Abstract
COVID-19 global pandemic affects health care and lifestyle worldwide, and its early detection is critical to control cases’ spreading and mortality. The actual leader diagnosis test is the Reverse transcription Polymerase chain reaction (RT-PCR), result times and cost of these tests are high, so other fast and accessible diagnostic tools are needed. Inspired by recent research that correlates the presence of COVID-19 to findings in Chest X-ray images, this papers’ approach uses existing deep learning models (VGG19 and U-Net) to process these images and classify them as positive or negative for COVID-19. The proposed system involves a preprocessing stage with lung segmentation, removing the surroundings which does not offer relevant information for the task and may produce biased results; after this initial stage comes the classification model trained under the transfer learning scheme; and finally, results analysis and interpretation via heat maps visualization. The best models achieved a detection accuracy of COVID-19 around 97%.
Keywords: COVID-19, Deep learning, Transfer learning, X-ray, Segmentation
Graphical abstract
1. Introduction
Coronavirus illness is a disease that comes from Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS). A novel coronavirus, COVID-19, is the infection caused by SARS-CoV-2 (Zhang, 2020). In December 2019, the first COVID-19 cases were reported in Wuhan city, Hubei province, China (Xu et al., 2020). World Health Organization (WHO) declared COVID-19 a pandemic (Ducharme, 2020) on March 11 2021, up to July 13 of 2021 there are 188,404,506 reported cases around the world, which have caused 4,059,220 deaths (Worldometer, 2020).
These diseases cause respiratory problems that can be treated without specialized medicine or equipment. Still, underlying medical issues such as diabetes, cancer, cardiovascular and respiratory illnesses can make this sickness worse (World Health Organization, 2020). Reverse transcription Polymerase chain reaction (RT-PCR), gene sequencing for respiratory or blood samples are now the main methods for COVID-19 detection (Wang et al., 2020). Other studies show that COVID-19 has similar pathologies presented in pneumonic illness, leaving chest pathologies visible in medical images. Research shows RT-PCR correlation with Chest CT (Ai et al., 2020), while others study its correlation with X-ray chest images (Kanne et al., 2020). Typical opacities or attenuation are the most common finding in these images, with ground-glass opacity in around 57% of cases (Kong & Agarwal, 2020). Even though expert radiologists can identify the visual patterns found in these images, considering monetary resources at low-level medical institutions and the ongoing increase of cases, this diagnostic process is quite impractical. Recent research in Artificial Intelligence (AI), especially in Deep Learning approaches, shows how these techniques applied to medical images performed well.
There are only a few large open access datasets of COVID-19 X-ray images; most of the published studies use as a foundation the COVID-19 Image Data Collection (Cohen et al., 2020), which was constructed with images from COVID-19 reports or articles, in collaboration with a radiologist to confirm pathologies in the pictures taken. Past approaches use different strategies to deal with small datasets such as transfer learning, data augmentation or combining different datasets, finding good results in papers as Civit-Masot et al. (2020) using a VGG16 with 86% accuracy; Ozturk et al. (2020) with a Dark Covid Net presents 87% accuracy classifying three classes in which is included Covid; Yoo et al. (2020) used a ResNet18 obtaining a 95% accuracy; Sethy et al. (2020) used a ResNet50 for a 95.33% accuracy, and Minaee et al. (2020) used Squeeze Net for a 95.45% accuracy; Panwar et al. (2020) achieved 97.62% using a nCovnet; Apostolopoulos and Mpesiana (2020) improved the results using a VGG19-MobileNet with a 97.8% accuracy, and finally higher results are found in Jain et al. (2020) using a ResNet101 with 98.95% and Khan et al. (2020) with a 99% accuracy using CoroNet a model based on an Xception.
This paper presents a new approach using existing Deep Learning models. It focuses on enhancing the preprocessing stage to obtain accurate and reliable results classifying COVID-19 from Chest X-ray images. The preprocessing step involves a network to filter the images based on the projection it is (lateral or frontal), some common operations such as normalization, standardization, and resizing to reduce data variability, which may hurt the performance of the classification models, and a segmentation model (U-Net) to extract the lung region which contains the relevant information, and discard the information of the surroundings that can produce misleading results (de Informática, 2020). Following the preprocessing stage comes the classification model (VGG16-19), using the transfer learning scheme that takes advantage of pre-trained weights from a much bigger dataset, such as ImageNet, and helps the training process of the network in performance and time to convergence. It is worth noting that the dataset used for this research is at least ten times bigger than the ones used in previous works. Finally, the visualization of heatmaps for different images provides helpful information about the regions of the images that contribute to the prediction of the network, which in ideal conditions should focus on the appearance of the lungs, backing the importance of lung segmentation in the preprocessing stage. After this section, the paper follows the next order: first, the Methodology applied for these approaches, followed by the experiments and results obtained, a discussion of the products, and lastly the conclusions.
2. Methodology
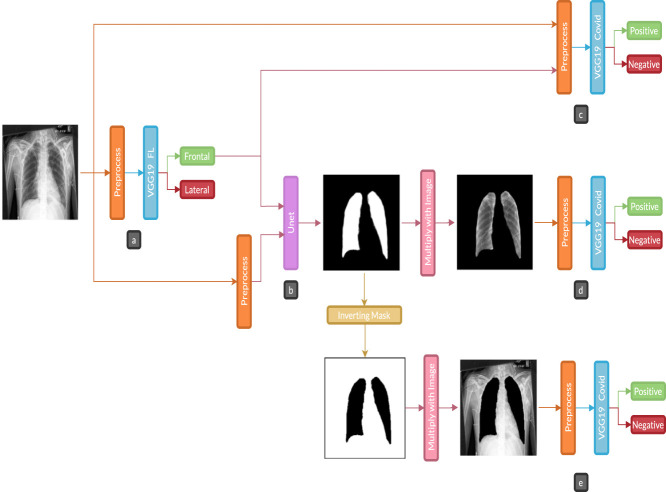
Our methodology consists of three main experiments to evaluate the performance of the models and assess the influence of the different stages of the process. Each experiment follows the workflow shown in Fig. 1. The difference between experiments is the dataset used. In all instances, the same images for COVID-19 positive cases were used. Meanwhile, three different datasets for negative cases were used. In that order, Experiment 1 and 2 consists of evaluating positive vs. negative cases datasets, and Experiment 3 involves Pre-COVID era images (images from -).
Fig. 1.
Experiment diagram: a is the first classification task, b is the lung segmentation task, c is a covid prediction with standard images, d is a covid prediction with only lungs part in the images, and e is covid prediction without lungs in images.. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.1. Datasets
A total of 9 Chest X-ray images datasets were used in different stages:
2.1.1. COVID-19 classification datasets
The following datasets were used to train the classification models: BIMCV-COVID19+(Vayá et al., 2020), BIMCV-COVID- (Medical Imaging Databank of the Valencia region BIMCV, 2020), and Spain Pre-COVID era dataset. These datasets were provided by the Medical Imaging Databank of the Valencia Region (BIMCV). Also, for comparing these processes with other previous works, we use another two databases. For positive cases, the COVID-19 Image Data Collection by Cohen et al. (2020), and negative cases compound by Normal, Viral Pneumonia and Bacterial Pneumonia database by Daniel Kermany et al. (2018), these last databases can be found (COVID-19 X rays, 2020).
2.1.2. Image projection filtering
The images from the COVID-19 datasets have a label corresponding to the image projection: frontal (posteroanterior and anteroposterior) and lateral. Upon manual inspection, several mismatched labels were found, affecting model performance, given the difference between the information available from the two views and that not every patient had both views available. In order to automate the process of filtering the images according to the projection, a classification model was trained on a subset of BIMCV-Padchest dataset (Bustos et al., 2020), with frontal images and lateral images. This model allowed us to filter the COVID-19 datasets efficiently and keep the frontal projection images that offer more information than lateral images.
Finally, to train COVID-19 classification models, the positive dataset (BIMCV-COVID19+), once separated, has 12,802 frontal images. In Experiment 1, images from BIMCV-COVID-dataset were used as negative cases, with 4610 frontal images. BIMCV-COVID — was not organized; also, some of the patients from this dataset were confirmed as COVID-19 positive in a posterior evaluation. Therefore, the models trained on this data could have a biased or unfavorable performance based on dataset size and false positives identified by radiologists. Experiment 2 used a curated version of BIMCV-COVID — for negative patients to avoid this bias, by eliminating patients’ images that correlate with the positive dataset, a total of 1370 images were excluded. Finally, Experiment 3 used a Pre-COVID dataset of images collected from European patients between and . There are 5469 images; this dataset was obtained from BIMCV, but it has not been published yet.
2.1.3. Lung segmentation
Three datasets were used to train the U-Net models for these segmentations: Montgomery dataset (Jaeger et al., 2020) with images, JSTR (Shiraishi et al., 2020) with , and NIH (Tang et al., 2020) with . Despite the apparent small amount of data, the quantity and variability of the images was enough to achieve a useful segmentation model.
2.2. Image separation
For the classification task, data were divided into a train (60%), validation (20%), and test (20%) partitions, following the clinical information to avoid images from the same subject in two different partitions, which could generate bias and overfitting in the models. Accordingly, the data distribution was as follows:
-
•
For the classification model to filter images based on the projection, the data was composed of frontal images, 1,150, , and for train, test, and validation partitions. In contrast, in the same partitions, the separation of lateral images was , , and images.
-
•
For the COVID-19 classification model, the positive cases dataset has 6475 images for train, 3454 for test, and 2873 for the validation set. Meanwhile, for the negative cases datasets, the BIMCV-COVID dataset is divided into 2342, 1228, and 1040 images for train, test, and validation. After the BIMCV-COVID-dataset was curated, there were 1645 images, , and for the train, test, and validation sets. Finally, the Pre-COVID era dataset was divided into 2803 images, 1401, and 1265 for the train, test, and validation sets.
-
•
For the COVID-19 comparison with previous works, the COVID cases dataset has images for train, for the test, and for the validation set. Meanwhile, for the negative cases datasets, Normal images are divided into 809 for training, 270 for test and validation sets. For Pneumonia, there are 2329 images for the train, 777 for the other two groups each.
The image quantity was considerably less for the segmentation task, so creating a test dataset was avoided, leaving the distribution of 80% ( images) for the train set and 20% ( images) for validation data.
2.3. Preprocessing
As the images come from several datasets with different image sizes and acquisition conditions, a preprocessing step is applied to reduce or remove effects on the performance of the models due to data variability. For instance, the BIMCV-Padchest dataset was collected all from the same hospital. In contrast, COVID-19 datasets have images mainly from the Valencian region in Spain, other parts of Spain, and other European countries. On the other hand, the Montgomery and NIH segmentation datasets come from US images, while JSRT is a Japanese dataset. In general, this implies that there were many types of X-ray devices used to take the images, with different technologies and resolutions. The preprocessing layer is shown orange in Fig. 1., it consists of three steps: resize all images to 224 × 224 pixels in one channel (grayscale). In the second step, Eq. (1) shows the normalization of datasets where represents the original images and , the normalized image. Finally, we standardized datasets according to Eq. (2), being the standardized image and the normalized image. When applying standardization to the validation and test sets, the mean and standard deviation (std) from the training set were used to unify the data distribution.
| (1) |
| (2) |
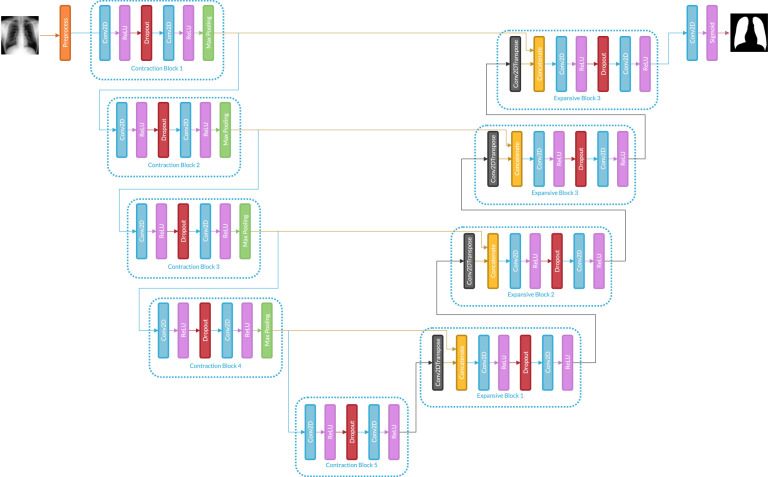
2.4. Segmentation
There are multiple ways to perform image segmentation; this paper uses a Deep Learning model based on U-Net architecture (Ronneberger et al., 2020). Previous articles show that U-Net architecture is accurate for the segmentation of medical images. This kind of model receives the target in the form of an image mask with ones (1) on the reconstruction area and zeros (0) on the rest; consequently, in a production setting, model input is an X-ray chest image, and the output is the predicted mask. Fig. 2. shows the structure of U-Net.
Fig. 2.
U-Net used for segmentation task.
For experimental purposes, we tested three different amounts of filters on convolutional layers to find the optimal for this task. The number of filters in contraction blocks are computed according to Eq. (3), where is the number of the initial filters, corresponds to the number of contraction blocks. Eq. (4) shows the number of filters for each block for Expansion blocks: is the number of filters at the last contraction block, and is the number of the corresponding expansion block. In the expansion block, the transposed convolution layer uses the same number of filters as convolutional layers.
| (3) |
| (4) |
The values used for were 16, 112, and 64, the models will be identified as U-Net 1,2, and 3, respectively.
2.4.1. Hyperparameters
Kernel size in convolutional layers is 3 × 3 with a kernel initialization he-normal and padding same. In Maxpooling layers, the pool size is 2 × 2, the Dropout rate in the first two Expansion and contraction blocks is 0.1, while in three and four of 0.2, and for contraction block five is 0.3. Transposed convolutional layers use kernel size of 2 × 2, strides of 2 × 2, and padding same. Finally, the last convolutional layer uses one filter and a kernel size of 1 × 1.
2.5. Classification
There are two classification tasks in this research, first to separate frontal and lateral Chest X-ray images, and the second one to distinguish COVID-19 positive cases from negative ones. For both tasks, VGG16 and VGG19 (Simonyan & Zisserman, 2020) Deep Learning models were used. The networks were trained using transfer learning (Bravo Ortíz et al., 2021) with pre-trained weights from the Imagenet dataset (Deng et al., 2020). These were trained, using millions of images to predict more than classes. The use of pre-trained models takes advantage of features learned on a larger dataset so that a new model converges faster and performs better on a smaller dataset (Aggarwal, 2020). Pre-trained models come from the Tensorflow+Keras library, these weights come from channels images, and the X-ray data comes in one channel. The following weights were used to convert the RGB values from channels to channel: Red 0.2989, Green 0.5870, and Blue 0.1140.
3. Experiments and results
Regarding Fig. 1, in part a, the dataset is filtered by a VGG19 model to find whether a Chest X-ray image is lateral or frontal. This network will be referred to as VGG19 FL to distinguish it from the other classification model. In part b, lung segmentation was performed with a U-Net model, only with the images that pass as frontal from the previous stage. A VGG19 classification model was used in parts c, d, and e, to predict COVID-19 positive and negative cases. For differentiation with the other VGG19 model, we use the name VGG19 Covid. In variation c, the datasets passed through the classification without lung segmentation. Variation d was using the segmented images, obtained by multiplying the predicted mask from part b and the original images, and passing them through the VGG19 Covid classifier; finally, in variation e, the mask from part b was inverted and applied to the original images to be passed through VGG19 Covid classifier; these three variations allowed us to assess the importance of the segmentation stage, by giving the model full or partial information and analyzing which part of the images contributes to the prediction.
3.1. VGG19 FL
To filter frontal and lateral images, a subset of samples from BIMCV-Padchest and BIMCV-COVID-datasets were labeled manually; experiments were performed using the VGG16 and VGG19 models with pre-trained weights from the Imagenet dataset. Table 1 shows the accuracy for these experiments leaving the best results in VGG19, making that model the one to be used for future parts in the Experiment diagram. Each model was trained for epochs with a batch size of .
Table 1.
Accuracy of part a models.
| Model | Train | Validation | Test |
|---|---|---|---|
| VGG16 | 0.9908 | 0.9803 | 0.9687 |
| VGG19 | 0.9973 | 0.9975 | 0.9937 |
3.2. U-Net
Lungs segmentation was performed with a U-Net model using a combination of three datasets. Three different models were applied, changing filter number in convolutional layers for each U-Net as shown in section 2.4 Segmentation. Table 2 shows Dice and Intersection over Union (IoU) metrics for evaluating segmentation tasks for each model. All networks were trained for epochs with a batch size of .
Table 2.
Dice coefficient an Interception over union (IoU) for U-Net models of part b.
| Model | Dice |
IoU |
||
|---|---|---|---|---|
| Train | Validation | Train | Validation | |
| U-Net 1 | 0.9869 | 0.9645 | 0.9609 | 0.9416 |
| U-Net 2 | 0.9828 | 0.9609 | 0.9520 | 0.9322 |
| U-Net 3 | 0.9867 | 0.9648 | 0.9591 | 0.904 |
Fig. 3 shows an example of image reconstruction for a specific type of image in BIMCV-COVID19+ dataset. Despite U-Net 1 achieving a higher value of IoU, some images are missing a lung portion. Instead, U-Net 3 reconstructs the lungs better in most cases. U-Net 3 was used for all future processes.
Fig. 3.
U-Net mask reconstructions of BIMCV COVID19 + dataset particular images.
3.3. VGG19 covid
For each of the three variations, the implementation for COVID case prediction was by selecting the best model between VGG16 and VGG19. For all of them, the model was trained for epochs with a batch size of .
3.3.1. Experiment 1
Table 3 shows the results of part c in which data has not segmentation applied. Meanwhile, Table 4 shows the results of part d and lung segmentation used over this data. Furthermore, Table 5 shows the results of part e in which segmentation masks were inverted and applied to images. For all Tables above, the models used were a VGG16 and a VGG19.
Table 3.
Accuracy of part c models in Experiment 1.
| Model | Train | Validation | Test |
|---|---|---|---|
| VGG16 | 0.9883 | 0.8898 | 0.8274 |
| VGG19 | 0.9863 | 0.9478 | 0.8996 |
Table 4.
Accuracy of part d models in Experiment 1.
| Model | Train | Validation | Test |
|---|---|---|---|
| VGG16 | 0.9835 | 0.9036 | 0.8767 |
| VGG19 | 0.9628 | 0.9379 | 0.9113 |
Table 5.
Accuracy of part e models in Experiment 1.
| Model | Train | Validation | Test |
|---|---|---|---|
| VGG16 | 0.9872 | 0.9366 | 0.8983 |
| VGG19 | 0.9954 | 0.9639 | 0.9538 |
Table 6 shows accuracy, sensitivity, specificity, and F1 score in the COVID label of parts c,d, and e with a threshold of 0.5.
Table 6.
Performance metrics of parts c, d , and e in COVID-19 label for Experiment 1.
| Part | Accuracy | Sensitivity | Specificity | F1 Score |
|---|---|---|---|---|
| c | 0.939 | 0.972 | 0.883 | 0.965 |
| d | 0.933 | 0.968 | 0.871 | 0.961 |
| e | 0.956 | 0.967 | 0.917 | 0.969 |
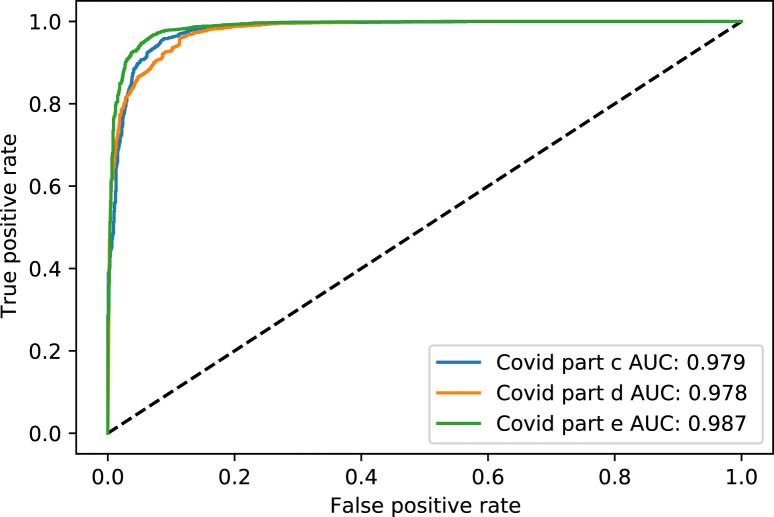
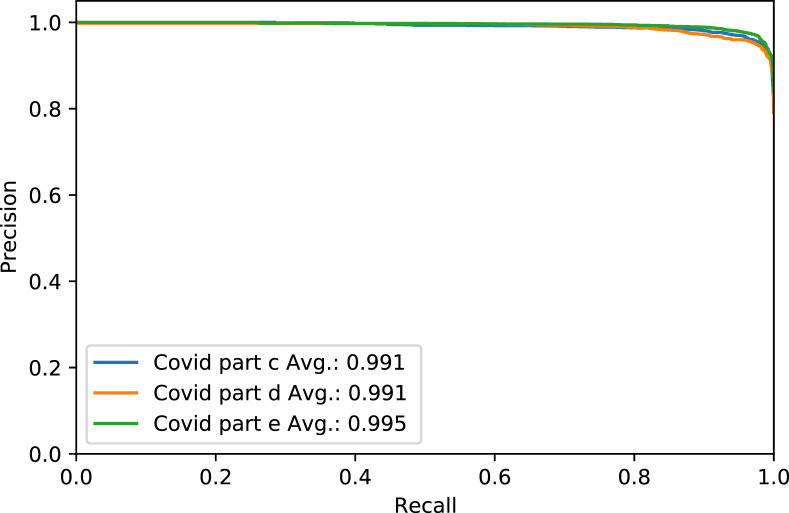
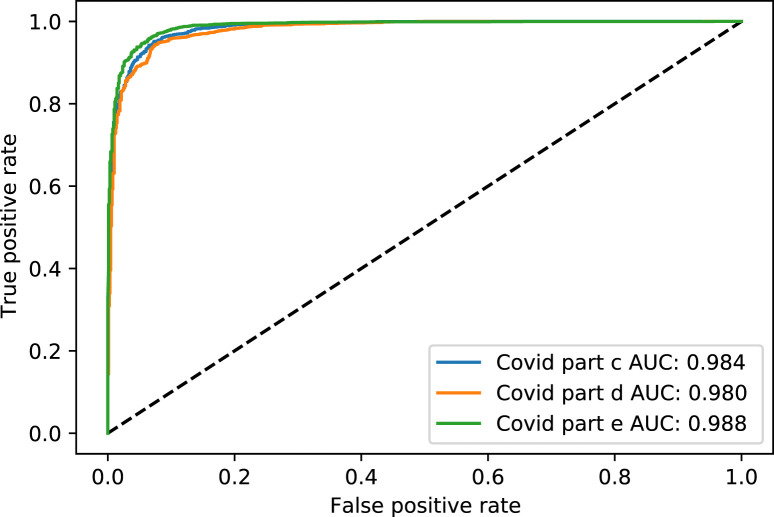
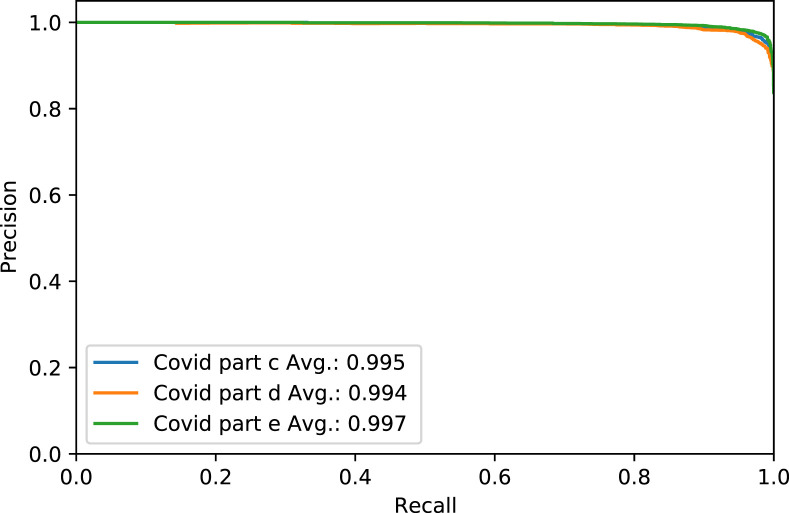
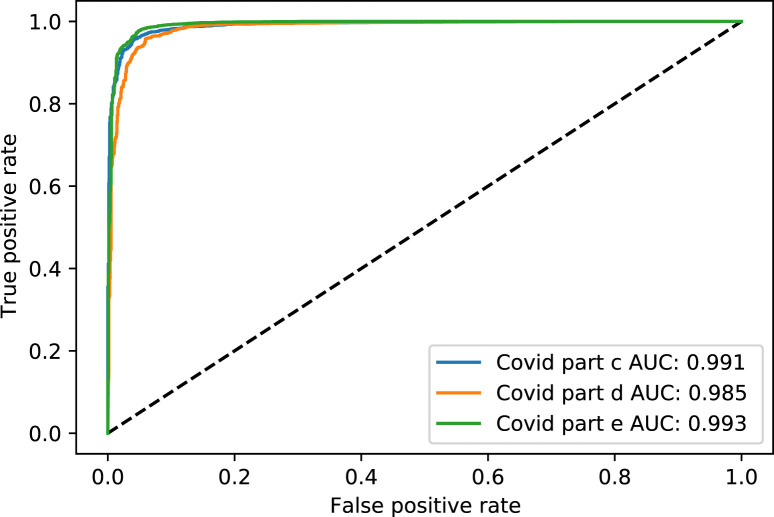
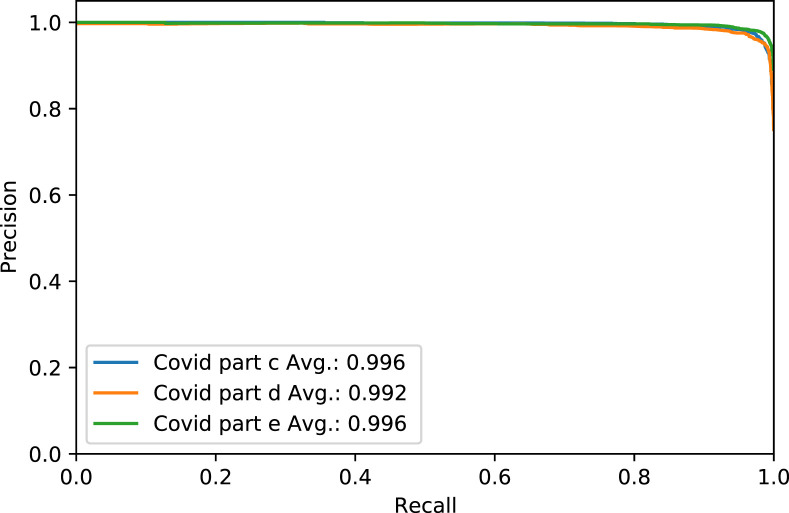
Fig. 4. shows the Receiver Operating Characteristic (ROC) curve of part c, d, and e for the rest of the thresholds, while Fig. 5. shows the same parts’ precision–recall curve for Experiment 1.
Fig. 4.
The ROC curve of COVID-19 test dataset in parts c, d and e, for Experiment 1.
Fig. 5.
The Precision–Recall curve of COVID-19 test dataset in parts c, d and e, for Experiment 1.
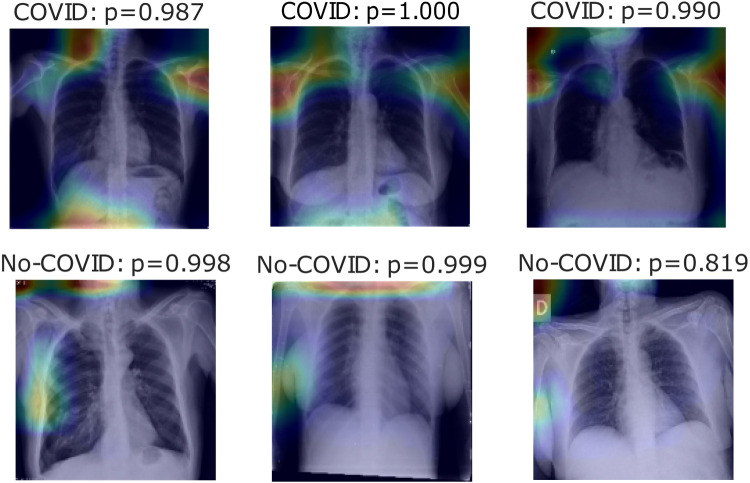
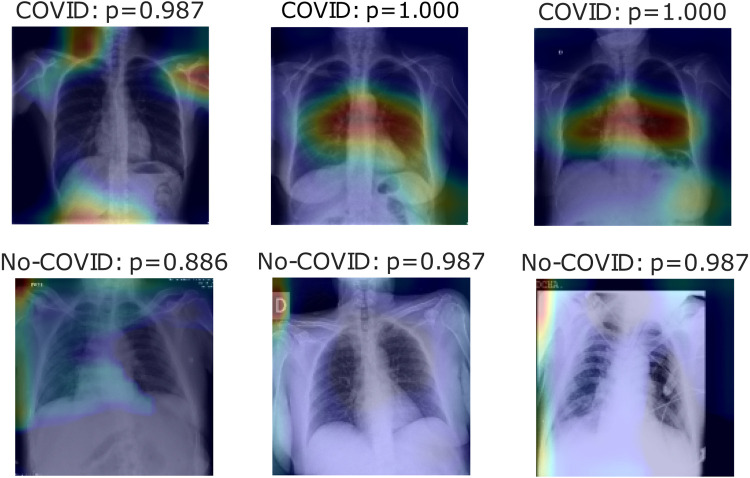
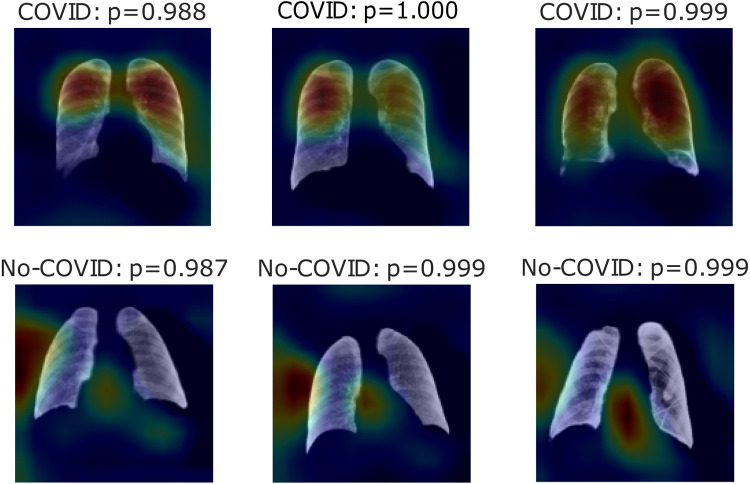
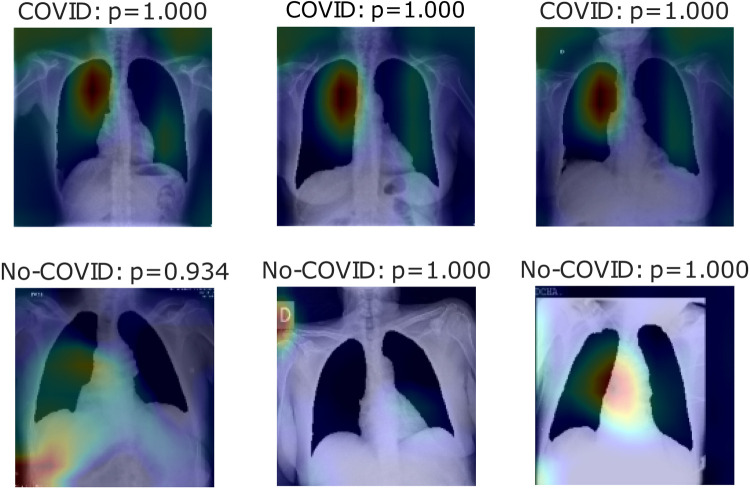
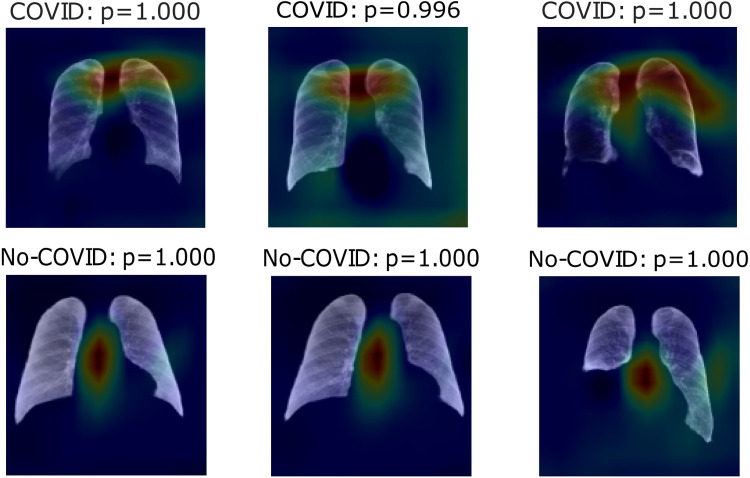
Fig. 6, Fig. 7, Fig. 8 show heatmaps of the last layers for some well predicted positive and negative cases in part c, d, and e, respectively, for Experiment 1.
Fig. 6.
Heatmaps of last layer in some images for part c experiment, for Experiment 1.
Fig. 7.
Heatmaps of last layer in some images for part d experiment, for Experiment 1.
Fig. 8.
Heatmaps of last layer in some images for part e experiment, for Experiment 1.
3.3.2. Experiment 2
Table 7 shows the results of part c. Currently, Table 8 shows the results of part d. Additionally, Table 9 shows the results of part e. For all of them, the models used were a VGG16 and a VGG19.
Table 7.
Accuracy of part c models in Experiment 2.
| Model | Train | Validation | Test |
|---|---|---|---|
| VGG16 | 0.9886 | 0.8970 | 0.8739 |
| VGG19 | 0.9743 | 0.9546 | 0.9241 |
Table 8.
Accuracy of part d models in Experiment 2.
| Model | Train | Validation | Test |
|---|---|---|---|
| VGG16 | 0.9774 | 0.8914 | 0.8705 |
| VGG19 | 0.9669 | 0.9359 | 0.9344 |
Table 9.
Accuracy of part e models in Experiment 2.
| Model | Train | Validation | Test |
|---|---|---|---|
| VGG16 | 0.9774 | 0.8914 | 0.8705 |
| VGG19 | 0.9669 | 0.9359 | 0.9344 |
Table 10 shows COVID label accuracy, sensitivity, specificity, and F1 score for parts c,d, and e with a threshold of 0.5 for Experiment 2.
Table 10.
Performance metrics of parts c, d , and e in COVID-19 label for Experiment 2.
| Part | Accuracy | Sensitivity | Specificity | F1 Score |
|---|---|---|---|---|
| c | 0.942 | 0.982 | 0.858 | 0.973 |
| d | 0.963 | 0.951 | 0.913 | 0.964 |
| e | 0.952 | 0.987 | 0.882 | 0.978 |
Fig. 9. shows the ROC curve of part c, d, and e for the rest of the thresholds. Meantime, Fig. 10. shows the precision–recall curve for the same parts, both figures for Experiment 2.
Fig. 9.
The ROC curve of COVID-19 test dataset in parts c, d and e, for Experiment 2.
Fig. 10.
The Precision–Recall curve of COVID-19 test dataset in parts c, d and e, for Experiment 2.
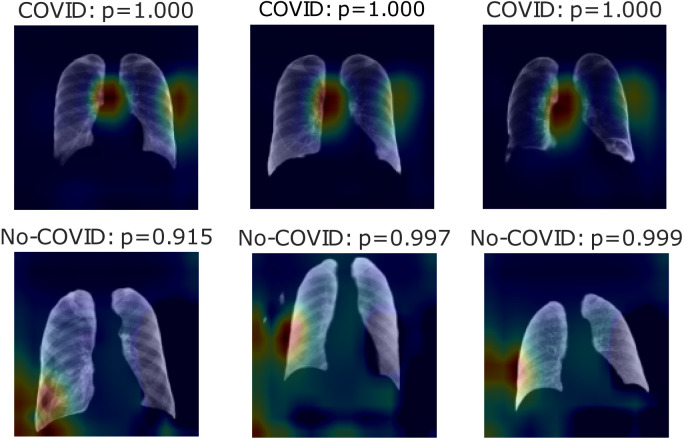
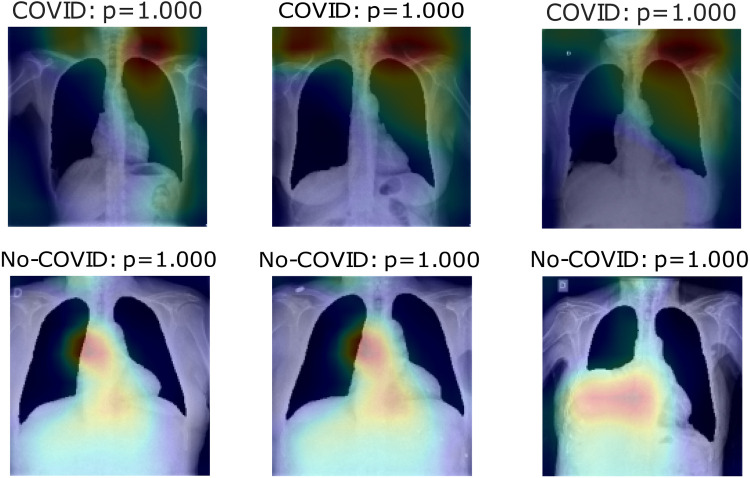
Fig. 11, Fig. 12, Fig. 13 show heatmaps of areas model set for prediction for both cases in parts c, d, and e, respectively, for Experiment 2.
Fig. 11.
Heatmaps of last layer in some images for part c experiment, for Experiment 2.
Fig. 12.
Heatmaps of last layer in some images for part d experiment, for Experiment 2.
Fig. 13.
Heatmaps of last layer in some images for part e experiment, for Experiment 2.
3.3.3. Experiment 3
Table 11, Table 12, Table 13 show the results of parts c, d, and e, respectively, for models VGG16 and VGG19.
Table 11.
Accuracy of part c models in Experiment 3.
| Model | Train | Validation | Test |
|---|---|---|---|
| VGG16 | 0.9927 | 0.8385 | 0.8447 |
| VGG19 | 0.9929 | 0.9645 | 0.8704 |
Table 12.
Accuracy of part d models in Experiment 3.
| Model | Train | Validation | Test |
|---|---|---|---|
| VGG16 | 0.9958 | 0.9376 | 0.9299 |
| VGG19 | 0.9937 | 0.9449 | 0.9363 |
Table 13.
Accuracy of part e models in Experiment 3.
| Model | Train | Validation | Test |
|---|---|---|---|
| VGG16 | 0.9495 | 0.9388 | 0.9118 |
| VGG19 | 0.9725 | 0.9690 | 0.9705 |
Table 14 presents COVID positive label accuracy, sensitivity, specificity, and F1 score metrics for parts c,d, and e with a threshold of 0.5 for Experiment 3.
Table 14.
Performance metrics of parts c, d , and e in COVID-19 label for Experiment 3.
| Part | Accuracy | Sensitivity | Specificity | F1 Score |
|---|---|---|---|---|
| c | 0.962 | 0.973 | 0.935 | 0.973 |
| d | 0.973 | 0.928 | 0.956 | 0.954 |
| e | 0.969 | 0.981 | 0.946 | 0.979 |
Fig. 14. shows the ROC curve of part c, d, and e for the COVID label for the rest of the thresholds. Fig. 15. shows the precision–recall curve for the same parts and case, both for Experiment 3.
Fig. 14.
The ROC curve of COVID-19 test dataset in parts c, d and e, for Experiment 3.
Fig. 15.
The Precision–Recall curve of COVID-19 test dataset in parts c, d and e, for Experiment 3.
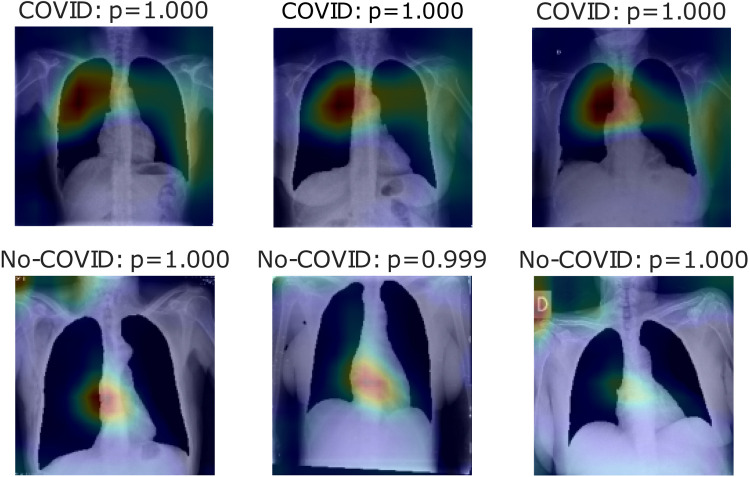
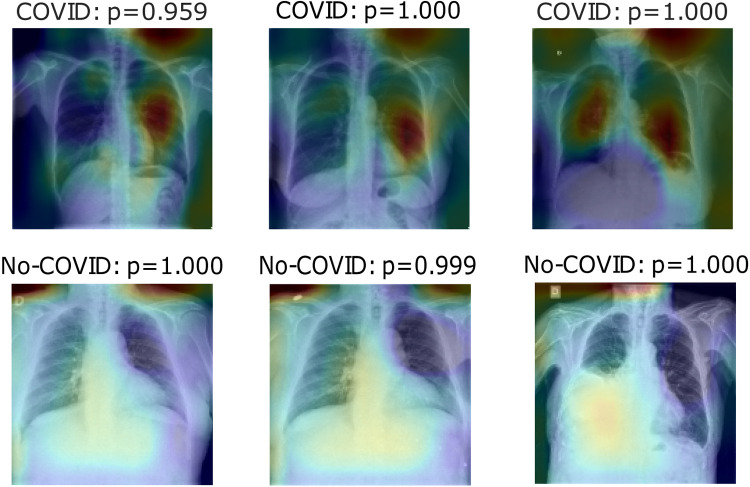
Fig. 16, Fig. 17, Fig. 18 show heatmaps predicted for correct predictions in COVID and No-COVID cases for parts c, d, and e, respectively, in Experiment 3.
Fig. 16.
Heatmaps of last layer in some images for part c experiment, for Experiment 3.
Fig. 17.
Heatmaps of last layer in some images for part d experiment, for Experiment 3.
Fig. 18.
Heatmaps of last layer in some images for part e experiment, for Experiment 3.
3.4. Results of comparison dataset
Table 15 presents COVID positive label accuracy, sensitivity, specificity, and F1 score metrics for parts c,d, and e with a threshold of 0.5 for comparison with previous models.
Table 15.
Performance metrics of parts c, d , and e in COVID-19 label for comparison dataset.
| Part | Accuracy | Sensitivity | Specificity | F1 Score |
|---|---|---|---|---|
| c | 0.991 | 0.995 | 0.986 | 0.966 |
| d | 0.993 | 0.971 | 0.996 | 0.965 |
| e | 0.993 | 0.986 | 0.996 | 0.973 |
3.5. Hardware and software
To develop this project, we used Python 3.8.1. All models were designed with TensorFlow 2.2.0 using the Keras library. We used Google Colaboratory for most of the experiments. In this case, Tensor Processor Unit (TPU) was used when possible; otherwise, we used the Graphic Processor Unit (GPU) depending on the Colaboratory assignation. RAM available in all instances was 12.72 GB. When Colaboratory was insufficient, we used a machine with Ubuntu 20.04 LTS as the operating system and a GeForce RTX Ti GPU, with GB to W. CUDA Version 11.0, and an AMD Ryzen -Core Processor, RAM with GB ( modules of GB with Mhz).
3.6. Future works
For more accurate results, we identified two main future work opportunities, first semantic segmentation of pathologies on lungs, especially the identification of Consolidation and Ground Glass Opacity pathologies. The second part consists of extending COVID datasets to generalize information and overcome the problems of using different image sources.
4. Discussion
For the classification tasks proposed in this research, the better results achieved were using the model VGG19. The first classification task was needed to filter data, as it was a real problem within the datasets, and as their size increases over time, the manual preprocessing becomes unmanageable. More than that is a powerful tool to prevent feeding images from lateral Chest X-ray to the model’s training process. It is appropriate to say that this classification does not avoid glitches from images different from frontal or lateral Chest X-ray ones.
For the previous study, by following experiment order is seen that for Experiment 1, first test accuracy, Table 4 shows better performance than Table 3, meaning segmentation works, but also Table 5 has better accuracy. In this case, lungs are out of images, meaning models use other image characteristics rather than lungs pathologies for classifying. As shown in Table 6, the COVID-19 positive label has higher accuracy for all parts. In general, these models use to mismatch more the negative cases than positive ones; ROC and Precision–Recall curves enhance Table 3, Table 4, Table 5 by showing that using a different threshold, the model predicts better with a continuous leading of part e. Heatmaps found attractively marked zones. Part c images see the model use any feature except the lungs for classifying, making these types not useful for other applications rather than this classification task. Meanwhile, part e commonly uses information in the lungs near surrounding or as COVID images in Fig. 8, also features in the removed lungs areas. Finally, in part d, Fig. 7 identifies similar zones in the lungs for classification. These experiments have correlation cases in the negative dataset related to the positive ones, so the model has difficulties recognizing those cases. Also, these can make mismatch predictions for outside dataset images. To solve those problems, correlation cases were taken out of the negative dataset and left Experiment 2.
Experiment 2 shows better classification results in almost all experiments than Experiment 1, except on part e. Accuracy in the COVID-19 positive label presents the same behavior as the previous experiment. In which the model used to mismatch more No-COVID patients, the Results in ROC and Precision–Recall curves are also enhanced compared to the last experiment. In these cases, the heatmaps show that the COVID images model initially focuses on lung information features. As the top-left image in Fig. 11 presents, these features are not used in all cases. Part e results are similar to the last experiment in which immediate surroundings and missing lung zones. In contrast to Experiment 1 results, part d heatmaps focus on the considerable lungs portion for prediction. As No-COVID images show, the model focuses on out of lung area images because there is no relevant information inside, meaning in these cases, the model sets on lungs pathologies.
Finally, Experiment 3 shows the Pre-COVID era incident in this model; the first three tables show different results from experiments 1 and 2 but with the same tendency. Hence, segmentation enhances classification but taking the lungs out presents even better results. Even accuracy for the COVID label is better in these cases in lung segmentation images. On the other hand, ROC and Precision–Recallcurves have better outcomes for part c and e experiments rather than part e, meaning that with a different threshold, it can perform better. Leaving the heatmaps as valuable information, the first experiment model uses inside the lungs information and outside info to represent bias noise testing in other images. Fig. 18 uses mainly information at the top-right of the pictures to classify COVID cases simultaneously for No-COVID use info near the lungs area. Finally, Fig. 17 shows information inside the lungs is taking into account for type COVID and outside of it for the contrary case.
In general, experiments demonstrate how segmentation helps the model focus on relevant information. As lung characteristics and distribution around datasets are different, the segmentation task provides information related to shape and size to the parts d and e. Hence, the mere fact of performing a segmentation of the lungs highlights relevant details of the images for better classification. Regarding part e, where models show unexpectedly high results, indicates how the information of the images surrounding the lungs alters the result by predicting the correct label for an image without even having the data used to perform a medical diagnosis. It is worth mentioning that the lung segmentation is not always perfect, leaving small lung regions in some cases. The segmentation model used for all experiments, U-Net 3, corresponds to the architecture presented in the original paper.
Finally, Table 16 shows the comparison of our development with other previous works.
Table 16.
Performance metrics of proposed method with other previous works using the comparison database.
| Model | Accuracy | F1 score | Recall | Precisión |
|---|---|---|---|---|
| Proposed Method | 99.06 | 99.06 | 99.06 | 99.07 |
| CoroNet (Khan et al., 2020) | 99 | 98.5 | 99.3 | 98.3 |
| VGG19 (Apostolopoulos & Mpesiana, 2020) | 98.75 | 93.06 | 92.85 | 93.27 |
As shown, the results are better using similar conditions. Meanwhile, it is a remarkable declaration that because the Cohen dataset was growing on the development of previous works, we have more images. The same happens with the Normal and Pneumonia dataset, plus choosing the images randomly. We can say it is not an ideal comparison.
5. Conclusions
This approach shows how existing models can be helpful for multiple tasks, especially if it is considered that the changed U-Net models do not have better performance. Also is shown how image noise can generate bias in the models. Most metrics show the images without segmentation as better for classifying COVID disease. Further analysis shows that even if metrics are better, these models are based on visible pathologies across lungs as clear evidence of COVID, so real accurate models must center on lungs parts for classifying. In this case, segmentation is needed for reliable results by reducing this bias. Transfer learning was vital for the results presented. As shown, classification models using this technique need between 20 and 30 epochs to converge, while segmentation models without transfer learning need about 200. Was presented a series of models to determine COVID-19 Disease in Chest X-ray images with a general accuracy of 92.72%, classifying COVID and NO-COVID images. Meanwhile, Only for the COVID label, the approach has a 95.63% accuracy in the test dataset for a threshold of 0.5. Changing the threshold shows an increase in the accuracy of models up to 98%.
The segmentation task shows a high probability of providing extra information to part d and e in all experiments, culminating in improved results by segmenting lungs and adding information combined with lungs surrounding noise. This noise is associated with cables, captured devices, patient’s age or gender, making images without lungs have more details for classifying in these cases. Either future application using models without lungs could have the highest chances of mislabeling images because of noise bias. Further investigation is required to segment pathologies identified by the expert radiologist to ensure any noise is a factor for bias. It is also essential to highlight that results presented do not necessarily mean the same performance in all datasets. For example, primary datasets come from European patients; other world patients may show minor data capture changes or pathologies, assuming a better classification is needed using worldwide datasets. In addition, separating the datasets by gender will provide more information on the model’s scope, as the soft tissues of the breast may hide parts of the lungs, and it is unknown whether this is considered a bias in the prediction of the model.
CRediT authorship contribution statement
Daniel Arias-Garzón: Data curation, Investigation, Software, Validation, Visualization, Writing – original draft. Jesús Alejandro Alzate-Grisales: Data curation, Investigation, Software, Supervision. Simon Orozco-Arias: Project administration, Supervision. Harold Brayan Arteaga-Arteaga: Formal analysis, Writing – review & editing. Mario Alejandro Bravo-Ortiz: Formal analysis, Writing – review & editing. Alejandro Mora-Rubio: Formal analysis, Writing – review & editing. Jose Manuel Saborit-Torres: Data curation, Conceptualization. Joaquim Ángel Montell Serrano: Data curation, Conceptualization. Maria de la Iglesia Vayá: Conceptualization, Methodology, Project administration, Resources, Supervision. Reinel Tabares-Soto: Conceptualization, Project administration, Methodology.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the Regional Ministry of Health of the Valencia Region, “The Medical Image Bank of the Valencian Community (BIMCV).” Part of the infrastructure used has been co-funded by the European Union through the Operational Program of the European Fund of Regional Development (FEDER) of the Valencian Community 2014-2020.
Funding
This work was supported by the National Ministery of technology and information (Minciencia) Colombia With the identification code 0831-2020.
Footnotes
The code (and data) in this article has been certified as Reproducible by Code Ocean: (https://codeocean.com/). More information on the Reproducibility Badge Initiative is available at https://www.elsevier.com/physical-sciences-and-engineering/computer-science/journals.
References
- Aggarwal C.C. 2020. Neural networks and deep learning; pp. 351–352. [DOI] [Google Scholar]
- Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest CT and RT-PCR testing for coronavirus disease 2019 (COVID-19) in China: A report of 1014 cases. Radiology. 2020;296(2):E32–E40. doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolopoulos I.D., Mpesiana T.A. Covid-19: automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020;43(2):635–640. doi: 10.1007/s13246-020-00865-4. arXiv:2003.11617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medical Imaging Databank of the Valencia region BIMCV . 2020. BIMCV-Covid19 – BIMCV. https://bimcv.cipf.es/bimcv-projects/bimcv-covid19/#1590859488150-148be708-c3f3. [Google Scholar]
- Bravo Ortíz M.A., Arteaga Arteaga H.B., Tabares Soto R., Padilla Buriticá J.I., Orozco-Arias S. Cervical cancer classification using convolutional neural networks, transfer learning and data augmentation. Revista EIA. 2021;18(35):1–12. doi: 10.24050/reia.v18i35.1462. https://revista.eia.edu.co/index.php/reveia/article/view/1462. [DOI] [Google Scholar]
- Bustos A., Pertusa A., Salinas J.M., de la Iglesia-Vayá M. PadChest: A large chest x-ray image dataset with multi-label annotated reports. Medical Image Analysis. 2020;66 doi: 10.1016/j.media.2020.101797. arXiv:1901.07441. [DOI] [PubMed] [Google Scholar]
- Civit-Masot J., Luna-Perejón F., Morales M.D., Civit A. Deep learning system for COVID-19 diagnosis aid using X-ray pulmonary images. Applied Sciences (Switzerland) 2020;10(13) doi: 10.3390/app10134640. [DOI] [Google Scholar]
- Cohen J.P., Morrison P., Dao L. 2020. COVID-19 image data collection. ArXiv, arXiv:2003.11597. [Google Scholar]
- COVID-19 X rays, . (2020). Kaggle. https://www.kaggle.com/andrewmvd/convid19-X-rays.
- Daniel Kermany A.S., Goldbaum M., Cai W., Anthony Lewis M., Xia H., Zhang Correspondence K. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172 doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- Deng J., Dong W., Socher R., Li L.-J., Li K., Fei-Fei L. ImageNet: A large-scale hierarchical image database. CVPR09. 2020;20(11) [Google Scholar]
- Ducharme J. 2020. The WHO just declared coronavirus COVID-19 a pandemic — time. https://time.com/5791661/who-coronavirus-pandemic-declaration/ [Google Scholar]
- de Informática I.T. Early detection in chest images informe de “in search for bias within the dataset”. ITI. 2020 [Google Scholar]
- Jaeger S., Candemir S., Antani S., Wáng Y.-X.J., Lu P.-X., Thoma G. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quantitative Imaging in Medicine and Surgery. 2020;4(6):475–477. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain G., Mittal D., Thakur D., Mittal M.K. A deep learning approach to detect Covid-19 coronavirus with X-Ray images. Biocybernetics and Biomedical Engineering. 2020;40(4):1391–1405. doi: 10.1016/j.bbe.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanne J.P., Little B.P., Chung J.H., Elicker B.M., Ketai L.H. Essentials for radiologists on COVID-19: An update—Radiology scientific expert panel. RSNA. 2020;78(May):1–15. doi: 10.1148/radiol.2020200527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.I., Shah J.L., Bhat M.M. CoroNet: A deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Computer Methods and Programs in Biomedicine. 2020;196 doi: 10.1016/j.cmpb.2020.105581. arXiv:2004.04931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W., Agarwal P.P. Chest imaging appearance of COVID-19 infection. Radiology: Cardiothoracic Imaging. 2020;2(1) doi: 10.1148/ryct.2020200028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minaee S., Kafieh R., Sonka M., Yazdani S., Jamalipour Soufi G. Deep-COVID: Predicting COVID-19 from chest X-ray images using deep transfer learning. Medical Image Analysis. 2020;65 doi: 10.1016/j.media.2020.101794. arXiv:2004.09363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Computers in Biology and Medicine. 2020;121(April) doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar H., Gupta P.K., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of COVID-19 in X-Rays using nCOVnet. Chaos, Solitons & Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger, O., Fischer, P., & Brox, T. (2020). U-net: Convolutional networks for biomedical image segmentation. in: Lecture notes in computer science (including subseries lecture notes in artificial intelligence and lecture notes in bioinformatics), Vol. 9351, pp. 234–241, arXiv:1505.04597, 10.1007/978-3-319-24574-4_28. [DOI]
- Sethy P.K., Behera S.K., Ratha P.K., Biswas P. Detection of coronavirus disease (COVID-19) based on deep features and support vector machine. International Journal of Mathematical, Engineering and Management Sciences. 2020;5(4):643–651. doi: 10.33889/IJMEMS.2020.5.4.052. [DOI] [Google Scholar]
- Shiraishi J., Katsuragawa S., Ikezoe J., Matsumoto T., Kobayashi T., Komatsu K.I., Matsui M., Fujita H., Kodera Y., Doi K. Development of a digital image database for chest radiographs with and without a lung nodule: Receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. American Journal of Roentgenology. 2020;174(1):71–74. doi: 10.2214/ajr.174.1.1740071. [DOI] [PubMed] [Google Scholar]
- Simonyan, K., & Zisserman, A. (2020). Very deep convolutional networks for large-scale image recognition. in: 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings, pp. 1–14, arXiv:1409.1556v6.
- Tang Y.B., Tang Y.X., Xiao J., Summers R.M. 2020. Xlsor: A robust and accurate lung segmentor on chest x-rays using criss-cross attention and customized radiorealistic abnormalities generation; pp. 457–467. ArXiv. [Google Scholar]
- Vayá M.d.l.I., Saborit J.M., Montell J.A., Pertusa A., Bustos A., Cazorla M., Galant J., Barber X., Orozco-Beltrán D., García-García F., Caparrós M., González G., Salinas J.M. 2020. BIMCV Covid-19+: a large annotated dataset of RX and CT images from COVID-19 patients; pp. 1–22. ArXiv, arXiv:2006.01174. [Google Scholar]
- Wang D., Hu B., Hu C., Zhu F., Liu X., Zhang J., Wang B., Xiang H., Cheng Z., Xiong Y., Zhao Y., Li Y., Wang X., Peng Z. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in wuhan, China. JAMA - Journal of the American Medical Association. 2020;323(11):1061–1069. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization . 2020. Coronavirus. https://www.who.int/health-topics/coronavirus#tab=tab_1. [Google Scholar]
- Worldometer . 2020. Coronavirus update (live): 55,912,871 cases and 1,342,598 deaths from COVID-19 virus pandemic - worldometer. https://www.worldometers.info/coronavirus/ [Google Scholar]
- Xu Y., Li X., Zhu B., Liang H., Fang C., Gong Y., Guo Q., Sun X., Zhao D., Shen J., Zhang H., Liu H., Xia H., Tang J., Zhang K., Gong S. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nature Medicine. 2020;26(4):502–505. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S.H., Geng H., Chiu T.L., Yu S.K., Cho D.C., Heo J., Choi M.S., Choi I.H., Cung Van C., Nhung N.V., Min B.J., Lee H. Deep learning-based decision-tree classifier for COVID-19 diagnosis from chest X-ray imaging. Frontiers in Medicine. 2020;7(July):1–8. doi: 10.3389/fmed.2020.00427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W. Imaging changes of severe COVID-19 pneumonia in advanced stage. Intensive Care Medicine. 2020;46(5):841–843. doi: 10.1007/s00134-020-05990-y. doi: 10.1007/s00134-020-05990-y. [DOI] [PMC free article] [PubMed] [Google Scholar]