Abstract
The evidence for pharmacogenetics has grown rapidly in recent decades. However, the strength of evidence required for the clinical implementation of pharmacogenetics is highly debated. Therefore, the purpose of this review is to summarize different perspectives on the evidence required for the clinical implementation of pharmacogenetics. First, we present two patient cases that demonstrate how knowledge of pharmacogenetic evidence affected their care. Then we summarize resources that curate pharmacogenetic evidence, types of evidence (with an emphasis on randomized controlled trials [RCT]) and their limitations, and different perspectives from implementers, clinicians, and patients. We compare pharmacogenetics to a historical example (i.e., the evidence required for the clinical implementation of pharmacokinetics/therapeutic drug monitoring), and we provide future perspectives on the evidence for pharmacogenetic panels and the need for more education in addition to evidence. While there are differences in the interpretation of pharmacogenetic evidence across resources, efforts for standardization are underway. Survey data illustrate the value of pharmacogenetic testing from the patient perspective, with their providers seen as key to ensuring maximum benefit from test results. However, clinicians and practice guidelines from medical societies often rely on RCT data to guide treatment decisions, which are not always feasible or ethical in pharmacogenetics. Thus, recognition of other types of evidence to support pharmacogenetic implementation is needed. Among pharmacogenetic implementers, consistent evidence of pharmacogenetic associations is deemed most critical. Ultimately, moving pharmacogenetics into practice will require consideration of multiple stakeholder perspectives, keeping particularly attuned to the voice of the ultimate stakeholder – the patient.
Keywords: pharmacogenetics, pharmacogenomics, evidence, clinical implementation, stakeholders, perspectives, randomized controlled trial
INTRODUCTION
Some medical centers have adopted pharmacogenetic testing into routine clinical care,(1–6) but these examples of pharmacogenetic testing in practice remain limited. This is in large part because of varying opinions on the level of evidence needed to support clinical implementation.(7–28) Specifically, some argue for randomized controlled trial (RCT) data demonstrating that pharmacogenetic testing improves health outcomes over the standard, non-genotype guided care, before supporting testing as part of clinical practice.(7, 24, 25) Others counter that pharmacogenetics is held to a higher standard than required for other patient specific factors (e.g. renal and liver function) routinely incorporated into prescribing decisions without RCT evidence.(8, 10, 26) For example, many commonly used drugs, such as metformin, angiotensin-converting enzyme inhibitors, and direct oral anticoagulants, are avoided or dose-adjusted based on serum creatinine level without RCT evidence demonstrating that improves patient outcomes.(29) Importantly, the voice of the patient, the ultimate stakeholder, is often lost in conversations about what evidence is important for informing pharmacogenomic implementation.
These different perspectives on pharmacogenetic evidence can dramatically influence patient care. Therefore we start this review by presenting two different patient cases that demonstrate how different perspectives on pharmacogenetic evidence influenced patient care. The first case focuses on a clinical molecular geneticist who was aware of evidence regarding CYP2D6 metabolic phenotype and tamoxifen efficacy, and she was aware of the controversy surrounding the evidence. Even though oncology clinical practice guidelines did not recommend CYP2D6 testing,(30, 31) she was aware of other literature (including CPIC guidelines)(32) showing less favorable survival statistics for CYP2D6 intermediate and poor metabolizers. When she was diagnosed with breast cancer, she used her knowledge to advocate for CYP2D6 testing and subsequent therapy change to avoid the possibility of unnecessary risk. Tamoxifen/CYP2D6 is an example of differing stances on clinical actionability evidence between CPIC, medical society guidelines, and in this case, a well-informed patient.
The second patient case demonstrates how different providers have varying perspectives on pharmacogenetic evidence, which played a role in their management of the patient. Other common themes demonstrated in these cases is that the patients learned about pharmacogenetic evidence from other resources than their providers, and the patients became their own advocates for their pharmacogenetic testing. Therefore it is critical that providers are also educated on pharmacogenetic evidence. Given the open question of the evidence required for the implementation of pharmacogenetics into clinical practice, after these patient cases we provide a broad overview of the resources curating pharmacogenetic evidence, the types of available evidence and their limitations, and then highlight different perspectives on pharmacogenetic evidence: historical, practice guidelines, implementers, clinicians, patients, and the future.
Patient Case 1
An ultrasound image of a tumor suddenly transformed a clinical molecular geneticist into a patient. The 55-year-old female was subsequently prescribed tamoxifen for treatment of estrogen-receptor positive, lymph node negative breast cancer (the patient had been taking oral contraceptives prior to her diagnosis and was therefore not known to be postmenopausal). As a geneticist, she was aware of the importance of cytochrome P450 (CYP) 2D6 in metabolism of tamoxifen to its major active metabolite, endoxifen, and of clinical studies showing an association between the poor metabolizer (PM) and intermediate metabolizer (IM) phenotypes and increased risk of breast cancer recurrence.(33, 34) After discussing relevant publications with her oncologist and sharing her concern about the possibility of unknowingly being in a genetically disadvantaged group regarding tamoxifen effectiveness, the clinician agreed to CYP2D6 testing. Testing was ordered through a commercial reference laboratory, with results revealing a *1/*4 genotype with gene duplication, and therefore the possibility of the IM phenotype. The oncologist agreed to switch treatment from tamoxifen to the aromatase inhibitor, anastrozole, which is not metabolized by CYP2D6 and has strong effectiveness for reducing the risk of recurrence and breast cancer mortality.(35) This therapy change is consistent with current CPIC guidelines for CYP2D6 and tamoxifen therapy.(32)
Patient Case 2
A 45-year-old female with hypertension, dyslipidemia, depression, and gastroesphogeal reflux disease (GERD) had a history of intolerance or inadequate response to multiple medications for her depression and GERD requiring frequent medication changes. The patient learned about pharmacogenetic testing through a patient support group website and requested testing through her primary care physician (PCP). She specifically relayed her hope that the testing results would help explain her unsuccessful treatment odyssey to date and point toward treatment most likely to improve her symptoms without causing side effects. The PCP was familiar with testing, having ordered it through a commercial vendor for previous patients and agreed that results would be useful for informing the patient’s therapy. Thus, testing was ordered, and on receiving the report, the PCP found that the patient was a CYP2C19 ultrarapid metabolizer. Therefore some of the patient’s current medications for treating depression and GERD could have decreased efficacy, and adjustments to her drug therapy may improve her symptoms. However, on sharing the results with the providers specifically managing the patient’s depression/anxiety and GERD, one provider placed the results in the medical chart without ever discussing them with the patient, and the other dismissed the report and its value all together. Therefore the patient’s pharmacogenetic test results were left unused, and their potential benefit was unexplored.
RESOURCES THAT CURATE PHARMACOGENETIC EVIDENCE
The FDA has incorporated genetic information into the labels for over 250 drugs, particularly when the impact of genotype on drug response is potentially serious or life-threatening.(36) In some cases, such as with clopidogrel, this information is in the form of a boxed warning, given the potentially serious implications of genotype for drug effectiveness.(37) For other drugs, this information may be included in other sections of the drug label, such as the clinical pharmacology, dosing and administration, or indications and usage. In early 2020, the FDA released a Table of Pharmacogenetic Associations,(38) listing medications and gene associations in three groups for which the data: a) support therapeutic management recommendations; b) indicate a potential impact on safety and response; and c) demonstrate a potential impact on pharmacokinetic properties only. The agency encourages public input regarding Table of Pharmacogenetic Associations and made some updates on March 18, 2021.
Pharmacogenetic evidence has also informed the development of clinical guidelines from organizations such as CPIC, the Dutch Pharmacogenomics Working Group (DPWG), the Canadian Pharmacogenomics Network for Drug Safety, and other pharmacogenetic expert groups on interpretation and translation of genotype results into prescribing decisions for numerous gene-drug pairs.(39–41) CPIC, DPWG, and the Pharmacogenomics Knowledgebase (PharmGKB®) use rigorous systems to grade levels of evidence for clinical actionability of gene-drug pairs. CPIC guidelines are developed for gene-drug pairs with strong evidence,(42) and as of early 2021, there were 25 CPIC guidelines. PharmGKB® has clinically annotated over 160 gene-drug pairs to date with high levels of evidence.(43) In reference to Case 1, the tamoxifen/CYP2D6 association has the highest level of evidence per CPIC (level A), DPWG (level 4), and PharmGKB® (level 1A). The evidence on the FDA-cleared tamoxifen label is considered actionable by PharmGKB®. The FDA label states that CYP2D6 poor metabolizers carrying two non-functional alleles exhibit significantly lower endoxifen plasma concentrations compared to patients carrying one or more fully functional alleles,(44) but it also states that the impact on the efficacy of tamoxifen is not well established.
There has been a push toward standardization in the field as a means to accelerate pharmacogenetic adoption,(45) and much progress has been made in this regard. This includes collaborative efforts by CPIC and the DPWG to standardize terms for pharmacogenetic test results and efforts by the Association of Molecular Pathology (AMP) to recommend which alleles to include in pharmacogenetic tests.(46) The PharmGKB® created a document cross-referencing the FDA Table of Pharmacogenetic Associations with CPIC gene-drug pairs and PharmGKB® annotations to illuminate differences.(47) Along the same lines, the Personalized Medicine Coalition (PMC) Pharmacogenomics Working Group is currently analyzing differences across these resources (personal communication). There are some gaps regarding pharmacogenetic information in FDA-approved prescribing information. For example, several drugs with CPIC level A gene drug designation have no pharmacogenetic information in their labeling. Differences in clinical pharmacogenetic recommendations among resources could be due to a variety reasons, including differences in the organizations’ methods of evidence evaluation. For example, unlike CPIC, the FDA does not specifically define the phenotype (e.g., alleles defining a poor metabolizer), nor does the FDA cite the evidence used to develop their recommendations. The American Society of Pharmacovigilance brought together a wide range of stakeholders, including patients, providers, industry, regulators, payers, and others to form the Standardizing Laboratory Practices in Pharmacogenomics (STRIPE) Collaborative Community in 2020. The goal of STRIPE is to harmonize pharmacogenetic testing-related standards, practices, and resources.(48) It is worth noting that differences among pharmacogenetic recommendations from different sources may never be resolved, especially because of differences in the organizations’ mission and approach. However a single standard of recommendations may not be necessary, as prescribers typically utilize a variety of information resources when making other types of prescribing decisions (e.g., UpToDate and Micromedex).
TYPES OF PHARMACOGENETIC EVIDENCE & THEIR LIMITATIONS
RCTs are the gold standard for evaluating the clinical utility of new interventions. Many RCTs have assessed the clinical utility of pharmacogenetic testing, by comparing outcomes of genotype-guided drug therapy versus the current standard of care (i.e., non-genotype guided drug therapy), and they are summarized in Table S1. Many of those pharmacogenetic RCTs have demonstrated improved outcomes with pharmacogenetic testing compared to usual care. RCTs have important limitations when applied specifically to pharmacogenetics. RCTs have been the gold standard for evaluating the clinical utility of new pharmacologic, surgical, or other interventions. However, they have not been typically used to evaluate the clinical utility of tailoring those interventions based on patient specific factors, such as patient age, renal function, and other laboratory values, including plasma drug concentrations. Patient specific factors such as those are routinely used to guide drug therapy in absence of RCT data showing that consideration of these factors improves outcomes. Rather, evidence that these variables influence the risk for adverse effects or likelihood of drug effectiveness is considered sufficient to support their consideration in practice. Moreover, only a subset of patients will carry the pharmacogenetic variant associated with an atypical drug response (i.e., reduced effectiveness or increased toxicity). Therefore, the RCT must be much larger than a typical drug trial (to account for the majority patients without the genetic variant), or a large number of patients must be screened in order to selectively enroll only those patients carrying the pharmacogenetic variant of interest. Sometimes the adverse event that the pharmacogenetic test is intended to prevent is rare, again requiring a very large sample size in order to provide sufficient power. RCTs in general cost millions of dollars and several years to complete, and thus adding those particular considerations for pharmacogenetic RCTs would increase the cost even more. The same limitations that apply to RCTs in general also apply to pharmacogenetic RCTs as well. For example, the generalizability of RCT results is usually limited, given the strict eligibility criteria for entry and lack of racial/ethnic diversity.(49)
In addition to limitations related to feasibility and generalizability, many argue that requiring a RCT for every gene-drug pair may also be unethical.(8–23, 26) In cases where a pharmacogenetic variant is associated with a life-threatening adverse drug effect (e.g. carbamazepine-induced severe cutaneous reactions in patients with a HLA*15:02 allele), it may be deemed unethical to randomize participants to the usual care arm. Because of these concerns, some advocate for alternative approaches to assessing outcomes with pharmacogenetic testing, such as pragmatic trials and observational studies or other types of evidence (e.g., pharmacokinetic data or case-control or cohort study designs), which may be supported by mechanistic or in vitro studies.(8–23, 26)
Examples of alternative approaches to evidence generation include pragmatic trials and observational studies of patients receiving testing as part of clinical care. Similar to RCTs, many pragmatic and observational studies to date have demonstrated improved outcomes with a pharmacogenetic-guided approach.(50–55) Data from additional pragmatic trials are forthcoming from efforts such as from the Implementing GeNomics In pracTicE (IGNITE) Pragmatic Trials Network (ClinicalTrials.gov Identifier: NCT04445792) and the Ubiquitous Pharmacogenomics Consortium.(4) Pragmatic studies are conducted in the context of clinical practice, and thus, provide an advantage over RCTs in that they are more generalizable. They are also generally less costly and more efficient to conduct. However, data from nonrandomized studies are prone to selection bias and confounding that cannot be completely mitigated through statistical approaches.
DIFFERENT PERSPECTIVES ON PHARMACOGENETIC EVIDENCE
Historical Perspective
Over 20 years ago, strikingly similar arguments over the level of evidence needed to support clinical implementation of pharmacogenetic testing occurred for the clinical implementation of pharmacokinetics (PK) and therapeutic drug monitoring (TDM).(56–61) Like pharmacogenetics, PK/TDM was a budding “new” field of pharmacology, transitioning from research applications to clinical implementation. Also like pharmacogenetics, some held the view that RCTs were the required level of evidence for the clinical implementation of PK/TDM, whereas others did not.(57, 58, 60) Just like for pharmacogenetics, most of the evidence supporting the clinical implementation of PK/TDM derived from in vitro and observational clinical studies, and many of the clinical studies had surrogate endpoints instead of clinical outcomes.(56) Some RCTs of PK/TDM showed significant benefit; some did not show significant benefit; and some even showed a significant negative impact on patient outcomes.(56)
TDM of aminoglycosides is a good example of how varying levels of evidence still led to eventual clinical implementation. Not all RCTs of TDM versus non-TDM guided aminoglycoside therapy showed significant benefit as far as clinical outcomes nor cost savings.(56) Regardless, TDM for aminoglycoside therapy is now a routine part of clinical care. The routine clinical implementation of PK/TDM occurred in the absence of consistent RCT level of evidence for every drug. Authors at the time reported similar challenges as far as the limited feasibility and ethicalness of performing PK/TDM guided RCTs.(59) Ultimately, the enthusiasm for RCTs of PK/TDM waned. Eventually, sponsors were no longer interested in funding RCTs of PK/TDM, and clinicians lost interest in those RCTs, because institutions began implementing PK/TDM on their own.(59) A similar situation seems to be occurring for pharmacogenetics, in which several institutions have begun implementing pharmacogenetics in the absence of RCT data.(6) To avoid repeating history, perhaps it is time to put to rest the general requirement of RCTs for pharmacogenetics as well.
Practice Guideline Perspective
Clinicians rely on practice guidelines from expert consensus panels to guide prescribing decisions. Guideline writing committees are charged with weighing and ranking the evidence, and the highest ranking is provided to RCT data. Case 1 is a good example of when a tightly held standard of requiring a RCT to demonstrate improved outcomes with genotype-guided therapy may not be in the best interest of patients. CYP2D6 testing in the context of tamoxifen is not currently recommended in the American Society of Clinical Oncology (ASCO) or National Comprehensive Cancer Network (NCCN) breast cancer guidelines,(30, 31) and CYP2D6 testing is not routinely offered by oncologists prior to tamoxifen prescribing. Negative findings from two secondary CYP2D6 analyses of previous RCTs (BIG 1–98 (62) and ATAC (63)) likely influenced the current ASCO and NCCN guidelines; however, these findings were found to be flawed. Multiple problems were identified in both studies, with serious genotyping errors in the BIG 1–98 study.(64, 65) Both studies used formalin tumor tissue for CYP2D6 genotyping, with erroneous genotype results due to loss of heterozygosity at the CYP2D6 locus.(65) A more recent trial (CYPTAM)(66) has also been criticized.(67) The initial papers showing CYP2D6 association with tamoxifen efficacy were in women with tamoxifen monotherapy,(68) while several negative studies have had treatment variability noise. When weighing evidence from studies, it is important to carefully evaluate the methodologies, patient populations, and treatments involved and focus on the appropriately designed studies.(64) A recent review reiterates that controversy continues in the oncology literature around CYP2D6 testing to predict response to tamoxifen,(69) as does a recent study showing no difference in endoxifen or 40H-tamoxifen levels between patients who had recurrent breast cancer and those who did not in a low dose trial.(70) The low dose trial differs from previous studies of patients using a standard dose of tamoxifen.
The practice guidelines for the antiplatelet drug clopidogrel, a prodrug activated through metabolism by CYP2C19, are also controversial. The American Heart Association and the American College of Cardiology (AHA/ACC) joint practice guidelines for cardiovascular disease management categorize recommendations, which range from class I (recommended; benefit clearly outweighs risk) to class III (not recommended; potentially harmful), based on the evidence and expert consensus opinion. Again, RCT data is ranked as the highest level of evidence. Prior to availability of RCT data, but with substantial and consistent data that CYP2C19 genotype is associated with risk for adverse cardiovascular outcomes after percutaneous coronary intervention (PCI), the guidelines provided a class IIb recommendation (benefit equal to or greater than risk) for genotyping in high risk patients.(71) A class III recommendation was provided for routine genotyping of all patients undergoing PCI.
RCT data on outcomes with CYP2C19-guided antiplatelet therapy have since emerged. One RCT showed a significant reduction in risk for bleeding, without compromising the risk for atherothrombotic events, with a genotype-guided approach versus universal use of potent P2Y12 inhibitors (e.g. ticagrelor or prasugrel).(72) Subsequently published guidelines by the European Society of Cardiology note these RCT data, but they still provided a class IIb recommendation. The European guidelines specifically state that testing may be considered in select patients, especially those “deemed unsuitable for potent platelet inhibition” (e.g. those at high bleeding risk), even though the RCT did not limit eligibility to such patients.
In a subsequent RCT of genotype-guided therapy, the comparator arm was universal clopidogrel.(73) The trial found a 34% relative risk reduction in adverse cardiovascular events with the genotype-guided approach without reaching statistical significance (p=0.06), likely because there were fewer cardiovascular events than anticipated. The trial also showed a significant reduction in pre-specified sensitivity analyses of cumulative events and a post-hoc analysis of events at 90 days in favor on genotype-guided therapy (p = 0.001). A meta-analysis of genotype-guided antiplatelet RCTs has recently been published.(74) The reduction of ischemic events in patients with coronary artery disease who predominantly underwent PCI by ticagrelor or prasugrel, in comparison with clopidogrel, was based primarily on the presence of CYP2C19 loss-of-function carrier status.(74)
It remains to be determined how these RCT data will influence future AHA/ACC guidelines. Will the guideline writing committee consider the multiple observational studies showing improved outcomes with CYP2C19-guided antiplatelet therapy in practice,(50, 51, 53) leading to an upgrade in the recommendation for genetic testing? Or conversely, will they downgrade the recommendation because one of the major RCTs just barely missed the definition of statistical significance? How will they consider the recent meta-analysis of RCTs? These are important questions, since how the AHA/ACC interprets this evidence may set the precedent for future pharmacogenetic recommendations in cardiology guidelines.
There are examples in which pharmacogenetic recommendations have been made in clinical practice guidelines based on lower levels of evidence than available for CYP2C19 and clopidogrel. The 2020 NCCN guidelines for the treatment of adult acute lymphoblastic leukemia (ALL) state “Determination of patient TPMT genotype using genomic DNA is recommended to optimize 6-MP [6-mercaptopurine] dosing…”.(75) The evidence supporting that recommendation is solely retrospective and observational.(76) To our knowledge, a RCT comparing genotype-guided thiopurine dosing versus non-genotype guided in patients with ALL has not been performed. RCTs of genotype-guided thiopurine dosing in inflammatory conditions have been conducted, but none of them showed that genotype-guided thiopurine dosing significantly improved outcomes.(77) Regardless, the American College of Gastroenterology clinical guideline for the management of Crohn’s disease in adults states “Thiopurine methyltransferase (TPMT) testing should be considered before initial use of azathioprine or 6-mercaptopurine to treat patients with Crohn’s disease (strong recommendation, low level of evidence).”(78) Another example is in the 2017 AHA/ACC/HRS guideline for the management of patients with syncope, which states “The response to beta-blockers depends on the genotype…”.(79) The data supporting that statement comes solely from two observational registries.(80) Nearly half of drug-gene pairs have differences in their guideline recommendations,(81) which demonstrates variable interpretations of the available evidence in practice guidelines.
Implementer Perspective
Implementers, such as from institutions that have already or are beginning to implement pharmacogenetic testing clinically, are faced with the challenge of finding the right amount and type of evidence needed to bring pharmacogenetics into clinical practice. This may mean going beyond medical society clinical guidelines and considering patient and provider needs. The NHGRI-funded Implementing GeNomics In pracTicE (IGNITE) and Electronic Medical Records and Genomics (eMERGE) networks both have pharmacogenetics working groups, and member institutions have experience in clinical implementation of pharmacogenetic testing.(3, 82) We performed a survey to gather their perspectives on different types of pharmacogenetic evidence. Survey responses were solicited from 2/20/2020 to 3/4/2020 via REDCap electronic data capture tool, hosted at Vanderbilt University Medical Center, with anonymous responses stored in the secure REDCap database.(83) The survey was approved by the institutional review board at the University of Florida as an exempt study. A total of 47 individuals from 21 institutions were invited to participate. Fifteen individuals responded from 13 unique institutions (32% response rate). Characteristics of the individuals and institutions are displayed in Table 1.
Table 1.
Characteristics of implementers of clinical pharmacogenetics (individuals and institutions) that completed the survey
| Individuals (n = 15) | |
| Type of Training* | n (%) |
| PhD | 2 (13%) |
| PharmD | 13 (87%) |
| Master’s Degree | 1 (7%) |
| Years Since Training Completed | |
| 0–5 | 6 (40%) |
| 6–10 | 4 (27%) |
| 11–15 | 3 (20%) |
| 16 or more | 2 (13%) |
| Pharmacogenetics Training* | |
| None | 2 (13%) |
| Fellowship | 9 (60%) |
| Master’s Degree | 1 (7%) |
| Residency | 6 (40%) |
| Seminars, Workshops, or CME | 5 (33%) |
| Online Training | 2 (13%) |
| Certificate Program | 1 (7%) |
| Institutions (n = 13) | |
| Type of Institution* | |
| Academic | 11 (85%) |
| Community | 3 (23%) |
| Pharmacogenetic Testing Offered | |
| Broad | 6 (46%) |
| Limited | 7 (54%) |
| None | 0 (0%) |
More than one answer allowed
CME = continuing medical education
In one set of questions, respondents were asked to rate, on a scale of 0 to 100, how essential, how easy to interpret, and how available six different types of evidence or guidance are: RCT data; real world evidence or observational studies testing clinical outcomes with pharmacogenomic testing; real world evidence or observational studies testing associations between genotype and drug response guidelines; pharmacogenetic guidelines (e.g. CPIC, DPWG); subspecialty guidelines and statements (e.g. American College of Obstetricians and Gynecologists, AHA/ACC); and FDA or other regulatory communications and agency drug labels (e.g. FDA label). Results indicate diversity of opinion for each type of data for every question (Figure 1). Particularly for ratings of how essential data are, RCT data, specialty guidelines, and FDA label guidance demonstrated wide ranges (range 10–80, 20–100, and 20–90, respectively). Pharmacogenetic guidelines received the highest scores regarding ease of interpretation (range 80–100) and availability (range 50–100).
Figures 1A-C:
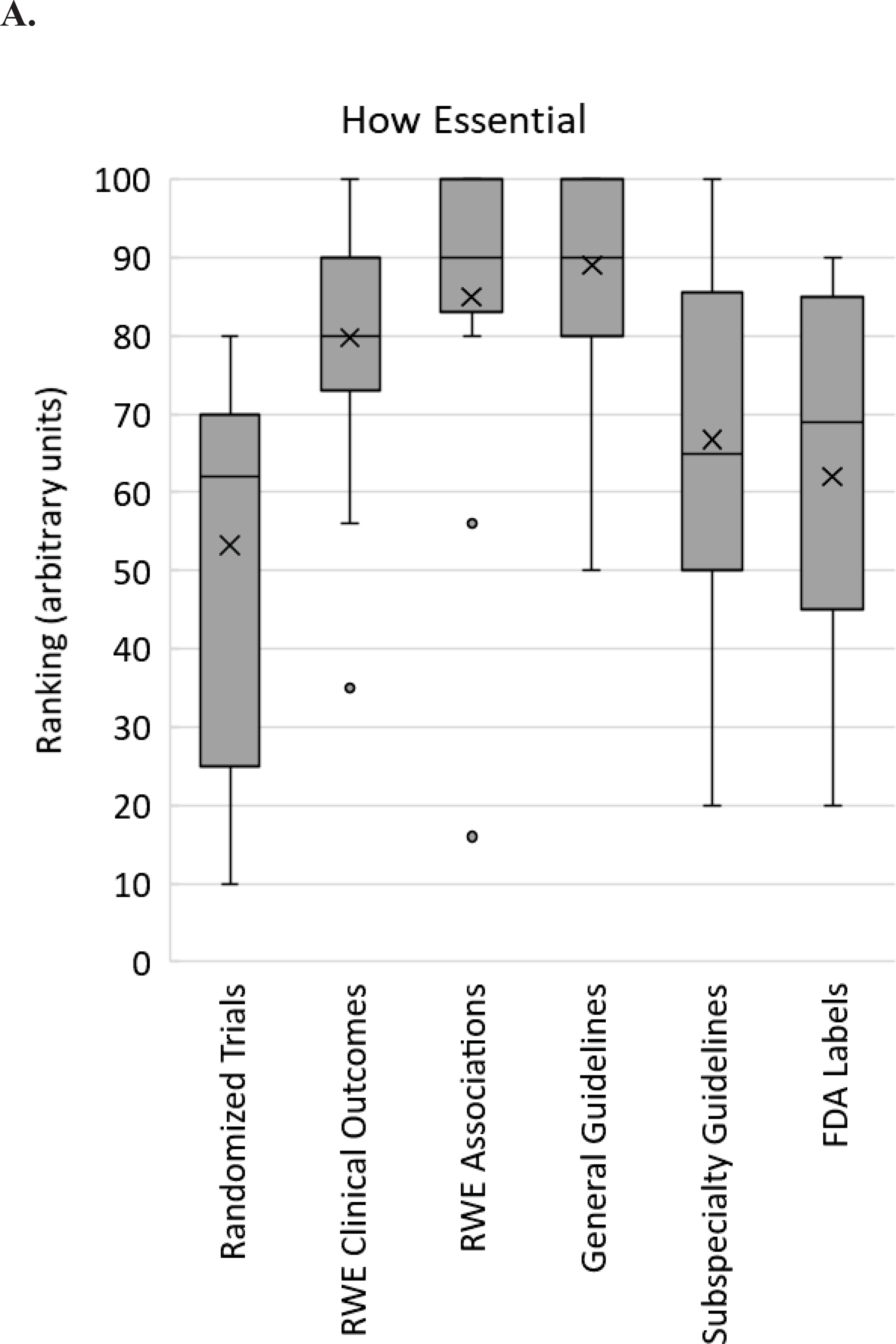
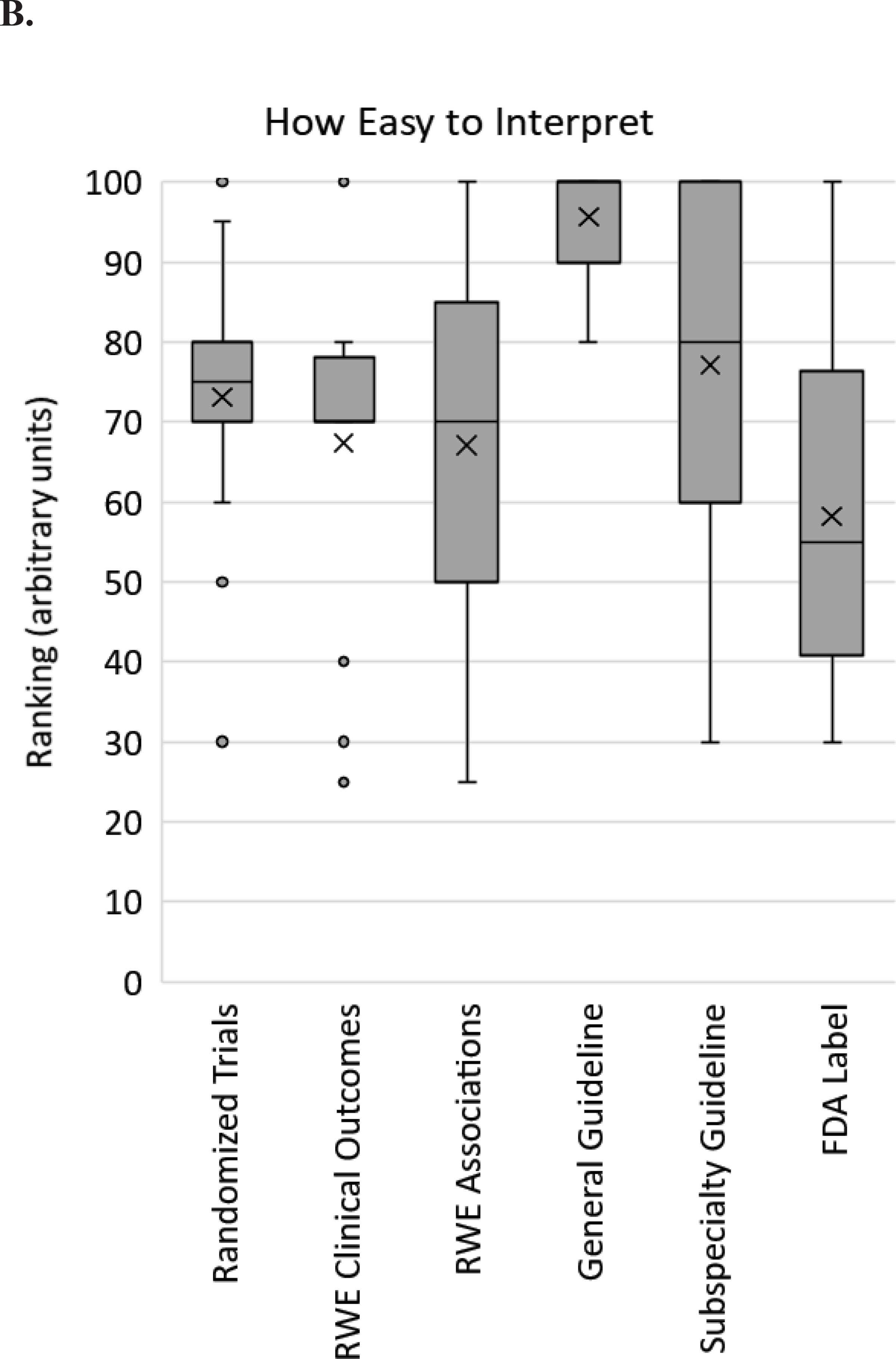
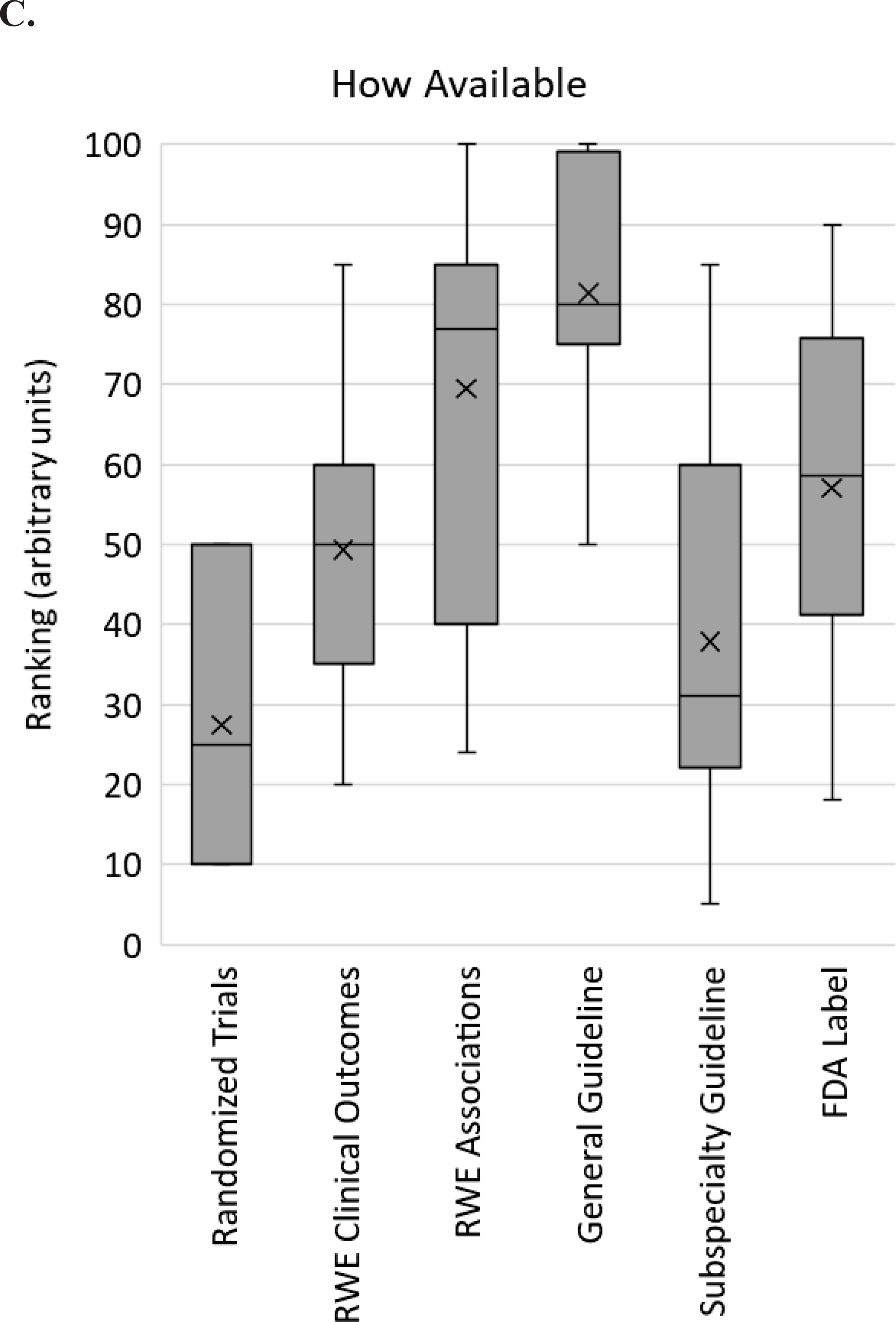
Survey respondent scores from implementers regarding pharmacogenomic evidence. For each of the 6 types of evidence listed on the X-axis, respondents were asked to rank how essential (A), how easy to interpret (B) and how available (C) that type of evidence is for clinical pharmacogenomic implementation on a scale from 0 to 100. The box and whisker plot shows the medians (horizontal lines within boxes), means (X), the interquartile ranges (boxes), the adjacent values (whiskers) and outliers (dots). RWE – real world evidence; FDA – Food and Drug Association.
We also asked in the survey, “What are the factors you consider when determining whether or not to implement pharmacogenomic testing?” Availability of CPIC or other guidelines were the most consistently endorsed factor (100% of respondents), with other factors less unanimously indicated (Figure 2). Thus, among experts in the field, opinions on evidence and resources essential to support pharmacogenetic implementation are highly variable.
Figure 2.
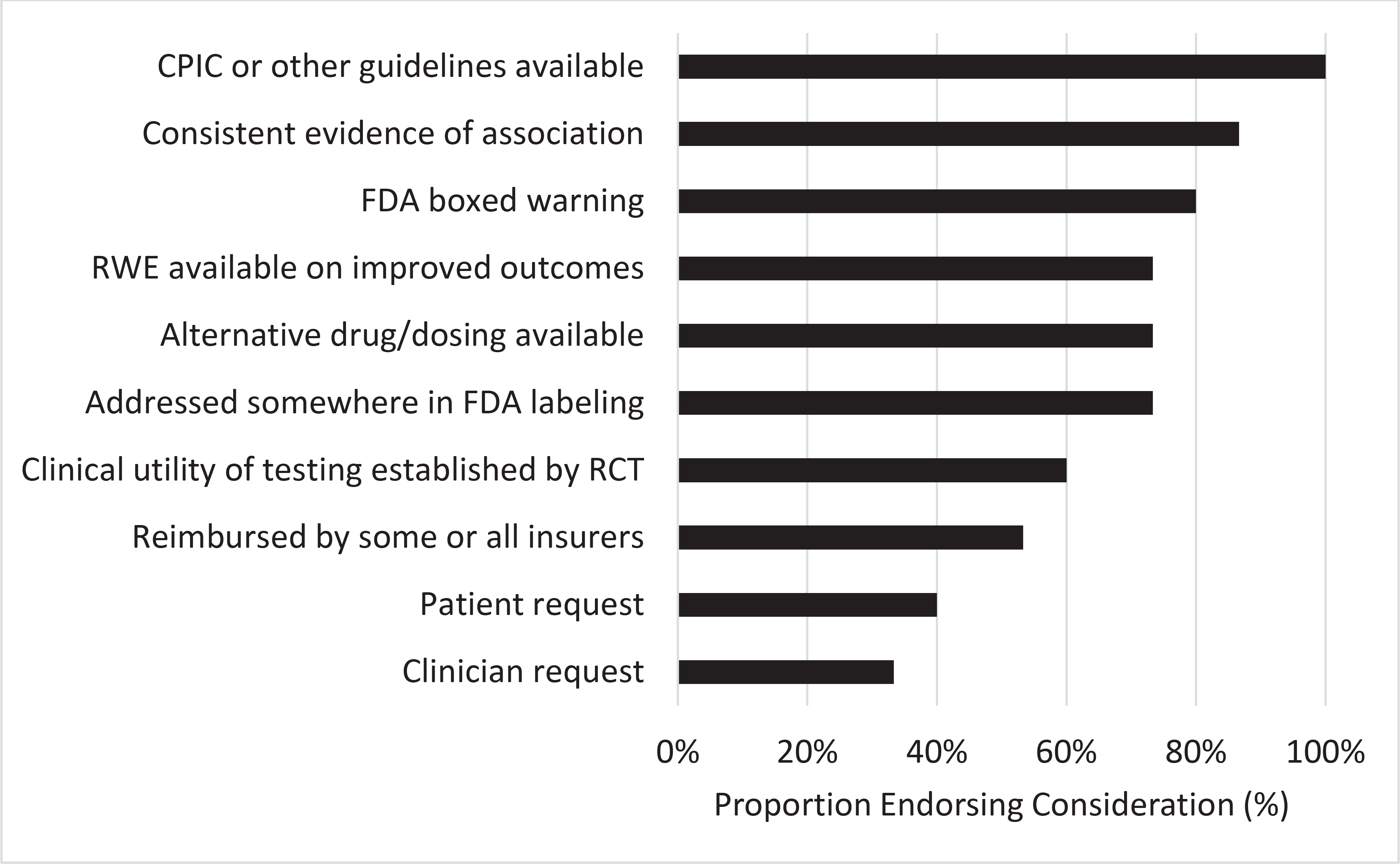
Factors considered by pharmacogenetic clinical implementers when determining whether or not to implement pharmacogenetic testing. Fifteen individuals from 13 different institutions were asked to rate their response on a scale from 0 to 100. CPIC = Clinical Pharmacogenetics Implementation Consortium; FDA = United States Food and Drug Administration; RWE = real world evidence
In addition to utilizing evidence to anchor their implementations in sound scientific data, implementers have an imperative to generate evidence for the use of pharmacogenetics in clinical practice, thereby ensuring the end-users of the implementation services, providers and patients, utilize pharmacogenetics in a responsible and prudent manner. Examining the patient and provider perspective of pharmacogenomic evidence illuminates the types of evidence implementers need to create.
Clinician Perspective
A problem with defining a single level of evidence for clinical implementation of pharmacogenetics is that every clinical scenario is unique, as demonstrated by the patient cases presented. The potential risks and benefits for pharmacogenetic-guided therapy vary greatly depending on the drug, the genomic signal, the alternative therapeutic strategies available, the indication for the drug, and many other variables, as displayed in Table 2. The balance of those risks and benefits should influence the level of evidence required for clinical pharmacogenetics, as has been argued by others.(10, 27, 28) All of these factors must be simultaneously considered and weighed in order to make rational, patient-centered decisions.
Table 2.
Variables that may affect the level of evidence required for the clinical implementation of pharmacogenetics
| Level of Evidence Potentially Required | ||
|---|---|---|
| Variable | Higher | Lower |
| Genetic test | -Results not already available -Not available in-house -Slow return of results -High cost |
-Results already available -Can be ordered in-house -Rapid return of results -Low cost |
| Genetic variant | -Rare -Low penetrance |
-Common -High penetrance |
| Drug | -Cost of drug (or alternative) is low -Rarely used -Alternatives available |
-Cost of drug (or alternative) is high -Commonly used -No alternatives available |
| Drug indication | -Mild -Rare |
-Severe -Common |
| Adverse Outcome | -Mild -Rare |
-Severe -Common |
| Setting | -Inpatient | -Outpatient |
| Patient | -Low risk for adverse outcome -Risk-averse |
-High risk for adverse outcome -Risk-taking |
| Provider | -Risk-averse | -Risk-taking |
| Specialty | -Cardiology | -Oncology -Pediatrics |
| Stakeholder | -Health system -Payers |
-Patients -Providers |
The clinical value of a test result drives the decision to test. The value is affected by many features including cost, variant frequency, clinical impact of outcomes, and strength of evidence. For a patient with no previous pharmacogenetic testing, a clinician’s first pharmacogenetic decision will be around whether to order testing. Two factors, actionability and cost, may contribute to a clinician’s decision making. Here, actionability refers to whether the results of the test will alter the management of the patient. If the evidence for the pharmacogenetic association is very weak, the clinical difference between genotypic groups is very small, and the variant tested for is exceedingly rare in the population, there may be little impetus for the clinician to order the test. On the other hand, testing for a rare variant with robust evidence for a severe, life-threatening toxicity (e.g., HLA variants and hypersensitivity to abacavir and carbamazepine, or TPMT/NUDT15 variants and myelosuppression from thiopurines) may be more quickly adopted into clinical practice. If there are two drugs with similar efficacy, but one has a higher cost (measured by financial cost, risk for toxicity, or need for ongoing monitoring), and a pharmacogenetic test can appropriately guide therapy, there may be higher uptake of the test.
Characteristics of the patient can also affect the acceptable level of evidence for a pharmacogenetic test for a provider. Lower levels of evidence may be required for patients with higher risk for the adverse clinical outcomes if not optimally treated because the potential benefit is greater. For example, lower levels of evidence for clopidogrel pharmacogenetic testing may be required for a patient who has multiple risk factors for adverse outcomes post-PCI (e.g., history of myocardial infarction, diabetes, chronic kidney disease), compared to a patient with fewer risk factors (e.g., stable coronary disease, non-diabetic, normal renal function). Clinicians may also seek context-specific evidence to support pharmacogenetic testing. For example, an interventional cardiologist who cares for patients in the cardiac catheterization laboratory and during their acute hospitalization may seek evidence for pharmacogenetic-guided antiplatelet therapy with endpoints relevant to time points when the patients are under his or her care (e.g., acute stent thrombosis). A general cardiologist managing patients’ chronic antiplatelet therapy may be attentive to endpoints far beyond the acute hospitalization (e.g., bleeding or need for revascularization). It also appears that the level of evidence for pharmacogenetics varies by the specialty within which the provider is practicing, e.g., cardiology, oncology, pediatrics.(80) Cardiology clinical practice guidelines, for example, usually have not supported recommendations for pharmacogenetic testing in the absence of RCT data, whereas oncology clinical practice guidelines have.(80) Lower levels of evidence may be acceptable in specialties like pediatrics, where providers are accustomed to adjusting doses based on patient-specific factors (including weight), using medications for off-label indications, and weighing evidence from a variety of study types, as large RCTs are uncommon for pediatric therapeutics. Thus, with all of these competing factors, it is difficult to define a single level of evidence for all clinical scenarios. A flexible standard for the required level of evidence for clinical implementation is rational.
Patient Perspective
As the ultimate benefactor of testing, the patient will likely be a key driver of moving pharmacogenetic discoveries into practice. Understanding patient needs from pharmacogenetics evidence is important, and like providers, is varied based on a multitude of factors. Different patients have different concerns about specific risks, and this needs to be incorporated into the decision making. Patients consider their own personal experience and perceived risk in clinical decisions, and biomarkers providing information on the likelihood of toxicity may influence their willingness to proceed with certain treatment choices.(84) The patient in case 1 was uniquely well-informed on the evidence that CYP2D6 influences plasma concentrations of the active tamoxifen metabolite, and that it may impact her risk for breast cancer recurrence. In her opinion, this evidence was impressive enough to warrant her request for testing.Unlike the patient in case 1 who had a genetic background, the patient in case 2 was less familiar with the evidence. Patients may hear about pharmacogenetic testing and request it from their providers. A thorough review of the patient’s medical history, including responses to current and previous medications, and determination of pharmacogenetic relevance of the medications, may be necessary to determine if pharmacogenetic testing is indeed warranted. Patients should be educated accordingly in order to ensure shared decision making between patient and provider. Pharmacogenetic clinics are emerging at some institutions where providers can refer patients to determine if testing may be beneficial, and if so, obtain consultation on how to best act on test results.(85) However the possibility that pharmacogenetics could help explain the patient’s responses to previous treatments, and identify the treatment regimen most likely to improve her symptoms with acceptable tolerability, was motivation enough for the patient to request testing.
Additional key insights into patient perspectives on pharmacogenetic evidence comes from focus groups and survey data. Cost is regularly cited as a barrier to patients completing pharmacogenomic testing.(86–89) Understanding the evidence behind the value and utility of pharmacogenomic testing is key to break through this barrier. A strong body of evidence is required to increase the insurance coverage of testing. In the last several years, there has been progress with reimbursement as United Healthcare and some Medicare local coverage determinations cover pharmacogenomic testing for patients meeting specific criteria.(90) Better reimbursement is expected to lead to reduced or no cost testing for patients, but it is still important for patients to understand the value and utility of testing in order to make informed decisions about whether to complete it. Patients have reported a wide range of value in pharmacogenetic evidence, including predicting effective therapy;(86) minimizing harm from wrong medications or doses;(91) increasing confidence in future medication decisions;(92) increasing compliance;(92, 93) and increasing trust in the health care system.(91) Several of these are evident in the patient cases presented. An outstanding question is whether the perceived value by patients is rooted in clear scientific data or just pseudoscience. Are RCTs required to answer this question? Implementers must continue to cultivate evidence to support these claims of value if patients are going to continue to be encouraged to believe in them.
The full benefit of testing may not be realized unless patients understand their results and implications of results for drug response. Programs have used reports, portals, and integrated models of pharmacogenetic delivery, well supported by a multidisciplinary team and pharmacogenetic experts(91, 92, 94) to help ensure patients receive and understand their results. Studies evaluating pharmacogenetic reports and portals have consistently found that graphic displays are better than tables or text at conveying pharmacogenetic information, but they are not perfect.(91, 94, 95) Regardless of how patients access their test results (e.g., reports and patient portals), patients believe their providers are key to receiving the most benefit from their pharmacogenomic results.(92, 94) More evidence is needed to determine the optimal method for returning results to patients and having them interact with the results through their lifetime. Patients are the ones who would ultimately benefit from pharmacogenetic testing. Therefore, while pharmacogenomic evidence from a patient perspective may be less about the drug-gene pairs, it is no less important to the future direction of the field.
THE FUTURE FOR PHARMACOGENETIC EVIDENCE
Evidence for Reactive, Individual Pharmacogenetic Tests Vs. Pre-Emptive Panels
Most of the currently available pharmacogenetic evidence focuses on individual drug-gene pairs (e.g. CYP2C19-clopidogrel; CYP2D6-tamoxifen). However recently, there has been a shift in interest regarding implementation, from reactive testing for individual drug-gene pairs to pre-emptive panels that cover multiple drug-gene pairs.(96, 97) Indeed, the cost of a dense pharmacogenetic genotyping array is now about the same as an individual pharmacogenetic test, and thus it is more cost efficient to order a pharmacogenetic panel instead of multiple individual pharmacogenetic tests.(26, 98) Additionally, with the cost of the sequencing decreasing, pharmacogenetics is increasingly being included in germline hereditary disease or population screening panels. Evidence from clinical implementation of pre-emptive pharmacogenetic panels is already showing tremendous potential. When as few as five gene-drug pairs were considered, a pre-emptive genotyping panel yielded actionable variants in >90% of patients at a single institution.(99) Implementation of the pre-emptive panel avoided the use of nearly 15,000 individual pharmacogenetic tests (if done reactively).(99) In 52,942 medical home patients in a single health system, it was estimated that nearly 400 adverse drug events could have been prevented by the implementation of that pre-emptive pharmacogenetic panel over the course of 5 years.(100) Recommendations based on the results of a pharmacogenetic panel resulted in an estimated $621 (USD) in annual savings per patient across a patient population on five or more medications.(101) An RCT assessing the clinical utility of pre-emptive pharmacogenetic panels, called the PREemptive Pharmacogenomic testing for prevention of Adverse drug REactions (PREPARE) study, is currently ongoing in Europe (as part of the Ubiquitous Pharmacogenomics Consortium [U-Pharmacogenetic]).(4) PREPARE is implementing pre-emptive genotyping of a panel of 50 variants in 13 pharmacogenes into clinical practice, in the context of a large prospective, international, block-randomized, controlled study (n = 8,100).
This cumulative evidence supporting all drug-gene pairs included on pharmacogenetic panels is developing, but it is important to note that the evidence supporting each individual gene-drug pair is still critical and the current barriers to panel testing. The content on pharmacogenetic panels varies widely (i.e., the specific genes and variants covered by the panel). (102)No two panels were the same in a comprehensive evaluation of several pharmacogenetic testing panels for 28 pharmacogenes(103)Though out of the scope of this paper for discussion, reimbursement is a significant barrier for pharmacogenetic panels, in particular. (90) Some companies and labs are offering pharmacogenetic test panels that include genes and variants with weak evidence for an association with drug response.(104) Therefore the evidence supporting each individual drug-gene pair informs many yet unanswered questions, such as which genes and variants should be included on a pharmacogenetic panel? Which results should be reported to the EHR? And which results from the panel have sufficient evidence to be used to guide patient care? Answers to these questions will evolve from ongoing discussions in the pharmacogenetics community. A single, standardized pharmacogenetic panel may not be the goal, but at least the genes and variants included on a pharmacogenetic panel should be supported by strong evidence.
Evidence Vs. Education
The clinical implementation of pharmacogenetics may be slowed as much by a lack of education as by the perception of a lack of evidence.(8) Would advocates for RCT data still argue for such data if they found themselves or a family member in the position of being a patient, and they were armed with the knowledge that a genotype is strongly and consistently associated with drug effectiveness or risk for an adverse effect? For example, what if a provider required PCI for coronary disease management and is prescribed clopidogrel afterwards? Knowing that 30% of the population carries a CYP2C19 no function allele, and that having a no function allele significantly increases the risk for clopidogrel failure, would he or she ask to be genotyped? Or would he or she refuse to be genotyped because one RCT (73) just barely missed statistical significance?
The cases presented at the beginning demonstrate the educational barrier on the part of the provider. Currently, many providers are not confident about interpreting pharmacogenetic test results, and they are not aware of the resources for pharmacogenetic evidence described above.(105) Referring to case 1, the switch from tamoxifen to anastrozole occurred for two key reasons. First, as a geneticist, the patient was knowledgeable about pharmacogenetics, and secondly, her physician was willing to embrace the scientific evidence presented to him by the patient and change the treatment plan accordingly. In case 2, the patient found that her primary care provider was experienced in pharmacogenetic testing, but the providers prescribing her relevant medications was not. Thus the pharmacogenetic test results went unused. Therefore, case 2 illustrates how even if pharmacogenetic tests results are readily available, eliminating the question about whether or not to order testing, there is significant diversity in the acceptance of pharmacogenetic evidence among physicians. Would the outcome of case 1 have been different had the patient not been knowledgeable about pharmacogenetics? Would the outcome of case 2 have been different had the providers been knowledgeable about the evidence supporting pharmacogenetic-guided prescribing decisions?
Data from a survey of approximately 10,000 US physicians suggest the answers to these questions is yes.(105) Specifically, the survey revealed that a large percentage of physicians (~98%) agreed that genetic variations may influence drug response, but few (~10%) felt adequately informed about pharmacogenetic testing. Only 29% reported receiving any pharmacogenetic education, and only 13% had ordered a test in the previous 6 months. Similarly, of 285 physicians surveyed across sites within the IGNITE Network, 70% believed that access to pharmacogenetic data would improve their ability to care for patients.(106) However only 30% responded they were confident in their ability to use the results, and only 32% said they could find or use reliable sources of pharmacogenetic information while caring for patients. Clinicians may have concerns about liability of testing for genetic variants with implications for multiple drugs. For example, a cardiologist may order a CYP2C19 genetic test to guide anti-platelet therapy, but the patient’s CYP2C19 phenotype may also affect responses to non-cardiovascular medications, such as antidepressants and proton pump inhibitors.
If the lack of pharmacogenetic education is more of a barrier than limited RCT evidence, then we need to focus our resources on education instead of further investment in RCT data. Efforts to include pharmacogenetics in the US medical school curriculum are important and underway. The PGx Dissemination Working Group is involved in the collaborative development of Pharmacogenetic continuing medical education (CME) materials with the Inter-Society Coordinating Committee for Practitioner Education in Genomics (ISCC) and the American Academy of Family Physicians (AAFP).(107) The goal of the PGx Dissemination Working Group is to raise awareness and educate about resources available for pharmacogenetic implementation such as CPIC guidelines, PharmGKB, the Pharmacogenomics Research Network (PGRN), and PharmVar, including also commentaries in journals, outreach to medical societies, letters to insurance companies, and through social media. Alignment of medical society practice guideline pharmacogenetic recommendations with these other resources is a critical step to enhance provider acceptance of the value of pharmacogenetic testing, (81) and to enhance the adoption into standard practice and availability for patients.
Conclusions
We reviewed multiple perspectives on the highly debated issue of the evidence required for the clinical implementation of pharmacogenetics. An overall summary of factors to consider in the clinical implementation of pharmacogenetic testing is presented in Figure 3. The perspective of the patient is often lost in this debate, and thus we presented two patient cases, in which the knowledge of pharmacogenetic evidence, on the part of both the provider and patient, affected their care. In our overview of the resources from organizations that curate pharmacogenetic evidence, such as CPIC, FDA, DPWG, and PharmGKB®, we point out that there are differences in the interpretation of the pharmacogenetic evidence across those resources. Efforts for standardization across these resources are underway. Our review of the types of pharmacogenetic evidence available included many RCTs of pharmacogenetic-guided therapy. However, the RCT design has many limitations when applied specifically to pharmacogenetics, and thus RCTs may not be necessary, appropriate, feasible, or ethical in pharmacogenetics. Similar arguments for RCT data for the clinical implementation of PK/TDM were reviewed in our historical perspective. However, similar to pharmacogenetics, institutions began routinely implementing PK/TDM in clinical practice in the absence of robust RCT data, and thus the arguments for RCT data supporting PK/TDM eventually waned.
Figure 3.
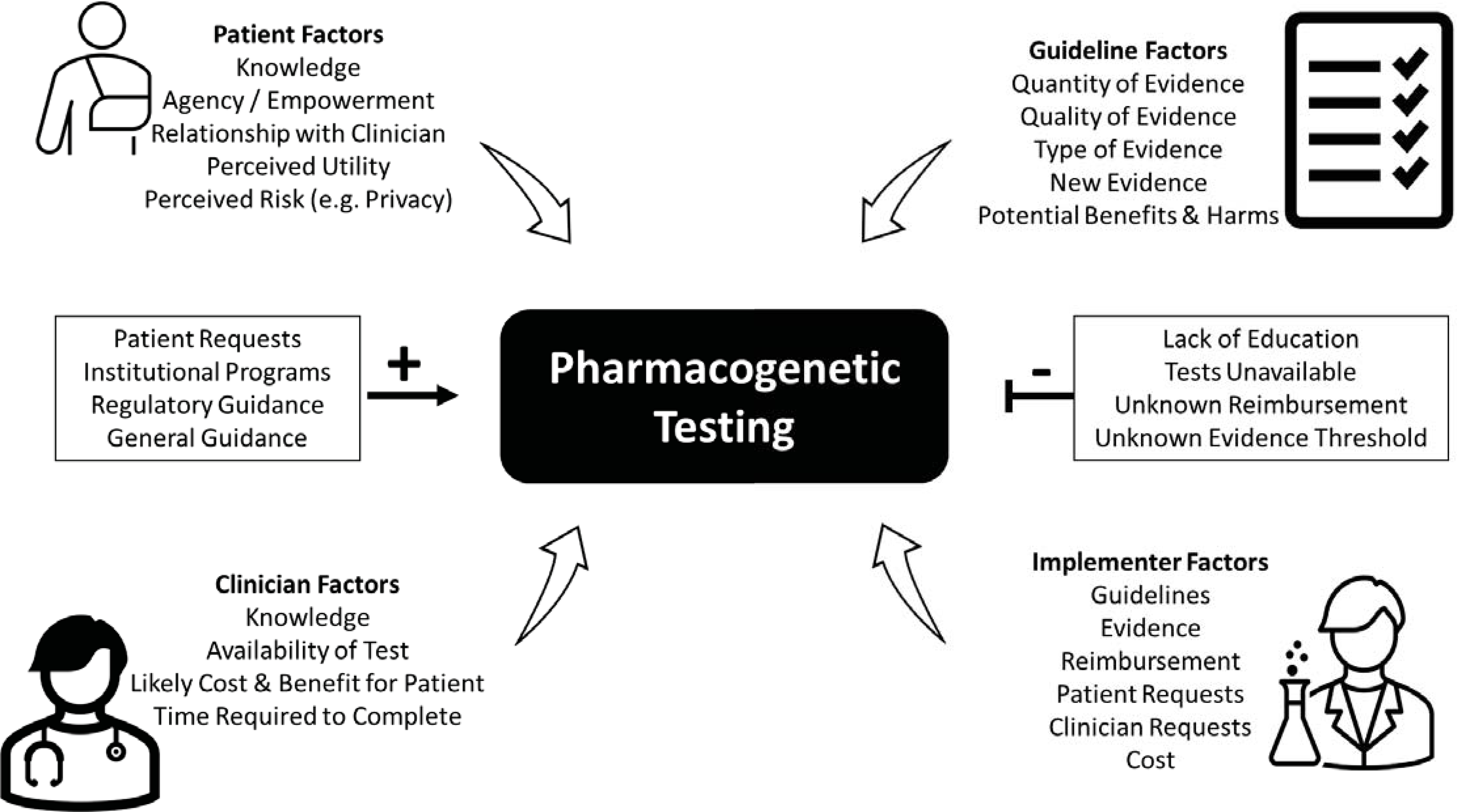
Factors to consider in the clinical implementation of pharmacogenetic testing. Plus sign indicates facilitators, and minus sign indicates barriers.
Pharmacogenetic recommendations in clinical practice guidelines from medical societies are different and controversial. That may explain why practice guidelines from medical societies were not one of the most common determinants of clinical implementation of pharmacogenetic tests in our small survey of implementers. The three most common determinants for current implementers of pharmacogenetics were the availability of CPIC guidelines, consistent evidence of pharmacogenetic associations, and an FDA boxed warning. From the clinician perspective, they must weigh a multitude of variables when making clinical decisions regarding pharmacogenetics, as a single level of evidence cannot be applied to every unique scenario. Examples of the many variables that clinicians must weigh include the severity of the potential clinical outcome, the individual patient’s risk factors, and the effect size and frequency of the genetic variant. Returning to the patient perspective, it is clear that patients also must weigh many variables in decisions on pharmacogenetic testing, especially the cost. Overall, patients believe that pharmacogenetic testing is valuable, and that their providers are key to receiving the most benefit from their pharmacogenomic results. Ultimately, the evidence for the pharmacogenetic tests described in the initially presented patient cases (i.e., tamoxifen and medications for treating depression and GERD) was not supported by the highest level of evidence (i.e., RCTs demonstrating clinical utility). Regardless, the evidence was sufficient for the patients to request pharmacogenetic testing from their providers.
Looking to the future of pharmacogenetics, it is important to recognize that clinical implementation of pharmacogenetics is shifting from individual, reactive tests to panel-based pre-emptive tests. Evidence for panels is showing early potential, and further efforts are underway to demonstrate the clinical utility of pharmacogenetic panels instead of just individual drug-gene pairs. The evidence for each individual gene-drug pair on a panel is still critical, as it will inform which genes and variants should be included in panels. In sum, to move pharmacogenetics into practice, a variety of stakeholders’ perspectives need to be considered, particularly keeping attuned to the voice of the ultimate stakeholder – the patient.
Supplementary Material
Funding Information:
JAL was supported by a NIH K08 award from NHLBI (K08 HL146990). LHC was partially supported by NIH grants U01 HG007269, UL1 TR001427, and R01 HL149752. SLV was partially supported by a Burroughs Wellcome Fund Innovation in Regulatory Science Award #1015006 and the NIH grant U01 HG010232.
Footnotes
Conflict of Interests: AKT is a Technical Director at Laboratory Corporation of America Holdings. All other authors declared no competing interests for this work.
SUPPLEMENTARY INFORMATION
REFERENCES
- (1).Dunnenberger HM et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol 55, 89–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Hicks JK et al. Implementation of Clinical Pharmacogenomics within a Large Health System: From Electronic Health Record Decision Support to Consultation Services. Pharmacotherapy 36, 940–8 (2016). [DOI] [PubMed] [Google Scholar]
- (3).Cavallari LH et al. The IGNITE Pharmacogenetics Working Group: An Opportunity for Building Evidence with Pharmacogenetic Implementation in a Real-World Setting. Clin Transl Sci 10, 143–6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).van der Wouden CH et al. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin Pharmacol Ther 101, 341–58 (2017). [DOI] [PubMed] [Google Scholar]
- (5).Petry N et al. Implementation of wide-scale pharmacogenetic testing in primary care. Pharmacogenomics 20, 903–13 (2019). [DOI] [PubMed] [Google Scholar]
- (6).Luzum JA et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: Outcomes and Metrics of Pharmacogenetic Implementations Across Diverse Healthcare Systems. Clinical pharmacology and therapeutics 102, 502–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Nissen SE Pharmacogenomics and clopidogrel: irrational exuberance? Jama 306, 2727–8 (2011). [DOI] [PubMed] [Google Scholar]
- (8).van der Wouden CH, Swen JJ, Samwald M, Mitropoulou C, Schwab M & Guchelaar HJ A brighter future for the implementation of pharmacogenomic testing. Eur J Hum Genet 24, 1658–60 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Pirmohamed M & Hughes DA Pharmacogenetic tests: the need for a level playing field. Nature reviews Drug discovery 12, 3–4 (2013). [DOI] [PubMed] [Google Scholar]
- (10).Khoury MJ Dealing with the evidence dilemma in genomics and personalized medicine. Clin Pharmacol Ther 87, 635–8 (2010). [DOI] [PubMed] [Google Scholar]
- (11).Frueh FW Back to the future: why randomized controlled trials cannot be the answer to pharmacogenomics and personalized medicine. Pharmacogenomics 10, 1077–81 (2009). [DOI] [PubMed] [Google Scholar]
- (12).Denny JC, Schildcrout JS, Pulley JM & Roden DM Response to “Doubt about the feasibility of preemptive genotyping”. Clin Pharmacol Ther 93, 234 (2013). [DOI] [PubMed] [Google Scholar]
- (13).Huddart R, Sangkuhl K, Whirl-Carrillo M & Klein TE Are Randomized Controlled Trials Necessary to Establish the Value of Implementing Pharmacogenomics in the Clinic? Clin Pharmacol Ther 106, 284–6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Bottorff MB, Bright DR & Kisor DF Commentary: Should Pharmacogenomic Evidence Be Considered in Clinical Decision Making? Focus on Select Cardiovascular Drugs. Pharmacotherapy 37, 1005–13 (2017). [DOI] [PubMed] [Google Scholar]
- (15).Drozda K & Pacanowski MA Clinical Trial Designs to Support Clinical Utility of Pharmacogenomic Testing. Pharmacotherapy 37, 1000–4 (2017). [DOI] [PubMed] [Google Scholar]
- (16).Caudle KE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Relling MV & Klein TE Evidence and resources to implement pharmacogenetic knowledge for precision medicine. Am J Health Syst Pharm 73, 1977–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Gillis NK & Innocenti F Evidence required to demonstrate clinical utility of pharmacogenetic testing: the debate continues. Clin Pharmacol Ther 96, 655–7 (2014). [DOI] [PubMed] [Google Scholar]
- (18).Janssens AC & Deverka PA Useless until proven effective: the clinical utility of preemptive pharmacogenetic testing. Clin Pharmacol Ther 96, 652–4 (2014). [DOI] [PubMed] [Google Scholar]
- (19).Ratain MJ & Johnson JA Meaningful use of pharmacogenetics. Clin Pharmacol Ther 96, 650–2 (2014). [DOI] [PubMed] [Google Scholar]
- (20).Altman RB Pharmacogenomics: “noninferiority” is sufficient for initial implementation. Clin Pharmacol Ther 89, 348–50 (2011). [DOI] [PubMed] [Google Scholar]
- (21).Evans BJ Establishing clinical utility of pharmacogenetic tests in the post-FDAAA era. Clin Pharmacol Ther 88, 749–51 (2010). [DOI] [PubMed] [Google Scholar]
- (22).Woodcock J The human genome and translational research: how much evidence is enough? Health affairs (Project Hope) 27, 1616–8 (2008). [DOI] [PubMed] [Google Scholar]
- (23).Zineh I & Lesko LJ Pharmacogenetics in medicine: barriers, critical factors and a framework for dialogue. Per Med 6, 359–61 (2009). [DOI] [PubMed] [Google Scholar]
- (24).Koch BC, van Schaik RH, van Gelder T, Mathijssen RH & Rotterdam Clinical Pharmacology-Pharmacogenetics, G. Doubt about the feasibility of preemptive genotyping. Clin Pharmacol Ther 93, 233 (2013). [DOI] [PubMed] [Google Scholar]
- (25).Wang B, Canestaro WJ & Choudhry NK Clinical evidence supporting pharmacogenomic biomarker testing provided in US Food and Drug Administration drug labels. JAMA Intern Med 174, 1938–44 (2014). [DOI] [PubMed] [Google Scholar]
- (26).Relling MV, Altman RB, Goetz MP & Evans WE Clinical implementation of pharmacogenomics: overcoming genetic exceptionalism. Lancet Oncol 11, 507–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Relling MV & Evans WE Pharmacogenomics in the clinic. Nature 526, 343–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Woodcock J Assessing the clinical utility of diagnostics used in drug therapy. Clin Pharmacol Ther 88, 765–73 (2010). [DOI] [PubMed] [Google Scholar]
- (29).Munar MY & Singh H Drug dosing adjustments in patients with chronic kidney disease. Am Fam Physician 75, 1487–96 (2007). [PubMed] [Google Scholar]
- (30).Harris LN et al. Use of Biomarkers to Guide Decisions on Adjuvant Systemic Therapy for Women With Early-Stage Invasive Breast Cancer: American Society of Clinical Oncology Clinical Practice Guideline. J Clin Oncol 34, 1134–50 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Breast Cancer. <https://www2.tri-kobe.org/nccn/guideline/breast/english/breast.pdf> (2020). [Google Scholar]
- (32).Goetz MP et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin Pharmacol Ther 103, 770–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Brauch H, Schroth W, Eichelbaum M, Schwab M, Harbeck N & in cooperation with the, A.G.O.T.C. Clinical Relevance of CYP2D6 Genetics for Tamoxifen Response in Breast Cancer. Breast Care (Basel) 3, 43–50 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Dean L Tamoxifen Therapy and CYP2D6 Genotype. In: Medical Genetics Summaries (eds. Pratt VM, Scott SA, Pirmohamed M, Esquivel B, Kane MS, Kattman BL et al.) 501–516 (Bethesda (MD), 2019). [Google Scholar]
- (35).Aromatase inhibitors versus tamoxifen in early breast cancer: patient-level meta-analysis of the randomised trials. Lancet 386, 1341–52 (2015). [DOI] [PubMed] [Google Scholar]
- (36).United States Food and Drug Administration. Table of Pharmacogenomic Biomarkers in Drug Labeling. <https://www.fda.gov/drugs/science-and-research-drugs/table-pharmacogenomic-biomarkers-drug-labeling> (2015).
- (37).United States Food and Drug Administration. Clopidogrel Prescribing Information. <https://www.accessdata.fda.gov/drugsatfda_docs/label/2021/020839s074lbl.pdf> (2017).
- (38).United States Food and Drug Administration. Table of Pharmacogenetic Associations. <https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations> (2020).
- (39).Relling MV & Klein TE CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 89, 464–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Swen JJ et al. Pharmacogenetics: from bench to byte--an update of guidelines. Clin Pharmacol Ther 89, 662–73 (2011). [DOI] [PubMed] [Google Scholar]
- (41).Ross CJ et al. The Canadian Pharmacogenomics Network for Drug Safety: a model for safety pharmacology. Thyroid : official journal of the American Thyroid Association 20, 681–7 (2010). [DOI] [PubMed] [Google Scholar]
- (42).Caudle KE et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab 15, 209–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hewett M et al. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res 30, 163–5 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).United States Food and Drug Administration. Prescribing Information for Tamoxifen. <https://www.accessdata.fda.gov/drugsatfda_docs/label/2018/021807s005lbl.pdf> (2018).
- (45).Caudle KE, Keeling NJ, Klein TE, Whirl-Carrillo M, Pratt VM & Hoffman JM Standardization can accelerate the adoption of pharmacogenomics: current status and the path forward. Pharmacogenomics 19, 847–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Association for Molecular Pathology. Association for Molecular Pathology Position Statement: Best Practices for Clinical Pharmacogenomic Testing. <https://www.amp.org/AMP/assets/File/position-statements/2019/Best_Practices_for_PGx_9_4_2019.pdf?pass=96> (2019).
- (47).PharmGKB. The PharmGKB Blog. <https://pharmgkb.blogspot.com/2020/05/pharmgkb-response-to-fda-table-for.html> (2020).
- (48).American Society of Pharmacovigilance. STRIPE Collaborative Community <https://www.stopadr.org/stripe> (2021). [Google Scholar]
- (49).Knepper TC & McLeod HL When will clinical trials finally reflect diversity? Nature 557, 157–9 (2018). [DOI] [PubMed] [Google Scholar]
- (50).Cavallari LH et al. Multisite Investigation of Outcomes With Implementation of CYP2C19 Genotype-Guided Antiplatelet Therapy After Percutaneous Coronary Intervention. JACC Cardiovasc Interv 11, 181–91 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Deiman BA et al. Reduced number of cardiovascular events and increased cost-effectiveness by genotype-guided antiplatelet therapy in patients undergoing percutaneous coronary interventions in the Netherlands. Neth Heart J 24, 589–99 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Smith DM et al. CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genet Med 21, 1842–50 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Hulot JS et al. Routine CYP2C19 Genotyping to Adjust Thienopyridine Treatment After Primary PCI for STEMI: Results of the GIANT Study. JACC Cardiovasc Interv 13, 621–30 (2020). [DOI] [PubMed] [Google Scholar]
- (54).Brunette CA et al. Pragmatic Trials in Genomic Medicine: The Integrating Pharmacogenetics In Clinical Care (I-PICC) Study. Clin Transl Sci 13, 381–90 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Oslin DW et al. Study design and implementation of the PRecision Medicine In MEntal health Care (PRIME Care) Trial. Contemp Clin Trials 101, 106247 (2021). [DOI] [PubMed] [Google Scholar]
- (56).Ensom MH, Davis GA, Cropp CD & Ensom RJ Clinical pharmacokinetics in the 21st century. Does the evidence support definitive outcomes? Clin Pharmacokinet 34, 265–79 (1998). [DOI] [PubMed] [Google Scholar]
- (57).McInnes GT The value of therapeutic drug monitoring to the practising physician--an hypothesis in need of testing. Br J Clin Pharmacol 27, 281–4 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Spector R, Park GD, Johnson GF & Vesell ES Therapeutic drug monitoring. Clin Pharmacol Ther 43, 345–53 (1988). [DOI] [PubMed] [Google Scholar]
- (59).Tonkin AL & Bochner F Therapeutic drug monitoring and patient outcome. A review of the issues. Clin Pharmacokinet 27, 169–74 (1994). [DOI] [PubMed] [Google Scholar]
- (60).Vozeh S Cost-effectiveness of therapeutic drug monitoring. Clin Pharmacokinet 13, 131–40 (1987). [DOI] [PubMed] [Google Scholar]
- (61).Watson ID & Thomson AH The value of therapeutic drug monitoring to the practising physician--an hypothesis needing sensible application. Br J Clin Pharmacol 28, 619–20 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Regan MM et al. CYP2D6 genotype and tamoxifen response in postmenopausal women with endocrine-responsive breast cancer: the breast international group 1–98 trial. J Natl Cancer Inst 104, 441–51 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Rae JM et al. CYP2D6 and UGT2B7 genotype and risk of recurrence in tamoxifen-treated breast cancer patients. J Natl Cancer Inst 104, 452–60 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (64).Goetz MP, Ratain M & Ingle JN Providing Balance in ASCO Clinical Practice Guidelines: CYP2D6 Genotyping and Tamoxifen Efficacy. J Clin Oncol 34, 3944–5 (2016). [DOI] [PubMed] [Google Scholar]
- (65).Johnson JA, Hamadeh IS & Langaee TY Loss of heterozygosity at the CYP2D6 locus in breast cancer: implications for tamoxifen pharmacogenetic studies. J Natl Cancer Inst 107, (2015). [DOI] [PubMed] [Google Scholar]
- (66).Sanchez-Spitman A et al. Tamoxifen Pharmacogenetics and Metabolism: Results From the Prospective CYPTAM Study. J Clin Oncol 37, 636–46 (2019). [DOI] [PubMed] [Google Scholar]
- (67).Goetz MP, Suman VJ, Nakamura Y, Kiyotani K, Jordan VC & Ingle JN Tamoxifen Metabolism and Breast Cancer Recurrence: A Question Unanswered by CYPTAM. J Clin Oncol 37, 1982–3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Schroth W et al. Association between CYP2D6 polymorphisms and outcomes among women with early stage breast cancer treated with tamoxifen. Jama 302, 1429–36 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Mulder TAM, de With M, Del Re M, Danesi R, Mathijssen RHJ & van Schaik RHN Clinical CYP2D6 Genotyping to Personalize Adjuvant Tamoxifen Treatment in ER-Positive Breast Cancer Patients: Current Status of a Controversy. Cancers (Basel) 13, 771 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).DeCensi A et al. Association of CYP2D6 genotype and tamoxifen metabolites with breast cancer recurrence in a low-dose trial. NPJ breast cancer 7, 34 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Levine GN et al. 2011 ACCF/AHA/SCAI Guideline for Percutaneous Coronary Intervention: a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines and the Society for Cardiovascular Angiography and Interventions. Circulation 124, e574–651 (2011). [DOI] [PubMed] [Google Scholar]
- (72).Claassens DMF et al. A Genotype-Guided Strategy for Oral P2Y12 Inhibitors in Primary PCI. N Engl J Med 381, 1621–31 (2019). [DOI] [PubMed] [Google Scholar]
- (73).Pereira NL et al. Effect of Genotype-Guided Oral P2Y12 Inhibitor Selection vs Conventional Clopidogrel Therapy on Ischemic Outcomes After Percutaneous Coronary Intervention: The TAILOR-PCI Randomized Clinical Trial. JAMA 324, 761–71 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Pereira NL et al. Effect of CYP2C19 Genotype on Ischemic Outcomes During Oral P2Y 12 Inhibitor Therapy: A Meta-Analysis. JACC: Cardiovascular Interventions 14, 739–750 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).National Comprehensive Cancer Network. Acute Lymphoblastic Leukemia. <https://www.nccn.org/professionals/physician_gls/pdf/all.pdf> (2021).
- (76).Relling MV et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Thiopurine Dosing Based on TPMT and NUDT15 Genotypes: 2018 Update. Clin Pharmacol Ther 105, 1095–105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Newman WG et al. A pragmatic randomized controlled trial of thiopurine methyltransferase genotyping prior to azathioprine treatment: the TARGET study. Pharmacogenomics 12, 815–26 (2011). [DOI] [PubMed] [Google Scholar]
- (78).Lichtenstein GR, Loftus EV, Isaacs KL, Regueiro MD, Gerson LB & Sands BE ACG Clinical Guideline: Management of Crohn’s Disease in Adults. The American journal of gastroenterology 113, 481–517 (2018). [DOI] [PubMed] [Google Scholar]
- (79).Shen WK et al. 2017 ACC/AHA/HRS Guideline for the Evaluation and Management of Patients With Syncope: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines and the Heart Rhythm Society. Circulation 136, e60–e122 (2017). [DOI] [PubMed] [Google Scholar]
- (80).Luzum JA & Cheung JC Does cardiology hold pharmacogenetics to an inconsistent standard? A comparison of evidence among recommendations. Pharmacogenomics 19, 1203–16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (81).Shugg T, Pasternak AL, London B & Luzum JA Prevalence and types of inconsistencies in clinical pharmacogenetic recommendations among major U.S. sources. NPJ Genom Med 5, 48 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (82).Rasmussen-Torvik LJ et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clin Pharmacol Ther 96, 482–9 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (83).Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N & Conde JG Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42, 377–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (84).Ballinger TJ et al. Discerning the clinical relevance of biomarkers in early stage breast cancer. Breast cancer research and treatment 164, 89–97 (2017). [DOI] [PubMed] [Google Scholar]
- (85).Arwood MJ et al. Design and Early Implementation Successes and Challenges of a Pharmacogenetics Consult Clinic. Journal of clinical medicine 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (86).Lee YM, Manzoor BS, Cavallari LH & Nutescu EA Facilitators and Barriers to the Adoption of Pharmacogenetic Testing in an Inner-City Population. Pharmacotherapy 38, 205–16 (2018). [DOI] [PubMed] [Google Scholar]
- (87).Mukherjee C, Sweet KM, Luzum JA, Abdel-Rasoul M, Christman MF & Kitzmiller JP Clinical pharmacogenomics: patient perspectives of pharmacogenomic testing and the incidence of actionable test results in a chronic disease cohort. Per Med 14, 383–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (88).Thornley T, Esquivel B, Wright DJ, Dop HVD, Kirkdale CL & Youssef E Implementation of a Pharmacogenomic Testing Service through Community Pharmacy in the Netherlands: Results from an Early Service Evaluation. Pharmacy (Basel) 9, 38 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (89).Bielinski SJ et al. Are patients willing to incur out-of-pocket costs for pharmacogenomic testing? Pharmacogenomics J 17, 1–3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (90).Empey PE, Pratt VM, Hoffman JM, Caudle KE & Klein TE Expanding evidence leads to new pharmacogenomics payer coverage. Genet Med, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (91).Holzer K et al. Hmong participants’ reactions to return of individual and community pharmacogenetic research results: “A positive light for our community”. J Community Genet 12, 53–65 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (92).Lemke AA et al. Patient perspectives following pharmacogenomics results disclosure in an integrated health system. Pharmacogenomics 19, 321–31 (2018). [DOI] [PubMed] [Google Scholar]
- (93).Christian C et al. Pharmacogenomic-Based Decision Support to Predict Adherence to Medications. Clin Pharmacol Ther 108, 368–76 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (94).Truong TM, Lipschultz E, Schierer E, Danahey K, Ratain MJ & O’Donnell PH Patient insights on features of an effective pharmacogenomics patient portal. Pharmacogenet Genomics 30, 191–200 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (95).Drelles K, Pilarski R, Manickam K, Shoben AB & Toland AE Impact of Previous Genetic Counseling and Objective Numeracy on Accurate Interpretation of a Pharmacogenetics Test Report. Public Health Genomics 24, 26–32 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (96).Roden DM et al. Benefit of Preemptive Pharmacogenetic Information on Clinical Outcome. Clin Pharmacol Ther 103, 787–94 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (97).Weitzel KW, Cavallari LH & Lesko LJ Preemptive Panel-Based Pharmacogenetic Testing: The Time is Now. Pharm Res 34, 1551–5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (98).Zhu Y et al. A model-based cost-effectiveness analysis of pharmacogenomic panel testing in cardiovascular disease management: preemptive, reactive, or none? Genet Med, 23, 461–470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (99).Van Driest SL et al. Clinically actionable genotypes among 10,000 patients with preemptive pharmacogenomic testing. Clin Pharmacol Ther 95, 423–31 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (100).Schildcrout JS et al. Optimizing drug outcomes through pharmacogenetics: a case for preemptive genotyping. Clin Pharmacol Ther 92, 235–42 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (101).Saldivar JS et al. Initial assessment of the benefits of implementing pharmacogenetics into the medical management of patients in a long-term care facility. Pharmgenomics Pers Med 9, 1–6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (102).Halverson CM, Pratt VM, Skaar TC & Schwartz PH Ending the pharmacogenomic gag rule: the imperative to report all results. Pharmacogenomics 22, 191–3 (2021). [DOI] [PubMed] [Google Scholar]
- (103).Pratt VM et al. Characterization of 137 Genomic DNA Reference Materials for 28 Pharmacogenetic Genes: A GeT-RM Collaborative Project. J Mol Diagn 18, 109–23 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (104).United States Food and Drug Administration. The FDA Warns Against the Use of Many Genetic Tests with Unapproved Claims to Predict Patient Response to Specific Medications: FDA Safety Communication. <https://www.fda.gov/medical-devices/safety-communications/fda-warns-against-use-many-genetic-tests-unapproved-claims-predict-patient-response-specific#actions> (2018).
- (105).Stanek EJ et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clinical pharmacology and therapeutics 91, 450–8 (2012). [DOI] [PubMed] [Google Scholar]
- (106).Obeng AO et al. Physician-reported benefits and barriers to clinical implementation of genomic medicine: a multi-site IGNITE-Network Survey. J Pers Med 8, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (107).Clinical Pharmacogenetics Implementation Consortium. PGx Dissemination Working Group <https://cpicpgx.org/dissemination/> (2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


