Abstract
Phytic acid is abundant in seeds, roots and stems of plants, it acts as an anti-nutrient in food and feed industry, since it affects the absorption of nutrients by humans and monogastric animals. Furthermore, phosphorus produced through its decomposition by microorganisms can cause environmental pollution. Phytase degrades phytic acid generating precursors of inositol that can be used in clinical practice; in addition, phytase treatment can minimize the anti-nutritional effect of phytic acid. The use of phytase synthesized from Bacillus is more advantageous due to its high activity. Additionally, its good heat resistance under neutral conditions greatly fills the gap of commercial utilization of acid phytase. In this review, we summarize the latest research results on Bacillus phytase, including its physiological and biochemical characteristics, molecular structure information, calcium effects on its catalytic activity and stability, its catalytic mechanism and molecular modification.
Keywords: Bacillus phytase, BPPhy, Physiological and biochemical characteristics, Molecular structure information, Catalytic mechanism, Molecular modification
Introduction
A very large amount of organic phosphorus in plants, soils, as well as in food and feed containing sorghum, glutinous rice, bran and other ingredients are in the form of phytic acid (also known as inositol hexaphosphate) (Jain et al. 2016). Phytic acid is known to be an anti-nutrient in the food and feed industry. In this regard, food and feed containing phytic acid cannot be properly absorbed by monogastric animals including humans, since phytic acid is able to chelate several minerals, so reducing the amount of bioavailable micronutrients. As regards feed, the amount of phytate phosphorus not absorbed and utilized by animals can cause phosphorus pollution in the environment. This hazard is particularly serious in areas with intensive animal husbandry (Balaban et al. 2016; Kumar et al. 2015a, b; Singh et al. 2018; Verma et al. 2016; Yang et al. 2019b). To reduce the anti-nutritional effect of phytic acid and to prevent environmental pollution, the application of genetic engineering technologies has allowed the production of phytase products employed in food and feed markets. Commercial phytase can effectively hydrolyze the phosphate of phytic acid, thereby reducing environmental pollution, and improving the utilization of phosphorus in monogastric animals (Farhat-Khemakhem et al. 2018). The inositol produced by the gradual decomposition of phytic acid by phytase has also great application value in feed, food, medicine and other industries, since inositol can be added to feed as a biological promoter for aquaculture and animal husbandry, and it can also be added to food as a nutritional fortifier. Remarkably, as a biologically active analog of vitamins B1 and H, inositol can be used to treat many chronic pathologies such as cardiovascular, cerebrovascular and liver diseases, as well as diabetes, and obesity (David et al. 2012; Dong 2015; Fan et al. 2019; Lu et al. 2018; Perelló et al. 2018; Shi 2017; Singh et al. 2018; Yang et al. 2019b).
The enzymes responsible for decomposing phytate in the fermentation process and their synthetic pathways are gradually being elucidated. However, limited to the understanding of the molecular level of these enzymes, the catalytic mechanism of Bacillus phytase, the regulation of its enzymatic activity, as well as its physiological functions are still unclear or controversial. Therefore, clarifying the biocatalytic mechanism through which Bacillus phytase degrades phytic acid to synthesize inositol and its phosphatidyl derivatives is of great significance in order to further improve food quality, to increase feed utilization and to promote environmental protection. Based on the existing researches, this review makes a brief summary of the research status of Bacillus phytase and provides reference for further research.
Phytase classification and related biochemical characteristics
Phytase is an extracellular enzyme belonging to the class of phosphomonoesterase, which can catalyze the hydrolysis of phytate to remove the phosphate group. Phytases are widely distributed in nature, as they were found in animals, plants and microorganisms (Kumar 2018; Rocky-Salimi et al. 2016; Vashishth et al. 2017). There are many types of phytases, their structures and properties are quite different, as well as their catalytic mechanisms According to their optimum pH, phytases can be divided into alkaline and acid phytases. Fungi, most bacteria and plants can secrete acid phytases, whereas a small number of bacteria and plants can produce neutral or alkaline phytases (Kai et al. 2015). There are various types of phytases, and also various sources. According to their catalytic mechanism, 3D structure and specific sequence properties, phytases are mainly divided into four categories: histidine acid phosphatase (HAPhy), cysteine phytase (CPhy), purple acid phosphatase (PAPhy) and β-propeller phytase (BPPhy) (Table 1). According to their consecutive phosphorylation sites, phytases are divided into 3-phosphatase, 5-phosphatase and 4/6-phosphatase, among them, BPPhy is the only phytase retaining enzyme activity under neutral or alkaline conditions (Chen et al. 2016; Greiner et al. 2007; Jain et al. 2016; Mullaney et al. 2007; Singh et al. 2018). BPPhy exerts several functions in different species, which are determined by its very different biochemical characteristics. BPPhy has good thermal stability, protease resistance and absolute substrate specificity (Jain et al. 2016; Lu et al. 2017; Reddy et al. 2015). BPPhy derived from Bacillus licheniformis can retain 80% of its enzyme activity after being treated at 95 °C for 10 min, and it displays almost no activity on phosphates other than those of phytate (Tye et al. 2002).
Table 1.
Classification and characteristics of phytases
| Source | Specific sequence properties | Active areas | Refs |
|---|---|---|---|
| Histidine acid phytases (HAPhy) | Conservative motif of the active center (RHGXRXP), conservative cysteine motif and CAP with the catalytic C-terminal dipeptide (HD) | pH 2.5–5.5 | (Ullah et al. 2011; Wang et al. 2004; Wyss et al. 1998) |
| Cysteine phytase (Cphy) | β-fold and α-helix domain and two α-helices near the C-terminal | pH 2.5–5.5 | (Kumar and Agrawal 2014; Piddington et al. 1993) |
| Purple acid phosphatase (PAPhy) | Motifs DXG, GDXXY, GNH [E / D], VXXH and GHXH | pH 2.5–5.5 | (Devillers et al. 2003; Kyrtopoulos et al. 1985) |
| β-propeller phytase (BPPhy) | P-loop HCXXGXXR and a WPD loop | Neutral or alkalescence | (Chan et al. 2006; Fang et al. 2018; Viader-Salvado et al. 2010) |
The catalytic mechanism of phytase is determined by its conserved sites; in particular, its active site, substrate binding site, ion binding site and other key sites form a unique "fingerprint" motif. These motifs can account for the tissue differences observed among different phytase groups (Balaban et al. 2016). Distinct phytase families have their own motifs and specific organizations, and the main types of important active amino acid residues reside in these motifs. Known eukaryotic and prokaryotic phytases have distinct motif structures, which might be related to changes in enzymatic functions during the evolutionary pathway of phytases (Fan et al. 2013).
HAPhy has an active site motif and a "two-step catalysis" mechanism, indeed, a conservative motif in the active center (RHGXRXP) and a conservative cysteine motif, related to the formation of disulfide bonds and responsible for thermal stability, are used for substrate binding or product release (Kostrewa et al.; Lee et al. 2003; Mullaney and Ullah 2005; Pramanik et al. 2018; Ullah and Dischinger 1993; Vohra and Satyanarayana 2003). CPhy contains two domains, i.e., a larger domain contains a β-sheet and three α-helices, the other domain is close to the C-terminus and contains two α-helices useful to promote substrate binding. PAPhy is usually a dimer containing some phosphomonoesterase motifs such as DXG, GDXXY, GNH [E/D], VXXH and GHXH, and including seven key amino acid residues for binding of metal ions (Klabunde et al. 1996; Olczak et al. 2003; Schenk et al. 2000). Animal PAPhy contains a binuclear metal center [Fe(III)Fe(II)], while plant PAPhy is mainly a homodimeric protein containing an iron ion Fe(III) that is connected to a zinc or manganese ion (Dionisio et al. 2007; Olczak et al. 2003). Alkaline PhyAsr (known as protein tyrosine phosphatase (PTP)-like phytase) is a kind of phytase from ruminants, it resides in their rumen and its sequence is very similar to that of other microbial phytases. Its active center contains a phosphate binding loop (P loop, HCXXGXXR) and a WPD loop, which form the substrate binding region (pocket) of tyrosine phosphatase (PTPs) (Chu et al. 2004; Puhl et al. 2007, 2008).
BPPhy exists in a variety of microorganisms including archaea(Becker et al. 2014), bacteria(Vieira Velloso et al. 2020), cyanobacteria(Teikari et al. 2015) and Arthrobotrys oligospora(Hou et al. 2020), BPPhy produced by Bacillus is also frequently reported. Differently from HAPs and PAPs, which display enzyme activity in an acidic environment (pH 2.5–5.5), the optimum pH for most BPPhys is neutral. According to literature data, PhyP of P. nyackensis MJ11 retains about 50% of its activity at pH 8.5, but has no activity at pH 9.0 (Huang et al. 2009); PhyS of S. oneidensis MR-1 is active under the pH range of 5.0–7.5, both enzymes are active and stable (Cheng and Lim 2006); PhyA115 of Janthinobacterium sp. TN115 shows the greatest activity at pH 8.5, and displays more than 60% of its activity in the pH range of 6.0–9.0 (Zhang et al. 2011). If compared with these BPPhys, BPPhy of Bacillus displays the same enzyme activity in the neutral or weak alkaline range, and it also exhibits high thermal stability (Choi et al. 2001; Kerovuo et al. 1998; Lu et al. 2014, 2017; Pal Roy et al. 2017; Powar and Jagannathan 1982; Shimizu 1992). Bacillus BPPhy has no conserved motif unique to acid phytases. Instead, BPPhys have a six-blade β-propeller structure (Kostrewa et al. 1997), and through the separation of each second phosphate group adjacent to the first one, phytic acid is stereospecifically dephosphorylated (Greiner et al. 2007). The catalytic reactions of these enzymes are uniquely dependent on the availability of calcium ions, since such enzymes have strict substrate specificity and can only hydrolyze calcium–phytate complexes (Fu et al. 2008). Optimal pH, molecular structure and catalytic mechanism of BPPhy are also different from acid phytases. BPPhys are resistant to papain, pancreatin and trypsin activity, but they are sensitive to pepsin, possibly because they are denatured at the low pH values necessary for pepsin activation (Lei et al. 2013). Although the structural characteristics, as well as the physical and chemical properties of BPPhys have been well studied, there are few and conflicting studies on their catalytic mechanism, which makes it difficult to analyze the degradation pathways of phytate complexes and to determine final products.
Enzymatic properties of Bacillus BPPhy
Environmental factors such as pH and temperature have a great impact on the activity of phytases. The enzymatic properties of neutral phytases of various Bacillus species are basically the same, since they have similar molecular weights and the optimal pH value for enzymatic reactions is about 7.0, which can effectively make up for the deficiency of acid phytases. Generally, phytases can maintain a high activity in a temperature range of 50–70 °C. In most cases, their activity is stable in the range of 45–60 °C, and it is easy to inactivate them by increasing the temperature. Bacillus phytase has a better thermal stability than that of acid phytases derived from fungi and E. coli. This advantage helps to prevent enzyme inactivation caused by high temperature during feed pelleting or expansion; hence, Bacillus phytase can be used as an ideal animal feed additive (Arastoo et al. 2019).
A variety of divalent metal ions can regulate the activity of Bacillus BPPhy; indeed, Ca2+ acts as an activator for BPPhy of Bacillus licheniformis and Bacillus subtilis, whereas their metal inhibitors include Cd2+, Mn2+, Cu2+, Ba2+, Hg2+, Zn2+, Co2+, Fe2+ and others. Metal ions inhibit BPPhy since the formation of metal–phytate complexes leads to a poor binding to active sites and to a scant substrate availability. These complexes may prevent substrate from contacting BPPhy resulting in a reduced enzyme activity (Kumar et al. 2014b; Reddy et al. 2015). Some acid phytase competitive inhibitors such as fluoride can competitively inhibit acid phytase, but they have no effect on BPPhy (Kim et al. 1999; Maenz et al. 1999; Powar and Jagannathan 1982). Interestingly, the activity of BPPhy can be inhibited by the chelating agent EDTA, whereas the activity of other types of phytase is unaffected; moreover, reducing agents do not affect BPPhy activity (Fasimoye et al. 2014; Kumar et al. 2014b; Pal Roy et al. 2017). Orthophosphate is a known competitive inhibitor of phytases; whereas, wolframate, molybdate and vanadate inhibit them by promoting the formation of complexes geometrically similar to the transition state [66]. Table 2 summarizes enzymatic properties of some Bacillus BPPhys.
Table 2.
Enzymatic properties of Bacillus BPPhys (Kumar et al. 2017)
| Source microbes | Strain | pH optima | Temp.optima ( °C) | pHa | Thermo stabilityb | Km (mM) /Vmax/ Kcat (s-1) | MW (kDa) | Refs |
|---|---|---|---|---|---|---|---|---|
| Bacillus sp. | YCJS | 6.0 | 50 | – | – | 0.95/15.3 U/– | 47.5 | (Yao et al. 2014) |
| B. subtilis | ARRMK33 | 7.0 | 55 | – | – | – | 42 | (Reddy et al. 2015) |
| B. subtilis (N) | US417 | 7.5 | 55 | 2.0–9.0 | 77 | – | 41 | (Farhat et al. 2008) |
| B. subtilis (R) | US417 | 7.5 | 55 | 3.0–9.0 | 90 | – | 41 | (Farhat-Khemakhem et al. 2012) |
| B. subtilis (R) | US417 | 7.5 | 55 | 3.0–9.0 | 100 | 0.45/–/64 | 43 | (Hmida-Sayari et al. 2014) |
| B. subtilis | B.S.46 | 7.3 | 56.5 | 6.0–10.0 | – | 2.05/–/– | ND | (Rocky-Salim et al. 2016) |
| Bacillus sp. | WYCQ02 | 7.5 | 55 | – | – | – | 53 | (Li et al. 2013) |
| B. licheniformis | – | 7.0 | 65 | 6.5–9.0 | 80 | – | 47 | (Tye et al. 2002) |
| B.licheniformis | PB-13 | 6.0–6.5 | 60 | 4.0–8.0 | – | 1.064/1.32U/mg/27.46 | ND | (Kumar et al. 2014b) |
| B. licheniformis | PFBL-03 | 6.0 | 55 | 4.0–7.5 | > 80 | 4.7/49.01 U/– | 36 | (Fasimoye et al. 2014) |
| B. licheniformis | ZJ-6 | 7.5 | 60 | 5.0–9.0 | 59.42 | – | 39 | (Wang et al. 2011) |
| B.amyloliquefaciens | US573 | 7.5 | 70 | 3.0–9.0 | – | 1.125/27.71U/mg/28.12 | 42 | (Boukhris et al. 2015) |
| B. licheniformis | – | 6.5–7.5 | 50 | 5.0–9.5 | – | – | 40–42 | (Dan et al. 2015) |
| B. amyloliquefacien | DS11 | 7.0–8.0 | 70 | – | – | 0.138/–/992.8 | 39 | (Shim and Oh 2012) |
| B. subtilis | 168 | 70 | 55 | – | 50 | 2.19/4.7/4965.7 | 44 | Chen et al. 2015) |
ND not determined
aStability range (> 60% activity)
b% residual activity after a heat treatment at 75/80 °C for 10 min
Notably, Calcium ions play an important role in maintaining conformational, catalytic activity and thermal stability of Bacillus BPPhys (Kim et al. 1998b). Various studies have confirmed that Ca2+ can increase thermal stability of Bacillus BPPhys, thereby preventing their thermal denaturation. Kerovuo et al. (Kerovuo et al. 2000a) observed a significant decrease in phytase activity after removing Ca2+ from the enzyme solution by dialysis. When activity of Bacillus DS11 phytase was tested in a solution containing 5 mM Ca2+, this enzyme displayed high thermal stability at 90 °C, and it still retained about 50% of its enzymatic activity even after a prolonged incubation of 10 min at 90 °C, but in the absence of the same Ca2+ concentration, phytase stability gradually decreased when incubation temperature exceeded 50 °C (Kim et al. 1998a). After adding 10 mM Ca2+ to a Bacillus KHU-10 phytase solution, this enzyme showed maximum activity at 60 °C (without further additions, it showed maximum activity at 40 °C), and it still retained 95% of its enzymatic activity after an incubation of 10 min at 60 °C (Choi et al. 2001). In addition, when adding 10 mM Ca2+, the optimum pH range of Bacillus KHU-10 phytase increased from 6.5–8.5 to 6.0–9.5 (Choi et al. 2001). Thermal stability of proteins depends on many structural features, including arrangement of peptide bonds and amino acid residues, content of α-helices, number of hydrogen bonds and salt bridges, presence of proline residues and other factors (Farhat-Khemakhem et al. 2012; Jang et al. 2019). To date, at least 13 different physical and chemical explanations have been proposed to explain enhanced thermostabilization of proteins, such as improved electrostatic interactions and shorter loops (Enrique et al. 1996). As yet, however, no single or preferred mode of thermostabilization has been found (Vogt et al. 1997).
To explore the molecular mechanism of the effect of Ca2+ on the thermal stability and catalytic activity, a large number of studies have been explained from the perspective of structural biology. Ha et al. research shows that binding of two calcium ions to high-affinity calcium binding sites results in a dramatic increase in enzyme thermostability by joining remote loop segments in the amino acid sequence (Ha et al. 2000). Zeng et al. confirmed that Ca9 (calcium binding site-9) is located on the outer surface of the enzyme, and it can interact with D220 and H226, as well as with multiple water molecules, thus speculating that Ca9 could be involved in the thermostability of the enzyme (Zeng et al. 2011). Oh et al. confirmed the identity of key amino acid residues involved in the binding of Ca2+ to substrate by single point mutation experiments at Y159, E211, E260, and D314 sites, which further confirmed the Ca2+-dependent catalytic activity of the phytase (Oh et al. 2001). Sejeong Shin et al. confirmed that mutation of the residues involved in the calcium chelation results in severe defects in the enzyme's activity. Therefore, they proposed that the phytase reaction is likely to proceed through a direct attack of the metal-bridging water molecule on the phosphorous atom of a substrate and the subsequent stabilization of the pentavalent transition state by the bound calcium ions (Shin et al. 2001). The above research shows that Ca2+ stimulates the activity of BPPhy in a concentration-dependent manner, mainly because Ca2+plays a key role in stabilizing the transition state in the catalytic process (Ha et al. 2000; Jang et al. 2018; Oh et al. 2001; Shin et al. 2001).
Molecular structure of Bacillus BPPhy
Recently, a considerable number of studies have focused on the nucleotide/amino acid sequence and structure of BPPhys to investigate their species diversity, as well as the key sites affecting their enzymatic properties. Kumar et al. analyzed 44 representative BPPhy protein sequences through in silico analysis showing that all the BPPhy conserved sites were located towards the N- and C-termini, and no cysteine was present in the full sequences (Kumar et al. 2014a). In addition, three main clusters were found by constructing evolutionary trees of BPPhy from different sources (Kumar et al. 2014a). A total of 10 motifs together with a 29-amino acid-long motif “DDPAIWVHPHDPEKSRIIGTNKKSGLAVY” were found in all the analyzed 44 protein sequences of BPPhys, these motifs can be used for diversity and expression analysis of BPPhys. A conservative sequence [D/A][STA]DDPA[I/V]W[I/V/L]T[N/D/L] was present at the N-terminus, followed by more than one NN[F/V]D[I/V/L] motif in all the analyzed sequences; in the highly conserved C-terminal "DG" sequence of BPPhy, an aspartic acid residue was considered to be important for catalysis, since it probably acts as a proton donor for the oxygen atom in order to cleave the phosphate monoester bond (Kumar et al. 2014a). In the second conserved motif (98–104) of BPPhys, the valine residue in position 100 was supposed to play a role in substrate binding during enzyme catalysis [62]. In silico analysis of Bacillus subtilis BPPhy revealed the presence of 6 binding sites, more in detail, Ca1, Ca2, Ca3 were responsible for the thermal stability of the enzyme; while Ca4, Ca5, and Ca6 were essential for its catalytic activity (Reddy et al. 2015).
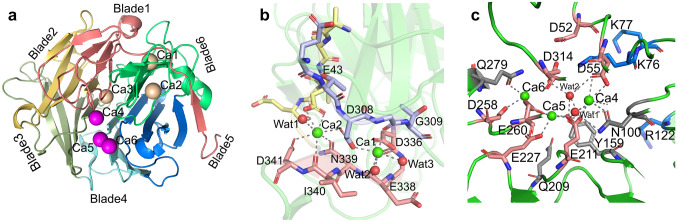
A crystal structure study led by Ha et al. (Ha et al. 2000) showed that the molecular structure of the thermostable phytase (TS-phy) phytase, including the amino acid residues 29 to 383 (the first 28 residues constitute a signal peptide), looks exactly like a propeller with 6 blades. At one end of this structure there is a crack suitable for substrate binding, while the other end is flat. In Table 3, some Bacillus phytases with known 3D structure are reported. In 2000, Ha et al. firstly analyzed the three-dimensional structure of the thermostable phytase of Bacillus amylolus (Fig. 1a). From a structural point of view, this enzyme has a round beta-propeller structure consisting of 6 anti-parallel beta sheet structures known as blades. Each blade has a highly curved sheet structure and consists of four–five anti-parallel β-sheets. They are connected to each other in a topologically identical manner, the fourth sheet of each leaf passes through the top of the molecule to connect to the first chain of the next leaf; the 1 + 3 layer combination of the sixth blade (C-end blade) and the extra β-layer, formed by the interaction of the N-terminal and the fifth layer, form together a "double clasp" structure. This latter further tightens the circular belt of the ring structure contributing to stabilize the structure of the enzyme; at the "top" of the structure there is a gap (Fig. 1a) (Fu et al. 2008; Ha et al. 2000).
Table 3.
Bacillus phytases with known 3D structure
Fig. 1.
The strip chart of TS-Phy (a); Stereoscopic view showing the coordinated geometry of the two calcium ions connecting the three rings, including the high-affinity calcium sites. The N-terminal ring is shown in yellow, the C-terminal ring in red, and the ring connecting the fifth and sixth blades in blue (b); calcium in a stereoscopic view of the low-affinity calcium site binding: calcium ion are shown in green, and oxygen atoms in red (c) (
modified from Ha et al. 2000)
The structure of TS-Phy contains two types of calcium connection sites, i.e., high-affinity calcium connection sites and low-affinity ones (Fig. 1a). There are three high-affinity calcium connection sites, in particular, two sites (Ca1, Ca2) are located at the periphery of the structure and one (Ca3) in the central channel (Ha et al. 2000). Ca1 and Ca2 sites are adjacent to the sixth leaf in the middle area of the "double clasp", and they form a dicalcium center through the bridge arms provided by the carboxyl group of Asp308. These two calcium ions play an important role in tightening the "double clasp"; Ca3 site is located in the middle of the central channel, and participates in the formation of an intricate hydrogen bond network that completely spans the channel (Fig. 1b) (Ha et al. 2000). Differential scanning calorimetry (DSC) showed that two of the high-affinity calcium connection sites (most likely Ca1 and Ca2) were responsible for increasing TS-Phy thermal stability (Ha et al. 2000). Furthermore, there are three low-affinity calcium connection sites (Ca4, Ca5, Ca6) located at the top of the structure (Fig. 1c). The connection of three calcium ions with Ca4, Ca5, and Ca6 can neutralize a total of six aspartic and glutamic acid residues around the "calcium cage", and induce the side chain involved in calcium ion chelation to change from a disordered state to an ordered one, while the conformation of the main chain remains unchanged. The cracks on the top of TS-Phy are mainly lined with negatively charged side chains. The occupation of calcium ions turns it into a favorable electrostatic environment, which facilitates the combination of phytate with the nearby amino acid residues Lys176, Lys77, Arg122 and Lys179, thus contributing to the formation of enzyme–substrate complexes and promoting enzyme catalysis (Fu et al. 2008; Ha et al. 2000). This theory was confirmed by studying the dependence on calcium ions of TS-Phy (Oh et al. 2001).
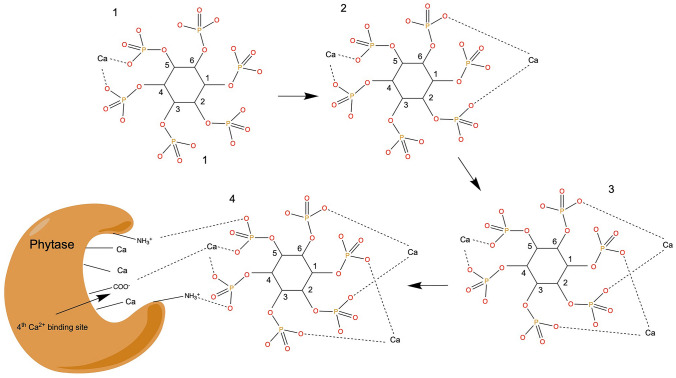
Kinetic studies showed that the enzyme activity of TS-Phy follows a fast and balanced orderly mechanism, in which the binding of Ca2+ to the active site is necessary for enzyme activation. TS-Phy can only hydrolyze the Ca2+–phytase complex; excessive free concentrations of both Ca2+ and phytic acid can competitively inhibit enzyme activity. Several point mutations found in TS-Phy have highlighted some key amino acid residues critical for binding to phytic acid: Y159, E211, E260 and D314. Isothermal titration calorimetry measurement of TS-Phy showed that the activity of this enzyme depends on the ionization of amino acid residues believed to be important for substrate binding. Based on their experiments, Oh et al. found that calcium is essential for activating this enzyme and its substrate. In particular, they highlighted that Ca2+ binds to paired oxo divalent bond of the phosphoryl group, and that binding occurs to the axial form (Fig. 2). Initially, the first Ca2+ ion connects to the phosphoryl group at positions 5 and 4/6, the second Ca2+ may connect to the phosphoryl group at positions 2 and 6, while the third Ca2+ may connect to the phosphoryl group at positions 1 and 3. The three Ca2+ ions combine with the Ca2+ binding site on the active site to form an ideal conformation and charge distribution, so that the Ca2+ of the Ca2+–phytic acid complex is connected to the fourth Ca2+ site of the active center, part of the negative charges of the phosphate group can interact with the amino groups of the lysine and arginine residues present in the active site (Oh et al. 2001).
Fig. 2.
Binding manner of substrate to the active site of phytase
The catalytic mechanism of Bacillus BPPhy
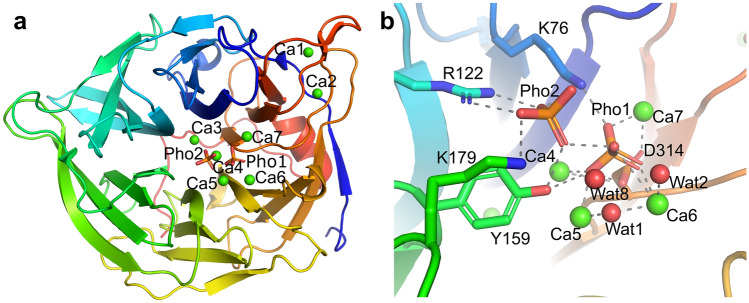
Phytase sequentially cleaves the phosphate group on the phytic acid molecule producing inositol triphosphate and inorganic phosphorus. However, the catalytic mechanism of phytase from different sources is not completely conserved. The catalytic mechanism of this enzyme was clearly understood by analyzing its crystal structure (Kumar et al. 2017). In 2001, Shin et al. attempted to describe the catalytic mechanism and properties of TS-Phy by analyzing the composite structure of Bacillus amyloliquefaciens TS-Phy including Ca2+ ions and inorganic phosphate. According to this structure, TS-Phy comprises seven calcium connection sites, two are located on the outside of the enzyme (Ca1, Ca2), one is in the center of the channel (Ca3), while the other four reside on the top (Ca4, Ca5, Ca6, Ca7) (Fig. 3a). The enzyme binds two phosphate ions (Pho1 and Pho2) at the top of its structure. Phosphate ions not only induce the connection of Ca7 by acting as coordinating arms, so promoting the connection of one more Ca2+ in Ca7, but they also induce side chain remodeling of a series of amino acids located near the connection site by supporting hydrogen bonds or ionic interactions. Among them, Pho1 is involved in the chelation of all the four Ca2+ ions and forms an octahedral coordination shell of Ca4–Ca7 by connecting the side chains of Asp52, Asp55, Tyr159, Glu211, Asp258, Glu260, Glu279, Asp314 and several water molecules. Hence, the connections favored by Pho1 are essential for the existence of such a structure. On the other hand, Pho2 can increase substrate binding affinity of the enzyme, but it does not participate in Ca2+ chelation (Fig. 3b) (Shin et al. 2001). According to enzyme activity assays, as well as to 2-phosphoinositide kinetic constant determination, TS-Phy can hydrolyze any phosphate on phytic acid, although substrate connection modeling experiments showed that two phosphates not structurally adjacent cannot overlap with Pho1 and Pho2. In brief, Shin et al. proposed a catalytic mechanism for TS-Phy, in detail, 1. The hydroxyl group of wat1, a bridging water molecule between Ca5 and CA6, attacks the phosphorus element of phytic acid occupying the pho1 site. The water molecules 2 and 8 (wat2 and wat8), along with Lys76, may provide protons as general acids (Fig. 3); 2. Only two adjacent phosphate groups can instantaneously occupy the cleavage site connecting Pho1 and the affinity site occupied by Pho2 (Fig. 4) (Shin et al. 2001).
Fig. 3.
Strip chart of the composite structure of Bacillus amyloliquefaciens TS-Phy with Ca2+ and inorganic phosphate (a). Stereoscopic view of the detailed atomic interactions with phosphate ions (b) (Shin et al. 2001)
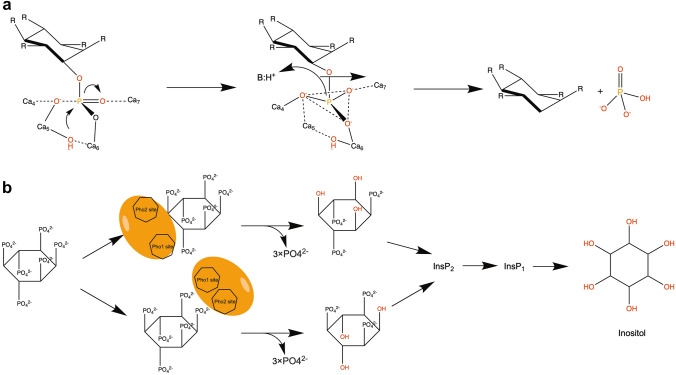
Fig. 4.
Possible enzyme mechanism of Bacillus amyloliquefaciens TS-Phy (a), and hydrolysis of phytic acid (b). a The hydroxide ion (Wat1) directly attacks the phosphorus atom of phytate phosphorus occupying the Pho1 site. Generally, the role of acid in catalysis can be provided by Wat2, Wat8 or Lys76. This catalysis seems to be severely affected in the hydrolysis of any non-adjacent phosphate group when the Pho2 site is empty. b Only two adjacent matrix phosphate groups can simultaneously occupy the "cleavage site" (Pho1 site) and the "affinity site" (Pho2 site) (Shin et al. 2001)
In 2011, Zeng et al. analyzed the three-dimensional structure of the complex of Bacillus subtilis BPPhy with divalent metal ions and inositol hexasulfate(Zeng et al. 2011). The structure analysis revealed four additional calcium binding sites (Ca8–Ca11). Ca8 is located at the top of the structure, It could interact with the side chains of D200 and D230 residues and indirectly interact with the third sulfate S3 of inositol hexasulfate through water molecules. So, Zeng et al. speculated that it could be involved in enzyme catalysis (Zeng et al. 2011). Moreover, the structure confirmed that Ca 9 was located on the outer surface of the enzyme, and it could interact with d220, h226, and several water molecules, which suggested that Ca 9 might be involved in the thermal stability of the enzyme. while Ca10 and Ca11 could be involved in crystal packaging. Since the connection between the active site of Bacillus subtilis BPPhy and the fourth sulfate radical of the substrate is stronger than other connections, Zeng hypothesized that BPPhy could preferentially hydrolyze the fourth phosphate radical (Zeng et al. 2011).
The hydrolysis pathway of phytic acid is still controversial. Shin et al. proposed for the first time that Bacillus BPPhy only cuts the phosphate groups on C3 and C6 of phytic acid, while Greiner and Kerovuo et al. proposed a dual pathway for enzymatic hydrolysis of phytic acid (Greiner 2002; Kerovuo et al. 2000b; Shin et al. 2001). Subsequently, Greiner used a combination of high-performance chromatography and kinetic studies demonstrating that phytic acid was catalyzed by BPPhy and then hydrolyzed into D-myo-inositol-1,2,4,5,6-pentakisphosphate (D-Ins(1,2,4,5,6)P5), D-myo-inositol-1,4,5,6-tetrakisphosphate (Ins(2,4,5,6)P4) and D-myo-Inositol-2,4,6-Triphosphate (Ins(2,4,6,)P3) (Greiner et al. 2007). In 2006, Oh et al. proposed that the degradation of phytic acid was carried out through the bidentate structure of P3–P4 of calcium phytate. Such a structure connects the two phosphate binding sites in the active structure of BPPhy, by promoting the hydrolysis of the phosphate group at position d-3 to produce Ins(1,2,4,5,6)P5. The hydrolysis of the phosphate groups at positions d-1 and d-5 also uses a similar mechanism to generate the final product Ins(2,4,6,)P3. In these cases, the bidentate structure cannot be formed, due to the lack of adjacent phosphate groups; therefore, the final product Ins(2,4,6,)P3 is no longer hydrolyzed by BPPhy (Oh et al. 2006).
Molecular modification of Bacillus BPPhy
Recently, molecular bioinformatics is an emerging research direction to study existing enzymes, especially when combined with site-directed mutagenesis and other molecular biology techniques able to modify enzymes to optimize their catalytic efficiency and enzymatic properties (Ni et al. 2019; Yang et al. 2019a). The current molecular modification strategies of enzymes include (i) rational design based on the structure and function of known enzymes, (ii) irrational design that does not require the knowledge of enzyme structure and function (such as directed evolution), and (iii) semi-rational design developed by combining the first two methods (such as screening of factor B based on protein extraction structure). The knowledge of enzyme mechanism, substrate binding, catalysis, as well as enzymatic properties are the main basis for rational design and transformation of enzymes (Li et al. 2019; Sutherland et al. 2016; Yang et al. 2019a; Yu et al. 2019).
Natural BPPhy has limited specific enzyme activity and thermal stability. In recent years, many studies were aimed to improve the specific activity and thermal stability of BPPhy through molecular modifications, which can make BPPhy more widely used in food and feed industry. Xu et al. conducted site-directed mutagenesis studies on DSM 1061 phytase derived from Bacillus amyloliquefaciens (Wang et al. 2015; Xu et al. 2014). They found that (i) the half-life of the D191E mutant was extended by 4.3 min at 85 °C if compared with that of the wild-type enzyme, although its catalytic efficiency dropped to 48.9% with respect to the wild-type enzyme; (ii) the D148E/H149R mutant had half-life similar to that of the wild-type, but remarkably its catalytic efficiency was 229% of that of the wild-type. Further studies found that both D148E and S197E mutants had improved catalytic activity and thermal stability; (iii) the Q67E/N68R double mutation also affected the thermal stability of the enzyme, indeed its catalytic efficiency was 93% of that of the wild-type enzyme, and the half-life was extended by 0.7 min. Using methods such as sequence alignment and homology modeling, Edward et al. found that the mutation of some amino acids into residues limiting conformational flexibility (any amino acid residue (Xaa) → Pro and Gly → Ala) could increase the catalytic activity of each mutant phytase (117A, G266A, H32P, S256P, K304P, K324P, S353P) (Tung et al. 2008). Tran et al. mutated the catalytic sites (P257R, E180N, E229V, S283R), as well as the active sites (K77R, K179R, E227S) on the surface of BPPhy to make its catalytic surface more positively charged (Tran et al. 2011). The related results showed that (i) K77R and K179R mutations reduced the specific activity of the enzyme, while the K77R/K179R double mutant showed a higher stability at pH values between 2.6 and 3.0; (ii) when compared with the optimal pH value of the wild-type phytase (6.0), the E227S mutant exhibited the best activity at pH 5.5, so displaying a higher stability under acidic conditions than the wild-type enzyme. The E227S mutant still retained most of its original activity (more than 80%) after three hours incubation at pH 2.6, whereas the wild-type phytase displayed only 40% of its original activity in similar conditions. Chen et al. firstly used engineered E. coli to study directed evolution of BPPhy of Bacillus subtilis 168, to obtain mutants (D24G/K70R/K111E/N121S) with increased activity (Chen et al. 2016). Such mutants were transformed into Bacillus subtilis 168, E. coli and Pichia pastoris for enzyme expression. Interestingly, the activity of these mutant enzymes expressed in Bacillus subtilis was higher than that of the enzymes expressed in E. coli and Pichia pastoris. Additionally, the D24G/K70R/K111E/N121S mutant showed higher activity than that of each single mutant. Recently, Zhang et al. added a disulfide bond (in the G197C/A358C variant) to the phytase (PhyBL) expressed by Bacillus licheniformis WHU, which increased the half-life of PhyBL at 60 °C by about 3.8 times compared to that of the wild type(Zhang et al. 2020).
At present, the target of modification of Bacillus phytase is generally the phytase selected in nature. However, in nature most enzymes occupy a small area in their protein sequence space, therefore, current molecular modifications are only aimed at areas with a small protein sequence space. In addition, since evolution is carried out through long mutations and natural selection after mutation, the sequence of natural proteins will not be evenly distributed in the entire sequence space, and the de novo design of enzymes can well make up for the above-mentioned defects (Lin 2018). The de novo design of a new enzyme is a new measure to achieve the modification of enzyme molecules. The calculation of quantum mechanics is used to simulate the conformational change process in the active site of the enzyme. The resulting conformation can be used to obtain a three-dimensional model of the ideal active site, also known as theoretical enzyme (Nanda and Koder 2010; Tantillo et al. 1998). Then, by using molecular dynamics simulation programs (RosettaMatch (Zanghellini et al. 2006), ORBIT (Bolon and Mayo 2002) or Scaffold Selection (Höcker 2009)) in order to find the optimal structure of the transition state and the catalytic group, the new enzyme structure can be simulated, and the evolution path from the initial design to the highly active new enzyme can be realized (Lassila et al. 2006; Meiler and Baker 2006). However, at present, there is no relevant report on the modification of phytase based on de novo design strategy, only lipase and other related de novo design reports have been reported, and the ideal results have been obtained. It is believed that there will be more research on the modification of phytase based on de novo design in the future.
Prospect
Phytase has a strong application prospect in the food industry, feed production and medicine. First of all, as an important additive for feed production, phytase can degrade phytate phosphorus in grain feed into inorganic phosphorus that can be absorbed by monogastric animals. This would increase the absorption of phosphorus by monogastric animals, thus reducing environmental phosphorus pollution. In addition, the catalyzed decomposition of phytic acid by phytase would facilitate the production of inositol, which could be used in the fields of feed, food and medicine. At present, both academia and industry are focusing on the research of acid phytase, but the defects of this kind of phytase are obvious: (i) in the aquaculture industry, the neutral digestive tract of fish affects the enzyme activity (ii) a large amount of heat is generated during food processing and feed crushing, and such a phytase has insufficient heat resistance, which also affects its enzyme activity. BPPhy overcomes the problems related to the low optimal pH value and the poor thermal stability of acid phytase, so it has important application value in food, feed and medicine industries.
Our knowledge of BPPhys began at the end of the last century. At present, research on BPPhys is in-depth, including their microbial source, three-dimensional structure, catalytic activity and substrate specificity, as well as the regulatory mechanism of their transcriptional expression, and their physiological functions in microorganisms. However, there are still many research challenges on this kind of enzymes; in particular, their catalytic mechanism is still controversial, their activity is low, and their thermal stability is still insufficient. These problems limit the application of BPPhys. The solution of these problems still depends on the screening of high-quality BPPhys in nature, the optimization of fermentation conditions, the determination of their three-dimensional structure and catalytic mechanism, the molecular modification of BPPhys to optimize their optimal pH value, thermal stability and specific enzyme activity, and even the de novo design of new BPPhys.
Author contributions
T.Z, Y.L. and R.C.: conception of the work. T.Z., X.Y., Z.Z. and Y.L.: collection of data, writing of manuscript. V.D. and R.C.: analysis of data.
Funding
This review was not funded by any organization.
Availability of data and material (data transparency)
All data and materials in this review are available.
Declarations
Conflict of interest
The authors declared that they have no conflicts of interest to this work. We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Code availability (software application or custom code)
All codes in this review are available.
Additional declarations for articles in life science journals that report the results of studies involving humans and/or animals
This review does not involve human and/or animal research results.
Ethics approval (include appropriate approvals or waivers)
This review does not involve human and/or animal research results.
Consent to participate (include appropriate statements)
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.
Consent for publication (include appropriate statements)
We confirm that all review authors agree to publish the review.
Footnotes
Ting Zhao and Xihao Yong authors contributed equally.
Contributor Information
Vincenza Dolce, Email: vincenza.dolce@unical.it.
Yuan Li, Email: liyuan@suse.edu.cn, Email: liyuan_italy@163.com.
Rosita Curcio, Email: rosita.curcio@unical.it.
References
- Arastoo BD, Maryam P, Zahra K. Characterization of a thermostable, acidic-phytase from Bacillus tequilensis Dm018; medium optimization by response surface methodology. Catal Lett. 2019;149:2961–2972. doi: 10.1007/s10562-019-02881-w. [DOI] [Google Scholar]
- Balaban NP, Suleimanova AD, Valeeva LR, Shakirov EV, Sharipova MR. Structural characteristics and catalytic mechanism of Bacillus beta-propeller phytases. Biochem Mosc. 2016;81:785–793. doi: 10.1134/S0006297916080010. [DOI] [PubMed] [Google Scholar]
- Becker EA, Seitzer PM, Tritt A, Larsen D, Krusor M, Yao AI, Wu D, Madern D, Eisen JA, Darling AE. Phylogenetically driven sequencing of extremely halophilic archaea reveals strategies for static and dynamic osmo-response. Plos Genetics. 2014;10:e1004784. doi: 10.1371/journal.pgen.1004784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolon DN, Mayo SL. Enzyme-like proteins by computational design. Proc Natl Acad Sci USA. 2002;98:14274–14279. doi: 10.1073/pnas.251555398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukhris I, Farhat-Khemakhem A, Blibech M, Bouchaala K, Chouayekh H. Characterization of an extremely salt-tolerant and thermostable phytase from Bacillus amyloliquefaciens US573. Int J Biol Macromol. 2015;80:581–587. doi: 10.1016/j.ijbiomac.2015.07.014. [DOI] [PubMed] [Google Scholar]
- Chan WL, Lung SC, Lim BL. Properties of beta-propeller phytase expressed in transgenic tobacco. Protein Expr Purif. 2006;46:100–106. doi: 10.1016/j.pep.2005.07.019. [DOI] [PubMed] [Google Scholar]
- Chen W, Ye L, Guo F, Lv Y, Yu H. Enhanced activity of an alkaline phytase from Bacillus subtilis 168 in acidic and neutral environments by directed evolution. Biochem Eng J. 2015;98:137–143. doi: 10.1016/j.bej.2015.02.021. [DOI] [Google Scholar]
- Chen W, Yu H, Ye L. Comparative study on different expression hosts for alkaline phytase engineered in Escherichia coli. Appl Biochem Biotechnol. 2016;179:997–1010. doi: 10.1007/s12010-016-2046-3. [DOI] [PubMed] [Google Scholar]
- Cheng C, Lim BL. Beta-propeller phytases in the aquatic environment. Arch Microbiol. 2006;185:1–13. doi: 10.1007/s00203-005-0080-6. [DOI] [PubMed] [Google Scholar]
- Choi YM, Joo SH, Kim JM. Purification and properties of extracellular phytase from Bacillus sp. KHU-10. J Protein Chem. 2001;20:287–292. doi: 10.1023/A:1010945416862. [DOI] [PubMed] [Google Scholar]
- Chu HM, Guo R-T, Lin T-W, Chou C-C, Shr H-L, Lai H-L, Tang T-Y, Cheng K-J, Selinger BL, Wang AH-J. Structures of selenomonas ruminantium phytase in complex with persulfated phytate: DSP phytase fold and mechanism for sequential substrate hydrolysis. Structure. 2004;12:2015–2024. doi: 10.1016/j.str.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Dan SK, Nandi A, Banerjee G, Ghosh P, Ray AK. Purification and characterization of extracellular phytase from Bacillus licheniformis isolated from fish gut. Proc Natl Acad Sci India. 2015;85:751–758. doi: 10.1007/s40011-015-0571-4. [DOI] [Google Scholar]
- David E, Ted P, Pamela F, Kathryn K, Barbara M, John A, Doug W, Darryl S. Analysis of free and total myo-inositol in foods, feeds, and infant formula by high-performance anion exchange chromatography with pulsed amperometric detection, including a novel total extraction using microwave-assisted acid hydrolysis and enzymatic trea. J AOAC Int. 2012;95:1469–1478. doi: 10.5740/jaoacint.12-028. [DOI] [PubMed] [Google Scholar]
- Devillers CH, Piper ME, Ballicora MA, Preiss J. Characterization of the branching patterns of glycogen branching enzyme truncated on the N-terminus. Arch Biochem Biophys. 2003;418:34–38. doi: 10.1016/s0003-9861(03)00341-2. [DOI] [PubMed] [Google Scholar]
- Dionisio G, Holm PB, Brinch-Pedersen H. Wheat (Triticum aestivum L.) and barley (Hordeum vulgare L.) multiple inositol polyphosphate phosphatases (MINPPs) are phytases expressed during grain filling and germination. Plant Biotechnol J. 2007;5:325–338. doi: 10.1111/j.1467-7652.2007.00244.x. [DOI] [PubMed] [Google Scholar]
- Dong S (2015) Effect and mechanism of inositol and D-chiro-inositol on alcoholic fatty liver rats. Master’s thesis, Tianjin University of Science & Technology, Tianjin
- Enrique Q, Perez-Pons JA, Angel MV. Analysis of protein conformational characteristics related to thermostability. Protein Eng. 1996;9:265–271. doi: 10.1093/protein/9.3.265. [DOI] [PubMed] [Google Scholar]
- Fan C-M, Wang Y-H, Zheng C-Y, Fu Y-F. Fingerprint motifs of phytases. Afr J Biotech. 2013;12:1138–1147. doi: 10.5897/AJB12.1279. [DOI] [Google Scholar]
- Fan D, Zhang N, Yang Y, Wen HL, Qin F, Liu H. Research on determination of inositol in food by microbiological method. Sci Technol Food Ind. 2019;40:192–200. doi: 10.13386/j.issn1002-0306.2019.16.032. [DOI] [Google Scholar]
- Fang L, Bei Z, Yong L, Ying S, Guo G, Yang P, Yao B, Guo S, Wei G. Sequencing and characterization of dual-domain β-propeller alkaline phytase. Acta Microbiol Sin. 2018;58:1582–1592. doi: 10.13343/j.cnki.wsxb.20170532. [DOI] [Google Scholar]
- Farhat A, Chouayekh H, Farhat MB, Bouchaala K, Bejar S. Gene cloning and characterization of a thermostable phytase from Bacillus subtilis US417 and assessment of its potential as a feed additive in comparison with a commercial enzyme. Mol Biotechnol. 2008;40:127–135. doi: 10.1007/s12033-008-9068-1. [DOI] [PubMed] [Google Scholar]
- Farhat-Khemakhem A, Ali MB, Boukhris I, Khemakhem B, Maguin E, Bejar S, Chouayekh H. Crucial role of Pro 257 in the thermostability of Bacillus phytases: Biochemical and structural investigation. Int J Biol Macromol. 2012;54:9–15. doi: 10.1016/j.ijbiomac.2012.11.020. [DOI] [PubMed] [Google Scholar]
- Farhat-Khemakhem A, Blibech M, Boukhris I, Makni M, Chouayekh H. Assessment of the potential of the multi-enzyme producer Bacillus amyloliquefaciens US573 as alternative feed additive. J Sci Food Agric. 2018;98:1208–1215. doi: 10.1002/jsfa.8574. [DOI] [PubMed] [Google Scholar]
- Fasimoye F, Olajuyigbe F, Sanni M. Purification and characterization of a thermostable extracellular phytase from Bacillus licheniformis PFBL-03. Prep Biochem Biotechnol. 2014;44:193–205. doi: 10.1080/10826068.2013.812565. [DOI] [PubMed] [Google Scholar]
- Fu S, Sun J, Qian L, Li Z. Bacillus phytases: present scenario and future perspectives. Appl Biochem Biotechnol. 2008;151:1–8. doi: 10.1007/s12010-008-8158-7. [DOI] [PubMed] [Google Scholar]
- Greiner R, Lim BL, Cheng C, Carlsson N-G. Pathway of phytate dephosphorylation by β-propeller phytases of different origins. Can J Microbiol. 2007;53:488–495. doi: 10.1139/W07-015. [DOI] [PubMed] [Google Scholar]
- Greiner R. Pathway of dephosphorylation ofmyo-inositol hexakisphosphate by phytases of legume seeds. J Agric Food Chem. 2002;50(23):6870. doi: 10.1021/jf025620t. [DOI] [PubMed] [Google Scholar]
- Ha NC, Oh BC, Shin S, Kim HJ, Oh TK, Kim YO, Choi KY, Oh BH. Crystal structures of a novel, thermostable phytase in partially and fully calcium-loaded states. Nat Str Biol. 2000;7:147–153. doi: 10.1038/72421. [DOI] [PubMed] [Google Scholar]
- Hmida-Sayari A, Elgharbi F, Farhat A, Rekik H, Blondeau K, Bejar S. Overexpression and biochemical characterization of a thermostable phytase from Bacillus subtilis US417 in pichia pastoris. Mol Biotechnol. 2014;56:839–848. doi: 10.1007/s12033-014-9764-y. [DOI] [PubMed] [Google Scholar]
- Höcker B. Automated scaffold selection for enzyme design. Proteins Str Funct Bioinf. 2009;77:74–83. doi: 10.1002/prot.22418. [DOI] [PubMed] [Google Scholar]
- Hou X, Shen Z, Li N, Kong X, Sheng K, Wang J, Wang Y. A novel fungal beta-propeller phytase from nematophagous Arthrobotrys oligospora: characterization and potential application in phosphorus and mineral release for feed processing. Microb Cell Fact. 2020;19(1):84. doi: 10.1186/s12934-020-01346-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Shao N, Wang Y, Luo H, Yang P, Zhou Z, Zhan Z, Yao B. A novel beta-propeller phytase from Pedobacter nyackensis MJ11 CGMCC 2503 with potential as an aquatic feed additive. Appl Microbiol Biotechnol. 2009;83:249–259. doi: 10.1007/s00253-008-1835-1. [DOI] [PubMed] [Google Scholar]
- Jain J, Sapna Singh B. Characteristics and biotechnological applications of bacterial phytases. Process Biochem. 2016;51:159–169. doi: 10.1016/j.procbio.2015.12.004. [DOI] [Google Scholar]
- Jang WJ, Lee JM, Park HD, Choi YB, Kong IS. N-terminal domain of the beta-propeller phytase of Pseudomonas sp. FB15 plays a role for retention of low-temperature activity and catalytic efficiency. Enzyme Microb Technol. 2018;117:84–90. doi: 10.1016/j.enzmictec.2018.06.008. [DOI] [PubMed] [Google Scholar]
- Jang WJ, Lee JM, Tawheed Hasan M, Kong IS. Fusion of the N-terminal domain of Pseudomonas sp. phytase with Bacillus sp. phytase and its effects on optimal temperature and catalytic efficiency. Enzyme Microb Technol. 2019;126:69–76. doi: 10.1016/j.enzmictec.2019.04.002. [DOI] [PubMed] [Google Scholar]
- Kai W, Wei Z, Shi W, Li S (2015) Study and application of Phytase. China Biotechnol 35: 85-93 10.13523/j.cb.20150913
- Kerovuo J, Lauraeus M, Nurminen P, Kalkkinen N, Apajalahti J. Isolation, characterization, molecular gene cloning, and sequencing of a novel phytase from Bacillus subtilis. Appl Environ Microbiol. 1998;64:2079–2085. doi: 10.1016/0269-7491(88)90045-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerovuo J, Lappalainen I, Reinikainen T. The metal dependence of Bacillus subtilis phytase. Biochem Biophys Res Commun. 2000;268:365–369. doi: 10.1006/bbrc.2000.2131. [DOI] [PubMed] [Google Scholar]
- Kerovuo J, Rouvinen J, Hatzack F. Analysis of myo-inositol hexakisphosphate hydrolysis by Bacillus phytase: indication of a novel reaction mechanism. Biochem J. 2000;352:623–628. doi: 10.1042/0264-6021:3520623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YO, Kim HK, Bae KS, Yu JH, Oh TK. Purification and properties of a thermostable phytase from Bacillus sp. DS11. Enzyme Microb Technol. 1998;22:2–7. doi: 10.1016/S0141-0229(97)00096-3. [DOI] [Google Scholar]
- Kim YO, Lee JK, Kim HK, Yu JH, Oh TK. Cloning of the thermostable phytase gene (phy) from Bacillus sp. DS11 and its overexpression in Escherichia coli. FEMS Microbiol Lett. 1998;162:185–191. doi: 10.1111/j.1574-6968.1998.tb12997.x. [DOI] [PubMed] [Google Scholar]
- Kim DH, Oh BC, Choi WC, Lee JK, Oh TK. Enzymatic evaluation of Bacillus amyloliquefaciens phytase as a feed additive. Biotechnol Let. 1999;21:925–927. doi: 10.1023/A:1005602717835. [DOI] [Google Scholar]
- Klabunde T, Strater N, Frohlich R, Witzel H, Krebs B. Mechanism of Fe(III) – Zn(II) purple acid phosphatase based on crystal structures. J Mol Biol. 1996;259:737–748. doi: 10.1107/S0108767396093841. [DOI] [PubMed] [Google Scholar]
- Kostrewa D, Grüninger-Leitch F, D’Arcy A, Broger C, Mitchell D, van Loon APGM. Crystal structure of phytase from Aspergillus ficuum at 2.5 Å resolution. Nature Str Biol. 1997;4:185–190. doi: 10.1038/nsb0397-185. [DOI] [PubMed] [Google Scholar]
- Kumar V, Agrawal S. Short communication: An insight into protein sequences of PTP-like cysteine phytases. Nusantara Biosci. 2014;6:102–106. doi: 10.13057/nusbiosci/n060116. [DOI] [Google Scholar]
- Kumar V, Sangwan P, Verma AK, Agrawal S. Molecular and biochemical characteristics of recombinant β-propeller phytase from Bacillus licheniformis strain PB-13 with potential application in aquafeed. Appl Biochem Biotechnol. 2014;173:646–659. doi: 10.1007/s12010-014-0871-9. [DOI] [PubMed] [Google Scholar]
- Kumar V, Singh G, Sangwan P, Verma AK, Agrawal S. Cloning, sequencing, and in silico analysis of β-propeller phytase Bacillus licheniformis strain PB-13. Biotechnol Res Int. 2014;2014:1–11. doi: 10.1155/2014/841353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Chanderman A, Singh S, Technology. Microbial production of phytases for combating environmental phosphate pollution and other diverse applications. Crit Rev Environ Sci. 2015;46:556–591. doi: 10.1080/10643389.2015.1131562. [DOI] [Google Scholar]
- Kumar V, Yadav AN, Verma P, Sangwan P, Saxena A, Kumar K, Singh B. beta-Propeller phytases: Diversity, catalytic attributes, current developments and potential biotechnological applications. Int J Biol Macromol. 2017;98:595–609. doi: 10.1016/j.ijbiomac.2017.01.134. [DOI] [PubMed] [Google Scholar]
- Kumar V, Singh D, Sangwan P, Gill PK (2015b) Management of environmental phosphorus pollution using phytases: current challenges and future prospects. In Applied environmental biotechnology: present scenario and future trends. pp 97–114. Springer
- Kumar V (2018) Phytase in animal feed, in Enzymes in Human and Animal Nutrition Principles and Perspectives (ed. N. Romano), pp 73–88, Academic Press
- Kyrtopoulos SA, Daskalakis G, Legakis NI, Konidaris N, Psarrou E, Bonatsos G, Golematis B, Lakiotis G, Bliouras N, Outram JR. Studies in gastric carcinogenesis. II. Absence of elevated concentrations of N-nitroso compounds in the gastric juice of Greek hypochlorhydric individuals. Carcinogenesis. 1985;6:1135–1140. doi: 10.1093/carcin/6.8.1135. [DOI] [PubMed] [Google Scholar]
- Lassila JK, Privett HK, Allen BD, Mayo SL. Combinatorial methods for small-molecule placement in computational enzyme design. Proc Natl Acad Sci USA. 2006;103:16710–16715. doi: 10.1073/pnas.0607691103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DC, Cottrill MA, Forsberg CW, Jia Z. Functional insights revealed by the crystal structures of Escherichia coli glucose-1-phosphatase. J Biol Chem. 2003;278:31412–31418. doi: 10.1074/jbc.M213154200. [DOI] [PubMed] [Google Scholar]
- Lei XG, Weaver JD, Mullaney E, Ullah AH, Azain MJ. Phytase, a new life for an "old" enzyme. Ann Rev Animal Biosci. 2013;1:283–309. doi: 10.1146/annurev-animal-031412-103717. [DOI] [PubMed] [Google Scholar]
- Li Z, Zhao A, Wang X, Jin X, Li J, Yu M. Cloning, overexpression, and functional characterization of a phytase from the genus Bacillus. J Mol Microbiol Biotechnol. 2013;23:193–202. doi: 10.1159/000347027. [DOI] [PubMed] [Google Scholar]
- Li J, Li X, Gai Y, Sun Y, Zhang D. Evolution of E. coli phytase for increased thermostability guided by rational parameters. J Microbiol Biotechnol. 2019;29:419–428. doi: 10.4014/jmb.1811.11017. [DOI] [PubMed] [Google Scholar]
- Lin GM (2018) Rational design and modification of lipase/esterase. Ph.D. Thesis, Huazhong University of Science and Technology, Wuhan
- Lu F, Guo G, Li Q, Feng D, Liu Y, Huang H, Yang P, Gao W, Yao B. Preparation, purification, crystallization and preliminary crystallographic analysis of dual-domain beta-propeller phytase from Bacillus sp. HJB17. Acta Crystallographica Sect F Str Biol Commun. 2014;70:1671–1674. doi: 10.1107/S2053230X14024388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu F, Zhang B, Liu Y, Song Y, Guo G, Feng D, Huang H, Yang P, Gao W, Guo S, Yao B. Crystallization and X-ray diffraction analysis of native and selenomethionine-substituted PhyH-DI from Bacillus sp. HJB17. Acta Crystallographica Sect FStr Biol Commun. 2017;73:607–611. doi: 10.1107/S2053230X17015102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y, Wang L, Teng F, Zhang J, Tao Y. Production of myo-inositol from glucose by a novel trienzymatic cascade of polyphosphate glucokinase, inositol 1-phosphate synthase and inositol monophosphatase. Enzyme Microb Technol. 2018;112:1–5. doi: 10.1016/j.enzmictec.2018.01.006. [DOI] [PubMed] [Google Scholar]
- Maenz DD, Engele-Schaan CM, Newkirk RW, Classen HL. The effect of minerals and mineral chelators on the formation of phytase-resistant and phytase-susceptible forms of phytic acid in solution and in a slurry of canola meal. Anim Feed Sci Technol. 1999;81:177–192. doi: 10.1016/S0377-8401(99)00085-1. [DOI] [Google Scholar]
- Meiler J, Baker D. ROSETTALIGAND: Protein–small molecule docking with full side-chain flexibility. Proteins Str Funct Bioinf. 2006;65:538–548. doi: 10.1002/prot.21086. [DOI] [PubMed] [Google Scholar]
- Mullaney EJ, Richardson AE, Turner BL. Inositol Phosphates: Linking Agriculture and the Environment. Wallingford: CABI; 2007. [Google Scholar]
- Mullaney EJ, Ullah AHJ. Conservation of cysteine residues in fungal histidine acid phytases. Biochem Biophys Res Commun. 2005;328:404–408. doi: 10.1016/j.bbrc.2004.12.181. [DOI] [PubMed] [Google Scholar]
- Nanda V, Koder R. Designing artificial enzymes by intuition and computation. Nat Chem. 2010;2:15–24. doi: 10.1038/nchem.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni D, Xu W, Zhu Y, Zhang W, Zhang T, Guang C, Mu W. Inulin and its enzymatic production by inulosucrase: Characteristics, structural features, molecular modifications and applications. Biotechnol Adv. 2019;37:306–318. doi: 10.1016/j.biotechadv.2019.01.002. [DOI] [PubMed] [Google Scholar]
- Oh B-C, Chang BS, Park K-H, Ha N-C, Kim H-K, Oh B-H, Oh T-K. Calcium-dependent catalytic activity of a novel phytase from Bacillus amyloliquefaciens DS11. Biochemistry. 2001;40:9669–9676. doi: 10.1021/bi010589u. [DOI] [PubMed] [Google Scholar]
- Oh B-C, Kim MH, Yun B-S, Choi W-C, Park S-C, Bae S-C, Oh T-K. Ca2+-inositol phosphate chelation mediates the substrate specificity of β-propeller phytase. Biochemistry. 2006;45:9531–9539. doi: 10.1021/bi0603118. [DOI] [PubMed] [Google Scholar]
- Olczak M, Morawiecka B, Watorek W. Plant purple acid phosphatases - Genes, structures and biological function. Acta Biochim Pol. 2003;50:1245–1256. doi: 10.18388/abp.2003_3648. [DOI] [PubMed] [Google Scholar]
- Pal Roy M, Datta S, Ghosh S. A novel extracellular low-temperature active phytase from Bacillus aryabhattai RS1 with potential application in plant growth. Biotechnol Prog. 2017;33:633–641. doi: 10.1002/btpr.2452. [DOI] [PubMed] [Google Scholar]
- Perelló J, Gómez M, Ferrer MD, Rodríguez NY, Salcedo C, Buades JM, Pérez MM, Torregrosa JV, Martín E, Maduell F. SNF472, a novel inhibitor of vascular calcification, could be administered during hemodialysis to attain potentially therapeutic phytate levels. J Nephrol. 2018;31:287–296. doi: 10.1007/s40620-018-0471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddington CS, Houston CS, Paloheimo M, Cantrell M, Miettinen-Oinonen A, Nevalainen H, Rambosek J. The cloning and sequencing of the genes encoding phytase (phy) and pH 2.5-optimum acid phosphatase (aph) from Aspergillus niger var. awamori. Gene. 1993;133:55–62. doi: 10.1016/0378-1119(93)90224-Q. [DOI] [PubMed] [Google Scholar]
- Powar VK, Jagannathan V. Purification and properties of phytate-specific phosphatase from Bacillus subtilis. J Bacteriol. 1982;151:1102–1108. doi: 10.1128/JB.151.3.1102-1108.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pramanik K, Kundu S, Banerjee S, Ghosh PK, Maiti TK. Computational-based structural, functional and phylogenetic analysis of Enterobacter phytases. 3 Biotech. 2018;8:262. doi: 10.1007/s13205-018-1287-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl AA, Gruninger RJ, Greiner R, Janzen TW, Mosimann SC, Selinger LB. Kinetic and structural analysis of a bacterial protein tyrosine phosphatase-like myo-inositol polyphosphatase. Protein Sci. 2007;16:1368–1378. doi: 10.1110/ps.062738307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhl AA, Greiner R, Selinger LB. A protein tyrosine phosphatase-like inositol polyphosphatase from Selenomonas ruminantium subsp. lactilytica has specificity for the 5-phosphate of myo-inositol hexakisphosphate. Int J Biochem Cell Biol. 2008;40:2053–2064. doi: 10.1016/j.biocel.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Reddy CS, Achary VMM, Manna M, Singh J, Kaul T, Reddy MK. Isolation and molecular characterization of thermostable phytase from Bacillus subtilis (BSPhyARRMK33) Appl Biochem Biotechnol. 2015;175:3058–3067. doi: 10.1007/s12010-015-1487-4. [DOI] [PubMed] [Google Scholar]
- Rocky-Salimi K, Hashemi M, Safari M, Mousivand M. A novel phytase characterized by thermostability and high pH tolerance from rice phyllosphere isolated Bacillus subtilis B.S.46. J Adv Res. 2016;7:381–390. doi: 10.1016/j.jare.2016.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk G, Guddat LW, Ge Y, Carrington LE, Jersey JD. Identification of mammalian-like purple acid phosphatases in a wide range of plants. Gene. 2000;250:117–125. doi: 10.1016/s0378-1119(00)00186-4. [DOI] [PubMed] [Google Scholar]
- Shi BQ (2017) The determination of phytic acid and its extraction process. Modern Food 10.16736/j.cnki.cn41-1434/ts.2017.15.027
- Shim J-H, Oh B-C. Characterization and application of calcium-dependent β-propeller phytase from Bacillus amyloliquefaciens DS11. J Agric Food Chem. 2012;60:7532–7537. doi: 10.1021/jf3022942. [DOI] [PubMed] [Google Scholar]
- Shimizu M. Purification and characterization of phytase from Bacillus suhtilis (natto) N–77. Biosci Biotechnol Biochem. 1992;56:1266–1269. doi: 10.1271/bbb.56.1266. [DOI] [Google Scholar]
- Shin S, Ha N-C, Oh B-C, Oh T-K, Oh B-H. Enzyme mechanism and catalytic property of β propeller phytase. Structure. 2001;9:851–858. doi: 10.1016/S0969-2126(01)00637-2. [DOI] [PubMed] [Google Scholar]
- Singh N, Kuhar S, Priya K, Jaryal R, Yadav R. Phytase: The Feed Enzyme, an Overview. In Advances in Animal Biotechnology and its Applications. Singapore: Springer; 2018. [Google Scholar]
- Sutherland JJ, Jolly RA, Goldstein KM, Stevens JL. Assessing concordance of drug-induced transcriptional response in rodent liver and cultured hepatocytes. PLoS Comput Biol. 2016;12:1–31. doi: 10.1371/journal.pcbi.1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantillo DJ, Chen J, Houk KN. Theozymes and compuzymes: theoretical models for biological catalysis. Curr Opin Chem Biol. 1998;2:743–750. doi: 10.1016/S1367-5931(98)80112-9. [DOI] [PubMed] [Google Scholar]
- Teikari J, Österholm J, Kopf M, Battchikova N, Wahlsten M, Aro EM, Hess WR, Sivonen K. Transcriptomic and proteomic profiling of Anabaena sp. strain 90 under inorganic phosphorus stress. Appl Environ Microbiol. 2015;81(15):5212–5222. doi: 10.1128/aem.01062-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TT, Mamo G, Búxo L, Le NN, Gaber Y, Mattiasson B, Hatti-Kaul R. Site-directed mutagenesis of an alkaline phytase: Influencing specificity, activity and stability in acidic milieu. Enzyme Microb Technol. 2011;49:177–182. doi: 10.1016/j.enzmictec.2011.05.012. [DOI] [PubMed] [Google Scholar]
- Tung ETK, Ma HW, Cheng C, Lim BL, Wong KB. Stabilization of beta-propeller phytase by introducing Xaa–>Pro and Gly–>Ala substitutions at consensus positions. Protein Pept Lett. 2008;15:297–299. doi: 10.2174/092986608783744216. [DOI] [PubMed] [Google Scholar]
- Tye A, Siu F, Leung T, Lim B. Molecular cloning and the biochemical characterization of two novel phytases from B. subtilis. Appl Microbiol Biotechnol. 2002;59:190–197. doi: 10.1007/s00706-010-0324-2. [DOI] [PubMed] [Google Scholar]
- Ullah AHJ, Dischinger HC. Aspergillus ficuum Phytase: complete primary structure elucidation by chemical sequencing. Biochem Biophys Res Commun. 1993;192:747–753. doi: 10.1006/bbrc.1993.1477. [DOI] [PubMed] [Google Scholar]
- Ullah AH, Sethumadhavan K, Mullaney EJ. Vanadate inhibition of fungal PhyA and bacterial AppA2 histidine acid phosphatases. J Agric Food Chem. 2011;59:1739–1743. doi: 10.1021/jf103783g. [DOI] [PubMed] [Google Scholar]
- Vashishth A, Ram S, Beniwal V. Cereal phytases and their importance in improvement of micronutrients bioavailability. Biotechnique. 2017;7:42. doi: 10.1007/s13205-017-0698-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma A, Singh VK, Gaur S. Computational based functional analysis of Bacillus phytases. Comput Biol Chem. 2016;60:53–58. doi: 10.1016/j.compbiolchem.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Viader-Salvado JM, Gallegos-Lopez JA, Carreon-Trevino JG, Castillo-Galvan M, Rojo-Dominguez A, Guerrero-Olazaran M. Design of thermostable beta-propeller phytases with activity over a broad range of pHs and their overproduction by Pichia pastoris. Appl Environ Microbiol. 2010;76:6423–6430. doi: 10.1128/AEM.00253-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira Velloso CC, de Oliveira CA, Gomes EA, Lana UGP, de Carvalho CG, Guimarães LJM, Pastina MM, de Sousa SM. Genome-guided insights of tropical Bacillus strains efficient in maize growth promotion. FEMS Microbiol Ecol. 2020 doi: 10.1093/femsec/fiaa157. [DOI] [PubMed] [Google Scholar]
- Vogt G, Woell S, Argos P. Protein thermal stability, hydrogen bonds, and ion pairs. J Mol Biol. 1997;269:631–643. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- Vohra A, Satyanarayana T. Phytases: microbial sources, production, purification, and potential biotechnological applications. Crit Rev Biotechnol. 2003;23:29–60. doi: 10.1080/713609297. [DOI] [PubMed] [Google Scholar]
- Wang XY, Meng FG, Zhou HM. The role of disulfide bonds in the conformational stability and catalytic activity of phytase. Biochem Cell Biol. 2004;82:329–334. doi: 10.1139/o03-082. [DOI] [PubMed] [Google Scholar]
- Wang Q, Fu SJ, Sun JY, Weng XY. Characterization of a thermostable alkaline phytase from Bacillus licheniformis ZJ-6 in Pichia pastoris. World J Microbiol Biotechnol. 2011;27:1247–1253. doi: 10.1007/s11274-010-0574-5. [DOI] [Google Scholar]
- Wang ZP, Xu W, Shao R, Wei P. Improvement of catalytic properties and structure-activity relationship of neutral phytase based on site-directed mutagenesis. Food Sci. 2015;36:118–123. [Google Scholar]
- Wyss M, Pasamontes L, Rémy R, Kohler J, Apgm VL. Comparison of the thermostability properties of three acid phosphatases from molds: Aspergillus fumigatus phytase, A. niger phytase, and A. niger PH 2.5 acid phosphatase. Appl Environ Microbiol. 1998;64:4446–4451. doi: 10.0000/PMID9797305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Wang ZP, Shao R. Site-directed mutagenesis of a neutral phytase from Bacillus amyloliquefaciens: influencing activity and stability. Adv Mater Res. 2014;1033–1034:271–278. doi: 10.4028/www.scientific.net/AMR.1033-1034.271. [DOI] [Google Scholar]
- Yang J, Lu YT, Zhao YY, Bai ZH, Ma Z. Site-directed mutation to improve the enzymatic activity of 5-carboxy-2-pentenoyl-CoA reductase for enhancing adipic acid biosynthesis. Enzyme Microb Technol. 2019;125:6–12. doi: 10.1016/j.enzmictec.2019.02.006. [DOI] [PubMed] [Google Scholar]
- Yang L, Tan HZ, Liu SB, Zhao JT, Huang HT, Chen L, Dong Y, Wang XQ, Wang FL, Feng JW. Research progress of phytic acid in feed ingredients. Cereal Feed Industry. 2019;383:53–57. doi: 10.7633/j.issn.1003-6202.2019.03.013. [DOI] [Google Scholar]
- Yao MZ, Lu WL, Chen TG, Wang W, Liang AH. Effect of metals ions on thermostable alkaline phytase from Bacillus subtilis YCJS isolated from soybean rhizosphere soil. Annals Microbiol. 2014;64:1123–1131. doi: 10.1007/s13213-013-0751-5. [DOI] [Google Scholar]
- Yu JM, Shi JQ, Zhang Y, Yu ZL. Molecular docking and site-directed mutagenesis of dichloromethane dehalogenase to improve enzyme activity for dichloromethane degradation. Appl Biochem Biotechnol. 2019;190:487–505. doi: 10.1007/s12010-019-03106-x. [DOI] [PubMed] [Google Scholar]
- Zanghellini A, Jiang L, Wollacott A, Cheng G, Meiler J, Althoff E, Röthlisberger D, Baker D. New algorithms and an in silico benchmark for computational enzyme design. Protein Sci. 2006;15:2785–2794. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng YF, Ko TP, Lai HL, Cheng YS, Wu THS, Ma YH, Chen CC, Yang CS, Cheng KJ, Huang CH. Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate. J Mol Biol. 2011;409:214–224. doi: 10.1016/j.jmb.2011.03.063. [DOI] [PubMed] [Google Scholar]
- Zhang R, Yang P, Huang H, Yuan T, Shi P, Meng K, Yao B. Molecular and biochemical characterization of a new alkaline β-propeller phytase from the insect symbiotic bacterium Janthinobacterium sp. TN115. Appl Microbiol Biotechnol. 2011;92:317–325. doi: 10.1007/s00253-011-3309-0. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yang J, Xie P, Gao Y, Bai J, Zhang C, Liu L, Wang Q, Gao X. Characterization of a thermostable phytase from Bacillus licheniformis WHU and further stabilization of the enzyme through disulfide bond engineering. Enzyme Microb Technol. 2020;142:109679. doi: 10.1016/j.enzmictec.2020.109679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and materials in this review are available.