Summary
The endoplasmic reticulum (ER) plays a central role in lipid homeostasis, but the role of individual ER subdomains in lipid biology has not been elucidated. WrappER is a curved wrapping type of rough-ER that establishes extensive contacts with almost every mitochondria of the hepatocyte in the mouse liver. Here, we describe a protocol for isolation of fractions enriched in wrappER-associated mitochondria from the mouse liver. We also provide techniques for assessing its quality by electron microscopy and biochemical/proteomic analysis.
For complete information on the use and execution of this protocol, please refer to Anastasia et al. (2021).
Subject areas: Cell Biology, Cell separation/fractionation, Metabolism, Protein Biochemistry
Graphical abstract
Highlights
-
•
Protocol for isolating fractions enriched in wrappER-associated mitochondria (WAM)
-
•
Companion protocol for isolating a control ER-enriched fraction
-
•
Procedure to validate WAM- and ER-enriched fractions by electron microscopy analysis
-
•
Procedure to validate WAM- and ER-enriched fractions by immunoblot analysis
The endoplasmic reticulum (ER) plays a central role in lipid homeostasis, but the role of individual ER subdomains in lipid biology has not been elucidated. WrappER is a curved wrapping type of rough-ER that establishes extensive contacts with almost every mitochondria of the hepatocyte in the mouse liver. Here, we describe a protocol for isolation of fractions enriched in wrappER-associated mitochondria from the mouse liver. We also provide techniques for assessing its quality by electron microscopy and biochemical/proteomic analysis.
Before you begin
Procedure to modify the tissue homogenizer
Timing: 30 min
CRITICAL: this protocol requires the use of a modified tissue grinder
-
1.
Take a new 5 mL Teflon Tissue Grinder Wheaton #358034.
-
2.
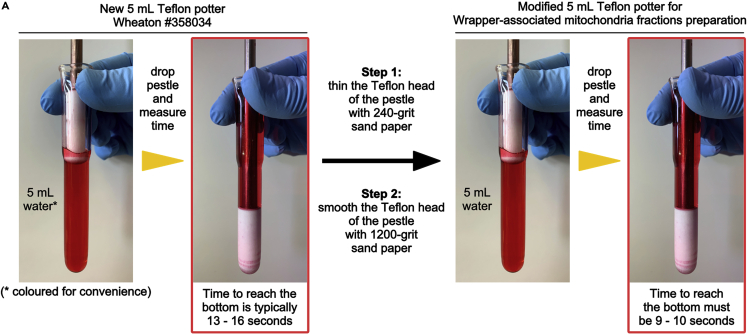
Fill the potter with 5 mL of water (Figure 1A).
-
3.
Hold the pestle as shown in Figure 1A.
-
4.
Drop the pestle and measure the time it takes to reach the bottom of the glass potter.
Note: typically, an unmodified 5 mL Teflon potter Wheaton #358034 takes 13–16 seconds to reach the bottom of the potter.
-
5.Increase the clearance between the Teflon pestle and the potter:
-
a.Thin the Teflon head of the pestle evenly with 240-grit sandpaper.
-
b.Smooth the Teflon head of the pestle with 1200-grit sandpaper.
-
a.
-
6.
Repeat step 4. The pestle should now take 9–10 s to reach the bottom of the potter (Figure 1A). If not, repeat steps 5 and 4 as long as necessary.
-
7.
The potter is now ready to be used.
Figure 1.
Procedure to modify the liver homogenizer
(A) Steps required to modify a Teflon Tissue Grinder Wheaton #358034. Note that in this figure red-colored water is used only for convenience, to better visualize the white potter Teflon head.
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| EGTA | Sigma-Aldrich | #E4378 |
| Halt Protease Inhibitor Cocktail (PIC) (100×) | Thermo Fisher Scientific | #78429 |
| Magnesium chloride hexahydrate | Sigma-Aldrich | #M9272 |
| Sucrose | Thermo Fisher Scientific | #BP220-212 |
| Trizma-base | Thermo Fisher Scientific | #T395 |
| Osmium tetroxide | Electron Microscopy Sciences | #19152 |
| Uranyl acetate | Electron Microscopy Sciences | #22400 |
| Propylene oxide | Electron Microscopy Sciences | #82320 |
| Lead nitrate | Electron Microscopy Sciences | #17900 |
| Sodium nitrate | Electron Microscopy Sciences | #21140 |
| Durcupan, single component A | Sigma-Aldrich | #44611 |
| Durcupan, single component B | Sigma-Aldrich | #44612 |
| Durcupan, single component C | Sigma-Aldrich | #44613 |
| Durcupan, single component D | Sigma-Aldrich | #44614 |
| Glutaraldehyde solution, 25% in H2O | Sigma-Aldrich | #G5882 |
| Calcium chloride, anhydrous | Thermo Fisher Scientific | #S25223 |
| Other | ||
| Teflon Dounce homogenizer 5 mL | Wheaton | #358034 |
| 30 mL Round-bottomed glass centrifuge tubes | Kimble | #45500-30 |
| Rotor adapter for 30 mL glass centrifuge tubes | Kimble | #45550-30 |
| Rotor adapter | Sigma-Aldrich | #EP022637266-2EA |
| Glass disposable Pasteur pipettes | VWR International | #14673-043 |
| Straight operating scissors: sharp/blunt blades | Stoelting Co | #52136-12P |
| Sandpaper 240 and 1200 grit | N/A | N/A |
| 15 mL and 50 mL Polypropylene Falcon tubes | N/A | N/A |
| 50 mL Pyrex glass beaker | N/A | N/A |
| 50 mL Plastic beaker | Electron Microscopy Sciences | #60968 |
| 2 mL Plastic transfer pipette | SARSTEDT | #86.1176.300 |
| Single Slot Copper grids | Electron Microscopy Sciences | #G2010-Cu |
| Biological samples | ||
| Adult male C57BL/6N mice (8 weeks of age) | Charles River | Stain code: 027 |
Materials and equipment
CRITICAL: All buffers should be freshly prepared on the morning of the experiment.
Buffer A (BA)
| Reagent | Stock concentration | Final concentration | Amount |
|---|---|---|---|
| Tris-HCl (pH 7.4) | 1 M | 10 mM | 500 μL |
| MgCl2 | 0.5 M | 1 mM | 100 μL |
| EGTA (pH 7.4) | 0.1 M | 0.1 mM | 50 μL |
| ddH2O | n/a | n/a | 50 mL |
| Total | 50 mL |
CRITICAL: Keep the BA at 20°C.
Buffer 10 (B10)
| Reagent | Stock concentration | Final concentration | Amount |
|---|---|---|---|
| Sucrose | n/a | 250 mM | 4.28 g |
| Tris-HCl (pH 7.4) | 1 M | 10 mM | 500 μL |
| MgCl2 | 0.5 M | 1 mM | 100 μL |
| EGTA (pH 7.4) | 0.1 M | 0.1 mM | 50 μL |
| ddH2O | n/a | n/a | 50 mL |
| Total | 50 mL |
CRITICAL: Keep the B10 always on ice.
Buffer 27 (B27)
| Reagent | Stock concentration | Final concentration | Amount |
|---|---|---|---|
| Sucrose | n/a | 27% (w/w) | 6.75 g |
| Buffer A | n/a | 73% (w/w) | 18.25 g |
| Total | 25 g |
CRITICAL: Keep the B27 at 20°C.
CaCl2 8 mM solution
| Reagent | Stock concentration | Final concentration | Amount |
|---|---|---|---|
| CaCl2 | 800 mM | 8 mM | 500 μL |
| ddH2O | n/a | n/a | 50 mL |
| Total | 50 mL |
CRITICAL: Keep the CaCl2 8 mM solution always on ice.
Step-by-step method details
Mouse liver WAM-enriched fraction preparation
Timing: 3 h
The following protocol describes the steps required to obtain WAM-enriched fractions from a mouse liver weighing between 1.0 and 1.4 grams.
Note: In our experience, this protocol allows the simultaneous processing of up to 3 livers, but no more.
CRITICAL: Do not clean glassware with detergents. Always thoroughly rinse all glassware with ddH2O before and after using them.
-
1.
Prepare all the buffers and cool down the centrifuge to 4°C.
Note: Use a fixed angle rotor type F34-6-38 for Eppendorf centrifuge 5800 (Figure 2A).
Figure 2.
Some of the equipment needed to isolate WAM-enriched mouse liver fractions
(A) Beaker containing mouse chopped liver in ice-cold B10 buffer.
(B) Motorized tissue homogenizer.
(C) Fixed angle rotor type F34-6-38 for Eppendorf centrifuge 5800 equipped with adapters for 50 mL Falcon tubes.
(D) 1 mL and 2 mL plastic transfer pipettes.
-
2.
To prepare B10PIC, transfer 11 mL of B10 to a 15mL-Falcon tube, then add 110 μL of protease inhibitor cocktail (PIC).
CRITICAL: Do not use a protease inhibitor cocktail containing EDTA, as chelating Mg++ interferes with retaining ribosomes attached to the ER membrane.
-
3.
Transfer 15 mL of B10 to a 50 mL-glass beaker. Keep it on ice.
-
4.
Euthanize the mouse, then incise the abdomen with surgical scissors to expose the liver. Excide the liver, discard the gallbladder, and transfer it to the 50 mL-glass beaker prepared in step 3.
-
5.
Use clean surgical scissors to cut the liver into small pieces (Figure 2B), then rinse three times with 15 mL of B10 to remove the blood.
Note: Let the small liver pieces decant, then remove most of the buffer by gently tilting the beaker. Use a P1000 pipette to remove the last few mL of B10.
-
6.
Add 4 mL of B10PIC to the chopped, rinsed liver and transfer it into the modified 5 mL Teflon Potter by tilting the beaker.
Note: Use surgical scissors or a wooden spatula to assist in the transfer of the small liver pieces.
Note: Do not fill the potter past the fill line. Depending on the size of the liver, it may be necessary to remove excess buffer with a P1000 before proceeding to the next step.
-
7.
Place the modified glass potter with the chopped liver inside a 50 mL Falcon tube containing ice.
Note: Keep the glass potter cold during liver homogenization.
-
8.Use an electric homogenizer (Figure 2C) to homogenize the liver by stroking up and down the potter.
-
a.Set the homogenizer speed to 1,600 rpm, then perform 16 strokes: 8 ups and 8 downs, 2 s per stroke; then pause for 30 s.
-
a.
-
9.
Repeat step 8a.
-
10.
Transfer the liver homogenate by tilting the glass potter into a 15 mL Falcon tube.
-
11.
Rinse the potter with the remaining 7 mL of cold B10PIC and add them to the liver homogenate. Mix gently by inverting the tube a few times and pour the homogenate into two 30 mL glass centrifuge tubes.
-
12.
Spin at 400 × g for 10 min at 4°C to remove nuclei and tissue debris.
Note: Use a fixed angle rotor type F34-6-38 for Eppendorf centrifuge 5800 (Figure 2A).
-
13.
Collect the supernatant (∼5 mL per tube) and transfer it to a clean 30 mL glass centrifuge tube.
CRITICAL: Stay away from the pellet: it contains unbroken cells and nuclei that would contaminate the WAM-enriched fraction.
Note: Use a P1000 pipette tip, cut ∼2 mm from the end to enlarge the opening.
-
14.
Repeat steps 12 and 13.
-
15.
Repeat step 12 one more time, collect the two supernatants and gently mix them into a 15 mL Falcon tube (the final volume should be ∼ 7.5 mL).
CRITICAL: Stay well away from the pellet.
Note: Use a P1000 pipette tip, cut ∼2 mm from the end to enlarge the opening.
Note: This is what, from here on, we call post-nuclear hepatic homogenate. You can set aside up to 500 μL and use it as a control in assays concurrent with biochemical validation of the WAM-enriched fraction.
-
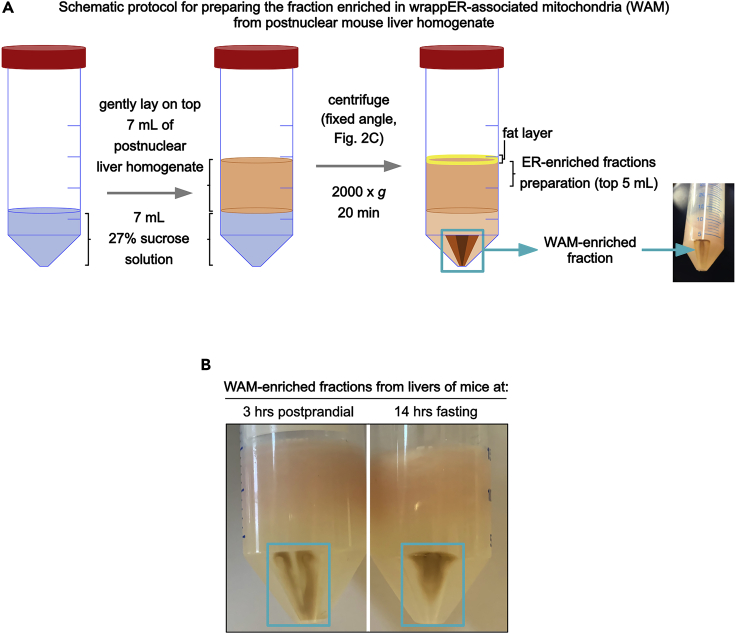
16.
Pour 7 mL of B27 (maintained at 20°C) into a 50 mL Falcon tube and carefully lay on top of it 7 mL of post-nuclear hepatic homogenate.
Note: Use a P1000 pipette tip, cut ∼2 mm from the end to enlarge the opening.
-
17.
Spin at 2,000 × g for 20 min at 4°C.
Note: Use a fixed angle rotor type F34-6-38 for Eppendorf centrifuge 5800 (Figure 2A).
-
18.
Collect the top 5 mL of the supernatant and use it to prepare the control ER-enriched fraction (see below “complementary protocol: preparation of ER-enriched fraction”).
-
19.Thoroughly remove the supernatant using a disposable glass Pasteur pipette connected to the vacuum line. The pellet is the WAM-enriched fraction (Figures 3A and 3B).
-
a.For omics analysis, resuspend the pellet in 500 μL B10PIC and store at −80°C.
-
b.For Electron Microscopy (EM) analysis, gently add 500 μL of B27PIC on top of the pellet. Using a paintbrush, gently detach it from the side of the tube; then, follow “Complementary Protocol: EM Analysis of WAM-Enriched Fractions”.
-
a.
CRITICAL: Step 19b goal is to obtain a flaky pellet, limiting its resuspension in the buffer.
Figure 3.
Schematic representation of the procedure used to prepare wrappER-mitochondria (WAM)-enriched fractions
(A) 7 mL of B27 are added into a 50 mL Falcon tube, on top of which 7 mL of post-nuclear liver homogenate are gently layered. The solution is centrifugated in a fixed rotor. The pellet is enriched in WAM.
(B) An image of the pellet enriched in WAM. Note that its appearance depends on the nutritional status of the mouse liver from which it was prepared.
Complementary protocol: Preparation of ER-enriched fraction
Timing: 1 h
The following procedure describes how to obtain ER-enriched fractions from WAM-depleted liver homogenates. They can be used per se or as a control to the proteomic/functional analysis of the WAM-enriched fractions. This protocol has been modified from (Hamilton et al., 1999).
-
20.
Starting from step 18, aliquot the supernatant into two 2 mL Eppendorf tubes.
Note: When collecting the supernatant in step 18, stay away from the fat layer that accumulates at the top and along the wall of the 50 mL Falcon tube.
-
21.
Spin at 12,000 × g for 10 min at 4°C to remove heavy organelles; collect the supernatants with a P1000 pipette tip and transfer them into two clean 2 mL Eppendorf tubes.
CRITICAL: Stay away from the pellet: it contains heavy organelles such as peroxisomes, lysosomes, mitochondria that would contaminate the ER-enriched fraction.
-
22.
Repeat step 21.
-
23.
Collect 1.5 mL of supernatant from each tube and gently mix them into a 50 mL Falcon tube. Gently add 22.5 mL of ice-cold CaCl2 8 mM solution.
-
24.
Incubate for 15 min at 4°C, with gentle rocking.
-
25.
Pour gently into two 30 mL glass tubes (12 mL/tube), then spin at 8,000 × g for 10 min at 4°C.
Note: Use a fixed angle rotor type F34-6-38 for Eppendorf centrifuge 5800.
-
26.Discard the supernatants; the pellet is the ER-enriched fraction.
-
a.For omics analysis, resuspend the pellet in 500 μL B10PIC and store at −80°C.
-
b.For Electron Microscopy (EM) analysis, gently add 500 μL of B27PIC on top of the pellet. Using a paintbrush, gently detach it from the side of the tube; then, follow “Complementary Protocol: EM Analysis of WAM-Enriched Fractions”.
-
a.
CRITICAL: Step 26b goal is to obtain a flaky pellet, limiting its resuspension in the buffer.
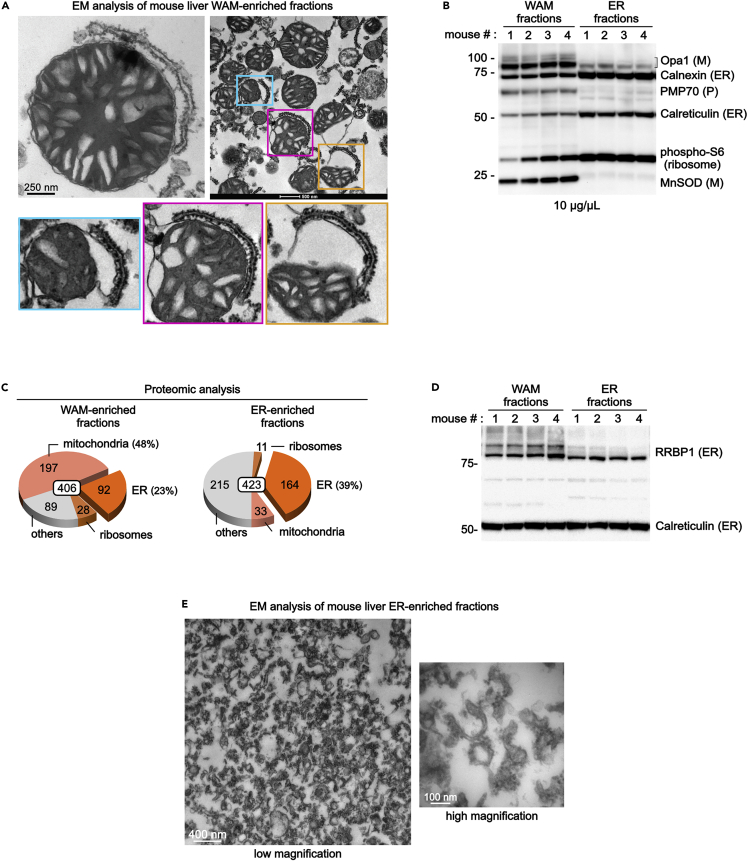
Note: Proteomic analysis of this fraction should indicate that ∼40% of the proteins are ER proteins (Figure 4C).
Figure 4.
Analysis of mouse liver WAM-enriched fractions
(A–D) The pellet shown in Figure 3B is analyzed by EM (A), immunoblot (B and D), and proteomic studies (C). Adapted from (Anastasia et al., 2021).
(E) EM analysis of the control mouse liver ER-enriched fractions.
Complementary protocol: EM analysis of WAM-enriched fractions
Timing: 3 days
Here we describe how to fix and embed WAM- and ER-enriched fractions for EM analysis, a reliable method for verifying their quality.
-
27.
Transfer the flaky pellet (step 19b or 26b) into a 2 mL Eppendorf tube and add 500 μL of B27PIC.
CRITICAL: Use a tube with a round, not conical, bottom.
CRITICAL: Use a P200 pipette tip, cut ∼5 mm from the end to enlarge the opening, and avoid resuspending the pellet. The pellet must remain visibly flaky.
-
28.
Spin at 3,000 × g for 10 min at 4°C. Remove and discard the supernatant.
-
29.
Gently add to the pellet 500 μL of B27 solution supplemented with 2% glutaraldehyde (freshly prepared), and incubate overnight (16 h) at 4°C.
CRITICAL: from this point on, the pellet must remain intact, possibly sticking against the wall of the Eppendorf tube. Through all the next steps, pay attention to always be gentle when adding and removing all various solutions; to this end, we recommend using plastic transfer pipettes (Figure 2D).
Note: Glutaraldehyde is highly toxic: work under a chemical hood.
-
30.
Remove the glutaraldehyde solution with a plastic transfer pipette, paying attention to leave the pellet undisturbed
-
31.
Rinse the pellet three times to eliminate any residual glutaraldehyde. To this end, gently apply onto the wall of the Eppendorf tube 700 μL of B27 solution (use a P1000 pipette).
-
32.
Incubate the pellet in a solution of 1% (w/v) osmium tetroxide in ddH2O for 30 min at 20°C, keeping the Eppendorf tube protected from light by wrapping it in aluminum foil.
Note: Osmium tetroxide is highly toxic: operate under a chemical hood.
-
33.During the incubation (step 32), reconstitute the Durcupan epoxy resin:
-
a.In a 50 mL plastic beaker placed on a scale, add 5 g of Durcupan component A, 5 g of Durcupan component B, 0.15 g of Durcupan component C, and 0.15 g of Durcupan component D.Note: Durcupan components are dense liquids: use plastic pipettes (Figure 2D).
-
b.Stir well with a wooden spatula until the resin changes color and becomes orange.Note: The reconstituted Durcupan epoxy resin is now ready to be used but it can be stored for up to 2 weeks at −20°C.
-
a.
-
34.
Rinse the pellet (step 32) three times to eliminate any residual osmium tetroxide. To this end, gently apply onto the wall of the Eppendorf tube 700 μL of ddH2O (use a P1000 pipette).
-
35.Pellet dehydration:Note: Use plastic transfer pipettes (Figure 2D) to add/remove the solutions used in this step.
-
a.Add 1 mL of 50% ethanol:water and incubate for 15 min. Discard ethanol solution.
-
b.Add 1 mL of 1% uranyl acetate in 70% ethanol:water and incubate for 40 min keeping the Eppendorf tube protected from light by wrapping it with aluminum foil. Remove and discard the uranyl acetate solution.
-
c.Add 1 mL of 90% ethanol:water and incubate for 15 min. Discard ethanol solution.
-
d.Add 1 mL of 100% ethanol and incubate for 15 min. Discard ethanol solution.
-
e.Repeat step 35d.
-
f.Add 1 mL of 100% propylene oxide to the pellet and incubate for 15 min. Discard propylene oxide.
-
g.Repeat step 35f.
 CRITICAL: Work rapidly as propylene oxide evaporates quickly at 20°C. The sample must not dry out when changing the solutions; perform all steps at 20°C.Note: Propylene oxide is highly toxic: operate under a chemical hood.
CRITICAL: Work rapidly as propylene oxide evaporates quickly at 20°C. The sample must not dry out when changing the solutions; perform all steps at 20°C.Note: Propylene oxide is highly toxic: operate under a chemical hood.
-
a.
-
36.Embed the pellet in the Durcupan epoxy resin:Note: Step 36b-d: leave the cap of the Eppendorf tube open.
-
a.Remove and discard the 100% propylene oxide and quickly embed the pellet in ∼500 μL of reconstituted Durcupan epoxy resin.Note: From this stage, the sample can be handled on the lab bench.
-
b.Incubate for 1 h at 37°C.
-
c.Remove as much epoxy resin as possible, leaving the pellet undisturbed and, ideally, attached to the side of the Eppendorf tube. Add 500 μL epoxy resin and move the sample in an oven at 58°C.
-
d.Incubate 48–72 h, to polymerize the reconstituted Durcupan epoxy resin.
-
a.
-
37.
Release the hardened epoxy resin block that contains the sample to analyze by EM; this can be done by cutting the 2 mL Eppendorf tube with a sharp blade or scissors.
-
38.
Cut the epoxy resin-embedded sample in ∼50 nm -thick slices using a Leica Ultramicrotome and place them on single-slot formvar coated copper grids (up to 5 slices/grid).
-
39.
Counterstain slices by incubating the grid for 5 min with Reynold’s lead citrate.
-
40.
The WAM and ER-enriched fractions can be now imaged under a transmission electron microscope (Figures 4A and 4E).
Expected outcomes
At the end of the protocol that yields the WAM-enriched fraction (step 19), the size and color of the pellet depend on the nutritional status of the animal when the liver was collected (Figure 3B). Regardless, at step 19a the expected protein concentration should be in the 5–10 μg/μL range (determined by Bradford assay).
Electron microscopy analysis is the most reliable method to verify that the protocol described here has been performed correctly (Figure 4A). EM analysis of WAM-enriched fractions should show that at least ∼75% of mitochondria are associated with wrappER (Figure 4A).
Immunoblot analysis of the WAM-enriched fraction (10 μg/lane) must reveal the enrichment of mitochondrial and ER proteins (membrane-anchored and soluble in the matrix/ER lumen); this can be done using post-nuclear liver homogenate as control (10 μg/lane; step 15). This analysis must also show enrichment of ribosomal proteins (Figure 4B). In contrast, contaminant lysosome, nucleus, plasma membrane, and Golgi proteins should be absent or minimally present. Note that peroxisomal proteins might be present because WAM-enriched fractions contain some wrappER-associated peroxisomes and peroxisome-wrappER-mitochondria complexes (our unpublished study).
Proteomic analysis of WAM-enriched fractions should show that ∼80% of the proteins are from mitochondria, ER, and ribosomes (Figure 4C) (Anastasia et al., 2021).
When compared to ER-enriched fractions (see above, step 20–26), the pattern of expression of the Rrbp1 protein should be as shown in Figure 4D (Anastasia et al., 2021).
This protocol is designed to retain the wrappER ribosomes and mRNAs, allowing to perform transcriptomic analysis. Such analyses should reveal a massive presence of ER protein transcripts and a minimal presence of nuclear-encoded mitochondrial protein transcripts (Anastasia et al., 2021); however, mitochondrial DNA-encoded transcripts should be highly expressed.
Limitations
Preparing WAM-enriched fractions from fatty livers can be problematic. The yield of WAM may diminish due to ineffective liver homogenate preparation and/or separation during the early phases of the protocol (steps 8 and 9). The presence of large lipid droplets may indeed cause wrappER shredding during sample homogenization. Therefore, any WAM isolation should be done by validating the integrity of this inter-organelle contact through EM analysis (step 27–40).
Recent studies from our laboratory have shown that the wrappER, in addition to mitochondria, also forms contacts with peroxisomes and that the protocol described here also yields wrappER-associated peroxisomes and peroxisome-wrappER-mitochondria (PEWM) complexes.
This protocol has been optimized for the isolation of WAM-enriched fractions from the mouse liver. We expect it to be equally effective in other model organisms, such as rats or fish, as long as the amount of liver used is not different from that given here.
Isolation of WAM-enriched fractions has not been attempted in other tissues and cultured cells, although we expect it to be possible in soft tissues such as the pancreas and thymus. As for cultured cells, the main hurdle is to adapt this protocol to a smaller scale and to modify the homogenization step.
Membrane-permeable molecules (including protease inhibitors) hurt the yield and structure of the WAM. Membrane-permeabilizing detergents must also be avoided, which is why indicated to clean the glassware by rinsing it with water.
Troubleshooting
Problem 1
Small pellet (step 19; Figure 3B). This typically means poor WAM yield, which can be caused either by poor liver homogenization (steps 8 and 9) or excessive organelle disruption/shredding (see problem 2).
Potential solution
Increase liver homogenization.
Start by chopping the liver into smaller pieces (step 5; Figure 2B); also, consider increasing the number of strokes (steps 8 and 9).
Problem 2
Small pellet (step 19; Figure 3B) and a) EM analysis shows either mitochondria without wrappER or broken mitochondria (step 40), or b) immunoblot shows low level of soluble mitochondria matrix markers (such as MnSOD, due to broken mitochondria).
Potential solution
Decrease liver homogenization.
In general, keep in mind that high hydraulic pressure destroys the integrity of any organelles, particularly in a viscous media. High hydraulic pressure is typically caused by pushing hard and fast the Teflon pestle into the glass potter containing the liver homogenate (step 8). Therefore, considering go slower in this step.
If hydraulic pressure is OK, then consider reducing the number of strokes to 10–12 (steps 8 and 9) and/or slow down the speed of the rotor to 1,200 rpm.
Consider also thinning a little more the Teflon head of the pestle (to 6–8 s), as this decreases hydraulic pressure too.
Excessive organelle shredding can also be caused by the presence of large masses of lipid droplets during homogenization (steps 8 and 9). Use 20% less liver without changing the volume of B10 solution (4 mL; step 6).
Problem 3
Electron microscopy analysis shows the presence of intact organelles such as, for example, lysosomes and peroxisomes (step 40).
Potential solution
Improve the homogenization described in steps 8 and 9. This requires increasing the number of strokes to 20–24. Also, increase the volume of B27 solution from 7 to 10 mL (step 16).
Problem 4
Immunoblot analysis of ER-enriched fractions detects membrane proteins from lysosomes, plasma membrane, peroxisomes, Golgi, etc (step 26a).
Potential solution
Decrease liver homogenization to limit the disruption of the organelles' integrity. Consider increasing the potter-pestle clearance (steps 5 and 6 of “procedure to modify the tissue homogenizer”) and/or reducing the number and speed of the strokes (steps 8 and 9).
Problem 5
Pellet breaks apart during steps 29–34 of “complementary protocol: EM analysis of WAM-enriched fractions”.
Potential solution
Centrifuge the sample at 6,000 × g for 10 min at 4°C.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Luca Pellegrini (luca.pellegrini@fmed.ulaval.ca).
Materials availability
This study did not generate new unique reagents.
Data and code availability
The published article (Anastasia et al., 2021) includes all datasets generated and analyzed during this study.
Acknowledgments
N.I. and I.A. are recipients of a PhD scholarship from the Centre Thématique de Recherche en Neurosciences (CTRN) and from Fonds de la recherche en santé du Québec (FRQS). This study was funded by grants from the Canadian Institutes of Health Research (CIHR, 201603PJT-365052) and the Natural Sciences and Engineering Research Council of Canada (NSERC, RGPIN-2017-06130) to L.P.
Author contributions
Conceptualization, N.I., I.A., and L.P.; investigation, N.I. and I.A.; writing – original draft, N.I., I.A., and L.P.; writing – review & editing, N.I. and L.P.; funding acquisition, L.P.; supervision, L.P.
Declaration of interests
The authors declare no competing interests.
Contributor Information
Nicolò Ilacqua, Email: nicolo.ilacqua.1@ulaval.ca.
Luca Pellegrini, Email: luca.pellegrini@fmed.ulaval.ca.
References
- Anastasia I., Ilacqua N., Raimondi A., Lemieux P., Ghandehari-Alavijeh R., Faure G., Mekhedov S.L., Williams K.J., Caicci F., Valle G. Mitochondria-rough-ER contacts in the liver regulate systemic lipid homeostasis. Cell Rep. 2021;34:108873. doi: 10.1016/j.celrep.2021.108873. [DOI] [PubMed] [Google Scholar]
- Hamilton R.L., Moorehouse A., Lear S.R., Wong J.S., Erickson S.K. A rapid calcium precipitation method of recovering large amounts of highly pure hepatocyte rough endoplasmic reticulum. J. Lipid Res. 1999;40:1140–1147. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The published article (Anastasia et al., 2021) includes all datasets generated and analyzed during this study.