Abstract
The association between the c.521T>C variant allele in SLCO1B1 (rs4149056) and simvastatin-induced myotoxicity was discovered over a decade ago; however, whether this relationship represents a class effect is still not fully known. The aim of this study was to investigate the relationship between rs4149056 genotype and statin-induced myotoxicity in patients taking atorvastatin and lovastatin. Study participants were from the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort. A total of 233 statin-induced myopathy + rhabdomyolysis cases met the criteria for inclusion and were matched to 2,342 controls. To validate the drug response phenotype, we replicated the previously-established association between rs4149056 genotype and simvastatin-induced myotoxicity. In particular, compared to homozygous T allele carriers, there was a significantly increased risk of simvastatin-induced myopathy + rhabdomyolysis in homozygous carriers of the C allele (CC vs TT, OR 4.6, 95% CI 1.58-11.9, p=2x10−3). For lovastatin users, homozygous carriers of the C allele were also at increased risk of statin-induced myopathy + rhabdomyolysis (CC vs TT, OR 4.5, 95% CI 1.68-10.8, p=1x10−3). In atorvastatin users, homozygous carriers of the C allele were twice as likely to experience statin-induced myopathy, though this association did not achieve statistical significance (CC vs TT, OR 2.0, 95% CI 0.44-6.59, p=0.3). In summary, our findings suggest that the association of rs4149056 with simvastatin-related myotoxicity may also extend to lovastatin. More data is needed to determine the extent of the association in atorvastatin users. Altogether, these data expand the evidence-base for informing guidelines of pharmacogenetic-based statin prescribing practices.
Keywords: Precision medicine, Statin, Pharmacogenetic, SLCO1B1, Genetic polymorphism
Introduction
Over 39 million adult Americans are on statin therapy due to its importance in primary and secondary prevention of cardiovascular events.1 Nevertheless, statin use comes with a risk of myotoxic adverse effects. Although clinical trial data show a substantial nocebo effect for statin-induced myotoxic adverse effects, observational studies report rates to be as high as 29%.2,3 Even myalgia, on the milder spectrum of myotoxicity, can have a major impact on everyday activities of patients as well as statin adherence.4 Additionally, rhabdomyolysis can have severe clinical consequences such as myoglobin-induced acute renal failure and death.5,6 Mortality rates have been reported as high as 10% in patients who develop rhabdomyolysis, with rates even higher in individuals with renal impairment.7 Importantly, myotoxic adverse effects of statins are dose-related; increased systemic exposure confers higher risk.8 Thus, factors impacting statin pharmacokinetics may play a role in statin-induced myotoxicity.
The SLCO1B1 gene on chromosome 12 encodes organic anion transporting polypeptide 1B1 (OATP1B1), which is responsible for hepatic uptake of statins.9,10 Polymorphisms in SLCO1B1 have been associated with marked elevations in plasma concentrations of simvastatin.11,12 A genetic substudy of the Study of the Effectiveness of Additional Reductions in Cholesterol and Homocysteine (SEARCH) trial found that the c.521T>C substitution in SLCO1B1 (rs4149056) was associated with an increased risk of simvastatin-induced myopathy.13 Another genetic substudy from the Heart Protection Study (HPS) found a similar association of rs4149056 and myopathy in patients taking simvastatin.13 In response to the mounting evidence for the association between rs4149056 and simvastatin-induced myopathy, the Clinical Pharmacogenetics Implementation Consortium (CPIC) has recommended with grade A evidence that patients with one or two copies of the allele be initiated on either a lower dose of simvastatin or an alternative statin.14
Despite the strong association between rs4149056 and the risk of simvastatin-induced myopathy, the data are less clear for other commonly prescribed statins.14 In particular, there remains conflicting evidence regarding the association between rs4149056 and atorvastatin-induced myotoxicity.15,16 In addition, no study has examined the association between rs4149056 and statin-induced myotoxicity in individuals taking lovastatin. Therefore, the objective of this study was to investigate the association between rs4149056 and statin-related myotoxicity outcomes for atorvastatin and lovastatin.
Methods
Data Source
All participants in the study were members of Kaiser Permanente Northern California (KPNC), an integrated health care delivery system. Participants were selected from the Genetic Epidemiology Research on Adult Health and Aging (GERA) cohort, a resource of KPNC members that links electronic health record (EHR), genome-wide variant, and demographic survey data. In accordance with the Kaiser Foundation Research Institute Institutional Review Board, informed consent was given by study participants. International Classification of Diseases 9th Revision (ICD-9) diagnosis codes, creatine kinase (CK) levels, fasting glucose levels, hemoglobin A1c levels, age, and dispensed medications from 1996 to 2018 were extracted from the EHR. ICD-9 codes included rhabdomyolysis (ICD-9 code 728.88), diabetes (ICD-9 code 250.xx) and myocardial infarction (ICD-9 codes 410.xx). Demographic survey data included height, weight, sex (reported as biologic sex), and race/ethnicity (self-reported). Race/ethnicity was categorized as one of four groups: Hispanic/Latino, African American, East Asian, or White/European/Other. Prescription medications evaluated included atorvastatin, amprenavir, atazanavir, cyclosporine, and diabetes medications (as previously defined)33, darunavir, fenofibrate, fosamprenavir, gemfibrozil, indinavir, lopinavir, lovastatin, nelfinavir, ritonavir, saquinavir, simvastatin, and tipranavir.
Study Population
GERA cohort participants who had received at least one prescription of simvastatin, lovastatin, or atorvastatin were included for analysis. Statin-induced myopathy exhibits a dose-dependent effect with a stronger correlation for milligram strength (compared to statin low-density lipoprotein cholesterol [LDL-C] lowering potency) when all statin types are considered together.17,18 Therefore, in order to enrich our study sample with a high frequency of statin-induced myopathy cases, we limited our sample to users who received statin therapy at a total daily dose ≥40 mg, regardless of type or LDL-C lowering potency. Total daily dose was calculated by multiplying tablet strength and daily frequency (tablets per day).19
Phenotype
We defined a case of statin-induced myopathy as a study participant with ≥1 CK level >5x the upper limit of normal (ULN) within six months after receiving a statin prescription.20 The ULN for CK utilized was 336units/L for males and 176units/L for females.21 CK levels reported within 7 days of myocardial infarction diagnosis were excluded from analysis. We defined a case of statin-induced rhabdomyolysis as a study participant with ≥1 diagnosis of rhabdomyolysis (via ICD-9 code) within six months after receiving a statin prescription. The large sample size provided a unique opportunity to analyze this rare and serious adverse event in the context of pharmacogenetics. Although there is no standardized definition for rhabdomyolysis, most clinical diagnostic criteria include acute renal impairment and CK greater than 10x ULN.22,23 To encompass these diagnostic elements, we defined rhabdomyolysis using an ICD-9 code, which has been previously utilized in epidemiologic research to accurately identify cases of rhabdomyolysis.24 Cases with a history of multiple rhabdomyolysis ICD-9 codes and/or CK levels >5x had the earliest of these events considered as the outcome for analysis. Cases with a dispensing history of any interacting non-statin medication within one year prior to the outcome were excluded from analysis. These interacting medications included amprenavir, atazanavir, cyclosporine, darunavir, fenofibrate, gemfibrozil, indinavir, nelfinavir, ritonavir, saquinavir, and tipranavir.7,25 Cases were then categorized into statin groups (atorvastatin, lovastatin, or simvastatin) based on the most recently prescribed statin type in the EHR directly prior to the instance of statin-induced myopathy or rhabdomyolysis. This statin prescription was considered the index statin prescription for cases. The primary outcome was the composite of statin-induced myopathy and statin-induced rhabdomyolysis (termed statin-induced myopathy + rhabdomyolysis). We set the secondary outcome as statin-induced rhabdomyolysis alone, since prior studies had not investigated this phenotype as defined. Study participants without a history of a primary or secondary outcome were identified as potential controls. From this potential control group, nine to eleven controls were identified for each case, matched based on age (within 5 years of case age), sex, statin type, and statin dose.26 Although controls had no history of the primary or secondary outcome throughout full follow-up, we considered the first statin dispensing record ≥40 mg daily dose as the index record for these participants. This is consistent with evidence suggesting that median time to onset for statin-induced muscle symptoms is 1 month after either initiation or upward dose titration of statin.27 Age, statin type, and statin dose for cases and controls were based on the index statin record.
Genotype
As previously described, DNA samples from study participants were genotyped on one of four Affymetrix Axiom arrays designed for individuals of East Asian (EAS), African American (AFR), Latino (LAT), and non-Hispanic white (EUR) race/ethnicity to maximize genome-wide coverage of common and less common variants.28 Study participants were categorized into one of three groups based on rs4149056 genotype: homozygous TT, heterozygous (TC) and homozygous CC.
Statistical Analyses
The primary analysis was determining the relationship between rs4149056 genotype and statin-induced myopathy + rhabdomyolysis using multivariate logistic regression. We first investigated the association between rs4149056 and simvastatin-induced myopathy + rhabdomyolysis, which served as a positive control for internal validation of our phenotype before conducting subsequent analyses in the other statin type users. Following analyses of each individual statin type, we also pre-specified to investigate the collective of all statins analyzed (atorvastatin, lovastatin, and simvastatin). Odds ratios (OR) were estimated along with 95% confidence intervals (CI) for the likelihood of developing statin-induced myopathy + rhabdomyolysis in relation to rs4149056 genotype. Pre-specified covariates for statin-induced myopathy + rhabdomyolysis included self-reported race/ethnicity and genetic ancestry eigenvectors.29,30 These genetic ancestry eigenvectors were previously generated through principal component analysis within each self-reported race/ethnicity (EUR, EAS, AFR, and LAT), and the first six eigenvectors were utilized as covariates.28 Among these covariates, only self-reported race was significantly associated with the outcome and included in the regression model.
We conducted two sensitivity analyses that incorporated more stringent matching criteria. Sensitivity analysis 1 matched on self-reported race/ethnicity in addition to criteria from the primary analysis (age, sex, statin type, and statin dose). 29,30 Sensitivity analysis 2 matched on obesity status and diabetes status at the time of index statin dispensing in addition to criteria from the primary analysis.31,32 Diabetes was defined using ICD-9 diagnosis codes, laboratory values, and prescription medications, as previously described.33 Study participants with a body mass index ≥30kg/m2 were considered obese.34
We conducted a secondary analysis, which was identical to the primary analysis except we used the secondary outcome of statin-induced rhabdomyolysis alone.
Power calculations were conducted a priori using QUANTO.35 Assuming a conservative effect size of 3.0 per C allele, minor allele frequency of 0.15, log additive mode of inheritance, and a conservative population risk of 5% for statin-induced myopathy based on prior studies, we calculated that at least 21 cases would be needed to have greater than 80% power to determine an association between rs4149056 and statin-induced myopathy.36
Minor allele (C allele) frequencies between cases and controls were also compared using χ2 test, which has been performed in prior pharmacogenetic studies.16,37 All analyses were done in R (verson.3.4.3, R Foundation for Statistical Computing) unless otherwise noted.38 P-values <0.05 were considered statistically significant.
Results
Primary Analysis
We identified 252 potential cases for the primary outcome; one case was excluded due to recent myocardial infarction and a further 18 cases were excluded due to dispensing history of an interacting non-statin medication. Subsequently, 233 cases were included for analysis and matched to 2,342 controls. Of these cases, 82 (35%) were taking simvastatin, 66 (28%) were taking atorvastatin, and 85 (37%) were taking lovastatin as their index statin. The corresponding matched control group consisted of 748 simvastatin users, 693 atorvastatin users, and 901 lovastatin users. The median duration of total statin use in day’s supply dispensed was 1,040 (interquartile range: 360-2,070) for controls and 1,000 (interquartile range: 400-1,742) for cases. Baseline characteristics are listed in Table 1. The genotyping call rate for rs4149056 was 100%. There was no deviation from Hardy-Weinberg equilibrium for controls within race/ethnicity groups (p>0.05).
Table 1.
Demographics for cases and controls of statin-induced myopathy + rhabdomyolysis by statin group (N = 2575)
| All Statins a | Simvastatin | Atorvastatin | Lovastatin | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases(n=233) | Controls(n=2342) | Cases(n=82) | Controls(n=748) | Cases(n=66) | Controls(n=693) | Cases(n=85) | Controls(n=901) | ||
| Age, median (IQR), y a | 76.5 (67.9-82.9) | 75.7 (67.8-81.4) | 75.3 (66.2-81.7) | 73.3 (65.1-80.6) | 77.1 (69.8-82.9) | 76.8 (70.2-81.5) | 75.8 (68.6-81.4) | 76.0 (68.5-82.2) | |
| Race/ethnicity, No. (%) b | |||||||||
| AFR | 17 (7.3) | 77 (3.3) | 5 (6.1) | 21 (2.8) | 4 (6.0) | 31 (4.5) | 8 (9.4) | 25 (2.8) | |
| EAS | 20 (8.6) | 136 (5.8) | 6 (7.3) | 41 (5.5) | 10 (15.2) | 36 (5.2) | 4 (4.7) | 59 (6.5) | |
| EUR | 168 (72.1) | 1965 (83.9) | 63 (76.8) | 627 (83.8) | 42 (63.6) | 571 (82.4) | 63 (74.1) | 767 (85.1) | |
| LAT | 28 (12.0) | 164 (7.0) | 8 (9.8) | 59 (7.9) | 10 (15.2) | 55 (7.9) | 10 (11.8) | 50 (5.6) | |
| Sex, No. (%) | |||||||||
| Male | 125 (53.6) | 1241 (53.0) | 53 (64.6) | 471 (63.0) | 27 (40.9) | 291 (42.0) | 45 (52.9) | 479 (53.2) | |
| Female | 108 (46.4) | 1101 (47.0) | 29 (35.4) | 277 (37.0) | 39 (59.1) | 402 (58.0) | 40 (47.1) | 422 (46.8) | |
| Statin Dose, No. | |||||||||
| 40mg | 154 | 1583 | 39 | 361 | 33 | 354 | 82 | 868 | |
| 80mg | 79 | 759 | 43 | 387 | 33 | 339 | 3 | 33 | |
Abbreviations: AFR, African American; EAS, East Asian; EUR, European/Caucasian/Other; LAT, Latino/Hispanic
Statin type and age were based on the dispense date of the index statin prescription.
Race/ethnicity was self-reported.
Among the 82 cases and 748 controls for simvastatin, both the heterozygous genotype (TC vs TT, OR 1.8, 95% CI 1.08-2.91, p=2x10−2, Figure 1) and the homozygous CC genotype (CC vs TT, OR 4.6, 95% CI 1.58-11.9, p=2x10−3) were associated with increased risk for the primary outcome compared to homozygous TT. There was a significant difference in the C allele frequency between cases (25%) and controls (15%, p=5x10−4, Table 2).
Figure 1. Impact of SLCO1B1 rs4149056 genotype on risk of statin-induced myopathy and rhabdomyolysis in simvastatin, lovastatin, and atorvastatin users.
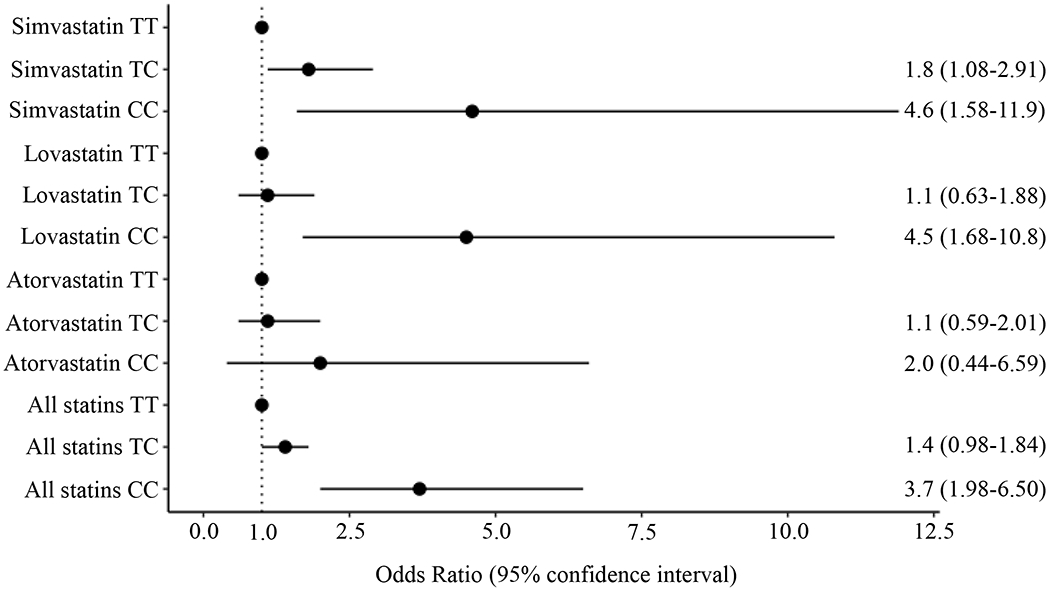
Odds ratios (ORs) and 95% confidence intervals for risk of statin-induced myopathy and rhabdomyolysis with rs4149056 genotype are shown by statin type. ORs for TC and CC genotype are compared to TT genotype. In the entire cohort of statin users, a significantly increased risk of statin-induced myopathy and rhabdomyolysis was observed in individuals with the CC genotype. In the subset of simvastatin and lovastatin users, CC genotype was associated with significantly increased risk of statin-induced myopathy and rhabdomyolysis. In addition, TC genotype conferred a significantly increased risk in simvastatin users. In contrast, for atorvastatin users, no statistically significant risk for statin-induced myopathy and rhabdomyolysis was identified.
Table 2.
Association between rs4149056 genotype and statin-induced myopathy + rhabdomyolysis by statin group
| Statin-type | Genotype | TT | TC | CC | MAF | χ2 p-valuea |
|---|---|---|---|---|---|---|
| All Statins | Cases | 153 | 64 | 16 | 0.21 | |
| Controls | 1722 | 569 | 51 | 0.14 | 1.80E-06 | |
| Simvastatin | Cases | 47 | 29 | 6 | 0.25 | |
| Controls | 537 | 195 | 16 | 0.15 | 4.55E-04 | |
| Lovastatin | Cases | 59 | 19 | 7 | 0.19 | |
| Controls | 671 | 210 | 20 | 0.14 | 8.32E-04 | |
| Atorvastatin | Cases | 47 | 16 | 3 | 0.17 | |
| Controls | 514 | 164 | 15 | 0.14 | 0.40 |
MAF, minor allele frequency
Differences in minor allele frequency between cases and controls for each statin group.
Analysis of the lovastatin subset (85 cases and 901 controls) showed a significantly increased risk for the primary outcome with the homozygous CC genotype (CC vs TT, OR 4.5, 95% CI 1.68-10.8, p=1x10−3). However, no association was found for the heterozygous genotype (TC vs TC, OR 1.1, 95% CI 0.63-1.88, p=0.7). The frequency of the C allele was significantly different between cases (19%) and controls (14%, p=8x10−4, Table 2).
Results from the atorvastatin subset, consisting of 66 cases and 693 controls, conferred no significant findings for the primary outcome in both the heterozygous (TC vs TT, OR 1.1, 95% CI 0.59-2.01, p=0.7) and the homozygous CC genotypes (CC vs TT, OR 2.0, 95% CI 0.44-6.59, p=0.3). There was no significant difference in the frequency of the C allele between cases (17%) and controls (14%, p=0.40, Table 2).
Across all statins analyzed, there was a significantly increased risk of statin-induced myopathy + rhabdomyolysis with the rs4149056 heterozygous genotype (TC vs TT, OR 1.4, 95% CI 1.02-1.92, p=0.03) and homozygous CC genotype (CC vs TT, OR 3.7, 95% CI 1.99-6.53, p=1x10−5; Figure 1) compared to homozygous TT. The frequency of the C allele was significantly different between cases (21%) and controls (14%, p=2x10−6, Table 2).
Results of the sensitivity analyses are presented in Table 3. Inclusion of matching criteria for race or obesity/diabetes status each yielded similar results to that of the primary analysis.
Table 3.
Odds ratios for statin-induced myopathy + rhabdomyolysis with rs4149056 genotype stratified by statin type using various matching criteria
| TC vs TT | CC vs TT | |||||
|---|---|---|---|---|---|---|
| Statin type | Analysis typea | Number of Cases/Controls | Odds ratio (95% confidence interval) | P-value | Odds ratio (95% confidence interval) | P-value |
| Simvastatin | Primary analysis | 82/748 | 1.78 (1.08-2.91) | 0.023 | 4.62 (1.58-11.9) | 0.003 |
| Sensitivity 1 | 82/229 | 1.82 (1.04-3.15) | 0.034 | 5.36 (1.47-21.7) | 0.012 | |
| Sensitivity 2 | 77/150 | 1.96 (1.06-3.62) | 0.031 | 5.09 (1.28-24.9) | 0.026 | |
| Lovastatin | Primary analysis | 85/901 | 1.11 (0.63-1.88) | 0.715 | 4.49 (1.68-10.8) | 0.001 |
| Sensitivity 1 | 85/251 | 1.09 (0.59-1.96) | 0.771 | 4.51 (1.39-15.7) | 0.013 | |
| Sensitivity 2 | 76/150 | 1.04 (0.52-2.03) | 0.904 | 5.02 (1.34-23.9) | 0.023 | |
| Atorvastatin | Primary analysis | 66/693 | 1.12 (0.59-2.01) | 0.716 | 1.99 (0.44-6.59) | 0.303 |
| Sensitivity 1 | 66/205 | 0.98 (0.49-1.84) | 0.940 | 2.38 (0.45-11.2) | 0.268 | |
| Sensitivity 2 | 61/121 | 1.25 (0.59-2.57) | 0.553 | 3.24 (0.52-25.3) | 0.206 | |
Controls for the primary analysis were matched based on age, sex, statin type, and statin dose. A series of sensitivity analyses were performed (Sensitivity 1 and Sensitivity 2) to determine the robustness of our findings. Sensitivity 1 matched based on age, sex, statin type, statin dose, and self-reported race/ethnicity. Sensitivity 2 matched based on age, sex, statin type, statin dose, obesity (body mass index ≥30kg/m2), and diabetes (a subset of participants with missing body mass index data were not included in Sensitivity 2).
Secondary Analysis
There were 193 cases of statin-induced rhabdomyolysis meeting the criteria for study inclusion and subsequently matched to 1,925 controls. Of the cases, 38 (20%) were taking atorvastatin, 78 (40%) were taking lovastatin, and 77 (40%) were taking simvastatin. The corresponding control group consisted of 407 atorvastatin users, 823 lovastatin users, and 695 simvastatin users. For all statins analyzed as a whole, the homozygous CC genotype conferred a significantly increased risk of statin-induced rhabdomyolysis (OR 3.7, 95% CI 1.97-6.75, p=2x10−5). There was no significantly increased risk associated with the heterozygous genotype (OR 1.3, 95% CI 0.94-1.89, p=0.1). Analysis by statin type yielded similar trends to that of the primary analysis (Figure 2, Table S1).
Figure 2. Impact of SLCO1B1 rs4149056 genotype on risk of statin-induced rhabdomyolysis in simvastatin, lovastatin, and atorvastatin users.
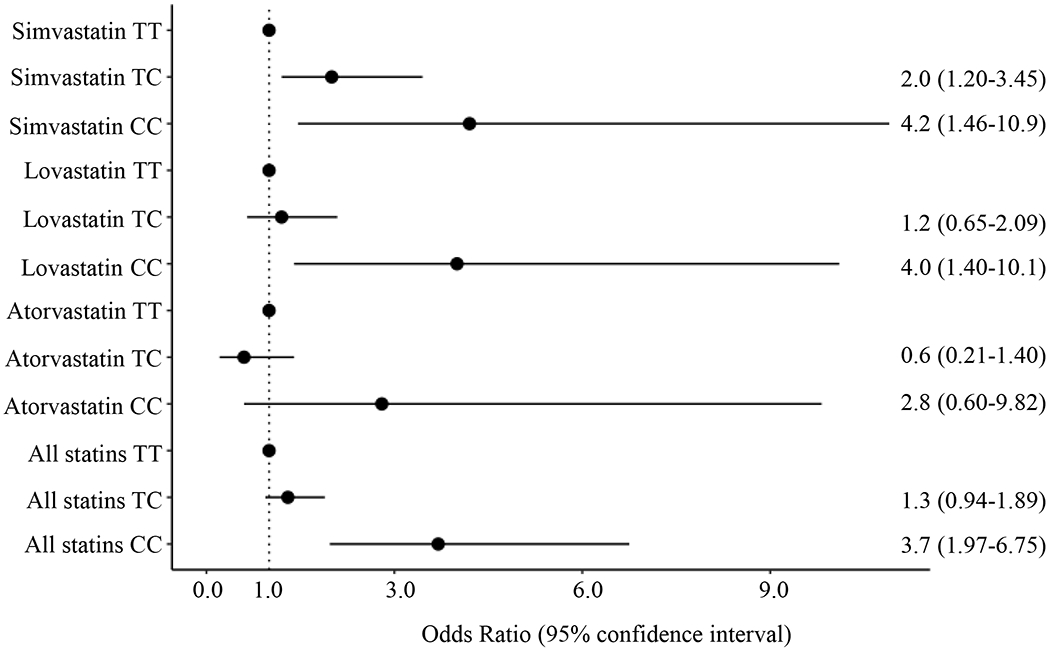
Odds ratios (ORs) and 95% confidence intervals for risk of statin-induced rhabdomyolysis with rs4149056 genotype are shown by statin type. ORs for TC and CC genotype are compared to TT genotype. In the entire cohort of statin users, homozygous recessive genotype conferred a significantly increased risk of statin-induced rhabdomyolysis. Analysis by statin type yielded similar trends to that of the primary analysis where CC genotype was associated with significantly increased risk of rhabdomyolysis in simvastatin and lovastatin users, but not in atorvastatin users.
Discussion
In this study, we sought to determine the relationship between rs4149056 genotype and the risk of statin-related myotoxicity outcomes for atorvastatin and lovastatin. We found that homozygous carriers of the C allele were at >4-fold increased risk of statin-induced myopathy + rhabdomyolysis among lovastatin users. This is the first study to investigate the association between this polymorphism and lovastatin-induced myotoxicity outcomes.
Based on substantial supporting evidence, the CPIC guidelines currently recommend that individuals with one or two copies of rs4149056 requiring statin therapy be initiated on low dose simvastatin (i.e., <40 mg daily dosing) or an alternative statin to reduce the risk of statin-induced myopathy.14 However, simvastatin is currently the only statin that has clearly outlined recommendations from CPIC. Thus, it is not fully known which alternative therapies are safe in carriers of the C allele at rs4149056.
The current findings in our subset of simvastatin users agree with the well-established association between simvastatin-induced myopathy and rs4149056 genotype.13–15 In particular, the ORs for statin-induced myopathy + rhabdomyolysis were 1.8 for heterozygous carriers of the C allele and 4.6 for homozygous carriers. These are lower than the ORs for statin-induced myopathy reported in the SEARCH study (4.5 and 16.9 for heterozygous and homozygous carriers of the C allele, respectively), but similar to the results from the Heart Protection Study Cohort (2.6 per C allele).13 Additionally, this is the first SLCO1B1 pharmacogenetic study of statin-related myotoxicity to use rhabdomyolysis as an endpoint; results mirrored statin-induced myopathy. Overall, our replication of the association between rs4149056 genotype and simvastatin-induced myopathy + rhabdomyolysis served as a positive control demonstrating the robustness of our study design before we applied this methodology to other statin types of interest.
To our knowledge, no previously published study has provided evidence of a clinical association between rs414056 and lovastatin-induced myopathy or rhabdomyolysis. Recently reported trends suggest that millions of lovastatin orders continue to be prescribed in the United States, thus clinicians could benefit from lovastatin pharmacogenetic prescribing guidance, especially for statin intolerant patients who may have to resort to this second-line statin option.39,40 We found that two copies of the C allele conferred a significantly increased risk for the primary outcome These data are in accord with pharmacokinetic analyses of single-dose lovastatin in which rs4149056 homozygous CC genotype yielded a nearly 3-fold higher lovastatin acid area under the plasma concentration time curve (AUC) compared to homozygous TT.41 Interestingly, we found in the current study that the increased risk of the primary outcome in CC genotype versus TT in lovastatin users was of a comparable magnitude to that of our simvastatin users. This is concordant with the similar chemical structure and pharmacokinetic characteristics between simvastatin and lovastatin.42,43
We identified no significant relationship between rs4149056 genotype and atorvastatin-induced myopathy + rhabdomyolysis. These findings contrast with the STRENGTH study, which identified a significantly increased risk of atorvastatin-induced myopathy in carriers of the C allele at rs4149056 (1.6 per allele).15 The broad definition of the primary endpoint in STRENGTH (a composite of drug discontinuation, myalgia, muscle cramping, and/or CK >3x ULN) may explain the differences in results compared to our study. However, our data parallel those of Brunham et al. (10 atorvastatin-induced myopathy cases) and Carr et al. (11 atorvastatin-induced myopathy cases), which also identified nonsignificant findings in atorvastatin users for risk of statin-induced myopathy despite observing positive results in simvastatin users.16,44 The varying degrees of myotoxicity induced by simvastatin, lovastatin, and atorvastatin in carriers of the C allele may be explained in part by differential selectivity among the statin types as substrates for hepatic uptake transporters. Within the SLCO family, OATP1B1 is the only hepatic transporter available for simvastatin and lovastatin. However, atorvastatin is a substrate for OATP1B1 along with other hepatic transporters of the SLCO family: OATP1A2, OATP1B3, and OATP2B1.45 This suggests that alternative pathways for hepatic uptake of atorvastatin could play a role in circumventing the markedly increased AUC and myotoxicity seen in simvastatin users carrying the C allele.13,46 In our data, there was a nonsignificant increased risk of statin-induced myopathy + rhabdomyolysis for the homozygous CC genotype (2.0 and 2.8 increased risk of atorvastatin-induced myopathy + rhabdomyolysis and rhabdomyolysis, respectively compared to homozygous TT), suggesting that rs4149056 genotype may have a weaker effect in atorvastatin users (compared to lovastatin and simvastatin users), but we were not powered to detect differences between statin types. Further studies with larger sample sizes (within a single cohort or as a meta-analysis of pooled studies) are needed to confirm this potential modest effect.
Our study has numerous strengths, including a relatively large sample size as well as a statin type (lovastatin) and myotoxicity endpoint (rhabdomyolysis alone) that has not been previously investigated. However, limitations must also be noted. First, this study could only account for a subset of all the potential statin-drug interactions that have the potential to impact results. However, interacting drugs of clinical significance (e.g. strong cytochrome P450 3A4 inhibitors) are readily detected by pharmacists in routine practice and are thus rarely co-dispensed with statins at the high doses we investigated. Second, there were fewer myopathy + rhabdomyolysis cases in atorvastatin users, which resulted in reduced power to detect significant associations compared to other statin types. This discrepancy may be explained, at least in part, by the fewer C allele carriers among the atorvastatin users. Indeed, there was a lower percentage of European descent participants receiving atorvastatin, an ancestry group with a higher C allele frequency relative to other populations.47 Nevertheless, with 66 total cases among atorvastatin users including 19 variant carriers, our study is among the largest to investigate rs4149056 genotype and atorvastatin-induced myotoxicity. Third, there is the potential that a proportion of our myopathy + rhabdomyolysis cases may be due to causes unrelated to statin therapy. However, our simvastatin results, which validate the well-established pharmacogenetic association with rs4149056, provide some confidence that the vast majority of our myotoxicity cases are truly statin-induced. Furthermore, our definition of rhabdomyolysis was based on a previously validated algorithm.24
In summary, our findings suggest that the association of rs4149056 with simvastatin-induced myotoxicity may also extend to lovastatin. More data is needed to determine the extent of the association in users of atorvastatin. Altogether, these novel data underscore the need for additional studies that evaluate the association between rs4149056 genotype and statin-related myotoxicity with the goal of more definitive pharmacogenetic-based statin prescribing guideline recommendations for atorvastatin, lovastatin, pitavastatin, pravastatin, and rosuvastatin.
Supplementary Material
Study Highlights .
What is the current knowledge on the topic?
Current literature suggests there is a strong association between the rs4149056 polymorphism in SLCO1B1 and simvastatin-induced myopathy. The Clinical Pharmacogenetics Implementation Consortium recommends that patients with the polymorphism be initiated on either a lower dose of simvastatin or an alternative statin.
What question did this study address?
Despite the strong association between rs4149056 and simvastatin-induced myopathy, there remains conflicting evidence with atorvastatin and no study with lovastatin users. The objective of this study was to investigate the association between rs4149056 and statin-related myotoxicity outcomes for atorvastatin and lovastatin.
What does this study add to our knowledge?
Our findings suggest that there is an association between rs4149056 and lovastatin-induced myopathy. We also explore the association between rs4149056 and statin-induced rhabdomyolysis, an endpoint that has not been previously studied in this setting.
How might this change clinical pharmacology or translational science?
Current national guidelines for pharmacogenetic-based statin prescribing only include recommendations for simvastatin. Our data provide rationale for expanding these guidelines to include lovastatin.
Funding
HHS | NIH | National Heart, Lung, and Blood Institute (NHLBI): Akinyemi Oni-Orisan K01 HL143109; HHS | NIH | National Institute of General Medical Sciences (NIGMS): Thomas (J) Hoffmann, Marisa (W) Medina, Carlos Iribarren, Ronald (M.) Krauss, Neil Risch, Akinyemi Oni-Orisan P50 GM115318; ASCPT Darrell Abernethy Early Stage Investigator Award: Akinyemi Oni-Orisan
Footnotes
Conflict of Interest/Disclosure
The authors declared no conflicts of interest.
Supplemental File:
1. Supplemental Material
References
- 1.Newman CB, Preiss D, Tobert JA, et al. Statin Safety and Associated Adverse Events: A Scientific Statement From the American Heart Association. Arterioscler Thromb Vasc Biol. 2019;39(2):e38–e81. doi: 10.1161/ATV.0000000000000073 [DOI] [PubMed] [Google Scholar]
- 2.Wood FA, Howard JP, Finegold JA, et al. N-of-1 Trial of a Statin, Placebo, or No Treatment to Assess Side Effects. N Engl J Med. 2020;383(22):2182–2184. doi: 10.1056/NEJMc2031173 [DOI] [PubMed] [Google Scholar]
- 3.Stroes ES, Thompson PD, Corsini A, et al. Statin-associated muscle symptoms: impact on statin therapy-European Atherosclerosis Society Consensus Panel Statement on Assessment, Aetiology and Management. Eur Heart J. 2015;36(17):1012–1022. doi: 10.1093/eurheartj/ehv043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruckert E, Hayem G, Dejager S, Yau C, Bégaud B. Mild to moderate muscular symptoms with high-dosage statin therapy in hyperlipidemic patients--the PRIMO study. Cardiovasc Drugs Ther. 2005;19(6):403–414. doi: 10.1007/s10557-005-5686-z [DOI] [PubMed] [Google Scholar]
- 5.Davidson MH, Clark JA, Glass LM, Kanumalla A. Statin safety: an appraisal from the adverse event reporting system. Am J Cardiol. 2006;97(8A):32C–43C. doi: 10.1016/j.amjcard.2005.12.008 [DOI] [PubMed] [Google Scholar]
- 6.Torres PA, Helmstetter JA, Kaye AM, Kaye AD. Rhabdomyolysis: pathogenesis, diagnosis, and treatment. Ochsner J. 2015;15(1):58–69. [PMC free article] [PubMed] [Google Scholar]
- 7.Zutt R, van der Kooi AJ, Linthorst GE, Wanders RJA, de Visser M. Rhabdomyolysis: review of the literature. Neuromuscul Disord NMD. 2014;24(8):651–659. doi: 10.1016/j.nmd.2014.05.005 [DOI] [PubMed] [Google Scholar]
- 8.De Angelis G The influence of statin characteristics on their safety and tolerability. Int J Clin Pract. 2004;58(10):945–955. doi: 10.1111/j.1368-5031.2004.00355.x [DOI] [PubMed] [Google Scholar]
- 9.Hsiang B, Zhu Y, Wang Z, et al. A novel human hepatic organic anion transporting polypeptide (OATP2). Identification of a liver-specific human organic anion transporting polypeptide and identification of rat and human hydroxymethylglutaryl-CoA reductase inhibitor transporters. J Biol Chem. 1999;274(52):37161–37168. doi: 10.1074/jbc.274.52.37161 [DOI] [PubMed] [Google Scholar]
- 10.Nakai D, Nakagomi R, Furuta Y, et al. Human liver-specific organic anion transporter, LST-1, mediates uptake of pravastatin by human hepatocytes. J Pharmacol Exp Ther. 2001;297(3):861–867. [PubMed] [Google Scholar]
- 11.Pasanen MK, Neuvonen M, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of simvastatin acid. Pharmacogenet Genomics. 2006;16(12):873–879. doi: 10.1097/01.fpc.0000230416.82349.90 [DOI] [PubMed] [Google Scholar]
- 12.Hirota T, Fujita Y, Ieiri I. An updated review of pharmacokinetic drug interactions and pharmacogenetics of statins. Expert Opin Drug Metab Toxicol. 2020;16(9):809–822. doi: 10.1080/17425255.2020.1801634 [DOI] [PubMed] [Google Scholar]
- 13.SEARCH Collaborative Group, Link E, Parish S, et al. SLCO1B1 variants and statin-induced myopathy--a genomewide study. N Engl J Med. 2008;359(8):789–799. doi: 10.1056/NEJMoa0801936 [DOI] [PubMed] [Google Scholar]
- 14.Ramsey LB, Johnson SG, Caudle KE, et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharmacol Ther. 2014;96(4):423–428. doi: 10.1038/clpt.2014.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voora D, Shah SH, Spasojevic I, et al. The SLCO1B1*5 genetic variant is associated with statin-induced side effects. J Am Coll Cardiol. 2009;54(17):1609–1616. doi: 10.1016/j.jacc.2009.04.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brunham LR, Lansberg PJ, Zhang L, et al. Differential effect of the rs4149056 variant in SLCO1B1 on myopathy associated with simvastatin and atorvastatin. Pharmacogenomics J. 2012;12(3):233–237. doi: 10.1038/tpj.2010.92 [DOI] [PubMed] [Google Scholar]
- 17.Jacobson TA. Statin safety: lessons from new drug applications for marketed statins. Am J Cardiol. 2006;97(8A):44C–51C. doi: 10.1016/j.amjcard.2005.12.009 [DOI] [PubMed] [Google Scholar]
- 18.Bays H Statin safety: an overview and assessment of the data--2005. Am J Cardiol. 2006;97(8A):6C–26C. doi: 10.1016/j.amjcard.2005.12.006 [DOI] [PubMed] [Google Scholar]
- 19.Oni-Orisan A, Hoffmann TJ, Ranatunga D, et al. Characterization of Statin Low-Density Lipoprotein Cholesterol Dose-Response Using Electronic Health Records in a Large Population-Based Cohort. Circ Genomic Precis Med. 2018;11(9):e002043. doi: 10.1161/CIRCGEN.117.002043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart A SLCO1B1 Polymorphisms and Statin-Induced Myopathy. PLoS Curr. 2013;5. doi: 10.1371/currents.eogt.d21e7f0c58463571bb0d9d3a19b82203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roger VL, Killian JM, Weston SA, et al. Redefinition of myocardial infarction: prospective evaluation in the community. Circulation. 2006;114(8):790–797. doi: 10.1161/CIRCULATIONAHA.106.627505 [DOI] [PubMed] [Google Scholar]
- 22.Zimmerman JL, Shen MC. Rhabdomyolysis. Chest. 2013;144(3):1058–1065. doi: 10.1378/chest.12-2016 [DOI] [PubMed] [Google Scholar]
- 23.Thompson PD, Panza G, Zaleski A, Taylor B. Statin-Associated Side Effects. J Am Coll Cardiol. 2016;67(20):2395–2410. doi: 10.1016/j.jacc.2016.02.071 [DOI] [PubMed] [Google Scholar]
- 24.Andrade SE, Graham DJ, Staffa JA, et al. Health plan administrative databases can efficiently identify serious myopathy and rhabdomyolysis. J Clin Epidemiol. 2005;58(2):171–174. doi: 10.1016/j.jclinepi.2004.10.004 [DOI] [PubMed] [Google Scholar]
- 25.Bellosta S, Corsini A. Statin drug interactions and related adverse reactions: an update. Expert Opin Drug Saf. 2018;17(1):25–37. doi: 10.1080/14740338.2018.1394455 [DOI] [PubMed] [Google Scholar]
- 26.Mancini GBJ, Baker S, Bergeron J, et al. Diagnosis, prevention, and management of statin adverse effects and intolerance: proceedings of a Canadian Working Group Consensus Conference. Can J Cardiol. 2011;27(5):635–662. doi: 10.1016/j.cjca.2011.05.007 [DOI] [PubMed] [Google Scholar]
- 27.Jacobson TA. Toward “pain-free” statin prescribing: clinical algorithm for diagnosis and management of myalgia. Mayo Clin Proc. 2008;83(6):687–700. doi: 10.4065/83.6.687 [DOI] [PubMed] [Google Scholar]
- 28.Banda Y, Kvale MN, Hoffmann TJ, et al. Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200(4):1285–1295. doi: 10.1534/genetics.115.178616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taha DA, De Moor CH, Barrett DA, Gershkovich P. Translational insight into statin-induced muscle toxicity: from cell culture to clinical studies. Transl Res J Lab Clin Med. 2014;164(2):85–109. doi: 10.1016/j.trsl.2014.01.013 [DOI] [PubMed] [Google Scholar]
- 30.Feng Q, Wilke RA, Baye TM. Individualized risk for statin-induced myopathy: current knowledge, emerging challenges and potential solutions. Pharmacogenomics. 2012;13(5):579–594. doi: 10.2217/pgs.12.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hopewell JC, Offer A, Haynes R, et al. Independent risk factors for simvastatin-related myopathy and relevance to different types of muscle symptom. Eur Heart J. 2020;41(35):3336–3342. doi: 10.1093/eurheartj/ehaa574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ochs-Balcom HM, Nguyen LM, Ma C, et al. Clinical features related to statin-associated muscle symptoms. Muscle Nerve. 2019;59(5):537–543. doi: 10.1002/mus.26397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karter AJ, Schillinger D, Adams AS, et al. Elevated rates of diabetes in Pacific Islanders and Asian subgroups: The Diabetes Study of Northern California (DISTANCE). Diabetes Care. 2013;36(3):574–579. doi: 10.2337/dc12-0722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Apovian CM. Obesity: definition, comorbidities, causes, and burden. Am J Manag Care. 2016;22(7 Suppl):s176–185. [PubMed] [Google Scholar]
- 35.Wj G Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med. 2002;21(1):35–50. doi: 10.1002/sim.973 [DOI] [PubMed] [Google Scholar]
- 36.Bitzur R, Cohen H, Kamari Y, Harats D. Intolerance to Statins: Mechanisms and Management. Diabetes Care. 2013;36(Supplement 2):S325–S330. doi: 10.2337/dcS13-2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pozhidaev IV, Boiko AS, Loonen AJM, et al. Association of Cholinergic Muscarinic M4 Receptor Gene Polymorphism with Schizophrenia. Appl Clin Genet. 2020;13:97–105. doi: 10.2147/TACG.S247174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.R Core Team (2017). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/ [Google Scholar]
- 39.Salami JA, Warraich H, Valero-Elizondo J, et al. National Trends in Statin Use and Expenditures in the US Adult Population From 2002 to 2013: Insights From the Medical Expenditure Panel Survey. JAMA Cardiol. 2017;2(1):56–65. doi: 10.1001/jamacardio.2016.4700 [DOI] [PubMed] [Google Scholar]
- 40.Grundy SM, Stone NJ, Bailey AL, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA Guideline on the Management of Blood Cholesterol: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol. 2019;73(24):e285–e350. doi: 10.1016/j.jacc.2018.11.003 [DOI] [PubMed] [Google Scholar]
- 41.Tornio A, Vakkilainen J, Neuvonen M, Backman JT, Neuvonen PJ, Niemi M. SLCO1B1 polymorphism markedly affects the pharmacokinetics of lovastatin acid. Pharmacogenet Genomics. 2015;25(8):382–387. doi: 10.1097/FPC.0000000000000148 [DOI] [PubMed] [Google Scholar]
- 42.Neuvonen PJ, Backman JT, Niemi M. Pharmacokinetic comparison of the potential over-the-counter statins simvastatin, lovastatin, fluvastatin and pravastatin. Clin Pharmacokinet. 2008;47(7):463–474. doi: 10.2165/00003088-200847070-00003 [DOI] [PubMed] [Google Scholar]
- 43.Ward NC, Watts GF, Eckel RH. Statin Toxicity. Circ Res. 2019;124(2):328–350. doi: 10.1161/CIRCRESAHA.118.312782 [DOI] [PubMed] [Google Scholar]
- 44.Carr DF, O’Meara H, Jorgensen AL, et al. SLCO1B1 genetic variant associated with statin-induced myopathy: a proof-of-concept study using the clinical practice research datalink. Clin Pharmacol Ther. 2013;94(6):695–701. doi: 10.1038/clpt.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodrigues AC. Efflux and uptake transporters as determinants of statin response. Expert Opin Drug Metab Toxicol. 2010;6(5):621–632. doi: 10.1517/17425251003713519 [DOI] [PubMed] [Google Scholar]
- 46.Niemi M Transporter pharmacogenetics and statin toxicity. Clin Pharmacol Ther. 2010;87(1):130–133. doi: 10.1038/clpt.2009.197 [DOI] [PubMed] [Google Scholar]
- 47.Ho RH, Choi L, Lee W, et al. Effect of drug transporter genotypes on pravastatin disposition in European- and African-American participants. Pharmacogenet Genomics. 2007;17(8):647–656. doi: 10.1097/FPC.0b013e3280ef698f [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


