Abstract
Purpose
Most radiomic studies use the features extracted from the manually drawn tumor contours for classification or survival prediction. However, large interobserver segmentation variations lead to inconsistent features and hence introduce more challenges in constructing robust prediction models. Here, we proposed an automatic workflow for glioblastoma (GBM) survival prediction based on multimodal magnetic resonance (MR) images.
Methods and Materials
Two hundred eighty-five patients with glioma (210 GBM, 75 low-grade glioma) were included. One hundred sixty-three of the patients with GBM had overall survival data. Every patient had 4 preoperative MR images and manually drawn tumor contours. A 3-dimensional convolutional neural network, VGG-Seg, was trained and validated using 122 patients with glioma for automatic GBM segmentation. The trained VGG-Seg was applied to the remaining 163 patients with GBM to generate their autosegmented tumor contours. The handcrafted and deep learning (DL)–based radiomic features were extracted from the autosegmented contours using explicitly designed algorithms and a pretrained convolutional neural network, respectively. One hundred sixty-three patients with GBM were randomly split into training (n = 122) and testing (n = 41) sets for survival analysis. Cox regression models were trained to construct the handcrafted and DL-based signatures. The prognostic powers of the 2 signatures were evaluated and compared.
Results
The VGG-Seg achieved a mean Dice coefficient of 0.86 across 163 patients with GBM for GBM segmentation. The handcrafted signature achieved a C-index of 0.64 (95% confidence interval, 0.55-0.73), whereas the DL-based signature achieved a C-index of 0.67 (95% confidence interval, 0.57-0.77). Unlike the handcrafted signature, the DL-based signature successfully stratified testing patients into 2 prognostically distinct groups.
Conclusions
The VGG-Seg generated accurate GBM contours from 4 MR images. The DL-based signature achieved a numerically higher C-index than the handcrafted signature and significant patient stratification. The proposed automatic workflow demonstrated the potential of improving patient stratification and survival prediction in patients with GBM.
Introduction
Glioma is the most common type of primary brain tumor in adults. It arises from glial cells, normally astrocytes and oligodendrocytes. According to the World Health Organization guideline, glioma can be classified into grade I to grade IV based on the histologic characteristics.1 Glioblastoma multiforme (GBM) is the most aggressive, grade IV, glioma. It accounts for 81% of malignant brain tumors.2 Despite extensive efforts, prognoses for patients with GBM remain dismal. The median overall survival (OS) is 14 to 16 months after diagnosis.3 The 5-year survival rate is below 5%.4 It is beneficial to build survival prediction models for assisting therapeutic decisions and disease management in patients with GBM.
Magnetic resonance imaging (MRI) is the preferred imaging modality for GBM diagnosis and monitoring. Radiomic features extracted from MR images using advanced mathematical algorithms may uncover tumor characteristics that fail to be appreciated by the naked eye. Many studies have investigated the association of MRI radiomic features with the survival outcomes of patients with GBM.5, 6, 7 However, radiomic features were extracted from the manually drawn tumor contours in these studies. Manual tumor segmentation is not only time-consuming but also sensitive to intraobserver and interobserver variabilities. These segmentation variations could result in many inconsistent radiomic features,8,9 which introduces more challenges in constructing robust prediction models.
Developing an automatic GBM segmentation model could eliminate the manual contour variations and enable an automatic survival prediction workflow. Convolutional neural networks (CNNs) have achieved state-of-the-art performance in medical image segmentation. Particularly, U-Net10 and fully convolutional network11 have been widely adopted. Shboul et al12 used an ensemble of the 2-dimensional (2D) U-Net and the 2D fully convolutional network for GBM segmentation followed by an XGBoost based regression model to achieve automatic GBM survival prediction. However, this study only investigated the handcrafted radiomic features that were extracted using explicitly designed algorithms. These features are normally low-level image features that are limited to current human knowledge. Another type of radiomic feature can be extracted using a pretrained CNN.13, 14, 15 We refer to these features as “deep learning (DL)-based features” in this study. These high-level features may have higher prognostic power than the handcrafted features.
In this study, we proposed an automatic workflow for GBM survival prediction based on 4 preoperative MR images. A novel 3D CNN, VGG-Seg, was proposed and trained for automatic GBM segmentation. The handcrafted and DL-based radiomic features were extracted from the autosegmented contours generated by the VGG-Seg and used to construct 2 separate Cox regression models for survival prediction. The prognostic powers of the constructed signatures were evaluated and compared. To our knowledge, this is the first paper to investigate the DL-based radiomic features for automatic GBM survival prediction.
Methods and Materials
Data set
Two hundred eighty-five patients with glioma were acquired from the Brain Tumor Segmentation 2018 challenge.16, 17, 18 Two-hundred and ten patients had GBM, and the remaining 75 patients had low-grade (grade II-III) glioma (LGG). Each patient had 4 preoperative MR images acquired. These included T1-weighted, contrast-enhanced T1-weighted, T2-weighted, and fluid-attenuated inversion recovery (FLAIR) MR images. Patient images were acquired with different clinical protocols and various scanners from multiple institutions. For each patient, MR images were coregistered, resampled to 1 mm3 resolution using linear interpolation, and skull-stripped.16,17 The final image dimension was 240 × 240 × 155. All patients had 3 ground truth tumor subregion labels (edema, enhancing tumor, and necrotic and nonenhancing tumor core) approved by experienced neuro-radiologists. OS data were available for 163 patients with GBM.
We applied the N4 bias correction algorithm on all images, except the FLAIR images, to remove low-frequency inhomogeneity.19 Each MR image was normalized to have zero mean and unit standard deviation in the brain voxels. Figure 1 shows the transverse slices of 4 preprocessed MR images and the corresponding tumor labels.
Fig. 1.
Transverse slices of preprocessed T1-weighted (T1w), contrast-enhanced T1-weighted (CE-T1w), T2-weighted (T2w), and fluid-attenuated inversion recovery (FLAIR) images along with the corresponding ground truth labels for edema, enhancing tumor, and necrotic and nonenhancing tumor core (NCR/NET) for a representative case.
VGG-Seg for automatic GBM segmentation
Figure 2 shows the architecture of the VGG-Seg proposed for automatic GBM segmentation. It contains 27 convolutional layers, forming an encoder and decoder architecture. The encoder network was constructed based on the VGG16 model20 that achieved accurate performance in object detection. Instance normalization layers21 and residual shortcuts22 were implemented to improve model performance. The VGG-Seg can be trained to perform an end-to-end mapping, converting the concatenation of 4 preprocessed images to 4 probability maps for 3 tumor subregion labels and background labels.
Fig. 2.
The overall VGG-Seg architecture. Four magnetic resonance (MR) images are concatenated and input into the VGG-Seg containing 27 convolutional layers. The model generates 4 probability maps. Each filled box represents a set of 4-dimensional (4D) feature maps, the numbers and dimensions of which are shown. The window size and the stride for convolutional, maxpooling, and deconvolutional layers are also presented. Abbreviations: Conv = convolutional layer; Deconv = deconvolutional layer; IN = instance normalization layer; Maxpool = maxpooling layer; ReLU = rectified linear unit.
In the model training stage, 122 patients without OS data were randomly split into a training set of 105 patients (75 patients with LGG and 30 patients with GBM) and a validation set of 17 patients with GBM. The Adam stochastic gradient descent method23 was used to minimize the multi-Dice loss:
| (1) |
where is the probability, after Softmax layers, of the voxel j being the label i; 4 labels are background label and 3 tumor subregion labels; is the ground truth label, 0 or 1, of the voxel j being the label i; and N is the voxel number. The validation set was used for tuning hyperparameters including the initial learning rate and the stopping epoch number. A batch size of 1 was used for model training.
The trained VGG-Seg was applied to the remaining 163 patients with GBM (all of whom had corresponding OS data) to generate their tumor subregion labels. The autosegmented tumor contour was acquired by merging the 3 predicted subregion labels. Model accuracy was evaluated using the Dice coefficient:
| (2) |
where and are the volumes of the ground truth tumor contour and autosegmented tumor contour, respectively.
Radiomic feature extraction
Handcrafted features
Using the PyRadiomics24 package (version 2.1.2) for all 163 patients with GBM, 1106 handcrafted features were extracted from 4 MR images. These features were extracted from the autosegmented tumor contour and contained 14 shape-based features, 72 first-order statistical features, 292 second-order statistical (textural) features, and 728 high-order statistical features. Shape-based features represented the shape characteristics of the tumor contour. First-order statistical features represented the characteristics of the tumor intensity distribution. Textural features were extracted based on gray level cooccurrence, gray level size zone, gray level run length, gray level dependence, and neighborhood gray-tone difference matrices. They represented the characteristics of the spatial intensity distributions. High-order statistic features were extracted from the images filtered using Laplacian of Gaussian filters.
DL-based features
Using a pretrained classification CNN VGG19 model,20 1472 DL-based features were extracted for all 163 patients with GBM in the testing set. We used a pretrained VGG19 that is available in the deep learning toolbox (version 12.0) from MATLAB (version 9.5, R2018b). It was trained on more than a million images from the ImageNet data set.25 Figure 3 shows the model architecture and feature extraction scheme. VGG19 contains 16 convolutional layers and 3 fully connected layers. Five max-pooling layers are used to achieve partial translational invariance, reduce model memory usage, and prevent overfitting. For each patient, we selected a square region of interest (ROI) from the transverse slice that had the largest tumor area. The size of the ROI was set as the maximum dimension of the tumor contour on the selected slice. We then resized the ROIs of FLAIR, T2-weighted, and contrast-enhanced T1-weighted MR images to 224 × 224 using bilinear interpolation, mapped the pixel intensity to the range (0–255) and concatenated them. The concatenation was input into the pretrained VGG19 for feature extraction. As shown in Figure 3, DL-based features were extracted by average-pooling the 5 feature maps after max-pooling layers. Each feature map generated a vector after average-pooling. Five feature vectors were first normalized with their Euclidean norms and then concatenated to form a single feature vector. DL-based features were acquired by normalizing the single feature vector with its Euclidean norm.
Fig. 3.
Deep learning (DL)–based feature extraction scheme using VGG19. VGG19 contains 16 convolutional layers, 5 max-pooling layers, and 3 fully connected layers. The average-pooling layers were used for extracting DL-based features. Feature maps and feature vectors after every layer are shown as cuboids and rectangles, respectively. The feature map depth and feature number are shown. A concatenation of fluid-attenuated inversion recovery (FLAIR), T2-weighted (T2w), and contrast-enhanced T1-weighted (CE-T1w) regions of interest (ROIs) was input into the pretrained VGG19 for feature extraction. By average-pooling along the spatial dimensions, 1472 DL-based features were extracted from max-pooling feature maps. Abbreviations: Conv = convolutional layer; ReLU = rectified linear unit.
Survival prediction model
The 163 patients with GBM with available OS data were randomly split into a training set of 122 patients and a testing set of 41 patients. Each feature was normalized using the mean and standard deviation of the training set. Because a large number of features may lead to overfitting, we preselected a subset of features having the highest univariate C-index. Higher C-index values indicate features with higher prognostic power. The Cox regression model with regularization was trained using the selected features to construct a radiomic signature for survival prediction in patients with GBM. The radiomic signature is a linear combination of the features weighted by the Cox regression model coefficients. We tested 3 regularization techniques: ridge, elastic net, and least absolute shrinkage and selection operator. The number of the preselected features, the regularization technique, and the corresponding regularization parameters were chosen with 5-fold cross-validation using the training set. Two Cox regression models were trained using either handcrafted features or DL-based features. The resulting radiomic signatures are referred to as the “handcrafted signature” and the “DL-based signature,” respectively.
The prognostic power of the 2 constructed radiomic signatures was evaluated using the C-index and the average areas under the receiver operating curves (AUCs) at different survival time points. A paired t test and DeLong tests were conducted to test the significance of the differences in the C-index and AUCs, respectively. A threshold on the radiomic signature can be set using the training set for patient stratification. We investigated 2 thresholds: 1 selected using the X-tile software26 and the other defined by the median signature value of the training patients. The X-tile software selected the optimal threshold by selecting the highest value of the data divisions. The chosen thresholds were then used to stratify the testing patients into high-risk and low-risk groups. Log-rank tests were conducted to test the difference between the 2 risk groups.
Results
OS statistics
The median and mean (standard deviation) of OS were 367.0 days and 416.5 (329.2) days in the training set, and 362.0 days and 442.1 (408.6) days in the testing set, respectively. A Mann-Whitney U test indicated that we cannot reject the null hypothesis that there was a difference in OS between 2 data sets (P = .83).
Tumor segmentation
The VGG-Seg was trained using an initial learning rate of 5 × 10−4 for 150 epochs. These hyperparameters resulted in the minimum validation loss. The Dice coefficients of the whole tumor contours for the training, validation, and testing sets are summarized in Table 1. The autosegmented contours achieved the Dice coefficient of 0.86 ± 0.09 on the whole tumor contour for 163 patients with GBM in the testing set.
Table 1.
Dice coefficients of the whole tumor contours for the training, validation, and testing sets
| Dice | Training (75 LGG and 30 GBM) |
Validation (17 GBM) |
Testing (163 GBM) |
|---|---|---|---|
| Whole tumor | 0.92 ± 0.03 | 0.90 ± 0.07 | 0.86 ± 0.09 |
Abbreviations: GBM = glioblastoma multiforme; LGG = low-grade glioma; SD = standard deviation.
Results were averaged and showed in (mean ± SD) format.
Survival prediction
Table 2 shows the optimal preselected feature number, regularization technique, and regularization parameter that achieved the best cross-validation result for each feature set.
Table 2.
Optimal regularization technique and hyperparameters selected by 5-fold cross-validation for each feature set
| Number of preselected features | Regularization technique | Regularization parameter (λ) | |
|---|---|---|---|
| Handcrafted features | 50 | Ridge | 3.439 |
| DL-based features | 80 | Ridge | 1.813 |
Abbreviation: DL = deep learning.
The handcrafted signature achieved a C-index of 0.64 (95% confidence intervals [CI], 0.55-0.73) on the testing set, whereas the DL-based signature achieved a C-index of 0.67 (95% CI, 0.57-0.77). A paired t test indicated that we could not reject the null hypothesis that there is no difference in C-index (P = .27).
Table S.1 shows the AUCs of the signatures, evaluated at the OS of 300 days and 450 days, of the testing set. The DL-based signature achieved numerically higher AUCs than the handcrafted signature. P values of DeLong tests were greater than .05.
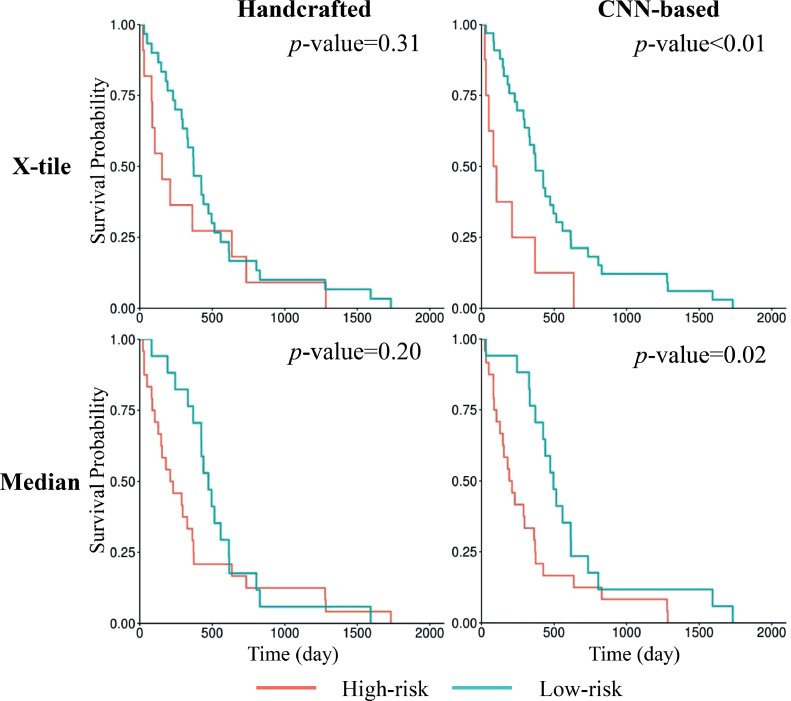
We split the testing patients into high-risk and low-risk groups based on signature thresholds. Figure 4 shows the Kaplan-Meier survival curves of the 2 risk groups. We cannot reject the null hypothesis that there was no difference between the risk groups, stratified by thresholding the handcrafted signature, and the patient OS (X-tile: P = .31; hazard ratio [HR], 1.44; 95% CI, 0.71-2.91; median: P = .20; HR, 1.51; 95% CI, 0.80-2.87). On the other hand, thresholds on the DL-based signature resulted in significant stratification of patients into 2 prognostically distinct groups (X-tile: P < .01; HR, 2.80; 95% CI, 1.26-6.24; median: P = .02; HR, 2.16; 95% CI, 1.12-4.17).
Fig. 4.
Kaplan-Meier survival curves of the testing patients. Patients were stratified into 2 risk groups based on thresholds of the handcrafted signature or the deep learning (DL)–based signature. The top row shows the stratification based on the threshold generated by X-tile software, and the bottom row shows the stratification based on the median signature value. P values of the corresponding log-rank tests are shown.
Discussion
In this paper, we proposed an automatic workflow for GBM survival prediction based on 4 preoperative MR images. The VGG-Seg was proposed and trained using 105 patients with glioma for automatically generating GBM contours from 4 MR images. The trained VGG-Seg was applied to 163 patients with GBM to generate their autosegmented tumor contours for survival analysis. We extracted handcrafted and DL-based radiomic features from the MR images using the autosegmented contours for these patients. Two Cox regression models were trained using the extracted features to construct the handcrafted and DL-based signatures for survival prediction.
The handcrafted signature achieved a C-index of 0.64, while the DL-based signature achieved a C-index of 0.67. The DL-based signature achieved numerically higher AUCs, evaluated at the OS of 300 days and 450 days, than the handcrafted signature. Additionally, the DL-based signature, unlike the handcrafted signature, resulted in prognostically distinct groups using either X-tile generated or median threshold. Shboul et al12 did not report the C-index but did report an accuracy of 0.52 in classifying patients with GBM into 3 survival outcome groups. However, DL-based radiomic features were not investigated in this study. It is also difficult to know whether significant patient stratification was achieved for testing patients with GBM in this study because log-rank tests were not conducted.
The VGG-Seg achieved accurate automatic GBM segmentation, with a mean Dice coefficient of 0.86 for the 163 patients with GBM. A study showed that the mean Dice coefficient between the whole tumor contours drawn by 2 experts based on multimodal MR images was 0.86.27 Recently, many studies have proposed novel 3D CNN architectures for improving glioma segmentation accuracy.28, 29, 30 The goal of this study was not to benchmark the best segmentation model but to develop an automatic workflow that can achieve accurate GBM survival prediction. Other automatic segmentation methods can be integrated into the proposed workflow but were not explored within the scope of this study. Potential future work includes selecting the best segmentation model and investigating whether more accurate autosegmented contours may result in a better survival prediction model.
We included 75 patients with LGG for training the VGG-Seg because we found that the VGG-Seg trained with both 75 patients with LGG and 30 patients with GBM achieved better performance than the VGG-Seg trained with 30 patients with GBM alone. This is expected, as LGG and GBM have a similar appearance in MR images. The VGG-Seg could generate 3 tumor subregion labels. However, the accuracy of segmenting subregion labels using the VGG-Seg was low, with the mean Dice coefficients of the tumor subregions smaller than 0.75. Hence, we decided to use the whole tumor contours for feature extraction.
Our study has several limitations. First, the number of patients is limited so we only investigated the transfer learning method for survival prediction. A CNN trained from scratch for survival prediction could directly learn useful features from MR images. However, it could be easily overfitted and hence require more patient data to achieve robust performance. Other methods like training an autoencoder for feature extraction would also be valuable to explore. Second, the information provided by the MR images may be limited and not powerful enough for achieving more accurate models. Future work could be done to include genomic features and investigate whether the combination of genomic and radiomic features could improve prediction performance. Third, we did not consider the treatment status of patients due to data scarcity. Integrating treatment status may help achieve better prediction performance and is worthy of investigation in the future.
Conclusions
We proposed an automatic workflow for GBM survival prediction based on 4 preoperative MR images. The proposed VGG-Seg generated accurate GBM contours. Our study showed that radiomic features, extracted from the autosegmented contours generated by the VGG-Seg, were associated with GBM OS. The DL-based radiomic signature resulted in a numerically higher C-index than the handcrafted signature and helped achieve significant patient stratification. Our automatic workflow based on DL-based radiomic features demonstrated the potential of improving patient stratification and survival prediction in patients with GBM.
Acknowledgments
The authors would like to acknowledge the BraTS 2018 challenge organizing committee.
Footnotes
Sources of support: This research is partially funded by Varian Master Research Grant.
Disclosures: Y.Y. reports personal fees from ViewRay, Inc, outside the submitted work; D.R. reports grants from Varian Medical Systems, Inc, during the conduct of the study; J.H.L. reports grants from Varian Medical Systems, Inc, during the conduct of the study.
Research data can be requested from https://ipp.cbica.upenn.edu/.
Supplementary material associated with this article can be found in the online version at https://doi.org/10.1016/j.adro.2021.100746.
Appendix. Supplementary materials
References
- 1.Louis DN, Perry A, Reifenberger G. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016;3:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 2.Ostrom QT, Bauchet L, Davis FG. The epidemiology of glioma in adults: A “state of the science” review. Neuro Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Domingo-Musibay E, Galanis E. What next for newly diagnosed glioblastoma? Futur Oncol. 2015;11:3273–3283. doi: 10.2217/fon.15.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tamimi AF, Juweid M. Epidemiology and outcome of glioblastoma. Glioblastoma. 2017:143–153. [PubMed] [Google Scholar]
- 5.Nicolasjilwan M, Hu Y, Yan C. Addition of MR imaging features and genetic biomarkers strengthens glioblastoma survival prediction in TCGA patients. J Neuroradiol. 2015;42:212–221. doi: 10.1016/j.neurad.2014.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanghani P, Ang BT, King NKK, Ren H. Regression based overall survival prediction of glioblastoma multiforme patients using a single discovery cohort of multi-institutional multi-channel MR images. Med Biol Eng Comput. 2019;57:1683–1691. doi: 10.1007/s11517-019-01986-z. [DOI] [PubMed] [Google Scholar]
- 7.Lao J, Chen Y, Li Z-C. A deep learning-based radiomics model for prediction of survival in glioblastoma multiforme. Sci Rep. 2017;7:10353. doi: 10.1038/s41598-017-10649-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavic M, Bogowicz M, Würms X. Influence of inter-observer delineation variability on radiomics stability in different tumor sites. Acta Oncol (Madr) 2018;57:1070–1074. doi: 10.1080/0284186X.2018.1445283. [DOI] [PubMed] [Google Scholar]
- 9.Fiset S, Welch ML, Weiss J. Repeatability and reproducibility of MRI-based radiomic features in cervical cancer. Radiother Oncol. 2019;135:107–114. doi: 10.1016/j.radonc.2019.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Ronneberger O, Fischer P, Brox T. Vol. 9351. Springer Verlag; Switzerland: 2015. U-net: Convolutional networks for biomedical image segmentation; pp. 234–241. (Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics)). [DOI] [Google Scholar]
- 11.Long J, Shelhamer E, Darrell T. Fully convolutional networks for semantic segmentation. Available at: https://people.eecs.berkeley.edu/~jonlong/long_shelhamer_fcn.pdf. Accessed September 11, 2018. [DOI] [PubMed]
- 12.Shboul ZA, Alam M, Vidyaratne L, Pei L, Elbakary MI, Iftekharuddin KM. Feature-guided deep radiomics for glioblastoma patient survival prediction. Front Neurosci. 2019;13:966. doi: 10.3389/fnins.2019.00966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antropova N, Huynh BQ, Giger ML. A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets. Med Phys. 2017;44:5162–5171. doi: 10.1002/mp.12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afshar P, Mohammadi A, Plataniotis KN, Oikonomou A, Benali H. From handcrafted to deep-learning-based cancer radiomics: Challenges and opportunities. IEEE Signal Process Mag. 2019;36:132–160. [Google Scholar]
- 15.Fu J, Zhong X, Li N. Deep learning-based radiomic features for improving neoadjuvant chemoradiation response prediction in locally advanced rectal cancer. Phys Med Biol. 2020;65 doi: 10.1088/1361-6560/ab7970. [DOI] [PubMed] [Google Scholar]
- 16.Menze BH, Jakab A, Bauer S. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) IEEE Trans Med Imaging. 2015;34:1993–2024. doi: 10.1109/TMI.2014.2377694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bakas S, Akbari H, Sotiras A. Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data. 2017;4 doi: 10.1038/sdata.2017.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakas S, Reyes M, Jakab A, et al. Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. Available at:http://arxiv.org/abs/1811.02629. Accessed October 23, 2019.
- 19.Tustison NJ, Avants BB, Cook PA. N4ITK: Improved N3 bias correction. IEEE Trans Med Imaging. 2010;29:1310–1320. doi: 10.1109/TMI.2010.2046908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv Prepr arXiv14091556. 2014.
- 21.Ulyanov D, Vedaldi A, Lempitsky V. Proceedings - 30th IEEE Conference on Computer Vision and Pattern Recognition, CVPR 2017. 2017-January. Institute of Electrical and Electronics Engineers Inc.; 2017. Improved texture networks: Maximizing quality and diversity in feed-forward stylization and texture synthesis; pp. 4105–4113. [Google Scholar]
- 22.He K, Zhang X, Ren S, Sun J. Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition. 2016-December. IEEE Computer Society; 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 23.Kingma D, Ba J. Adam: A method for stochastic optimization. arXiv Prepr arXiv14126980. 2014.
- 24.van Griethuysen JJM, Fedorov A, Parmar C. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77:e104–e107. doi: 10.1158/0008-5472.CAN-17-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng J, Dong W, Socher R, Li L-J, Kai L, Li F-F. ImageNet: A large-scale hierarchical image database. IEEE. 2010:248–255. [Google Scholar]
- 26.Camp RL, Dolled-Filhart M, Rimm DL. X-tile: A new bio-informatics tool for biomarker assessment and outcome-based cut-point optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. [DOI] [PubMed] [Google Scholar]
- 27.Porz N, Bauer S, Pica A. Multi-modal glioblastoma segmentation: Man versus machine. PLoS One. 2014;9:e96873. doi: 10.1371/journal.pone.0096873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ghosal P, Reddy S, Sai C, Pandey V, Chakraborty J, Nandi D. TENCON 2019 - 2019 IEEE Region 10 Conference (TENCON) 2019. A deep adaptive convolutional network for brain tumor segmentation from multimodal MR images; pp. 1065–1070. [Google Scholar]
- 29.Myronenko A. Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics) Vol. 11384. Springer Verlag; Switzerland: 2019. 3D MRI brain tumor segmentation using autoencoder regularization; pp. 311–320. LNCS. [Google Scholar]
- 30.Fu J, Singhrao K, Qi XS, Yang Y, Ruan D, Lewis JH. Three-dimensional multipath DenseNet for improving automatic segmentation of glioblastoma on pre-operative multimodal MR images. Med Phys. 2021;418:2859–2866. doi: 10.1002/mp.14800. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.