SUMMARY
The accurate identification and quantitation of RNA isoforms present in the cancer transcriptome is key for analyses ranging from the inference of the impacts of somatic variants to pathway analysis to biomarker development and subtype discovery. The ICGC-TCGA DREAM Somatic Mutation Calling in RNA (SMCRNA) challenge was a crowd-sourced effort to benchmark methods for RNA isoform quantification and fusion detection from bulk cancer RNA sequencing (RNA-seq) data. It concluded in 2018 with a comparison of 77 fusion detection entries and 65 isoform quantification entries on 51 synthetic tumors and 32 cell lines with spiked-in fusion constructs. We report the entries used to build this benchmark, the leaderboard results, and the experimental features associated with the accurate prediction of RNA species. This challenge required submissions to be in the form of containerized workflows, meaning each of the entries described is easily reusable through CWL and Docker containers at https://github.com/SMC-RNA-challenge. A record of this paper’s transparent peer review process is included in the supplemental information.
In brief
The SMC-RNA Challenge benchmarked isoform quantification and fusion detection methods. Challenge participants submitted CWL workflows made up of containerized methods to the challenge administrators who then ran the code on held-out samples never available to contestants. For the Fusion Detection sub-challenge, Arriba and STAR-Fusion were identified as top performers.
INTRODUCTION
While only a small fraction of the genome encodes proteins, the majority is either transcribed or has putative regulatory functions, with the consequence that cellular functions are extensively regulated at the RNA level. The regulation of RNA, and its dramatic dysregulation in cancer cells, occurs in multiple ways. RNA abundances of certain spliced products may be altered and these have served as the basis for clinically important prognostic biomarkers. RNA sequencing (RNA-seq) uses sequencing techniques to detect and quantify specific RNA isoforms. These isoforms can derive from the same gene but differ in many ways, including through alternative splicing, by germline or somatic variation on any allele, or through the generation of novel fusion transcripts. The raw read counts from an RNA-seq study can be used to estimate transcript abundances, and from it elucidate other biologically relevant information. Traditional protocols for RNA-seq involve reverse transcription into cDNA, which is then sequenced using high-throughput technologies, such as Illumina HiSeq, Roche 454, or PacBio (Metzker, 2010). After sequencing, reads can be assembled de novo, aligned to a reference genome, or aligned to a reference transcriptome. Some key challenges in RNA-seq include biases occurring in RNA fragmentation, cDNA fragmentation, and library preparation, in addition to, potential polymerase chain reaction (PCR) artifacts that skew estimated abundances and possible alignment to multiple locations in a reference genome (Han et al., 2015). Many of these same artifacts remain for the more recent task of interpreting RNAs from individual cells (i.e., with single-cell RNA-seq platforms). Due to these and other influences, methods for detecting and quantifying transcriptional isoforms and fusion products remains an important task.
Genomic rearrangements in cancer cells produce fusion transcripts, which may give rise to protein products not present in normal cells. These can serve as robust diagnostic markers, e.g., TMPRSS2-ERG in prostate cancer (Tomlins et al., 2008) or drug targets, e.g., SET-NUP214 in acute T-lymphoblastic leukemia (Mohseni et al., 2018). Ongoing research efforts are beginning to unveil the potential clinical relevance of aberrant processing of RNA in cancer, such as defects in alternative splicing. An assortment of computational methods is needed to fully document the transcriptomic differences between tumor cells and their normal counterparts. Cataloging the “alterome” of tumors by fully characterizing their RNA landscapes will expand our understanding of cancer mechanisms, provide new biomarkers, and reveal possible new RNA-based therapeutics, improving personalized patient treatment.
Gene fusions occur when two genes are joined through a DNA translocation, interstitial deletion, or chromosomal inversion. Trans-splicing events can also occur in which two transcripts are fused (Zaphiropoulos, 2011). Gene fusions often have an important role in the initial steps of tumorigenesis. Specifically, gene fusions have been found to be the driver mutations in neoplasia and have been linked to various tumor subtypes. An increasing number of gene fusions are being recognized as important diagnostic and prognostic parameters in malignant hematological disorders and childhood sarcomas. Reviews have estimated that gene fusions occur in all malignancies and that 16.5% of human cancer cases harbor at least one driving RNA fusion event (Gao et al., 2018).
Isoforms are alternative combinations of exons combined into a transcript formed from splicing during post-transcriptional processing. Dysregulation of alternative splicing occurs in every one of the hallmarks of cancer (Hanahan and Weinberg, 2000, 2011). Modifications in splicing may occur due to mutations of cisacting splicing elements, trans-acting regulators, and microRNAs. Moreover, the switch from one isoform to another in cancer cells leads to functional consequences and measurable differences in patient outcomes, especially when observed in multiple tumor types (Vitting-Seerup and Sandelin, 2017).
The goal of the ICGC-TCGA DREAM SMC-RNA Challenge was to use a crowd-based competition to identify optimal method(s) for quantifying isoforms and detecting mRNA fusions from RNA-seq data. Several methods have been developed to detect and quantify cancer-associated RNA species abundance. It is not clear which methods are best used and in what contexts. However, the evaluations published in these studies may suffer from the well-known “self-assessment trap,” as the benchmarking includes one of the tools developed by the evaluators. The challenge we describe evaluated workflows composed of one or more methods for two separate sub-challenges using an objective approach. The Fusion Detection sub-challenge measured performance in detecting cancer-associated fusions at any expression level while the Isoform Quantification sub-challenge measured performance in predicting the relative level of each transcript across samples. For each sub-challenge, an unbiased assessment was conducted by using a combination of computationally simulated in silico RNA sequences as well as experimentally generated in vitro RNAs. All submissions were run by the challenge administrators so that contestants never had access to evaluation datasets. Participants submitted to the administrators their trained model including their workflows (composed of one or more methods), parameters, and environments needed for execution using Docker and Common Workflow Language (CWL) definitions. The challenge ran for seven months, with 221 participants comprising of 17 teams that submitted 65 entries for the Isoform Quantification sub-challenge and 82 for the Fusion Detection sub-challenge. Participants were able to submit up to three trained models to include in the evaluation for the leaderboards. Submissions were run on the Institute for Systems Biology Cancer Genomics Cloud with resulting predictions stored and evaluated. Performance metrics of the evaluated submissions were used to generate leaderboards for each round of the challenge. Notably, because the administrators had access to all of the results, we were able to investigate possible explanations leading to algorithm failure.
For the Fusion Detection sub-challenge, two methods—Arriba (https://github.com/suhrig/arriba) and STAR-Fusion (Haas et al., 2017)—outperformed all others submitted. Both of these align the transcriptome using STAR (Dobin et al., 2013) and use “chimeric reads” as the basis for identifying potential fusion junction sites. Further, both emphasize the importance of using filters to detect bona fide fusions from myriad background fusions. In these analyses, junction coverage and abundance were the most important influences upon false negatives, while GC content and the total number of alternate gene isoforms contributed most to false positives. For the Isoform Quantification sub-challenge, no submissions outperformed standard approaches used to initialize the leaderboard. We found methods that had the most error in distinguishing between isoforms for a few genes when spiked-in levels differed by 2-fold compared with 5-fold.
RESULTS
The SMC-RNA challenge included two sub-challenges: Fusion Detection and Isoform Quantification. For these sub-challenges, in silico simulated and in vitro-derived spike-in datasets were designed for use in evaluating entries (Figure 1). To generate simulated data, a custom pipeline called rnaseqSim was created to simulate RNA-seq reads that mimic several realistic aspects of biology and current technology such as uneven read coverage across a transcript, the insert size distribution, GC content biases, and the presence of possibly different haplotypes produced from a diploid genome (STAR methods, isoform and fusion simulation pipeline). The final test set contained an in vitro benchmark of 6 cell lines with 5 replicates each, with varying cell line backgrounds, transcript or fusion construct spiked-ins, and spike-in concentrations (Table S7 and STAR methods, spike-in fusion construction, benchmark transcript selection). The data and various quality estimates are available on Synapse(https://www.synapse.org/Synapse:syn22344794). The spike-in design varied in complexity across samples and included multiple isoforms from the same gene (from 1 up to 3) as well as different levels of the transcripts and number of fusion events (Table S8 and STAR methods, spike-in fusion construction, and benchmark transcript selection). Participants were required to submit two components for the challenge: a Docker image encapsulating their code, executables, and environment, and a CWL workflow to define the steps and parameters for running their algorithms. The resulting output of each entry also had to meet the format specifications published on the challenge website. For both sub-challenges, participants were allowed to submit any number of entries but were restricted to selecting up to three entries for scoring on the official leaderboard. Participants could choose to have these three entries be based on different algorithms or optimize the same algorithm with different trained parameters. The performances of entries for both the Fusion Detection and the Isoform Quantification sub-challenges were benchmarked against constructs spiked into the cell line-based samples. For the Fusion Detection sub-challenge, we identified two entries, using the methods Arriba and StarFusion, that were better than all others. For the Isoform Quantification sub-challenge, there was no top entry and none of the participant submissions outperformed the challenge organizer based submissions.
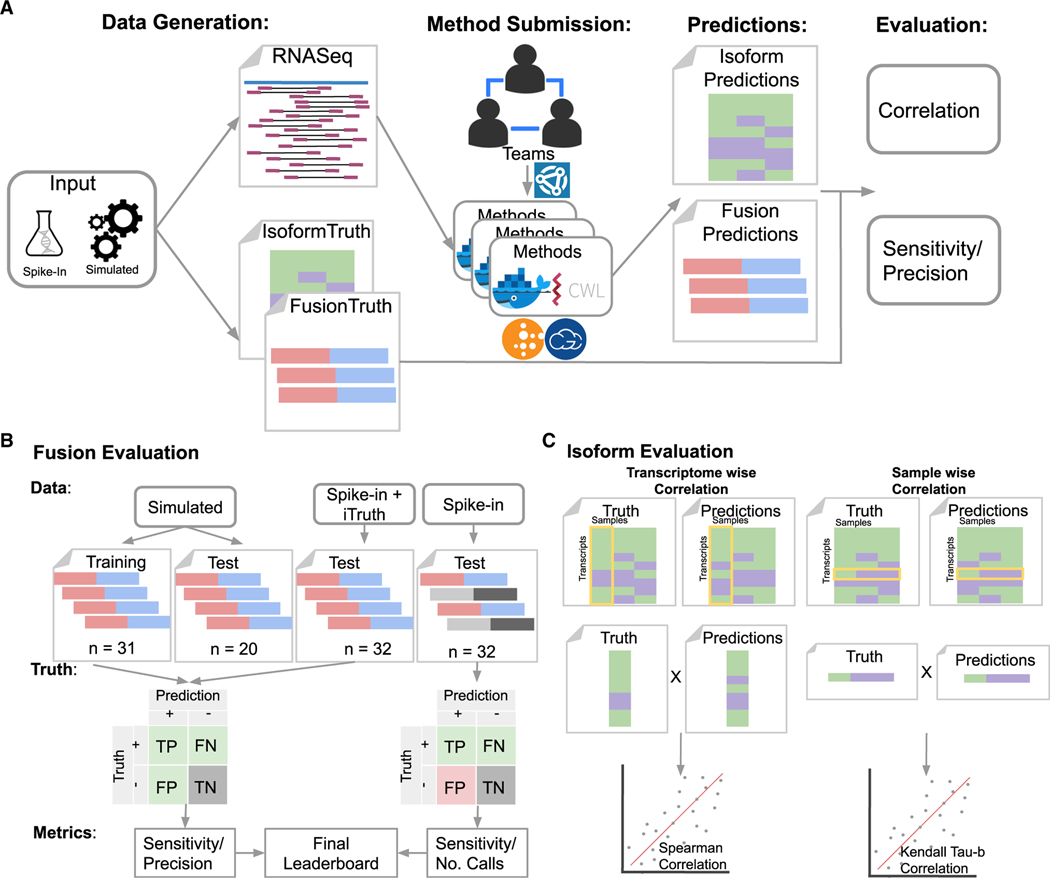
Figure 1. Overview of the challenge.
(A–C) The challenge generated simulated (or in silico) and spike-in datasets represented as RNA-seq reads (FastQ files) and ground truth. Challenge participants could submit entries (i.e., CWL workflows and Docker images) as individuals or teams using Synapse. Submitted entries were run on the FastQ files using cloud-based compute resources to generate predictions. The resulting predictions were evaluated based on statistical performance measurements. Evaluation of the Fusion Detection sub-challenge (B) used four types of input datasets to calculate sensitivity and either precision or the total number of fusion calls. Datasets where the fusion genes are known are represented as red (5’ donor) and blue (3’ acceptor), and datasets where unknown fusion genes may exist are represented as light and dark gray. The confusion matrix displays the known (green), unknown (red), and irrelevant (gray) parameters used to calculate the subsequent statistical metrics. Evaluation of the isoform quantification sub-challenge (C) used two metrics for evaluating the correlation of predictions to the truth. The transcriptome-wise evaluation compared predictions and truth in a single sample across all transcripts using a Spearman correlation. The sample-wise evaluation compared predictions and truth for a single transcript across multiple sample replicates using Kendall’s tau-β.
Fusion detection sub-challenge results
The Fusion Detection sub-challenge evaluation received 77 entries, of which the organizers were able to execute and evaluate 35 (Table S1). The majority of entries failed due to an ill-formed submission (submission error, 26 entries), followed by the packaged code running into problems during execution (workflow error, 13 entries), and lastly, a few cases produced output that was unable to be properly evaluated (evaluation error, 3 entries). Of the successful 35 entries, 17 represented valid entries after restricting submissions to allow up to three from any one team as specified by the challenge rules. Fusion detection entry workflows were often composed of two steps to first align sequence reads followed by fusion detection and calling. Examples of commonly used alignment methods included STAR and GSNAP and fusion detection methods included STAR-Fusion, STAR-SEQR, Arriba, FusionRnadt, and Hera (Table S1 includes the full list of methods submitted).
Two different datasets were created to evaluate entries, a computationally simulated dataset and an experimentally generated set using spike-ins (Figure 1B). The simulated dataset was used to evaluate entries in the preliminary rounds. The simulated data were generated with the program rnaseqSim (https://github.com/Sage-Bionetworks/rnaseqSim) that created reads from computationally constructed fusions. On average, the simulated tumor samples contained 39 fusions per transcriptome, ranging from 3 to 100 to test how callers reacted to various levels of signal. This number is in line with those reported for several popular cell lines (Picco et al., 2019).
A second evaluation dataset of spiked-in fusions was used for the final assessment of entries. The spike-in data were created in the lab using a predefined series of 18 fusion products, formed between arbitrarily selected gene partners. The RNA from each of 6 different cell lines was aliquoted into 5 replicates, 4 of which were spiked with designed quantities of synthetic fusion RNA. Lung, ALL, prostate, and breast cancer cell lines were used. Each fusion was introduced at an amount of either 0, 5, 25, or 50 pg. The 5th replicate was spiked only with 20-μL nuclease-free water to act as a negative control. Three technical replicates were made for one of the HCC1143 cell line’s spike-in designs by splitting the cell line’s RNA into three aliquots prior to adding the same spike-in mixture to each.
The spike-ins provide a basis for evaluating the methods. On the one hand, if a method fails to detect a fusion construct known to be added at a particular level in a sample, we call this event a fusion false negative (FFN). On the other hand, methods that report on a fusion that was not spiked into a sample, but are nonetheless reported by a method, are labeled as a fusion false positive (FFP). Notably, FFPs could result from the existence of transcripts in the cell line’s background. For such naturally occurring transcripts, we expect many (or all) of the methods to detect their presence. Thus, we introduced a correction to the evaluation that extends the truth set (see discussion of i-Truth below). As far as FFNs, we found that a majority of the fusions included in the spike-in experiment were detected at similar rates, with only three fusion constructs missed by more than half of the methods. We also looked into extreme cases to determine whether any properties influence detection difficulty. One such extreme case is the designed fusion of interleukin-15 (IL-15) and IL-21 that was never detected by any of the methods. The construct appears to have been synthesized correctly as we verified by manual inspection the presence of junction-spanning reads. We suspect that the homology between IL-15 and IL-21 lead to detection failures. Either the mapping step misaligned the relevant junction-spanning reads, or the methods themselves filtered these reads out (ironically, methods often exclude junctions spanning homologous genes to remove a major source of mapping misalignment noise). We next discuss our approach to systematically evaluate the accuracy of the methods using the spike-in designs that include estimates of both types of errors, FFNs, and FFPs.
On the one hand, it is straightforward to estimate the sensitivity of an entry as the fraction of spike-in controls reported. On the other hand, it is not as obvious how to estimate precision or specificity due to the possibility that true fusions exist outside the spike-in set because any naturally occurring fusions present in the cell lines would also be detected by contesting algorithms. One approach would be to use a long-read technology that could detect the native constructs. We found that current read depths of a Nanopore-based long-read approach were insufficient to accurately detect the presence of fusions. Inspired by recent work in the area (Ahsen et al., 2018), we instead estimated a set of “imputed truth” (i-Truth) fusions from the calls made by the entries (STAR methods, imputing an extended truth dataset for fusion evaluation). Along with spiked-in controls, predicted fusions were considered as positives for evaluation if several callers detected them in the replicates of the same cell line background. To this end, a “meta caller” was created to combine the submitted predictions into a consensus score, made up of the proportion of callers voting in favor of the presence of a particular fusion in a specific sample. If the consensus score exceeded a critical threshold, then a fusion event was considered as good as truth and included in the i-Truth set. Assuming the i-Truth contains bona fide fusions, the recall of the entries should be similar to when they are run on the actual truth, i.e., on the spike-ins. Using this reasoning, we set the critical threshold such that the recall measured using the i-Truth matched the recall measured using the spike-ins (STAR methods, imputing an extended truth dataset for fusion evaluation). This produced an i-Truth set containing 48 predicted fusions, ranging from 2 up to 17 fusions in every cell line (Tables S8 and S12). This set offered a notable increase in the number of events to gauge entry performance compared with using the spike-ins alone.
Including even a small proportion of erroneous events as truth could detrimentally affect the ultimate ranking of entries. We, therefore, estimated the accuracy of the i-Truth by querying several cancer-specific fusion databases including the Broad’s cancer cell line encyclopedia (CCLE) database for the presence of the i-Truth fusions in cancer cell lines as well as several other databases documenting fusions in normal tissue including the GTEx dataset of fusions in normal tissue to confirm their absence in non-cancerous tissue (Table S8). Remarkably, 28 (61%) of the i-Truth fusions had evidence for the existence of the exact breakpoint in the predicted cell line based on the CCLE collection. Of the remaining i-Truth fusions, another 10 (22%) had evidence that the 3’ and 5’ partner genes participated in a fusion in the same cell line, albeit with different breakpoints. The remaining 10 (22%) had no evidence of either breakpoint being present in the CCLE collection. Of these 10, 6 were found to have partial matches in either the ChimerSeq or TumorFusion guanosine diphosphate (GDP) collection. Thus, altogether, 44 out of the original 48 (92%) had either an exact or inexact match in existing fusion databases. Reassuringly, none of the i-Truth fusions were found recorded in normal tissue databases, reflecting their cancer specificity. Encouraged by the documented existence for all of the i-Truth fusions, each of the predicted events was combined with the spike-ins to create an extended truth set.
To create the final leaderboard, all submitted entries were ranked by their F1 scores based on their performance predicting fusions included in the extended truth benchmark (spike-ins plus i-Truth; Figure 1B). F1, an average of precision and recall, was chosen because limiting the number of extraneous predictions is just as important as predicting known fusions since only a few options can be considered in cancer treatment due to factors like time and cost. Two of the submitted entries/methods emerged as the overall winners of this sub-challenge, Arriba (F1 = 0.73) followed by STAR-Fusion (F1 = 0.70) (Table S3). These winners were followed by other lower-ranking entries that were found to be statistically lower in score based on bootstrap resampling (STAR methods). The third highest ranked entry was a variation of the STAR-Fusion method (F1 = 0.63), followed by fusioncatcher (F1 = 0.58) contributed by this challenge’s organizers, then STAR-SEQR (F1 = 0.47).
Features influencing the accuracy of fusion detection
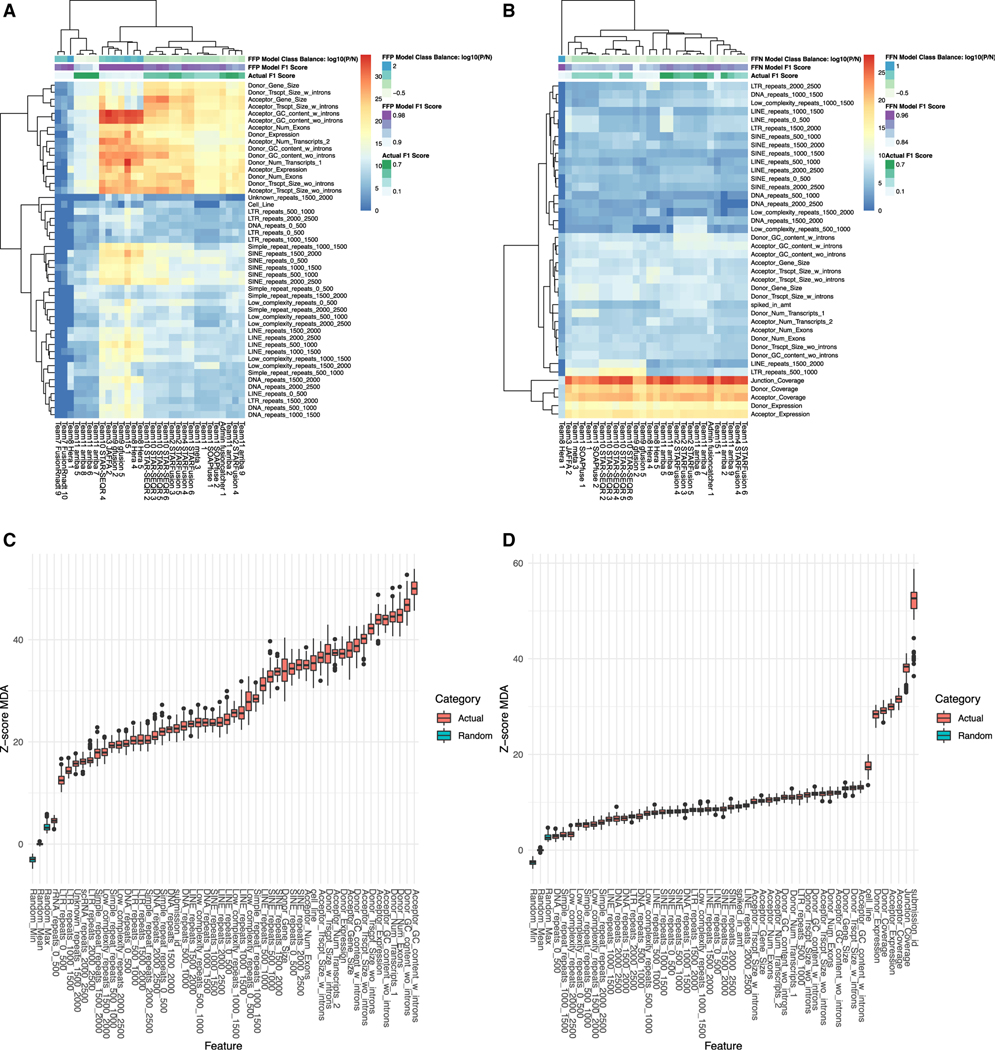
To determine what factors influence entries to incorrectly call fusion events, we created a fusion feature importance pipeline, similar to what was done for the ICGC/TCGA DREAM SMCDNA challenge (Lee et al., 2018). We collected 128 genomic features related to each predicted fusion event, including gene length, transcript length, distance from the breakpoint to repeats, and the abundance for each fusion partner. To identify features predictive of error across cell lines and entries, the cell line and submission identifier were also included as features to account for those covariates. The full list of features is recorded in Table S9. Next, we built a random forest (RF) classification model to predict FFPs from each submission. In other words, the RF model was trained to select features that predict when an entry erroneously calls a fusion event when no such event was present according to the extended truth. We built a second RF model to select features that predict FFNs; i.e., the RF predicts when an entry fails to detect a spiked-in fusion construct. To quantify feature importance for each of our classification models, we applied the Boruta feature selection algorithm to the RF models (Figures 2A and 2B) (Degenhardt et al., 2019; Kursa and Rudnicki, 2010). Boruta determines feature relevance by comparing the original importance with the importance achievable at random, estimated using permuted versions of a feature, and progressively eliminates insignificant features to stabilize a test statistic. An accurate FFP model was obtained that achieved an out-of-bag error rate of 0.26% (see resource table for links to SMC-RNA-Eval code). The FFN model had lower, but still respectable, accuracy due to fewer observations, achieving an error rate of 7.64%. The Boruta algorithm revealed that the number of transcripts and GC content were the most important features for determining FFPs among all fusion prediction methods whereas submission identifier, coverage across the junction and abundance were the top features for FFNs (Figures 2C and 2D). We speculate that the informative GC feature could reflect the presence of low complexity repeats influencing hybridization efficiency or alignment problems in the area of the predicted fusions. Further analysis for FFNs revealed a marked decrease in coverage and abundance as additional top features (Figure S1A).
Figure 2. Boruta feature importance analysis across by fusion submissions.
(A–D) A heatmap showing results from performing the Boruta algorithm on each submission’s false-positive fusion events (A) and false-negative fusion events (B). Each cell in the heatmap represents the Z score mean decrease in accuracy. Higher Z scores are in red and represent more important features. Rows are the fusion submission names and columns are the features. Only features that had a mean value greater than Boruta’s shadow maximum value are shown. Boxplots showing results from performing the Boruta algorithm on all Fusion Detection sub-challenge submissions. (C) is the importance analysis against false positives and (D) is against the false negatives. The y axis represents the Z score MDA and features are across the x axis. The red plots are the Z scores of the actual features and blue are Boruta’s shadow features, which are considered the randomized background features. Only features that performed better (p < 0.05) than the random features are shown in this plot.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
|
| ||
| SpikeIn sequencing Data | https://www.synapse.org/Synapse:syn22344794 | N/A |
|
| ||
| Experimental Models: Cell Lines | ||
|
| ||
| PC-3 | Joe W. Gray Lab | N/A |
| Jurkat I 9.2 | Joe W. Gray Lab | N/A |
| HCC1143 | Joe W. Gray Lab | N/A |
| LNCapFGC | Joe W. Gray Lab | N/A |
| PC-9 | Joe W. Gray Lab | N/A |
| A549 | Joe W. Gray Lab | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| Fusion Spike-in DNA templates | Integrated DNA Technologies | N/A |
|
| ||
| Software and Algorithms | ||
|
| ||
| rnaseqSim | https://github.com/Sage-Bionetworks/rnaseqSim | N/A |
| SMC-RNA-Eval | https://github.com/smc-rna-challenge/SMC-RNA-Eval | N/A |
Isoform quantification sub-challenge results
For the Isoform Quantification sub-challenge, we received 65 submissions, of which 32 were successfully executed to completion through the leaderboard evaluation pipeline. Of the 32 that were successful, 16 were included in the final leaderboard (Table S4). Entries failed for several reasons including submissions of the wrong format (23 Submission Errors), incompatibility with the runtime system based on the CWL (8 workflow errors), and two that were runnable but produced ill-formatted output for evaluation (Table S2).
A diverse set of algorithms were submitted to the challenge representing two major classes of isoform quantification approaches: alignment-based workflows (e.g., STAR and RSEM) and hashing-based workflows (e.g., Kallisto and Salmon). Common components of the entries (i.e., workflows) submitted for the Isoform Quantification sub-challenge included STAR (Dobin et al., 2013), Kallisto (Bray et al., 2016), Salmon (Patro et al., 2017), Hera, RSEM (Li and Dewey, 2011), GSNAP(Wu et al., 2016), eXpress (Roberts and Pachter, 2013), Cufflinks, and Flux-Capacitor. We considered transcriptome-wise and sample-wise evaluation of the results (Figure 1C and STAR methods, Evaluating Isoform Quantification). Transcriptome-wise correlation (TWC) measures the degree to which the levels of a transcript relative to other transcripts in the same sample match the known set. On the other hand, sample-wise correlation (SWC) measures how well the level of a transcript matches relative to the same transcript across different samples. TWC reflects the ability of an entry to estimate dominant splice forms from others while SWC measures the accuracy for use in differential abundance analysis when sample subgroups are compared. An evaluation using the computationally simulated data found that the top-performing entry was based on RSEM using TWC as a measure (Figure S2D). Similar results were obtained when SWC was used (data not shown). However, since the simulation program itself invokes RSEM to generate FASTQ reads, this result could indicate a systematic bias and not reflect the accuracy of entries when run on real tumors.
For this reason, we compared entries using a spike-in dataset and a non-parametric comparison of submissions (STAR methods, benchmark transcript selection). Submissions were evaluated against a set of 20 synthetic tumors and a panel of six cell lines with 18 native transcripts spiked in at different levels. As was done for fusions, the same six cell lines were used to introduce four different spike-in designs plus a negative control (no spike-in), and a technical replicate was created for one of the HCC1143 designs. The transcripts were selected from genes exhibiting expression levels at or below that of background across a mix of breast cancer cell lines and tumors.
For technical reasons related to manipulating spike-ins, we used SWC for the evaluation because the relationship between the abundance spiked into the number of sequenced reads scripts spiked in at the same concentrations may not show a could differ from one gene to the next, adding an appreciable comparable number of reads due to sequencing efficiencies amount of noise to a calculation of TWC. For example, two tran- that may vary from transcript to transcript (e.g., potentially, though not necessarily, due to causes such as GC content or differing hybridization efficiencies of the probes). On the other hand, results from our pilot studies suggest that the relationship could be much more comparable for a particular transcript from one sample to the next (data not shown).
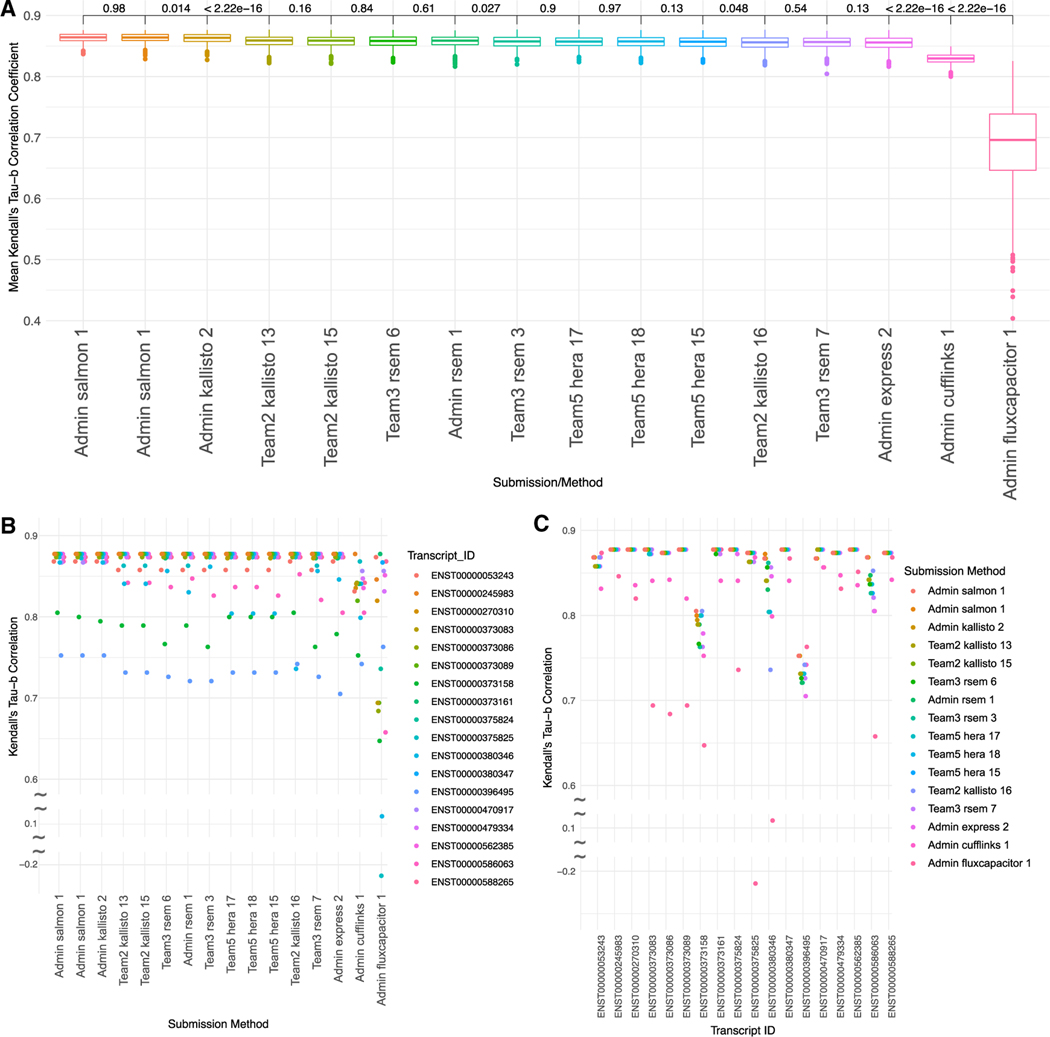
To this end, we calculated Kendall’s Tau-β correlation for each transcript for each submission that measured the agreement of ranking between the predicted and actual levels across all of the cell lines. The final Kendall’s Tau-β score (KTBS) for a submission was then determined by taking an average across all of these transcript-specific correlation values. Standard deviations for each entry were obtained by creating bootstrap replicates (see STAR methods).
Despite the range of different methods included in the benchmark, the spike-in based evaluation failed to identify a clearly superior entry. The top 14 entries, covering pipelines including Kallisto, Salmon, RSEM, Hera, and Express, all had statistically indistinguishable scores within a span of 8×10−3 of each other, with a standard deviation across all submissions of 4.16 × 10−2. The two top-performing entries were based on Salmon and submitted by the challenge organizers followed by another organizer-submitted version of Kallisto, scoring only slightly worse (p = 0.043) (Figure 3A). Of the entries submitted by challenge participants, the best performing entry was based on Kallisto, followed closely by RSEM and Hera. Entries submitted by the challenge organizers were not considered for deciding the challenge winner. However, because of the lack of separation between the top participant-submitted entries evaluated using spike-in controls, no winner was declared for the Isoform Quantification sub-challenge.
Figure 3. Isoform abundance Kendall Tau-β correlation coefficient bootstrap.
(A–C) Ranking of methods based on their performance in predicting isoform levels as measured by 1,000 bootstrap replicates of the Kendall Tau-β score (KTBS) (see STAR methods). The x axis represents the submissions and the y axis the KTBS. Each boxplot represents the 1,000 mean Tau-β scores for each bootstrap. Results of the Student’s t test for closely ranked submissions shown between boxplots. Values greater than 0.05 were considered as ties between submissions. (B and C) Kendall’s tau-β correlation by transcript and submission method. Plots show Kendall’s tau-β correlation coefficient for each transcript with Submission ID across the x axis (B) or transcript across the x axis (C). The color corresponds to the feature in the legend.
Features influencing the accuracy of isoform detection
Although no entries emerged as a leading approach for this sub-challenge, we investigated the influence of various aspects of the data on calling accuracy, to determine whether particular callers might be more accurate under certain circumstances. First, we attempted to identify any genomic features influencing the abundance estimates of the entries. We investigated transcript length, gene size, number of exons, and GC content, but did not find any correlation with the rankings of transcripts among the entries (data not shown). We note that while we expect transcript size and exon count to be inversely related to the accuracy, the spike-in design used in the challenge was likely too simplistic to reveal such dependencies.
Next, we analyzed, which spike-in quantities were misordered as part of the discordant pairs influencing the KTBS (STAR methods, Evaluating Isoform Quantification). Transcripts were spiked at 0, 5, 25, and 50 pg (see “spike-in fusion construction” section in STAR methods). Interestingly, there was an overwhelming majority of incorrectly predicted orderings between the 25- and 50-pg pairs, not only across transcripts (Figure S2B) but also across cell line pairs (Figure S2C). The entries may have more difficulty in quantifying the 25:50 comparison either because the difference is merely doubled, whereas all of the other pairs have at least a 5-fold difference, or the spike-in amount is already saturated at the 25-pg level. For the other relative pairwise rankings—0:50, 0:25, and 5:50—there were no incorrect pairs among any of the entries. Since there was an obvious bias toward the incorrect 25:50 pairs in only a few transcripts, we re-ran the ranking after removing those pairs and we found the ranking to be even less discerning between submissions. In fact, most submissions tied for first place with only submissions using Kallisto and Cufflinks coming in last.
DISCUSSION
The winning submissions for the Fusion Detection sub-challenge, based on Arriba and STAR-Fusion, implement several strategies that may contribute to their superior performance over other approaches. Both entries make use of filtering strategies to eliminate potentially thousands of artifacts from true fusions among the chimeric reads found in RNA-seq alignments. Arriba identifies three types of false positives: alignment artifacts, in vitro-generated artifacts, and benign transcripts, which are erroneously classified as aberrant due to the incomplete annotation of genes. Alignment artifacts are mediated by sequence homology in the genome, causing reads to be mapped to the wrong locus, or by regions posing challenges to short-read aligners, such as homopolymers, tandem repeats, and loci subject to somatic hypermutation. By discarding reads with low sequence complexity, an excessive number of mismatches, or segments aligning to homologous genes, Arriba eliminates such spurious alignments. A substantial amount of artifactual chimeric fragments are produced in vitro during reverse transcription (Houseley and Tollervey, 2010) and amplified by the PCR step of library preparation. These artifacts are effectively reduced by ignoring PCR duplicates and by requiring a higher number of supporting reads with an increasing level of a gene’s background noise, estimated as the total number of fusion candidates involving that gene. Many benign transcripts are not annotated by available gene models, including circular RNAs, trans-splicing, read-through fusions, and alternative promoters. Such transcripts give rise to chimeric reads, which are hard to distinguish from reads originating from aberrant transcripts and may thus lead to false-positive fusion predictions. Such benign transcripts are discarded by Arriba with the help of a blacklist trained on samples from normal tissue. While false-positive filtering certainly helped top-performing entries, we found that maintaining sensitivity was just as important to maintain accuracy. STAR-Fusion applies similar ideas, but with some small differences. For dealing with likely mismappings, reads with low complexity and paralogous sequences are excluded. To deal with the PCR artifact issue, STAR-Fusion requires more evidence, quantified by the number of supporting reads, for breakpoints that fail to match reference annotation splice sites. In the version of STAR-Fusion used for the competition, read mappings that anchored to regions of transcripts that matched the repbase repeat library (Bao et al., 2015) were excluded. STAR-Fusion uses a filter to remove “promiscuous” fusion calls. These calls are characterized when a fusion gene partner “A” has multiple partners, e.g., “A–B”, “A–C”, and “A–D”. Finally, STAR-Fusion utilizes a blacklist of “red herrings,” including fusions recurrently seen in normal data sets, which could be the result of trans-splicing or other artifact-producing processes. Various additional elements are considered for removal as part of filtering steps for these top two methods (see Table S10).
If filtering out false positives is truly what separates the performance of entries, then one would expect the variability in false-positive calls across these entries to be higher than for true-positive calls. For example, in the extreme case that all entries find the same fusions but differ in the number of false positives, they would have the same sensitivity, i.e., zero standard deviation in the true positive rate (TPR) but a non-zero standard deviation in the false discovery rate (FDR). In fact, we do find this trend among the top-performing entries—where top entries are defined as the nine submissions with F1 at least 0.25—in which the standard deviation of the TPR is 0.106 and nearly twice as high for the FDR, 0.201 (Table S3). For example, the ninth versus the first report a similar number of true positives (448 versus 423, respectively) while the ninth called four times as many total fusions (2,465 versus 619). However, when considering all of the submissions, the variability is much more comparable and the relationship reversed, with the TPR standard deviation was calculated to be 0.310 and the FDR standard deviation was calculated to be 0.265. This suggests that the top-performing entries distinguish themselves from poorer-performing entries by maintaining both high sensitivity and precision. Whereas, when considering the top-performers among themselves, additional improvements were obtained by controlling the FDR possibly due to the benefits of the employed filtering strategies.
The challenge utilized the “Model to Data” approach (Ellrott et al., 2019; Guinney and Saez-Rodriguez, 2018), where participants produced and shipped a functional prediction model to the challenge organizers that could be run on held-out data. There are many advantages to this setup beyond avoiding the transfer of large data files. Notably, participants never saw the final testing data set. Instead, the organizers provided simulated training datasets to allow participants to run their model, check their compatibility of output, estimate performance, and make adjustments as needed. Administrators ran containerized workflows on behalf of participants that specified all parameters needed for execution and thus all data remained protected. For example, the same set up could be used to preserve patient privacy in those cases where the evaluation data contain such sensitive information. All entries ranked on leaderboards are reproducible, rerunnable, and able to be distributed to the community for further analysis. For example, we expect subsequent efforts to create better fusion detectors may come from the investigation of “wisdom of crowds” ensembles (Marbach et al., 2012) that combine the strengths of the methods. The portability has allowed the top-performing fusion methods to be adapted into the NCI’s genomic data commons (GDC) workflow system and deployed across several large datasets. Methods profiled by this benchmarking effort were used to generate fusion calls on the NCI’s TARGET dataset and were included in release 25.0 of the GDC dataset. Future work datasets profiled with these methods will also include the BeatAML and CPTAC cohorts.
Recent systematic comparisons have been performed to evaluate RNA-seq analysis methods (Kanitz et al., 2015; Teng et al., 2016; Zhang et al., 2017). Kumar et al. (2016) conducted an impartial survey of 12 different methods based on their accuracy, length of execution time, and memory requirements. Zhang et al. (2017) and Kanitz et al. compare several methods on isoform detection and find accuracy dependent on gene complexity (e.g., the number of transcripts or exons), read depth, and alignment method. Similar to our findings, both reviews report that the majority of methods perform similarly well and that a difference in accuracy across methods was only seen at higher transcript complexities when genes had more than 1–5 transcripts. Krantiz et al., then goes on to explore different memory and computational efficiency considerations, which was not a focus of our study. Teng et al. (2016) investigate data preprocessing and metrics for method comparison. They advise against using correlations on raw levels due to non-normality, which inspired our use of the non-parametric Tau-βeta correlations in this study. Several methods were not included in the challenge because they were not submitted by competing teams. The challenge administrators augmented the submissions with additional methods, however, a number of programs were excluded due to being either outdated or failing to pass sanity checks, producing thousands of fusion calls per sample. Additionally, a number of methods have been developed since the time of running the competition (see Table S1). Even so, recent reviews suggest our survey of methods here reflect those that are most competitive (Haas et al., 2019).
Our review here has included several tools, the use of spike-ins for an unbiased assessment of sensitivity, an objective evaluation framework in which the administrators have run submitted methods to generate all predictions, and a statistical procedure to infer background fusions to accurately measure precision. In addition to providing an evaluation of methods, our work contributes a tool for simulating RNA isoforms and fusions, a new benchmark dataset against which forthcoming methods can be compared, and all of the tested methods in standardized workflows for re-execution, which should facilitate further progress in this area of study. As part of our benchmark, we employed a computational simulation that can create a cancer transcriptome that includes alternative isoform levels as well as novel fusions. The simulator is available as an open-source repository and the full details of its design are described in a companion manuscript (unpublished data). However, while the in silico benchmarking provides a valuable assessment, an in vitro analysis was also used to avoid any evaluation biases among methods that use overlapping computational strategies with the simulator as well as to assess any issues in detecting RNA species stemming from laboratory and sequencing effects. For this reason, we synthetically constructed isoforms and fusion transcripts that were introduced into cell line backgrounds. The constructs were added at pre-specified quantities of 0, 5, 25, and 50 pg. While the spike-in design provided valuable information to rank fusion detection entries, we failed to elucidate a meaningful ranking of entries for the Isoform Quantification sub-challenge. All entries were able to perfectly identify higher from lower transcript levels between all comparisons except the two highest levels (e.g., 25 pg compared with 50 pg). We speculate that either the tested methods were not accurate enough to predict the 2-fold relative difference between the 25 and 50-pg quantities or the transcripts that were ultimately sequenced did not reflect the input quantities either due to saturation or internal cellular degradation that both effectively equalized the concentrations of these two spike-in levels. An important follow-up investigation could include an additional spike-in level among the array of levels tested here. For example, the use of an additional 10 pg could have helped assess methods in their ability to distinguish in the 2- to 2.5-fold range of resolution. It is our theory that the methodology to estimate transcript abundance may have plateaued or that the challenge design itself lacked critical resolution to discriminate among methods. Additional experiments including higher quantities of spike-ins (25 to 50 pg range) also would help further elucidate the issue.
For the Fusion Detection sub-challenge, the spike-ins were effective for assessing the sensitivity of the submitted entries. However, there is an issue in estimating the precision of the entries because fusions were added to cell lines that may express their own background fusions. Thus, methods predicting the presence of such background fusions would be improperly penalized in a precision assessment. We, therefore, attempted to estimate the background fusions in a number of ways, first using long-read sequencing approaches that each failed for different reasons (STAR methods, attempts to assess background transcripts with long read sequencing). To compensate, we introduced a computational strategy to infer fusions present in the background from submitted predictions. We reasoned that in such cases the fusions would be predicted in multiple designs that included the same cell line (i.e., the background set of fusions should be the same or very similar), multiple submitted entries would predict such cases, and they would also be detected by more accurate entries. We computationally determined a set of fusions called the imputed truth (i-Truth) that were added to the spike-in truth set (see resource table for links to SMC-RNA-Eval code). The i-Truth contributed an additional 48 high-confidence fusion calls for the final evaluation. Follow-up validation revealed that 43 out of 48 of the i-Truth constructs were supported by one or more sources of external evidence. Thus, even in the absence of a ground truth orthogonal set, the procedure and results of this challenge establish a computational strategy highly effective for unbiased assessment of methods that could be applied more broadly to an additional set of problems beyond RNA-seq analysis.
The detection of RNA species is becoming an increasingly important diagnostic tool in the analysis of cancer samples, with multi-gene transcript abundance panels used for prognosis and prediction of response to therapy, and fusion transcripts used for diagnosis and prediction of treatment efficacy. These applications continue to expand, and an improved understanding of the ways in which the cancer transcriptome is dysregulated has the potential for basic, translational, and clinical applications in essentially every cancer type. Key applications will include refining tumor subtypes and their differentiation status, mapping clonal complexity, illuminating the role of the microenvironment, pinpointing the state and function of immune cells, linking transcriptomic biomarkers to targeted treatments, and understanding the differential activity of specific driver mutations. It remains unclear what sequencing and computational approaches will have sufficient accuracy to identify transcript variants and estimate their abundances for routine clinical use. Our results suggest that additional work is needed to identify fusions in complex samples. For example, the sensitivity for detecting the smallest quantities of a fusion in this challenge (5 pg) were 82% ± 8% compared with 88.0 ± 9% for 25 pgand 88.7 ± 12% for 50 pg (see Table S11). The top caller suffered the same drop in sensitivity for the lowest spike-in level, achieving 93.6% for the 5-pg spike-in compared with 98.9% (for 25 pg) and 100% (for 50 pg). If we assume that the 25 pg levels reflect the typical expression level of a fusion in a relatively pure tumor sample, then the 5-pg quantities reflect fusions expressed at 5-fold lower levels or those expressed at the same level in only one out of the five cells sequenced due to normal contamination or tumor subclonal heterogeneity. While suitable for routine cases, current methods would lead to a large number of missed calls for subclonal variants or in samples with large amounts of normal tissue admixture. For example, at these estimated sensitivity levels, the best method is expected to miss 1 out of every 15 to 16 cases of a driving fusion if present in one out of every five subclones or if it is expressed at lower levels. On the other hand, for applications in which relative transcript abundances are used to calculate signature scores, such as the well-known PAM50 breast cancer subtypes, methods provide accurate quantitation. In conclusion, we identified, benchmarked, and made available in a standardized containerized format a suite of tools for estimating key features of the altered cancer transcriptome that should further the applicability of RNA’s use in patient care.
CONSORTIA
The participants of the SMC-RNA Challenge are Hongjiu Zhang, Yifan Wang, Yuanfang Guan, Cu Nguyen, Christopher Sugai, Alokkumar Jha, Jing Woei LI, and Alexander Dobin.
FEATURE IMPORTANCE ANALYSIS
Random forest models were created using R’s randomForest function, version 4.6–14, which implements Breiman’s random forest algorithm (based on Breiman and Cutler’s original Fortran code) (https://cran.r-project.org/web/packages/randomForest/randomForest.pdf).All parameters were left as default except for the number of trees which was set to 100. The feature importance analysis was performed using the the Boruta R package with default parameters, version 6.0.0 (https://www.jstatsoft.org/article/view/v036i11).
STAR★METHODS
Detailed methods are provided in the online version of this paper and include the following:
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kyle Ellrott (ellrott@ohsu.edu)
Materials availability
This study did not generate new materials.
Data and code availability
SpikeIn Sequencing Data has been deposited at Synapse.org and is publicly available under the accession numbers: syn22344794.
Challenge workflows have been deposited at Github.org and is available under https://github.com/smc-rna-challenge
rnaseqSim original code is publicly available at https://github.com/Sage-Bionetworks/rnaseqSim
The scripts used to generate the figures reported in this paper are available at https://github.com/smc-rna-challenge/SMC-RNA-Eval
Any additional information required to reproduce this work is available from the Lead Contact.
METHOD DETAILS
Isoform and fusion simulation pipeline
The simulated benchmark was constructed using 32 training and 20 test datasets (Table S5). The datasets varied in the number of simulated fusion events ranging between 3 and 111 events. Other parameters that varied between datasets included read depth (50100 million reads), insert size (150 or 200 base pairs), coverage bias, and the abundance of individual transcripts.
These genomes were created using a new simulator called rnaseqSim (the source code can be found at https://github.com/Sage-Bionetworks/rnaseqSim ).
Fusion transcripts were simulated by randomly selecting two protein coding transcripts (using Ensembl v75 annotation). For each of the selected transcripts, a random number of exons are used to generate the fusion. If the transcript was selected as the donor, then the number of exons incorporated are counted from the beginning of the transcript. Conversely, if the transcript was selected as the acceptor, then the number of exons incorporated are counted from the end of the transcript. Selected transcripts are fused only at the exon-intron boundaries. Using the exon coordinates for each selected transcript, a synthetic fusion sequence is generated using the GRCh37.75 genome and biopython. A reference index is generated for the synthetic fusion sequence using RSEM v1.2.31 with the STAR 2.4.2a aligner.
For simulating isoform abundance, a diploid genome was synthetically designed to capture allele-specific SNPs and haplotypes during read generation. First, the GRCh37 genome build (Homo sapiens GRCh37.75) and GTF annotation (Homo sapiens Ensembl v75) were duplicated and chromosomes were labeled to distinguish the two sets of haploid chromosomes. bcftools consensus was then used to introduce phased SNPs found in the Genome in a Bottle into each set of haploid chromosomes. The diploid genome sequence and annotations were then used to generate a reference index with STAR v2.4.2a. Isoform abundance was simulated using abundance data originating from prostate cancer samples (Chen et al., 2019) and select TCGA samples including:
TCGA-GBM: TCGA-26–5139-01A-01R-1850–01
TCGA-LUAD: TCGA-44–6775-01A-11R-1858–07
TCGA-LUSC: TCGA-21–1082-01A-01R-0692–07
TCGA-OV: TCGA-24–1467-01A-01R-1566–13
TCGA-BRCA: TCGA-BH-A1F8–11B-21R-A13Q-07
TCGA-BLCA: TCGA-H4-A2HQ-01A-11R-A180–07
Abundance profiles were estimated for each sample using RSEM v1.2.31. Each profile was adjusted by adding noise, modeled using a gamma distribution, to a subset of transcript selected using a binomial distribution. Synthetic fusion transcripts were incorporated into the expression. An abundance is randomly assigned to the fusion transcript, such that its value was greater than the overall median transcript abundance. Abundances are then normalized to sum to 1 million, i.e. TPMs (Li and Dewey, 2011; Wagner et al., 2012). During this normalization step, donor transcripts were removed. The abundance assigned to the fusion transcript was then divided (randomly following a uniform distribution) between the original donor transcript and the fusion transcript. Abundances were then allocated to one of two alleles in the diploid GTF annotation (previously described) using a uniform distribution to model allelic expression. Finally, RSEM v1.2.31 was used to simulate the generation of the FASTQ sequence reads.
Simulated tumor workflow deployment
Each entry was submitted as a defined workflow written using the Common Workflow Language (CWL v1.0). Source code for the algorithm and any dependencies needed for installing or running the algorithm were built into a Docker image by the participant. Workflow descriptions for all entries are available at: https://github.com/smc-rna-challenge and Docker images are available from https://quay.io/organization/smc-rna-challenge. All workflows were provided the GRCh37.75 genome assembly and annotation as reference files. If additional reference files were required for the workflow, participants were allowed to upload files to synapse and link to those files using a synapse ID.
Deployment of workflows was done using ISB cloud resources (Google Compute Engine). Most entries for the Isoform Quantification sub-challenge were provided a virtual machine with 4 vCPUs and 15 GB of RAM (n1-standard-4) while most entries for the Fusion Detection sub-challenge were provided a virtual machine with 16 vCPUs and 60 GB of RAM (n1-standard-16). For some entries, the default resources were not sufficient to run, in which case, a virtual machine with more resources was provided (maximum 16 vCPUS and 104 GB RAM, n1-highmem-16). All entries were provided a 400 GB persistent disk and a time limit of 35 hours to complete running of the workflow.
A virtual machine was created for each workflow being run on a given test dataset. The CWL workflow, docker image, default reference files, participant-provided reference files, and test datasets were pulled down onto the VM. A JSON file was generated to point to all necessary input files for the workflow. The CWL workflows were run with cwltool v1.0.20161007181528. Output files generated by the CWL workflows were stored in a Google Bucket for evaluation.
Spike-in fusion construction
For the fusion constructs, genes were randomly selected, with an eye only toward the likelihood of successful PCR during library preparation and a total length of less than 1kb. The Invitrogen GeneArt Gene Synthesis service built DNA constructs from our provided sequences, and the RNA spike-in material was generated using the NEB HiScribe T7 Quick high Yield RNA Synthesis Kit. Low abundances of these genes were observed post sequencing (Figure S3E). Additionally, to verify the presence of the spike-in products in the short read sequencing, we inspected the alignment of the reads using IGV to confirm reads mapped as expected across the junctions. We did this by including the constructed fusion sequences in the reference genome. This allowed the alignment algorithm to easily identify reads from the fusion constructs and verify that indeed hundreds of reads exist in the cell line files.
The spike-in constructs were programmed according to the design in Table S8. Some genes had multiple isoforms (up to 3). The transcript complexity (from 1 up to 3 per gene) varied across the samples. In addition, a range of fusions, from 7 up to 18, were included in the samples.
Benchmark transcript selection
To ensure the accuracy of our technical spike-in approach, we elected to assemble a list of transcripts known to be non-expressors in a suitable cell line. To this end, we assembled a cohort of normal breast tissue RNA-seq data from GTEx (GTEx Consortium, 2013), Fantom5 (Abugessaisa et al., 2017), Illumina Body Map (Petryszak et al., 2016) and TCGA (Nawy, 2018) to establish a baseline for all transcripts. To be considered a non-expressing transcript in normal breast tissue, transcripts were filtered to include only those with FPKM <= 0.5 and total expression of the corresponding gene with FPKM <= 0.9, where data was available for >= 80% of the samples (lincRNAs were excluded from consideration). Transcripts retained following this filtration of the normal data were confirmed to also be non-expressing in both the JWGray Breast Cancer Cell Line Panel (Neve et al., 2006) and in the TCGA BRCA RNA-seq data (Neve et al., 2006).
Criteria for transcript selection:
We selected non-expressing genes in breast cancer cell lines, with individual transcript expression of FPKM <=0.5 and overall gene expression of with FPKM < 0.9
- We selected genes with 3–5 transcripts, of which one or more of the following structural variations were present in at least one of those transcripts:
- Alternate 5’ UTR
- Alternate 3’ UTR
- Cassette exon
- Retained intron
- Alternate transcription start site
- Alternate stop codon
Transcripts with length >0.5kbp and < 1kbp
Isoforms selected for benchmarking were converted to spike-in RNA in identical fashion to that of the synthetic fusion set discussed above.
Final benchmark collection: 20 transcripts from 6 different genes, 3–5 transcripts/gene, plus 40 additional transcripts selected for construction of the 20 synthetic fusions (alternate splicing not taken into account for fusion).
As proof-of-concept we could accurately detect proportional increases in “expression” between different spike amounts, 5 replicates of MDA-MB-415 (breast adenocarcinoma metastasis) RNA was spiked with 5pg, 25pg, and 125pg and sequenced. Following this test run, we were satisfied with our ability to measure proportional “differential expression” between the spike amounts. Further examination of this BCCL exploratory analysis indicated 5pg to be closest to endogenous expression of most transcripts in these cell lines while 125pg was excessively high, so the spike amounts were adjusted to 5pg, 25pg, and 50pg.
For the benchmark experiment, we chose six cell lines for use in the challenge. These include A549 (lung carcinoma), HCC1143 (breast primary ductal carcinoma), Jurkat I 9.2 (acute T-lymphoblastic leukemia), LNCaP clone FGC (prostate carcinoma metastasis), PC-3 (prostate adenocarcinoma metastasis), and PC9 (non-small cell lung carcinoma). Cell lines were grown to subconfluency in RPMI media supplemented with 10% FBS.
From the original 40 selected transcripts and synthetic fusions, we selected 36 by removing the two highest and the two lowest expressing transcripts/fusions (including one failed construct), then randomly assigning each to one of six evenly populated spike-in groups. . Minor modifications to the random assignment were made to ensure no group contained more than one transcript from the same gene. The six cell lines were each divided into five aliquots. Four of these were each spiked with the transcripts/fusions from two of the six spike-in groups (12 constructs per replicate), attempting to randomize the groups per cell line as much as possible in order to minimize the pairing of any two groups within the same replicate. (Table S6). The fifth aliquot remained unadulterated. Finally, each sample aliquot underwent RNA sequencing.
RNA preparation and sequencing
RNA was isolated from cell lines using a Zymo Research Quick-RNA Kit following manufacturer’s instructions. Extracted RNA samples were divided into 5 aliquots (1 ug each) per cell line and spiked with different amounts of transcript and fusion constructs (Table S7). Library preparation for RNA-Seq was performed using the Agilent SureSelect Strand-Specific RNA Library Prep Kit. Samples were sequenced at the OHSU Massively Parallel Sequencing Shared Resource (MPSSR) core facility using the Illumina NextSeq500 for 2×100 cycles. The results of the sequencing have been uploaded to Synapse under syn22344794.
QUANTIFICATION AND STATISTICAL ANALYSIS
Attempts to assess background transcripts with long read sequencing
Because spike-ins were added to established cell lines that contained their own background transcripts, we attempted to estimate the fusions present in the background by sequencing the cell lines using three different approaches based on long read sequencing data. First, we attempted direct long-read sequencing on the LNCaP and A549 cell lines, using MinION nanopore sequencing. We performed direct sequencing of poly-A RNA from A549 cell line which yielded 293,813 reads. We also performed nanopore sequencing of cDNA from A549 poly-A RNA, which yielded 281,319 reads. While there were fusions detected in the existing reads, the read depth was insufficient to conclusively rule out background or technical artifacts, and the large amount of sample RNA that would be required prevented further analysis of the matched samples used for spike-in studies. Second, we performed indirect long read sequencing to estimate the background. We obtained long-read sequencing of the LNCap cell line using the IsoSeq protocol, paired to matched short-read sequencing. Integrated analysis of both the long- and short-read data to call fusions using IDP-Fusion (Weirather et al., 2015), resulted in one high confidence (supported by both short/long reads) fusion (chr5:95234564-/chr5:135587632+; KIAA0825-PCBD2). We compared the results of the hybrid short/long read fusion detection results with our own short read paired-end sequencing data for the LNCap cell and found no fusion calls for this exact fusion (same breakpoints or same donor/acceptor genes). We did observe 2 entries call 2 different fusions involving PCBD2 as the donor gene. This sample provided a comparison point, but would not take into account any fusion events that could have occurred in the passages that separated the two aliquots. Finally, we attempted to estimate false-positive rates using the Genome in a Bottle (GIAB) sample as a null model. To our knowledge, given the available transcriptomic data, no fusions have been detected for the SRR5665260 GIAB sample. Consistent with this expectation, we ran the IDP-Fusion caller (Weirather et al., 2015) using both the short reads and long reads from the sample and indeed found no identifiable fusions. Therefore, if any entries predicted fusions in this sample we could assume they represented false positives. We ran contestant entries using the short reads of the GIAB sample. A number of the entries failed to run on these new samples. In total, 19 of the entries were able to run on the GIAB short read data. The number of fusions predicted by entries ranged from 0 to 208 with an average of 56 (median 22) fusions called. In summary, the results on long read sequencing either provided too little sequencing depth to base fusion predictions or the results were inconclusive due to issues with entries failing to run.
Imputing an extended truth dataset for fusion evaluation
From the design of the experiment, three factors enabled deeper analysis of these potential native fusions. First, contestants had contributed a wide distribution of workflows composed of different detection methods and filtering options. Second, the same cell lines had been used multiple times across separate spike-in experiments that created a set of biological replicates. Third, the spike-in panel provided an estimate of the sensitivity of different entries as well as any potential meta-calling entry.
Given these factors, we created an imputed truth set (i-Truth) for each cell line made up of the known spike-ins and those predicted to be in the background based on a meta-caller created from the consensus of submitted entries. The first step in creating the meta-caller was to eliminate entries that were too similar, to remove the bias of having multiple, near identical methods over-influence what is interpreted as truth. Second, we removed entries that fell below a sensitivity cutoff, in this case a true positive rate of 0.6, which is the approximate sensitivity found when running callers on the spike-ins. This meta-calling was done at the cell line level, aggregating the calls across multiple spike-in experiments. This means the meta-calling approach would be unable to detect the spike-ins, which would only occur in a fraction of the biological replicates, but the background native fusions would be common across the replicates. Each i-Truth was based on an agreement cutoff, with the total number of agreeing calls across all the entries and all the biological replicates. Fifty percent agreement could come from half of the entries agreeing on a call across all the biological replicates, or all the entries agreeing across half of the replicates. Across the six cell lines, we used a threshold of two or more entries or replicates being in agreement, which would yield a total of 30,031 potential fusion junction breakpoints. With five replicates per cell line and ten representative callers, there were 50 potential callsets.
We expect background fusions to be consistently predicted across these callsets but do not know a good agreement level across the callsets to set for a priori detection. Higher levels of required agreement decrease the total number of calls and increase the requirement that the calls be based on a wide variety of supporting methods and evidence. To identify an agreement threshold, we utilized the determined rate of recall seen in the spike-in set using the representative set of callers, which was 0.726. By using various agreement levels, we could create a new i-Truth set and evaluate both sensitivity and specificity. The total number predicted i-Truth sites vary as a function of the confidence level, measures as k out of the 50 callsets that predicted a site’s existence (Figure S1B). Conversely, as more low confidence sites are added to the i-Truth the average recall rate of entries decreases. At its lowest setting, an agreement level of 4%, the recall rate of the meta-caller is 5%, as the agreement rate increases, the total number of new sites added to the i-Truth decreases and the recall rate increases. For example, out of the 30,031 breakpoints, 289 of them were predicted by at least 10 out of 50 call sets (20%). By increasing the agreement rate to 50%, the recall rate of the meta-caller approached a recall rate of 0.736, similar to the recall rate seen in the spike-in data set. Using this threshold of agreement, 48 additional possible RNA fusions were predicted across the cell lines and used as the i-Truth set and added to the synthetic constructs. We used this extended set for evaluating the sensitivity and precision of individual entries.
Imputed truth fusion validation
We collected database reports and literature support for the 48 i-Truth fusions to determine prior predictions or validation for each. In searching for previous observations of these 48 fusions, 44 had some level of breakpoint support. Of these, 28 were exact matches in both the donor and acceptor breakpoints as well as occurring in the correct cell line or tumor type, as per either the Broad Cancer Cell Line Encyclopedia (Ghandi et al., 2019; Panigrahi et al., 2018), a unique database which accumulates and reports fusion support data from multiple databases at once. At least 3 fusions have been experimentally validated in previous literature (Guo et al., 2016; Maher et al., 2009; Winters et al., 2018). While the contents of these databases would have been generated using source material parallel to the sequencing used in the benchmark, these databases are likely generated using the same algorithm. The summary of this analysis can be found in Table S9. The spreadsheet covers results from both the CCLE and from FusionHub, which compiled reports from the following databases and/or methods for predicting and reporting fusions:
18Cancers [EC] - FusionCatcher
Babiceanu Dataset [BD] - SOAPfuse
ChimerKB [KB] - Fusion database with FISH, SangerSeq, or RT-PCR validation
ChimerSeq [CS] - PRADA, FusionScan, TopHat-Fusion, ChiTaRS
ChimerPub [CP] - PubMed text mining
ChiTaRS-2.1 [CH] - Database of chimeric transcripts
FusionCancer [FC] - Tophat2, FusionMap, SOAPfuse, chimerascan
Klijn Database [KD] - GSNAP
Known Fusions [KF] - FusionCatcher fusions from literature
Literature [LT] - Known fusions compiled from literature
Prostate Dataset [PD] - Tophat2
Tumor Fusions GDP [TF] – PRADA
1000 Genomes [TG] - FusionCatcher for 1000 Genome project
GTEx [GX] - FusionAnnotator & FusionCatcher on normal tissue
Non Tumor Cells [NT] - FusionCatcher for non-tumor cell
Evaluating Isoform Quantification
The Isoform Quantification sub-challenge was evaluated using the Kendall’s Tau-β correlation coefficient. Simulated data was initially evaluated using Spearman correlation coefficient as the input model data for the simulator was TPM quantities, fully described and in the same dimension as the results data being produced by the submissions. However, the spike-in data was much more sparse, with 18 separate isoforms spiked in at three different concentrations. Additionally the truth data from the experiment involved picograms of spike-in material, a much different metric than the results. For this reason, the scoring against the spike-in set was done using Kendall’s tau coefficient to evaluate rank based correlation.
For synthetically generated samples, in the Isoform Quantification sub-challenge, the abundance, in the form of Transcripts Per Million (TPM) is a known input into the simulator. Calculation of a Spearman correlation coefficient of the TPM outputs for the entries could be fully calculated. However, evaluation of the isoform abundance in the spike-ins is confounded by two factors: 1) the input quantities are much more sparse and 2) the units of measurement are not linearly associated to the output units. The full spike-in experiment was developed by spiking in pairs of transcripts across 6 different cell lines, which causes a much more sparse set of possible points for evaluation. Secondly, the inputs to the spike-in system were in picograms of material spiked into the system. We have demonstrated correlation of spike-in quantity to TPM (shown in Figure S2E), but this is for the same transcript across multiple samples. Each transcript will have a different coefficient that connects the spike-in amount in picograms to the output TPMs. This means that direct comparison between different transcripts in a single sample in the TPM space could be distorted by this mix of coefficients. Thus, for evaluating isoform quantity in the spike-in set, we evaluated the predicted abundance level of a single transcript across multiple samples.
In order to evaluate an entry’s ability to determine isoform abundance in the spike-in samples, we calculated a Kendall’s Tau-β Score (KTBS), by first calculating separate Kendall Tau-βeta correlations for each transcript (across replicates and cell lines). The Kendall’s Tau-β correlation thus compares the agreement of the abundance ordering between the predictions and the truth for one transcript. Importantly, the Kendall Tau-β statistic makes adjustment for ties, which do exist in our truth set.
The KTBS for each entry was then defined as the mean of the Kendall Tau-β correlations computed for each of the 18 transcripts. (Figure 3B). We then ranked the submissions by KTBS (Figure 3C). Because this ranking resulted in close scores among many entries, we performed a leave-one-out cross-validation and bootstrap ranking (Figure 3A). The leave-one-out procedure was performed by setting aside one transcript from the KTBS calculation to ensure that any one transcript did not unduly influence the ranking of any given method. The final ranking of the leave-one-out procedure was based on the average rank of the method across 18 folds. In order to more finely compare methods with similar accuracies, we performed a bootstrap procedure. To do this, for each method, , we drew bootstrap samples from the 18 transcript Kendall Tau-β correlations 1000 times. We then ranked the methods by the mean of each Tau-β score distribution. This allowed us to estimate and significantly compare the mean and variance of the closely ranked methods. We also confirmed that there was no bias between genes by calculating a Tau-β correlation score within the 3 transcripts per gene and the 5 replicates among cell lines (Figure S2A).
Supplementary Material
Highlights.
The SMC-RNA Challenge benchmarked isoform quantification and fusion detection methods
These methods were benchmarked using both in silico and in vitro datasets
Methods were captured using reproducible computing methods, including docker and CWL
The best methods have been incorporated into the NCI’s Genomic Data Commons
ACKNOWLEDGMENTS
The authors would like to acknowledge the work done by Alison Tang and input from Dr. Angela Brooks for working with long-read RNA data, Chris Boniface for his work as a lab consultant, and Dr. Joe Gray for the contribution of the cell lines used for the benchmark dataset creation. The authors would like to acknowledge the support of the National Cancer Institute. This includes funding to UCSC from the ITCR (grant R01CA180778), Oregon Health & Science University (U24CA210957, U24CA143799, and HHSN261200800001E), UCLA (P30CA016042), and Sage Bionetworks (5U24CA209923).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.cels.2021.05.021.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Abugessaisa I, Noguchi S, Carninci P, and Kasukawa T. (2017). The FANTOM5 computation ecosystem: genomic information hub for promoters and active enhancers. Methods Mol. Biol. 1611, 199–217. [DOI] [PubMed] [Google Scholar]
- Ahsen ME, Vogel R, and Stolovitzky G. (2018). Unsupervised evaluation and weighted aggregation of ranked predictions. J. Mach. Learn. Res. 20, 1–40. [Google Scholar]
- Bao W, Kojima KK, and Kohany O. (2015). Repbase Update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, and Pachter L. (2016). Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527. [DOI] [PubMed] [Google Scholar]
- Chen S, Huang V, Xu X, Livingstone J, Soares F, Jeon J, Zeng Y, Hua JT, Petricca J, Guo H, et al. (2019). Widespread and functional RNA circularization in localized prostate. Cancer Cell 176, 831–843. [DOI] [PubMed] [Google Scholar]
- Degenhardt F, Seifert S, and Szymczak S. (2019). Evaluation of variable selection methods for random forests and omics data sets. Brief. Bioinform. 20, 492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellrott K, Buchanan A, Creason A, Mason M, Schaffter T, Hoff B, Eddy J, Chilton JM, Yu T, Stuart JM, et al. (2019). Reproducible biomedical benchmarking in the cloud: lessons from crowd-sourced data challenges. Genome Biol. 20, 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Liang WW, Foltz SM, Mutharasu G, Jayasinghe RG, Cao S, Liao WW, Reynolds SM, Wyczalkowski MA, Yao L, et al. (2018). Driver fusions and their implications in the development and treatment of human cancers. Cell Rep. 23, 227–238.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER 3rd, Barretina J, Gelfand ET, Bielski CM, Li H, et al. (2019). Next-generation characterization of the cancer cell line encyclopedia. Nature 569, 503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium GTEx (2013). The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J, and Saez-Rodriguez J. (2018). Alternative models for sharing confidential biomedical data. Nat. Biotechnol. 36, 391–392. [DOI] [PubMed] [Google Scholar]
- Guo T, Gaykalova DA, Considine M, Wheelan S, Pallavajjala A, Bishop JA, Westra WH, Ideker T, Koch WM, Khan Z, et al. (2016). Characterization of functionally active gene fusions in human papillomavirus related oropharyngeal squamous cell carcinoma. Int. J. Cancer 139, 373–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Dobin A, Li B, Stransky N, Pochet N, and Regev A. (2019). Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 20, 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Dobin A, Stransky N, Li B, Yang X, Tickle T, Bankapur A, Ganote C, Doak TG, Pochet N, et al. (2017). STAR-fusion: fast and accurate fusion transcript detection from RNA-Seq. bioRxiv. 10.1101/120295. [DOI] [Google Scholar]
- Han Y, Gao S, Muegge K, Zhang W, and Zhou B. (2015). Advanced applications of RNA sequencing and challenges. Bioinform. Biol. Insights 9, 29–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2000). The hallmarks of cancer. Cell 100, 57–70. [DOI] [PubMed] [Google Scholar]
- Hanahan D, and Weinberg RA (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Houseley J, and Tollervey D. (2010). Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS One 5, e12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanitz A, Gypas F, Gruber AJ, Gruber AR, Martin G, and Zavolan M. (2015). Comparative assessment of methods for the computational inference of transcript isoform abundance from RNA-seq data. Genome Biol. 16, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Vo AD, Qin F, and Li H. (2016). Comparative assessment of methods for the fusion transcripts detection from RNA-Seq data. Sci. Rep. 6, 21597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursa MB, and Rudnicki WR (2010). Feature selection with the Boruta package. J. Stat. Soft. 36, 1–13. [Google Scholar]
- Lee AY, Ewing AD, Ellrott K, Hu Y, Houlahan KE, Bare JC, Espiritu SMG, Huang V, Dang K, Chong Z, et al. (2018). Combining accurate tumor genome simulation with crowdsourcing to benchmark somatic structural variant detection. Genome Biol 19, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, and Dewey CN (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12, 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, and Chinnaiyan AM (2009). Transcriptome sequencing to detect gene fusions in cancer. Nature 458, 97–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbach D, Costello JC, Küffner R, Vega NM, Prill RJ, Camacho DM, Allison KR, DREAM5 Consortium, Kellis M, Collins JJ, Collins JJ, et al. (2012). Wisdom of crowds for robust gene network inference. Nat. Methods 9, 796–804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzker ML (2010). Sequencing technologies - the next generation. Nat. Rev. Genet. 11, 31–46. [DOI] [PubMed] [Google Scholar]
- Mohseni M, Uludag H, and Brandwein JM (2018). Advances in biology of acute lymphoblastic leukemia (ALL) and therapeutic implications. Am. J. Blood Res. 8, 29–56. [PMC free article] [PubMed] [Google Scholar]
- Nawy T. (2018). A pan-cancer atlas. Nat. Methods 15, 407. [DOI] [PubMed] [Google Scholar]
- Neve RM, Chin K, Fridlyand J, Yeh J, Baehner FL, Fevr T, Clark L, Bayani N, Coppe JP, Tong F, et al. (2006). A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 10, 515–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panigrahi P, Jere A, and Anamika K. (2018). FusionHub: A unified web platform for annotation and visualization of gene fusion events in human cancer. PLoS One 13, e0196588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T, Füllgrabe A, Fuentes AM-P, Jupp S, Koskinen S, et al. (2016). Expression Atlas update–an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 44, D746–D752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picco G, Chen ED, Alonso LG, Behan FM, Gonçalves E, Bignell G, Matchan A, Fu B, Banerjee R, Anderson E, et al. (2019). Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat. Commun. 10, 2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, and Pachter L. (2013). Streaming fragment assignment for real-time analysis of sequencing experiments. Nat. Methods 10, 71–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng M, Love MI, Davis CA, Djebali S, Dobin A, Graveley BR, Li S, Mason CE, Olson S, Pervouchine D, et al. (2016). A benchmark for RNA-seq quantification pipelines. Genome Biol. 17, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, et al. (2008). Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia 10, 177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitting-Seerup K, and Sandelin A. (2017). The landscape of isoform switches in human cancers. Mol. Cancer Res. 15, 1206–1220. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Kin K, and Lynch VJ (2012). Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci. 131, 281–285. [DOI] [PubMed] [Google Scholar]
- Weirather JL, Afshar PT, Clark TA, Tseng E, Powers LS, Underwood JG, Zabner J, Korlach J, Wong WH, and Au KF (2015). Characterization of fusion genes and the significantly expressed fusion isoforms in breast cancer by hybrid sequencing. Nucleic Acids Res. 43, e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winters JL, Davila JI, McDonald AM, Nair AA, Fadra N, Wehrs RN, Thomas BC, Balcom JR, Jin L, Wu X, et al. (2018). Development and verification of an RNA sequencing (RNA-Seq) assay for the detection of gene fusions in tumors. J. Mol. Diagn. 20, 495–511. [DOI] [PubMed] [Google Scholar]
- Wu TD, Reeder J, Lawrence M, Becker G, and Brauer MJ (2016). GMAP and GSNAP for genomic sequence alignment: enhancements to speed, accuracy, and functionality. Methods Mol. Biol. 1418, 283–334. [DOI] [PubMed] [Google Scholar]
- Zaphiropoulos PG (2011). Trans-splicing in higher eukaryotes: implications for cancer development? Front. Genet. 2, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang B, Lin LL, and Zhao S. (2017). Evaluation and comparison of computational tools for RNA-seq isoform quantification. BMC Genomics 18, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
SpikeIn Sequencing Data has been deposited at Synapse.org and is publicly available under the accession numbers: syn22344794.
Challenge workflows have been deposited at Github.org and is available under https://github.com/smc-rna-challenge
rnaseqSim original code is publicly available at https://github.com/Sage-Bionetworks/rnaseqSim
The scripts used to generate the figures reported in this paper are available at https://github.com/smc-rna-challenge/SMC-RNA-Eval
Any additional information required to reproduce this work is available from the Lead Contact.