Abstract
In a previous study, we identified a 117 base severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) sequence in the human genome with 94.6% identity. The sequence was in chromosome 1p within an intronic region of the netrin G1 (NTNG1) gene. The sequence matched a sequence in the SARS-CoV-2 Orf1b gene in non-structural protein 14 (NSP14), which is an exonuclease and NSP15, an endoribonuclease. In the current study we compared the human genome with other viral genomes to determine some of the characteristics of human sequences found in the latter. Most of the viruses had human sequences, but they were short. Hepatitis A and St Louis encephalitis had human sequences that were longer than the 117 base SARS-Cov-2 sequence, but they were in non-coding regions of the human genome. The SARS-Cov-2 sequence was the only long sequence found in a human gene (NTNG1). The related coronaviruses SARS-Cov had a 41 BP human sequence on chromosome 3 that was not part of a human gene, and MERS had no human sequence. The 117 base SARS-CoV-2 human sequence is relatively close to the viral spike sequence, separated only by NSP16, a 904 base sequence. The mechanism for SARS-CoV-2 infection is the binding of the virus spike protein to the membrane-bound form of angiotensin-converting enzyme 2 (ACE2) and internalization of the complex by the host cell. We have no explanation for the NSP14 and NSP15 SARS-Cov-2 sequences we observed here or how they might relate to infectiousness. Further studies are warranted.
Keywords: COVID-19, ORF1b gene, NTNG1 gene, the UCSC Genome Browser
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a positive-sense single-stranded RNA virus (1). In January 2020, SARS-CoV-2 was identified as the cause of an outbreak of viral pneumonia in Wuhan, PR China. The disease, COVID-19, quickly spread worldwide. In the first three months after COVID-19 appeared nearly 1 million people were infected and 50,000 died. The genome of SARS-CoV-2 is less than 30,000 bases, whereas the human genome is over 3 billion. SARS-CoV-2 genes have been identified for 29 proteins, which carry out a range of functions from making copies of the virus to suppressing the body’s immune responses.
SARS-CoV-2 is related to two other coronaviruses, Middle East respiratory syndrome (MERS)-CoV and SARS-CoV. Both are much less infectious than SARS-CoV-2. MERS is a viral respiratory disease that was first reported in Saudi Arabia in September 2012 and has since spread to 27 countries. Humans infected with MERS coronavirus (MERS-CoV) develop severe acute respiratory illness, including fever, cough, and shortness of breath. From its emergence through January 2020, the World Health Organization (WHO) has confirmed 2,519 MERS cases and 866 deaths (about 1 in 3). Among all reported human cases, about 80% have occurred in Saudi Arabia. Only two people in the United States tested positive for MERS-CoV, both of whom recovered. They were healthcare providers who lived in Saudi Arabia, where they likely were infected before traveling to the U.S., according to the US Centers for Disease Control and Prevention (CDC).
SARS-CoV can also cause a severe viral respiratory illness. SARS was first identified in Asia in February 2003, though cases were subsequently traced to November 2002. SARS rapidly spread to 26 countries before being contained after about four months. More than 8,000 people contracted SARS and 774 died. Since 2004, there have been no reported SARS cases. Research evidence suggests that SARS-CoV and MERS-CoV originated in bats, and it is likely that SARS-CoV-2 did as well. SARS-CoV spread from infected civets to people, while MERS-CoV spread from infected dromedary camels to people.
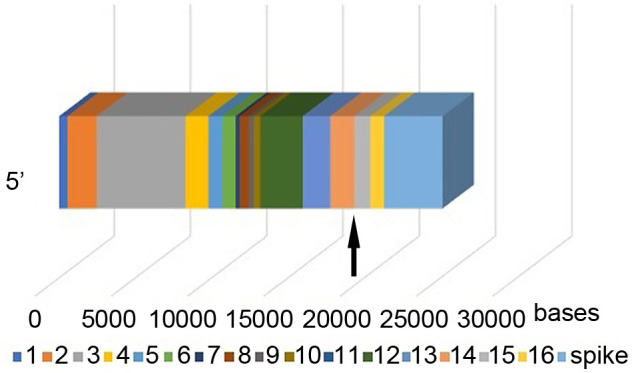
SARS-CoV strains have 2 Orf1 (open reading frame) genes, Orf1a and Orf1b. The 16 Orf1ab non-structural proteins (NSPs) are directly involved in viral replication. 5 of the NSPs, NSP12 – NSP16, are on Orf1b (Figure 1). In a previous study, we identified a 117 base SARS-CoV-2 sequence in the human genome with 94.6% identity. The sequence was in chromosome 1p within an intronic region of the netrin G1 (NTNG1) gene. The sequence matched a sequence in the SARS-CoV-2 Orf1b gene (2). In the current study we compared the human genome with other viral genomes to determine some of the characteristics of human sequences found in the latter.
Figure 1. Orf1ab genome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), showing the 16 non-structural proteins (NSPs) and the viral spike. The human sequence is within NSP 14 and NSP 15 (arrow). The human sequence is separated from the spike by NSP 16, a small sequence of 904 bases. The mechanism for SARS-CoV-2 infection is the binding of the virus spike protein to the membrane-bound form of angiotensin-converting enzyme 2 (ACE2) and internalization of the complex by the host cell. [Figure originally appeared in (2)].
We utilized the UCSC Genome Browser, an on-line genome browser at the University of California, Santa Cruz (UCSC). The browser is an interactive website offering access to genome sequence data from a variety of vertebrate and invertebrate species and major model organisms, integrated with a large collection of aligned annotations. The Genome Browser Database, browsing tools, downloadable data files, and documentation are all accessible on the UCSC Genome Bioinformatics website (https://genome.ucsc.edu) (3).
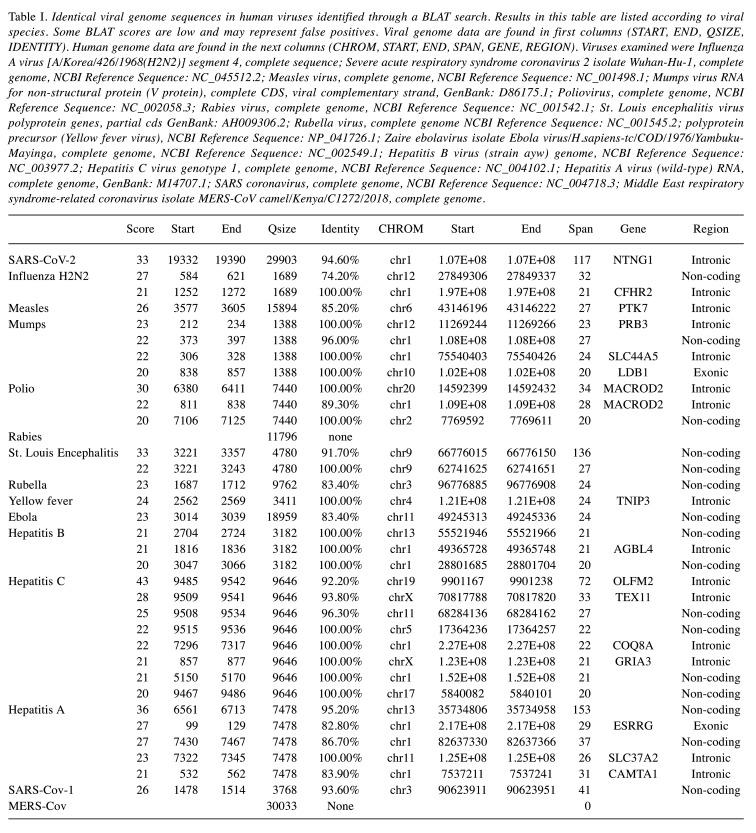
To compare viral genomes to the human genome we used BLAT, the Blast-Like Alignment Tool of the UCSC Genome Browser (3). BLAT can align a user sequence of 25 bases or more to the genome. Because some level of mismatch is tolerated, cross-species alignments may be performed provided the species have not diverged too far from each other; this capability previously allowed comparison of the Mouse Mammary Tumor Virus genome to the human genome (4). BLAT calculates a percent identity score to indicate differences between sequences without a perfect match (i.e. without 100% identity). The differences include mismatches and gaps (5). A BLAT search returns a list of results that are ordered in decreasing order based on the score (5). The results are presented in Table I. Most of the viruses had human sequences, but they were short. For example, three polio sequences were 34 bases, 24 bases, and 20 bases (6). Hepatitis A and St Louis encephalitis had human sequences that were longer than the 117 base SARS-Cov-2 sequence, but they were in non-coding regions of the human genome. The SARS-Cov-2 sequence was the only long sequence found in a human gene (NTNG1). Human NTNG1 encodes a preproprotein that is processed into a secreted protein containing eukaryotic growth factor (EGF)-like domains. This protein acts to guide axon growth during neuronal development. Polymorphisms in this gene may be associated with schizophrenia (7). The related coronaviruses SARS-Cov had a 41 BP human sequence on chromosome 3 that was not part of a human gene, and MERS had no human sequence.
Table I. Identical viral genome sequences in human viruses identified through a BLAT search. Results in this table are listed according to viral species. Some BLAT scores are low and may represent false positives. Viral genome data are found in first columns (START, END, QSIZE, IDENTITY). Human genome data are found in the next columns (CHROM, START, END, SPAN, GENE, REGION). Viruses examined were Influenza A virus [A/Korea/426/1968(H2N2)] segment 4, complete sequence; Severe acute respiratory syndrome coronavirus 2 isolate Wuhan-Hu-1, complete genome, NCBI Reference Sequence: NC_045512.2; Measles virus, complete genome, NCBI Reference Sequence: NC_001498.1; Mumps virus RNA for non-structural protein (V protein), complete CDS, viral complementary strand, GenBank: D86175.1; Poliovirus, complete genome, NCBI Reference Sequence: NC_002058.3; Rabies virus, complete genome, NCBI Reference Sequence: NC_001542.1; St. Louis encephalitis virus polyprotein genes, partial cds GenBank: AH009306.2; Rubella virus, complete genome NCBI Reference Sequence: NC_001545.2; polyprotein precursor (Yellow fever virus), NCBI Reference Sequence: NP_041726.1; Zaire ebolavirus isolate Ebola virus/H.sapiens-tc/COD/1976/Yambuku-Mayinga, complete genome, NCBI Reference Sequence: NC_002549.1; Hepatitis B virus (strain ayw) genome, NCBI Reference Sequence: NC_003977.2; Hepatitis C virus genotype 1, complete genome, NCBI Reference Sequence: NC_004102.1; Hepatitis A virus (wild-type) RNA, complete genome, GenBank: M14707.1; SARS coronavirus, complete genome, NCBI Reference Sequence: NC_004718.3; Middle East respiratory syndrome-related coronavirus isolate MERS-CoV camel/Kenya/C1272/2018, complete genome.
Eight percent of DNA in the human genome comes from human endogenous retroviruses (HERV), and some human diseases have been attributed to this DNA. HERV sequences have occasionally been adapted by the human body to serve a useful purpose, such as in the placenta, where they may safeguard fetal-maternal tolerance (8). However, MERS, SARS-CoV, and SARS-CoV-2 are not retroviruses. Short segments of non-retroviral genomes have been found within the human genome. We are unaware of such a long non-retroviral sequence in the human genome.
The SARS-CoV-2 human sequence lies within the non-structural protein 14 (NSP14), an exonuclease (9) and non-structural protein 15 (NSP15), an endoribonuclease (10). As NSP12 duplicates the coronavirus genome, it sometimes adds an incorrect base to the new copy. NSP14 cuts out these errors, so that the correct base can be added instead. NSP15 protein cuts residual virus RNA segments to evade the infected cell’s antiviral defenses.
The 117 base SARS-CoV-2 human sequence is quite close to the viral spike sequence, separated only by NSP16, a 904 base sequence (Figure 1). Human cells have antiviral proteins that identify viral RNA and shred it. NSP16 protein works with NSP10 to camouflage the viral genes and protect them. The mechanism for SARS-CoV-2 infection is the binding of the virus spike protein to the membrane-bound form of angiotensin-converting enzyme 2 (ACE2) and internalization of the complex by the host cell (11).
We have no explanation for the NSP14-NSP15-SARS-Cov-2 sequence we observed here or how it might relate to infectiousness. Further studies are warranted.
Conflicts of Interest
There are no conflicts of interest.
Authors’ Contributions
Dr. Lehrer and Dr. Rheinstein contributed equally to the conception, data analysis, and writing of this article.
References
- 1.Khan S, Siddique R, Shereen MA, Ali A, Liu J, Bai Q, Bashir N, Xue M. The emergence of a novel coronavirus (Sars-CoV-2), their biology and therapeutic options. J Clin Microbiol. 2020 doi: 10.1128/JCM.00187-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lehrer S, Rheinstein P. Sars-CoV-2 orf1b gene sequence in the ntng1 gene on human chromosome 1. In Vivo. 2020;34(3) doi: 10.21873/invivo.11953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuhn RM, Haussler D, Kent WJ. The ucsc genome browser and associated tools. Brief Bioinform. 2013;14(2):144–161. doi: 10.1093/bib/bbs038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lehrer S, Rheinstein PH. Mouse mammary tumor viral env sequences are not present in the human genome but are present in breast tumors and normal breast tissues. Virus Res. 2019;266:43–47. doi: 10.1016/j.virusres.2019.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhagwat M, Young L, Robison RR. Using blat to find sequence similarity in closely related genomes. Curr Protoc Bioinformatics Chapter. 2012;10:Unit10. doi: 10.1002/0471250953.bi1008s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lehrer S, Rheinstein PH. Three poliovirus sequences in the human genome associated with colorectal cancer. Cancer Genomics Proteomics. 2019;16(1):65–70. doi: 10.21873/cgp.20112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilcox JA, Quadri S. Replication of ntng1 association in schizophrenia. Psychiatr Genet. 2014;24(6):266–268. doi: 10.1097/YPG.0000000000000061. [DOI] [PubMed] [Google Scholar]
- 8.Kurth R, Bannert N. Beneficial and detrimental effects of human endogenous retroviruses. Int J Cancer. 2010;126(2):306–314. doi: 10.1002/ijc.24902. [DOI] [PubMed] [Google Scholar]
- 9.Shannon A, Le NT, Selisko B, Eydoux C, Alvarez K, Guillemot JC, Decroly E, Peersen O, Ferron F, Canard B. Remdesivir and sars-cov-2: Structural requirements at both nsp12 rdrp and nsp14 exonuclease active-sites. Antiviral Res. 2020;178:104793. doi: 10.1016/j.antiviral.2020.104793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y, Jedrzejczak R, Maltseva NI, Wilamowski M, Endres M, Godzik A, Michalska K, Joachimiak A. Crystal structure of nsp15 endoribonuclease nendou from sars-cov-2. Protein Sci. 2020 doi: 10.1002/pro.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.South AM, Diz DI, Chappell MC. Covid-19, ace2, and the cardiovascular consequences. Am J Physiol Heart Circ Physiol. 2020;318(5):H1084–H1090. doi: 10.1152/ajpheart.00217.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]