Abstract
SARS-CoV-2 virions are composed of structural proteins, but during virus infection, an additional 30 proteins could be expressed according to putative open reading frames (ORFs) of the viral genome. Some of these additional proteins modulate cellular processes through direct interactions, their truncations can affect disease pathogenesis and they can also serve as antigenic targets for more specific serology. In addition to structural proteins, the ORF1a/b polyprotein and accessory proteins can stimulate antibody responses during infection. Antibodies that target non-structural proteins can impact viral infection, through Fc mediated effector functions, through interactions during virus entry, fusion, replication and egress within infected cells. Characterization of the serological responses to additional proteins, provides a snapshot of the ‘antibody landscape’, which includes the antibody magnitude, antigenic specificity and informs the biological relevance of SARS-CoV-2 proteins.
Current Opinion in Virology 2021, 50:139–146
This review comes from a themed issue on Viral immunology
Edited by Matteo Iannacone and Antonio Bertoletti
For complete overview about the section, refer “Viral immunology”
Available online 20th August 2021
https://doi.org/10.1016/j.coviro.2021.08.006
1879-6257/© 2021 The Author(s). Published by Elsevier B.V. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
SARS-CoV-2 proteins for diverse functions
The COVID-19 pandemic has upturned the world and caused incalculable losses. Since the emergence of SARS-CoV-2 in December of 2019, scientists have scrambled to characterize the novel virus, in terms of its replication, disease pathogenesis and immune responses to develop targeted therapies, diagnostics and vaccines. Characterization of the antibody response to SARS-CoV-2 helps to reveal which antigens are expressed during infection and the dynamic relationship between antibody response magnitude and viral protein expression can be used to develop sensitive serological testing and insights of virus replication. Therefore, the specificity and magnitude of the antibody landscape generated during SARS-CoV-2 infection is the focus of this review.
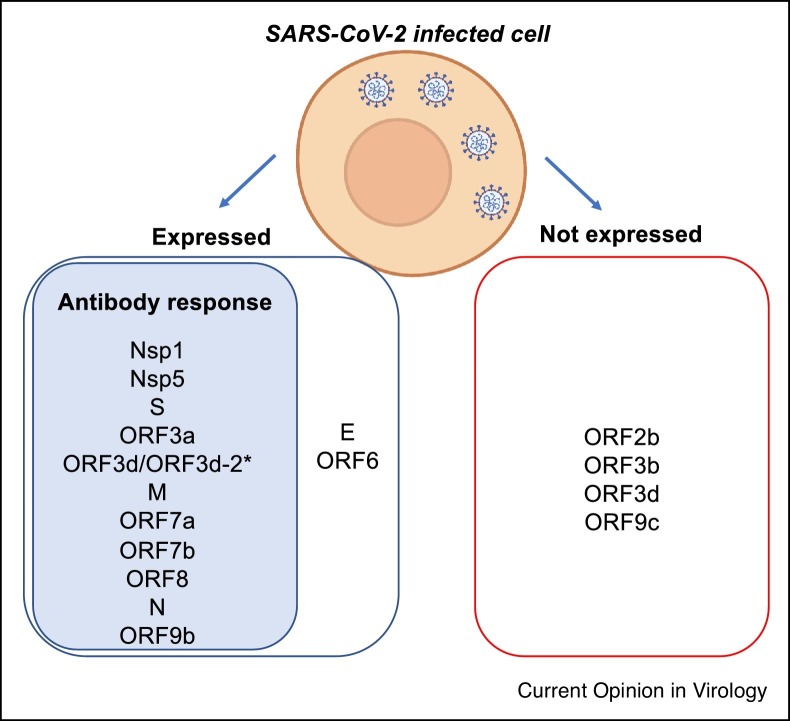
SARS-CoV-2 has at least four structural proteins: Spike (S), Envelope (E), Membrane (M) and Nucleocapsid (N). In addition, SARS-CoV-2 genome encodes for around 30 putative non-structural and accessory proteins [1] (Table 1 ). The open reading frame (ORF) 1a/b encodes for a large polyprotein that is proteolytically cleaved into at least 16 non-structural proteins (NSP1-16). The NSPs are mainly involved in the replication machinery of the virus (NSP3, NSP4, NSP5, NSP6, NSP7, NSP8, NSP9, NSP10, NSP12, NSP13, NSP14, NSP15, NSP16) but take also part in the evasion of the host immune response (NSP1, NSP15 notably) [2]. At the 3′ end of the SARS-CoV-2 genome are additional ORFs S, 3a, 3c, E, M, 6, 7a, 7b, 8, N and 9b which are translated and functional; ORFs 2b and 3d-2 which are translated but not functional and ORFs 3b, 3d, 9c and 10 which appear not to be translated to biologically meaningful levels (Table 1, Figure 1 ) [3•]. As the pandemic has progressed and alternate reading frame products have been defined, nomenclature of the ORF3 has been updated [3•], for clarity the ORF3b we previously referred to [4] is actually now referred to as ORF3d (which is not expressed) and also contains the isoform ORF3d-2, that is expressed, which we believe elicits an antibody response detected by the full ORF3d antigen [1] (Figure 1). Therefore, study of the antibody landscape can be linked to studies of protein function, interactomes and evolutionary rates to determine functional expression.
Table 1.
SARS-CoV-2 viral proteins their function in viral lifecycle, host response for immune evasion, antibody response and mutations in variants of concern
| Protein function | Antibody response | Mutations in VoCc | ||||||
|---|---|---|---|---|---|---|---|---|
| Viral protein | Viral | Host immune evasion | Alpha | Beta | Gamma | Delta | ||
| B1.1.7 | B1.351 | P1 | B.1.617.2 | |||||
| ORF1a/b | Nsp1 | N/A | IFN antagonist, promotes host mRNA degradation | Y | T1001I, A1708D, I2230T, SGF 3675-3677del | T85I, H295Y, K837N, T1456X, N1457X, K90R, S106del, G107del, F108del, D323del, P378X, A379X, D484X, G485X, G486X, C487X, T806X, Q196X | synT733C, synC2749T, S1188L, K1795Q, 11288-11296del, synC12778T, synC13860T, E5665D | |
| Nsp2 | Modulation of host cell survival | N/A | N/A | |||||
| Nsp3 | N-terminal Cleavage of viral polyprotein | Blocks host innate immune response (IFN antagonist, inhibition of NF-kB signalling) | N/A | P1469S, P822L, A1711V, A488S | ||||
| Nsp4 | Assembly of DMVs (with nsp3) | N/A | N/A | V167L, T492I, A446V | ||||
| Nsp5 | C-terminal Cleavage of viral polyprotein | N/A | Yb | |||||
| Nsp6 | Autophagosome induction from host ER, IFN antagonist | N/A | T77A, V149A,T181I | |||||
| Nsp7 | Forms complex with nsp8 may act as primase | N/A | N/A | |||||
| Nsp8 | Forms complex with nsp7 may act as primase | N/A | N/A | |||||
| Nsp9 | RNA binding protein | N/A | N/A | |||||
| Nsp10 | Stimulates exoribonuclease and methyltransferase activity of nsp14 and 16 | N/A | N/A | |||||
| Nsp11 | N/A | N/A | N/A | |||||
| Nsp12 | Replication and transcription of viral genome | N/A | N/A | P323L, G671S | ||||
| Nsp13 | Helicase essential for unwinding of nucleic strands for translation | IFN antagonist | N/A | P77L | ||||
| Nsp14 | Proofreading exoribonuclease for RNA replication | IFN antagonist | N/A | A394V | ||||
| Nsp15 | Uridylate-specific endoribonuclease at 3′ and 5′ end | IFN antagonist | N/A | |||||
| Nsp16 | Cap methylation of mRNA | Immune evasion | N/A | |||||
| Spike | Cell attachment by interaction with host hACE2 receptor | Neutralisation | Y | HV 69-70del, Y144del, N501Y, A570D, P681H, T716I, S982A, D1118H | D80A, D215G, L242del, A243del, L244del, K417N, E484K, N501Y, D614G, A701V | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, H655Y, T1027I | T19R, V70F, T95I, G142D, E156del, F157del, R158G, A222V, W258L, K417N, L452R, T478K, D614G, P681R, D950N, R158del | |
| Antibody-dependent enhancement of infectivity by antibodies binding NTD | ||||||||
| ORF2ba | N/A | N/A | N/A | |||||
| ORF3a | Homotetrameric ion channel formation that may modulate virus release | Fibrinogen upregulation in lung epithelial cells, induces apoptosis, IFN antagonist, block autophagy | Y | Q57H, W131C, S171L | S26L | |||
| ORF3ba | N/A | IFN antagonist | N/A | Deleted | ||||
| ORF3c | N/A | IFN antagonist | N/A | |||||
| ORF3da | N/A | IFN antagonist | Y | |||||
| Envelope | Viral morphogenesis and assembly | Induction of apoptosis and IL-1B overproduction | N | P71L | ||||
| Membrane | Component of viral envelope and morphogenesis | N/A | Y | T175M, I82T | ||||
| ORF6 | Disruption of cell nuclear import complex formation and retention of import factors at ER/golgi | IFN antagonist | N | |||||
| ORF7a | Tetherin activation to circumvent virus budding blockade | IFN antagonist | Y | V82A, T120I | ||||
| ORF7b | Structural component of virion | N/A | Y | T401I | ||||
| ORF8 | N/A | IFN antagonist, MHCI downregulation | Y | Q27stop, R52I, Y73C | I121L | E92K | ||
| Nucleocapsid | Packaging of vRNA into helical ribonucleocapsid, fundamental role in virion assembly by interacting with M protein | IFN antagonist | Y | D3L, S235F | A90S, T205I | P80R | D377Y, R203M, G215C, R203K | |
| ORF9b | N/A | N/A | Yb | |||||
| ORF9ca | N/A | N/A | N/A | |||||
| ORF10a | N/A | N/A | N | |||||
Not translated into functional/biologically relevant proteins.
Yes antibody response based on protein microarray Jiang et al. Nat Comms 2020.
Mutations were found using EpiCoV-GISAID CoVsurver as of August 10th 2021.
Figure 1.
SARS-CoV-2 viral proteins with confirmed expression and immunogenic antibody response. *As ORF3d and ORF3d-2 are in frame (see Jungeris et al. Nat Comms 2021), it is likely that the ORF3d antibody responses identified are against ORF3d-2.
Recently, a large scale interactome study of the proteins of SARS-CoV-2 infected human lung cell line revealed the systems level host proteins that directly interact with viral proteins [5••] and diverse roles of the accessory proteins and NSPs were found for immunomodulation, cellular trafficking and metabolism among many more cellular processes, whilst the S and N proteins had more limited functions within infected cells.
The most prominent immunomodulation effect studied to date has been type-I interferon antagonism which is mediated by NSP1, NSP6, NSP13, NSP14 and NSP15 of ORF1a/b polyprotein [6,7•], as well as ORF3d [8], ORF6 [9], ORF7a [10], ORF8 [9] and N [11] (Table 1). Moreover, ORF7a transmembrane protein [12] has been shown to inhibit Tetherin activation circumventing blockade of viral budding [13] and ORF3a has been shown to induce apoptosis in vitro through activation of the caspase-8 pathway [14] and to block the mechanism of autophagy [15]. The accessory proteins therefore mediate immune evasion through a multitude of pathways and appear to be essential in COVID-19 pathogenesis.
This is further supported by the positive selection of ORF1a/b, ORF3a and ORF8 genes which drives the evolution of SARS-CoV-2 [16]. Accessory proteins are also subject to mutations, to a lesser extent than S and N (Table 1), but have led to the emergence of variants which are highly transmissible and can modulate disease severity. Notably, ORF8 Q27stop mutation in Alpha VoC (B1.1.7 lineage) is associated with the surge in cases in the United Kingdom, and ORF8 mutations are found in other VoC at different positions (Beta: I121L, Gamma: E92K) that may modulate function (Table 1). ORF8 is a glycoprotein which can downregulate MHC-I [17] which may impact CD8+ T cell responses, and could play an important role in viral fitness and pathogenesis as its deletion led to reduced COVID-19 severity [18]. However, its involvement, location and function within the virus life cycle remain to be determined. Another accessory protein which has been prone to deletions is ORF3b (and ORF3c, 3d, 3d-2), prominently due to a Q57H mutation in ORF3a (in VoC Beta, B1.351 lineage, Table 1) which leads to its truncation and loss of antagonism function of ORF3b [19]. Important roles are emerging for the ORF1a/b polyprotein and accessory proteins in SARS-CoV-2, highlighting their essential role in viral pathogenesis and replication, hence the need for further investigation into their location and function within the virus and its life cycle.
Most vaccines in use against COVID-19 target the S protein to elicit neutralizing antibodies to block infection [20], as the S protein contains the receptor binding domain (RBD) which is critical for viral entry [21]. Whilst the N protein is the most abundant protein within infected cells and the virion, and is a target of routine serological tests like S. The sensitivity and specificity of S and N-based serological testing is not as effective in children [22•] and wanes with time considerably post infection, with only 36% of S antibody levels and 7% N antibody levels remaining at 1 year post infection [23]. Inactivated vaccines that contain structural proteins, such as CoronaVac which is widely distributed to some countries by China, and S-based vaccines (as an mRNA, recombinant protein, or as an adenovirus encoding for the S gene) form the majority of vaccines in use but have limited or no content of SARS-CoV-2 accessory and non-structural proteins, therefore alternate serological measures exclusive of S and N could detect current or past infection in vaccinated subjects. Furthermore, a similar issue to determine vaccine efficacy variants within the S have been undermined by reduced sensitivity of current serological tests. Protein domain-specific antigens may improve the sensitivity of N testing [24], but the same issue of waning and whole virion inactivated vaccines remains. Therefore, confirmatory serological testing with additional serological makers of SARS-CoV-2 infection will be useful when vaccine use is widespread, but waning immunity from vaccines may require further diagnosis of subsequent infection.
SARS-CoV-2 antibody landscape
Upon the initiation of an adaptive immune response against a pathogen, antibody targets are selected depending on their physical properties, their expression and the host’s immune repertoire diversity. For SARS-CoV-2, antibodies to the S protein that are neutralizing were largely studied early in the pandemic by several teams [25], but the spectrum of the antibody landscape was first described by our team in April 2020 [4,26]. Since then, others have also confirmed that COVID-19 induces robust antibody responses to epitopes throughout the SARS-CoV-2 proteome to define cross-reactive B cells and accurate serology [27•], identify unique epitopes by virscan [28], and the proteome response by microarray [29•]. Antibody landscapes can also refer to the viral strain diversity recognized by the humoral response, for example influenza-specific antibodies to hemagglutinin antigens over time with infection and vaccination [30, 31, 32, 33].
To characterize the SARS-CoV-2 antibody landscape we employed an unbiased and quantitative approach by luciferase immunoprecipitation system (LIPS) [34•] to assess the serological response to a panel of 15 SARS-CoV-2 antigens representing the structural (S, N, M and E, and S subunits), non-structural (representative NSP1) and accessory viral (ORF3a, 3b, 6, 7a, 7b, 8 and 10) proteins in SARS-CoV-2 infected adults and children, including asymptomatic cases and longitudinal samples compared to pre-pandemic negative controls. Antibody responses were detected in COVID-19 patients with various sensitivity levels allowing the establishment of the hierarchy of relative responses to SARS-CoV-2 proteins as N >> ORF8 >> ORF3b > ORF7a proteins. Furthermore, antibody responses were not mounted to the ORF10, ORF6, S2 (uncleaved Spike S2 domain) and E proteins in infected adults (Table 1, Figure 1), which questions the level of expression and immunogenicity of these proteins during infection, or their cross-reactivity to existing antibody responses from past common cold CoV infections.
Prior immunity did not impact the magnitude of ORF8 and ORF3b due to minimal cross-reactivity of these responses with HCoVs [4]. ORF8 is not encoded by other endemic common cold corona viruses, and only found in some sarbecoronaviruses (bat coronaviruses and SARS-CoV), where it has the smallest sequence homology of all genes [1]. Whilst only the NL63 common cold α-coronavirus, has an ORF3 protein which shares minimal homology with the SARS-CoV-2 derived ORF3a and ORF3b/c/d, making cross-reactivity unlikely. We found the combined use of ORF3b and ORF8 was a highly sensitive and specific serological marker of infection, as early as 5 days post infection and long term up to 6 months after infection.
This is consistent with the concept of ‘dating’ time estimates of infection based on the relative magnitude of antibodies to different antigens, as waning for different specificities can result in their unequal decline [35] and has been adopted by other fields such as malaria [36], Ebola [37], and influenza infection [38]. However, so far, combined antigen testing and the ‘dating’ of recent infection using S, RBD and N has wide ranges in estimates [23] which may undermine its utility without incorporation of additional more specific antigens like ORF8 and ORF3b. Therefore, commercial protein production should progress from the limited available accessory antigens and structural S and N, to improve the research efforts and tools widely available on the role of alternate targets of SARS-CoV-2.
The antibody landscape was markedly different in SARS-CoV-2 infected children compared to infected adults [39]. The antibody landscape in children was lower in magnitude toward S and N proteins which may undermine current serological tests in their diagnosis, whereas infected children had elevated responses to E and ORF6 proteins compared to infected adults. Furthermore, there was an increased proportion of the landscape contributed to by accessory proteins in infected children. Overall, these results suggested a different turnover of viral proteins or a different virus life cycle and pathogenesis within infected children and may account for the mild symptomatology of their disease. There was no difference between symptomatic (mild cases) and asymptomatic infections for their landscapes irrespective of age. However rare severe pediatric cases like MIS-C were not included in the study but have been shown by others to make a reduced antibody breadth for S and N antigens [22•].
Other reports also highlight the importance of antibody responses to non-structural and accessory proteins in SARS-CoV-2 patients. Jiang et al. constructed a SARS-CoV-2 proteome microarray containing 18 SARS-CoV-2 proteins and applied it to the characterization of the IgG and IgM antibodies responses in the sera from 29 convalescent patients [29•]. Besides responses to N and S1 proteins, patients mounted significant antibody responses to ORF9b and NSP5. The panel of this microarray protein differs from our LIPS protein panel, as it included 10 NSPs (versus one in the LIPS panel) but only 3 accessory ORFs (versus 7 in the LIPS panel), and therefore the results of both studies are complementary. Yet, whilst patients mounted a substantial response to ORF9b and NSP5, these antibodies were only detected in 44.8% and 10.3% of the patients, showing that they are not suitable antigenic targets for diagnostic purposes, to the contrary of ORF8 and ORF3b that showed high diagnostic performances. Across the antigenic targets of SARS-CoV-2, others report that besides S and N, the M structural protein also represent a major antibody target [27•]. Using a peptide microarray mapping Heffron et al., show that one epitope of the M protein achieved accurate diagnosis and could represent an interesting serology tool complementary to current serological tests.
Immune function of antibodies to alternate targets
Apart from their role for serology diagnostic, antibodies to alternate targets could play a role at diverse levels of the infection. In addition to the broad spectrum of antigenic targets, antibody class (IgG, IgA, IgM) brings another level of diversity of the antibody landscape as it impacts the antibody half-life and tissue distribution (e.g. presence of antibodies at mucosal entry sites or in the systemic circulation), along with effector functions for immune modulation.
Whilst T cells are well characterized to recognise both structural and non-structural/accessory proteins due to MHC presentation during infection, and have a large proportion of SARS-CoV-2 epitopes to non-structural proteins [40,41], the magnitude and function of antibodies towards non-structural non-strcuctural proteins is less understood. Non-structural and accessory proteins are likely exposed to the immune system for immune priming during lysis of infected cells after virus replication, leading to rupture characteristic of the cytopathic effect, either by necrosis, apoptosis or pyroptosis, and denuding of the epithelium during virus infection. In influenza A virus infection, evidence of cell surface expression of some viral antigens, such as Nucleoprotein has been reported [42], similarly some SARS-CoV-2 accessory proteins could be incorporated in virions and presented on the cell surface.
In SARS-CoV-2, ORF8 responses over time significantly correlated with S antibodies in longitudinal samples [4], suggesting the possibility of surface expression, secretion or incorporation into virus particles of ORF8. The recent demonstration of the interaction of ORF8 to the IL17RA extra-cellular receptor also supports the possible secretion of ORF8 [43,44]. Long term antigen retention of the N protein in gut biopsies months after recovery from infection may be pivotal in the retention and maturation of the memory B cell response, and in the high levels and persistence of N antibodies [45••]. The longevity of other viral antigens is not currently known as only N-specific antigen detection has been determined.
The B cell response [46•,47] to other viral antigens including, S, N and ORF8, shows that memory B cells are enriched following infection with greater levels of somatic hypermutation (SHM) of all specificities tested revealing a maturation of these cells and the initiation of a germinal center reaction [46•]. However, this is not evident during acute infection [48], where there is a presence of highly polyclonal B cell populations that have class switched in some cases, but with little or no SHM [49,50] and S-specific memory B cells with SHM appear progressively at later timepoints of infection [51]. Moreover in severe cases a significant proportion of S-specific B cells were derived from HCoV cross-reactivity [46•] or new de novo responses were made [52]. Similarly, memory B cells to N and ORF8 showed a pronounced maturation over time, however monoclonal antibodies to these internal targets do not confer protection in vivo in mice [46•].
As antibodies specific for accessory proteins are non-neutralizing and do not directly block infection through their Fab, they may mediate other functions either by blocking the virus replication or the virus interaction with host proteins, or by engaging their Fc with receptors on immune cells. Immune effector functions may include Antibody-dependent cellular cytotoxicity (ADCC), Antibody-dependent complement deposition (ADCD) and Antibody-dependent cellular phagocytosis (ADCP) or antibody-dependent enhancement (ADE). In SARS-CoV-2 infection, systems serology studies have recently suggested that qualitative changes to antibody Fc were induced, though the antigenic targets studied were limited to S and N [53]. N-specific effector functions imbalanced with S-specific effector functions have coincided with fatal COVID-19 [54], whereby S-specific ADCP and ADCD responses were found in survivors, whilst N-specific ADCD was increased in fatal cases.
Non-neutralizing antibodies may also inadvertently mediate antibody-dependent enhancement (ADE), as shown in severe patients in which divalent antibodies binding to S-NTD allowed for a conformational change of the spike protein resulting in enhanced infectivity [55]. This could possibly also occur for other S-specific antibodies outside of the RBD or that bind with a lower affinity, such as the variants of concern Gamma (P.1) and Alpha (B.1.351) which have the S-N501Y mutation leading to incomplete neutralization [56]. However, although ADE did occur leading to enhanced virus infection [55,57], replication is not apparent in primary macrophages [58], however modulation of the myeloid compartment is associated with disease outcomes [59] and immune activation [60].
Antibodies that target non-neutralising antigens may also mediate their effect by directly interfering with virus replication by targeting the Replicase machinery formed by the NSPs [2] (such as NSP12 which is responsible for replication and transcription of viral genome [61]), viral entry at fusion (such as S2′ specific antibodies), virus budding (such as ORF7a) [2]. Therefore, antibody specificity and effector functions leading to immune cell recruitment can drive clinical outcomes. However, the antibody effector functions of non-structural and accessory proteins remain uncharacterized currently and warrants further studies, especially as variants of concern, such as Beta (B1.1.7), Alpha (B1.351), Gamma (P.1) and Delta (B1.617) have alterations within these key proteins (Table 1).
The utility of antibodies to non-neutralising and internal proteins is gaining traction with a growing body of work for other virus infections as diagnosis, treatment and vaccines. Antibody landscapes can be used to detect spillover infection [62], whereby viruses that have limited protein tools, such as bat orthoreoviruses, can have their entire genomes cloned, expressed and screened for antibody binding in population serosurveillance. Unbiased and quantitative approaches, like LIPS [34•], microarray [29•], virscan [28] or cell-based ELISA [25,63] for serosurveillance will be useful tools in the post-COVID-19 future to highlight and identify key immunogenic targets of novel viruses.
Conflict of interest statement
A Hachim, N Kavian, and SA Valkenburg have filed an IDF (US 63/016,898) for the use of ORF8 and ORF3b as diagnostics of SARS-CoV-2 infection.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This study was partly supported by the Theme based Research Grants Scheme (T11-712/19-N), Health and Medical Research Fund (HMRF COVID-190115 and COVID-190126). Figure 1 was partly made with Biorender.
References
- 1.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., et al. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.V’Kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Jungreis I., Sealfon R., Kellis M. SARS-CoV-2 gene content and COVID-19 mutation impact by comparing 44 Sarbecovirus genomes. Nat Commun. 2021;12 doi: 10.1038/s41467-021-22905-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper assessed genetic stability of SARS-CoV-2 and related viruses to determine adaptive changes to determine epitopes and expressed proteins.
- 4.Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., et al. ORF8 and ORF3b antibodies are accurate serological markers of early and late SARS-CoV-2 infection. Nat Immunol. 2020;21:1293–1301. doi: 10.1038/s41590-020-0773-7. [DOI] [PubMed] [Google Scholar]
- 5••.Stukalov A., Girault V., Grass V., Karayel O., Bergant V., Urban C., et al. Multilevel proteomics reveals host perturbations by SARS-CoV-2 and SARS-CoV. Nature. 2021;594:246–252. doi: 10.1038/s41586-021-03493-4. [DOI] [PubMed] [Google Scholar]; This paper defines the transcriptome, proteome, ubiquitinome and phosphoproteome of SARS-CoV-2 in an in vitro cell model to define the comprehensive cellular networks and targets for rational drug design.
- 6.Xia H., Shi P.Y. Antagonism of type I interferon by severe acute respiratory syndrome coronavirus 2. J Interferon Cytokine Res. 2020;40:543–548. doi: 10.1089/jir.2020.0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Yuen C.K., Lam J.Y., Wong W.M., Mak L.F., Wang X., Chu H., et al. SARS-CoV-2 nsp13, nsp14, nsp15 and orf6 function as potent interferon antagonists. Emerg Microbes Infect. 2020;9:1418–1428. doi: 10.1080/22221751.2020.1780953. [DOI] [PMC free article] [PubMed] [Google Scholar]; Interferon antagonism is a major viral evasion mechanism of SARS-CoV-2 and the entire proteome of SARS-CoV-2 is screened in this study to define the Nsps and ORF6 as IFN antagonists.
- 8.Konno Y., Kimura I., Uriu K., Fukushi M., Irie T., Koyanagi Y., et al. SARS-CoV-2 ORF3b is a potent interferon antagonist whose activity is increased by a naturally occurring elongation variant. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li J.Y., Liao C.H., Wang Q., Tan Y.J., Luo R., Qiu Y., et al. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020;286 doi: 10.1016/j.virusres.2020.198074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cao Z., Xia H., Rajsbaum R., Xia X., Wang H., Shi P.Y. Ubiquitination of SARS-CoV-2 ORF7a promotes antagonism of interferon response. Cell Mol Immunol. 2021;18:746–748. doi: 10.1038/s41423-020-00603-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mu J., Fang Y., Yang Q., Shu T., Wang A., Huang M., et al. SARS-CoV-2 N protein antagonizes type I interferon signaling by suppressing phosphorylation and nuclear translocation of STAT1 and STAT2. Cell Discov. 2020;6 doi: 10.1038/s41421-020-00208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arya R., Kumari S., Pandey B., Mistry H., Bihani S.C., Das A., et al. Structural insights into SARS-CoV-2 proteins. J Mol Biol. 2021;433 doi: 10.1016/j.jmb.2020.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taylor J.K., Coleman C.M., Postel S., Sisk J.M., Bernbaum J.G., Venkataraman T., et al. Severe acute respiratory syndrome coronavirus ORF7a inhibits bone marrow stromal antigen 2 virion tethering through a novel mechanism of glycosylation interference. J Virol. 2015;89:11820–11833. doi: 10.1128/JVI.02274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ren Y., Shu T., Wu D., Mu J., Wang C., Huang M., et al. The ORF3a protein of SARS-CoV-2 induces apoptosis in cells. Cell Mol Immunol. 2020;17:881–883. doi: 10.1038/s41423-020-0485-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miao G., Zhao H., Li Y., Ji M., Chen Y., Shi Y., et al. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev Cell. 2021;56:427–442.e5. doi: 10.1016/j.devcel.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Velazquez-Salinas L., Zarate S., Eberl S., Gladue D.P., Novella I., Borca M.V. Positive selection of ORF1ab, ORF3a, and ORF8 genes drives the early evolutionary trends of SARS-CoV-2 during the 2020 COVID-19 pandemic. Front Microbiol. 2020;11 doi: 10.3389/fmicb.2020.550674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y., Chen Y., Li Y., Huang F., Luo B., Yuan Y., et al. The ORF8 protein of SARS-CoV-2 mediates immune evasion through down-regulating MHC-Iota. Proc Natl Acad Sci U S A. 2021;118 doi: 10.1073/pnas.2024202118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Young B.E., Fong S.W., Chan Y.H., Mak T.M., Ang L.W., Anderson D.E., et al. Effects of a major deletion in the SARS-CoV-2 genome on the severity of infection and the inflammatory response: an observational cohort study. Lancet. 2020;396:603–611. doi: 10.1016/S0140-6736(20)31757-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam J.Y., Yuen C.K., Ip J.D., Wong W.M., To K.K., Yuen K.Y., et al. Loss of orf3b in the circulating SARS-CoV-2 strains. Emerg Microbes Infect. 2020;9:2685–2696. doi: 10.1080/22221751.2020.1852892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections-more than just the common cold. JAMA. 2020;323:707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 21.Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., et al. A highly conserved cryptic epitope in the receptor-binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22•.Weisberg S.P., Connors T.J., Zhu Y., Baldwin M.R., Lin W.H., Wontakal S., et al. Distinct antibody responses to SARS-CoV-2 in children and adults across the COVID-19 clinical spectrum. Nat Immunol. 2021;22:25–31. doi: 10.1038/s41590-020-00826-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; MIS-C in children is an extremely rare and severe by product following SARS-CoV-2 infection. The S and N IgG and IgM response is compared in this study across the clinical spectrum. Children had reduced responses compared to adults regardless of MIS-C.
- 23.Pelleau S., Woudenberg T., Rosado J., Donnadieu F., Garcia L., Obadia T., et al. Serological reconstruction of COVID-19 epidemics through analysis of antibody kinetics to SARS-CoV-2 proteins. medRxiv. 2021 doi: 10.1101/2021.03.04.21252532. [DOI] [Google Scholar]
- 24.Wu C., Qavi A.J., Hachim A., Kavian N., Cole A.R., Moyle A.B., et al. Characterization of SARS-CoV-2 N protein reveals multiple functional consequences of the C-terminal domain. iScience. 2021;24:102681. doi: 10.1016/j.isci.2021.102681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perera R.A., Mok C.K., Tsang O.T., Lv H., Ko R.L., Wu N.C., et al. Serological assays for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), March 2020. Euro Surveill. 2020;25 doi: 10.2807/1560-7917.ES.2020.25.16.2000421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hachim A., Kavian N., Cohen C.A., Chin A.W.H., Chu D.K.W., Mok C.K.P., et al. Beyond the Spike: identification of viral targets of the antibody responses to SARS-CoV-2 in COVID-19 patients. medRxiv. 2020 doi: 10.1101/2020.04.30.20085670. [DOI] [Google Scholar]
- 27•.Heffron A.S., McIlwain S.J., Amjadi M.F., Baker D.A., Khullar S., Armbrust T., et al. The landscape of antibody binding in SARS-CoV-2 infection. PLoS Biol. 2021;19:e3001265. doi: 10.1371/journal.pbio.3001265. [DOI] [PMC free article] [PubMed] [Google Scholar]; Using an ultra dense peptide microarray the antibody landscape following SARS-CoV-2 infection is used to define serological targets and B cell epitopes.
- 28.Shrock E., Fujimura E., Kula T., Timms R.T., Lee I.H., Leng Y., et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020;370 doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29•.Jiang H.W., Li Y., Zhang H.N., Wang W., Yang X., Qi H., et al. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat Commun. 2020;11 doi: 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; The antibody landscape following SARS-CoV-2 infection was assessed by proteome microarray and correlated with clinical biomarkers (LDH and lymphocyte percentage), unique targets were defined (ORF9b and NSP5) but at low sensitivity.
- 30.Dugan H.L., Guthmiller J.J., Arevalo P., Huang M., Chen Y.Q., Neu K.E., et al. Preexisting immunity shapes distinct antibody landscapes after influenza virus infection and vaccination in humans. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.abd3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jang H., Ross T.M. Hemagglutination Inhibition (HAI) antibody landscapes after vaccination with H7Nx virus like particles. PLoS One. 2021;16 doi: 10.1371/journal.pone.0246613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Z.N., Liu F., Gross F.L., Kim L., Ferdinands J., Carney P., et al. Antibody landscape analysis following influenza vaccination and natural infection in humans with a high-throughput multiplex influenza antibody detection assay. mBio. 2021;12 doi: 10.1128/mBio.02808-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fonville J.M., Wilks S.H., James S.L., Fox A., Ventresca M., Aban M., et al. Antibody landscapes after influenza virus infection or vaccination. Science. 2014;346:996–1000. doi: 10.1126/science.1256427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34•.Burbelo P.D., Ching K.H., Klimavicz C.M., Iadarola M.J. Antibody profiling by Luciferase Immunoprecipitation Systems (LIPS) J Vis Exp (JoVE) 2009;32 doi: 10.3791/1549. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper is a Methods paper that details the technique of the Luciferase Immunoprecipitation System that allows the discovery of new antigens.
- 35.Rosado J., Pelleau S., Cockram C., Merkling S.H., Nekkab N., Demeret C., et al. Multiplex assays for the identification of serological signatures of SARS-CoV-2 infection: an antibody-based diagnostic and machine learning study. Lancet Microbe. 2021;2:e60–e69. doi: 10.1016/S2666-5247(20)30197-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosado J., White M.T., Longley R.J., Lacerda M., Monteiro W., Brewster J., et al. Heterogeneity in response to serological exposure markers of recent Plasmodium vivax infections in contrasting epidemiological contexts. PLoS Negl Trop Dis. 2021;15 doi: 10.1371/journal.pntd.0009165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Halfmann P.J., Eisfeld A.J., Watanabe T., Maemura T., Yamashita M., Fukuyama S., et al. Serological analysis of Ebola virus survivors and close contacts in Sierra Leone: a cross-sectional study. PLoS Negl Trop Dis. 2019;13 doi: 10.1371/journal.pntd.0007654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson A.H., Mahic M., Savic M., Tunheim G., Hungnes O., Trogstad L., et al. Detection of anti-NS1 antibodies after pandemic influenza exposure: evaluation of a serological method for distinguishing H1N1pdm09 infected from vaccinated cases. Influenza Other Respir Viruses. 2020;14:294–301. doi: 10.1111/irv.12712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hachim A., Gu H., Kavian O., Kwan M.Y.W., Chan W.-H., Yau Y.S., et al. The SARS-CoV-2 antibody landscape is lower in magnitude for structural proteins, diversified for accessory proteins and stable long-term in children. medRxiv. 2021 doi: 10.1101/2021.01.03.21249180. [DOI] [Google Scholar]
- 40.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yewdell J.W., Frank E., Gerhard W. Expression of influenza A virus internal antigens on the surface of infected P815 cells. J Immunol. 1981;126:1814–1819. [PubMed] [Google Scholar]
- 43.Gordon D.E., Hiatt J., Bouhaddou M., Rezelj V.V., Ulferts S., Braberg H., et al. Comparative host-coronavirus protein interaction networks reveal pan-viral disease mechanisms. Science. 2020;370 doi: 10.1126/science.abe9403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin X., Fu B., Yin S., Li Z., Liu H., Zhang H., et al. ORF8 contributes to cytokine storm during SARS-CoV-2 infection by activating IL-17 pathway. iScience. 2021;24 doi: 10.1016/j.isci.2021.102293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45••.Gaebler C., Wang Z., Lorenzi J.C.C., et al. Evolution of antibody immunity to SARS-CoV-2. Nature. 2021;591:639–644. doi: 10.1038/s41586-021-03207-w. [DOI] [PMC free article] [PubMed] [Google Scholar]; Long term memory B cell stability is a key immune correlate for recall at re-infection, and was defined in this study as reassuringly stable due to long term antigen retention in tissue biopsies.
- 46•.Dugan H.L., Stamper C.T., Li L., Changrob S., Asby N.W., Halfmann P.J., et al. Profiling B cell immunodominance after SARS-CoV-2 infection reveals antibody evolution to non-neutralizing viral targets. Immunity. 2021;54:1290–1303.e7. doi: 10.1016/j.immuni.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper uses high through put B cell sequencing the define the receptor sequences and somatic hypermutation rates of protein-specific responses.
- 47.Guthmiller J.J., Stovicek O., Wang J., Changrob S., Li L., Halfmann P., et al. SARS-CoV-2 infection severity is linked to superior humoral immunity against the spike. mBio. 2021;12 doi: 10.1128/mBio.02940-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roltgen K., Boyd S.D. Antibody and B cell responses to SARS-CoV-2 infection and vaccination. Cell Host Microbe. 2021;29:1063–1075. doi: 10.1016/j.chom.2021.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nielsen S.C.A., Yang F., Jackson K.J.L., Hoh R.A., Roltgen K., Jean G.H., et al. Human B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Cell Host Microbe. 2020;28:516–525.e5. doi: 10.1016/j.chom.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuri-Cervantes L., Pampena M.B., Meng W., Rosenfeld A.M., Ittner C.A.G., Weisman A.R., et al. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abd7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakharkar M., Rappazzo C.G., Wieland-Alter W.F., Hsieh C.L., Wrapp D., Esterman E.S., et al. Prolonged evolution of the human B cell response to SARS-CoV-2 infection. Sci Immunol. 2021;6 doi: 10.1126/sciimmunol.abg6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim S.I., Noh J., Kim S., Choi Y., Yoo D.K., Lee Y., et al. Stereotypic neutralizing VH antibodies against SARS-CoV-2 spike protein receptor binding domain in patients with COVID-19 and healthy individuals. Sci Transl Med. 2021;13 doi: 10.1126/scitranslmed.abd6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Selva K.J., van de Sandt C.E., Lemke M.M., Lee C.Y., Shoffner S.K., Chua B.Y., et al. Systems serology detects functionally distinct coronavirus antibody features in children and elderly. Nat Commun. 2021;12 doi: 10.1038/s41467-021-22236-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., et al. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu Y., Soh W.T., Kishikawa J.-i, Hirose M., Nakayama E.E., Li S., et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell. 2021;184:3452–3466.e18. doi: 10.1016/j.cell.2021.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hoffmann M., Arora P., Gross R., Seidel A., Hornich B.F., Hahn A.S., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grant R.A., Morales-Nebreda L., Markov N.S., Swaminathan S., Querrey M., Guzman E.R., et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hui K.P.Y., Cheung M.C., Perera R., Ng K.C., Bui C.H.T., Ho J.C.W., et al. Tropism, replication competence, and innate immune responses of the coronavirus SARS-CoV-2 in human respiratory tract and conjunctiva: an analysis in ex-vivo and in-vitro cultures. Lancet Respir Med. 2020;8:687–695. doi: 10.1016/S2213-2600(20)30193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schulte-Schrepping J., Reusch N., Paclik D., Bassler K., Schlickeiser S., Zhang B., et al. Severe COVID-19 is marked by a dysregulated myeloid cell compartment. Cell. 2020;182:1419–1440.e23. doi: 10.1016/j.cell.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ferreira A.C., Soares V.C., de Azevedo-Quintanilha I.G., Dias S., Fintelman-Rodrigues N., Sacramento C.Q., et al. SARS-CoV-2 engages inflammasome and pyroptosis in human primary monocytes. Cell Death Discov. 2021;7 doi: 10.1038/s41420-021-00428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kirchdoerfer R.N., Ward A.B. Structure of the SARS-CoV nsp12 polymerase bound to nsp7 and nsp8 co-factors. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10280-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Uehara A., Tan C.W., Mani S., Chua K.B., Leo Y.S., Anderson D.E., et al. Serological evidence of human infection by bat orthoreovirus in Singapore. J Med Virol. 2019;91:707–710. doi: 10.1002/jmv.25355. [DOI] [PubMed] [Google Scholar]
- 63.Chen Y.Q., Wohlbold T.J., Zheng N.Y., Huang M., Huang Y., Neu K.E., et al. Influenza infection in humans induces broadly cross-reactive and protective neuraminidase-reactive antibodies. Cell. 2018;173:417–429.e10. doi: 10.1016/j.cell.2018.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]