Abstract
Background and Aims
Nonalcoholic steatohepatitis is rapidly becoming the leading cause of liver failure and indication for liver transplantation. Hepatic inflammation is a key feature of NASH but the immune pathways involved in this process are poorly understood. B lymphocytes are cells of the adaptive immune system that are critical regulators of immune responses. However, the role of B cells in the pathogenesis of NASH and the potential mechanisms leading to their activation in the liver are unclear.
Approach and Results
In this study, we report that NASH livers accumulate B cells with elevated pro‐inflammatory cytokine secretion and antigen‐presentation ability. Single‐cell and bulk RNA sequencing of intrahepatic B cells from mice with NASH unveiled a transcriptional landscape that reflects their pro‐inflammatory function. Accordingly, B‐cell deficiency ameliorated NASH progression, and adoptively transferring B cells from NASH livers recapitulates the disease. Mechanistically, B‐cell activation during NASH involves signaling through the innate adaptor myeloid differentiation primary response protein 88 (MyD88) as B cell–specific deletion of MyD88 reduced hepatic T cell–mediated inflammation and fibrosis, but not steatosis. In addition, activation of intrahepatic B cells implicates B cell–receptor signaling, delineating a synergy between innate and adaptive mechanisms of antigen recognition. Furthermore, fecal microbiota transplantation of human NAFLD gut microbiotas into recipient mice promoted the progression of NASH by increasing the accumulation and activation of intrahepatic B cells, suggesting that gut microbial factors drive the pathogenic function of B cells during NASH.
Conclusion
Our findings reveal that a gut microbiota–driven activation of intrahepatic B cells leads to hepatic inflammation and fibrosis during the progression of NASH through innate and adaptive immune mechanisms.
Abbreviations
- Acta2
smooth muscle α actin
- ALT
alanine transaminase
- AST
aspartate transaminase
- AUC
area under the curve
- BAFF‐R
B‐cell activating factor receptor
- BCR
B‐cell receptor
- B‐MYD
B cell–specific deletion of MyD88
- CM
central memory
- Col1a1
collagen type 1 alpha 1 chain
- CyTOF
cytometry by time of flight
- DC
dendritic cell
- DEG
differentially expressed gene
- EM
effector memory
- FMT
fecal microbiota transfer
- GFP
green fluorescent protein
- GTT
glucose tolerance test
- H&E
hematoxylin and eosin
- HFHC
high‐fat high‐carbohydrate
- IFN
interferon
- IPA
Ingenuity Pathway Analysis
- ITT
insulin tolerance test
- KC
Kupffer cell
- LPS
lipopolysaccharide
- mAb
monoclonal antibody
- Mmp2
matrix metalloproteinase
- MHC
major histocompatibility complex
- MO
monocytes
- MyD88
myeloid differentiation primary response protein 88
- Mφ
macrophages
- NAS
NAFLD activity score
- NCD
normal chow diet
- NK
natural killer cell
- NKT
natural killer T
- PMA
phorbol 12‐myristate 13‐acetate
- PTT
pyruvate tolerance test
- RNA‐seq
RNA sequencing
- RT‐PCR
real‐time polymerase chain reaction
- Th1
T helper 1
- Timp1
tissue inhibitor of metalloproteinase‐1
- TLR
toll‐like receptor
- WT
wild‐type
NAFLD is estimated to affect 30% of the population and is now recognized as the most prevalent chronic liver disease worldwide.( 1 ) The disease covers a wide spectrum of liver pathology, ranging from simple lipid accumulation to the development of NASH, defined by hepatic steatosis, local inflammation, hepatocellular injury, and fibrosis.( 2 ) NASH‐associated inflammation is driven by innate and adaptive immune mechanisms comprising macrophages, dendritic cells, neutrophils, and lymphocytes.( 3 ) Recent single‐cell transcriptome analyses have uncovered the heterogeneity of intrahepatic immune populations during NASH, particularly that of hepatic macrophages.( 4, 5 ) Although hepatic inflammation is a key event in the progression of steatosis to the more serious NASH, its triggers are less clear.( 6 ) Alterations in the gut microbiota associate with the severity of NAFLD( 7 ) in part due to the increased translocation of bacterial antigens into the liver.( 8 ) However, the mechanisms involved in the recognition of these antigens and the downstream immune effector functions remain unclear.
B lymphocytes can promote inflammatory disease through secretion of pro‐inflammatory cytokines, modulation of neighboring immune cells, and differentiation into autoreactive antibody‐secreting cells.( 9 ) Obesity‐associated metabolic disease is characterized by an accumulation of pro‐inflammatory B2 B cells in the visceral adipose tissue,( 10 ) a decrease of anti‐inflammatory B1a cells in the peritoneum,( 11 ) and a reduction in tolerogenic IgA+ B cells in the gut.( 12 ) In NAFLD, B cells have been shown to accumulate in the liver of mice with steatosis where they express increased pro‐inflammatory cytokines.( 13 ) Furthermore, depletion of B2 cells ameliorates hepatic inflammation and fibrosis in mice fed a methionine and choline‐deficient diet, which induces liver inflammation but causes severe weight loss.( 14 ) However, a precise role for intrahepatic B cells in obesity‐associated NASH, the mechanisms regulating their activation, and their contribution to disease relative to other inflammatory cells remain unclear.
Here, we demonstrate that pro‐inflammatory B cells accumulate in the NASH liver. B cell–deficient mice fed a NASH‐inducing diet showed improved hepatic insulin sensitivity, inflammation, and fibrosis. B cell–specific depletion of myeloid differentiation primary response 88 (MyD88) ameliorated NASH progression, suggesting that B‐cell activation is mediated by this canonical adaptor downstream of toll‐like receptor (TLR) signaling. To our surprise, B‐cell responses in NASH also involve B cell–receptor (BCR) signaling, suggesting that B cells integrate innate and adaptive immune mechanisms of activation. Finally, we show that fecal microbiota transplantation (FMT) of human NAFLD gut microbiotas into recipient mice increased the accumulation and activation of B cells in the liver. Overall, our findings demonstrate a critical role for hepatic B cells in driving NASH progression.
Experimental Procedures
Experimental Animals
Wild‐type (WT) C57BL/6J, μMT,( 15 ) Cd19cre,( 16 ) and Myd88fl ( 17 ) mice on the C57BL/6J background were purchased from Jackson Laboratory (Bar Harbor, ME). Nur77GFP mice( 18 ) were gifted by Kristin A. Hoqquist (University of Minnesota). At 6 weeks of age, mice received a normal chow diet (NCD) or high‐fat high‐carbohydrate diet (HFHC) (40%kcal palm oil, 20%kcal fructose, and 2% cholesterol) supplemented with 42 g/L of carbohydrates in the drinking water (55% fructose, 45% sucrose; Sigma‐Aldrich, St. Louis, MO).( 19 ) Additional details are found in the Supporting Information. Animal experiments followed the Guide for the Care and Use of Laboratory Animals and were approved by the University of Minnesota Institutional Animal Care and Use Committee.
Single‐Cell RNA Sequencing
Cells were barcoded with Totalseq‐A hashtag antibodies (Biolegend, San Diego, CA) for multiplexing. Samples were loaded into a 10x Genomics chip (Pleasanton, CA) and sequenced using a Novaseq S4 chip (Illumina, San Diego, CA). Analyses were performed using Seurat, and visualization of clusters was enabled using uniform manifold approximation and projection. Differential expression testing was performed using the Wilcoxon rank‐sum test. For a detailed description, see the Supporting Information.
Human Specimens
Fecal microbiota samples were collected from participants of the Minnesota Microbiota Therapeutics Donor Program and the Cincinnati Children’s Medical Center for NAFLD. Samples were obtained with institutional review board approval. Donors included a lean individual (body mass < 85th percentile) and 3 obese patients with diagnosed NAFLD (Supporting Table S2). For the assessment of B cells in human liver, we used specimens from a cohort of patients with NAFLD obtained with approval by the Research Ethics Board at the University Health Network. Consent was collected by the collaborators as approved by IRB. For more information, see the Supporting Information.
Fecal Microbiota Transplantation
Mice received a regimen of systemic and nonabsorbable antibiotics in the drinking water to improve engraftment before FMT.( 20 ) Mice received a single oral gavage with 100 μL of fecal material from either a lean or NAFLD human donor. DNA was extracted from mouse and human stool samples. For a description of microbiome sequencing, see the Supporting Information.
Statistical Analysis
Statistical significance between means was determined with an unpaired Student t test using GraphPad Prism 8.3 (La Jolla, CA). Spearman’s rank test was used to determine correlation. Data are presented as means ± SEM. Statistical significance was set to 5% and denoted by *P < 0.05, **P < 0.01, and ***P < 0.001.
Results
Pro‐inflammatory B Cells Accumulate in the NASH Liver
To determine the role of intrahepatic B cells in NASH progression, we fed WT mice an HFHC diet that induces human‐relevant features of NASH( 19 ) for up to 20 weeks. Compared with a NCD diet, HFHC feeding increased body weight, liver weight, hepatic triglycerides, and worsened insulin resistance as indicated by glucose (GTT), insulin (ITT), and pyruvate (PTT) tolerance tests (Supporting Fig. S1A). At 15 weeks of dietary intervention, HFHC‐fed mice showed signs of hepatic fibrosis (Supporting Fig. S1B). We isolated the immune cells from the livers of HFHC‐fed mice and performed cytometry by time of flight (CyTOF) to identify the major immune subsets by unsupervised clustering and high‐dimensional single‐cell analysis (Fig. 1A). Perfusion of the liver removed circulatory immune cells (Supporting Fig. S1C). Quantification of CyTOF data showed that CD19+ B220+ B cells comprise between 20% and 30% of all CD45+ immune cells in the liver, regardless of the diet (Supporting Fig. S1D). Compared with NCD controls, the livers of HFHC‐fed mice had an increasing number of monocytes and macrophages as early as 5 weeks, followed by B cells, natural killer (NK) cells, and dendritic cells (DCs) at 10 weeks, and T cells at 15 weeks of dietary intervention (Fig. 1B). Given that HFHC‐fed mice show all features of NASH after 15 weeks of HFHC feeding, we characterized B‐cell subsets at this time point. Regardless of diet, B2 cells constituted most intrahepatic B cells, and only a small proportion of B1b cells were detected (Fig. 1C). Compared with NCD, HFHC‐fed mice showed an increased frequency of B2 cells and a reduced percentage of B1b cells (Fig. 1C). As previously shown,( 21 ) most intrahepatic B2 cells were IgM+ IgD+, resembling splenic B2 cells (Supporting Fig. S1E). To assess B‐cell cytokine production, we determined the intracellular expression of TNF‐α and IL‐6 in B cells after ex vivo stimulation with phorbol 12‐myristate 13‐acetate (PMA) or several TLR agonists. HFHC feeding resulted in an increased number of B cells expressing TNF‐α and IL‐6, following PMA and lipopolysaccharide (LPS) stimulation, respectively (Fig. 1D and Supporting Fig. S1F). To measure the secreted amounts of these cytokines relative to other inflammatory cells, we purified intrahepatic B cells, neutrophils, and macrophages (Supporting Fig. S1G) and measured their secreted TNF‐α and IL‐6 in the supernatants after 5 days of culture with LPS. Macrophages released the highest amounts of TNF‐α and IL‐6, followed by neutrophils and B cells. All cell types released increased amounts of IL‐6 and TNF‐α in HFHC mice, compared with NCD controls (Fig. 1E). B cells from HFHC mice also showed up‐regulation of cell‐surface major histocompatibility complex (MHC) class I and II and CD86, suggesting increased activation and antigen‐presenting capacity (Fig. 1F). To establish the relevance of intrahepatic B cells in human disease, we assessed the infiltration of CD19+ B cells in liver biopsies from a cohort of patients with NAFLD and detected a positive correlation between their NAFLD activity score (NAS) and intrahepatic B cell accumulation (Supporting Results). Together, these results show that activated B cells, with an increased capacity to release pro‐inflammatory cytokines and present antigens, accumulate in the liver during NASH.
FIG. 1.
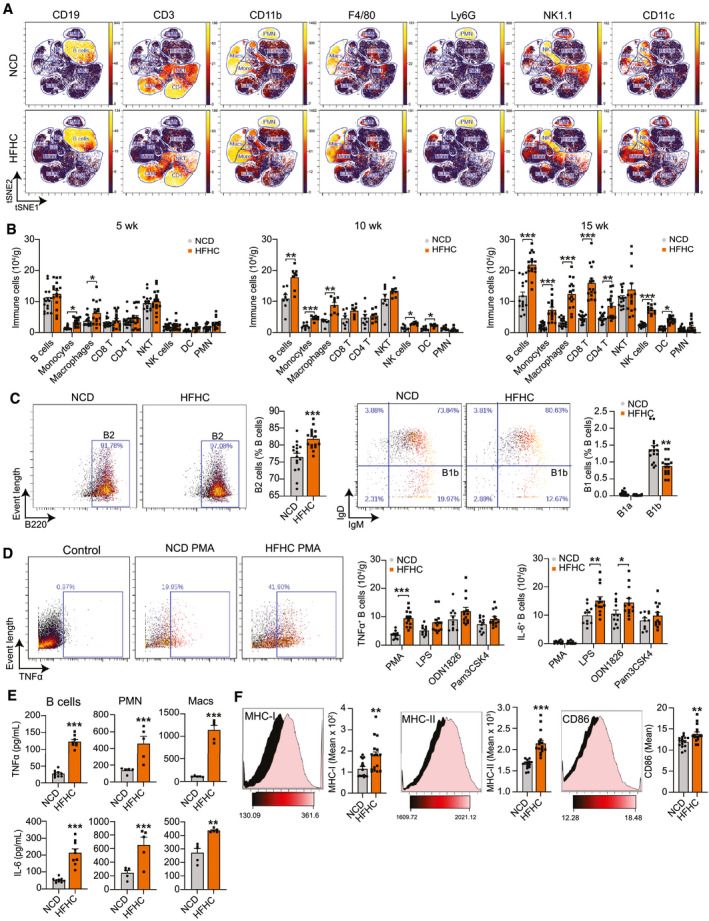
Pro‐Inflammatory B cells accumulate in the liver of mice with NASH. (A) Representative viSNE plots from CyTOF data showing unsupervised clustering and expression of CD19, CD3, CD11b, F4/80, Ly6G, NK1.1, and CD11c by intrahepatic immune cells from male mice fed either a NCD or HFHC diet for 15 weeks. Gates were drawn based on the expression of commonly used markers to distinguish B cells, monocytes, macrophages, CD8 and CD4 T cells, NKT cells, NK cells, DCs, and polymorphonuclear neutrophils. (B) Quantification of immune cell subsets from CyTOF data as in (A) in mice fed either an NCD or HFHC for 5 (n = 15 mice per group), 10 (n = 8 mice per group), or 15 weeks (n = 16 mice per group). (C) Representative CyTOF plot showing B2 cells (CD3− NK1.1− CD19+ CD23+ CD5− B220hi, far left) and their quantification expressed as a percentage of total B cells (middle left). Representative CyTOF plot showing B1b cells (CD3− NK1.1− CD19+ CD23− CD5− B220lo IgM+ IgD−, middle right) and quantification of B1a and B1b cells expressed as a percentage of total B cells (far right). Mice were fed either an NCD or HFHC for 15 weeks (n = 16 mice per group). (D) Representative CyTOF plot showing TNFα+ B cells gated from CD3− NK1.1− CD19+ B220+ cells in unstimulated control and PMA conditions (left) and quantification of TNF‐α+ (middle) and IL‐6+ (right) B cells after a 5‐hour stimulation with PMA or the TLR agonists LPS (TLR4), ODN1826 (TLR9), and Pam3CSK4 (TLR1 and 2) in mice fed either an NCD or HFHC for 15 weeks (n = 12 mice per group). (E) Quantification of TNF‐α and IL‐6 secreted by purified intrahepatic B cells (n = 9 wells per group, 2 pooled mice per well), polymorphonuclear neutrophils (n = 5 wells per group, 2 pooled mice per well), and macrophages (n = 5 wells per group, 2 pooled mice per well) from mice fed either an NCD or HFHC for 15 weeks. (F) Representative CyTOF histograms and quantification of mean intensity showing the B‐cell expression of MHC‐I (left), MHC‐II (middle), and CD86 (right) in mice fed either an NCD or HFHC for 15 weeks (n = 16 mice per group). Data correspond to at least three independent experiments and are presented as mean ± SEM. Statistical significance is denoted by *P < 0.05, **P < 0.01, and ***P < 0.001. See also Supporting Fig. S1. Abbreviation: PMN, polymorphonuclear neutrophil.
Intrahepatic B Cells Display a Pro‐inflammatory Gene Profile During NASH
To better understand how NASH alters the transcriptomic profile of B cells, we performed single‐cell RNA sequencing (RNA‐seq) of immune cells from NCD and HFHC livers. Unsupervised clustering of cells revealed 24 clusters of CD45+ immune cells (Fig. 2A). The identity of these clusters was determined using SingleR( 22 ) and confirmed by the differential expression of established immune subset marker genes (Supporting Fig. S2A). We found transcriptionally distinct clusters of B cells, monocytes, macrophages, natural killer T (NKT) cells, neutrophils, T cells, NK cells, plasmacytoid DCs, innate lymphoid cells, and conventional DCs (Fig. 2A and Supporting File S1). HFHC livers showed an increased percentage of clusters 2, 3, and 4 (monocytes), 5 (monocyte‐derived macrophages), 19 (neutrophils), 9 (CD8 T cells), and 17 (DCs), as well as a decreased percentage of clusters 11 and 13 (B cells), 7 (Kupffer cells [KCs]), 0 (NKT cells), and 14 (plasmacytoid DCs) (Fig. 2A). We focused on the gene‐expression profiles of B‐cell clusters and determined that the largest B‐cell subpopulation (cluster 1) consists of mature B cells expressing Sell, Fcer2a, H2‐aa, and Ighd (Fig. 2B). Cluster 11 were immature B cells expressing Vpreb3, Fam129C, and enriched for Ms4a1 (CD20) and Tnfsrf13c (B‐cell activating factor receptor [BAFF‐R]),( 23, 24, 25 ) while cluster 13 was high in mitochondrial genes (Fig. 2B). Next, we analyzed the differential expression of genes within each cluster between NCD and HFHC conditions (Supporting File S2). After combining B‐cell clusters, we detected 63 differentially expressed genes (DEGs) between NCD and HFHC livers (Supporting Table S3) that showed increased expression of the inflammatory genes Il‐1b, S100a8, Cxcl2, Cxcr4, and H2‐aa in HFHC B cells (Fig. 2C). Overall, these findings highlight the heterogeneity of the intrahepatic immune cell landscape and its transition toward an inflammatory phenotype in NASH. To improve our ability to detect B‐cell DEGs between NCD and HFHC conditions, we also performed bulk RNA‐seq analysis of purified B cells from the livers of NCD or HFHC mice. Multidimensional scaling showed a substantial separation between B cells from NCD and HFHC mice and a high congruency between biological replicates in the HFHC and some diversity in the NCD group (Fig. 2D, left). When plotting the top 500 variance genes in a hierarchical heatmap, differential clustering of NCD and HFHC B cells was evident (Fig. 2D, right). Gene‐expression analysis revealed 1,179 DEGs, including 570 up‐regulated and 609 down‐regulated in the HFHC B cells. Consistent with our single‐cell analysis, several pro‐inflammatory genes were up‐regulated in HFHC B cells (Fig. 2E and Supporting Fig. S2B). To interpret the biological meaning of these data, we used Ingenuity Pathway Analysis (IPA) to detect gene‐expression patterns. The IPA regulator effects tool predicted the activation of upstream regulator molecules involved in inflammation (TNF, interferon [IFN]‐γ , IL‐2, CD44, and IL‐18) and TLR signaling (MyD88, TLR4, IFN regulatory factor), as well as the inhibition of the anti‐inflammatory regulators IL‐10 receptor alpha and suppressor of cytokine signaling 1 (Fig. 2F, left). The most activated inflammation pathways included LPS/IL‐1 inhibition of retinoid X receptor function, acute phase response signaling, and role of pattern recognition receptors (Fig. 2F, right). These findings are consistent with our functional analyses of intrahepatic B cells showing increased activation and inflammatory phenotype in HFHC‐fed mice.
FIG. 2.
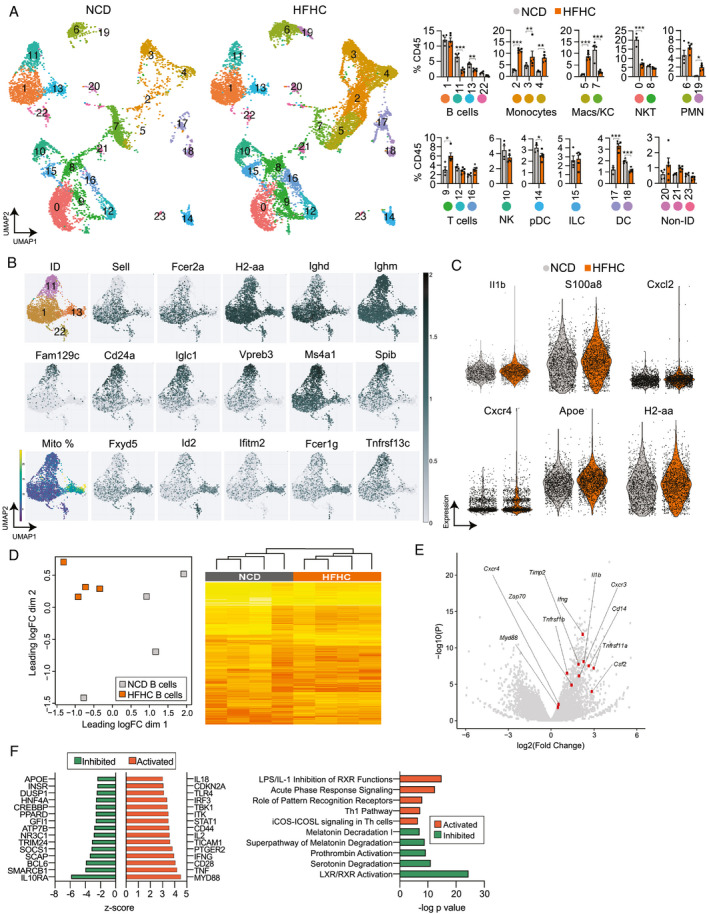
Intrahepatic B cells display a pro‐inflammatory gene profile during NASH. (A‐C) Single‐cell RNA‐seq of intrahepatic immune cells from WT mice fed either a NCD (n = 4) or HFHC (n = 5) for 15 weeks. After quality control and de‐multiplexing, we identified 18,700 unique single cells including 6,972 and 11,728 cells from NCD and HFHC livers, respectively. (A) Uniform manifold approximation and projection of single cells clustered in 24 unique clusters with identification of immune cell identity. NCD‐derived and HFHC‐derived cells were plotted separately to visualize their abundance (left) and relative abundance of each cluster of cells (right) in NCD (n = 4) and HFHC (n = 5) conditions. (B) Identification of B cell clusters based on the expression of the mature (cluster 1: Sell, Fcer2a, H2‐aa, and Ighd) and immature (cluster 11: Fam129c, Cd24a, Iglc1, Ms4a1, and Spib) B‐cell genes. Cluster 13 was enriched in mitochondrial genes and Fxyd, Id2, Ifitm2, Fcer1g, and Tnfrsf13c. (C) Violin plots showing the expression of inflammatory (Il‐1b, S100a8, Cxcl2, Cxcr4, and Apoe) and activation genes (H2‐aa), which were differentially expressed (adj. P < 0.05) between NCD and HFHC B‐cell clusters. (D‐F) B cells were purified from the liver of WT mice fed either an NCD or HFHC for 15 weeks, and their gene expression assessed by bulk RNA‐seq (n = 4 mice per group). (D) Multidimensional scaling plot based on normalized RNA‐seq gene‐expression data showing the pattern of proximities among groups (left) and hierarchical heatmap showing expression changes for the top 500 variance genes in intrahepatic B cells (right). (E) Volcano plot representation of gene‐expression analysis showing normalized fold change (x‐axis) and P value (y‐axis), highlighting selected pro‐inflammatory and pro‐fibrotic genes in intrahepatic B cells. (F) Top predicted upstream regulators, either activated (positive Z‐score) or inhibited (negative Z‐score) from IPA of differentially expressed genes in HFHC B cells, relative to NCD B cells (left) and top canonical pathways based on log‐transformed P values, either activated (positive Z‐score) or inhibited (negative Z‐score) from IPA of differentially expressed genes in HFHC B cells, relative to NCD B cells from mice (right). See also Supporting Fig. S2, Table S3, and Files S1 and S2. Abbreviations: Apoe, Apolipoprotein E; Cxcl2, Chemokine (C‐X‐C motif) ligand 2; Cxcl4, Chemokine (C‐X‐C motif) ligand 4; CD24a, Cluster of differentiation 24; Fam129c, B cell novel protein 1 / Family with sequence similarity 129 member C; Fcer1g, High affinity immunoglobulin epsilon receptor subunit gamma; Fcer2a, Fc receptor, IgE, low affinity II, alpha polypeptide; Fxyd, Fxyd protein; H2‐aa, Histocompatabiltiy 2, class II antigen A, alpha; ID, Identified; Id2, DNA‐binding protein inhibitor ID‐2; Ifitm2, Interferon induced transmembrane protein 2; Ighd, Immunoglobulin heavy constant delta; Iglc1, Immunoglobulin lambda constant 1; IL‐1b, Interleukin 1 beta; ILC, innate lymphoid cell; LXR, liver X receptor; Macs, macrophages; Ms4a1, B‐lymphocyte antigen CD20; pDC, plasmacytoid dendritic cell; RXR, retinoid X receptor; S100a8, S100 calcium‐binding protein A8; Sell, L‐selectin; Spib, Transcription factor Spi‐B; Tnfsrsf13c, B‐cell activating factor receptor; UMAP, uniform manifold approximation and projection.
B‐Cell Deficiency Ameliorates Inflammation and Fibrogenesis During NASH
To determine whether B cells play a direct role in the pathogenesis of NASH, we assessed NASH progression in 15‐week HFHC‐fed B cell–deficient μMT mice.( 15 ) Despite no differences in body weight (Supporting Fig. S3A), HFHC μMT mice showed an improved GTT (Supporting Fig. S3B), ITT (Supporting Fig. S3C), and PTT (Fig. 3A), compared with WT littermates. No differences were detected in liver weight (Fig. 3B, left) and liver triglycerides (Fig. 3B, middle), suggesting that metabolic improvements were independent of changes in steatosis. Hematoxylin and eosin (H&E) staining of liver sections and NAS scoring confirmed similar steatosis but revealed a lower total NAS score in μMT mice (Fig. 3B, right) driven by a lower inflammation component (Supporting Fig. S3D), compared with WT controls. In agreement, we found a substantial reduction in the hepatic expression of pro‐inflammatory genes in μMT mice (Fig. 3C, left). Compared with WT controls, HFHC μMT mice lacked intrahepatic B cells but showed no alteration in the total number of T cells, monocytes, macrophages, DCs, NK cells, and neutrophils (Fig. 3C, middle and right). As B cells can activate T cells through antigen presentation, we examined the frequencies of naïve, central memory (CM), and effector memory (EM) T cells and their expression of IFN‐γ and TNF‐α in HFHC μMT mice. Compared with WT controls, livers from HFHC μMT mice had decreased percentages of EM CD4 and CD8 T cells (Fig. 3D) and IFN‐γ+ CD4 T cells (Fig. 3E, left), but no differences in TNF‐α+ CD4 and CD8 T cells (Fig. 3E, right). HFHC μMT mice also showed markedly decreased fibrosis, as determined by trichrome staining‐based quantification of collagen deposition (Fig. 3F, left), lower expression of fibrogenesis genes (Fig. 3F, middle), and reduced serum levels of alanine transaminase (ALT) and aspartate transaminase (AST) (Fig. 3F, right), confirming that B cells promote liver injury during NASH. Importantly, no differences in metabolic tests (Supporting Fig. S3E) and intrahepatic immune cell frequencies (Supporting Fig. S3F) were detected between healthy NCD‐fed WT and μMT mice, suggesting that B cells cause metabolic and inflammatory disturbances in the setting of HFHC feeding.
FIG. 3.
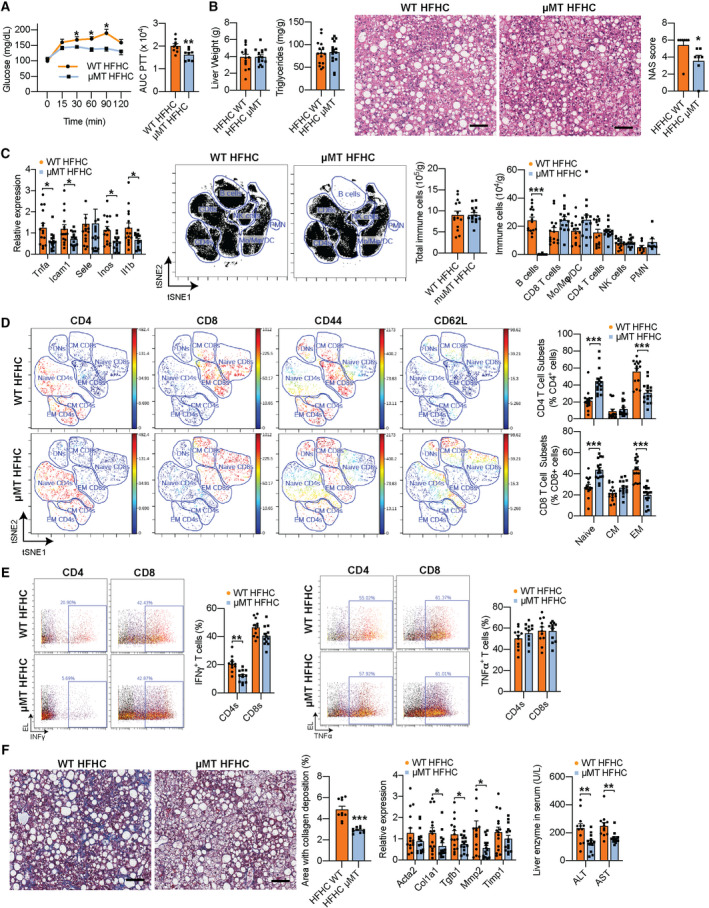
B‐cell deficiency ameliorates inflammation and fibrogenesis during NASH. (A‐F) WT and littermate μMT mice were fed the NASH‐inducing, HFHC diet for 15 weeks. (A) Pyruvate tolerance test (left) with corresponding areas under the curve (AUCs) (right; n = 12 mice per group). (B) Liver weight (far left; n = 15 mice per group), triglyceride content (middle left; n = 15 mice per group), and representative H&E liver stain (middle right; ×200 magnification; scale bar = 100 μm) with corresponding total NAS scores of H&E histology sections (far right, n = 7 mice per group). (C) Real‐time polymerase chain reaction gene‐expression analysis of liver pro‐inflammatory genes (far left; Tnfa, Icam1, Sele, Inos, and Il1b; n = 15 mice per group), representative viSNE plots from CyTOF data (middle left) showing unsupervised, uncolored clustering of intrahepatic B cells, monocytes, macrophages, DCs, CD4 and CD8 T cells, and NK cells with corresponding total immune cell count (middle right), and quantification of immune cell subsets (far right; n = 15 mice per group). (D) Representative viSNE plots from CyTOF data showing clustering of intrahepatic T cells with colored expression of CD4, CD8, CD44, and CD62L and gating of double‐negative T cells, naïve, EM, and CM CD4 and CD8 T cells (left), and quantification of naïve, CM, and EM CD4 and CD8 T cells (right; n = 15 mice per group). (E) Representative CyTOF plots and quantification of IFN‐γ+ CD4 and CD8 T cells (left) and TNF‐α+ CD4 and CD8 T cells (right) after a 5‐hour stimulation with PMA (n = 11 mice per group). (F) Representative trichrome liver stain (far left; ×200 magnification; scale bar = 100 μm) with quantification of the area with collagen deposition (middle left; n = 10 mice per group), RT‐PCR gene‐expression analysis of liver pro‐fibrotic genes (middle right; Acta2, Col1a1, Tgfb1, Mmp2, and Timp1; n = 15 mice per group), and serum ALT/AST levels (far right; n = 12 mice per group). Data correspond to at least three independent experiments and are presented as mean ± SEM. Statistical significance is denoted by *P < 0.05, **P < 0.01, and ***P < 0.001. See also Supporting Fig. S3. Abbreviations: Acta2, smooth muscle α actin; Col1a1, collagen type 1 alpha 1 chain; Mo, monocytes; Mφ, macrophages; RT‐PCR, real‐time polymerase chain reaction; Timp1, tissue inhibitor of metalloproteinase‐1.
Intrahepatic B Cells Stimulate EM and T Helper 1 Responses In NASH
Having established that genetic deficiency of B cells reduces T‐cell activation, we hypothesized that antibody‐mediated depletion of B cells can revert the generation of EM T cells during NASH. A single dose of monoclonal anti‐CD20 antibody (CD20 mAb) depleted most of the B cells in the liver, blood, spleen, and adipose tissue for at least 3 weeks (Supporting Fig. S4A). Thus, we injected three doses of CD20 mAb or isotype control into HFHC‐fed mice at weeks 6, 9, and 12 of dietary treatment and determined their effects at week 15. The CD20 mAb did not affect body and liver weight (Fig. 4A) and depleted mice of intrahepatic B cells without affecting other immune cell types (Fig. 4B). Similar to the genetic deficiency in μMT mice, CD20 mAb–mediated depletion of B cells decreased the frequency of EM CD4 and EM CD8 (Fig. 4C) T cells in the liver, confirming a role for B cells in promoting T‐cell activation during NASH.
FIG. 4.
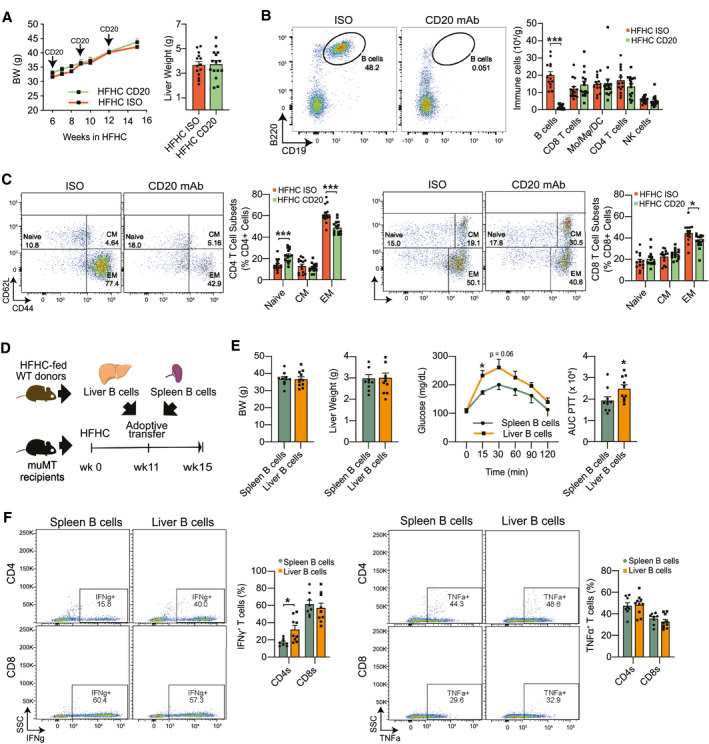
Intrahepatic B cells stimulate effector memory and Th1 responses in NASH. (A‐C) WT mice were injected with three doses of either a CD20 mAb or isotype control at 6, 9, and 12 weeks after initiation of HFHC feeding. Experiments were performed 15 weeks after the initiation of HFHC feeding. (A) Body (left) and liver weight (right; n = 15 mice per group). (B) Representative flow cytometry plot showing CD19+ B220+ B cells (left) and quantification of intrahepatic B cells, Mo, Mφ, DCs, CD4 and CD8 T cells, and NK cells (right; n = 15 mice per group). (C) Representative flow cytometry plot and quantification of naïve, EM, and CM CD4 T cells (left; n = 15 mice per group) and CD8 T cells (right, n = 15 mice per group). (D‐F) B cells (1 × 106 cells) were purified from either the spleen or liver of WT donor mice fed the HFHC diet for 15 weeks and adoptively transferred via intraperitoneal injection into μMT recipient mice fed the HFHC diet for 11 weeks. Experiments were performed 4 weeks after adoptive transfer. (D) Adoptive transfer experimental design. (E) Body (far left) and liver weight (middle left; n = 10 mice per group). PTT (middle right) with corresponding AUCs (far right; n = 10 mice per group). (F) Representative flow cytometry plot and quantification of IFN‐γ+ (left) and TNF‐α+ (right) CD4 and CD8 T cells after a 5‐hour stimulation with PMA (n = 8 mice per group). Data correspond to at least three independent experiments and are presented as mean ± SEM. Statistical significance is denoted by *P < 0.05, **P < 0.01, and ***P < 0.001. See also Supporting Fig. S4. Abbreviations: BW, body weight; ISO, isotype control; SSC, side scatter; wk, week.
To distinguish between the effects of intrahepatic and systemic B cells on T‐cell activation, we reconstituted HFHC‐fed μMT recipients with B cells from either the spleen or liver of HFHC‐fed WT donor mice (Fig. 4D). Despite no differences in body (Fig. 4E, left) and liver weights (Fig. 4E, middle), recipients of liver B cells showed worsened responses to a GTT (Supporting Fig. S4B), ITT (Supporting Fig. S4C), and PTT (Fig. 4E, right), compared with recipients of spleen B cells. The worsened metabolic parameters in recipients of liver B cells were accompanied by an increased frequency of IFN‐γ+ CD4 T cells, although no changes were detected in the frequency of TNF‐α+ CD4, or CD8 T cells (Fig. 4F). These findings show that while B‐cell deficiency prevents the accumulation of intrahepatic IFN‐γ+ CD4 T cells, reconstituting μMT mice with NASH‐associated liver B cells reverts this effect.
Cell‐Intrinsic MyD88 Mediates the Pro‐inflammatory Function of B Cells in NASH
Our bulk RNA‐seq analysis identified MyD88 as the top upstream regulator of pro‐inflammatory genes in liver B cells from HFHC mice (Supporting Fig. S5A). To investigate whether B cell–intrinsic MyD88 signaling influences NASH progression, we used mice with a B cell–specific deletion of MyD88 (B‐MYD) and investigated their responses to 15 weeks of HFHC feeding. HFHC‐fed B‐MYD mice showed no differences in body weight but displayed improved responses to a GTT, ITT (Supporting Fig. S5B), and PTT (Fig. 5A). Compared with WT controls, HFHC‐fed B‐MYD mice showed a similar liver weight (Fig. 5B, left) and triglyceride content (Fig. 5B, middle). H&E staining and corresponding NAS scoring showed no differences in steatosis but a lower total NAS score in B‐MYD mice (Fig. 5B, right) due to a lower inflammation NAS component (Supporting Fig. S5C). We detected no differences in the total number and subset immune cell populations between HFHC‐fed B‐MYD and WT mice (Fig. 5C). Nevertheless, intrahepatic B cells from HFHC‐fed B‐MYD mice showed reduced expression of cell‐surface MHC‐I and MHC‐II (Fig. 5D and Supporting Fig. S5D), suggesting a lower ability to present antigens due to reduced TLR‐mediated activation.( 26 ) Consistently, HFHC B‐MYD mice had a lower frequency of EM CD4 and CD8 T cells (Fig. 5E, top), as well as a decrease in the percentage of IFN‐γ+ CD4 T cells but not TNF‐α+ T cells (Fig. 5E, bottom). Most importantly, HFHC B‐MYD mice showed decreased collagen deposition (Fig. 5F, top), expression of fibrogenesis genes, and serum levels of ALT and AST (Fig. 5F, bottom), suggesting that MyD88 is required for B cells to promote liver inflammation, fibrosis, and injury during NASH. No differences were detected in metabolic parameters (Supporting Fig. S5E) and intrahepatic immune cell composition (Supporting Fig. S5F) in healthy NCD‐fed WT and B‐MYD mice, suggesting that B cell–intrinsic MyD88 signaling does not influence disease in basal conditions.
FIG. 5.
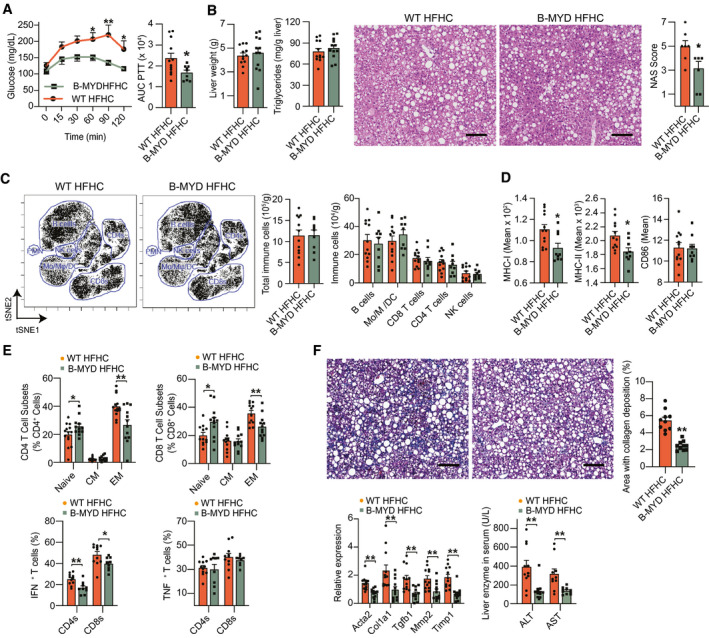
Cell‐intrinsic MyD88 mediates the inflammatory function of B cells in NASH. (A‐F) WT and littermate B‐MYD mice were fed the NASH‐inducing HFHC diet for 15 weeks. (A) PTT (left) with corresponding AUCs (right; n = 12 mice per group). (B) Liver weight (far left), liver triglyceride content (middle left; n = 12 mice per group), and representative H&E liver stain (middle right; ×200 magnification; scale bar = 100 μm) with corresponding total NAS scores (far right; n = 7 mice per group). (C) Representative viSNE plots from CyTOF data showing unsupervised, uncolored clustering of intrahepatic B cells, Mo, Mφ, DCs, CD4 and CD8 T cells, and NK cells (left), total immune cell count (middle), and quantification of immune cell subsets from CyTOF data (right; n = 12 mice per group). (D) Mean intensity of B‐cell expression of MHC‐I (left), MHC‐II (middle), and CD86 (right; n = 12 mice per group). (E) Frequency of naïve, EM, and CM CD4 (top left) and CD8 (top right) T cells determined by flow cytometry (n = 12 mice per group). Frequency of IFN‐γ+ (bottom left) and TNF‐α+(bottom right) CD4 and CD8 T cells after a 5‐hour stimulation with PMA (n = 10 mice per group). (F) Representative trichrome liver stain (top left; ×200 magnification; scale bar = 100 μm) and quantification of the area with collagen deposition (top right; n = 10 mice per group). RT‐PCR gene‐expression analysis of liver pro‐fibrotic genes (bottom left; Acta2, Col1a1, Tgfb1, Mmp2, and Timp1; n = 12 mice per group) and serum ALT (bottom right; n = 11 mice per group) and AST (n = 10 mice per group) levels. Data correspond to at least three independent experiments and are presented as mean ± SEM. Statistical significance is denoted by *P < 0.05, **P < 0.01, and ***P < 0.001. See also Supporting Fig. S5.
Intrahepatic B Cells Are Activated Through BCR Stimulation in NASH
We next determined whether intrahepatic B‐cell activation during NASH involves direct BCR stimulation using a reporter mouse in which BCR signaling induces green fluorescent protein (GFP) expression under the control of the Nur77 gene.( 18 ) We fed Nur77‐GFP mice an NCD or the HFHC diet for 15 weeks and determined the proportions of B cells expressing GFP and CD69 in the liver, colon, and spleen by flow cytometry. B cells from the liver of Nur77‐GFP mice expressed a basal level of GFP that was consistently above the WT background (Fig. 6A), as previously described.( 18 ) Compared with NCD controls, HFHC Nur77‐GFP mice had a substantial increase in the frequency and number of GFP+ CD69− intrahepatic B cells (Fig. 6A), indicating that activation during NASH involves preferential BCR stimulation in the liver. We determined the number of GFP‐expressing B cells in the colon lamina propria, as antigen‐specific BCR responses against gut microbes and self‐antigens have been shown in B cells from this compartment.( 27 ) Accordingly, our data show that HFHC Nur77‐GFP mice have increased GFP+ colonic lamina propria B cells (Fig. 6B). In contrast, no differences were detected in the frequency and number of GFP+ B cells from the spleen (Fig. 6C). The levels of Nur77 expression, which correlate with the strength of stimulus,( 18 ) increased in B cells from the liver (Fig. 6D), lamina propria (Fig. 6E), but not the spleen (Fig. 6F). As Nur77‐GFP transgenes are also specific for T‐cell receptors, we determined the GFP expression in T cells in mice with NASH. Compared with NCD controls, HFHC Nur77‐GFP mice showed increased frequency and expression intensity of GFP+ CD4 and CD8 T cells from the liver and colon, but not the spleen (Supporting Fig. S6). These data suggest that B cells and T cells perceive stronger antigen receptor signaling during NASH.
FIG. 6.
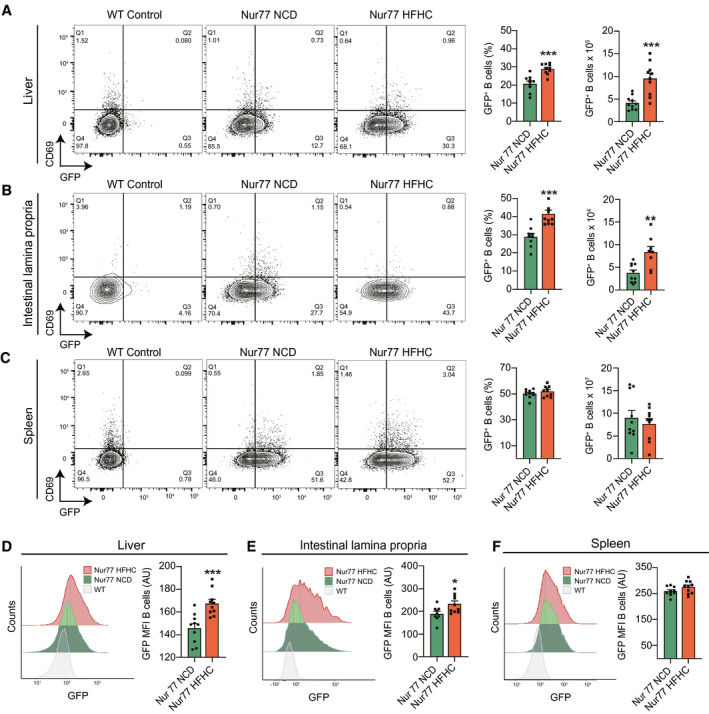
Intrahepatic B cells are activated through BCR in NASH. (A‐F) Nur77‐GFP mice were fed either a NCD or the HFHC diet for 15 weeks. WT mice were used as internal controls of GFP expression. (A) Representative flow cytometry plots (left), quantification (middle), and total number (right) of intrahepatic GFP+ CD69− B cells (n = 10 mice per group). (B) Representative flow cytometry plots (left), quantification (middle), and total number (right) of colonic lamina propria GFP+ CD69− B cells (n = 10 mice per group). (C) Representative flow cytometry plots (left), quantification (middle), and total number (right) of splenic GFP+ CD69− B cells (n = 10 mice per group). (D) Representative histograms (left) and mean fluorescence intensity quantification (right) of GFP in intrahepatic B cells (n = 10 mice per group). (E) Representative histograms (left) and mean fluorescence intensity quantification (right) of GFP in colon lamina propria B cells (n = 10 mice per group). (F) Representative histograms (left) and mean fluorescence intensity quantification (right) of GFP in splenic B cells (n = 10 mice per group). Data correspond to at least three independent experiments and are presented as mean ± SEM. Statistical significance is denoted by *P < 0.05, **P < 0.01, and ***P < 0.001. See also Supporting Fig. S6.
Intestinal‐Derived Microbial Factors Promote Intrahepatic B‐Cell Activation
To investigate whether bacterial products resulting from NAFLD‐associated dysbiosis promote intrahepatic B‐cell activation, we used a model of FMT consisting of antibiotic treatment of mice followed by oral gavage with human donor microbiota.( 20 ) NCD‐fed mice were treated with antibiotics to disrupt their indigenous gut microbiota before the FMT from either a healthy lean donor or a patient with NAFLD (donor A). The antibiotic treatment alone did not result in dehydration or weight loss (Supporting Fig. S7A). Compared with untreated mice, antibiotic treatment in the FMT groups resulted in a marked decrease in microbial diversity at the time of FMT (Fig. 7A left), suggesting a successful disruption of the indigenous microbiota. Characterization of fecal communities showed a depletion of the predominant phyla Bacteroidetes and Firmicutes in antibiotic‐treated mice at the time of FMT, which recovered by day 7 following FMT (Fig. 7A, right). Consistent with the composition of the donor, NAFLD microbiota recipients showed a decreased abundance of Firmicutes for at least 8 weeks, while Bacteroidetes and Proteobacteria increased during the first week following the FMT (Fig. 7B [left] and Supporting Fig. S7B). The NAFLD donor had an increased abundance of Bacteroides, a genus independently associated with NASH progression in patients with NAFLD.( 7 ) The similarity in genera between the human donors and their recipient mice peaked during the first week after the FMT and persisted for at least 5 weeks (Fig. 7B, right). Having validated our FMT strategy, we determined the effects of FMT of microbiotas from a total of 3 donors with NAFLD on the NASH progression of independent cohorts of recipient mice. Compared with the healthy FMT, recipients of the NAFLD microbiotas showed slightly increased liver weight, triglycerides, and H&E‐based steatosis and total NAS score 10 weeks after FMT (Fig. 7C). Mice that received the NAFLD FMT had increased number of intrahepatic B cells (Fig. 7D) with increased expression of antigen‐presenting and costimulatory molecules (Fig. 7E). In contrast, no differences were detected in the number of B cells in the spleen (Supporting Fig. S7C) and their expression of antigen‐presenting and costimulatory molecules (Supporting Fig. S7D). NAFLD FMT recipients showed slightly increased fibrosis, as determined by trichrome staining with quantification of collagen deposition (Fig. 7F, left), hepatic gene expression of Tgfb1 and matrix metalloproteinase (Mmp2) (Fig. 7F, middle), and serum AST (Fig. 7F, right). These data suggest that NAFLD‐related changes in the intestinal microbiota are sufficient to induce B‐cell accumulation and activation in the liver, while transmitting aspects of liver pathology from the human donor to the recipient mice.
FIG. 7.
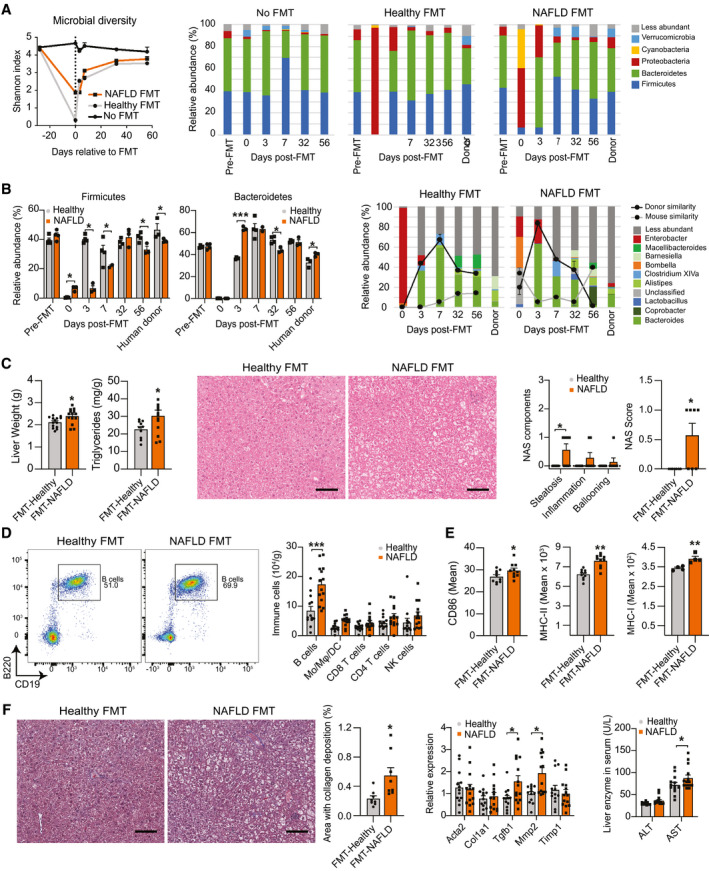
Intestinal‐derived microbial factors promote intrahepatic B‐cell activation. (A‐F) WT mice fed a NCD were left untreated (No FMT) or received antibiotics in the drinking water for 21 days followed by a single oral gavage with fecal material from a healthy lean (healthy FMT) or obese with NAFLD (NAFLD FMT) human donors. Fecal pellets from one cohort of recipient mice were collected for microbial analysis before antibiotic treatment (Pre‐FMT) and at 0, 3, 7, 32, and 56 days relative to the FMT. (A) Microbiota diversity determined by average Shannon indices in bacterial communities (left) and relative abundance of predominant phyla in fecal communities (right) of healthy FMT (n = 3 mice) and NAFLD FMT (donor A; n = 4 mice) donors and recipient mice at different time points relative to the FMT. (B) Relative abundance of Firmicutes and Bacteroidetes (left) and of predominant genera (right) in fecal communities of healthy FMT (n = 3 mice) and NAFLD FMT (donor A; n = 4 mice) donors and recipient mice at different time points relative to the FMT. The donor and mouse similarity at each time point were determined by SourceTracker analysis. The samples collected before FMT were used as the reference for mouse similarity analysis. (C) Liver weight and triglyceride content (left) of healthy FMT (n = 13 mice) and NAFLD FMT recipients (n = 14 mice) 10 weeks after FMT. Representative H&E liver stain (middle; ×200 magnification; scale bar = 100 μm) with individual component and total NAS score (right) of healthy and NAFLD FMT recipients (n = 7 mice per group) 10 weeks after FMT. (D) Representative flow cytometry plot showing CD19+ B220+ B cells (left), and total number of intrahepatic B cells, Mo, Mφ, DCs, CD4 and CD8 T cells, and NK cells (right) in healthy FMT (n = 13 mice) and NAFLD FMT recipients (n = 14 mice) 10 weeks after FMT. (E) Mean intensity of intrahepatic B‐cell expression of CD86 (left; n = 10 mice per group), MHC‐II (middle; n = 10 mice per group), and MHC‐I (right; n = 4 mice per group) in healthy FMT and NAFLD FMT recipient mice 10 weeks after FMT. (F) Representative trichrome liver stain (×200 magnification; scale bar = 100 μm) with quantification of the area with collagen deposition (left; n = 8 mice per group), RT‐PCR gene‐expression analysis of liver pro‐fibrotic genes (middle; Acta2, Col1a1, Tgfb1, Mmp2, and Timp1) of healthy FMT (n = 13) and NAFLD FMT (n = 14) recipient mice 10 weeks after FMT, and serum ALT and AST levels (right) of healthy FMT (n = 13) and NAFLD FMT (n = 14) recipient mice 10 weeks after FMT. Data shown in (E) and (F) represent three independent mouse experiments using three different NAFLD human donors (A, B, and C). Data are presented as mean ± SEM. Statistical significance is denoted by *P < 0.05, **P < 0.01, and ***P < 0.001. See also Supporting Fig. S7.
Discussion
We have identified intrahepatic B‐cell accumulation and activation as an important mechanism that contributes to the pathogenesis of NASH by promoting hepatic glucose intolerance, inflammation, and fibrosis. Compared with macrophages and neutrophils, we found that B cells secrete less inflammatory cytokines IL‐6 and TNF‐α on a per‐cell basis. However, we demonstrate that B cells are among the most abundant immune cells in the liver, suggesting a substantial contribution to hepatic inflammation in NASH. Previous research has shown that CD4, CD8, and NKT cells promote NASH through increased release of IL‐17, IFN‐γ, and LIGHT( 28, 29, 30, 31 ) in a process facilitated by antigen‐presenting cells such as DCs and macrophages.( 28, 32 ) Our data expand on this paradigm and suggest that B cells are strong inducers of T‐cell responses in NASH, as shown in other inflammatory diseases.( 33 ) Our single‐cell analysis provides insights into the cellular heterogeneity of the liver, showing remarkable alterations of the immune cellular landscape during NASH. In agreement with previous reports,( 5, 34 ) we found that NASH livers show a dramatic accumulation of monocytes and monocyte‐derived macrophages and a loss of KCs. Notably, our data revealed four distinct B‐cell populations, with the major cluster comprised of mature B cells expressing increased inflammatory genes during NASH. Unexpectedly, we identified a cluster of immature B cells in the liver that decreased in relative abundance in the NASH liver. This subset of B cells expressed higher levels of the gene encoding for BAFF‐R, suggesting that BAFF may promote the maturation and activation of B cells in NASH.( 14 ) Future research is needed to investigate the functional relevance of these B‐cell subsets in NASH.
Although TLR4 activation in NASH has been attributed to KCs and hepatocytes,( 35 ) B cells can respond to pathogen‐derived ligands through TLR4 signaling.( 36 ) Our bulk RNA‐seq data unveiled that B cells display an innate‐like gene signature with increased expression of Il‐1b, Csf2, and Ccl2 during NASH. A similar “innate” function has been described for B cells that secrete CSF‐2 following engagement of TLRs during bacterial sepsis,( 37 ) and IL‐1β‐producing adipose B cells that can respond to gut bacterial antigens in aging.( 38 ) In our study, B cells from NASH livers showed expression of the pro‐fibrotic genes Tgfb1 and Timp2, and B cell–deficient mice had decreased fibrosis. Indeed, B cells have been shown to promote fibrosis in a model of acute liver injury.( 21, 39 ) Such pro‐fibrotic function of B cells can be attributed to IL‐6 and instigation of T helper 1 (Th1) responses, which disrupt extracellular matrix turnover in chronic inflammation,( 40 ) activation of hepatic stellate cells through the production of TGF‐β1,( 41 ) or inhibition of extracellular matrix degradation through TIMP‐2.( 42 ) Our study revealed that the net effect of B‐cell depletion is the amelioration of NASH, providing evidence that B‐cell activation in the liver is causative toward NASH and not a bystander effect. We observed that B cells in NASH livers stimulate the generation of EM CD4 and CD8 T cells, which promotes inflammation during NAFLD.( 31 ) Our finding that intrahepatic, but not splenic, B cells promote Th1 responses suggests that the B‐cell inflammatory phenotype may result from the local activation and is not a systemic event. This is consistent with studies showing that inflamed tissues such as the obese adipose tissue alters the function of tissue‐resident, but not systemic, B cells.( 10 ) As NASH pathogenesis involves the activity of multiple immune cell types, the potential crosstalk among B cells, macrophages, neutrophils, DCs, and NKT cells remains to be determined.
Mechanistically, we show that MyD88 signaling is an intrinsic mechanism by which B cells may respond to endogenous or exogenous antigens during NASH. This finding is consistent with previous research suggesting that MyD88 signaling is required for the pro‐fibrotic role of B cells in a carbon tetrachloride model of liver injury.( 39 ) TLR4 ligands can activate B cells through the TLR4–MyD88 pathway, leading to stimulation of the NF‐κB.( 43 ) By taking advantage of the Nur77 reporter mouse, our study raises the prospect that antigen‐specific mechanisms drive the B‐cell activation during NASH. One possibility is that B‐cell activation during NASH occurs simultaneously through the MyD88 and BCR signaling pathways, as antigens such as LPS can dually engage the TLR4 and BCR.( 44 ) Our data suggest that gut‐derived microbial antigens may serve as ligands that activate intrahepatic B cells during NASH. Gut‐derived microbes can influence the immune system directly or through their secretion of metabolites such as short‐chain fatty acids.( 45 ) Thus, bacterial metabolites draining into the liver during NASH may influence intrahepatic B‐cell function through MyD88‐dependent or independent pathways. In addition to exogenous ligands, endogenous DNA‐containing antigens released from dying cells can activate B cells through TLR9.( 46 ) Indeed, mitochondrial DNA released from lipid‐laden hepatocytes is elevated in humans and mice with NASH and can promote disease in a TLR9‐dependent manner.( 47 ) Finally, our quantification of intrahepatic B cells in a cohort of patients with NAFLD provides evidence of B‐cell infiltration in human NASH, in agreement with previous reports.( 14, 48 ) Future research should aim to understand the changes in B‐cell infiltration at different stages of human NASH and the functional tissue spatial organization of immune cells. In summary, this study demonstrates an important role for intrahepatic B cells in promoting NASH in a process involving MyD88 and BCR signaling. Notably, we show that gut‐derived bacterial products associated with a dysbiotic NAFLD microbiota promote the accumulation and activation of intrahepatic B cells.
Author Contributions
X.S.R., F.B, and S.K. conceived the study and designed the experiments. X.S.R., F.B, and S.K. interpreted results, generated figures and tables, and wrote the manuscript. F.B, S.K., G.F., H.W., K.D., S.R., P.P., and X.S.R. performed the experiments. A.H. performed the analysis of single‐cell RNA‐seq data. M.M. provided human stool samples for FMT experiments. T.K. and C.S. performed the FMT experiments and analyzed the microbiota sequencing data. D.A.W., S.W., and O.A. analyzed and provided data from the human liver specimens. A.K., K.A.H., and C.S. provided feedback and supervised aspects of the study. X.S.R. obtained funding for, supervised, and led the overall execution of the study.
Supporting information
Supplementary Material
File S1
File S2
Acknowledgment
The authors thank the staff from the Research Animal Resources, University Flow Cytometry Resource, Mass Cytometry Facility, Genomics Center, and the Clinical and Translational Science Institute at the University of Minnesota for their assistance. Particularly, we thank Juan Abrahante (Genomics Center) for his contribution to the RNA‐seq analysis. We also thank Seokwon Jo for his assistance with mouse studies.
Supported by the National Institute of Diabetes and Digestive and Kidney Diseases (DK122056 to X.S.R.), the American Association of Immunologists (Careers in Immunology Fellowship to X.S.R), and the Canadian Liver Foundation and the Canadian Institutes of Health Research (FDN‐148385 to D.A.W.).
Potential conflict of interest: Nothing to report.
References
Author names in bold designate shared co‐first authorship.
- 1.Younossi Z, Anstee QM, Marietti M, Hardy T, Henry L, Eslam M, et al. Global burden of NAFLD and NASH: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol 2018;15:11‐20. [DOI] [PubMed] [Google Scholar]
- 2.Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology 2010;52:1836‐1846. [DOI] [PubMed] [Google Scholar]
- 3.Parthasarathy G, Revelo X, Malhi H. Pathogenesis of nonalcoholic steatohepatitis: an overview. Hepatol Commun 2020;4:478‐492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xiong X, Kuang H, Ansari S, Liu T, Gong J, Wang S, et al. Landscape of intercellular crosstalk in healthy and NASH liver revealed by single‐cell secretome gene analysis. Mol Cell 2019;75:644‐660.e645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Remmerie A, Martens L, Thoné T, Castoldi A, Seurinck R, Pavie B, et al. Osteopontin expression identifies a subset of recruited macrophages distinct from Kupffer cells in the fatty liver. Immunity 2020;53:641‐657.e614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schuster S, Cabrera D, Arrese M, Feldstein AE. Triggering and resolution of inflammation in NASH. Nat Rev Gastroenterol Hepatol 2018;15:349‐364. [DOI] [PubMed] [Google Scholar]
- 7.Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo‐Perez F, et al. The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 2016;63:764‐775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cani PD, Amar J, Iglesias MA, Poggi M, Knauf C, Bastelica D, et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 2007;56:1761‐1772. [DOI] [PubMed] [Google Scholar]
- 9.Hofmann K, Clauder AK, Manz RA. Targeting B cells and plasma cells in autoimmune diseases. Front Immunol 2018;9:835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Winer DA, Winer S, Shen L, Wadia PP, Yantha J, Paltser G, et al. B cells promote insulin resistance through modulation of T cells and production of pathogenic IgG antibodies. Nat Med 2011;17:610‐617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shen L, Chng MH, Alonso MN, Yuan R, Winer DA, Engleman EG. B‐1a lymphocytes attenuate insulin resistance. Diabetes 2015;64:593‐603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luck H, Khan S, Kim JH, Copeland JK, Revelo XS, Tsai S, et al. Gut‐associated IgA+ immune cells regulate obesity‐related insulin resistance. Nat Commun 2019;10:3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F, Jiang WW, Li X, Qiu XY, Wu Z, Chi YJ, et al. Role of intrahepatic B cells in non‐alcoholic fatty liver disease by secreting pro‐inflammatory cytokines and regulating intrahepatic T cells. J Dig Dis 2016;17:464‐474. [DOI] [PubMed] [Google Scholar]
- 14.Bruzzì S, Sutti S, Giudici G, Burlone ME, Ramavath NN, Toscani A, et al. B2‐Lymphocyte responses to oxidative stress‐derived antigens contribute to the evolution of nonalcoholic fatty liver disease (NAFLD). Free Radic Biol Med 2018;124:249‐259. [DOI] [PubMed] [Google Scholar]
- 15.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell‐deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature 1991;350:423‐426. [DOI] [PubMed] [Google Scholar]
- 16.Rickert RC, Roes J, Rajewsky K. B lymphocyte‐specific, Cre‐mediated mutagenesis in mice. Nucleic Acids Res 1997;25:1317‐1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hou B, Reizis B, DeFranco AL. Toll‐like receptors activate innate and adaptive immunity by using dendritic cell‐intrinsic and ‐extrinsic mechanisms. Immunity 2008;29:272‐282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moran AE, Holzapfel KL, Xing Y, Cunningham NR, Maltzman JS, Punt J, et al. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J Exp Med 2011;208:1279‐1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohli R, Kirby M, Xanthakos SA, Softic S, Feldstein AE, Saxena V, et al. High‐fructose, medium chain trans fat diet induces liver fibrosis and elevates plasma coenzyme Q9 in a novel murine model of obesity and nonalcoholic steatohepatitis. Hepatology 2010;52:934‐944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Staley C, Kaiser T, Beura LK, Hamilton MJ, Weingarden AR, Bobr A, et al. Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome 2017;5:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Novobrantseva TI, Majeau GR, Amatucci A, Kogan S, Brenner I, Casola S, et al. Attenuated liver fibrosis in the absence of B cells. J Clin Invest 2005;115:3072‐3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aran D, Looney AP, Liu L, Wu E, Fong V, Hsu A, et al. Reference‐based analysis of lung single‐cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019;20:163‐172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kleiman E, Salyakina D, De Heusch M, Hoek KL, Llanes JM, Castro I, et al. Distinct transcriptomic features are associated with transitional and mature B‐cell populations in the mouse spleen. Front Immunol 2015;6:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohnishi K, Takemori T. Molecular components and assembly of mu.surrogate light chain complexes in pre‐B cell lines. J Biol Chem 1994;269:28347‐28353. [PubMed] [Google Scholar]
- 25.Hong R, Lai N, Ouchida R, Xiong E, Zhou Y, Min Q, et al. The B cell novel protein 1 (BCNP1) regulates BCR signaling and B cell apoptosis. Eur J Immunol 2019;49:911‐917. [DOI] [PubMed] [Google Scholar]
- 26.Barr TA, Brown S, Ryan G, Zhao J, Gray D. TLR‐mediated stimulation of APC: distinct cytokine responses of B cells and dendritic cells. Eur J Immunol 2007;37:3040‐3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Benckert J, Schmolka N, Kreschel C, Zoller MJ, Sturm A, Wiedenmann B, et al. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen‐specific. J Clin Invest 2011;121:1946‐1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maricic I, Marrero I, Eguchi A, Nakamura R, Johnson CD, Dasgupta S, et al. Differential activation of hepatic invariant NKT cell subsets plays a key role in progression of nonalcoholic steatohepatitis. J Immunol 2018;201:3017‐3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rau M, Schilling A‐K, Meertens J, Hering I, Weiss J, Jurowich C, et al. Progression from nonalcoholic fatty liver to nonalcoholic steatohepatitis is marked by a higher frequency of Th17 cells in the liver and an increased Th17/resting regulatory T cell ratio in peripheral blood and in the liver. J Immunol 2016;196:97‐105. [DOI] [PubMed] [Google Scholar]
- 30.Wolf M, Adili A, Piotrowitz K, Abdullah Z, Boege Y, Stemmer K, et al. Metabolic activation of intrahepatic CD8+ T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross‐talk with hepatocytes. Cancer Cell 2014;26:549‐564. [DOI] [PubMed] [Google Scholar]
- 31.Ghazarian M, Revelo XS, Nøhr MK, Luck H, Zeng K, Lei H, et al. Type I interferon responses drive intrahepatic T cells to promote metabolic syndrome. Sci Immunol 2017;2:eaai7616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas JT, Vonghia L, Mogilenko DA, Verrijken AN, Molendi‐Coste O, Fleury S, et al. Transcriptional network analysis implicates altered hepatic immune function in NASH development and resolution. Nat Metab 2019;604‐614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan J, Harvey BP, Gee RJ, Shlomchik MJ, Mamula MJ. B cells drive early T cell autoimmunity in vivo prior to dendritic cell‐mediated autoantigen presentation. J Immunol 2006;177:4481‐4487. [DOI] [PubMed] [Google Scholar]
- 34.MacParland SA, Liu JC, Ma X‐Z, Innes BT, Bartczak AM, Gage BK, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun 2018;9:4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rivera CA, Adegboyega P, van Rooijen N, Tagalicud A, Allman M, Wallace M. Toll‐like receptor‐4 signaling and Kupffer cells play pivotal roles in the pathogenesis of non‐alcoholic steatohepatitis. J Hepatol 2007;47:571‐579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrio L, Saez de Guinoa J, Carrasco YR. TLR4 signaling shapes B cell dynamics via MyD88‐dependent pathways and Rac GTPases. J Immunol 2013;191:3867‐3875. [DOI] [PubMed] [Google Scholar]
- 37.Rauch PJ, Chudnovskiy A, Robbins CS, Weber GF, Etzrodt M, Hilgendorf I, et al. Innate response activator B cells protect against microbial sepsis. Science 2012;335:597‐601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Camell CD, Günther P, Lee A, Goldberg EL, Spadaro O, Youm Y‐H, et al. Aging induces an Nlrp3 inflammasome‐dependent expansion of adipose B cells that impairs metabolic homeostasis. Cell Metab 2019;30:1024‐1039.e1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thapa M, Chinnadurai R, Velazquez VM, Tedesco D, Elrod E, Han J‐H, et al. Liver fibrosis occurs through dysregulation of MyD88‐dependent innate B‐cell activity. Hepatology 2015;61:2067‐2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fielding C, Jones G, McLoughlin R, McLeod L, Hammond V, Uceda J, et al. Interleukin‐6 signaling drives fibrosis in unresolved inflammation. Immunity 2014;40:40‐50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xu L, Hui AY, Albanis E, Arthur MJ, O'Byrne SM, Blaner WS, et al. Human hepatic stellate cell lines, LX‐1 and LX‐2: new tools for analysis of hepatic fibrosis. Gut 2005;54:142‐151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brew K, Nagase H. The tissue inhibitors of metalloproteinases (TIMPs): an ancient family with structural and functional diversity. Biochim Biophys Acta 2010;1803:55‐71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schweighoffer E, Nys J, Vanes L, Smithers N, Tybulewicz VLJ. TLR4 signals in B lymphocytes are transduced via the B cell antigen receptor and SYK. J Exp Med 2017;214:1269‐1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pone EJ, Zhang J, Mai T, White CA, Li G, Sakakura JK, et al. BCR‐signalling synergizes with TLR‐signalling for induction of AID and immunoglobulin class‐switching through the non‐canonical NF‐κB pathway. Nat Commun 2012;3:767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 2016;20:202‐214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sindhava VJ, Oropallo MA, Moody K, Naradikian M, Higdon LE, Zhou L, et al. A TLR9‐dependent checkpoint governs B cell responses to DNA‐containing antigens. J Clin Invest 2017;127:1651‐1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Garcia‐Martinez I, Santoro N, Chen Y, Hoque R, Ouyang X, Caprio S, et al. Hepatocyte mitochondrial DNA drives nonalcoholic steatohepatitis by activation of TLR9. J Clin Invest 2016;126:859‐864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gadd VL, Skoien R, Powell EE, Fagan KJ, Winterford C, Horsfall L, et al. The portal inflammatory infiltrate and ductular reaction in human nonalcoholic fatty liver disease. Hepatology 2014;59:1393‐1405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
File S1
File S2


