J. M.(2016). Antifungal activity of the ribosome-inactivating protein BE27 from sugar beet(Beta vulgaris L.) against the green mould Penicillium digitatum . Mol Plant Pathol 17(2): 261-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Endo Y. and Tsurugi K.(1988). The RNA N-glycosidase activity of ricin A-chain. The characteristics of the enzymatic activity of ricin A-chain with ribosomes and with rRNA. J Biol Chem 263(18): 8735-8739. [PubMed] [Google Scholar]
- 4.Iglesias R., Citores L., Di Maro A. and Ferreras J. M.(2015). Biological activities of the antiviral protein BE27 from sugar beet(Beta vulgaris L.) . Planta 241(2): 421-433. [DOI] [PubMed] [Google Scholar]
- 5.Iglesias R., Citores L., Ragucci S., Russo R., Di Maro A. and Ferreras J. M.(2016). Biological and antipathogenic activities of ribosome-inactivating proteins from Phytolacca dioica L . Biochim Biophys Acta 1860(6): 1256-1264. [DOI] [PubMed] [Google Scholar]
- 6.Pelham H. R. and Jackson R. J.(1976). An efficient mRNA-dependent translation system from reticulocyte lysates. Eur J Biochem 67(1): 247-256. [DOI] [PubMed] [Google Scholar]
- 7.Sallustio S. and Stanley P.(1990). Isolation of Chinese hamster ovary ribosomal mutants differentially resistant to ricin, abrin, and modeccin. J Biol Chem 265(1): 582-588. [PubMed] [Google Scholar]