Abstract
Primary somatosensory neurons, whose cell bodies reside in the dorsal root ganglion (DRG) and trigeminal ganglion, are specialized to transmit sensory information from the periphery to the central nervous system. Our molecular understanding of peripheral sensory neurons has been limited by both their heterogeneity and low abundance compared with non-neuronal cell types in sensory ganglia. We describe a protocol to isolate nuclei from mouse DRGs using iodixanol density gradient centrifugation, which enriches for neuronal nuclei while still sampling non-neuronal cells such as satellite glia and Schwann cells. This protocol is compatible with a range of downstream applications such as single-nucleus transcriptional and epigenomic assays.
Keywords: Dorsal root ganglion, Sensory neurons, Single-nucleus RNA-seq, Single-nucleus ATAC-seq, Gene regulation, Epigenomics
Background
Gene expression is dynamically regulated in dorsal root ganglion (DRG) cells after peripheral nerve injury, a process important for axonal regeneration and the neuronal hyperexcitability that contributes to neuropathic pain. Previous studies have analyzed gene expression and chromatin structure from bulk DRG tissues ( Perkins et al., 2014 ; Chandran et al., 2016 ), but their interpretation is complicated by the cellular heterogeneity of the DRG.
Single-cell/single-nucleus genomics has become an important tool for studying molecular features within distinct peripheral sensory neuronal subtypes ( Usoskin et al., 2015 ; Nguyen et al., 2017 and 2019; Zeisel et al., 2018 ; Sharma et al., 2020 ). While single-cell transcriptional profiling from sensory ganglia appears to provide greater transcriptional diversity than single-nucleus transcriptional profiling, a key advantage of analyzing nuclei is that it can be applied to both fresh and archived frozen specimens. This offers flexibility in sample preparation and greater access to human samples. Nuclei isolation is also associated with induction of immediate early genes contrary to whole-cell isolation because it is faster, avoids cytoplasmic signaling cascades, and is conducted entirely at 4°C. These features make single-nucleus analyses particularly attractive for studying changes that occur rapidly after a stimulus.
Here, we describe a density gradient centrifugation protocol for purifying nuclei from frozen mouse DRG samples, which is compatible with droplet-based single-nucleus genomic analyses ( Renthal et al., 2020 ). This protocol, which is based on previous protocols for extracting mouse and human CNS nuclei ( Mo et al., 2015 ; Renthal et al., 2018 ), yields a greater fraction of neuronal nuclei (40-50% neurons as measured by single-nucleus RNA sequencing) as compared with commonly used non-gradient dissociation methods (5-10% neurons) ( Habib et al., 2017 ; Avraham et al., 2020 ; Slyper et al., 2020 ), while still sampling a broad range of non-neuronal DRG cell types.
Materials and Reagents
1.5-ml centrifuge tubes (Eppendorf, catalog number: 022431021)
5-ml centrifuge tubes (Eppendorf, catalog number: 0030108310)
50-ml conical centrifuge tubes (Falcon, catalog number: 352098)
Cell strainers, 40-µm (BD Falcon, catalog number: 352340)
Ultracentrifuge tubes (Beckman Coulter, catalog number: 344059)
Laboratory film (Parafilm, catalog number: PM-996)
Freshly dissected or freshly frozen mouse DRG tissue
RNase inhibitor (Promega, catalog number: N2611)
60% Iodixanol solution (Sigma, catalog number: D1556)
IGEPAL CA-630 (Sigma, catalog number: I8896)
Nuclease-free water (Invitrogen, catalog number: 4387936)
35% Bovine Serum Albumin solution (BSA; Sigma, catalog number: A7979-50ML)
Sucrose (Millipore, catalog number: 573113)
KCl (Thermo Fisher Scientific, catalog number: AM9640G)
MgCl2 (Thermo Fisher Scientific, catalog number: AM9530G)
Tricine (Sigma, catalog number: T0377)
KOH (Sigma, catalog number: 221465)
Actinomycin (Sigma Aldrich, catalog number: A9415)
PBS (Thermo Fisher Scientific, catalog number: 10010023)
Trypan Blue (Fisher Scientific, catalog number: 15-250-061)
(Optional) Nuclei buffer (10× genomics, catalog number: PN-2000207)
Stock buffer HB (see Recipes)
Diluent buffer (see Recipes)
Equipment
Ultracentrifuge (Beckman Coulter, model: Optima L-90K)
Ultracentrifuge swinging-bucket rotor (Beckman Coulter, model: SW 41 Ti)
Centrifuge with swinging-bucket rotors (Thermo Scientific, model: 75004261)
7-ml Dounce homogenizer (Thermo Fisher Scientific, model: 06435A)
Tissue-Tearor (Biospec Products Inc., model: 985370-04)
Hemocytometer (Hausser Scientific, 1492)
Procedure
-
Preparation
Prepare fresh DRGs from experimental animals ( Lin et al., 2018 ). Alternatively, DRG samples can be prepared in advance and stored at -80°C.
-
Prepare the fresh buffers listed below and maintain them at 4°C (buffer volumes shown below are for one DRG sample).
Note: It is important to vortex any solutions containing iodixanol well every time before use.
7.5 ml Working Solution (50% iodixanol): 6.25 ml 60% iodixanol solution + 1.25 ml Diluent.
1.25 ml 30% iodixanol: 750 μl Working Solution + 500 μl Stock Buffer HB + 1.5 μl 35% BSA + 2 μl RNase inhibitor.
1.25 ml 40% iodixanol: 1 ml Working Solution + 250 μl Stock Buffer HB + 1.5 μl 35% BSA + 2 μl RNase inhibitor.
5 ml Working Buffer HB: 5 ml Stock Buffer HB + 6 μl 35% BSA + 7.5 μl RNase inhibitor.
400 μl 5% IGEPAL CA-630: 20 μl IGEPAL CA-630 + 380 μl Working Buffer HB.
-
Pre-cool centrifuge, ultracentrifuge, and rotor to 4°C. All centrifugations should be done at 4°C.
Note: Using swing-bucket rotors is recommended to improve the final nuclei recovery.
-
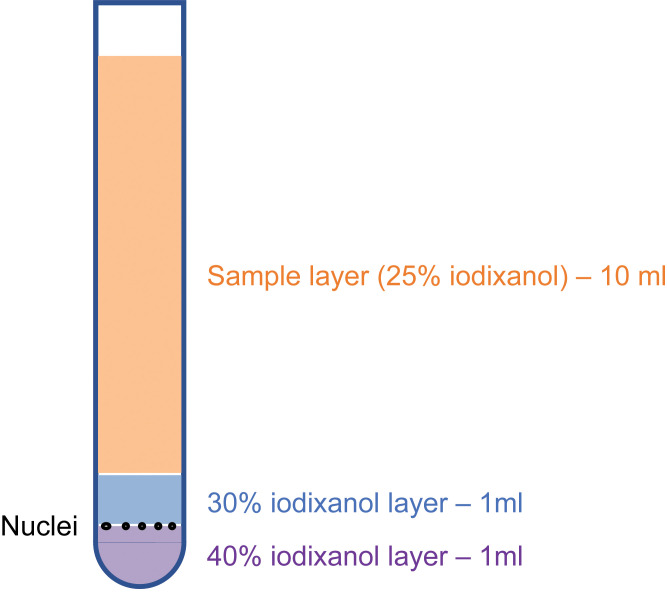
Prepare one ultracentrifuge tube for each DRG sample. In each tube, prepare the density gradient solution by layering 1 ml 40% iodixanol at the bottom and then slowly adding 1 ml 30% iodixanol on top of it, without disturbing the interface. Pre-cool tubes on ice.
Note: While preparing the density gradient solution, pipette just above the 40% layer against the wall, allowing it to form two distinct layers. Try again if you do not see two distinct layers after sitting for 1 min. Before placing the tube on ice, pre-indent the ice with another tube to avoid disrupting the density gradient layers.
-
Lysis
Transfer each DRG sample to a 5-ml centrifuge tube. Prepare one DRG sample at a time.
Add 4 ml Working Buffer HB to the 7-ml Dounce homogenizer and pre-cool the homogenizer on ice.
Add 1 ml ice-cold Working Buffer HB to the DRG sample and place the tube on ice for 15-30 s until the DRGs sink.
Homogenize the DRG sample with a Tissue-Tearor using the low setting (< level four out of seven levels for Tissue-Tearor 985370-04) for about 5 s. Visually inspect the sample and confirm that the tissue is homogenized.
Transfer the homogenate to the pre-cooled 7-ml Dounce homogenizer.
Further homogenize the tissue by douncing with a tight pestle for 10-12 strokes.
Add 320 μl 5% IGEPAL CA-630 and dounce with a tight pestle for five additional strokes. Dounce slowly to avoid bubbles.
Filter the homogenate through a 40-μm cell strainer into a 50-ml conical tube.
-
Add 5 ml recently vortexed Working Solution to the lysed sample to reach a final volume of 10 ml. Gently pipette up and down to mix the solution.
Note: The iodixanol concentration in the lysed sample is 25%.
-
Place the 50-ml conical tube containing 10 ml lysed sample on ice. Repeat Steps B1-B9 for each additional DRG sample.
Note: Lysed samples can be placed on ice for as long as 30 min before proceeding to Procedure C. It is important to remove undissociated myelin and fully clean the Tissue-Tearor and 7-ml Dounce homogenizer by rinsing with Stock Buffer HB several times between samples.
-
Gradient formation and centrifugation
Using a 1-ml pipette tip, slowly pipette each of the lysed samples onto the 30% iodixanol layer of the density gradient solution prepared in Step A3.
Balance the weight of the samples with nuclease-free water prior to centrifugation. Seal the top of each ultracentrifuge tube with parafilm.
Ultracentrifuge samples at 10,000 × g (equivalent to 7,600 rpm with SW 41 Ti rotor) for 18 min at 4°C.
After ultracentrifugation, white particles that contain the desired nuclei may be visible at the interface between 30% and 40% iodixanol if there is sufficient starting material (Figure 1).
Discard ~10 ml of the top layer to facilitate access to the interface between 30% and 40% iodixanol.
Place a 1-ml pipette tip at the interface between the 30% and 40% iodixanol layers. While moving the pipette tip around the layer, slowly aspirate 400 μl nuclei sample just above the 40% iodixanol layer and transfer it to a new 1.5-ml centrifuge.
-
Dilution and resuspension
Note: This step may vary based on the downstream analyses. Here, we demonstrate the sample preparation for inDrops single-cell RNA sequencing (Zilioniset et al., 2017) and 10× Genomics assays such as the Single Cell Gene Expression Assay and Single Cell Assay for Transposase-accessible Chromatin (ATAC). As an optional step, fluorescence-activated cell sorting can be used to further remove cellular debris and/or purify genetically labeled cell populations.
-
inDrops
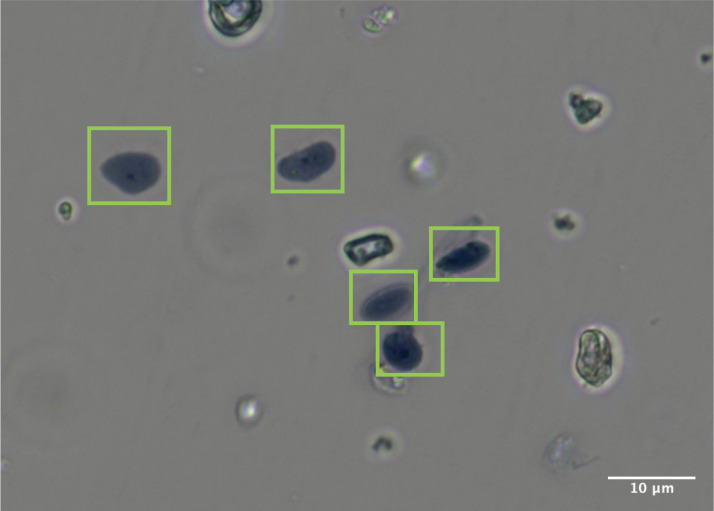
Take 10 μl nuclei sample, stain it with Trypan Blue, and count nuclei using a hemocytometer (Figure 2).
Dilute nuclei with 30% iodixanol to 80,000 nuclei/ml.
Proceed to inDrops.
Note: Some DRG nuclei have elongated shapes in this preparation.
-
10× Genomics Single Cell assays
-
Single Cell Gene Expression
To dilute the iodixanol, add 4 ml PBS with 0.05% BSA to the nuclei sample. Gently pipette up and down to mix the solution.
Centrifuge at 500 × g for 5 min. Remove supernatant.
Centrifuge at 500 × g for 2 min. Remove excess supernatant.
Resuspend the pellet in 45.5 μl PBS with 0.05% BSA.
Take 2 μl sample, stain it with Trypan Blue, and count nuclei using a hemocytometer (Figure 2).
Calculate the input volume of nuclei sample based on the Cell Suspension Volume Calculator Table in the 10× Genomics Single Cell Gene Expression User Guide.
Proceed with the 10× Genomics Single Cell Gene Expression Protocol.
-
Single Cell ATAC
-
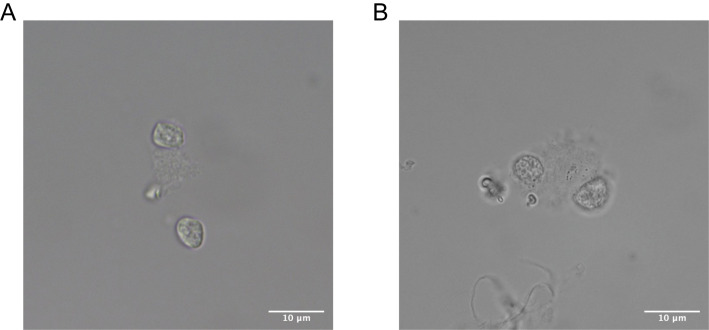
Take 10 μl sample and visualize the nuclear membrane at 60× magnification to ensure no blebbing (Figure 3).
Note: Samples with blebbing nuclei are not recommended for use.
To dilute the iodixanol, add 4 ml PBS with 0.05% BSA to the nuclei sample. Gently pipette up and down to mix the solution.
Centrifuge at 500 × g for 5 min. Remove supernatant.
Centrifuge at 500 × g for 2 min. Remove excess supernatant.
Resuspend the pellet in 7 μl 1× nuclei buffer.
Take 2 μl sample, stain it with Trypan Blue, and count nuclei using a hemocytometer (Figure 2).
Dilute the sample to the desired loading concentration based on the Nuclei Concentration Guidelines in the 10× Genomics Single Cell ATAC User Guide.
Proceed with the 10× Genomics Single Cell ATAC Protocol.
-
-
-
Figure 1. Illustration of iodixanol layers and nuclei positions after ultracentrifugation.
Figure 2. DRG nuclei sample stained with 0.2% Trypan Blue.
Image was taken at 60× magnification. The particles highlighted with green rectangles are intact nuclei.
Figure 3. Representative images of mouse DRG nuclei at 60× magnification.
(A). High-quality intact nuclei show well-resolved edges. (B). Low-quality nuclei show evidence of blebbing.
Recipes
-
Stock buffer HB
0.25 M sucrose
25 mM KCl
5 mM MgCl2
20 mM Tricine-KOH, pH 7.8
5 mg/ml actinomycin
Sterilize using a 0.2-μm filter and store at 4°C for up to 1 month
-
Diluent buffer
150 mM KCl
30 mM MgCl2
120 mM Tricine-KOH, pH 7.8
Sterilize using a 0.2-μm filter and store at 4°C for up to 1 month
Acknowledgments
W.R. is supported by the National Institute of Neurological Disorders and Stroke (NINDS) K08NS101064, the Burroughs Wellcome Fund, and the Migraine Research Foundation. C.J.W. is supported by NINDS R35NS105076-01, the DARPA Panacea program (HR0011-19-2-0022), and Adelson Medical Research Foundation.
Competing interest
L.Y. and W.R. have no competing interests. I.T. is an employee at Regeneron. C.J.W. is a founder of Nocion Therapeutics and QurAlis.
Citation
Readers should cite both the Bio-protocol article and the original research article where this protocol was used.
References
- 1.Chandran V., Coppola G., Nawabi H., Omura T., Versano R., Huebner E. A., Zhang A., Costigan M., Yekkirala A., Barrett L., Blesch A., Michaelevski I., Davis-Turak J., Gao F., Langfelder P., Horvath S., He Z., Benowitz L., Fainzilber M., Tuszynski M., Woolf C.J. and Geschwind D. H.(2016). A Systems-Level Analysis of the Peripheral Nerve Intrinsic Axonal Growth Program. Neuron 89: 956-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perkins J. R. Antunes-Martins A., Calvo M., Grist J., Rust W., Schmid R., Hildebrandt T., Kohl M., Orengo C., McMahon S. B. and Bennett D. L.(2014). A comparison of RNA-seq and exon arrays for whole genome transcription profiling of the L5 spinal nerve transection model of neuropathic pain in the rat. Mol Pain 10: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Usoskin D., Furlan A., Islam S., Abdo H., Lönnerberg P., Lou D., Hjerling-Leffler J., Haeggström J., Kharchenko O., Kharchenko P. V., Linnarsson S. and Ernfors P.(2015). Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci 18: 145-153. [DOI] [PubMed] [Google Scholar]
- 4.Zeisel A., Hochgerner H., Lönnerberg P., Johnsson A., Memic F., van der Zwan J., Häring M., Braun E., Borm L. E., La Manno G., Codeluppi S., Furlan A., Lee K., Skene N., Harris K. D., Hjerling-Leffler J., Arenas E., Ernfors P., Marklund U. and Linnarsson S.(2018). Molecular Architecture of the Mouse Nervous System. Cell 174: 999–1014..e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharma N., Flaherty K., Lezgiyeva K., Wagner D. E., Klein A.M. and Ginty D. D.(2020). The emergence of transcriptional identity in somatosensory neurons. Nature 577: 392-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen M. Q., Le Pichon C. E. and Ryba N.(2019). Stereotyped transcriptomic transformation of somatosensory neurons in response to injury. Elife 8: e49679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen M. Q., Wu Y., Bonilla L. S., von Buchholtz L. J. and Ryba N. J. P.(2017). Diversity amongst trigeminal neurons revealed by high throughput single cell sequencing. PLoS One 12: e0185543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Renthal W., Tochitsky I., Yang L., Cheng Y. C., Li E., Kawaguchi R., Geschwind D. H. and Woolf C. J.(2020). Transcriptional Reprogramming of Distinct Peripheral Sensory Neuron Subtypes after Axonal Injury. Neuron 108: 128–144..e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renthal W., Boxer L. D., Hrvatin S., Li E., Silberfeld A., Nagy M. A., Griffith E. C., Vierbuchen T. and Greenberg M. E.(2018). Characterization of human mosaic Rett syndrome brain tissue by single-nucleus RNA sequencing. Nat Neurosci 21: 1670-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mo A., Mukamel E. A., Davis F. P., Luo C., Henry G. L., Picard S., Urich M. A., Nery J.R., Sejnowski T. J., Lister R., Eddy S. R., Ecker J. R. and Nathans J.(2015). Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86: 1369-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slyper M., Porter C. B. M., Ashenberg O., Waldman J., Drokhlyansky E., Wakiro I. and Smillie C. et al.(2020). A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med 26: 792-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Habib N., Avraham-Davidi I., Basu A., Burks T., Shekhar K., Hofree M., Choudhury S. R., Aguet F., Gelfand E., Ardlie K., Weitz D. A., Rozenblatt-Rosen O., Zhang F. and Regev A.(2017). Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14: 955-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Avraham O., Deng P. Y., Jones S., Kuruvilla R., Semenkovich C. F., Klyachko V. A. and Cavalli V.(2020). Satellite glial cells promote regenerative growth in sensory neurons. Nat Commun 11: 4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Y. T. and Chen J. C.(2018). Dorsal Root Ganglia Isolation and Primary Culture to Study Neurotransmitter Release. J Vis Exp 140: e57569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zilioniset R., Nainys J., Veres A., Savova V., Zemmour D., Klein A.M. and Mazutis L.(2017). Single-cell barcoding and sequencing using droplet microfluidics. Nat Protoc 12: 44-73. [DOI] [PubMed] [Google Scholar]