Abstract
The pancreas is a heavily innervated organ, but pancreatic innervation can be challenging to comprehensively assess using conventional histological methods. However, recent advances in whole-mount tissue clearing and 3D rendering techniques have allowed detailed reconstructions of pancreatic innervation. Optical clearing is used to enhance tissue transparency and reduce light scattering, thus eliminating the need to section the tissue. Here, we describe a modified version of the optical tissue clearing protocol iDISCO+ (immunolabeling-enabled three-dimensional imaging of solvent-cleared organs) optimized for pancreatic innervation and endocrine markers. The protocol takes 13-19 days, depending on tissue size. In addition, we include protocols for imaging using light sheet and confocal microscopes and for 3D segmentation of pancreatic innervation and endocrine cells using Imaris.
Keywords: Pancreas, Innervation, iDISCO, Clearing, Light sheet
Background
The pancreas and islets of Langerhans are heavily innervated by the autonomic nervous system and sensory nerves (Ahren, 2000; Rodriguez- Diaz et al., 2011 ; Rodriguez-Diaz and Caicedo, 2014). The autonomic nervous system consists of the sympathetic and parasympathetic nervous systems. The sympathetic nervous system is activated by decreased blood glucose and is essential in initiating the counter-regulatory responses to hypoglycemia (Ahren and Holst, 2001). The parasympathetic nervous system mediates the cephalic phase insulin secretion and is activated in response to feeding (Louis-Sylvestre, 1978; Strubbe and Steffens, 1993; Teff and Townsend, 1999). The roles of sensory innervation in pancreatic function remain poorly understood, but sensory islet innervation is implicated in hormone release and glucose homeostasis (Ahren, 2000). Parasympathetic intrapancreatic ganglia, consisting of aggregates of neuronal cell bodies, are scattered throughout the pancreas and act as neuronal networks that integrate intrinsic and external nerve inputs to modulate pancreatic functions ( Li et al., 2019 ). Metabolic pathologies, including type 1 diabetes (T1D) and type 2 diabetes (T2D), remodel the pancreatic environment in mice and humans, including islet morphology and endocrine and exocrine innervation ( Giannulis et al., 2014 ; Mundinger et al., 2016 ; Lundberg et al., 2017 ; Butterworth et al., 2018 ; Alvarsson et al., 2020 ). However, the mechanisms behind the remodeling of pancreatic innervation in diabetes, and the functional consequences, remain poorly understood.
The pancreas is a highly heterogeneous organ, which in the mouse macroscopically consists of three diffuse lobes: the duodenal, splenic, and gastric lobes, separated by adipose, lymphatic, and connective tissue ( Dolenšek et al., 2015 ). Pancreatic innervation is difficult to comprehensively assess using conventional histological methods due to its filamentous structure and the long-range projections of pancreatic nerves ( Fasanella et al., 2007 ). Therefore, there is a need for 3-dimensional high-resolution organ-wide pancreatic imaging techniques. Recent advances in optical clearing techniques, including immunolabeling-enabled three-dimensional imaging of solvent-cleared organs (iDISCO+) ( Renier et al., 2016 ), allow the visualization of intact organs, including dispersed neuronal networks, in 3D (Video 1). The purpose of optical clearing is to render tissue transparent by reducing light scattering and absorption and by equilibrating its refractory index with that of an imaging medium, thus allowing full penetration of light during imaging and eliminating the need to section the tissue. This approach readily allows detailed analyses of tissue architecture, spatial relationships between structures, and regional differences in three dimensions. This provides an ideal approach to study the architecture of innervation across the pancreas. Here, we describe a modified version of iDISCO+ optimized for pancreatic innervation ( Alvarsson et al., 2020 ), which takes 13-19 days to complete depending on sample size (Figure 1). We also describe basic protocols for imaging optically cleared pancreata using light sheet microscopy and confocal microscopy and protocols for the segmentation of 3-dimensional pancreatic structures.
Video 1. Cleared mouse pancreas with insulin and NF200 immunolabeling and 3D segmentation.
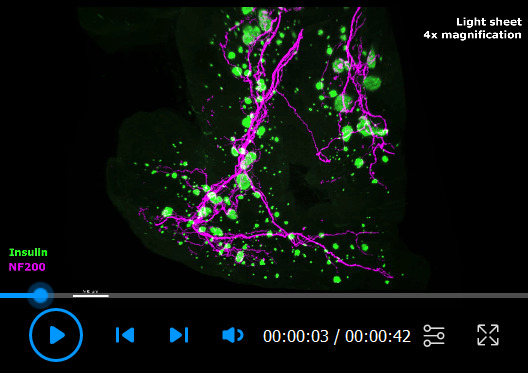
Figure 1. Flowchart of the protocol.
Materials and Reagents
-
Materials
5-ml Eppendorf tubes (Eppendorf, catalog number: 0030119401)
2-ml Eppendorf tubes (Eppendorf, catalog number: 022363352)
1.5-ml Eppendorf tubes (Eppendorf, catalog number: 022364111)
0.5-ml Eppendorf tubes (Eppendorf, catalog number: 022363611)
Glass-bottomed μ-slides (Ibidi, catalog number: 80827)
Glass-bottomed μ-dishes (Ibidi, catalog number: 81158)
Neoprene gloves (Ansell, catalog number: 25-101)
Polystyrene Petri dishes (Sigma-Aldrich, catalog number: P5731)
-
Reagents
16% paraformaldehyde (Electron Microscopy Sciences, catalog number: 15710-S)
Heparin sodium salt from porcine intestinal mucosa (Sigma-Aldrich, catalog number: H3393)
Dichloromethane (Sigma-Aldrich, catalog number: 270997)
Dibenzyl ether (Sigma-Aldrich, catalog number: 108014)
Dimethyl sulfoxide (Fisher Scientific, catalog number: D128-500)
Triton X-100 (Sigma-Aldrich, catalog number: T8787)
Glycine (Sigma-Aldrich, catalog number: G7126)
Double distilled H2O
Tween-20 (Sigma-Aldrich, catalog number: P9416)
Sodium azide (Sigma-Aldrich, catalog number: 58032)
Hydrogen peroxide solution (30%) (Sigma-Aldrich, catalog number: 216763)
Normal donkey serum (Jackson ImmunoResearch, catalog number: 017-000-121)
Methanol (Sigma-Aldrich, catalog number: 322415-2L)
Agarose (Invitrogen, catalog number: 16520-050)
PBS (Fisher Scientific, catalog number: BP2944100)
Forane (Isoflurane, USP) (Baxter, Deerfield, IL, USA)
4% PFA buffer (see Recipes)
10% Sodium azide (NaN3) stock (see Recipes)
Modified PTxwH (see Recipes)
Permeabilization buffer (see Recipes)
Blocking buffer (see Recipes)
Equipment
Fine tweezers (Dumont tweezers, WPI, catalog numbers: 14098 [#5]; 500234 [#5B])
Iris scissor (WPI, catalog number: 501758)
Blunt tweezers (e.g., Dressing Forceps 12.5 cm; WPI, catalog number: 501217)
Single edge razor blade or scalpel (e.g., Single edge razor blade #12, Titan, Seattle, WA, USA; Disposable scalpel #20, WPI, catalog number: 500352)
Cyanoacrylate glue (e.g., Gorilla super glue, Loctite super glue, Starbond EM-02, Krazy glue all purpose)
Absorbent pads (e.g., Medline, catalog number: DDPAD1724Z)
Peristaltic pump (Gilson, catalog number: F155005)
Dissection microscope (United Scope LLC, model: SM-2T-6WB-V331)
Nutating shaker (Benchmark Scientific, model: B3D5000)
Incubator (37°C, no CO2 required) with space for a shaker or rotary mixer inside, or with a built-in shaker/mixer
Centrifuge (no refrigeration required) (Labnet International, model: C2400)
Standard household microwave (e.g., Westinghouse, model: WCM660W)
Light sheet microscope (Ultramicroscope II, LaVision Biotec, Bielefeld, Germany, or similar instrument)
Confocal microscope (Inverted Zeiss LSM, Carl Zeiss AG, Oberkochen, Germany, or similar instrument equipped with standard 10× objective, red and far-red filters)
Image analysis computer (Suggested specifications: Motherboard: Gigabyte X299 Designare EX; CPU: Intel Core i7 7800X 3.5GHz Six Core 8.25MB 140W; RAM: 32GB; Video Card: Nvidia Quadro P4000 8GB; Hard drive: 512GB SSD plus 1TB SSD; Power supply: 850W; Cooling: Noctua NH-U12DX i4 CPU Cooling and Arctic MX-2 thermal compound paste; Operating system: Windows 10 64-bit)
Software
Image analysis software for 3D segmenting (e.g., Imaris, Bitplane, Zürich, Switzerland)
Spreadsheet software (e.g., Excel, Microsoft, Redmond, WA, USA)
Data analysis software (e.g., GraphPad Prism, GraphPad Software Inc., San Diego, CA, USA)
Procedure
-
Sample collection and preparation
Mouse tissue
All animal experiments must be performed according to an approved institutional animal protocol.
Note: Paraformaldehyde (PFA) and its fumes are toxic. Always consult the safety data sheet before handling and use the recommended personal protective equipment (including gloves, lab coat, and face protection). Handle PFA inside a fume hood, and collect all waste for proper disposal in accordance with the institutional requirements. During perfusion, high-absorbency disposable pads can be placed inside the hood to collect blood and PFA.
Anesthetize the mouse with isoflurane (3-5% for induction and 1-2% for maintenance). Proceed when the mouse is fully anesthetized, as indicated by the absence of a response to toe pinching.
Perform a transcardial perfusion ( Wu et al., 2021 ) with ~20 ml cold heparinized phosphate-buffered saline (PBS) (10-20 units/ml), using a peristaltic pump (flow rate ~6 ml/min), until the blood is removed from the tissue and the fluid exiting the right atrium is clear.
-
Continue perfusing with ~20 ml ice-cold 4% PFA until the body is stiff.
Note: A good perfusion is critical to the quality of the experiment. To ensure a good perfusion, it is essential to remove as much blood as possible. The addition of heparin to the PBS is important to prevent blood clot formation. When perfusing with heparinized PBS, observe the color of the liver and continue perfusing with heparinized PBS for 2-3 additional minutes after the liver becomes pale before switching to PFA to ensure that blood has been removed from the pancreas.
-
Dissect the pancreas immediately, taking care not to damage the stomach and intestines (Figure 2). Place the dissected pancreas into a Petri dish containing cold PBS and remove adipose and mesenteric tissue under a dissection microscope.
Note: Mesenteric adipose tissue looks pale and glistening under a dissection microscope and can easily be pulled away from the pancreatic tissue with blunt tweezers. It can also be carefully cut away with small scissors.
For easier anatomical assessment, the pancreas can be divided into anatomical subregions (duodenal, splenic, or gastric) before proceeding with the clearing protocol (Figure 2). Alternatively, for antibody evaluation, the tissue can be sectioned into smaller pieces (2-3 mm3).
-
Postfix pancreata overnight in 4% PFA at 4°C with shaking (~25 RPM on a nutating shaker).
Note: Use a ratio of 5:1 PFA to tissue volume in an Eppendorf tube (for sizing, see Table 1). Prolonged fixation can increase tissue autofluorescence.
Wash samples 3 times in PBS for 30 min.
Place the sample in a Petri dish with PBS, and use fine tweezers to remove hair and lint from the sample under a dissection microscope.
-
Proceed immediately with sample delipidation (Part B).
Note: If necessary, samples can be stored in PBS with 0.02% sodium azide (NaN3) at 4°C for a few weeks, but this may impact the quality of the experiment.
Human tissue- Fix samples in 4% PFA immediately upon acquisition. Leave overnight in 4% PFA at 4°C with shaking (~25 RPM).
-
Proceed with the protocol for mouse tissue starting at Step A7 (PBS × 3).Note: Since human tissue will not be perfused, its quality may be more variable than that of rodent tissue.
-
Sample pretreatment (delipidation, bleaching, and permeabilization)
Note: All steps with shaking should be performed at 25 RPM on a nutating shaker.
-
Transfer samples to tubes of adequate size (Table 1) with the sample ideally taking up no more than 25% of the volume of the tube, and label each tube carefully using a scalpel or razor blade. Colored polypropylene tubes can be used to separate samples.
Note: Permanent markers are easily removed by methanol.
-
Dehydrate samples in a gradient of methanol in H2O (20, 40, 60, 80, and 100%). Incubate in each concentration for 1 h at RT with shaking. Always fill tubes completely to avoid sample oxidation.
Note: Methanol is toxic, flammable, and volatile. Always handle methanol inside a fume hood and collect all waste for proper disposal according to the institutional requirements.
Wash samples in 100% methanol for 1 h at RT with shaking.
-
Wash samples in 100% dichloromethane (DCM) for 1 h at RT with shaking to remove hydrophobic lipids.
Note: Handle DCM inside a fume hood and use only with compatible materials (e.g., polypropylene). Use double nitrile gloves or gloves compatible with DCM (e.g., neoprene). Collect all waste for proper disposal according to the institutional requirements.
Wash samples in 100% methanol for 3 × 30 min at RT with shaking.
-
Bleach samples in 5% H2O2 in methanol overnight at 4°C without shaking to reduce tissue autofluorescence. Keep samples protected from light.
Note: H2O2 may degrade with shaking, light, and heat exposure.
Rehydrate samples in a reversed gradient of methanol (80, 60, 40, and 20%). Incubate in each concentration for 30 min at RT with shaking.
Wash samples in permeabilization buffer for 1 h at RT with shaking.
Incubate samples in permeabilization buffer overnight at RT with shaking.
Wash samples in PTxwH for 3 × 30 min at RT with shaking.
-
Proceed immediately with immunolabeling.
Note: If necessary, delipidated samples can be stored in PTxwH with 0.02% NaN3 at 4°C for a few weeks. Prolonged storage may impair staining efficiency.
-
-
Immunolabeling
Primary and secondary antibodies that have been validated for use in pancreatic tissue with this protocol are listed in Table 2.
Block samples in blocking buffer overnight at 37°C with shaking.
Prepare primary antibodies by diluting them in PTxwH, mixing on a shaker for 10 min, and centrifuging at 16,000 × g for 10 min. The volume prepared depends on the tube and sample size (Table 1). Transfer the spun-down antibody solution to the sample tube, leaving ~10 μl at the bottom of the antibody tube to remove precipitates. Add 3% NDS to the sample. If necessary, add extra PTxwH to the sample to fill the tube all the way up.
-
Incubate samples with primary antibodies for 3-6 days, depending on sample size, at 37°C with shaking. Protect samples from light.
Note: Add 0.01% NaN3 to prevent microbial growth and protect the antibodies from degrading during extended incubation times. An incubation time of 6 days is recommended for whole mouse pancreatic tissue and large hemipancreata to ensure complete and even antibody diffusion (see “Notes” section).
Wash samples with PTxwH 5 times over 1 day at RT, with the final wash overnight.
-
Prepare secondary antibodies by diluting them in PTxwH, mixing them on a shaker for 10 min, and centrifuging them at 16,000 × g for 10 min. The volume prepared depends on the tube and sample size (Table 1). Transfer the spun-down antibody solution to the sample tube, leaving ~10 μl at the bottom of the antibody tube to remove precipitates. Add 3% NDS to the sample. If necessary, add extra PTxwH to the sample to fill the tube all the way up.
Note: It is recommended to use red or far-red secondary antibodies. Due to tissue autofluorescence, GFP-tagged secondary antibodies can only be used for markers with very high expression, and it is not recommended to use secondary antibodies with shorter wavelengths. Near-infrared secondary antibodies can also be used for highly expressed markers but may require prolonged exposure times during imaging.
-
Incubate samples with secondary antibodies for 3-6 days, depending on sample size, at 37°C with shaking. Protect samples from light.
Note: Add 0.01% NaN3 to prevent microbial growth and protect the antibodies from degradation during extended incubation times.
Wash samples with PTxwH 5 times over 1 day at RT, with the final wash overnight.
Wash samples with PBS 5 times over 1 day at RT, with the final wash overnight.
-
Embed pancreatic samples in 1% low melting point agarose in PBS. Heat the agarose/PBS in a microwave until melted, then pour into a Petri dish to cool. When the temperature of the agarose falls below 37°C (this can be measured with a thermometer or by hand), place the pancreas carefully in the liquid agarose using tweezers, taking care to position it properly and cover it in agarose with no air bubbles. Low melting point agarose starts to solidify at 25°C, but this process can be sped up by placing the Petri dish in the fridge. Use a razor blade or scalpel to cut the agarose to size after solidifying.
Note: Embedding is necessary for light sheet imaging but may not be necessary for confocal imaging of small pancreatic sections in an Ibidi μ-slide or glass-bottomed Petri dish.
-
Tissue clearing
Note: Protect samples from light during all steps.
-
Dehydrate samples in a gradient of methanol/H2O (20, 40, 60, 80, and 100%); each step is for 1 h at RT with shaking. The samples can optionally be left overnight in methanol at RT.
Note: Methanol is toxic, flammable, and volatile. Always handle inside a fume hood and collect all waste for proper disposal.
Wash samples in 100% methanol 3 times for at least 30 min each at RT with shaking.
-
Wash samples in 100% DCM 3 times for 30 min each at RT with shaking.
Note: Handle DCM inside a fume hood and be careful to only use it with compatible materials (e.g., polypropylene). Use double nitrile gloves or gloves compatible with DCM (e.g., neoprene). Collect all waste for proper disposal.
-
Transfer samples into dibenzyl ether (DBE) for clearing (Figure 3). Fill tubes with DBE to the top to avoid any air being left in the tube, which can cause sample oxidation. Store samples in DBE, protected from light.
Note: Samples in DBE can be stored at RT, but storage at 4°C may preserve fluorescence for longer. The melting point of DBE is 1.5-3.5°C, so samples stored in a fridge may freeze. Handle DBE in a fume hood and collect any waste for proper disposal according to the institutional requirements.
-
-
Imaging
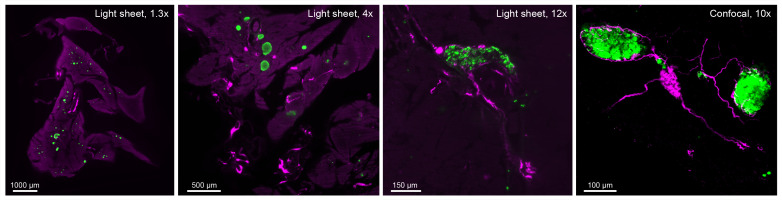
A light sheet microscope (LaVision Ultramicroscope II or similar) is recommended for whole tissue and hemipancreata. A 1.3× or 2× objective is suitable for whole-tissue mounts, whereas a 4× or 12× objective is recommended for fine innervation. An inverted confocal microscope (Zeiss LSM or similar) equipped with a 10× objective is recommended for imaging small pancreatic sections, particularly when a higher resolution is needed, such as for analyses of nerve contacts or pancreatic ganglia. Figure 4 shows the differences in magnification between the systems and objectives.
Light sheet imaging
Note: Always consult the microscopy core facility and/or specific microscope user manual for details on operation.
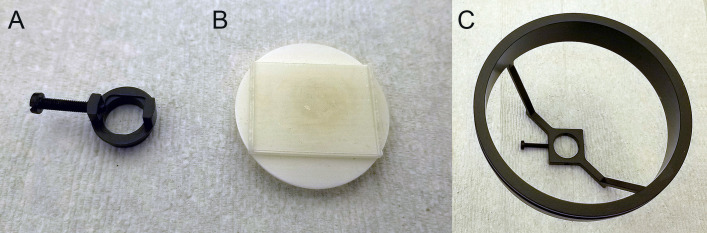
In a fume hood, remove the sample from its storage vial with blunt tweezers and attach it to a correctly sized sample holder (Figure 5A).
Large, agarose-embedded samples may need to be secured to a flat sample holder with cyanoacrylate glue (Figure 5B).
Assemble the sample holder into the sample mount, which is then submerged in DBE in the cuvette of the light sheet microscope (Figure 5C). Take care not to spill DBE on the instrument, and clean any spills immediately with methanol. Replace gloves contaminated with DBE before handling any instrument or equipment to avoid damage.
Orient the sample mount in the cuvette to ensure that the sample is perpendicular to the light sheet.
Move the sample using the joystick until the light sheet illuminates the sample, then lower the objective to focus the sample. If using a dipping objective, move it until it touches the surface of the DBE, then lower it slowly to prevent accidentally hitting the sample or holder. Adjust the display settings to properly view the sample while focusing.
In the light sheet microscope software (ImSpectorPro), select which light sheet(s) to use (this will depend on the specific instrument; consult with the imaging core) and adjust the width of the light sheet so that it evenly illuminates the entire sample. Select the lasers and adjust the focus for each channel individually if using multiple wavelengths. Adjust the laser power and exposure time to ensure a non-saturated signal.
-
Save your user settings to ensure consistency between imaging sessions.
Note: Always use the same imaging settings for samples that will be statistically compared.
For whole-mount imaging, acquire z-stacked optical sections at 1.3× or 2× magnification with the lowest zoom setting and a step size of 3-5 μm. This resolution is sufficient to image insulin+ islets and large filamentous structures such as nerve trunks, ducts, and blood vessels.
-
Acquire z-stacked optical sections at 4× or 12× magnification with dynamic horizontal focus and a step size of 2-5 μm to generate images suitable for assessing finer structures, such as individual α cells, β cells, fine filamentous innervation, fine ducts, blood capillaries, and intrapancreatic ganglia. For analyses of ganglia, which can consist of as few as two neurons, it is important to use sufficient magnification to confidently identify individual cell bodies (Figure 4).
Tiled mosaic images can be acquired at 4× and 12× with 20% overlap and stitched using the plugin TeraStitcher (Bria and Iannello, 2012). However, generating mosaic images can be time consuming and result in very large files that require more processing power to analyze. An alternative to mosaic images is to acquire 1-3 separate z-stacks at 4× or 12× magnification from each sample, segment and analyze them separately, and pool the results.
Note: Factors that affect the light sheet acquisition time are exposure time, dynamic horizontal focus (number of steps per image), and step size. The step size will also affect the final file size. The exposure time can be shortened by increasing the laser power; however, an increased laser power increases the risk of sample bleaching, so only increase the laser power immediately before imaging.
Confocal imaging
Note: If performing light sheet and confocal imaging of the same samples, do the confocal imaging last due to the increased risk of sample bleaching. Always consult the microscopy core facility and/or microscope user manual for details on operation.- Remove the sample from the storage vial using fine blunt tweezers and place it in a glass-bottomed μ-slide or a μ-dish, avoiding air bubbles between the sample and the glass.
- Check the integrity of the chamber/slide before and after imaging to ensure that DBE is not leaking, which can damage the microscope.
- Acquire z-stacked optical sections with a standard 10× objective and a step size of 5-10 μm for larger structures such as islets and ganglia, or a step size <5 μm for finer details such as nerve contacts. Note: Due to the shorter working distance, a standard 20× objective can only be used for imaging structures that are close to the sample surface. For higher magnification, it is recommended to use an objective with a long working distance.
Figure 2. Mouse pancreas with surrounding organs and anatomical subregions indicated.
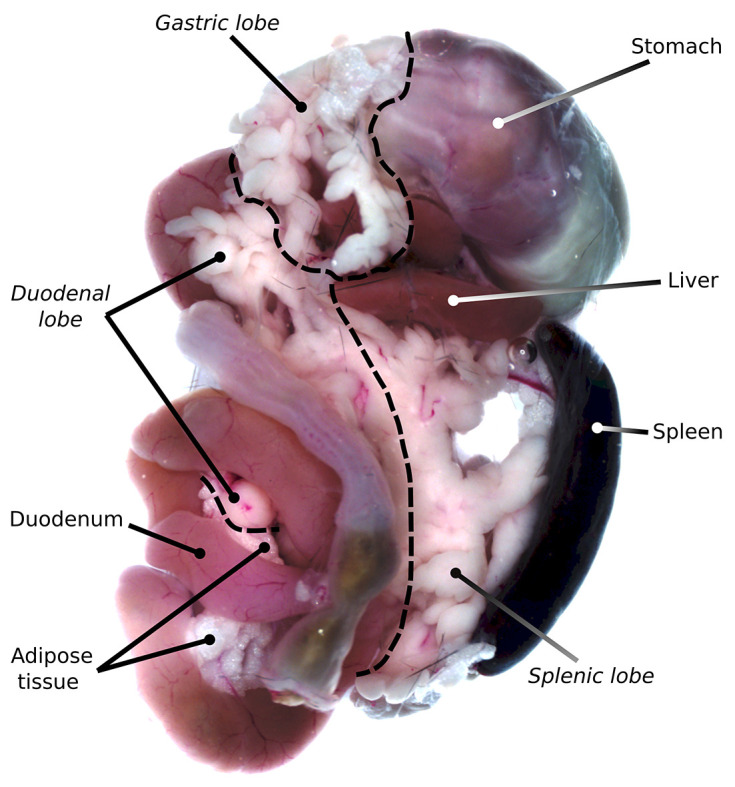
Table 1. List of tube sizes and usage.
| Tube size | Usage | |
|---|---|---|
| 5-ml | Postfixation, washing, re-/dehydration, and clearing of large samples (e.g., intact pancreata or large hemi-pancreata) | |
| 2-ml | Postfixation, washing, re-/dehydration, and clearing of small or intermediate samples (e.g., small hemi-pancreata) | Antibody incubation of large samples (e.g., intact pancreata or large hemi-pancreata) |
| 1.5-ml | Postfixation, washing, re-/dehydration, and clearing of small samples (e.g., pancreatic sections a few mm in diameter) | Antibody incubation of intermediate samples (e.g., small hemi-pancreata) |
| 0.5-ml | Antibody incubation of small samples (e.g., pancreatic sections a few mm in diameter) | |
Table 2. Antibodies validated for iDISCO+ in the pancreas.
Adapted from Alvarsson et al. (2020) .
| PRIMARY ANTIBODIES FOR ENDOCRINE MARKERS | ||||||
|---|---|---|---|---|---|---|
| Target | Dilution | Host species | Manufacturer | Product number |
RRID number |
|
| Glucagon | 1:200 | Rabbit | Cell Signaling Technology | 2760 | AB_659831 | |
| Glucagon | 1:2000-1:5000 | Mouse | Sigma-Aldrich | G2654 | AB_259852 | |
| Insulin | 1:1000 | Guinea pig | Dako/Agilent | A0564 | AB_10013624 | |
| Insulin | 1:500 | Rat | R&D Systems | MAB1417 | AB_2126533 | |
| Somatostatin | 1:1000 | Goat | Santa Cruz | SC7819 | AB_2302603 | |
| PRIMARY ANTIBODIES FOR NEURONAL MARKERS | ||||||
| Neurofilament 200 kDA | 1:500 | Rabbit | Sigma-Aldrich | N4142 | AB_477272 | |
| Synapsin | 1:500 | Rabbit | Cell Signaling Technology | 5297 | AB_2616578 | |
| TRPV1 | 1:500 | Rabbit | Alomone labs | ACC-030 | AB_2313819 | |
| Tyrosine Hydroxylase | 1:500 | Rabbit | Millipore | AB152 | AB_390204 | |
| Vesicular Acetylcholine Transporter | 1:500 | Rabbit | Synaptic Systems | 139 103 | AB_887864 | |
| MISCELLANEOUS PRIMARY ANTIBODIES | ||||||
| Mucin 1 | 1:200 | Armenian hamster | Thermo FisherScientific | MA5-11202 | AB_11000874 | |
| Green Fluorescent Protein | 1:2000 | Chicken | Aves | GFP-1010 | AB_2307313 | |
| Red Fluorescent Protein | 1:2000 | Rabbit | Rockland | 600-401-379 | AB_2209751 | |
| HA-tag | 1:500 | Rabbit | Cell Signaling | 3724 | AB_1549585 | |
| SECONDARY ANTIBODIES | ||||||
| Target | Conjugate | Dilution | Host species | Manufacturer |
Product number |
RRID number |
| Armenian Hamster | Alexa Fluor® 594 | 1:500 | Goat | Jackson ImmunoResearch | 127-585-099 | AB_2338998 |
| Goat | Alexa Fluor® 546 | 1:500 | Donkey | Thermo Fisher Scientific | A-11056 | AB_2534103 |
| Guinea pig | Alexa Fluor® 647 | 1:500 | Donkey | Jackson ImmunoResearch | 706-605-148 | AB_2340476 |
|
Mouse |
Alexa Fluor® Plus 647 | 1:500 | Donkey | Invitrogen | A32787 | AB_2762830 |
| Rabbit | Alexa Fluor® 647 | 1:500 | Donkey | Jackson ImmunoResearch | 711-605-152 | AB_2492288 |
| Rabbit | Alexa Fluor® 594 | 1:500 | Donkey | Invitrogen | A-21207 | AB_141637 |
| Rabbit | Alexa Fluor® 546 | 1:500 | Donkey | Thermo Fisher Scientific | A10040 | AB_2534016 |
| Rat | DyLight® 550 | 1:500 | Donkey | Thermo Fisher Scientific | SA5-10027 | AB_2556607 |
| Rat | Alexa Fluor® 488 | 1:500 | Donkey | Thermo Fisher Scientific | A-21208 | AB_25357794 |
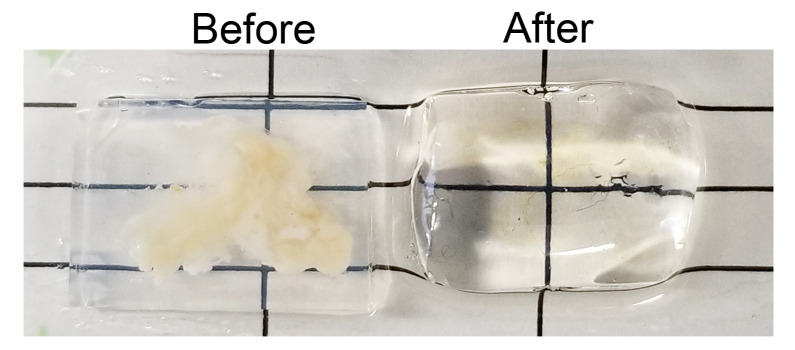
Figure 3. Optical clearing.
Agarose-embedded mouse pancreata before (left) and after (right) being subjected to the optical clearing protocol.
Figure 4. System comparisons.
Single z-stack images captured with a light sheet microscope (LaVision Ultramacroscope II) equipped with a 1.3× objective, a 4× objective, and a 12× objective (hemipancreata), and an inverted confocal microscope equipped with a 10× objective (small pancreatic section).
Figure 5. Sample mounting for light sheet microscopy.
Sample holder for smaller agarose-embedded samples, where the sample is fixed with a screw (A). Sample holder for large agarose-embedded samples, where the sample is fixed with glue (B). The sample holder is then attached to the sample mount and fixed with a screw for submerging in DBE (C).
Data analysis
Image processing and analysis
Note: Use a 3D image processing software to generate 3D images from acquired z-stacks and to perform volume and distance analyses. The protocol below is adapted for image analysis in Imaris, but note that there are other commercial and open-source image analysis programs that can be used to a similar end. Additional online video tutorials and webinars on using the Imaris software can be found athttps://imaris.oxinst.com/tutorialsandhttps://imaris.oxinst.com/homeschool.
Move the acquired z-stack image to the image analysis computer and make sure to always keep these data backed up. Files generated by light sheet microscopy are large (10-200 GB per image), so make a plan for long-term data storage.
Import z-stacks into Imaris by adding the image folder as an “Observe Folder,” and double click on the z-stack to automatically generate a compressed 3D image.
Use the Creation Wizard to create digital surfaces covering the signal of interest in each channel (outlined in detail below). These surfaces will be used to extract measurements specific for each channel, such as volume and signal intensity.
Segmentation
-
Segmentation of insulin+ islets:
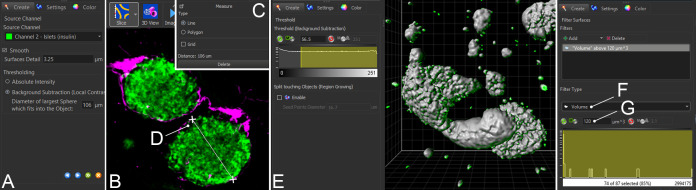
Create a digital surface by clicking the blue “surface” button in the top left menu. In the Creation Wizard, choose to analyze the entire image or a Region of Interest (ROI). In the next step, select the correct channel, smoothing factor, and “Background Subtraction” (Figure 6A).
-
The default smoothing factors can usually be used for analysis of islets.
Note: Threshold values may have to be modified depending on sample quality and computing power, but should ideally be kept the same between analyses. A smaller smoothing factor will give finer detail but will require more memory to process than a larger smoothing factor.
For “Background Subtraction,” the threshold factor should correspond to the diameter of the largest islet in each sample. This value can be obtained by switching to “Slice” view under the “View” menu (Figure 6B). Scroll through the entire z stack to identify the largest islet, and use the “Measure” tool in the top right corner to obtain the diameter (Figure 6C, 5D).
-
In the next step, set the detection threshold, such that it adequately covers insulin+ islets and β cells without covering any unwanted fluorescence (Figure 6E). Toggle the surface on and off to compare the volume coverage with the islet channel, making sure that no rendered volumes appear too large or to inadequately cover dimmer structures.
Note: Since large datasets may vary in their insulin intensity, also within the same sample, the detection threshold may have to be adjusted for each sample to allow for optimal islet detection. For this reason, it is very important to keep the display adjustment similar between samples. The display adjustment does not change the intensity output values but alters how the intensity is displayed on the screen, which in turn will make islets look dimmer/smaller/larger and therefore may affect the thresholding when setting it manually. If necessary due to high variability in insulin intensity, several ROIs can be created to cover islets of different intensities as part of separate islet surfaces, which then can be merged into one islet surface upon completion using the “Edit” tool.
-
Antibody precipitates are common artefacts that appear as tiny, bright, and sometimes elongated dots or stars, often situated along the surface of the sample. To remove antibody precipitates from the islet surface, volumes below 120 μm3 (the approximate size of one β cell) can be excluded from the final surface using the “Filter” function in the Creation Wizard. Under “Filter Type,” select “Volume” (Figure 6F) and exclude objects with volumes below 120 μm3 (Figure 6G). This function can also be used to remove islets below a certain size, such as proto-islets consisting of only a few β cells, from the final analysis. Such exclusion criteria should ideally be established before analysis and applied equally across all samples.
Note: Artifacts can only be removed from the analysis by removing the volume covering the artifact. Clipping planes and slicing tools only remove the volume from the field of view but not from the analysis.
-
Segmentation of nerves:
It is generally recommended to use a threshold factor corresponding to the width of the widest nerve trunk (e.g., 12 μm). However, for filamentous structures, lower threshold values may give better digital surface coverage depending on the specific characteristics of the neuronal marker used, including the diameter of the finest nerve fibers.
-
Likewise, the smoothing factor needs to be optimized depending on the specific neuronal marker (e.g., 3.5-1 μm).
Note: A smaller smoothing factor will give finer detail but will require more memory to process than a larger smoothing factor.
-
When staining for tyrosine hydroxylase (TH), the presence of TH+ β cells may confound measurements of TH+ pancreatic innervation (Iturriza and Thibault, 1993; Persson-Sjögren et al., 2002). For optimal detection of TH+ nerves, TH+ β cells have to be manually removed from the final TH+ nerve surface. This can be done by excluding volumes below 120 μm3 residing within insulin+ islets and overlapping with insulin staining. Once the TH+ nerve surface is created, highlight it and click the pencil icon (“Edit”) in the bottom left part of the screen. Select “circle” and use the mouse wheel to select the size of the mouse pointer. Zoom in over islets and use the pointer to select the volumes corresponding to β cells. Press the “delete” key to remove these volumes from the final surface.
Note: Only TH+ β cells that do not touch nerves can be removed using this method.
-
Segmentation of endocrine innervation:
This is done by masking the innervation channel to remove the background and then masking it again using the islet volume to remove innervation outside islets, thus generating a new channel, where the remaining signal is from innervation inside islets.
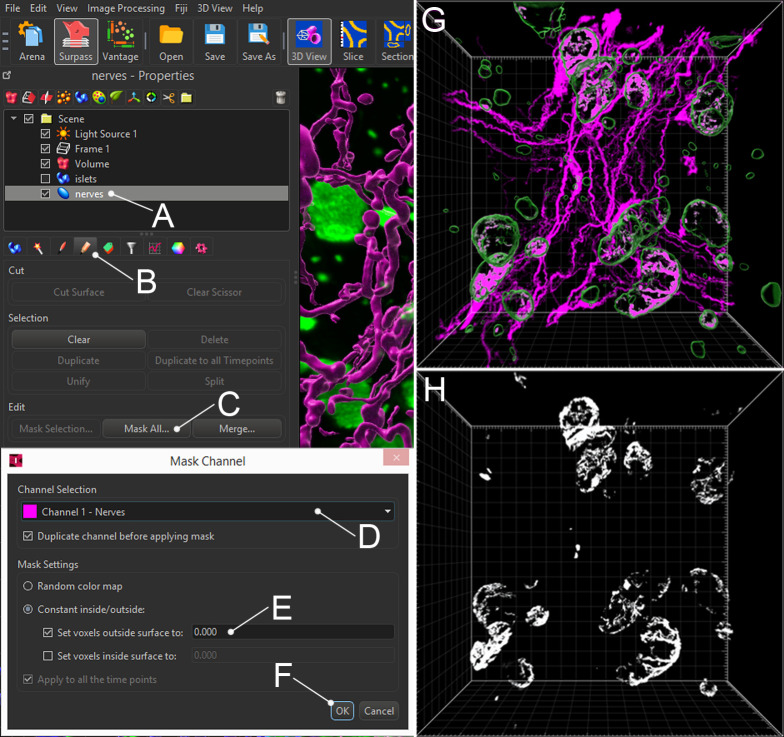
Select the innervation surface (Figure 7A) and click the pencil (“Edit”) icon (Figure 7B). Click “Mask All” under “Edit” (Figure 7C). In the pop-up box, select the innervation channel (Figure 7D) and “Set voxels outside surface to: 0.000” (Figure 7E), then click “OK (Figure 7F)”. Imaris will now create a new channel “Masked [innervation channel name],” containing innervation with no background.
Select the islet surface (Figure 7G), click “Edit,” and then “Mask all.” In the pop-up box, select the newly created masked innervation channel and “Set voxels outside surface to: 0.000”, then click “OK.” Imaris will now create a new channel, where the signal represents innervation inside islets with no background (Figure 7H).
Create a new surface based on this new channel using the same settings as when segmenting the nerves in step 2.
-
Segmentation of ganglia:
Use images of sufficient magnification to confidently identify cell bodies.
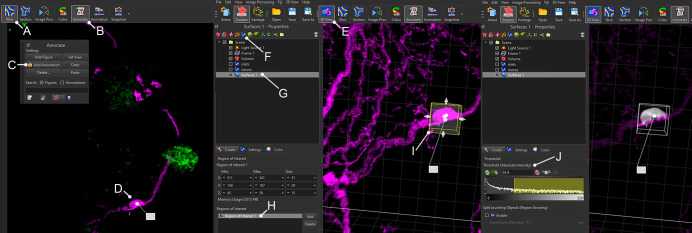
Use the “Slice” view to identify ganglia (Figure 8A), and label them using the “Annotate” tool under “Edit” (Figure 8B-D).
Switch to 3D view (Figure 8E) and use the creation wizard to manually create one ROI around each individual ganglion (Figure 8F-I). This will generate digital surfaces specifically covering ganglionic cell bodies, but not nerve fibers.
Set the threshold to fully cover the cell bodies of the ganglion, and proceed with segmentation as above (Figure 8J).
-
Segmentation of the total sample volume:
For each sample undergoing volumetric analysis, create a surface covering the total volume of the sample using the channel with the highest background signal. This value is required to adjust for variations in total sample volume.
Figure 6. Segmentation of islets of Langerhans.
Screenshots from Imaris showing the key steps for analyzing insulin+ islets, which include setting up the smoothing (A), background subtraction (B) with identification of the largest diameter (C, D), thresholding (E), and filtering based on volume (F, G).
Figure 7. Segmentation of endocrine innervation in Imaris.
Screenshots from Imaris showing the key steps for analyzing innervation inside endocrine cells. This is done by masking the nerve channel twice; once using the “nerve” surface to remove all background from the nerve channel (A-F) and a second time using the “islets” surface (green outlines) to remove all nerve signals outside islets (G), thus generating a new channel, where the signal only corresponds to innervation inside islets (H).
Figure 8. Segmentation of intrapancreatic ganglia in Imaris.
Screenshots from Imaris showing the key steps in segmenting pancreatic ganglia. “Slice” view (A) can be used with the “Annotate” tool (B, C) to identify ganglia in 2D (D). In 3D view, create a new surface (F, G) with one ROI covering each individual ganglion (H, I). Set the threshold to fully cover the cell bodies of the ganglion (J).
Distance calculations
The purpose of distance calculations is to identify surfaces that are closely associated, suggesting physiological interactions. Non-synaptic autonomic neurotransmission occurs over distances up to 2 μm, whereas hormones and cytokines can act over longer distances (Burnstock, 2008).
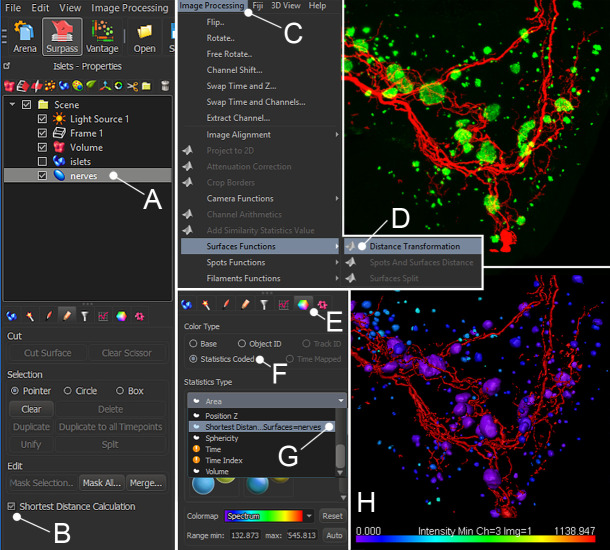
In Imaris 9.5 and newer versions, distance measurements can be obtained using the “Shortest Distance Calculation” function in the creation wizard when creating a surface (Figure 9A and B). This will generate a new parameter called “shortest distance to surface=[name]” when exporting the data.
If using Imaris 9.3 or older, the Imaris “Distance Transformation” XTension function can be used to calculate the distance of each islet surface from the innervation surface. To use this function, select the final innervation surface (Figure 9A), then select “Distance Transformation” under “Image Processing” (Figure 9C) → “Surface Functions” (Figure 9D). Run the plugin and select the “Outside Surfaces object” option. This will generate a new channel, where the intensity is based on the distance from the selected surface. Distances from islets to nerves should be reported as the intensity minimum of the distance transformation channel (intensity 0 = islet touching nerve) for each islet surface to the nerve surface as calculated by the distance transformation operation.
To visually display the distance from one surface to another, select a surface and click “Color” (Figure 9E). Set the color type to “Statistics Coded” (Figure 9F), and under “Statistics Type” select “Shortest Distance to Surface[name]” (Figure 9G-H). If the distance measurements were done using the “Distance Transformation” XTension, the surface should instead be statistics coded for the minimum intensity of the distance transformation channel.
-
In images with sufficient magnification, digital surfaces can be created to cover nerves and individual α or β cells, and the distance between nerves and individual cells can be calculated using the principles outlined in steps 1-3. A nerve/cell distance of 0 indicates a nerve contact.
Note: This method does not allow for quantitation of the number of contacts per endocrine cell.
Figure 9. Distance calculations in Imaris.
The shortest distance between a selected surface and any other surface can be obtained in Imaris using the “Shortest Distance Calculation” (A, B) or using the “Distance Transformation” XTension function (C, D). To make a color overlay that illustrates the distance to nerves, highlight the “islets” surface and click “Color” (E), select “Statistics Coded” (F), and under “Statistics Type” select “Shortest distance to surface, Surface=nerves” (G). H shows pancreatic islets and innervation without surfaces (upper panel) and with an “islets” surface color-coded for distance to the “nerves” surface (lower panel).
Data export and processing
Export data as Excel spreadsheets by highlighting each segmented surface and clicking the export button.
Create a summarizing Excel document to keep the exported data organized.
-
In Excel:
Calculate surface volumes as percentages of the total sample volume to adjust for differences in sample size.
Calculate endocrine nerve volume as a percentage of the total islet volume in each sample.
Use the mean or median intensity values to calculate insulin intensity.
For surfaces consisting of multiple volumes (e.g., insulin), Imaris will generate a unique ID number for each volume, which will be displayed under “ID” in the exported Excel sheet. This ID number can be used to correlate different parameters such as volume, intensity, and distance.
Notes
This section lists problems that may occur with this protocol and how to best mitigate them.
-
The sample does not clear in DBE during the final step of the protocol.
This can happen when there is some water remaining in the sample during the final steps, and it is more likely to happen in large agarose-embedded samples. Reverse the clearing steps until the sample is back in 100% methanol. Wash a couple of additional times in 100% methanol to ensure that all DBE and water are removed, and optionally leave the sample in 100% methanol overnight. Proceed with the clearing protocol. If the sample is large, it can help to prolong the DCM washes.
If the agarose has stiffened and warped, rehydrate the sample completely, carefully remove the agarose using fine tweezers, and re-embed and dehydrate the sample again with the modifications suggested above.
-
The cleared sample looks blurry under the light sheet microscope.
This can either be because of poor clearing (see above), a poor perfusion (blood remaining in the tissue), lipids remaining in the sample (e.g., failure to remove mesenteric adipose tissue completely), or because the samples have been stored for too long before clearing.
This can also happen if the light sheet microscope is not set up properly. Move the horizontal focus to verify that it is focused, and adjust the sheetmotor calibration if needed.
-
The antibody is staining the surface of the sample.
Poor antibody penetration occurs with some primary antibodies and can to some extent be mitigated by increasing the antibody incubation time. Decreasing the antibody concentration may also increase antibody penetration. If the problem persists, try to find an alternative antibody. Antibodies with smaller molecular weights (e.g., nanobodies) may penetrate tissue better; however, the availability of primary detection nanobodies is currently limited.
-
The antibody does not stain the center of the sample.
This can be seen as a result of the antibody being depleted, usually when targeting an abundantly expressed antigen, and can be avoided by increasing the antibody concentration.
-
The antibody does not appear to stain the sample at all.
This could be due to poor tissue quality (e.g., poor clearing, poor perfusion, the presence of lipids, or the sample having been stored for too long before clearing). In this case, the sample may look slightly blurry under the light sheet.
If the sample looks crisp under the light sheet, this suggests a problem with the antibody rather than with the sample. This could be due to the primary antibody not being compatible with the protocol. We recommend testing the antibody on thick sections and/or small pieces of tissue before attempting whole-mount experiments. Use the manufacturer’s recommended dilution for immunohistochemistry as the starting dilution.
This could be caused by a weak antibody signal-to-background ratio. It is recommended to use the red or far-red channels to minimize the effects of tissue autofluorescence. Moreover, directly conjugated antibodies tend to yield weak signals; thus, it is recommended to use secondary antibodies to amplify the signal.
-
The antibody is precipitating.
Some antibodies are more prone to precipitating, and this may also be a sign of an antibody being compromised. Spinning the antibodies down before adding them to the sample or filtering them through a 0.2-μm syringe filter can to some extent mitigate the problem. If possible, find an alternative antibody.
Depending on the amount of precipitate, it is often possible to filter away precipitates during the image analysis step.
-
3D rendering reveals hair/fibers in the GFP channel.
Remove hair and fibers manually under a dissection microscope before embedding the sample.
Avoid using the GFP channel due to high levels of autofluorescence in the cleared samples.
This type of artifact can often be removed easily from 3D analysis using the “Edit” tool.
-
I want to add another antibody to an already cleared sample.
It is possible to reverse the clearing in order to add another antibody. Run the sample through a reversed version of the clearing protocol until the sample is back in PTxwH. Remove any agarose and repeat the protocol beginning with the primary antibody incubation step. Protect the sample from light to preserve any existing fluorophores. Since imaging and general handling may compromise the integrity of existing antibodies, it is not recommended to use restained samples for analysis.
-
The image analysis generates unexpectedly high/low islet counts/volumes.
Check that there are no precipitates or other artifacts in the islet channel that could confound the analysis. Precipitates are sometimes easier to identify under the “Slice” view.
Check that the total sample volume is calculated correctly by checking that the volume only covers pancreatic tissue. If the surrounding adipose tissue is not removed completely, it may lead to an overestimation of the total volume. Look for the presence of islets, and go over the whole sample in 2D view. If the sample contains non-pancreatic tissue, create a more defined ROI when creating a surface for the total sample volume.
Recipes
-
Heparinized PBS (500 ml, 20 units per ml)
500 ml dH2O
PBS tablets
10,000 units heparin sodium salt (e.g., add 55.6 mg ≥180 USP units/mg heparin)
-
4% PFA buffer (100 ml)
75 ml H2O
25 ml 16% PFA
0.5 PBS tablets
Note: Prepare fresh and keep on ice during perfusion. PFA and its fumes are toxic. Always consult the safety data sheet before handling and use the recommended personal protective equipment (including gloves, lab coat, and face protection). Handle PFA inside a fume hood and collect all waste for proper disposal in accordance with the institutional requirements.
-
10% sodium azide (NaN3) stock (100 ml)
100 ml H2O
10 g NaN3
Note: NaN3 is highly toxic, readily penetrates the skin, and can react with metal. Measure NaN3 powder inside a fume hood and do not use a metal spatula. Always handle >5% NaN3 stock solutions inside a fume hood. Never pour waste buffers containing NaN3 down the drain. Collect NaN3 waste for proper disposal. Store at 2-8°C.
-
Modified PTxwH (1000 ml)
994 ml H2O
5 PBS tablets
1 ml Tween-20 (0.1%)
5 ml Triton X-100 (0.5%)
2 mg heparin
0.1 g NaN3 (optional)
Note: This modified PTxwH uses higher concentrations of detergent as compared with the original PTxwH recipe (Renier et al., 2016) for optimized staining of pancreatic innervation. Store at RT.
-
Permeabilization buffer (100 ml)
100 ml PTxwH
5 ml DMSO (5%)
2.25 g glycine (0.3 M)
Note: Store at RT, protecting from light.
-
Blocking buffer
PTxwH
3% normal donkey serum
Note: Prepare fresh as needed.
Acknowledgments
This protocol was adapted from the previous study published by Alvarsson et al. (2020). A.A. was supported by a senior postdoctoral fellowship from the Charles H. Revson Foundation (grant no. 18-25), a fellowship from Sweden-America Foundation (Ernst O Eks fond), and a postdoctoral scholarship from the Swedish Society for Medical Research (SSMF). M.J.G and A.A. are supported by the Department of Defense Discovery Award (# W81XWH-20-1-0156). This work was also supported by the American Diabetes Association Pathway to Stop Diabetes Grant ADA #1-17-ACE-31 and, in part, by grants from the NIH (R01NS097184, OT2OD024912, and R01DK124461), Department of Defense (W81XWH-20-1-0345), and the Alexander and Alexandrine Sinsheimer Scholar Award. This work was supported in part by a Mindich Child Health and Development Institute Pilot and Feasibility Grant and by a Mount Sinai Distinguished Scholar award. Microscopy and image analysis were performed at the Microscopy CoRE at the Icahn School of Medicine at Mount Sinai.
Competing interests
S.A.S. is a named inventor of the intellectual property, “Compositions and Methods to Modulate Cell Activity,” and is a co-founder of, consults for, and has equity in the private company Redpin Therapeutics (preclinical stage gene therapy company developing neuromodulation technologies). The authors declare that they have no other competing interests.
Ethics
All protocols were approved by the Institutional Animal Care and Use Committee.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Ahren B.(2000). Autonomic regulation of islet hormone secretion--implications for health and disease. Diabetologia 43(4): 393-410. [DOI] [PubMed] [Google Scholar]
- 2.Ahren B. and Holst J. J.(2001). The cephalic insulin response to meal ingestion in humans is dependent on both cholinergic and noncholinergic mechanisms and is important for postprandial glycemia. Diabetes 50(5): 1030-1038. [DOI] [PubMed] [Google Scholar]
- 3.Alvarsson A., Jimenez-Gonzalez M., Li R., Rosselot C., Tzavaras N., Wu Z., Stewart A. F., Garcia-Ocana A. and Stanley S. A.(2020). A 3D atlas of the dynamic and regional variation of pancreatic innervation in diabetes. Sci Adv 6(41): eaaz9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bria A. and Iannello G.(2012). TeraStitcher- a tool for fast automatic 3D-stitching of teravoxel-sized microscopy images. BMC Bioinformatics 13: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burnstock G. J.(2008). Non-synaptic transmission at autonomic neuroeffector junctions. Neurochemistry International 52(1-2): 14-25. [DOI] [PubMed] [Google Scholar]
- 6.Butterworth E. A., Nasif L. H., Nasif K., Carty K. N., Mathews C. E., Atkinson M. A., Gerling I. C. and Campbell-Thompson M.(2018). Sympathetic innervation of human alpha cells. Diabetes 67(Supplement 1), 1799-P. [Google Scholar]
- 7.Dolenšek J., Rupnik M. S. and Stožer A. J.(2015). Structural similarities and differences between the human and the mouse pancreas. Islets 7(1): e1024405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giannulis I., Mondini E., Cinti F., Frontini A., Murano I., Barazzoni R., Barbatelli G., Accili D. and Cinti S.(2014). Increased density of inhibitory noradrenergic parenchymal nerve fibers in hypertrophic islets of Langerhans of obese mice. Nutr Metab Cardiovasc Dis 24(4): 384-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fasanella K. E., Christianson J. A., Chanthaphavong R. S. and Davis B. M.(2008). Distribution and neurochemical identification of pancreatic afferents in the mouse. J Comp Neurol 509:42-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iturriza F. C. and Thibault J.(1993). Immunohistochemical investigation of tyrosine-hydroxylase in the islets of Langerhans of adult mice, rats and guinea pigs. Neuroendocrinology 57(3): 476-480. [DOI] [PubMed] [Google Scholar]
- 11.Li W., Yu G., Liu Y. and Sha L.(2019). Intrapancreatic Ganglia and Neural Regulation of Pancreatic Endocrine Secretion. Front Neurosci 13: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Louis-Sylvestre J.(1978). Relationship between two stages of prandial insulin release in rats. Am J Physiol 235(2): E103-111. [DOI] [PubMed] [Google Scholar]
- 13.Lundberg M., Lindqvist A., Wierup N., Krogvold L., Dahl-Jørgensen K. and Skog O. J. P. o. (2017). The density of parasympathetic axons is reduced in the exocrine pancreas of individuals recently diagnosed with type 1 diabetes. PLoS ONE 12(6): e0179911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mundinger T. O., Mei Q., Foulis A. K., Fligner C. L., Hull R. L. and Taborsky G. J., Jr (2016). Human Type 1 Diabetes Is Characterized by an Early, Marked, Sustained, and Islet-Selective Loss of Sympathetic Nerves. Diabetes 65(8): 2322-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Persson-Sjogren S., Forsgren S. and Taljedal I. B.(2002). Tyrosine hydroxylase in mouse pancreatic islet cells, in situ and after syngeneic transplantation to kidney. Histol Histopathol 17(1): 113-121. [DOI] [PubMed] [Google Scholar]
- 16.Renier N., Adams E. L., Kirst C., Wu Z., Azevedo R., Kohl J., Autry A. E., Kadiri L., Umadevi Venkataraju K., Zhou Y., Wang V. X., Tang C. Y., Olsen O., Dulac C., Osten P. and Tessier-Lavigne M.(2016). Mapping of Brain Activity by Automated Volume Analysis of Immediate Early Genes. Cell 165(7): 1789-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rodriguez-Diaz R. and Caicedo A.(2014). Neural control of the endocrine pancreas. Best Pract Res Clin Endocrinol Metab 28(5): 745-756. [DOI] [PubMed] [Google Scholar]
- 18.Rodriguez-Diaz R., Abdulreda M. H., Formoso A. L., Gans I., Ricordi C., Berggren P. O. and Caicedo A.(2011). Innervation patterns of autonomic axons in the human endocrine pancreas. Cell Metab 14(1): 45-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Strubbe J. H. and Steffens A. B.(1993). Neural control of insulin secretion. Horm Metab Res 25(10): 507-512. [DOI] [PubMed] [Google Scholar]
- 20.Teff K. L. and Townsend R. R.(1999). Early phase insulin infusion and muscarinic blockade in obese and lean subjects. Am J Physiol 277(1): R198-208. [DOI] [PubMed] [Google Scholar]
- 21.Wu J., y. Cai, Wu X., Ying Y., Tai Y. and He M.(2021). Transcardiac Perfusion of the Mouse for Brain Tissue Dissection and Fixation. Bio-protocol 11(5): e3988. [DOI] [PMC free article] [PubMed] [Google Scholar]