Abstract
Immunotherapies play critical roles in cancer treatment. However, given that only a few patients respond to immune checkpoint blockades and other immunotherapeutic strategies, more novel technologies are needed to decipher the complicated interplay between tumor cells and the components of the tumor immune microenvironment (TIME). Tumor immunomics refers to the integrated study of the TIME using immunogenomics, immunoproteomics, immune-bioinformatics, and other multi-omics data reflecting the immune states of tumors, which has relied on the rapid development of next-generation sequencing. High-throughput genomic and transcriptomic data may be utilized for calculating the abundance of immune cells and predicting tumor antigens, referring to immunogenomics. However, as bulk sequencing represents the average characteristics of a heterogeneous cell population, it fails to distinguish distinct cell subtypes. Single-cell-based technologies enable better dissection of the TIME through precise immune cell subpopulation and spatial architecture investigations. In addition, radiomics and digital pathology-based deep learning models largely contribute to research on cancer immunity. These artificial intelligence technologies have performed well in predicting response to immunotherapy, with profound significance in cancer therapy. In this review, we briefly summarize conventional and state-of-the-art technologies in the field of immunogenomics, single-cell and artificial intelligence, and present prospects for future research.
Subject terms: Tumour immunology, Tumour immunology
Introduction
Tumor cells exist with nearby cells in sophisticated community, which strongly affects how tumor cells grow, behave and communicate with other cells.1,2 Among these cells, immune cells are critical players, and many studies have proven that crosstalk between tumor cells and immune cells is bidirectional. Indeed, immune cells both promote and inhibit carcinogenesis, tumor progression, metastasis, and recurrence. Therefore, here we focus on the tumor immune microenvironment (TIME).2,3 And accordingly, promoting the transition from a pro-tumor to an anti-tumor effect to maximize the efficacy of anti-tumor immunity is a main goal of immunotherapy.4,5 Recent tumor immunotherapy strategies, such as immune checkpoint blockades (ICBs), cancer vaccines, and adoptive cell transfer (ACT) therapy, have shown unprecedented clinical efficacy.6–12 Nevertheless, in the face of therapeutic resistance and adverse effects, among others, their applications are hindered by the incomplete understanding of tumor immunity.
Despite achieving great advancements in exploring the mechanism of tumor-immune interplay, traditional techniques, such as western blotting (WB), coimmunoprecipitation (Co-IP), and real-time quantitative polymerase chain reaction (RT-qPCR), cannot provide a thorough landscape of the TIME. There is an urgent need for novel methods to characterize tumor immunological features in detail. Applying high-throughput technologies, such as genomics, transcriptomics, proteomics, epigenomics, cytomics, and informatics, to comprehensively understand tumor immunity has emerged as a brand-new discipline, i.e., tumor immunomics, providing novel insights for researchers.13,14 Next-generation sequencing (NGS) technologies greatly promote the development of immunogenomics, an important branch of immunomics. Furthermore, single-cell sequencing and artificial intelligence (AI) have ushered in a new epoch of tumor immunity in recent years. Due to the tremendous development of tumor immunology and bioinformatics, an increasing number of technologies and potential clinical implications are a matter of great concern.
In this review, we discuss the technological advances and clinical implications of immunomics in tumors to date, especially in the field of immunogenomics, single-cell, and AI.
Brief introduction to the TIME
Over the past years, knowledge of tumors has undergone metamorphosis due to innumerous researchers’ efforts to achieve progress against tumors. The definition of tumors has also evolved from the mere aggregation of tumor cells to a complex organ-like structure composed of tumor cells, immune cells, fibroblasts, vascular endothelial cells, and other stromal cells in communities.15–17 Encompassing all structures in the organ, such as immune infiltration, vascular vessels, the extracellular matrix, etc. the tumor surrounding, which is also called the tumor microenvironment (TME), has been one of the hottest research topics in oncology.18,19 With the development of tumor immunity, the immune context of the TME, i.e., TIME, has been proven to play a decisive role in carcinogenesis, tumor progression, metastasis, recurrence, and potential therapeutic targets making it being the focus of our review.20,21
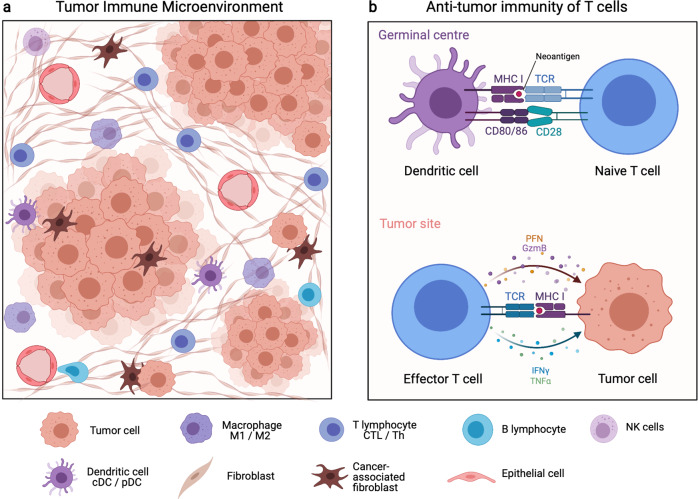
There are two main categories for the compositions of the TIME, i.e., immune cells and secreted factors, such as cytokines, chemokines, and growth factors. Regarding the former, the TIME contains extremely diverse subsets of immune cells, including T lymphocytes, B lymphocytes, natural killer (NK) cells, macrophages, dendritic cells (DCs), granulocytes, and myeloid-derived suppressor cells (MDSCs), among others.22,23 Normally, T cells, B cells, NK cells, and macrophages help inhibit tumor growth, while MDSCs and regulatory T cells (Tregs) tend to suppress anti-tumor immunity.23,24 However, available studies have confirmed that given the complex interactions with tumor cells, the specific role of immune cells could dynamically change and even become the exact opposite. For example, the anti-tumor function of CD8 + T cells may be inhibited via the exhaustion of T cells, and after CTLA-4 blockade in glycolysis-low tumors, the functional destabilization of Treg cells towards interferon-γ-producing cells may promote anti-tumor immunity.25
In summary, innumerable immune cell types and even different functional states of specific immune cell types may produce the opposite effect on anti-tumor immunity (Fig. 1). Thus, it is not wise to explore tumor immunity in a reductionistic way. With the aid of state-of-art bioinformatics technologies, to a great extent, researchers could characterize tumor immunological features systematically and provide more information to enhance our understanding of tumor immunity.
Fig. 1.
Components and interactions of the tumor immune microenvironment. a Cellular compositions of the tumor immune microenvironment. b Brief illustration of cell–cell interaction in anti-tumor immunity. cDC conventional dendritic cells, CTL cytotoxic T lymphocyte, Gzm granzyme, IFN interferon, MHC major histocompatibility complex, NK cells natural killer cells, pDC plasmacytoid dendritic cells, PFN perforin, TCR T cell receptor, Th T helper cell, TNF tumor necrosis factor
Immunomics technologies in the NGS era—immunogenomics
Over the last two decades, NGS, including whole-genome sequencing (WGS), whole-exome sequencing (WES), and RNA sequencing (RNA-seq), has been successfully developed and applied to obtain whole-genome information in humans. Compared to Sanger sequencing, NGS generates high-throughput genomic and transcriptomic data, laying a foundation for research investigating the multi-step immune response. Studies utilizing immunogenomics in the NGS era not only provide a global view of the immune cell compositions of the TIME through bioinformatic algorithms but also identify immunogenic proteins by abnormal peptide prediction, human leukocyte antigen (HLA) typing, and major histocompatibility complex (MHC)-peptide binding affinity prediction.
Quantification of immune cells in the TIME
The TIME comprises various immunocytes. For the quantification of tumor immune cell components in the TIME, conventional methods, such as flow cytometry and immunohistochemistry (IHC), are impractical for massive profiling because of their high cost and low tissue availability. With the rapid development of NGS, in silico analysis has become an alternative approach to address this issue. Considering the high cellular heterogeneity, gene expression profiles are very different among the different immune cell types and could represent immune cell types to a certain extent. Thus, we are able to estimate the abundance of dozens of immune cell types through NGS data, which have also been validated as reliable. The sources of these analyses are mainly DNA and RNA sequencing, especially the latter. Regarding RNA-seq data, which we mainly discuss, the rationales of the computational methods are mainly classified into gene set enrichment analysis (GSEA) and deconvolution26 (Tables 1–2).
Table 1.
Computational tools used in tumor immunogenomics with high-throughput next-generation sequencing data
| Tool | Characteristics | URL | Year | Ref. |
|---|---|---|---|---|
| Quantification of immune cells in TIME | ||||
| CIBERSORT | Based on linear support vector regression, deconvolution from microarray data, and known gene expression profiling set. | https://cibersort.stanford.edu/ | 2015 | 30 |
| CIBERSORTx | Expanding data source to single-cell RNA-seq. | https://cibersortx.stanford.edu/ | 2019 | 34 |
| DeconRNASeq | Constrained least square regression model validated with RNA-seq data from five human tissues | http://bioconductor.org/packages | 2013 | 273 |
| EPIC | Based on constrained least square, incorporates a non-negative condition into deconvolution. | https://gfellerlab.shinyapps.io/EPIC_1-1/ | 2017 | 274 |
| ESTIMATE | Generating a stromal score and immune score to reflect the tumor purity based on ssGSEA | https://sourceforge.net/projects/estimateproject/ | 2013 | 27 |
| FARDEEP | Based on adaptive least trimmed square, FARDEEP removes outliers and outputs the absolute quantification of cell types | https://github.com/YuningHao/FARDEEP.git | 2019 | 32 |
| MCP-counter | The score is the geometric mean of the expression level of cell-specific genes, implying the absolute abundance of immune cell types among samples | http://github.com/ebecht/MCPcounter | 2016 | 29 |
| MuSiC | Deconvolution of bulk sequencing data based on cell type-specific gene expression reference from single-cell RNA-seq | https://github.com/xuranw/ MuSiC | 2019 | 33 |
| NITUMD | A semi-supervised nonnegative matrix factorization framework with a trichotomous signature matrix | https://github.com/tdw1221/NITUMID | 2020 | 275 |
| PERT | A non-negative maximum likelihood-based method applied to fresh human umbilical cord blood samples | – | 2012 | 276 |
| quanTIseq | Quantification of ten different immune cell types and other uncharacterized cells based on RNA-seq data | http://icbi.at/quantiseq | 2019 | 277 |
| TIMER | Immune cells are estimated via transcriptomic data and the correlations among the immunological, genomic, and clinical features were established. | https://cistrome.shinyapps.io/timer/ | 2016 | 278 |
| xCell | Spillover compensation is used to separate cell types with high correlation | http://xcell.ucsf.edu/ | 2017 | 28 |
| Prediction of mutated proteins | ||||
| CN-Learn | Machine-learning framework integrating calls from multiple CNV detection algorithms and learning to accurately identify true CNVs | https://github.com/girirajanlab/CN_Learn | 2019 | 180 |
| deepSNV | Detecting and quantifying sub-clonal SNVs in mixed populations even for low-frequency variants | http://www.bioconductor.org | 2012 | 181 |
| DeepVariant | SNP and small-indel variant caller using deep neural networks in aligned NGS read data | https://github.com/google/deepvariant/ | 2018 | 182 |
| EBCall | Discriminating somatic mutations from sequencing errors with both moderate and low allele frequencies | https://github.com/friend1ws/EBCall | 2013 | 61 |
| GATK | Industry standard for identifying SNPs and indels via analyzing WES, WGS, and RNA-seq data | https://gatk.broadinstitute.org/hc/en-us | 2010 | 51 |
| LoFreq | Modeling sequencing run-specific error rates to accurately call variants occurring in <0.05% of a population | http://sourceforge.net/projects/lofreq/ | 2012 | 53 |
| MuTect2 | Sensitive detection of somatic point mutations especially in low-allelic-fraction events | https://software.broadinstitute.org/cancer/cga/mutect | 2013 | 52 |
| Platypus | Using local de novo assembly to generate candidate variants, including SNPs, indels, and complex polymorphisms | http://www.well.ox.ac.uk/platypus | 2014 | 279 |
| PyroHMMsnp | Realigning read sequences around homopolymers and inferring the underlying genotype by using a Bayesian approach | https://github.com/homopolymer/PyroTools/ | 2013 | 280 |
| SAMtools | Variant caller utilizing the post-processing alignments in the SAM/BAM format | http://samtools.sourceforge.net | 2009 | 62 |
| SCcaller | Firm foundation for standardized somatic-mutation analysis in single-cell genomics based on single-cell multiple displacement amplification (SCMDA) | https://github.com/biosinodx/SCcaller/ | 2017 | 281 |
| SomaticSeq | Somatic mutation detection pipeline used to produce highly accurate somatic mutation calls for both SNVs and small INDELs | http://bioinform.github.io/somaticseq/ | 2015 | 282 |
| SomaticSniper | Calling of somatic SNPs and indels from matched tumor–normal NGS data | http://gmt.genome.wustl.edu/packages/somatic-sniper/ | 2011 | 56 |
| Strelka2 | Fast and accurate caller of germline and somatic variants based on Strelka | https://github.com/Illumina/strelka | 2018 | 58 |
| VarDict | Variant caller of SNV, MNV, INDELs, and SVs, enabling ultra-deep sequencing | https://github.com/AstraZeneca-NGS/VarDict | 2016 | 60 |
| VarScan2 | Detection of somatic mutations and CNVs in exome data from tumor–normal pairs | http://varscan.sourceforge.net | 2012 | 55 |
| HLA typing | ||||
| HISAT2 | Graph-based genome alignment and genotyping, also applied for DNA fingerprinting | https://github.com/DaehwanKimLab/hisat2 | 2019 | 84 |
| HLA-HD | Extraction of six-digit resolution HLA-I and HLA-II from NGS data | https://www.genome.med.kyoto-u.ac.jp/HLA-HD/ | 2017 | 77 |
| HLA-miner | HLA-I and HLA-II typing directly from non-targeted RNA-seq, WGS and WES data | http://www.bcgsc.ca/platform/bioinfo/software/hlaminer | 2012 | 76 |
| HLAProfiler | K-mer profile-based method for HLA calling in RNA-seq data for both rare and common HLA alleles at two-field precision | https://github.com/ExpressionAnalysis/HLAProfiler | 2017 | 283 |
| HLAreporter | Extraction of HLA-I and HLA-II from NGS data at four-digit resolution | http://paed.hku.hk/genome/ | 2015 | 81 |
| HLAscan | Determination of HLA type across the whole-genome, exome, and target sequences | http://www.genomekorea.com/display/tools/HLA_SCAN | 2017 | 284 |
| HLAssign | First highly automated open-source HLA-typing method for NGS data to three-field resolution | https://www.ikmb.uni-kiel.de/resources/download-tools/software/hlassign | 2015 | 285 |
| HLA-VBseq | Genotyping of HLA alleles at an 8-digit resolution from WGS data without the need of prior knowledge regarding the HLA loci | http://nagasakilab.csml.org/hla | 2015 | 79 |
| Kourami | Graph-guided assembly technique used to provide highly classical HLA typing | https://github.com/Kingsford-Group/kourami | 2018 | 85 |
| Optitype | Genotyping of major and minor HLA-I alleles from RNA-seq, WGS, and WES data not specifically enriched for the HLA cluster | http://github.com/FRED-2/OptiType | 2014 | 78 |
| PHLAT | High-accuracy genotyping of HLA-I and HLA-II alleles from RNA-seq, WGS, WES, and targeted sequencing at a four-digit resolution | https://sites.google.com/site/phlatfortype | 2014 | 83 |
| Polysolver | High-precision HLA typing of WES data even relatively low-coverage WES data, and subsequent mutation detection | http://www.broadinstitute.org/cancer/cga/polysolver | 2015 | 82 |
| seq2HLA | Using standard RNA-Seq reads as input to determine the HLA-I and HLA-II types and expression at a four-digit resolution | https://github.com/TRON-Bioinformatics/seq2HLA | 2012 | 70 |
| SNP2HLA | Imputing four-digit classical alleles and amino acid polymorphisms at class I and class II loci | http://faculty.washington.edu/browning/beagle/beagle.html | 2013 | 80 |
| Prediction of antigen-MHC binding affinity | ||||
| ACME | Pan-specific peptide–MHC class I binding prediction through attention-based deep neural networks | https://github.com/HYsxe/ACME | 2019 | 286 |
| MHCAttnNet | MHC-peptide binding prediction of MHC alleles classes I and II using an attention-based deep neural model | https://github.com/gopuvenkat/MHCAttnNet | 2020 | 287 |
| MHCflurry | Open-source class I MHC binding affinity prediction, using mass spectrometry datasets for model selection and showing competitive accuracy | https://github.com/openvax/mhcflurry | 2018 | 98 |
| MHCSeqNet | Open-source deep neural network model for universal MHC binding prediction, accepting peptides of any length | https://github.com/cmbcu/MHCSeqNet | 2019 | 288 |
| NetMHC | High accuracy prediction of pMHC binding affinity to human and non-human MHC-I molecules based on ANN and PSSMs | http://www.cbs.dtu.dk/ services/NetMHC | 2008 | 93 |
| NetMHCII | High accuracy prediction of pMHC binding affinity to human and non-human MHC-II molecules based on ANN and PSSMs | http://www.cbs.dtu.dk/services/NetMHCII | 2018 | 289 |
| NetMHCIIpan | Pan-specific version of netMHCII | http://www.cbs.dtu.dk/services/NetMHCIIpan | 2020 | 102 |
| NetMHCpan | Pan-specific version of netMHC | http://www.cbs.dtu.dk/services/NetMHCpan | 2020 | 102 |
| PSSMHCpan |
PSSM based software for predicting class I peptide-HLA binding affinity |
https://github.com/BGI2016/PSSMHCpan | 2017 | 94 |
| PUFFIN | Deep residual network-based computational approach that quantifies uncertainty in pMHC affinity prediction | http://github.com/gifford-lab/PUFFIN | 2019 | 290 |
ANN artificial neural network, CNVs copy number variations, HLA human leukocyte antigen, INDELs insertions and deletions, NGS next-generation sequencing, MNVs multiple-nucleotide variants, PSSM position-specific scoring matrix, RNA-seq RNA sequencing, SNP single-nucleotide polymorphism, SNVs single nucleotide variations, SVs structural variants, TIME tumor immune microenvironment, WES whole-exome sequencing, WGS whole-genome sequencing
Table 2.
Strengths and weaknesses of immune cells quantification algorithms
| Algorithm | Category | Strengths | Weakness |
|---|---|---|---|
| ESTIMATE | G | Available for tumor purity and global immune status | Only a stromal score and an immune score are output. The information is limited27 |
| xCell | G | Available for inference of 64 immune and stromal cell |
The definitions of the cell subtypes are sometimes not clear Accuracy of prediction of some cell types is uncertain26 |
| MCP-counter | G |
Available for inference of fibroblasts and endothelial cells Available for an absolute quantification of specific cell population across samples Available for between-sample comparison |
Relatively less cell types included in the inference (8 types) |
| CIBERSORT | D |
Available for inference of 22 immune cell subtypes Available for between-cell-type comparison |
Relative proportion of distinct cell types in a single sample Trained on microarray rather than RNA-seq data30 |
| EPIC | D |
Available for inference of fibroblasts, endothelial cells, and uncharacterized cells Enabling inference of tumor purity from uncharacterized cell proportion Available for both between-sample and between-cell-type comparison |
Only 6 immune cell types available Not available for discrimination of cell types with transcriptional similarity |
| quanTIseq | D |
Available for inference of 10 immune cell subtypes Available for both between-sample and between-cell-type comparison |
Not available for quantification of stromal cells (e.g., cancer-associated fibroblasts) |
| TIMER | D | A user-friendly analytic web tool for cancer immunology research |
Only 6 immune cell types and no stromal cells available Relative proportion of distinct cell types in a single sample |
| CIBERSORTx | D | Adopting a more convincing gene expression reference from single-cell sequencing | Suitability for some tumor types needs further validation |
| MuSiC | D |
Adopting a more convincing gene expression reference from single-cell sequencing Available for tissues with intensively correlated cell types |
Suitability for some tumor types needs further validation Not available for TPM data as input33 |
| FARDEEP | D |
A robust machine learning tool eliminating outliers in the dataset Suitable for deconvolution of noisy datasets |
Different signature matrix should be adopted according to the type of gene expression data32 |
G GSEA-based method, D deconvolution method
In general, the representative GSEA-based algorithms include ESTIMATE, xCell, and MCP-counter. Based on the gene signature and single sample GSEA (ssGSEA), the ESTIMATE algorithm provides an immune score and a stromal score to represent the proportion and distribution of immune cells and stromal cells.27 The ESTIMATE score can differentiate the tumor and stomal components but cannot distinguish specific immune cell types. xCell is another ssGSEA-based method that obtains gene sets to characterize distinct cell types from multiple RNA-seq and microarray-based data sources, increasing the robustness to avoid noise disturbances. Compared with ESTIMATE, xCell uses a spillover compensation correction to better distinguish among cell types with close relationships and high similarity.28 MCP-counter generates an abundance score for each TIME cell population (including not only immune cells but also endothelial cells and fibroblasts) in every single sample based on the geometric mean of marker gene expression levels.29 For the sake of accuracy, a common characteristic of GSEA-based methods is the need for a specific gene set for each immunocyte subpopulation of interest.
The deconvolution of cell components is a reverse process of the convolution of cell subtypes in bulk tissues based on gene expression signatures. The deconvolution-based tools include DeconRNASeq, PERT, CIBERSORT, TIMER, EPIC, quanTIseq, and deconf.26 CIBERSORT, which is among the most popular algorithms based on deconvolution, utilizes linear support vector regression and a gene expression signature matrix to characterize immune infiltrating components.30 QuanTIseq is designed specifically for RNA-seq data, and the analysis pipeline comprises raw RNA-seq data preprocessing, gene expression quantification, and constrained least squares regression-based deconvolution. Remarkably, using this method, integrated image information from hematoxylin-eosin (H&E)-, IHC-, and immunofluorescence (IF)-stained slides is utilized to complement gene expression deconvolution, enabling immune profiling of the absolute cell fraction and unique immune cell densities.31 Recently, more novel deconvolution-based algorithms have been developed. For example, FARDEEP focuses on significant issue such that the deconvolution accuracy is influenced by outlier contamination of gene expression, which has not been addressed by previous algorithms.32 FARDEEP relies on the least trimmed square (LTS) to construct a robust model suitable for datasets with heavy-tailed noise. MuSiC considers cross-subject and cross-cell consistency and leverages cross-subject single-cell RNA sequencing (scRNA-seq) to generate cell type-specific gene sets for the deconvolution analysis of bulk RNA-seq data.33 However, the sensitivity and specificity of the newly developed algorithms mentioned above require more validation. Currently, ESTIMATE, CIBERSORT, and MCP-counter remain the most commonly used methods for determining immune components, and CIBERSORT has recently been updated to CIBERSORTx to fit single-cell sequencing data.34
These immunogenomics technologies are widely used to elaborate the global immune infiltration characteristics of specific cancer types. In recent multiomics studies, xCell was applied to portray the immune landscape of clear cell renal cell carcinoma, lung adenocarcinoma, and head and neck squamous cell carcinoma.35–37 Remarkably, an immune landscape was interpreted with 10000 tumors of 33 cancer types compiled in TCGA. Thorsson and colleagues divided the cancer-immune status into six distinct clusters, and CIBERSORT was used to dissect the composition of immune cells in each immune subtype.38 In addition, these algorithms were applied to compare the TIME composition of two or more groups of patients with distinct pathological features, therapeutic strategies, and treatment responses. Using CIBERSORT, Gil Del Alcazar et al.39 uncovered the difference in the infiltrating T cell subpopulation between breast ductal carcinoma in situ (DCIS) and breast invasive ductal carcinomas (IDCs). CD8 + T cells were enriched in DCIS, whereas Tregs and CD4 + T helper cells were more infiltrated in IDC. Wheeler et al. analyzed the components of the TME of hepatocellular carcinoma (HCC). These authors found that compared to normal adjacent tissue, HCC tissue was more likely to accommodate immunosuppressive cells. This research portrayed an immune evasion microenvironment and supported evidence suggesting that ICBs might be feasible in HCC patients with moderate to high levels of immune infiltration.40
Although RNA-seq data have been widely used as input resources for deconvolution, the instability of RNA molecules affects the accuracy of results obtained under chemical agent fixation. DNA molecules are more stable, and DNA methylation is highly cell-type specific, rendering DNA methylation a potential surrogate in TIME deconvolution. Cell composition dissection based on DNA methylation from blood samples has been reported, such as methylCIBERSORT and MethylResolver. The accuracy of methylCIBERSORT has been validated in immune infiltration analysis, and its clinical implications in both head and neck squamous cell carcinoma and pediatric central nervous system tumors have been presented.41 By adopting an LTS regression as described above using the FARDEEP algorithm, MethylResolver fabricates a methylation signature compendium of the leukocyte population and accomplishes relative quantification of immune infiltrates and evaluation of tumor purity.42
Notably, the combination of NGS data and bioinformatics algorithms could roughly differentiate immune cell types. Nevertheless, immune cells in the TIME comprise numerous subtypes with different properties and biological functions. Consequently, single-cell technologies should be developed to identify these cell subtypes at a higher resolution.
Identification of tumor antigens
Genomic-level mutations, transcriptomic-level mutations, and proteomic-level alternations can cause the expression of abnormal proteins, i.e., tumor antigens, which can be recognized by immune cells and trigger the anti-tumor immune response.43–45 Among these antigens, viral antigens, cancer germline antigens, and neoantigens (tumor-specific antigens resulting from somatic DNA alterations) have relatively high tumoral specificity and, thus, have become the main tumor vaccine targets.7,45–49 Consequently, we mainly discuss the identification of these tumor-specific antigens, particularly neoantigens. According to the process of antigen recognition, immunogenomics technologies perform in silico analysis to predict abnormal peptides, perform HLA typing and predict MHC-peptide binding affinity, which is greatly helpful and necessary for the identification of tumor antigens (Table 1).
Prediction of abnormal peptides from WES, WGS, or RNA-seq data
Somatic DNA mutations, including single-nucleotide variants (SNVs) and small insertions and deletions (INDELs), account for the major sources of abnormal proteins.45 Given recent systematic reviews concerning variant detection tools, we provide a summary of several standard tools and briefly discuss future perspectives.50
Currently, Genome Analysis Toolkit (GATK) is the industry standard used to identify SNVs and INDELs by analyzing WES, WGS, and RNA-seq data. Its scope is also expanding to cover copy number variations (CNVs) and structural variations (SVs).50,51 In the case of variants with a low allele frequency (normally allelic fractions as low as 0.1 and below), LoFreq and MuTect have higher sensitivities with a similar specificity and may be a better choice; the latter, which applies a Bayesian classifier, has been used more widely.52–54 In contrast, VarScan2 and SomaticSniper require higher allele fractions to guarantee sufficiently high sensitivity.55–57 Regarding liquid tumor analysis, Strelka2 introduces a normal sample contamination model to improve the variant calling accuracy and functions fairly well in the computing cost.58,59 In addition, VarScan, FreeBayes, Samtools, Vardict, and EBCall are valuable for identifying tumor antigens.55,60–62
However, regarding false positivity or false negativity, none of these tools is satisfactory in all aspects. Therefore, the scientific community has not established gold standards for calling variants.63,64 How to optimize the present tools and design a versatile and efficient variant caller to better discriminate true variants from sequencing errors is worthy of further research.65 By integrating VarScan, GATK, Pindel, BreakDancer, Strelka, and Genome STRiP in a large web interface, the Genome Variant Investigation Platform (GenomeVIP) provides a new method and has been used in large data projects, such as TCGA PanCanAtlas, to provide high-confidence annotated somatic, germline, and de novo variants of potential biological significance.66,67 Furthermore, it is advisable to select more than two tools to predict abnormal proteins in practice.
HLA typing
Abnormal peptides need to bind HLA to assist recognition by the T cell receptor (TCR) to elicit an immune response. HLA genes are the most polymorphic genes in the human genome and comprise three major gene loci for class I (A, B, and C) and three for class II (DP, DQ, and DR).68–71 Different HLAs have distinct binding affinities to abnormal proteins. Thus, crucial for antigen recognition, predicting HLA typing is essential for the identification of tumor antigens.70,72
After a long development period, limited by their efficiency and reliability, serological and cellular typing methods have been gradually replaced by DNA typing methods.73,74 Although real-time polymerase chain reaction (PCR) and sequencing-based methods have been the standard HLA typing methods, their low throughput limits their wide application.75 In particular, the tools used for HLA typing in the era of NGS have dramatically changed the field. HLA-miner and Seq2HLA are two of the early tools used for HLA typing from NGS data, massively circumventing the time and cost at that time.70,76 Subsequently, great efforts have been achieved to improve HLA typing performance in terms of both accuracy and resolution. PHLAT, HLAreporter, SNP2HLA, HLA-HD, Optitype and HLA-VBSeq perform fairly well at a four-digit, six-digit, and eight-digit resolution in different cancers.77–83 Notably, among these tools, Polysolver enables high-precision HLA typing and is among the currently accepted standard tools using low-coverage WES data, particularly when applied to cancer-associated somatic mutations.82 Graph-guided genotyping tools used to perform highly classical HLA typing, such as Kourami and HISAT2, provide a new perspective to improve the efficacy of typing.84,85 However, considering the complexity of the HLA types, we still expect independent benchmarking studies and more tools to be presented.
Prediction of antigen-MHC binding affinity
In addition to identifying abnormal peptides and HLA typing, antigen-MHC binding affinity is the next focus of tumor antigen prediction.86,87 Human MHC molecules are divided into the following three subtypes: Class I, Class II, and Class III. Class I MHC molecules (MHC-Is) are expressed by all nucleated cells and present intracellular peptides, such as viral and tumor antigens, to CD8 + T cells to elicit an immune response. In addition, expressed on professional antigen-processing cells (APCs), such as DCs, macrophages, and B cells, class II MHC molecules (MHC-II) present exogenous peptides to activate CD4 + T cells.88,89 Despite substantial research on MHC-I and tumor immunotherapy, recent studies have shown that tumor-specific MHC-II molecules are also associated with favorable outcomes in patients with cancer.90 MHC IIIs are not markers on the cell surface and are not discussed here.
Compared with MHC-II molecules, MHC-I molecules bind shorter peptides between 8 and 11 amino acids.50 Based on artificial neural network (ANN) training methods and position-specific scoring matrix (PSSM), many peptide-MHC-I (pMHC-I) binding affinity prediction tools, such as the currently widely used tools, NetMHC and NetMHCpan, have been developed.91–93 Moreover, even without ANN training, the PSSM-based software called PSSMHCpan could accurately and efficiently predict the pMHC-I binding affinity. After analyzing a 10-fold cross-validation of a training database containing 87 HLA alleles and another independent dataset, Li et al. claimed that PSSMHCpan may be superior to other currently available methods; however, this finding requires verification in further research.94 Currently, the industry standard for predicting the pMHC-I binding affinity is NetMHCpan-4.1, though the number of candidate tumor antigens that could be identified by specific T lymphocytes remains low.95–97 Using mass spectrometry datasets for model selection, MHCflurry provides another choice in addition to tools based on ANNs, which have been validated to show competitive accuracy.98 With the development of AI, an increasing number of tools based on deep neural networks are also promising for improving the current situation, which we would discuss in the following section of the review.
The process of the formation of peptide-MHC-II (pMHC-II) is similar to that of pMHC-I, but it usually binds longer peptides up to 30 amino acids. Furthermore, the impressive diversity of the length of MHC-II-binding peptides and the “openness” of the peptide-binding groove of HLA class II, which permits the binding of a highly degenerate set of peptides, both hinder the development of competitive predictive tools.99,100 Therefore, the prediction of pMHC-II affinity is more challenging, and naturally, the number of available pMHC-II binding affinity prediction methods is far less than pMHC-I, such as CONSENSUS, ProPred, MixMHC2pred, MHCnuggets, NetMHCII, and NetMHCIIpan.101–105 As frequent updates, NetMHCIIpan may be the priority for researchers depending on its competitive performance. Nielsen et al. used the epitope dataset described by Reynisson et al.106 for independent validation and found that NetMHCIIpan-4.0 is much better than the other tools. However, research gaps still persist in the prediction of antigen-MHC II affinity, representing a pause in the development of the prediction of tumor antigens.
Although varying in principles, intended uses, and input and output formats, these tools are not perfect in all aspects, such as sensitivity, accuracy, and availability. Much work is needed to optimize the current tools to better predict tumor antigens to assist with follow-up vaccine design.
Considering the above information, immunogenomics technologies in the NGS era allow researchers to take full advantage of and comprehensively understand sequencing data. On the one hand, considering that conventional tools used to calculate the content of immune cells, such as flow cytometry and IHC, could only quantify a few cellular subtypes, immunogenomics technologies represented by CIBERSORT and ESTIMATE could simultaneously quantity dozens of immune cell types at a relatively lower cost with considerable convenience. On the other hand, with the development of the abovementioned sequencing technology and bioinformatic algorithms, researchers could extract maximal meaning from sequencing data to correlate genetic abnormalities with anti-tumor immunity to make predictions regarding tumor antigens, enabling the design of tumor vaccines.107 Thus, in the NGS era, the technological advances of immunogenomics greatly promote the development of tumor immunity research.
Immunomics in the single-cell era
Although studies using NGS technologies to investigate tumor immunity have greatly promoted the development of oncology, the deficiencies of bulk sequencing have gradually emerged. Performed with RNA (or DNA) extracted from tissue or large cell populations, bulk sequencing may result in a dilution of the signal below the lower detection limit and average out individual cellular expression patterns, masking the reaction of a single cell.108–111 In addition to intra-tumoral heterogeneity (ITH) and the dramatic diversity of immune cells, numerous significant biological phenomena may be obscured by bulk sequencing in the exploration of tumor immunity.
Until recently, technological breakthroughs in single-cell-related approaches revolutionized our understanding of tumor immunity and transitioned the research level from the bulk level to the single-cell level.112–115 In addition to immune cells and tumor cells in the TIME, all cells in the TIME are highly heterogeneous and have unique gene expression profiles and membrane protein expression. We can utilize sequencing technologies and antigen-antibody combination reactions to reflect the features of a single cell. Here, we mainly discuss several technologies applied to the tumor immune cell repertoire and TIME spatial architecture20,116–118 (Table 3).
Table 3.
Comparison of immunomics technologies at the single-cell level
| Technology | Spatial | Strengths | Weaknesses |
|---|---|---|---|
| H&E | √ |
Simple intelligible protocol Lower cost and less time Impressive preservation of tissue morphology |
Lack of specific markers Only morphological features and basophilic or eosinophilic information available |
| mIHC&IF | √ |
Highly specific marker Detailed information regarding the abundance, distribution and localization of certain substances |
Spectral overlap Limited simultaneously detectable markers Time-consuming and labor intensive |
| Flow cytometry |
Affordable and fast Machinery available in most institutes More tools available for analysis Could perform cell sorting |
Spectral overlap Fluorescent spill-over Targets need to be selected carefully (biased) |
|
| CyTOF |
More simultaneously detectable markers Higher accuracy without spectral overlap |
Costly (both the machine and antibodies) Slower processing speed and lower sensitivity Targets need to be carefully selected (biased) |
|
| Spectral flow cytometry |
Compatible with flow cytometry (both the machine and antibodies) Greatly eliminates confounding factors |
Targets need to be carefully selected (biased) | |
| Single-cell seq |
Unbiased Parallel multi-omics analysis Generation of new hypotheses |
Limited to nearly 10,000 cells Limited sequencing depth/coverage Costly, time-consuming and labor intensive |
|
| CODEX | √ |
Higher accuracy and specificity Detection of over 50 markers in a single slide |
Affected by the tissue quality Accumulative structural changes Costly, time-consuming and labor intensive |
| IMC | √ |
At near-optical resolution Could be applied to biobanked tissues More simultaneously detectable markers |
Lack of suitable commercial antibodies for use Comparatively lower rate of image acquisition Limited extent to which slides can be scanned Costly and only available in high-end facilities |
| MIBI-TOF | √ |
High accuracy at near-optical resolution Could be applied to biobanked tissue Indefinitely stable samples More simultaneously detectable markers |
Lack of suitable commercial antibodies for use Comparatively lower rate of image acquisition Limited extent to which slides can be scanned Costly and only available in high-end facilities |
| Spatial transcriptomics | √ | Visualization and quantitative analysis of the transcriptome with spatial resolution | Small-niche but not real single-cell sequencing Comparatively low resolution |
| Slide-seq | √ |
High spatial resolution High scalability to large tissue volumes Lower cost and better accessibility |
Small-niche but not real single-cell sequencing Not suitable for analyzing multiple sections Confined to transcriptomics data |
| HDST | √ |
Higher spatial resolution than Slide-seq High scalability to large tissue volumes Lower cost and better accessibility |
Small-niche but not real single-cell sequencing Not suitable for analyzing multiple sections Confined to transcriptomics data |
| DBiT-seq | √ |
Unbiased High spatial resolution multi-omics seq Compatible with different tissues High accessibility and operability |
Small-niche but not real single-cell sequencing Existence of a theoretical limit of the pixel size |
| ZipSeq | √ |
Provides a complete map of live tissues May integrate with multimodal measurements |
Confined to transcriptomics data Costly and only available in few facilities |
CODEX codetection by indexing, CyTOF cytometry by time-of-light, DBiT-seq deterministic barcoding in tissue for spatial omics sequencing, HDST high-definition spatial transcriptome, H&E hematoxylin-eosin, mIHC multiplex immunohistochemistry, mIF multiplex immunofluorescence, IMC imaging mass cytometry, MIBI-TOF multiplexed ion beam imaging by time-of-flight
Single-cell-based tumor immune cell repertoire
As a highly complex whole, the biological behaviors of tumors, including carcinogenesis, tumor progression, metastasis, recurrence, and response to therapy, all depend on the crosstalk between tumor cells and the surrounding cells in the TIME, especially the immune stromal elements.22,119 Therefore, characterizing the TIME and determining the cellular components could be highly beneficial for tumor immunity studies.
Protein-based single-cell analysis—THE KNOWN UNKNOWN
Polychromatic flow cytometry
Based on the physical characteristics and proteins expressed on the cell surface or within cells that are relatively unique to each cell type, flow cytometry could identify and quantify various cell types utilizing fluorescent dye-conjugated antibodies.120 Flow cytometry has emerged as a core tool in medical research, particularly regarding tumor immune cells.121 The power of multiparametric analysis to discriminate functionally and physically distinct subsets of immune cells has driven flow cytometry to the routinely used 8-parameter flow cytometer. In addition, coupled with technological advances, the design, and implementation of instruments that could measure more parameters (including fluorescent colors and physical parameters) are and could be realized, such as 30- and 50-parameter flow cytometers.122,123 The more parameters that can be measured by flow cytometry, the more information that can be attained from the same sample for further advanced analysis (this also enhances the difficulty of analysis and decreases the accuracy, which we would discuss below). Technological development is confined to not only improving the number of measurable parameters but also better analyzing the existing data. For example, more computational tools for preprocessing, population identification (e.g., FlowJo, FCS Express, WinMDI, and CytoPaint), clustering (e.g., DensVM, kmeans, and mclust), visualization (e.g., flowViz, ggCyto, RchyOptimyx, SPADE, Citrus, and t-stochastic neighbor embedding (t-SNE)), and sorting (e.g., fluorescence-activated cell sorting (FACS)) are available.124–126 However, when deciding how to optimize flow cytometry, researchers are often faced with the following dilemma: more measurable parameters with a lower accuracy or a higher accuracy with limited measurable parameters, particularly due to the overlap between the emission spectra of fluorochromes. Thus, to some extent, these disadvantages limit the application and further development of flow cytometry.
Cytometry by time-of-flight
Mass cytometry, which is a recent innovation in this field and is also termed cytometry by time-of-flight (CyTOF), combines flow cytometry with mass spectrometry and bridges the gap.122,127 Compared with traditional flow cytometry, mass cytometry labels antibodies with metal isotopes instead of fluorophores and then quantifies the signal using a time-of-flight detector, which detects at least 40 parameters and avoids the problem of spectral overlap. CyTOF has been validated as an accurate approach for performing high-dimensional analyses of tumor tissues for exploratory immune profiling and biomarker discovery.128,129 Chevrier et al.130 applied mass cytometry to successfully depict an in-depth atlas of the TIME in clear renal cell carcinoma and correlated immune compositions with clinical features, which has great clinical significance and could guide follow-up studies. Another interesting study performed by Friebel et al. creatively showed that the immune response to cancer in the brain is shaped by the cancer type. Using CyTOF, the TIME of patients with primary brain tumors and brain metastases could be mapped and differentiated according to the heterogeneous composition of tissue-resident and invading immune cells, facilitating the proper design of follow-up targeted immunotherapy strategies.131
Although mass cytometry theoretically allows us to detect at most 100 parameters per cell, the processing speed and throughput are limited by ion flight. After being atomized and ionized, cells are completely destroyed during preprocessing, rendering follow-up cell sorting applications infeasible.122 In addition, regarding measuring certain low-expressed molecular features, CyTOF may be inappropriate because of its low sensitivity.127
Spectral flow cytometry
Spectral flow cytometry is another recent technological advance that promotes the efficacy of conventional flow cytometry. Differing from mass cytometry, spectral flow cytometry still labels antibodies with fluorescent dyes but replaces classical optics and detectors with dispersive optics and novel detectors that measure the full emission spectrum.132 Based on the same principle, conventional flow cytometry and spectral flow cytometry maintain fairly good compatibility, particularly regarding the availability of commercial antibodies, but better eliminate confounding factors, such as spectral overlap, to improve efficiency. Along with the development of compensation technologies, spectral flow cytometry has the potential to replace polychromatic flow cytometry.133
Flow cytometry, mass cytometry, and spectral flow cytometry all base on binding a specific label with the corresponding cellular subgroup and identifying that label, indicating that the targets must be determined before sample acquisition. Thus, the initial targets limit the information obtained from these technologies, seriously diluting the creativeness of research findings.127 We believe that we can only find “THE KNOWN UNKNOWN” via these technologies. In addition, during the actual process, the expense, processing speed, and operability should all be carefully considered. For example, although mass cytometry can avoid the problem of spectral overlap, the cost of specific detectors and access to the required commercial antibodies could render the technique impractical. Finally, we believe that these three technologies are based on the expression of proteins, which may provide a relatively narrow view of the single-cell repertoire, particularly in the era of multi-omics, and urgent innovations are needed.
Single-cell RNA sequencing—THE UNKNOWN UNKNOWN
Fortunately, the advent of single-cell sequencing has driven the single-cell area to new heights. Based on NGS, single-cell sequencing can be divided into the following two main steps: single-cell separation and single-cell analysis.134 Single-cell separation, which is also called single-cell isolation, plays an indispensable role in single-cell studies, including FACS, laser microdissection, manual cell picking, random seeding/dilution, and microfluidics/lab-on-a-chip devices.135 Regarding single-cell analysis, genomic, transcriptomic (mainly), proteomic, and even metabolomic profiles of a single cell are unquestionable research priorities.136–138 No longer limited by predetermined targets as flow cytometry, an individual cell can be sequenced using the standard NGS protocol to obtain unbiased multi-omics profiling that can be used to identify “THE UNKNOWN UNKNOWN”.
Currently, the application of scRNA-seq is relatively more mature than other methods, to be our focus here. Zeisel et al. revealed cell types in the mouse cortex and hippocampus by scRNA-seq, which is a finding considered a groundbreaking discovery.139,140 Subsequently, research applying scRNA-seq to depict the TIME began worldwide. Tirosh et al.141 unraveled the ecosystem of metastatic melanoma by scRNA-seq to provide insight with implications for both targeted and immune therapies. Moreover, in human triple-negative breast cancer (TNBC), the combination of single-cell DNA and RNA sequencing also helped depict the evolutionary trajectories of chemoresistance, which provided further directions for therapies.142 Thus, the prospects of single-cell sequencing technologies are promising and deserve further investigation considering their scientific merit and clinical significance. Since the specific experimental protocols of single-cell sequencing have been reviewed in detail recently, we do not list them again but discuss their advantages and disadvantages in discriminating cellular components.143–145
Commonly, the technical noise resulting from the amplification of trace materials remains the most significant challenge. Regarding other drawbacks, considering scRNA-seq, the whole workflow contains the following five basic steps: single-cell sample preparation, whole-genome or transcriptome amplification, library preparation, sequencing, and data analysis. How to isolate a single cell and maintain its biological activity, how to address the vast technical noise introduced by amplification and improve sensitivity, how to obtain the highest amounts of measurable genes at the lowest price, and how to more efficiently analyze the data greatly raise the threshold for single-cell sequencing and limit its widespread use. Although currently, all technologies compromise coverage, sensitivity or throughput to some extent, we are still optimistic regarding the development of single-cell sequencing and expect more benchmarking studies in the future.
Approaches used to identify the TIME spatial architecture
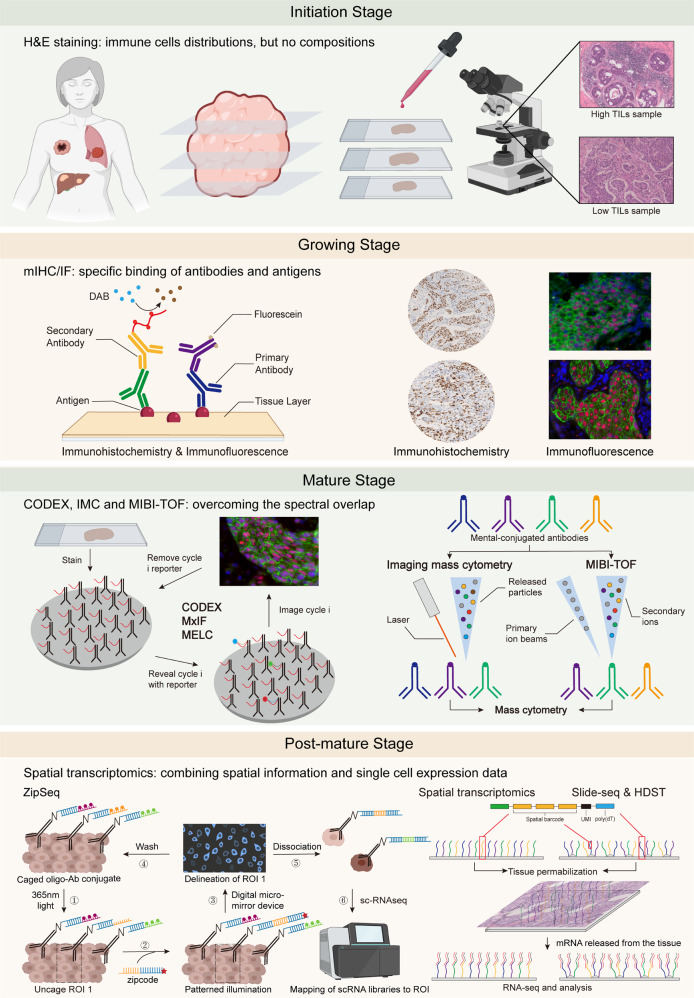
Studies have increasingly found that not only the components of the TIME but also the spatial architectures significantly influence anti-tumor immunity.146 Given that single-cell isolation is necessary for flow cytometry and single-cell sequencing, none of the single-cell technologies mentioned above can be applied to studies investigating spatial architecture. Thus, we briefly introduce several recent single-cell level spatial technologies. According to the principles, we divide the development of TIME spatial architecture approaches into the following four stages: initiation or emerging stage, growing stage, mature stage, and postmature stage (Fig. 2).
Fig. 2.
Development of single-cell spatial technologies: from germination to maturity. (1) Initiation stage: H&E staining, a conventional but significant method that clearly demonstrates the cellular and tissue structure but underperforms in the discrimination of immune cells. (2) Growing stage: The specific binding of antibodies and antigens drove the spatial technologies to a new height as represented by IHC and IF. In addition, multiplex IHC/IF technologies allow the detection of multiple markers simultaneously on a single slice, improving our understanding of the TIME spatial architecture. (3) Mature stage: Given that the spectral overlap limits the further application of mIHC/IF, utilizing dye cycling is a main optimization strategy in which only two or three antibodies are imaged by fluorescence microscopy in each cycle. Then, the fluorophores are cleaved and washed, and this cycle is repeated until all antibodies are imaged, such as CODEX, MxIF, and MELC. Also, IMC and MIBI-TOF utilize mental-conjugated antibodies to eliminate confounding factors, such as spectral overlap, and are also promising. (4) Postmature stage: Combining high-resolution spatial information with single-cell expression data, spatial transcriptomics, slide-seq, HDST, etc. explore brand-new ideas for the characterization of the spatial architecture. CODEX codetection by indexing, HDST high-definition spatial transcriptome, H&E hematoxylin-eosin, IF immunofluorescence, IHC immunohistochemistry, IMC imaging mass cytometry, MELC multiepitope ligand cartography, MIBI-TOF multiplexed ion beam imaging by time-of-flight, MxIF multiplexed fluorescence microscopy method
Initiation stage: H&E-staining
In the initiation stage, a microscopic analysis of the tissue components of an H&E-stained tumor sample slide allows pathologists to clearly differentiate the alkaliphilic nucleus and acidophilic cytoplasm of cells, providing an image of the spatial architecture.147,148 However, without specific markers, we can only empirically divide cells into several large subgroups, such as parenchyma cells, fibroblasts, muscle cells, and inflammatory cells, which is not suitable for characterizing the spatial architecture of the TIME.
Growing stage: mIHC & mIF
Then, at the growing stage, the development of immunological markers markedly improved TIME spatial architecture approaches. IHC and IF utilize fluorescent dye- or enzyme reporter-labeled antibodies targeted against certain antigens in specific cells to more precisely discriminate cell types.149,150 Multiplex immunohistochemistry/immunofluorescence (mIHC/IF) enables the simultaneous detection of multiple markers on a single slice, improving our understanding of the TIME spatial architecture. Unfortunately, similar to flow cytometry, mIHC/IF remains limited by spectral overlap.
Mature stage: CODEX, IMC, and MIBI-TOF
Entering the mature stage, codetection by indexing (CODEX), which is a multiplexed cytometric imaging approach, replaces fluorescent dyes or enzyme reporters with designed specific barcodes comprising a unique oligonucleotide sequence. The fluorescent dNTP analogs and in situ polymerization-based indexing procedure help provide an image of the slice. Interestingly, cells are stained with a mixture of all tagged antibodies simultaneously, but only two or three antibodies are imaged by fluorescence microscopy at each cycle. Then, the fluorophores are cleaved and washed, and the cycle is repeated until all antibodies are imaged. Using computational tools, all antibodies are visualized to reconstruct the multiparameter image.151 Hence, the accuracy of CODEX is higher than that of mIHC/IF to minimize spectral overlap. With the help of CODEX, Goltsev et al.151 observed many previously uncharacterized splenic cell-interaction dynamics in fresh-frozen spleen tissues from animals with systemic autoimmune disease, which is promising for enabling the systemic characterization of tissue architecture. Regarding cancer, the application of CODEX also enabled Schürch et al.146 to identify conserved, distinct cellular neighborhoods (CNs) and explore their correlation with clinical outcomes, re-engineering them to be compatible with formalin-fixed, paraffin-embedded (FFPE) tissue and tissue microarrays.
The multiplexed fluorescence microscopy method (MxIF) and multiepitope ligand cartography (MELC) are two other technologies that use dye cycling analogous to that in CODEX, allowing the detection of at most 100 antigens in a single sample.152–154 In contrast, MxIF is superior to MELC because it can provide a quantitative, single-cell, and subcellular characterization of multiple analytes in FFPE tissue and integrate histological staining with DNA fluorescence in situ hybridization (FISH) to unambiguously compare identical regions in the same sample.154 However, the characteristic feature of these technologies also results in a disadvantage, as follows: repeated elution and imaging could change the antigenicity of the target specimen and may cost too much time and money.
In addition, imaging mass cytometry (IMC) is another expansion of mass cytometry that is perhaps similar to the combination of IHC and mass cytometry. IMC uses laser ablation to generate particles that are carried to the mass cytometer by inserting gas and then yields a high-resolution picture of the region of interest on the slide.155 Notably, IMC preserves the antigen specificity and can simultaneously provide the spatially resolved analysis of 32 proteins.156 On this basis, Damond et al.157 presented a new mechanism of type I diabetes progression as follows: the loss of β cell markers and recruitment of cytotoxic T cells and T helper cells precede β cell destruction. Regarding tumors, Fisher and colleagues unraveled the spatial architecture of classic Hodgkin lymphoma to correlate LAG3 3-expressing Tr1-type Treg cells with MHC-II-negative Hodgkin lymphoma.158
In addition, based on analogous but more complex principles, matrix-assisted laser desorption/ionization (MALDI) mass cytometry can directly identify the distributions of proteins, lipids, metabolites, and drugs with a higher accuracy and sensitivity but lower sample requirements to identify large molecules rather than the TIME spatial architecture.159–161 Further application of laser ablation coupled with inductively coupled plasma mass spectrometry (LA-ICP-MS) is also limited by the laser spot size, analysis speed, and sensitivity but may be combined with IHC, which we do not discuss in detail here.156,162
Notably, the onset of multiplexed ion beam imaging by time-of-flight (MIBI-TOF) had an impact on this field. Compared with mIHC/IF, MIBI-TOF utilizes secondary ion mass spectrometry to image antibodies tagged with mental isotopes and can analyze at most 100 targets simultaneously with high accuracy, low spectral overlap, and no need for channel compensation.163 For example, a structured TIME in TNBC characterized by in situ expression of 36 proteins covering identity, function, and immune regulation at subcellular resolution in 41 TNBC patients has been revealed using MIBI-TOF.164 In 2019, a purpose-built mass spectrometer for MIBI analysis was also designed to further promote the application of MIBI-TOF.165 Combined with CyTOF, MIBI-TOF helped researchers to draw a single-cell metabolic profile of cytotoxic T cells.166 In addition, MIBI-TOF and IMC can both be applied to FFPE tissue sections to perform a retrospective analysis of patient cohorts whose outcome is known.
Post-mature stage: spatial transcriptomics
Finally, the development of spatial technologies has entered the post-mature stage. Recently, the post-genomics era started with an increasing number of sequenced model organisms and further decreases in cost. How to correlate high-resolution spatial information with single-cell expression data and whether single-cell sequencing technology can be utilized to characterize spatial architectures have remained hurdles for a long time in this era. Fortunately, spatial transcriptomics has emerged to address this issue. Making the best use of NGS, similar to spatial transcriptomics, slide-seq and high-definition spatial transcriptome (HDST) utilize a monolayer of spatially barcoded beads on a glass slide to capture mRNAs released from tissue placed on top to demonstrate spatial transcriptome mapping at the cellular level (which could be reduced to 2 µm for HDST).166–171 Furthermore, not confined to transcriptomics data, a recent innovation involving a microfluidic-based method, deterministic barcoding in tissue for spatial omics sequencing (DBiT-seq), enables the realization of high-spatial-resolution multi-omics sequencing in FFPE slides, revolutionizing a range of research fields.172 Notably, ZipSeq can label live cells in intact tissues with unique illumination and photocaged oligonucleotide “zipcodes” and then lyse tissues into individual cells for RNA-seq to broaden the scope of research.173,174 Using these technologies to match clustered regions with individual cells, a spatial landscape of the transcriptome can be generated. However, depending on the designed bead decoding or deterministic barcoding, the number of detected genes is limited; therefore, real spatial transcriptome sequencing has not been realized but is warranted.
Using scRNA-seq data and in situ hybridization patterns as the input, Seurat, which is a spatial map technique, uses a series of sophisticated models to infer the original spatial location of a single cell. This R package (Seurat v3) can accurately localize cellular subpopulations and has been developed into one of the standard tools that have been validated in zebrafish (Danio rerio).175,176 Andersson et al.177 also developed a model-based probabilistic method that performs guided deconvolution of mixed expression profiles to integrate scRNA-seq and spatial transcriptomics data and then spatially map cell types.
The field of single-cell spatial transcriptomics is greatly expanding, and the number of correlative technologies is exploding. Nonetheless, most technologies do not perform NGS or characterize the spatial architecture completely at the single-cell level but rather have tiny pixel sizes (10 µm, even 2 µm). These technologies still perform bulk analyses only for a smaller mixture of cells, possibly representing the greatest challenge faced by current single-cell spatial technologies. Thus, more single-cell technologies and related benchmarking studies are still expected for a long time.
Immunomics and AI
With the development of computer technology, AI, i.e., an intelligence demonstrated by machines that mimic the cognitive functions performed by the human mind, such as learning and making decisions, has been applied all to various fields worldwide.178 In medicine, scientists and clinicians are paying extensive attention to the applications of machine learning or even more advanced deep learning in disease diagnosis, prognosis, and therapeutic response prediction.179 Regarding tumor immunity, AI assists clinicians in better analyzing tumor immunological features associated with the TIME and response to immunotherapy. The technological advances of AI in cancer immunity research principally involve the following aspects: (1) attenuating the workload of the manual recognition of immune infiltration on pathological slides; (2) offering an alternative technology to recognize immune cell subpopulations and spatial architectures that can be hardly distinguished by the human eye; and (3) providing a non-invasive approach to predict specific patient characteristics of the TIME and response to immunotherapy. The major theory of AI in cancer immunity research harnesses high-dimensional features or a black-box operation program to deeply excavate the characteristics of patients’ intra-tumoral immune infiltration.
Tumor antigen prediction with deep learning methods
Concise and accurate tumor antigen prediction is necessary for the investigation and fabrication of personalized tumor vaccines. An important problem resulting in a relatively high false-positive rate is that the current tools predicting antigen presentation are mostly trained by in vitro binding affinity data, thus ignoring other factors, such as gene expression, proteasome cleavage, and transporters associated with antigen processing (TAP) transportation. Considering the aforementioned factors, a robust neoantigen prediction model that comprises reliable training data and an advanced algorithm framework is necessary.
The first step to deciphering tumor antigens is to predict abnormal peptides. In addition to the multiple developed algorithms identifying SNVs, a recently designed CN-learn tool has been designed to detect CNVs, exhibiting favorable performance.180–182 Regarding HLA typing, Bulik et al.183 generated a large integrated dataset including HLA types and HLA peptides from various types of cancer tissues and published data that could be used to train the full mass spectrometry deep learning model EDGE, which has been validated in non-small-cell lung cancer (NSCLC) patients. Two promising computational deep learning methods, MARIA and MixMHC2pred, were recently introduced, greatly increasing the MHC-II prediction accuracy. MARIA is trained using not only in vitro binding affinity data but also naturally presented MHC-II ligand detected by liquid chromatography-tandem mass cytometry (LS-MS/MS) and gene expression levels and conducts a recurrent neural network (RNN) to output a presentation score.184 Racle et al.103 developed a motif deconvolution algorithm, i.e., Modec, to train the deep learning MHC-II peptide predictor MixMHC2pred. These two deep learning methods outperformed the previously prevalent tool NetMHCIIpan, and the neoantigens predicted by both programs have been proven to stimulate responsive CD4 + T cells.
Radiomics in tumor immunity
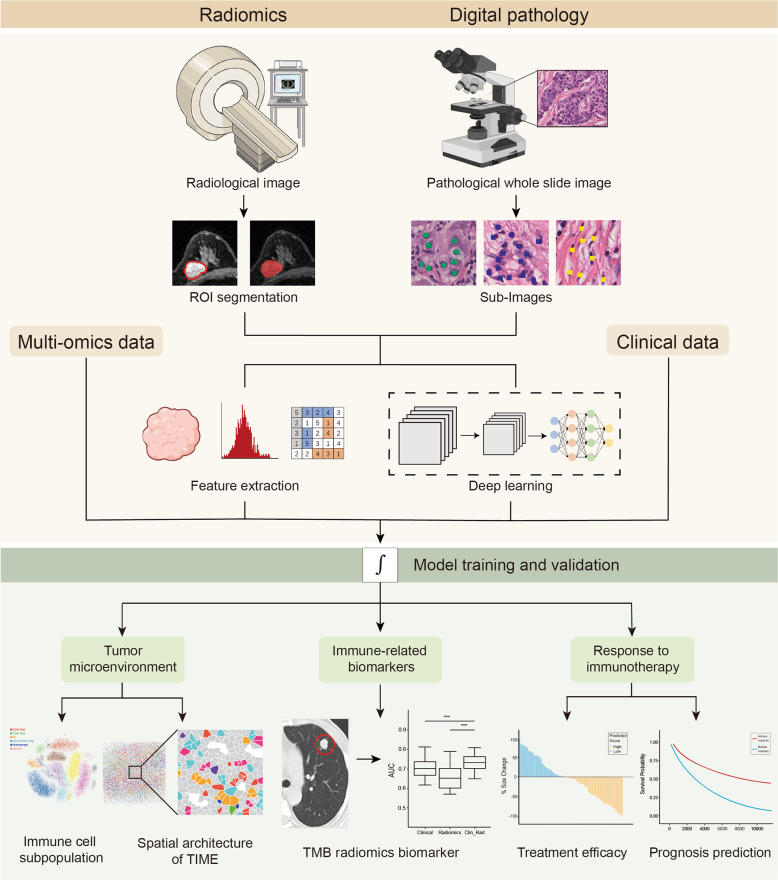
With the development of AI in medical imageology, imaging is far beyond simply a picture but a large scale of digital data. The quantitative and qualitative features extracted from regions of interest (ROIs, usually containing tumor sites) characterize tumor biological behavior and can be correlated with clinical outcomes. This process of analyzing imaging data using AI technology is radiomics.185 To the best of our knowledge, radiomics technology applied to tumor immunity is mainly used to identify biomarkers reflecting immune infiltration and predict the therapeutic response of ICB-treated patients (Fig. 3).
Fig. 3.
Radiomics and computational pathology in tumor immunity exploration. Radiological and pathological image-derived omics data enable the investigation of the tumor immune microenvironment (TIME) and response to immunotherapy. For a raw radiology image, regions of interest, generally representing the tumor lesion area, are segmented, while a pathological image is divided into numerous sub-images. Two methods can be applied to analyze these high-dimensional data. First, features, including but not limited to statistical features, tumor volume features, and texture features, are extracted and analyzed by professional clinicians. Alternatively, images are input into a convolutional neural network (CNN). After a complicated deep learning process, robust models are output. These radiomics or digital pathologic models could finally be established to evaluate or predict the immune index, which can be divided into three aspects. First, TIME dissection encompasses distinct immune cell subset classification and TIME spatial architecture characterization, resembling single-cell technologies and CODEX, respectively. Second, immune-related biomarkers, such as the tumor mutation burden (TMB), could be predicted. Third, response to immunotherapy and clinical outcomes could be predicted. ROI regions of interest, TIME tumor immune microenvironment, TMB tumor mutation burden
First, radiomics provides a non-invasive method to estimate immune-related biomarkers, such as the cytolytic activity score (CytAct) predicted by the deep learning of fluorodeoxyglucose positron emission tomography (FDG-PET)186 and the ImmunoScore of gastric cancer predicted by radiomic features.187,188 A tumor mutational burden radiomics biomarker (TMBRB) was also developed and outperformed the current clinical models in dividing NSCLC patients into high and low tumor mutation burden (TMB) patients who have different clinical outcomes.189 Interestingly, researchers have compared the alterations in the radiomic texture (DelRADx) between baseline and post-treatment CT imaging to discriminate responders from nonresponders. In particular, the relationship among DelRADx, the tumor-infiltrating lymphocytes (TILs) density, and programmed cell death ligand 1 (PD-L1) expression provides a reasonable interpretation for predicting clinical outcomes via radiomics approaches.190
On the other hand, radiomics applications in immunotherapy start with a radiomics signature of CD8 + T cell output by a machine learning model. Imaging-related features and RNA-seq from patients in the MOSCATO clinical trial were input as the training set. This radiomics signature was confirmed to be a biomarker of the response to immunotherapy in validation cohorts. Compared with establishing connections between radiomics features and clinical responses via T cell infiltration, a radiomics biomarker was directly trained and validated using images and clinical data.191 This biomarker was more effective than the lesion volume in predicting the immunotherapy response and overall survival.192
During ICB administration, some atypical responses have been reported. One is hyperprogression (HP), which represents an unexpected accelerated progression after immunotherapy is initiated.193,194 Due to its poor prognosis, predictive biomarkers are desperately required. Textural characteristics and novel quantitative vessel tortuosity features were integrated to distinguish HPs from responders and nonresponders.195 Another response is pseudoprogression, which is defined as an increase in tumor size or newly identified lesion after treatment is initiated before a decrease in tumor size is observed. This phenomenon is due to an inflammatory pseudotumor formed by lymphocyte infiltration.193,194 By comparing blood, volume, and radiomics models alone or combined, a multimodality approach combining the blood biomarker LDH and radiomics features best-predicted pseudoprogression (AUC = 0.82).196
Altogether, radiomics technology enables the identification of changes in tumors during an early stage, stratifying patients’ sensitivity to immunotherapy and predicting their clinical outcomes via a noninvasive method. However, since most current studies are retrospective studies, these results still need to be validated in larger cohorts and prospective studies.
Computational pathology in tumor immunity
Distinct from radiologists, pathologists are devoted to identifying histological alterations from a more microscopic perspective. H&E staining, IHC, and IF help pathologists differentiate among distinct cell populations. AI in pathology, or so-called digital pathology, provides novel insight into exploring the interaction between immune cells and tumor cells and the connection among key behaviors of cancer biology via computational analyses.
CNN-based deep learning models have been established to explore the quantification and spatial distribution of tumor infiltrating immune cells on H&E or IHC staining slides.197–200 In a recent study, AbdulJabbar et al. developed a deep learning framework to profile the spatial architecture of the TIME, revealing that heterogeneity in immune infiltration exists in different samples from identical patients and that prognosis depends on the number of “immune-cold” regions. In addition, evolutionary patterns, clonal neoantigens, and antigen presentation are associated with TIL distribution and the spatial complexity of the TIME.201 Furthermore, predictive models that incorporate the immune cell composition and spatial organization correlate with cancer prognosis, which several groups have proven in colorectal cancer, HCC, and melanoma202–205 (Fig. 3).
Similar to radiomics, digital pathology combined with deep learning excavates invisible information from images; however, the latter enables us to comprehend the TIME on a cellular or molecular level. Consistent with the high-dimensional imaging technology IMC and MIBI-TOF,156,163,165 digital pathology could be a promising approach for investigating the TIME structure and the relationship between cancer biology and therapy. More importantly, deep learning of computational pathology is a paradigm of large-scale detection, i.e., AI can analyze numerous pathological slides simultaneously. Moreover, AI maps analytical results to original slides, which provides a better visualization performance.
In this section, we focus on the AI-based excavation of medical imaging and pathological slides in the era of cancer immunity and immunotherapy. The last decade has observed great achievements in radiomics in clinical practice. In particular, radiomics exhibits potential in predicting immune infiltration and the response to immunotherapy. Although the processes of feature extraction and model training are mainly conducted manually in the current stage, we envision that in the near future, deep learning approaches in medical imaging will be highlighted in research investigating the TIME. In comparison, digital pathology primarily adopts a deep learning approach to dissect the spatial architecture. Although pathological slides involve invasive examination, they can provide more detailed immune information than imaging; thus, radiomics and digital pathology complement each other in the study of the immune microenvironment.
Applications of immunomics in tumor immunotherapy
Major categories of cancer immunotherapy
Cancer immunotherapy is mainly classified into the following six categories: oncolytic viruses, cytokine therapy, antibody-based therapy, ICBs, ACT, and cancer vaccines. (1) Oncolytic viruses. Oncolytic viruses are genetically modified viruses that enable tumor cells to attack and stimulate the immune system simultaneously. Recently, due to progress in genetic engineering, an oncolytic virus, i.e., talimogene laherparepvec (T-VEC), has been proven to benefit advanced melanoma patients and was approved by the Food and Drug Administration (FDA).206 (2) Cytokine therapy. As messengers in communication between immune cells and crucial orchestrating factors in the immune system, cytokines also have the potential to restrict tumor growth.207 Interleukin 2 (IL-2) was approved by the FDA for metastatic melanoma and kidney cancer as an immunotherapy regimen.208 Interferon (IFN) and tumor necrosis factor (TNF) are also regarded as cytokines with potential cancer therapeutic effects. (3) Antibody-based therapy. Monoclonal antibodies were attached to the surface marker of tumor cells and, thus, triggered an enlarged immune response or impeded signal transduction in tumor cells. At the end of the 20th century, rituximab was approved by the FDA for the treatment of non-Hodgkin’s lymphoma. Rituximab binds the CD20 molecule on immature B lymphocytes, guiding NK cells to eradicate these abnormal monoclonal tumor cells.209 (4) Immune checkpoint blockades. Immune checkpoint refers to negative costimulatory molecules expressed on immune cells and tumor cells. In the immune system, the interaction of checkpoint molecules partially offsets positive costimulatory signals to prevent the excess activation of the immune response, which is utilized by tricky tumor cells to render them capable of immune evasion.210–212 Consequently, blockades of such checkpoints reinforce anti-tumor immunity and yield durable therapy responses in cancer patients. (5) ACT. ACT involves the genetic modification of autologous lymphocytes to strengthen anti-tumor activity and reinfusion to the patient’s body.213 Engineering TCRs and chimeric antigen receptors (CARs) are the two types of antigen receptors designed to be expressed on T cells expanding ex vivo, redirecting T cells toward tumor cells specifically.214 (6) Cancer vaccines. Distinct from the prevention effect of conventional antimicrobial vaccines, cancer vaccines trigger the immune system to eradicate preexisting tumor cells. Effective components of cancer vaccines consist of DNAs, RNAs, proteins, and cells (e.g., tumor cells or DCs).215 Cell-based vaccines are classified into autologous and allogenic cell vaccines.
Immunomics technologies: a milestone of immunotherapy
Principle of immunomics application in cancer immunotherapy
Immunotherapy has been one of the most important therapeutic approaches in addition to surgery, chemotherapy, and radiotherapy in multiple types of cancers. Tremendous benefits have been provided to cancer patients with this promising treatment option. Nevertheless, large numbers of patients show less response to immunotherapy. We must address two crucial missions. First, it is necessary to identify novel biomarkers to discriminate responders from non-responders to ICBs. Second, it is essential to authenticate effective targets for engineering T cells and cancer vaccines.
Immunomics technologies offer considerable insight into the microenvironment of tumors to facilitate achieving the two goals above. First, prospective biomarkers of ICBs could be identified by bioinformatics algorithms and single-cell-based technologies. With transcriptomic data, researchers enumerate the immune cell composition in the TIME and estimate the tumor purity with GSEA-based or cell deconvolution-based algorithms, such as ESTIMATE, CIBERSORT, and MCP-counter. Recent years have marked the rapid development of the identification of membrane molecules at a single-cell resolution. In addition to conventional techniques, such as IHC and flow cytometry, techniques, such as CyTOF and single-cell sequencing, permit the identification of more unraveled prognosis- or ICB efficacy-related immune cell subpopulations. Furthermore, promising techniques, such as IMC, CODEX, and MIBI-TOF, offer not only therapeutically significant cell populations but also the relative spatial distribution of distinct immune cells and tumor cells, which are potential targets or biomarkers. Radiomics technology is also able to predict the immune infiltration status in multiple cancer types and patient responses to immunotherapy.
Second, neoantigen prediction via bioinformatic algorithms and AI enables the identification of effective targets of adoptive cell therapy and cancer vaccines. Initially, HLA typing was inferred from genomics and transcriptomic data, and candidate neoantigens were predicted by mutation information and MHC-peptide binding affinity. After experimental validation (i.e., mass spectrometry, ELISpot, and MHC tetramers), selection and prioritization, ultimately determined neoantigens are utilized to generate neoantigen vaccines or neoantigen-targeted engineered T cells.216
Identifying biomarkers of ICBs for patient stratification
Although ICB is undoubtably a milestone of tumor therapy, only a proportion of patients benefit from it. Thus, therapeutic biomarkers are needed to stratify patients into sensitive and non-sensitive to ICB and guide precision medicine. These aforementioned technologies have remarkably promoted the identification of ICB-related biomarkers.
As a target of ICB, the PD-L1 expression level detected by IHC was the first discovered prediction biomarker,217 but several clinical trials have revealed moderate efficacy of ICB in patients with high PD-L1 expression.218 Other biomarkers are urgently required to fill this gap. Promising biomarkers are roughly classified into the following two categories: tumor cell-related biomarkers and immune cell-related biomarkers.
In 2014, investigators first connected TMB with the clinical survival of patients accepting CTLA-4 inhibitor therapy through WES. Subsequently, other retrospective studies also proved that high TMB correlates with a durable clinical benefit.219–221 Translational analyses using clinical trial cohorts of immunologically “cold” metastatic castration-resistant prostate cancer demonstrated that higher TMB is related to a better prognosis after nivolumab plus ipilimumab combination administration.222 Regarding the approaches used to assess TMB, due to the high cost and complicacy of WES, two surrogate NGS panels, i.e., FoundationOne CDx (F1CDx) and MSKCC Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT), were approved by the FDA and validated by several prospective studies of multiple cancers.221
On the other hand, immune cell infiltrations, particularly TILs, play a pivotal role in the immune response. Among the determinants of the anti-tumor response of immune cells, counts, phenotypes and the spatial architecture are the three most highlighted.22,223 Initially, quantified by IHC or flow cytometry, the density of TILs was used to reflect the intensity of the anti-tumor response.224,225 Substantial studies have proven that the intensity of TILs strongly correlates with the ICB response and clinical outcomes.224 Furthermore, according to the number of TILs and their proximity to tumor cells, the TIME can be divided into immune-inflamed, immune-excluded, and immune-desert, which explicitly determine the response to immunotherapy and have better application.226
Nonetheless, a considerable proportion of TILs are only a bystander without cytolytic effects on tumor cells.227 To discover more ideal therapeutic and prognostic biomarkers, single-cell sequencing was used to identify more immune cell subpopulations. It has been found that TCF7 + memory-like T cells improved the clinical outcomes of melanoma patients with anti-PD1 treatment, and stem-like TCF1 + PD1 + T cells were confirmed to be conducive to tumor control in response to ICB.228,229 More therapeutic and prognosis-related T cell subsets and functional status were identified.230–233 CyTOF was performed to compare the TIME of pre- and post-ICB-administered advanced melanoma patients. Krieg et al.234 identified CD14 + CD16 − HLA-DRhi monocytes for the prediction of the response to anti-PD-1 therapy. Furthermore, Helmink et al.235 leveraged CyTOF and single-cell sequencing to reveal a distinctive B cell functional status and tertiary lymphoid structure localization in a melanoma neoadjuvant ICB clinical trial cohort. In addition to adaptive immune cells, new subtypes of innate immune cells, such as macrophages, DCs, and innate lymphoid cells, were also classified by single-cell transcriptome analyses and demonstrated to influence anti-tumor immunity and prognosis236–238 (Table 4).
Table 4.
Clinical significance of immunomics technologies
| Category | Representative technique | Example of clinical relevance | Cancer type | Ref. |
|---|---|---|---|---|
| Bulk sequencing | Abnormal peptides prediction | Developing tumor neoantigen vaccines | Melanoma | 262,291 |
| HLA typing | Glioblastoma | 267 | ||
| MHC-antigen binding affinity | Melanoma, NSCLC | 292 | ||
| Conventional staining on pathological slides |
Immunohistochemistry Immunofluorescence |
Detecting immunotherapy biomarkers, such as PD-L1 and TILs | Multiple cancer types | 293,294 |
| Single-cell technologies | CyTOF | Identifying multiple prognosis-correlated T cell and macrophage phenotypes | Renal cell carcinoma | 130 |
| Revealing immune cell heterogeneity between glioma and brain metastases | Brain cancer | 131 | ||
| Discovering distinct liver TIME driving resistance to immunotherapy | Liver metastatic cancer | 295 | ||
| MIBI-TOF | Dissecting the spatial architecture of the TIME as a promising immunotherapy biomarker | Triple-negative breast cancer | 164 | |
| Single-cell transcriptomics | ILCregs indicate a poor prognosis | Colorectal cancer | 238 | |
| TNFRSF9 + Treg cells refer to a poor prognosis | Lung adenocarcinoma | 231 | ||
| CLEC9A + DC represents a better clinical outcome | Nasopharyngeal carcinoma | 237 | ||
| TCF1-PD1 + T cells correlate to sensitive to immunotherapy | Melanoma | 229 | ||
| CD11b + F4/80+ macrophages lead to resistance to immunotherapy | Liver metastatic cancer | 295 | ||
| CCL22 + cDC1 cells are related to sensitivity to CD40 agonist therapy | Colon cancer | 236 | ||
| Cytotoxic CD4 + T cells serve as a biomarker of responders of anti-PD-L1 treatment | Bladder cancer | 232 | ||
| Artificial intelligence | Radiomics | Radiomics signature predicts immunotherapy biomarker “CytAct” | Lung adenocarcinoma | 186 |
| Radiomics signature predicts immunotherapy biomarker TMB | NSCLC | 189 | ||
| Radiomics signature predicts chemotherapy biomarker “ImmunoScore“ | gastric cancer | 187 | ||
| Radiomics signature predicts response to immunotherapy | Multiple cancer types | 191 | ||
| Digital pathology | Discovering the relationship between the number of immune cold regions and tumor relapse risk | Lung adenocarcinoma | 201 |
cDC conventional dendritic cell, CytAct cytolytic activity score, CyTOF cytometry by time-of-light, DC dendritic cell, HLA human leukocyte antigen, ILCreg regulatory innate lymphoid cell, MHC major histocompatibility complex, MIBI-TOF multiplexed ion beam imaging by time-of-flight, NSCLC non-small cell lung cancer, TMB tumor mutation burden, Treg regulatory T cell
In addition to the components of immune cells in the TIME, the spatial organization largely influences the anti-tumor efficacy of immunotherapy. Recently, CODEX was used to image distinct cell subtypes from low-risk and high-risk colorectal cancer patients. Through a computational analysis, researchers established a CN model and then revealed different functional states in CNs and communication networks between CNs, representing the spatial heterogeneity of the TIME and correlates to clinical outcome.146 From another perspective, deep learning models have been used to analyze digital pathological slides to elucidate the spatial heterogeneity of tumor antigen presentation and tumor evolution.201 In breast cancer, researchers designed an IMC panel that enables 35 biomarkers to be labeled simultaneously, thus revealing breast cancer and connecting heterogeneity with clinical outcomes.239 While these emerging technologies are not mature enough, a landscape will be portrayed, and the TIME organization could be watched even from a higher dimensional perspective in the future (Table 4).
Predicting neoantigens for ACT therapy
ACT is an immunotherapy approach in which genetically modified or expanded autologous or allogeneic T cells are reinfused into patients to enhance anti-tumor immunity.240 Immunogenomics primarily functions in the identification of ideal tumor antigens in ACT therapy. Specifically, once patient NGS data are obtained, it is feasible to enter an ACT-targeted tumor antigen prediction pipeline comprising abnormal peptide prediction, HLA typing, antigen-MHC binding affinity, and neoantigen prioritization.
As the first approach of ACT, engineering TCR T cells construct tumor antigen-specific TCRs to recognize tumor-associated antigens (TAAs), such as MAGE, NY-ESO-1, or more ideal target neoantigens defined by immunogenomics data by WES and RNA-seq.241 chimeric antigen receptor T cells (CAR-T cells) are another approach in which T cells are armored by CAR dominantly composed of a single-chain variable fragment (scFv) from a monoclonal antibody. In contrast to TCR-engineered T cells, CAR-T cells recognize tumor antigens with an MHC-independent pattern, directly identifying and combining targeting surface molecules expressed on tumor cells.242 Although successful in multiple hematopoietic malignancies, the benefit of CAR-T cells in solid tumors has not been forthcoming.243–245
Clinical trials have been conducted to demonstrate the antitumor response of TAA-specific T cells produced by TCR engineering in synovial sarcoma, melanoma, and colorectal cancers.246–248 Currently, neoantigen-specific TCR-engineered T cells have not been applied clinically at bedside. However, it is gratifying that several case reports have shown the efficacy of T-cell recognition against tumor neoantigens predicted by immunogenomics in colorectal cancer, breast cancers, and cholangiocarcinoma.249–251 Researchers cocultured T cells with neoantigen-armored APCs and T cells to identify neoantigen-activated T cells and reinfused them back into the body. Tran et al. conducted WGS of a sample from a metastatic cholangiocarcinoma patient to identify 26 somatic mutations. Tandem minigenes composed of the mutated genes were transcribed and transfected into autologous APCs, after which the neoantigen-presenting APCs were cocultured with patient-derived TILs, eventually identifying antigen-specific CD4 + Vb22+ T cell clones, which induced regression of epithelial cancer.251
Resulting from the difficulty in isolating TILs from tumor sites, peripheral blood neoantigen-recognizing T cells were isolated and proved to be identical to TILs in the immunological process.252 Thus, WES along with neoantigen T cell isolation has become a promising approach for promoting noninvasive cancer therapeutic strategies.
However, conventional neoantigen selection based on autologous APC and T cell coculture is limited by its low throughput, high cost, and time-consuming attributes. To eliminate these barriers, more high-throughput immunogenic neoantigen detection technologies have been developed. Li et al. established a trogocytosis-based platform in which surface marker proteins transfer from APCs to T cells when TCR and pMHC combine. Therefore, ideal neoantigens could be identified by analyzing marker protein-positive cells.253 Coincidently, another cell-based platform utilizing signaling and antigen-presenting bifunctional receptors was established for neoantigen identification.254 In these cases, cell lines expressing the predicted neoantigens and TCR replaced patient-derived APCs and lymphocytes and realized high-throughput neoantigen selection as burgeoning immunogenomics technologies.
Selecting neoantigens for personalized cancer vaccines
Since William Coley discovered that bacterial toxins elicited body immunity to attack tumor cells, cancer vaccines have received attention.255,256 Subsequently, the discovery of TAA paved the way for further investigations of tumor-specific vaccine therapeutics.257,258 However, targeting TAAs likely harms normal cells by autoimmunity, and anti-tumor immunity is insufficient since T cells experience negative selection in the thymus against autoantigens.259–261 Personalized “neoantigens” from tumor mutations are more appropriate for effective vaccine design, and personalized vaccines have achieved favorable efficacy in several clinical trials, such as GAPVAC-10 and IVAC MUTANOME.262–269
Immunogenomics approaches have been widely applied in vaccine development in clinical research. In general, neoantigens used to generate personalized vaccines are identified by analyzing WES and RNA-seq of tumor and normal tissues and predicting effective epitopes via algorithms, such as NetMHCpan. Through this method, for instance, four in six high-risk melanoma patients accepting vaccination were free of recurrence in 25 months, while the other two patients received ICB after recurrence and had a complete response. Furthermore, ex vivo immunological experiments indicated that polyfunctional CD4 + T cells and CD8 + T cells were stimulated by 60% and 16% neoantigens, respectively.262 Similarly, neoantigen vaccines have shown efficacy in phase Ib clinical trials of glioblastoma. Single-cell TCR analysis also suggests that antigen-specific T cells are stimulated and distributed in intracranial tumor lesions.267
Similar to ACT, the crucial parameter of tumor vaccine development is ideal neoantigen identification. Considerable efforts have been exerted to develop immunogenomics technology to improve the neoantigen prediction accuracy and prioritize immunogenic neoepitope selection pipelines. In a recent study, Wells et al.270 compiled all neoantigen prediction and selection methods and provided a brand-new candidate determination pipeline incorporating 14 immunogenic features of MHC presentation and T cell recognition. This study lays a solid foundation for promoting the efficacy of tumor vaccines and adoptive cell therapy.
Conclusions and future directions
It is patently obvious that with the giant leap of emergent technologies in the realm of immunomics, we are now able to dissect tumor immunity at an unprecedented depth (Fig. 4). In this review, we present a picture of conventional and state-of-the-art technologies in tumor immunology along with prospects for clinical application as a reference for researchers.
Fig. 4.
A landscape of immunomics: developmental tendency and future direction. a The timeline of immunomics technologies. b Historical development trajectory and future prospective of tumor immunomics. CODEX codetection by indexing, CyTOF cytometry by time-of-light, ESTIMATE estimation of stromal and immune cells in malignant tumors using expression data, GATK Genome Analysis Toolkit, GenomeVIP Genome Variant Investigation Platform, HDST high-definition spatial transcriptome, IMC imaging mass cytometry, MCP-counter microenvironment cell populations-counter, MIBI-TOF multiplexed ion beam imaging by time-of-flight, mIF multiplex immunofluorescence, mIHC multiplex immunohistochemistry
In the era of bulk sequencing, methods for estimating tumor immune cells, mainly including computational algorithms, such as CIBERSORT and MCP-counter, allow us to better explore the individual infiltration pattern of tumor immune cells. Furthermore, comprising the prediction of abnormal peptides, HLA typing, and prediction of tumor antigen-MHC binding affinity, the use of immunogenomics technologies to predict tumor antigens has demonstrated credible efficacy in both preclinical and clinical studies, as represented by personalized tumor vaccines and ACT.
Moreover, it is wise to explore tumor immunity at the single-cell level considering the high diversity of immune cell subtypes and ITH. With the development of single-cell immune-related technologies, from flow cytometry and spectral flow cytometry to CyTOF, the single-cell tumor immune atlas should assist with immune cell subgroup classification to decipher components of the TIME. Regarding spatial architecture, using H&E, IHC/IF, MIBI-TOF, or spatially resolved transcriptomics, which is a crowned method of 2020, provide a high-resolution visualization of the TIME.271
The advent of AI also provides a new direction for the development of immunomics. Radiological and pathological image-derived omics data enable the characterization of the TIME to predict the prognosis and response to immunotherapy, indicating the potential of clinical applications with noninvasive or minimally invasive methods.
As immunomics technologies flourish, several issues should be considered for sustainable development. First, although numerous methods for quality control and improvement of the algorithm principle have been implemented, the efficacy of these technologies can be improved. In particular, regarding the prediction of tumor antigens, single-cell sequencing, and spatially resolved transcriptomics, technical noise and confounding factors hamper subsequent analyses. Second, more cost-effective, accessible, and automated technologies are expected to emerge to revolutionize the development of the discipline. Third, we also expect that researchers will fully use existing technologies to explore tumor immunity and promote clinical transformation. Utilizing advanced technologies to analyze samples from clinical trials may be a practical solution. For example, Grasso et al.272 showed that an increase in T cell infiltration and downstream IFN-γ signaling drive clinical responses by analyzing the CheckMate 038 study using technologies, such as NGS and immune cell quantitation, representing the regeneration of immunogenomics in the NGS era. Studies investigating tumor immunotherapy, such as tumor vaccines and ACT, should be promoted. Finally, it is necessary to develop more cancer type-specific technologies. Currently, some technologies are indeed appropriate and perform well in specific tumor types. For example, spatial single-cell technologies are suitable for solid tumors because the spatial architecture of the TIME is not involved in hematological malignancies. TCR-T cell therapy is mainly applied in melanoma, and CAR-T cell therapy performs better in hematological malignancies such as leukemia and lymphoma; neoantigen prediction technologies are suitable for these cancer types. However, as discussed above, tumor type-specific technologies are confined to hematological/solid malignancies or immune “hot” tumors in the current stage. We anticipate that further cancer type-specific technologies will emerge based on the distinctive characteristics of each cancer, greatly contributing to the development of precision oncology.
Although there is much to be accomplished, immunomics is likely to dominate the field of future tumor immunology, and its clinical value will undoubtedly dramatically promote the development of this discipline, in the field of immunogenomics, single-cell, and AI.
Acknowledgements
This work was supported by grants from the National Key Research and Development Project of China (2020YFA0112304), the National Natural Science Foundation of China (81922048, 81874112, 82002792), the Program of Shanghai Academic/Technology Research Leader (20XD1421100), the Shanghai Key Laboratory of Breast Cancer (ZDSYS2101), the Shanghai Key Clinical Specialty of Oncology (shslczdzk02001), the Shenkang Three Year Program for Clinical Research (SK2020) and Shanghai Sailing Program (20YF1408600). The funders had no role in the study design, data collection, and analysis, decision to publish, or preparation of the manuscript. And the figures are created with Biorender.com.
Author contributions
Y.-Z.J., Z.-M.S., Y.X., and Y.X. designed and finalized the study. Y.X. and G.-H.S. wrote the paper. Y.X., G.-H.S., Y.X., and D.M. revised the paper. All authors approved the final version submitted.
Consent for publication
The content of this manuscript has not been previously published and is not under consideration for publication elsewhere. All authors are aware of and agree to the content of the paper and are listed as coauthors of the paper.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Ying Xu, Guan-Hua Su
Contributor Information
Yi Xiao, Email: yixiao11@fudan.edu.cn.
Zhi-Ming Shao, Email: zhimingshao@yahoo.com.
Yi-Zhou Jiang, Email: yizhoujiang@fudan.edu.cn.
References
- 1.Anderson NM, Simon MC. The tumor microenvironment. Curr. Biol. 2020;30:R921–R925. doi: 10.1016/j.cub.2020.06.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat. Rev. Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei X, et al. Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Lett. 2020;470:126–133. doi: 10.1016/j.canlet.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 4.Chen DS, Mellman I. Oncology meets immunology: the cancer-immunity cycle. Immunity. 2013;39:1–10. doi: 10.1016/j.immuni.2013.07.012. [DOI] [PubMed] [Google Scholar]
- 5.Mellman I, Coukos G, Dranoff G. Cancer immunotherapy comes of age. Nature. 2011;480:480–489. doi: 10.1038/nature10673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zou W, Wolchok JD, Chen L. PD-L1 (B7-H1) and PD-1 pathway blockade for cancer therapy: mechanisms, response biomarkers, and combinations. Sci. Transl. Med. 2016;8:328rv324. doi: 10.1126/scitranslmed.aad7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Goedegebuure SP, Gillanders WE. Preclinical and clinical development of neoantigen vaccines. Ann. Oncol. 2017;28:xii11–xii17. doi: 10.1093/annonc/mdx681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer. 2012;12:252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yamamoto TN, Kishton RJ, Restifo NP. Developing neoantigen-targeted T cell–based treatments for solid tumors. Nat. Med. 2019;25:1488–1499. doi: 10.1038/s41591-019-0596-y. [DOI] [PubMed] [Google Scholar]
- 10.Butterfield LH. Cancer vaccines. BMJ. 2015;350:h988. doi: 10.1136/bmj.h988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Majzner RG, Mackall CL. Tumor antigen escape from CAR T-cell therapy. Cancer Discov. 2018;8:1219–1226. doi: 10.1158/2159-8290.CD-18-0442. [DOI] [PubMed] [Google Scholar]
- 12.Chandran SS, Klebanoff CA. T cell receptor-based cancer immunotherapy: emerging efficacy and pathways of resistance. Immunol. Rev. 2019;290:127–147. doi: 10.1111/imr.12772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klysik J. Concept of immunomics: a new frontier in the battle for gene function? Acta Biotheor. 2001;49:191–202. doi: 10.1023/A:1011901410166. [DOI] [PubMed] [Google Scholar]
- 14.Tay SH, et al. Immunomics in pediatric rheumatic diseases. Front. Med. 2019;6:111. doi: 10.3389/fmed.2019.00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hui L, Chen Y. Tumor microenvironment: sanctuary of the devil. Cancer Lett. 2015;368:7–13. doi: 10.1016/j.canlet.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 16.DeBerardinis RJ. Tumor microenvironment, metabolism, and immunotherapy. N. Engl. J. Med. 2020;382:869–871. doi: 10.1056/NEJMcibr1914890. [DOI] [PubMed] [Google Scholar]
- 17.Vitale I, et al. Macrophages and metabolism in the tumor microenvironment. Cell Metab. 2019;30:36–50. doi: 10.1016/j.cmet.2019.06.001. [DOI] [PubMed] [Google Scholar]
- 18.Meurette O, Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell. 2018;34:536–548. doi: 10.1016/j.ccell.2018.07.009. [DOI] [PubMed] [Google Scholar]
- 19.van de Wall S, et al. Sialoglycans and siglecs can shape the tumor immune microenvironment. Trends Immunol. 2020;41:274–285. doi: 10.1016/j.it.2020.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Binnewies M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat. Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang TY, et al. Advantages of targeting the tumor immune microenvironment over blocking immune checkpoint in cancer immunotherapy. Signal Transduct. Tar. 2021;6:1–13. doi: 10.1038/s41392-020-00451-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Labani-Motlagh A, Ashja-Mahdavi M, Loskog A. The tumor microenvironment: a milieu hindering and obstructing antitumor immune responses. Front. Immunol. 2020;11:940. doi: 10.3389/fimmu.2020.00940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pitt JM, et al. Targeting the tumor microenvironment: removing obstruction to anticancer immune responses and immunotherapy. Ann. Oncol. 2016;27:1482–1492. doi: 10.1093/annonc/mdw168. [DOI] [PubMed] [Google Scholar]
- 25.Zappasodi R, et al. CTLA-4 blockade drives loss of Treg stability in glycolysis-low tumours. Nature. 2021;591:652–658. doi: 10.1038/s41586-021-03326-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finotello F, Trajanoski Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol. Immunother. 2018;67:1031–1040. doi: 10.1007/s00262-018-2150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshihara K, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013;4:2612. doi: 10.1038/ncomms3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220. doi: 10.1186/s13059-017-1349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becht E, et al. Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016;17:218. doi: 10.1186/s13059-016-1070-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods. 2015;12:453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Plattner C, Finotello F, Rieder D. Deconvoluting tumor-infiltrating immune cells from RNA-seq data using quanTIseq. Methods Enzymol. 2020;636:261–285. doi: 10.1016/bs.mie.2019.05.056. [DOI] [PubMed] [Google Scholar]
- 32.Hao Y, et al. Fast and robust deconvolution of tumor infiltrating lymphocyte from expression profiles using least trimmed squares. PLoS Comput. Biol. 2019;15:e1006976. doi: 10.1371/journal.pcbi.1006976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X, et al. Bulk tissue cell type deconvolution with multi-subject single-cell expression reference. Nat. Commun. 2019;10:380. doi: 10.1038/s41467-018-08023-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Newman AM, et al. Determining cell type abundance and expression from bulk tissues with digital cytometry. Nat. Biotechnol. 2019;37:773–782. doi: 10.1038/s41587-019-0114-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Clark DJ, et al. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell. 2019;179:964–983.e931. doi: 10.1016/j.cell.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gillette MA, et al. Proteogenomic CHaracterization Reveals Therapeutic Vulnerabilities in Lung Adenocarcinoma. Cell. 2020;182:200–225.e235. doi: 10.1016/j.cell.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C, et al. Proteogenomic insights into the biology and treatment of HPV-negative head and neck squamous cell carcinoma. Cancer Cell. 2021;39:361–379.e316. doi: 10.1016/j.ccell.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thorsson V, et al. The Immune Landscape of Cancer. Immunity. 2018;48:812–830. doi: 10.1016/j.immuni.2018.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gil Del Alcazar CR, et al. Immune escape in breast cancer during in situ to invasive carcinoma transition. Cancer Discov. 2017;7:1098–1115. doi: 10.1158/2159-8290.CD-17-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cancer Genome Atlas Research Network. Electronic address, w. b. e. & Cancer Genome Atlas Research, N. Comprehensive and integrative genomic characterization of hepatocellular carcinoma. Cell169, 1327–1341.e1323 (2017). [DOI] [PMC free article] [PubMed]
- 41.Chakravarthy A, et al. Pan-cancer deconvolution of tumour composition using DNA methylation. Nat. Commun. 2018;9:3220. doi: 10.1038/s41467-018-05570-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arneson D, Yang X, Wang K. MethylResolver-a method for deconvoluting bulk DNA methylation profiles into known and unknown cell contents. Commun. Biol. 2020;3:422. doi: 10.1038/s42003-020-01146-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jiang T, et al. Tumor neoantigens: from basic research to clinical applications. J. Hematol. Oncol. 2019;12:93. doi: 10.1186/s13045-019-0787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krassowski M, et al. ActiveDriverDB: human disease mutations and genome variation in post-translational modification sites of proteins. Nucleic Acids Res. 2018;46:D901–D910. doi: 10.1093/nar/gkx973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou C, Zhu C, Liu Q. Toward in silico identification of tumor neoantigens in immunotherapy. Trends Mol. Med. 2019;25:980–992. doi: 10.1016/j.molmed.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 46.Ehx G, Perreault C. Discovery and characterization of actionable tumor antigens. Genome Med. 2019;11:29. doi: 10.1186/s13073-019-0642-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T. Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat. Rev. Cancer. 2014;14:135–146. doi: 10.1038/nrc3670. [DOI] [PubMed] [Google Scholar]
- 48.Gubin MM, Artyomov MN, Mardis ER, Schreiber RD. Tumor neoantigens: building a framework for personalized cancer immunotherapy. J. Clin. Invest. 2015;125:3413–3421. doi: 10.1172/JCI80008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paschen A, Eichmuller S, Schadendorf D. Identification of tumor antigens and T-cell epitopes, and its clinical application. Cancer Immunol. Immunother. 2004;53:196–203. doi: 10.1007/s00262-003-0479-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hackl H, Charoentong P, Finotello F, Trajanoski Z. Computational genomics tools for dissecting tumour-immune cell interactions. Nat. Rev. Genet. 2016;17:441–458. doi: 10.1038/nrg.2016.67. [DOI] [PubMed] [Google Scholar]
- 51.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wilm A, et al. LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40:11189–11201. doi: 10.1093/nar/gks918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koboldt DC, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2012;22:568–576. doi: 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Larson DE, et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics. 2012;28:311–317. doi: 10.1093/bioinformatics/btr665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang Q, et al. Detecting somatic point mutations in cancer genome sequencing data: a comparison of mutation callers. Genome Med. 2013;5:91. doi: 10.1186/gm495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim S, et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat. Methods. 2018;15:591–594. doi: 10.1038/s41592-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 59.Saunders CT, et al. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics. 2012;28:1811–1817. doi: 10.1093/bioinformatics/bts271. [DOI] [PubMed] [Google Scholar]
- 60.Lai Z, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016;44:e108–e108. doi: 10.1093/nar/gkw227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shiraishi Y, et al. An empirical Bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res. 2013;41:e89. doi: 10.1093/nar/gkt126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.O’Rawe J, et al. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Med. 2013;5:28. doi: 10.1186/gm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bian X, et al. Comparing the performance of selected variant callers using synthetic data and genome segmentation. BMC Bioinforma. 2018;19:429. doi: 10.1186/s12859-018-2440-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Do Valle ÍF, et al. Optimized pipeline of MuTect and GATK tools to improve the detection of somatic single nucleotide polymorphisms in whole-exome sequencing data. BMC Bioinforma. 2016;17:27–35. doi: 10.1186/s12859-016-1190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mashl RJ, et al. GenomeVIP: a cloud platform for genomic variant discovery and interpretation. Genome Res. 2017;27:1450–1459. doi: 10.1101/gr.211656.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Huang KL, et al. Pathogenic germline variants in 10,389 adult cancers. Cell. 2018;173:355–370. doi: 10.1016/j.cell.2018.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Consortium, T. G. P. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Williams TM. Human leukocyte antigen gene polymorphism and the histocompatibility laboratory. J. Mol. Diagnostics. 2001;3:98–104. doi: 10.1016/S1525-1578(10)60658-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boegel S, et al. HLA typing from RNA-Seq sequence reads. Genome Med. 2012;4:102. doi: 10.1186/gm403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Marsh SGE. Nomenclature for factors of the HLA system, update April, May, and June 2020. HLA. 2020;96:384–412. doi: 10.1111/tan.14000. [DOI] [PubMed] [Google Scholar]
- 72.Birnbaum ME, et al. Deconstructing the peptide-MHC specificity of T cell recognition. Cell. 2014;157:1073–1087. doi: 10.1016/j.cell.2014.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Opelz G, et al. Survival of DNA HLA-DR typed and matched cadaver kidney transplants. The Collaborative Transplant Study. Lancet. 1991;338:461–463. doi: 10.1016/0140-6736(91)90540-6. [DOI] [PubMed] [Google Scholar]
- 74.Erlich HA, Opelz G, Hansen J. HLA DNA Typing and Transplantation. Immunity. 2001;14:347–356. doi: 10.1016/S1074-7613(01)00115-7. [DOI] [PubMed] [Google Scholar]
- 75.Bontadini A. HLA techniques: typing and antibody detection in the laboratory of immunogenetics. Methods. 2012;56:471–476. doi: 10.1016/j.ymeth.2012.03.025. [DOI] [PubMed] [Google Scholar]
- 76.Warren RL, et al. Derivation of HLA types from shotgun sequence datasets. Genome Med. 2012;4:95. doi: 10.1186/gm396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kawaguchi S, et al. HLA‐HD: an accurate HLA typing algorithm for next‐generation sequencing data. Hum. Mutat. 2017;38:788–797. doi: 10.1002/humu.23230. [DOI] [PubMed] [Google Scholar]
- 78.Szolek A, et al. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30:3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nariai N, et al. HLA-VBSeq: accurate HLA typing at full resolution from whole-genome sequencing data. BMC Genomics. 2015;16:S7. doi: 10.1186/1471-2164-16-S2-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jia X, et al. Imputing amino acid polymorphisms in human leukocyte antigens. PLoS ONE. 2013;8:e64683. doi: 10.1371/journal.pone.0064683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang Y, et al. HLAreporter: a tool for HLA typing from next generation sequencing data. Genome Med. 2015;7:25. doi: 10.1186/s13073-015-0145-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shukla SA, et al. Comprehensive analysis of cancer-associated somatic mutations in class I HLA genes. Nat. Biotechnol. 2015;33:1152–1158. doi: 10.1038/nbt.3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bai Y, et al. Inference of high resolution HLA types using genome-wide RNA or DNA sequencing reads. BMC Genomics. 2014;15:325. doi: 10.1186/1471-2164-15-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim D, et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lee H, Kingsford C. Kourami: graph-guided assembly for novel human leukocyte antigen allele discovery. Genome Biol. 2018;19:16. doi: 10.1186/s13059-018-1388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Rock KL, Reits E, Neefjes J. Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol. 2016;37:724–737. doi: 10.1016/j.it.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wieczorek M, et al. Major histocompatibility complex (MHC) class I and MHC class II proteins: conformational plasticity in antigen presentation. Front. Immunol. 2017;8:292. doi: 10.3389/fimmu.2017.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Neefjes J, Jongsma MLM, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat. Rev. Immunol. 2011;11:823–836. doi: 10.1038/nri3084. [DOI] [PubMed] [Google Scholar]
- 89.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat. Rev. Genet. 2010;11:47–59. doi: 10.1038/nrg2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Axelrod ML, Cook RS, Johnson DB, Balko JM. Biological consequences of MHC-II expression by tumor cells in cancer. Clin. Cancer Res. 2019;25:2392–2402. doi: 10.1158/1078-0432.CCR-18-3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Desrichard A, Snyder A, Chan TA. Cancer neoantigens and applications for immunotherapy. Clin. Cancer Res. 2016;22:807–812. doi: 10.1158/1078-0432.CCR-14-3175. [DOI] [PubMed] [Google Scholar]
- 92.Jurtz V, et al. NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J. Immunol. 2017;199:3360–3368. doi: 10.4049/jimmunol.1700893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lundegaard C, et al. NetMHC-3.0: accurate web accessible predictions of human, mouse and monkey MHC class I affinities for peptides of length 8–11. Nucleic Acids Res. 2008;36:W509–W512. doi: 10.1093/nar/gkn202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu G, et al. PSSMHCpan: a novel PSSM-based software for predicting class I peptide-HLA binding affinity. Gigascience. 2017;6:1–11. doi: 10.1093/gigascience/gix017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rappazzo CG, Huisman BD, Birnbaum ME. Repertoire-scale determination of class II MHC peptide binding via yeast display improves antigen prediction. Nat. Commun. 2020;11:4414. doi: 10.1038/s41467-020-18204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Castro A, et al. Strength of immune selection in tumors varies with sex and age. Nat. Commun. 2020;11:4128. doi: 10.1038/s41467-020-17981-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kristensen VN. The antigenicity of the tumor cell - context matters. N. Engl. J. Med. 2017;376:491–493. doi: 10.1056/NEJMcibr1613793. [DOI] [PubMed] [Google Scholar]
- 98.O’Donnell TJ, et al. MHCflurry: open-source class I MHC binding affinity prediction. Cell Syst. 2018;7:129–132.e124. doi: 10.1016/j.cels.2018.05.014. [DOI] [PubMed] [Google Scholar]
- 99.Roudko V, Greenbaum B, Bhardwaj N. Computational prediction and validation of tumor-associated neoantigens. Front. Immunol. 2020;11:27. doi: 10.3389/fimmu.2020.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nielsen M, Lund O, Buus S, Lundegaard C. MHC class II epitope predictive algorithms. Immunology. 2010;130:319–328. doi: 10.1111/j.1365-2567.2010.03268.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Singh H, Raghava GP. ProPred: prediction of HLA-DR binding sites. Bioinformatics. 2001;17:1236–1237. doi: 10.1093/bioinformatics/17.12.1236. [DOI] [PubMed] [Google Scholar]
- 102.Reynisson B, et al. NetMHCpan-4.1 and NetMHCIIpan-4.0: improved predictions of MHC antigen presentation by concurrent motif deconvolution and integration of MS MHC eluted ligand data. Nucleic Acids Res. 2020;48:W449–W454. doi: 10.1093/nar/gkaa379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Racle J, et al. Robust prediction of HLA class II epitopes by deep motif deconvolution of immunopeptidomes. Nat. Biotechnol. 2019;37:1283–1286. doi: 10.1038/s41587-019-0289-6. [DOI] [PubMed] [Google Scholar]
- 104.Shao XM, et al. High-throughput prediction of MHC class I and II neoantigens with MHCnuggets. Cancer Immunol. Res. 2020;8:396–408. doi: 10.1158/2326-6066.CIR-19-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Moutaftsi M, et al. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 106.Reynisson B, et al. Improved prediction of MHC II antigen presentation through integration and motif deconvolution of mass spectrometry MHC eluted ligand data. J. Proteome Res. 2020;19:2304–2315. doi: 10.1021/acs.jproteome.9b00874. [DOI] [PubMed] [Google Scholar]
- 107.Koch CM, et al. A Beginner’s Guide to Analysis of RNA Sequencing Data. Am. J. Respir. Cell Mol. Biol. 2018;59:145–157. doi: 10.1165/rcmb.2017-0430TR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rosenfeld N. Gene regulation at the single-cell level. Science. 2005;307:1962–1965. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- 109.Olsen TK, Baryawno N. Introduction to single-cell RNA sequencing. Curr. Protoc. Mol. Biol. 2018;122:e57. doi: 10.1002/cpmb.57. [DOI] [PubMed] [Google Scholar]
- 110.Eberwine J, et al. Quantitative biology of single neurons. J. R. Soc. Interface. 2012;9:3165–3183. doi: 10.1098/rsif.2012.0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Savas P, et al. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 2018;24:986–993. doi: 10.1038/s41591-018-0078-7. [DOI] [PubMed] [Google Scholar]
- 112.Shapiro E, Biezuner T, Linnarsson S. Single-cell sequencing-based technologies will revolutionize whole-organism science. Nat. Rev. Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 113.Li H, et al. Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat. Genet. 2017;49:708–718. doi: 10.1038/ng.3818. [DOI] [PubMed] [Google Scholar]
- 114.Azizi E, et al. Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. 2018;174:1293–1308.e1236. doi: 10.1016/j.cell.2018.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chung W, et al. Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat. Commun. 2017;8:15081. doi: 10.1038/ncomms15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Potter SS. Single-cell RNA sequencing for the study of development, physiology and disease. Nat. Rev. Nephrol. 2018;14:479–492. doi: 10.1038/s41581-018-0021-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Stegle O, Teichmann SA, Marioni JC. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 2015;16:133–145. doi: 10.1038/nrg3833. [DOI] [PubMed] [Google Scholar]
- 118.Kolodziejczyk AA, et al. The technology and biology of single-cell RNA sequencing. Mol. Cell. 2015;58:610–620. doi: 10.1016/j.molcel.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 119.Liu CC, Steen CB, Newman AM. Computational approaches for characterizing the tumor immune microenvironment. Immunology. 2019;158:70–84. doi: 10.1111/imm.13101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Adan A, et al. Flow cytometry: basic principles and applications. Crit. Rev. Biotechnol. 2017;37:163–176. doi: 10.3109/07388551.2015.1128876. [DOI] [PubMed] [Google Scholar]
- 121.Robinson JP, Roederer M. HISTORY OF. Sci. Flow. Cytom. strikes gold. Sci. 2015;350:739–740. doi: 10.1126/science.aad6770. [DOI] [PubMed] [Google Scholar]
- 122.Saeys Y, Van Gassen S, Lambrecht BN. Computational flow cytometry: helping to make sense of high-dimensional immunology data. Nat. Rev. Immunol. 2016;16:449–462. doi: 10.1038/nri.2016.56. [DOI] [PubMed] [Google Scholar]
- 123.Perfetto SP, Chattopadhyay PK, Roederer M. Seventeen-colour flow cytometry: unravelling the immune system. Nat. Rev. Immunol. 2004;4:648–655. doi: 10.1038/nri1416. [DOI] [PubMed] [Google Scholar]
- 124.Vazquez J, Ong IM, Stanic AK. Single-cell technologies in reproductive immunology. Am. J. Reprod. Immunol. 2019;82:e13157. doi: 10.1111/aji.13157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Montante S, Brinkman RR. Flow cytometry data analysis: recent tools and algorithms. Int. J. Lab. Hematol. 2019;41:56–62. doi: 10.1111/ijlh.13016. [DOI] [PubMed] [Google Scholar]
- 126.O’Neill K, Aghaeepour N, Spidlen J, Brinkman R. Flow cytometry bioinformatics. PLoS Comput. Biol. 2013;9:e1003365. doi: 10.1371/journal.pcbi.1003365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Spitzer H, Matthew, Nolan PGarry. Mass cytometry: single cells, many features. Cell. 2016;165:780–791. doi: 10.1016/j.cell.2016.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Gadalla R, et al. Validation of CyTOF against flow cytometry for immunological studies and monitoring of human cancer clinical trials. Front. Oncol. 2019;9:415. doi: 10.3389/fonc.2019.00415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hartmann FJ, Bendall SC. Immune monitoring using mass cytometry and related high-dimensional imaging approaches. Nat. Rev. Rheumatol. 2020;16:87–99. doi: 10.1038/s41584-019-0338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chevrier S, et al. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169:736–749.e718. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Friebel E, et al. Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell. 2020;181:1626–1642.e1620. doi: 10.1016/j.cell.2020.04.055. [DOI] [PubMed] [Google Scholar]
- 132.Nolan, J. P. & Condello, D. Spectral flow cytometry. Curr. Protoc. Cytom. 10.1002/0471142956.cy0127s63 (2013). [DOI] [PMC free article] [PubMed]
- 133.Robinson JP. Spectral flow cytometry-Quo vadimus? Cytom. A. 2019;95:823–824. doi: 10.1002/cyto.a.23779. [DOI] [PubMed] [Google Scholar]
- 134.Liang S-B, Fu L-W. Application of single-cell technology in cancer research. Biotechnol. Adv. 2017;35:443–449. doi: 10.1016/j.biotechadv.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 135.Gross A, et al. Technologies for single-cell isolation. Int. J. Mol. Sci. 2015;16:16897–16919. doi: 10.3390/ijms160816897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Artyomov MN, Van den Bossche J. Immunometabolism in the single-cell era. Cell Metab. 2020;32:710–725. doi: 10.1016/j.cmet.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Brennecke P, et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods. 2013;10:1093–1095. doi: 10.1038/nmeth.2645. [DOI] [PubMed] [Google Scholar]
- 138.Jia C, et al. Accounting for technical noise in differential expression analysis of single-cell RNA sequencing data. Nucleic Acids Res. 2017;45:10978–10988. doi: 10.1093/nar/gkx754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Jaitin DA, et al. Massively parallel single-cell RNA-seq for marker-free decomposition of tissues into cell types. Science. 2014;343:776–779. doi: 10.1126/science.1247651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zeisel A, et al. Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science. 2015;347:1138–1142. doi: 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- 141.Tirosh I, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 2016;352:189–196. doi: 10.1126/science.aad0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Kim C, et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018;173:879–893.e183. doi: 10.1016/j.cell.2018.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Grun D, van Oudenaarden A. Design and analysis of single-cell sequencing experiments. Cell. 2015;163:799–810. doi: 10.1016/j.cell.2015.10.039. [DOI] [PubMed] [Google Scholar]
- 144.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat. Rev. Immunol. 2018;18:35–45. doi: 10.1038/nri.2017.76. [DOI] [PubMed] [Google Scholar]
- 145.Ziegenhain C, et al. Comparative analysis of single-cell RNA sequencing methods. Mol. Cell. 2017;65:631–643.e634. doi: 10.1016/j.molcel.2017.01.023. [DOI] [PubMed] [Google Scholar]
- 146.Schurch CM, et al. Coordinated cellular neighborhoods orchestrate antitumoral immunity at the colorectal cancer invasive front. Cell. 2020;182:1341–1359.e1319. doi: 10.1016/j.cell.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Azevedo Tosta TA, de Faria PR, Neves LA, do Nascimento MZ. Computational normalization of H&E-stained histological images: progress, challenges and future potential. Artif. Intell. Med. 2019;95:118–132. doi: 10.1016/j.artmed.2018.10.004. [DOI] [PubMed] [Google Scholar]
- 148.Romano LA, Pedrosa VF. Re-claiming H&E: back to the future. Postgrad. Med. J. 2020;96:58. doi: 10.1136/postgradmedj-2019-136955. [DOI] [PubMed] [Google Scholar]
- 149.Tan WCC, et al. Overview of multiplex immunohistochemistry/immunofluorescence techniques in the era of cancer immunotherapy. Cancer Commun. 2020;40:135–153. doi: 10.1002/cac2.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Magaki S, et al. An introduction to the performance of immunohistochemistry. Methods Mol. Biol. 2019;1897:289–298. doi: 10.1007/978-1-4939-8935-5_25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Goltsev Y, et al. Deep profiling of mouse splenic architecture with CODEX multiplexed imaging. Cell. 2018;174:968–981.e915. doi: 10.1016/j.cell.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Schubert W, et al. Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat. Biotechnol. 2006;24:1270–1278. doi: 10.1038/nbt1250. [DOI] [PubMed] [Google Scholar]
- 153.Friedenberger M, Bode M, Krusche A, Schubert W. Fluorescence detection of protein clusters in individual cells and tissue sections by using toponome imaging system: sample preparation and measuring procedures. Nat. Protoc. 2007;2:2285–2294. doi: 10.1038/nprot.2007.320. [DOI] [PubMed] [Google Scholar]
- 154.Gerdes MJ, et al. Highly multiplexed single-cell analysis of formalin-fixed, paraffin-embedded cancer tissue. Proc. Natl Acad. Sci. USA. 2013;110:11982–11987. doi: 10.1073/pnas.1300136110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Chang Q, et al. Imaging mass cytometry. Cytom. A. 2017;91:160–169. doi: 10.1002/cyto.a.23053. [DOI] [PubMed] [Google Scholar]
- 156.Giesen C, et al. Highly multiplexed imaging of tumor tissues with subcellular resolution by mass cytometry. Nat. Methods. 2014;11:417–422. doi: 10.1038/nmeth.2869. [DOI] [PubMed] [Google Scholar]
- 157.Damond N, et al. A map of human type 1 diabetes progression by imaging mass cytometry. Cell Metab. 2019;29:755–768. doi: 10.1016/j.cmet.2018.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Fisher DAC, Oh ST. Unraveling the architecture of classic hodgkin lymphoma one cell at a time. Cancer Discov. 2020;10:342–344. doi: 10.1158/2159-8290.CD-19-1538. [DOI] [PubMed] [Google Scholar]
- 159.Cornett DS, Reyzer ML, Chaurand P, Caprioli RM. MALDI imaging mass spectrometry: molecular snapshots of biochemical systems. Nat. Methods. 2007;4:828–833. doi: 10.1038/nmeth1094. [DOI] [PubMed] [Google Scholar]
- 160.Schober Y, Guenther S, Spengler B, Rompp A. Single cell matrix-assisted laser desorption/ionization mass spectrometry imaging. Anal. Chem. 2012;84:6293–6297. doi: 10.1021/ac301337h. [DOI] [PubMed] [Google Scholar]
- 161.Neumann EK, Comi TJ, Rubakhin SS, Sweedler JV. Lipid heterogeneity between astrocytes and neurons revealed by single-cell MALDI-MS combined with immunocytochemical classification. Angew. Chem.-Int. Ed. 2019;58:5910–5914. doi: 10.1002/anie.201812892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fernandez B. Elemental and molecular imaging by LA-ICP-MS. Anal. Bioanal. Chem. 2019;411:547–548. doi: 10.1007/s00216-018-1523-7. [DOI] [PubMed] [Google Scholar]
- 163.Angelo M, et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 2014;20:436–442. doi: 10.1038/nm.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Keren L, et al. A structured tumor-immune microenvironment in triple negative breast cancer revealed by multiplexed ion beam imaging. Cell. 2018;174:1373–1387.e1319. doi: 10.1016/j.cell.2018.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Keren L, et al. MIBI-TOF: a multiplexed imaging platform relates cellular phenotypes and tissue structure. Sci. Adv. 2019;5:eaax5851. doi: 10.1126/sciadv.aax5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hartmann FJ, et al. Single-cell metabolic profiling of human cytotoxic T cells. Nat. Biotechnol. 2020;39:186–197. doi: 10.1038/s41587-020-0651-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vickovic S, et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods. 2019;16:987–990. doi: 10.1038/s41592-019-0548-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rodriques SG, et al. Slide-seq: A scalable technology for measuring genome-wide expression at high spatial resolution. Science. 2019;363:1463–1467. doi: 10.1126/science.aaw1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chelvanambi S, et al. Slide-seq for spatially mapping gene expression. metabolic syndrome exacerbates group 2 pulmonary hypertension, and NAD metabolism is influenced by tissue origin. Am. J. Respir. Cell Mol. Biol. 2020;62:112–114. doi: 10.1165/rcmb.2019-0333RO. [DOI] [PubMed] [Google Scholar]
- 170.Stahl PL, et al. Visualization and analysis of gene expression in tissue sections by spatial transcriptomics. Science. 2016;353:78–82. doi: 10.1126/science.aaf2403. [DOI] [PubMed] [Google Scholar]
- 171.Burgess DJ. Spatial transcriptomics coming of age. Nat. Rev. Genet. 2019;20:317. doi: 10.1038/s41576-019-0129-z. [DOI] [PubMed] [Google Scholar]
- 172.Liu Y, et al. High-spatial-resolution multi-omics sequencing via deterministic barcoding in tissue. Cell. 2020;183:1665–1681. doi: 10.1016/j.cell.2020.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hu KH, et al. ZipSeq: barcoding for real-time mapping of single cell transcriptomes. Nat. Methods. 2020;17:833–843. doi: 10.1038/s41592-020-0880-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Krummel M, et al. Visualizing cancer. Cancer Cell. 2020;38:753–756. doi: 10.1016/j.ccell.2020.11.014. [DOI] [PubMed] [Google Scholar]
- 175.Satija R, et al. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 2015;33:495–502. doi: 10.1038/nbt.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Andersson A, et al. Single-cell and spatial transcriptomics enables probabilistic inference of cell type topography. Commun. Biol. 2020;3:565. doi: 10.1038/s42003-020-01247-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Yu KH, Beam AL, Kohane IS. Artificial intelligence in healthcare. Nat. Biomed. Eng. 2018;2:719–731. doi: 10.1038/s41551-018-0305-z. [DOI] [PubMed] [Google Scholar]
- 179.Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N. Engl. J. Med. 2019;380:1347–1358. doi: 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 180.Pounraja VK, et al. A machine-learning approach for accurate detection of copy number variants from exome sequencing. Genome Res. 2019;29:1134–1143. doi: 10.1101/gr.245928.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gerstung M, et al. Reliable detection of subclonal single-nucleotide variants in tumour cell populations. Nat. Commun. 2012;3:811. doi: 10.1038/ncomms1814. [DOI] [PubMed] [Google Scholar]
- 182.Poplin R, et al. A universal SNP and small-indel variant caller using deep neural networks. Nat. Biotechnol. 2018;36:983–987. doi: 10.1038/nbt.4235. [DOI] [PubMed] [Google Scholar]
- 183.Bulik-Sullivan B, et al. Deep learning using tumor HLA peptide mass spectrometry datasets improves neoantigen identification. Nat. Biotechnol. 2018;37:55–63. doi: 10.1038/nbt.4313. [DOI] [PubMed] [Google Scholar]
- 184.Chen B, et al. Predicting HLA class II antigen presentation through integrated deep learning. Nat. Biotechnol. 2019;37:1332–1343. doi: 10.1038/s41587-019-0280-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Gillies RJ, Kinahan PE, Hricak H. Radiomics: Images Are More than Pictures, They Are Data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Park C, et al. Tumor immune profiles noninvasively estimated by FDG PET with deep learning correlate with immunotherapy response in lung adenocarcinoma. Theranostics. 2020;10:10838–10848. doi: 10.7150/thno.50283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Jiang Y, et al. Noninvasive imaging evaluation of tumor immune microenvironment to predict outcomes in gastric cancer. Ann. Oncol. 2020;32:578–578. doi: 10.1016/j.annonc.2021.01.064. [DOI] [PubMed] [Google Scholar]
- 188.Jiang Y, et al. ImmunoScore Signature: A Prognostic and Predictive Tool in Gastric Cancer. Ann. Surg. 2018;267:504–513. doi: 10.1097/SLA.0000000000002116. [DOI] [PubMed] [Google Scholar]
- 189.He, B. et al. Predicting response to immunotherapy in advanced non-small-cell lung cancer using tumor mutational burden radiomic biomarker. J. Immunother. Cancer.10.1136/jitc-2020-000550 (2020). [DOI] [PMC free article] [PubMed]
- 190.Khorrami M, et al. Changes in CT radiomic features associated with lymphocyte distribution predict overall survival and response to immunotherapy in non-small cell lung cancer. Cancer Immunol. Res. 2020;8:108–119. doi: 10.1158/2326-6066.CIR-19-0476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sun R, et al. A radiomics approach to assess tumour-infiltrating CD8 cells and response to anti-PD-1 or anti-PD-L1 immunotherapy: an imaging biomarker, retrospective multicohort study. Lancet Oncol. 2018;19:1180–1191. doi: 10.1016/S1470-2045(18)30413-3. [DOI] [PubMed] [Google Scholar]
- 192.Trebeschi S, et al. Predicting response to cancer immunotherapy using noninvasive radiomic biomarkers. Ann. Oncol. 2019;30:998–1004. doi: 10.1093/annonc/mdz108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Nishino M, Hatabu H, Hodi FS. Imaging of cancer immunotherapy: current approaches and future directions. Radiology. 2019;290:9–22. doi: 10.1148/radiol.2018181349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Garcia-Figueiras, R. et al. Assessing immunotherapy with functional and molecular imaging and radiomics. Radiographics 200070, (2020). [DOI] [PubMed]
- 195.Vaidya, P. et al. Novel, non-invasive imaging approach to identify patients with advanced non-small cell lung cancer at risk of hyperprogressive disease with immune checkpoint blockade. J Immunother. Cancer.10.1136/jitc-2020-001343 (2020). [DOI] [PMC free article] [PubMed]
- 196.Basler L, et al. Radiomics, tumor volume, and blood biomarkers for early prediction of pseudoprogression in patients with metastatic melanoma treated with immune checkpoint inhibition. Clin. Cancer Res. 2020;26:4414–4425. doi: 10.1158/1078-0432.CCR-20-0020. [DOI] [PubMed] [Google Scholar]
- 197.Saltz J, et al. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23:181–193.e187. doi: 10.1016/j.celrep.2018.03.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lu Z, et al. Deep-learning-based characterization of tumor-infiltrating lymphocytes in breast cancers from histopathology images and multiomics data. JCO Clin. Cancer Inform. 2020;4:480–490. doi: 10.1200/CCI.19.00126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Krijgsman D, et al. Quantitative whole slide assessment of tumor-infiltrating CD8-positive lymphocytes in ER-positive breast cancer in relation to clinical outcome. IEEE J. Biomed. Health Inform. 2020;25:381–392. doi: 10.1109/JBHI.2020.3003475. [DOI] [PubMed] [Google Scholar]
- 200.Klauschen F, et al. Scoring of tumor-infiltrating lymphocytes: From visual estimation to machine learning. Semin Cancer Biol. 2018;52:151–157. doi: 10.1016/j.semcancer.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 201.AbdulJabbar K, et al. Geospatial immune variability illuminates differential evolution of lung adenocarcinoma. Nat. Med. 2020;26:1054–1062. doi: 10.1038/s41591-020-0900-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Kather JN, et al. Predicting survival from colorectal cancer histology slides using deep learning: a retrospective multicenter study. PLoS Med. 2019;16:e1002730. doi: 10.1371/journal.pmed.1002730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Shi JY, et al. Exploring prognostic indicators in the pathological images of hepatocellular carcinoma based on deep learning. Gut. 2020;70:951–961. doi: 10.1136/gutjnl-2020-320930. [DOI] [PubMed] [Google Scholar]
- 204.Vayrynen JP, et al. Prognostic significance of immune cell populations identified by machine learning in colorectal cancer using routine hematoxylin and eosin-stained sections. Clin. Cancer Res. 2020;26:4326–4338. doi: 10.1158/1078-0432.CCR-20-0071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Zormpas-Petridis K, et al. Superpixel-based conditional random fields (SuperCRF): incorporating global and local context for enhanced deep learning in melanoma histopathology. Front. Oncol. 2019;9:1045. doi: 10.3389/fonc.2019.01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Andtbacka RH, et al. Talimogene Laherparepvec improves durable response rate in patients with advanced melanoma. J. Clin. Oncol. 2015;33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 207.Waldmann, T. A. Cytokines in cancer immunotherapy. Cold Spring Harb. Perspect. Biol. 10.1101/cshperspect.a028472 (2018). [DOI] [PMC free article] [PubMed]
- 208.Abbott M, Ustoyev Y. Cancer and the immune system: the history and background of immunotherapy. Semin Oncol. Nurs. 2019;35:150923. doi: 10.1016/j.soncn.2019.08.002. [DOI] [PubMed] [Google Scholar]
- 209.Rudnicka D, et al. Rituximab causes a polarization of B cells that augments its therapeutic function in NK-cell-mediated antibody-dependent cellular cytotoxicity. Blood. 2013;121:4694–4702. doi: 10.1182/blood-2013-02-482570. [DOI] [PubMed] [Google Scholar]
- 210.Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell. 2015;27:450–461. doi: 10.1016/j.ccell.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Postow MA, Callahan MK, Wolchok JD. Immune checkpoint blockade in cancer therapy. J. Clin. Oncol. 2015;33:1974–1982. doi: 10.1200/JCO.2014.59.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov. 2018;8:1069–1086. doi: 10.1158/2159-8290.CD-18-0367. [DOI] [PubMed] [Google Scholar]
- 213.Rosenberg SA, et al. Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat. Rev. Cancer. 2008;8:299–308. doi: 10.1038/nrc2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Klebanoff CA, Rosenberg SA, Restifo NP. Prospects for gene-engineered T cell immunotherapy for solid cancers. Nat. Med. 2016;22:26–36. doi: 10.1038/nm.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Zhang Y, Zhang Z. The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol. Immunol. 2020;17:807–821. doi: 10.1038/s41423-020-0488-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.De Mattos-Arruda L, et al. Neoantigen prediction and computational perspectives towards clinical benefit: recommendations from the ESMO Precision Medicine Working Group. Ann. Oncol. 2020;31:978–990. doi: 10.1016/j.annonc.2020.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Topalian SL, et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012;366:2443–2454. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sunshine J, Taube JM. PD-1/PD-L1 inhibitors. Curr. Opin. Pharmacol. 2015;23:32–38. doi: 10.1016/j.coph.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Carbone DP, et al. First-line nivolumab in stage IV or recurrent non-small-cell lung cancer. N. Engl. J. Med. 2017;376:2415–2426. doi: 10.1056/NEJMoa1613493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Rizvi NA, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Samstein RM, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019;51:202–206. doi: 10.1038/s41588-018-0312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Sharma P, et al. Nivolumab plus ipilimumab for metastatic castration-resistant prostate cancer: preliminary analysis of patients in the CheckMate 650 Trial. Cancer Cell. 2020;38:489–499.e483. doi: 10.1016/j.ccell.2020.08.007. [DOI] [PubMed] [Google Scholar]
- 223.Tsujikawa T, et al. Prognostic significance of spatial immune profiles in human solid cancers. Cancer Sci. 2020;111:3426–3434. doi: 10.1111/cas.14591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Pages F, et al. International validation of the consensus Immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet. 2018;391:2128–2139. doi: 10.1016/S0140-6736(18)30789-X. [DOI] [PubMed] [Google Scholar]
- 225.Kumagai S, et al. The PD-1 expression balance between effector and regulatory T cells predicts the clinical efficacy of PD-1 blockade therapies. Nat. Immunol. 2020;21:1346–1358. doi: 10.1038/s41590-020-0769-3. [DOI] [PubMed] [Google Scholar]
- 226.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541:321–330. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 227.Simoni Y, et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature. 2018;557:575–579. doi: 10.1038/s41586-018-0130-2. [DOI] [PubMed] [Google Scholar]
- 228.Sade-Feldman M, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175:998–1013. doi: 10.1016/j.cell.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Siddiqui I, et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T cells with stem-like properties promote tumor control in response to vaccination and checkpoint blockade immunotherapy. Immunity. 2019;50:195–211.e110. doi: 10.1016/j.immuni.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 230.Yost KE, et al. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 2019;25:1251–1259. doi: 10.1038/s41591-019-0522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Guo X, et al. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 2018;24:978–985. doi: 10.1038/s41591-018-0045-3. [DOI] [PubMed] [Google Scholar]
- 232.Oh DY, et al. Intratumoral CD4(+) T cells mediate anti-tumor cytotoxicity in human bladder cancer. Cell. 2020;181:1612–1625.e1613. doi: 10.1016/j.cell.2020.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Zheng C, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356.e1316. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 234.Krieg C, et al. High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat. Med. 2018;24:144–153. doi: 10.1038/nm.4466. [DOI] [PubMed] [Google Scholar]
- 235.Helmink BA, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature. 2020;577:549–555. doi: 10.1038/s41586-019-1922-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zhang L, et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020;181:442–459.e429. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 237.Chen YP, et al. Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res. 2020;30:1024–1042. doi: 10.1038/s41422-020-0374-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Wang S, et al. Transdifferentiation of tumor infiltrating innate lymphoid cells during progression of colorectal cancer. Cell Res. 2020;30:610–622. doi: 10.1038/s41422-020-0312-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Jackson HW, et al. The single-cell pathology landscape of breast cancer. Nature. 2020;578:615–620. doi: 10.1038/s41586-019-1876-x. [DOI] [PubMed] [Google Scholar]
- 240.Rosenberg SA, Restifo NP. Adoptive cell transfer as personalized immunotherapy for human cancer. Science. 2015;348:62–68. doi: 10.1126/science.aaa4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ping Y, Liu C, Zhang Y. T-cell receptor-engineered T cells for cancer treatment: current status and future directions. Protein Cell. 2018;9:254–266. doi: 10.1007/s13238-016-0367-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.June CH, et al. CAR T cell immunotherapy for human cancer. Science. 2018;359:1361–1365. doi: 10.1126/science.aar6711. [DOI] [PubMed] [Google Scholar]
- 243.Park JH, et al. Long-term follow-up of CD19 CAR therapy in acute lymphoblastic leukemia. N. Engl. J. Med. 2018;378:449–459. doi: 10.1056/NEJMoa1709919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Neelapu SS, et al. Axicabtagene Ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N. Engl. J. Med. 2017;377:2531–2544. doi: 10.1056/NEJMoa1707447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Rafiq S, Hackett CS, Brentjens RJ. Engineering strategies to overcome the current roadblocks in CAR T cell therapy. Nat. Rev. Clin. Oncol. 2020;17:147–167. doi: 10.1038/s41571-019-0297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Robbins PF, et al. Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J. Clin. Oncol. 2011;29:917–924. doi: 10.1200/JCO.2010.32.2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Robbins PF, et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin. Cancer Res. 2015;21:1019–1027. doi: 10.1158/1078-0432.CCR-14-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Parkhurst MR, et al. T cells targeting carcinoembryonic antigen can mediate regression of metastatic colorectal cancer but induce severe transient colitis. Mol. Ther. 2011;19:620–626. doi: 10.1038/mt.2010.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Zacharakis N, et al. Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat. Med. 2018;24:724–730. doi: 10.1038/s41591-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Tran E, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med. 2016;375:2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tran E, et al. Cancer immunotherapy based on mutation-specific CD4+ T cells in a patient with epithelial cancer. Science. 2014;344:641–645. doi: 10.1126/science.1251102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Gros A, et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med. 2016;22:433–438. doi: 10.1038/nm.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Li G, et al. T cell antigen discovery via trogocytosis. Nat. Methods. 2019;16:183–190. doi: 10.1038/s41592-018-0305-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Joglekar AV, et al. T cell antigen discovery via signaling and antigen-presenting bifunctional receptors. Nat. Methods. 2019;16:191–198. doi: 10.1038/s41592-018-0304-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Coley WB. The treatment of inoperable sarcoma by bacterial toxins (the mixed toxins of the Streptococcus erysipelas and the Bacillus prodigiosus) Proc. R. Soc. Med. 1910;3:1–48. doi: 10.1177/003591571000301601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Coley, W. B. The treatment of malignant tumors by repeated inoculations of erysipelas. With a report of ten original cases. 1893. Clin. Orthop. Relat. Res. 3–11 (1991). [PubMed]
- 257.van der Bruggen P, et al. A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science. 1991;254:1643–1647. doi: 10.1126/science.1840703. [DOI] [PubMed] [Google Scholar]
- 258.Chen YT, et al. A testicular antigen aberrantly expressed in human cancers detected by autologous antibody screening. Proc. Natl Acad. Sci. USA. 1997;94:1914–1918. doi: 10.1073/pnas.94.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Dudley ME, et al. Cancer regression and autoimmunity in patients after clonal repopulation with antitumor lymphocytes. Science. 2002;298:850–854. doi: 10.1126/science.1076514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Theobald M, et al. Tolerance to p53 byA2.1-restricted cytotoxic T lymphocytes. J. Exp. Med. 1997;185:833–841. doi: 10.1084/jem.185.5.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Thaxton JE, Li Z. To affinity and beyond: harnessing the T cell receptor for cancer immunotherapy. Hum. Vaccin Immunother. 2014;10:3313–3321. doi: 10.4161/21645515.2014.973314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ott PA, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547:217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Sahin U, et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 264.Carreno BM, et al. Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science. 2015;348:803–808. doi: 10.1126/science.aaa3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Hu Z, Ott PA, Wu CJ. Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat. Rev. Immunol. 2018;18:168–182. doi: 10.1038/nri.2017.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Hilf N, et al. Actively personalized vaccination trial for newly diagnosed glioblastoma. Nature. 2019;565:240–245. doi: 10.1038/s41586-018-0810-y. [DOI] [PubMed] [Google Scholar]
- 267.Keskin DB, et al. Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature. 2019;565:234–239. doi: 10.1038/s41586-018-0792-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sahin U, Tureci O. Personalized vaccines for cancer immunotherapy. Science. 2018;359:1355–1360. doi: 10.1126/science.aar7112. [DOI] [PubMed] [Google Scholar]
- 269.Fang Y, et al. A pan-cancer clinical study of personalized neoantigen vaccine monotherapy in treating patients with various types of advanced solid tumors. Clin. Cancer Res. 2020;26:4511–4520. doi: 10.1158/1078-0432.CCR-19-2881. [DOI] [PubMed] [Google Scholar]
- 270.Wells DK, et al. Key parameters of tumor epitope immunogenicity revealed through a consortium approach improve neoantigen prediction. Cell. 2020;183:818–834.e813. doi: 10.1016/j.cell.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Marx V. Method of the Year: spatially resolved transcriptomics. Nat. Methods. 2021;18:9–14. doi: 10.1038/s41592-020-01033-y. [DOI] [PubMed] [Google Scholar]
- 272.Grasso CS, et al. Conserved interferon-gamma signaling drives clinical response to immune checkpoint blockade therapy in melanoma. Cancer Cell. 2020;38:500–515.e503. doi: 10.1016/j.ccell.2020.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Gong T, Szustakowski JD. DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics. 2013;29:1083–1085. doi: 10.1093/bioinformatics/btt090. [DOI] [PubMed] [Google Scholar]
- 274.Racle, J. et al. Simultaneous enumeration of cancer and immune cell types from bulk tumor gene expression data. Elife.10.7554/eLife.26476 (2017). [DOI] [PMC free article] [PubMed]
- 275.Tang D, Park S, Zhao H. NITUMID: Nonnegative matrix factorization-based Immune-TUmor MIcroenvironment Deconvolution. Bioinformatics. 2020;36:1344–1350. doi: 10.1093/bioinformatics/btz748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Qiao W, et al. PERT: a method for expression deconvolution of human blood samples from varied microenvironmental and developmental conditions. PLoS Comput. Biol. 2012;8:e1002838. doi: 10.1371/journal.pcbi.1002838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Finotello F, et al. Molecular and pharmacological modulators of the tumor immune contexture revealed by deconvolution of RNA-seq data. Genome Med. 2019;11:34. doi: 10.1186/s13073-019-0638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Li B, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17:174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Rimmer A, et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat. Genet. 2014;46:912–918. doi: 10.1038/ng.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zeng F, Jiang R, Chen T. PyroHMMsnp: an SNP caller for Ion Torrent and 454 sequencing data. Nucleic Acids Res. 2013;41:e136. doi: 10.1093/nar/gkt372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Dong X, et al. Accurate identification of single-nucleotide variants in whole-genome-amplified single cells. Nat. Methods. 2017;14:491–493. doi: 10.1038/nmeth.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Fang LT, et al. An ensemble approach to accurately detect somatic mutations using SomaticSeq. Genome Biol. 2015;16:197. doi: 10.1186/s13059-015-0758-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Buchkovich ML, et al. HLAProfiler utilizes k-mer profiles to improve HLA calling accuracy for rare and common alleles in RNA-seq data. Genome Med. 2017;9:86. doi: 10.1186/s13073-017-0473-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Ka S, et al. HLAscan: genotyping of the HLA region using next-generation sequencing data. BMC Bioinforma. 2017;18:258. doi: 10.1186/s12859-017-1671-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Wittig M, et al. Development of a high-resolution NGS-based HLA-typing and analysis pipeline. Nucleic Acids Res. 2015;43:e70. doi: 10.1093/nar/gkv184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Hu Y, et al. ACME: pan-specific peptide-MHC class I binding prediction through attention-based deep neural networks. Bioinformatics. 2019;35:4946–4954. doi: 10.1093/bioinformatics/btz427. [DOI] [PubMed] [Google Scholar]
- 287.Venkatesh G, Grover A, Srinivasaraghavan G, Rao S. MHCAttnNet: predicting MHC-peptide bindings for MHC alleles classes I and II using an attention-based deep neural model. Bioinformatics. 2020;36:i399–i406. doi: 10.1093/bioinformatics/btaa479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Phloyphisut P, Pornputtapong N, Sriswasdi S, Chuangsuwanich E. MHCSeqNet: a deep neural network model for universal MHC binding prediction. BMC Bioinforma. 2019;20:270. doi: 10.1186/s12859-019-2892-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Jensen KK, et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology. 2018;154:394–406. doi: 10.1111/imm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zeng H, Gifford DK. Quantification of uncertainty in peptide-MHC binding prediction improves high-affinity peptide selection for therapeutic design. Cell Syst. 2019;9:159–166.e153. doi: 10.1016/j.cels.2019.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Hu Z, et al. Personal neoantigen vaccines induce persistent memory T cell responses and epitope spreading in patients with melanoma. Nat. Med. 2021;27:515–525. doi: 10.1038/s41591-020-01206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Ott PA, et al. A phase Ib trial of personalized neoantigen therapy plus anti-PD-1 in patients with advanced melanoma, non-small cell lung cancer, or bladder cancer. Cell. 2020;183:347–362.e324. doi: 10.1016/j.cell.2020.08.053. [DOI] [PubMed] [Google Scholar]
- 293.Davis AA, Patel VG. The role of PD-L1 expression as a predictive biomarker: an analysis of all US Food and Drug Administration (FDA) approvals of immune checkpoint inhibitors. J. Immunother. Cancer. 2019;7:278. doi: 10.1186/s40425-019-0768-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Savic Prince S, Bubendorf L. Predictive potential and need for standardization of PD-L1 immunohistochemistry. Virchows Arch. 2019;474:475–484. doi: 10.1007/s00428-018-2445-7. [DOI] [PubMed] [Google Scholar]
- 295.Yu J, et al. Liver metastasis restrains immunotherapy efficacy via macrophage-mediated T cell elimination. Nat. Med. 2021;27:152–164. doi: 10.1038/s41591-020-1131-x. [DOI] [PMC free article] [PubMed] [Google Scholar]