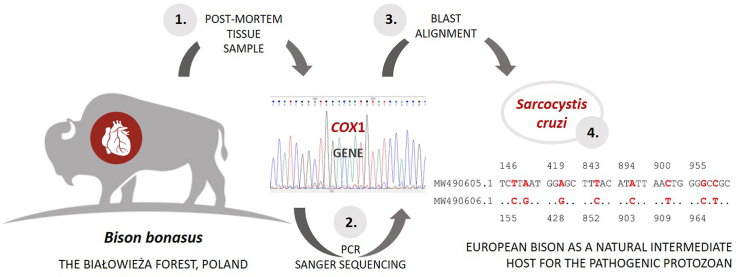
Abstract
European bison are susceptible to a range of pathogens which may influence their health, and hence, to ensure their protection, it is essential to provide effective monitoring of potential exposure. This study presents the first molecular confirmation of Sarcocystis cruzi infection in European bison based on PCR amplification of the cytochrome c oxidase subunit I (cox1) gene. A sample of heart tissue taken from one fifteen-year-old European bison cow was examined by light microscopy for the presence of heart sarcocysts. The genomic DNA isolated from any identified sarcocysts was subjected to PCR to amplify cox1 gene sequences, and the obtained amplicons were sequenced by Sanger dideoxy sequencing. Two partial cox1 sequences were obtained; they were identified as S. cruzi and deposited in the GenBank™ database under the accession numbers MW490605 and MW490606. BLAST analysis found them to demonstrate the closest similarity to S. levinei (MH255771-MH255779 and KU247874-KU247884), sharing an identity of 93.14–93.8 %. This is the first report to identify sarcocysts isolated from heart tissue of infected European bison living in the Białowieża forest to species level using cox1 analysis. Our findings confirm that the European bison is a natural intermediate host for S. cruzi. As such, coordinators of future conservation programmes should consider the impact of these diseases on reintroduced animals.
Keywords: Sarcocystis cruzi, European bison, cox1 gene
Graphical abstract
Highlights
-
•
Molecular identification targeting cox1 gene confirmed S. cruzi infection in European bison.
-
•
The pathogenic species of Sarcocystis in endangered bison from Białowieża forest.
-
•
Bison bonasus as a natural intermediate host for Sarcocystis cruzi.
1. Introduction
The European bison (Bison bonasus bonasus L.), the largest herbivorous animal in Europe, is protected by the international and national laws. Historically, the species was distributed across the breadth of Europe, from west to east. After being made extinct in the wild at the beginning of the 20th Century, the European bison has been successfully recovered in two distinct genetic lines derived from only twelve and seven captive founders (Druet et al., 2020). In 1966, the European bison was classified as an endangered species in the International Union for the Conservation of Nature and Natural Resources (IUCN) Red List of Threatened Species.
The largest existing population, comprising 800 individuals, divided between one group maintained in captivity and another group living in the wild (Olech and Perzanowski, 2016), inhabits the Białowieża Forest, the last ancient primeval woodland in Europe (Raczyński, 2019). This population plays a particularly important role in the protection and restitution program: many of the Białowieża bison are transported to other locations in Poland and other European countries, including Spain, Denmark, Germany, Slovakia and Sweden, to create new free-living and captive herds.
Due to their low genetic heterogeneity resulting from inbreeding depression, and the consequent depletion of variation in the genes responsible for immunity against parasites, modern-day European bison demonstrate decreased resistance to infections and higher vulnerability to pathogens (Karbowiak et al., 2014); such susceptibility may negatively impact their individual health and prevent genetically valuable individuals from breeding. Furthermore, some parasite infections can lead to abortion (Bień et al., 2010; Moskwa et al., 2017).
Sarcocystis species are cyst-forming sporozoan parasites with an obligatory two-host life cycle: sexual development and oocyst formation occurs in the intestinal mucosa of their definitive host, followed by asexual multiplication in the vascular endothelial cells and striated muscle cells (sarcocyst stage) of the intermediate host (Dubey et al., 1980a).
The occurrence of Sarcocystis spp. in European bison (Bison bonasus) was first recorded in the heart muscles of animals from the Białowieża Forest in histopathological studies by Szwejkowski (1954), however, the parasite was not classified to the species level.
Several studies have since demonstrated the competence of American bison as hosts for Sarcocystis cruzi (Dubey, 1980; Fayer et al., 1982). In addition to S. cruzi, infections by S. hominis and S. hirsuta have been detected in European bison, both captive and free ranging animals (Odening et al., 1994, 1995; Pyziel and Demiaszkiewicz, 2009). It is important to note that these three species mainly use cattle as intermediate hosts.
The purpose of the present study was to obtain a molecular identification of the sarcocysts isolated from the heart muscles of a European bison living in the Białowieża Forest.
2. Materials and methods
2.1. Animal and source of samples
The Białowieża Forest, the last ancient primeval woodland in Europe, is a contiguous forest complex covering 1500 km2. The western part of the complex, covering 650 km2, is located in Poland, and the eastern part, covering 875 km2, lies within Belarus. The forest extends for 55 km from east to west and 51 km from north to south. Its area lies within the coordinates 23°31’ −24°21′ E and 52°29’ −52°37’ N (Sokołowski, 2004).
The study was performed on a European bison shot in the autumn of 2019 in the Białowieża Forest, Poland. A sample of heart tissue (24 g of weight) obtained from a 15-year-old European bison cow, was divided into smaller parts, gently broken up in physiological saline solution using a blender, and the resulting suspension was filtered through gauze. The resulting sediments were collected and eight sarcocysts were found. The basic morphology (shape, size) of any cysts present were examined in wet mounts and collected under an inverted Olympus IX50 microscope fitted with a camera. The sarcocyst was individually preserved in sterile H2O in Eppendorf tubes for molecular investigation and stored at − 20 °C (Cabaj et al., 2020).
2.2. DNA isolation and amplification of cox1 gene
Genomic DNA was extracted from the sarcocysts with a Nucleospin Tissue DNA extraction kit (Macherey-Nagel, Germany). The isolation procedure was performed according to the manufacturer's protocol. The PCR amplification of the cox1 gene was carried out with primers SF1 5′- ATG GCG TAC AAC AAT CAT AAA GAA -3′ and SR8D 5′- CAT TGC CCA TDA CTA CGC C-3′; the amplicon product was expected to be 1072 bp in length (Gjerde, 2013).
PCR was performed in a final 25 μl volume consisting of 0.5 μM of each primer, 12.5 μl DreamTaq PCR Master Mix (2x) (Thermo Scientific™), 3 μl DNA and nuclease-free water.
The cycling conditions began with one cycle at 95 °C for 5 min, followed by 35 cycles at 94 °C for 45 s, 52.1 °C for 60 s, 72 °C for 80 s, and ending with one cycle at 72 °C for 10 min.
The PCR products were separated by electrophoresis in 1.5 % agarose gels and stained with GelRed (Nucleic Acid Gel Stain, Biotium). The selected PCR products were purified using.
Clean-up Product Purification Kits (A&A Biotechnology, Poland) according to the manufacturer's instructions. DNA concentration was estimated using a NanoDrop ND-1000 Spectrophotometer (NanoDrop Technologies, USA).
2.3. Sequence analysis
The purified PCR fragments were subjected to Sanger dideoxy sequencing in Genomed S.A. Warsaw, Poland. The obtained data were processed in Unipro UGENE and aligned to the sequences of closely-related organisms deposited in GenBank™ using the BLAST tool. Nucleotide sequence data reported in this paper have been deposited in the GenBank™ database under the accession numbers: MW490605.1, MW490606.1.
2.4. Phylogenetic analysis
Our partial cox1 gene sequences were subjected to phylogenetic analysis using MEGA X software (Kumar et al., 2018). The analysis was performed against 7 existing partial sequences previously obtained from different isolates of Sarcocystis cruzi from cattle, and 29 sequences from Sarcocystis species identified in Bovidae and Cervidae. All sequences were truncated slightly at both ends to preserve the homologous nucleotide positions for further analysis.
The final dataset for the phylogenetic trees comprised 979 positions. A codon-based ClustalW multiple alignment of all sequences was performed in MEGA X. Maximum Parsimony trees were obtained using the Subtree-Pruning-Regrafting (SPR) algorithm. The phylogeny was tested with the bootstrap method using 1000 bootstrap replications. All codon positions were used.
3. Results
3.1. Partial cox1 gene analysis
The cox1 gene sequences were amplified by PCR using the genomic DNA isolated from the heart sarcocysts as templates. All the obtained amplicons were submitted to sequencing. Six out of eight isolates of cysts resulted in poor quality reads, two sequences were identified in BLAST as Sarcocystis cruzi and submitted to GenBank database under accession numbers MW490605 and MW490606. The sequences differed from each other at eight out of 978 nucleotide positions (98.97 % identity) as shown on Fig. 1. Both were identified as belonging to S. cruzi on the basis of their high sequence identity with most previous cox1 gene sequence of this species: MW490605 with 99.9 to 99.0 % identity, and MW490606 with 99.9 to 98.3 % identity. BLAST analysis found the MW490605 sequence shares an identity of 93.38–93.8 % with the partial cox1 gene from Sarcocystis levinei (MH255771-MH255779 and KU247874-KU247884), followed by Sarcocystis pilosa (MT070670-MT070677) with 90.9–91.31 % identity. Similarly, the MW490606 sequence shared 93.14–93.55 % identity with S. levinei, and 90.59–90.8 % with S. pilosa.
Fig. 1.
Overview of consistent nucleotide differences between two cox1 gene sequences of S. cruzi MW490605 and MW490606. Numbers above and below the sequences refer to nucleotide positions in the two GenBank sequences used in the comparison. Positions where the two sequences were identical are signified with Dots (.).
3.2. Phylogenetic analysis
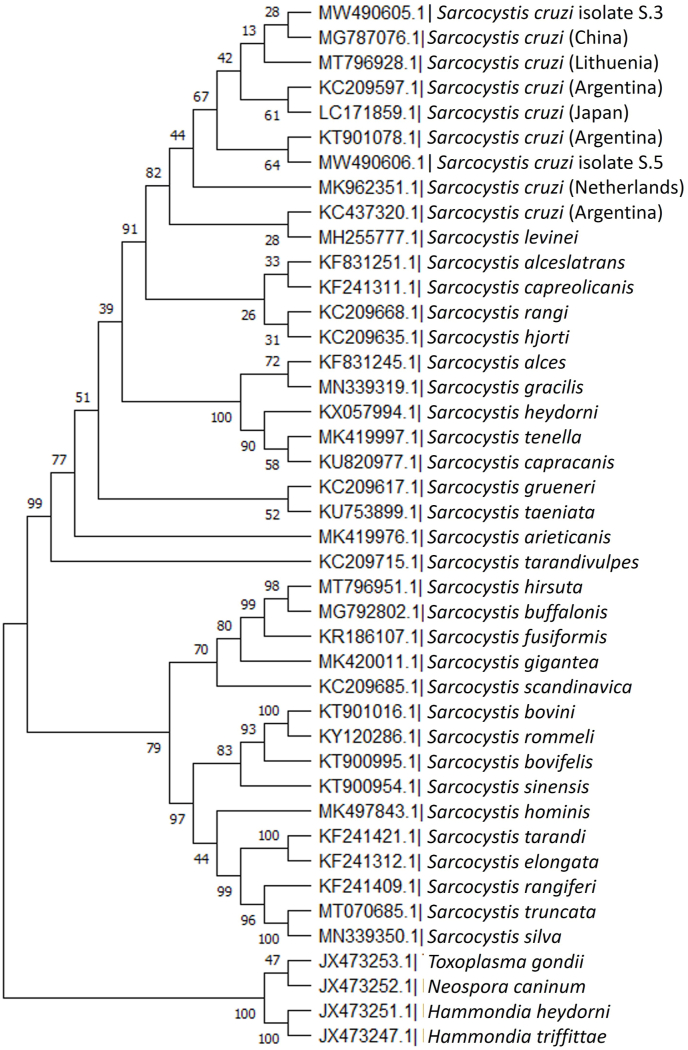
The partial cox1 gene sequences of Sarcocystis cruzi sarcocysts collected from the heart muscle of Bison bonasus were phylogenetically compared with various S. cruzi cox1 gene sequences from cattle (Bos taurus) previously deposited in GenBank, and sequences from Sarcocystis species identified in Bovidae and Cervidae; these sequences were selected for analysis on the basis of the initial BLAST alignment. Any records with product lengths shorter than 989 bp were rejected. Detailed information about the accession numbers, hosts and countries of origin of the selected S. cruzi sequences are presented in Table S1.
The phylogenetic analysis (Fig. 2), placed S. cruzi isolates S.3 and S.5 in separate clades with relationships that were not well resolved. MW490605 was a sister to a sequence from a China Bos taurus isolate (MG787076), and this clade formed a sister clade to Lithuanian isolate. The relationships of the three isolates with respect to each other were not clearly resolved. MW490606 was closely related with the isolate from Bos taurus host from the Argentina (KT901078); the pair formed a sister clade to the sequence from Argentina. All sequences of S. cruzi were placed, with near-maximum support, as a monophyletic sister group to a S. levinei sequence from a water buffalo (MH255777). Both taxa were sisters to sequences from Cervidae: S. alceslatrans from moose, S. rangi from reindeer, S. hjorti from red deer and S. capreolicanis from roe deer.
Fig. 2.
Phylogenetic tree for selected Sarcocystis cruzi isolates and selected members of Sarcocystis species found in Bovidae based on partial sequences of cox1 using the maximum parsimony method. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches. GenBank accession numbers are listed next to the taxon names. The country of origin of S. cruzi isolates are listed in brackets.
4. Discussion
The story of the European bison is an excellent example of the successful restitution of a species which had previously become extinct in the wild through many years of efforts by conservationists (Olech and Perzanowski, 2016). Today, its population continues to grow, and in 2019, the total number of individuals globally exceeded 8000. In Białowieża Forest, the current bison population consists of 800 individuals (Raczyński, 2019). However, as the spatial distribution of these populations is unfavourable in most countries, their management is becoming a growing challenge for conservationists.
As their population densities increase, so do the health risks for the European bison, and diseases have a significant influence on their condition and demography (Scott, 1988). This is particularly true for the reintroduced bison, as they are extremely susceptible to infection by apicomplexan parasites (Cabaj et al., 2005; Bień et al., 2010; Moskwa et al., 2017).
The presence of sarcocysts has been confirmed in American bison (Bison bison L) living wild in North America (Dubey, 1980a, 1980b; Fayer et al., 1982), and in those kept in Central European zoos (Ippen et al., 1974; Henne et al., 1977).
Until now, Sarcocystis spp. in the European bison from Białowieża Forest have been identified exclusively on the basis of light microscope/ultrastructural observations and histopathological changes during Sarcocystis infection; however, the invasive species have never been identified to species (Piusiński et al., 1996).
In the present study, PCR amplification of the mitochondrial cytochrome c oxidase subunit-I gene (cox1) was used to identify sarcocysts isolated from European bison, with the results confirming the presence of Sarcocystis cruzi: the most pathogenic species of Sarcocystis in cattle, which can cause retarded growth and fatalities in cows (Dubey et al., 2015). Our findings represent the first cox1 gene based molecular report of infection by S. cruzi in European bison (Bison bonasus bonasus L.) and of the host competence of this bovid species. Due to the close systematic relationship between European bison and cattle (Bos taurus), with both being members of the Family Bovidae, it is reasonable to assume that the two species will also demonstrate similar clinical symptoms of sarcocystosis.
Our findings indicate that our identified sequences, MW490605 and MW490606, obtained from the genomic DNA of sarcocysts isolated from European bison, are similar to sequences of the cox1 genes of S. cruzi sarcocysts isolated from cattle.
In a previous study, the amplification of bradyzoite 18S rRNA fragments, and their subsequent phylogenetic analyses, found a sequence isolated from wood bison in Alaska (KP640133) to have great similarity with two others from S. cruzi isolated previously from water buffalo and cattle in China: AF176932 and AF176934, respectively (Yang et al., 2001); in addition, the KP640133 sequence demonstrated 98 % homology with another sequence deposited in GenBank for Sarcocystis cruzi, 18S ribosomal RNA gene, fragment 1, partial sequence isolated from European bison (AF006480) (Calero-Barnal et al., 2015; Fischer and Odening, 1998). Nevertheless, Sarcocystis species cannot be unambiguously distinguished based on 18S rRNA gene alone, as evidenced by Gjerde et al. (2016), who demonstrate the superiority of the cox1 gene over ribosomal rRNA genes as tools for identification.
Our results are in line with previous findings by Pyziel and Demiaszkiewicz (2009), where sarcocysts from the heart, oesophagus and diaphragm of European bison from the Białowieża Forest were morphologically identified as S. cruzi. The authors note the presence of a sarcocyst wall, which was thin (1 mm up to 1.2 mm) and smooth in all cases, with villar protrusions, on the surface of the cysts. In addition, they report that the size of the sarcocysts varied depending on the muscle sample from which they were isolated: the longest and the most slender sarcocysts were found in the diaphragm, with those from the oesophagus being slightly smaller and those from heart muscle tissue being the smallest (Pyziel and Demiaszkiewicz, 2009).
Odening et al. (1994) also report the presence of sarcocysts in a wisent (Bison bonasus) which was born and kept in Germany. Light microscope and TEM examination revealed the presence of sarcocysts consistent with those of S. cruzi, S. hirsuta and S. hominis. All three species are typical for cattle (Odening et al., 1994).
Despite sharing 98.97 % of their nucleotides, the S. cruzi partial cox1 gene sequences from the Bison bonasus identified in our study were placed in separate clades by the phylogenetic analysis. Both clades included S. cruzi isolates obtained from Bos taurus. The identified MW490605 sequence was closely related to isolates from Lithuania and Argentina, whereas MW490606 was similar to those from the Netherlands, Japan and Argentina. S. cruzi has been the most frequently-detected species in cattle in Belgium and the Netherlands in recent years (Hoeve-Bakker et al., 2019; Zeng et al., 2021), with it being identified in 68 of 104 samples in the Netherlands, and 113 out of 200 in Belgium based on 18s rDNA and cox1 fragment analysis. The widespread distribution of S. cruzi in cattle in Europe may account for the invasion of European bison.
The S. cruzi sequences identified herein demonstrate a close relationship with S. levinei (MH255777) from water buffalo. Similar findings were obtained by Gjerde et al. (2016) in a comparison of different S. cruzi and S. levinei sequences from cattle and water buffalo isolates, identified through the cox1 marker; the identified sequences (Gjerde et al., 2016) were partly or fully similar to each other.
The relatedness presented on our phylogenetic tree based on the limited number of isolates examined from Bison bonansus. Hence, there is a great need to continue research on higher amount of isolates/clones of Sarcocystis spp. from European bison, what could disclose more variability in the cox1 gene, and thus allow a more precise idea of the relationships between them and other Sarcocystis species isolated from Bovidae and Cervidae. Previous studies suggest that the most common species of parasite in European bison living in Białowieża Forest is S. cruzi, and considering the large range of definitive hosts (e.g. foxes, wolves) occupying the same niche as bison, this may well be the case. Furthermore, the occurrence of Sarcocystis species in wild ruminants can increase the prevalence of infection in the carnivorous definitive host and thus support its transfer to other wild and domestic herbivores which shares the same habitat and presents particular Sarcocystis spp infection specificity.
5. Conclusion
Sarcocystis cruzi was identified in the heart tissue of infected European bison living in Białowieża Forest. This is the first report to confirm a species level identification of sarcocysts in European bison using cox1 analysis. The importance of parasitic diseases and their impact on reintroduced animals should be taken into account in conservation and reintroduction programmes.
CRediT authorship contribution statement
Władysław Cabaj: Conceptualization, Methodology, Resources, Writing – original draft, Writing – review & editing, Supervision. Sylwia Grzelak: Validation, Investigation, Data curation, Writing – review & editing. Bożena Moskwa: Writing – original draft, Writing – review & editing. Justyna Bień-Kalinowska: Methodology, Validation, Investigation, Writing – original draft, Writing – review & editing.
Funding
This research was funded by The Witold Stefański Institute of Parasitology Polish Academy of Sciences financial resources.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijppaw.2021.07.010.
Contributor Information
Władysław Cabaj, Email: cabajw@twarda.pan.pl.
Sylwia Grzelak, Email: sylwia.grzelak@twarda.pan.pl.
Bożena Moskwa, Email: moskwa@twarda.pan.pl.
Justyna Bień-Kalinowska, Email: jkalinowska@twarda.pan.pl.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Bień J., Moskwa B., Cabaj W. In vitro isolation and identification of the first Neospora caninum isolate from European bison (Bison bonasus bonasus L.) Vet. Parasitol. 2010;29:200–205. doi: 10.1016/j.vetpar.2010.06.038. https://doi: 10.1016/j.vetpar.2010.06.038 [DOI] [PubMed] [Google Scholar]
- Cabaj W., Bień Kalinowska J., Goździk K., Basałaj K., Steiner Bogdaszewska Ż., Bogdaszewski M., Moskwa B. Molecular identification of sarcocysts from tissue of fallow deer (Dama dama) farmed in the open pasture system based on ssu rRNA gene. Acta Parasitol. 2020;65:354–360. doi: 10.2478/s11686-019-00159-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabaj W., Moskwa B., Pastusiak K., Gill J. Antibodies to Neospora caninum in the blood of European bison (Bison bonasus bonasus L.) living in Poland. Vet. Parasitol. 2005;128:63–68. doi: 10.1016/j.vetpar.2004.09.033. https://doi: 10.1016/j.vetpar.2004.09.033 [DOI] [PubMed] [Google Scholar]
- Calero-Bernal R., Vermaa S.K., Seatonb C.T., Sinnett D., Ball E., Dunams D., Rosenthal B.M., Dubey J.P. Sarcocystis cruzi infection in wood bison (Bison bison athabascae) Vet. Parasitol. 2015;210:102–105. doi: 10.1016/j.vetpar.2015.03.007. [DOI] [PubMed] [Google Scholar]
- Druet T., Oleński K., Flori F., Bertrand F.B., Olech W., Tokarska M., Kaminski S., Gautier M. Genomic footprints of recovery in the European Bison. J. Hered. 2020;111:194–203. doi: 10.1093/jhered/esaa002. https://doi: 10.1093/jhered/esaa002 [DOI] [PubMed] [Google Scholar]
- Dubey J.P. Coyote as a final host for Sarcocystis species of goats, sheep, cattle, elk, bison, and moose in Montana. Am. J. Vet. Res. 1980;41:1227–1229. [PubMed] [Google Scholar]
- Dubey J.P. Sarcocystis species in moose (Alces alces), bison (Bison, bison), and pronghorn (Antilocapra americana) in Montana. Am. J. Vet. Res. 1980;41:2063–2065. [PubMed] [Google Scholar]
- Dubey J.P., Calero-Bernal R., Rosenthal B.M., Speer C.A., Fayer R. CRC Press; Boca Raton, FL: 2015. Sarcocystosis of Animals and Humans. [Google Scholar]
- Fayer R., Dubey J.P., Leek R.G. Infectivity of Sarcocystis spp. from bison, elk, moose, and cattle for cattle via sporocysts from coyotes. J. Parasitol. 1982;68:681–685. [PubMed] [Google Scholar]
- Fischer S., Odening K. Characterization of bovine Sarcocystis species by analysis of their 18S ribosomal DNA sequences. J. Parasitol. 1998;84:50–54. https://doi: 10.2307/3284529 [PubMed] [Google Scholar]
- Gjerde B. Phylogenetic relationships among Sarcocystis species in cervids, cattle and sheep inferred from the mitochondrial cytochrome c oxidase subunit I gene. Int. J. Parasitol. 2013;3:579–591. doi: 10.1016/j.ijpara.2013.02.004. https://doi: 10.1016/j.ijpara.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Gjerde B., Hilali M., Abbas I.E. Molecular differentiation of Sarcocystis buffalonis and Sarcocystis levinei in water buffaloes (Bubalus bubalis) from Sarcocystis hirsuta and Sarcocystis cruzi in cattle (Bos taurus) Parasitol. Res. 2016;115:2459–2471. doi: 10.1007/s00436-016-4998-1. [DOI] [PubMed] [Google Scholar]
- Henne D., Ippen R., Blazek K. Zum Sarkosporidienbefall der muskulatur von Zoo- und Wildtieren. Varhandl. Ber. Int. Symp. Erkrank. Zootiere (Berlin). 1977;19:251–257. [Google Scholar]
- Hoeve-Bakker B.J.A., van der Giessen J.W.B., Franssen F.F.J. Molecular identification targeting cox1 and 18S genes confirms the high prevalence of Sarcocystis spp. in cattle in The Netherlands. Int. J. Parasitol. 2019;49:859–866. doi: 10.1016/j.ijpara.2019.05.008. [DOI] [PubMed] [Google Scholar]
- Ippen R., Blazek K., Henne D., Kotrly A. Ein Beitrag zur Sarkosporidiose der Zoo- und Wildtiere. Verhandl. Ber. Int. Sym. Erkrank. Zootiere (Berlin). 1974;16:315–321. [Google Scholar]
- Karbowiak G., Demiaszkiewicz A.W., Pyziel A., Wita I., Moskwa B., Werszko J., Bień J., Goździk K., Lachowicz J., Cabaj W. The parasitic fauna of the European bison (Bison bonasus) (Linnaeus, 1758) and their impact on the conservation. Part 1. The summarising list of parasites noted. Acta Parasitol. 2014;59:363–371. doi: 10.2478/s11686-014-0252-0. https://doi:10.2478/s11686-014-0252-0 [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskwa B., Bień J., Kornacka A., Cybulska A., Goździk K., Krzysiak M.K., Reiterova K., Cabaj W. First Toxoplasma gondii isolate from an aborted foetus of European bison (Bison bonasus bonasus L.) Parasitol. Res. 2017;116:2457–2461. doi: 10.1007/s00436-017-5549-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odening K., Wesemeier H.H., Walter G., Bockhardt J. The wisent (Bison bonasus, Bovidae) as an intermediate host of three Sarcocystis species (Apicomplexa: Sarcocystidae) of cattle. Folia Parasitol. 1994;41:115–121. [PubMed] [Google Scholar]
- Odening K., Wesemeier H.H., Walter G., Bockhardt J. On the morphological diagnostics and host specificity of the Sarcocystis species of some domesticated and wild Bovini (cattle, banteng and bison) Appl. Parasitol. 1995;36:161–178. [PubMed] [Google Scholar]
- Olech W., Perzanowski K. Changes of size and structure of world population of European bison in years 2000–2015. Eur. Bison Conserv.. Newsletter. 2016;9:5–10. [Google Scholar]
- Piusiński W., Malicka E., Bielecki W., Osińska B., Lenartowicz-Kubrat Z. Zmiany patomorfologiczne u żubrów w Puszczy Białowieskiej. Med. Weter. 1996;52:386–388. [Google Scholar]
- Pyziel A.M., Demiaszkiewicz A. Sarcocystis cruzi (Protozoa: Apicomplexa: Sarcocystiidae) infection in European bison (Bison bonasus) from Białowieża forest, Poland. Wiad. Parazytol. 2009;55:31–34. [PubMed] [Google Scholar]
- Raczyński J. European Bison pedigree book 2019. Białowieża National Park, Scott, M.E., 1988 the impact of infection and disease on animal populations: implications for conservation biology. Conserv. Biol. 2019;2:40–56. doi: 10.1111/j.1523-1739.1988.tb00334.x. [DOI] [Google Scholar]
- Sokołowski A.W. Centrum Informacyjne Lasów Państwowych; Warszawa: 2004. Lasy Puszczy Białowieskiej; pp. 1–363. (in Polish) [Google Scholar]
- Szwejkowski H. vol. 30. Gdańsk; 1954. Sarcocystis w mięśniu sercowym żubra (Bison bonasus Boj.) w Polsce; pp. 118–119. (IV Congres of the Polish Parasitologica Society). 10.-01.11.1954, (in Polish) [Google Scholar]
- Yang Z.Q., Zuo Y.X., Yao Y.G., Chen X.W., Yang G.C., Zhang Y.P. Analysis of the 18S rRNA genes of Sarcocystis species suggests that the morphologically similar organisms from cattle and water buffalo should be considered the same species. Mol. Biochem. Parasitol. 2001;115:283–288. doi: 10.1016/s0166-6851(01)00283-3. [DOI] [PubMed] [Google Scholar]
- Zeng H., Van Damme I., Kabi T.W., Šoba B., Gabriël S. Sarcocystis species in bovine carcasses from a Belgian abattoir: a cross-sectional study. Parasites Vectors. 2021;14:271. doi: 10.1186/s13071-021-04788-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.