Abstract
Wastewater-based epidemiology (WBE) was utilized to monitor SARS-CoV-2 RNA in sewage collected from manholes specific to individual student dormitories (dorms) at the University of Arizona in the fall semester of 2020, which led to successful identification and reduction of SARS-CoV-2 transmission events. Positive wastewater samples triggered clinical testing of residents within that dorm; thus, SARS-CoV-2 infected individuals were identified regardless of symptom expression. This current study examined clinical testing data to determine the abundance of asymptomatic versus symptomatic cases in these defined communities. Nasal and nasopharyngeal swab samples processed via antigen and PCR tests indicated that 79.2% of SARS-CoV-2 infections were asymptomatic, and only 20.8% of positive cases reported COVID-19 symptoms at the time of testing. Clinical data was paired with corresponding wastewater virus concentrations, which enabled calculation of viral shedding rates in feces per infected person. Mean shedding rates averaged from positive wastewater samples across all dorms were 7.30 ± 0.67 log10 genome copies per gram of feces (gc/g-feces) based on the N1 gene. Quantification of SARS-CoV-2 fecal shedding rates from infected individuals has been the critical missing component necessary for WBE models to measure and predict SARS-CoV-2 infection prevalence in communities. The findings from this study can be utilized to create models that can be used to inform public health prevention and response actions.
Keywords: SARS-CoV-2, Fecal shedding, Asymptomatic, SARS-CoV-2 infection, Public health response, Wastewater-based epidemiology, COVID-19, Dormitories
Graphical abstract
1. Introduction
SARS-CoV-2 infections may not conclude with current vaccination efforts alone. Some scientists conjecture that the virus’ evolution may require long-term monitoring and more intensive public health measures to control transmission of new variants. Thus, additional public health surveillance strategies should be implemented to prevent and respond to outbreaks; wastewater based epidemiology (WBE) may be one of those tools.
Human wastewater (i.e., sewage) may be among the first indications of aggregated, population-based SARS-CoV-2 infections in a defined community (Medema et al., 2020b), followed by healthcare facility reported number of infections, hospitalizations, and ultimately deaths. WBE is an effective tool that has been demonstrated to monitor concentrations of SARS-CoV-2 RNA in wastewater as an indicator to survey the SARS-CoV-2 infection prevalence at a population level (Medema et al., 2020b). Scientists have adapted results from these surveys to track community infection dynamics (Peccia et al., 2020) and guide targeted public health response actions (Betancourt et al., 2021). Therefore, fecal shedding of SARS-CoV-2 may be utilized as a biomarker to estimate SARS-CoV-2 infections in a population.
Viral shedding estimates of SARS-CoV-2 from infected individuals have strong implications for the effectiveness of WBE to determine disease prevalence and guide public health interventions. Estimations of disease prevalence is one tool which should be paired with other public health interventions including clinical testing, public outreach and education, outbreak prevention and response action, and multidisciplinary collaboration (Lundy et al., 2021). Although SARS-CoV-2 has been demonstrated to shed in the upper respiratory tract, the lower respiratory tract, feces, urine and blood serum (Cevik et al., 2020), WBE leverages RNA shedding of SARS-CoV-2 in feces. Infected individuals shed virus into sewage via feces (Cevik et al., 2020), or urine (Brönimann et al., 2020), both of which can then be used to monitor population-level infection changes. Although SARS-CoV-2 has been isolated in urine of a COVID-19 patient (Sun et al., 2020), the incidence of the virus in urine is reported to be low (Brönimann et al., 2020; Morone et al., 2020); whereas, SARS-CoV-2 particles have been isolated in larger concentration from feces (Wang et al., 2020; Xiao et al., 2020; Zhang et al., 2020). Further, SARS-CoV-2 RNA has been recovered even in the absence of intact virus isolation from stool samples (Wölfel et al., 2020), demonstrating the sensitivity of molecular methods applied for WBE.
During the early stages of the pandemic in the Netherlands, SARS-CoV-2 was detected in sewage six days before initial cases were reported (Medema et al., 2020a). Mulitple jurisdictions around the world have monitored concentrations of SARS-CoV-2 RNA in sewage samples to quantify the total number of infected individuals in the community that excreted the virus into wastewater (Ahmed et al., 2020; Chavarria-Miró et al., 2021; Curtis et al., 2020). More recently, the University of Arizona (UArizona) used WBE, dovetailed with targeted clinical testing, to prevent COVID-19 outbreaks in student dormitories (dorms). In particular, 91 wastewater samples containing SARS-CoV-2 RNA provided early-warning that at least one infected individual was present in the community (Betancourt et al., 2021). The UArizona case study highlights the effectiveness of WBE to detect the presence of SARS-CoV-2 infected individuals in a defined community and to ultimately contain outbreaks by triggering public health response actions. Although WBE has been utilized to raise the alert of SARS-CoV-2 presence with a dichotomous threshold, quantifying the number of SARS-CoV-2 infected individuals is limited in the absence of accurate fecal shedding rate estimates.
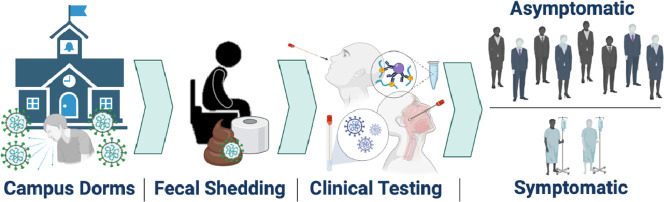
This research aims to estimate SARS-CoV-2 fecal shedding rates in infected university students by aggregating data from positive wastewater samples with numbers of clinically positive, symptomatic and asymptomatic individuals in defined communities. To that end, the Campus Re-entry study at the UArizona in Fall 2020 (Betancourt et al., 2021) provided a unique setting to calculate mean shedding rates for defined communities composed of young adults (18-20 years old). Specifically, 13 student dorms in UArizona served as a case study to test the utility of WBE to monitor SARS-CoV-2 levels in sewage and initiate public health action based upon those results. Positive SARS-CoV-2 detection in wastewater samples from any given dorm led to point prevalence antigen and/or PCR testing for dormitory residents regardless of symptomatic expression. This study determines the proportion of COVID-19 infections that were asymptomatic versus symptomatic and pairs these clinical data with wastewater results to extrapolate the fecal shedding rate of SARS-CoV-2 in infected university students. Findings from this study may act as a benchmark to assess SARS-CoV-2 prevalence in a population and inform public health prevention and response actions.
2. Methods
2.1. Dormitory sites
In total, 13 student dorms (Dorm A–M) at UArizona main campus were monitored throughout the Fall 2020 Semester (August 17–November 20). Data from the final weeks of classes and exams following the Thanksgiving break (November 21–December 17) are not included in this analysis, given students were advised to complete the semester virtually and not return to campus. Each wastewater sample was collected from sewer manholes specific to each dorm's effluent prior to convergence with other sewer pipelines; thus, all wastewater samples were specific to the defined communities living in each dorm. However, two sets of dorms (Dorm E and F; Dorm H and I) required sampling from a single sewer manhole distal from the mixing of wastewater from the two dorms but proximal to convergence with other pipelines. The dorms in each set are juxtaposed and were considered a ‘combined dorm’ for all public health response actions (i.e., clinical testing interventions) and data analysis. Dorms varied in infrastructure and resident occupancy (Table 1 ). Due to the pandemic, UArizona limited dorm room occupancy to a maximum of two persons.
Table 1.
Meta-data for student dormitories.
| Dorm | Fall 2020 occupancya | Capacity | Room type | Bathrooms |
|---|---|---|---|---|
| A | 611 | 722 | Single, double | Community, all-gender |
| B | 342 | 400 | Single, double | Community |
| C | 123 | 181 | Single, double, tripleb | Community |
| D | 623 | 731 | Single, double, suite-style | Suite |
| E | 206 | 300 | Single, double | Community, all-gender |
| F | 56 | 60 | Single | Community |
| G | 231 | 300 | Apartments | In-room (1 per bedroom) |
| H | 195 | 238 | Single, double | Community, all-gender |
| I | 181 | 238 | Single, double | Community, all-gender |
| J | 424 | 482 | Single, double | Community |
| K | 328 | 369 | Double | Community, all-gender |
| L | 76 | 106 | Single, double, tripleb | Community, sink-in-room |
| M | 132 | 152 | Single, double | Community |
Occupancy varied throughout the Fall 2020 semester, so the highest numbers of residents at any given time throughout the semester are shown.
Due to the pandemic, UArizona limited rooms to a maximum of two occupants. No triple-occupant rooms were offered during the time of this analysis.
2.2. Wastewater sampling and analysis
Wastewater samples from each dorm were collected between 9:00 am and 10:30 am, then analyzed for SARS-CoV-2 RNA at least twice per week. One set of dorms was monitored on Monday/Wednesday/Friday (Dorms A, B, C, D, G, L), while a second set was monitored Tuesday/Thursday/Saturday (Dorms E-F, H-I, J, K, M). Grab samples were collected inside sewer manholes with sterile Nalgene® bottles that were affixed to extension poles, which allowed collection at depths of 5 to 15 ft. Sewer pipes were concrete, spherical, and 6 in (15.2 cm) diameter. Velocity (ft3/s) of wastewater within the sewer was measured using a Global Water FP 211 flow probe (Global Water, College Station, TX). Multiple readings of the minimum and maximum flow rates (gallons per minute) over a one-minute period were averaged and recorded daily. Flow rates were calculated using Manning's equation for partially full pipes (Akgiray, 2005). For all wastewater samples, sewage height and depth were estimated to be approximately 5.08 cm, based upon the height of the fluid on the propellor sensor since exact measurements were infeasible inside manholes. The sewage depth was never observed to be above the propellor sensor and a velocity reading could not be recorded if wastewater height was not near the top of the propellor (5.08 cm height). Statistical methods were used to estimate flow rates for samples for which velocity could not be measured due to site obstruction or the flow probe not being available (see Section 2.6).
Grab samples collected at the same time of each sampling day were considered adequate based on a prior multiple sampling event. In that case, multiple samples were collected at a particular site over a 30-min period, and analysis indicated virtually identical virus concentrations (5.49 ± 4.17 log10 gc/L) (Betancourt et al., 2021). This suggests that virus particles disperse upon entering the piping and remain in the sewer for extended periods of time. Samples were stored on ice and travel time to the WEST Center laboratory was under 30 min. Wastewater processing and analysis for SARS-CoV-2 RNA followed procedures previously described (Betancourt et al., 2021). Samples were tested for the virus using the United States (U.S.) Centers for Disease Control and Prevention (CDC) RT-PCR assays that target regions of the nCoV nucleocapsid gene (N1; Table S1) (Research use only kit, Integrated DNA Technologies, Coralville, IA). Real-time PCR for the detection of N1 gene was performed on a LightCycler 480 Instrument II (Roche Diagnostics) with the LightCycler® Multiplex RNA Virus Master (Roche Diagnostics) (Table S2). Detection of N2 gene was not considered, as the assay efficiency (0.65) was unacceptable and did not generate reliable data (Table S2). The average percent recovery (10.9%) was estimated based on 23 matrix spike samples with human coronavirus (HCoV) 229E (data not shown). However, virus concentrations were not adjusted because percent recoveries were unavailable for all 319 study samples.
2.3. Clinical testing data
Positive detection of SARS-CoV-2 RNA (via the N1 gene region) in wastewater acted as the leading indicator for the presence of SARS-CoV-2 infections within the dorm communities. Results were immediately communicated to the UA Task Force and Campus Re-Entry Working Groups, which planned and conducted clinical testing of residents (Betancourt et al., 2021).
Clinical tests for COVID-19 diagnosis were performed via antigen testing from nasal swab samples and RT-PCR from nasopharyngeal swab samples (Betancourt et al., 2021). Analytical performance characteristics for the antigen test were 96% sensitivity and 100% specificity (Sofia SARS Antigen FIA, Quidel, San Diego, CA, USA). Performance of the RT-PCR tests were determined by the University of Arizona Genetics Core for Clinical Services (CDC 2019-nCoV RT-PCR Diagnostic Panel) with a limit of detection at 150 viral copies/reaction, or 30 viral copies/μl of sample.
Clinical testing data was obtained from the University Campus Health Services (CHS) and the Test All Test Smart (TATS) program. Symptomatic and asymptomatic cases in the study were tracked based on location and program where clinical tests were conducted. Individuals with COVID-19 symptoms who sought clinical testing were tested through CHS. Clinical testing of non-self-reporting (i.e., asymptomatic and sub-clinical) students and employees was conducted through the TATS program that runs several testing locations on campus and mobilizes pop-up sites as needed (i.e., targeted clinical testing at dorms with positive wastewater detection of SARS-CoV-2). A negative antigen test at CHS, where patients were exhibiting symptoms, was followed by PCR testing to confirm results; data was de-duplicated in situations when both testing methods were used to confirm test results (Betancourt et al., 2021). According to UArizona public health and safety guidelines, students who tested positive for COVID-19 and completed isolation protocols were exempt from clinical testing for 90 days, which was a duration longer than the remainder of the semester. Students who had not tested positive for COVID-19 in the previous 90 days were required to participate in clincal testing via TATS and CHS programs. The percent of eligible students who agreed to participate in clinical testing was reported (Table S3). In some cases, the percent testing numbers over 100% because some individuals who were not required to undergo clinical testing may have volunteered regardless; however, this occurred infrequently.
To ensure compliance with the Human Subjects Protection Program (HSPP), the use of clinical data was reviewed and approved by a UArizona Institutional Review Board (IRB).
2.4. Alignment of wastewater and clinical data
To estimate the number of SARS-CoV-2 infected individuals who contributed to a single positive wastewater sample, a 6-day range of clinical data was considered. Positive clinical cases from the day before, day-of, and four days after sampling were included in the count of infected individuals contributing to viral shedding. The rationale for this approach is:
-
1)
Residual virus—shed from individuals testing positive for COVID-19 the day prior to sampling—may be detected in wastewater the following day. Previous sampling indicates that virus can persist for extended periods of time and be detected even when an estimated 1000 gal of wastewater has continued to flow through a sewer system (Betancourt et al., 2021).
-
2)
Individuals infected with SARS-CoV-2 may shed virus into wastewater prior to showing symptoms and/or being identified as a clinical case. The viral load of SARS-CoV-2 appears to peak in the upper respiratory tract of infected individuals within the first week of infection (Cevik et al., 2020) and the median incubation period for COVID-19 is estimated to be approximately five days (Lauer et al., 2020).
Following a positive wastewater sample, clinical testing was performed on nearly all residents living in the dorm via the TATS program (Betancourt et al., 2021). Individuals that tested positive were removed from the dorm and transferred to another location for isolation. However, some individuals in the early stages of infection may have tested negative on the day of wastewater testing, and tested positive after subsequently reaching peak viral load of SARS-CoV-2 at a later date. It was also possible that students were shedding virus in feces, but tested negative by TATS within the 24-h period after the positive wastewater signal. Additionally, the full dorm community was not always available on the first day of targeted testing; some were tested a day or more after a positive wastewater detection. Therefore, clinical data for 4 days following positive wastewater samples was included in the count of infected individuals since these individuals likely shed and contributed virus to the wastewater samples for several days.
Although infected individuals can shed virus several weeks after infection (Cevik et al., 2020), the 6-day window for calculating shedding rates was appropriate since wastewater testing was performed at least twice per week and all infected individuals that tested positive within the 3-4 day window between samples were removed into isolation prior to the next sampling event. Also, individuals were assumed to be at peak shedding for the first 6 days in which an infected person could be identified via UArizona WBE protocols.
2.5. Viral shedding rate estimation
The fecal shedding rate of SARS-CoV-2 RNA per gram of feces from an infected individual was enumerated based on known concentrations of viral RNA in positive wastewater samples and the fact that the total numbers of infected individuals contributing to the total virus load in the samples were also known. Equations from previous reports using viral loads in wastewater to estimate the number of infected individuals in a community (Ahmed et al., 2020; Chavarria-Miró et al., 2021; Curtis et al., 2020) were modified so that the fecal shedding rate of viral RNA per infected person could be quantified. The fecal shedding rate (FS) defined as genome copies per gram-feces (gc/g-feces) was calculated as follows:
| (1) |
where VC is the virus concentration (genome copies/L) in the wastewater sample, Q is the flow rate (gpm) of wastewater in the sewer manhole at time of sample collection, f is the conversion factor between gallons and liters, h is the conversion factor between minutes and days, G is the average mass of stool produced per person per day (Curtis et al., 2020; Rose et al., 2015), and I is total number of infected individuals contributing to the wastewater sample based on the 6-day range for clinical data (see Section 2.5). Confidence intervals were calculated based on the standard deviations between sample calculations.
Fecal shedding rates were based on the N1 gene concentration. Overall, the average percent recovery rate was 10.9% based on 23 matrix spike samples. However, wastewater virus concentrations were not adjusted as we did not have percent recoveries for all 319 samples.
2.6. Statistical analysis
Statistical analyses were performed in Microsoft Excel (version 16.47.1, 2021) and RStudio ((RStudio, 2020) RStudio: Integrated Development for R. RStudio, PBC, Boston, MA. http://www.rstudio.com/). Multiple imputation for measurement error (MIME) was utilized to estimate flow rates for missing data when the flow probe was unavailable.
2.7. Sensitivity analyses
Sensitivity analyses were conducted to explore how three variables within the fecal shedding rate estimation equation (Eq. (1)) may have influenced results: 1) depth of wastewater in the pipeline, 2) viral RNA concentration in wastewater (VC), and 3) proportion of infected individuals who shed SARS-CoV-2 in feces. Potential measurement error of sewage depth may have biased actual flow rate estimations (refer to Section 2.2) and subsequent calculations in shedding rates. Flow rates, originally calculated with the assumption of a 5.08 cm depth (29% of pipe filled) were resestimated using 2.54 cm (11% of pipe filled), 3.81 cm (20% of pipe filled), 6.35 cm (39% of pipe filled), 7.62 cm (50% of pipe filled), and 15.24 cm (100% of pipe filled).
Measurement errors in the molecular detection of viral RNA concentrations may be attributed due to several factors (e.g., assay efficiency and inhibition). Mean virus concentrations detected in wastewater were increased and decreased by one log to determine its influence on the calculated mean fecal shedding rate.
Lastly, not all infected individuals may shed SARS-CoV-2 particles in feces. Therefore, fecal shedding rates were calculated based on different proportions (i.e. 50% vs. 100%) of infected individuals who shed viral particles/RNA in feces.
3. Results
3.1. Asymptomatic and symptomatic clinical COVID-19 cases
Asymptomatic and symptomatic cases of COVID-19 were differentiated based on clinical testing program – TATS for asymptomatic and CHS for symptomatic (see Section 2.4). For all dates in the study period, a total of 711 clinical cases were reported among the 13 unique dorms. Clinical data was combined for dorms with converged sewage systems (Dorm E-F and Dorm H-I). Across all dorms, 148 symptomatic cases and 563 asymptomatic cases were reported; thus, 79.2% of SARS-CoV-2 infections were asymptomatic and only 20.8% were symptomatic (Table 2 ).
Table 2.
Clinical positives for COVID-19 in wastewater-monitored dorms, 8/17/20-11/20/20.
| Dorm | Clinical |
Symptomatic (CHS) |
Asymptomatic (TATS)a |
||
|---|---|---|---|---|---|
| Positives | Total | Percentb | Total | Percentb | |
| A | 164 | 35 | 21.3% | 129 | 78.7% |
| B | 92 | 16 | 17.4% | 76 | 82.6% |
| C | 10 | 3 | 30.0% | 7 | 70.0% |
| D | 171 | 34 | 19.9% | 137 | 80.1% |
| E & F | 16 | 8 | 50.0% | 8 | 50.0% |
| G | 2 | 0 | 0.0% | 2 | 100.0% |
| H & I | 74 | 17 | 23.0% | 57 | 77.0% |
| J | 111 | 16 | 14.4% | 95 | 85.6% |
| K | 66 | 15 | 22.7% | 51 | 77.3% |
| L | 0 | 0 | – | 0 | – |
| M | 5 | 4 | 80.0% | 1 | 20.0% |
| Total | 711 | 148 | 20.8% | 563 | 79.2% |
Asymptomatic cases may also include mildly symptomatic individuals that did not self-report.
Percent of total new clinical positive cases of infection.
Examining specific dorm communities, reported clinical cases ranged from zero in Dorm L to 171 in Dorm D (Table 2). Dorms G and M had the fewest reported infections, as well as the highest and lowest asymptomatic rates. With limited data (two infections in Dorm G and five infections in Dorm M), the asymptomatic rate was calculated as 100% for Dorm G and 20% for Dorm M. Combined Dorm E & F was the only dorm to report equal number of symptomatic and asymptomatic cases (Table 2). Seven of the eleven monitored communities had asymptomatic rates between 70 and 85% (Table 2). Two dorms (E & F and M) had asymptomatic rates at 50% or below, while Dorm G was the only to report 100% of new cases as asymptomatic (Table 2).
3.2. Virus fecal shedding estimation
In total, 74 samples in the study period were positive for the SARS-CoV-2 N1 gene and 246 samples resulted in no detection of the virus (Table S3). Clinical and wastewater data were aggregated (see Section 2.5) to calculate the viral shedding rate per infected person (see Section 2.6). One sample was omitted because it did not meet MIQE guidelines (Bustin et al., 2009). Fourteen other samples were omitted due to zero reported cases of infection within the 6-day range for clinical data (Table S3). This situation may have arisen when it was not possible to test all individuals following a positive wastewater detection (Table S3). The majority (68 out of 81) of positive wastewater samples were concordant with new reported cases of infections within this 6-day period. Negative wastewater samples were not included in viral shedding estimations due to no detection of SARS-CoV-2. Negative samples did not trigger a response action to conduct clinical testing on residents; thus, there is limited clinical data for the days that wastewater was negative. Overall, 59 total positive wastewater samples were aligned with clinical data to extrapolate the fecal shedding rates of SARS-CoV-2 N1 gene.
The average N1 shedding rate per infected person was calculated to be 7.30 ± 0.67 log10 gc/g-feces, considering all positive wastewater samples (n = 59) across all dorms (Table 3 ). While there is likely to be shedding rate differences between specific individuals, analysis of a large number of individuals within a community provides a more precise shedding rate. The median was 6.64 log10 gc/g-feces with a full range 5.74 - 8.28 log10 gc/g-feces (Table 3). Within specific defined communities, Dorm K had the highest average shedding rate at 7.73 ± 0.89 (n = 12), while Dorm C had the lowest average at 6.53 ± 0.14 log10 gc/g-feces (n = 2). The widest range of shedding rates was found in Dorm K with a minimum at 5.80 and a maximum at 8.28 log10 gc/g-feces.
Table 3.
Fecal shedding rates of SARS-CoV-2 RNA extrapolated from Uarizona dorm WBE.
| Dorma | Avg. | Stdev | Med. | Min. | Max. | n |
|---|---|---|---|---|---|---|
| A | 6.90 | 0.60 | 6.36 | 5.78 | 7.66 | 13 |
| B | 7.15 | 0.49 | 7.21 | 6.21 | 7.40 | 5 |
| C | 7.27 | 0.14 | 7.26 | 7.16 | 7.35 | 2 |
| E & F | 6.66 | 0.12 | 6.65 | 6.57 | 6.73 | 2 |
| H & I | 7.15 | 0.68 | 6.65 | 5.74 | 7.71 | 12 |
| J | 7.08 | 0.56 | 6.55 | 5.98 | 7.69 | 13 |
| K | 7.73 | 0.89 | 6.68 | 5.80 | 8.28 | 12 |
| Total | 7.30 | 0.67 | 6.64 | 5.74 | 8.28 | 59 |
All values are presented in log10 genome copies per gram of feces (log10 gc/g-feces), except for n (number of samples). Avg., average; Stdev, standard deviation; Med., median; Min., minimum; Max., maximum; n, number of samples.
Shedding rates could not be calculated from Dorms D, L, or M due to no positive wastewater samples associated with a new reported case(s) of infection or clinical positive test.
3.3. Sensivity analysis for virus fecal shedding estimation
Several sensitivity analyses were conducted on equation variables to test the impact of possible measurement error on fecal shedding rates of SARS-CoV-2 particles. Sensitivity analysis showed that altering sewage depth between 11 and 50% of the pipe filled (2.54 cm, 3.81 cm, 5.08 cm, 6.35 cm, and 7.62 cm) did not result in significant changes in the computed shedding rates (Table S4). One exception is when the pipe was completely filled (15.24 cm) (Table S4). Of note, sample could not be collected if the sewage depth was less than 2.54 cm (11% pipe filled) and no instance occurred where the sewage height was observed to be over half the diameter (7.62 cm).
Upon adjusting the viral RNA concentration by one log, fecal shedding rates were proportionally altered (Table S5). Across all dorms, virus RNA in wastewater was detected at 4.77 ± 0.59 log10 gc/L (min. 3.39; max. 6.06), which resulted in a mean 7.30 ± 0.67 log10 gc/g-fecal shedding rate.
Lastly, no significant difference in calculated fecal shedding rates was observed when considering the proportion (50% vs. 100%) of infected individuals who shed viral particles/RNA in feces (Table S6).
4. Discussion
4.1. Estimation of asymptomatic cases as a percent of total cases
The total number of asymptomatic and symptomatic cases of COVID-19 were discerned in this study based on clinical testing programs: TATS for asymptomatic and CHS for symptomatic (see Section 2.4). Of the total 711 cases of COVID-19 that were reported via positive antigen and/or PCR clinical tests, the vast majority (79.2%) were identified through the TATS testing program and were therefore considered asymptomatic (Table S3). However, it is important to note that follow-up data on students was unavailable to distinguish between asymptomatic and pre-symptomatic cases. This high rate of asymptomatic cases may in part be due to the younger and likely healthier population surveyed in this study based upon their university enrollment and dormitory setting. However, findings suggest that the amount of people infected with SARs-CoV-2 in the U.S. may be vastly underestimated, which could have downstream effects for determining the immune population and herd immunity thresholds.
4.2. Viral shedding estimation
Fecal shedding rates of SARS-CoV-2 RNA were estimated from dorm wastewater samples using a modified equation from previous reports (Ahmed et al., 2020; Chavarria-Miró et al., 2021; Curtis et al., 2020). This calculation accounts for the number of infected people within a defined community, the amount of fecal material excreted, the shedding rate of infected individuals, the flow rate of the wastewater, and the viral load within the wastewater. Of these parameters, the fecal shedding rate of SARS-CoV-2 from infected individuals is the least understood. Until this research, shedding rate estimations were derived from limited results that indicated a large variance across a small number of individuals (Wölfel et al., 2020). Shedding rates were further extrapolated from the 90th percentile of this limited dataset and used to calculate the number of infected individuals contributing to viral loads in wastewater (Ahmed et al., 2020; Chavarria-Miró et al., 2021; Curtis et al., 2020). However, models and predictions based on these percentile shedding rates are likely to be inaccurate without pairing wastewater results with clinical data.
In this study, wastewater effluent from student dorms with defined and enumerated communities was sampled and assayed for SARS-CoV-2 RNA. Wastewater samples that tested positive for SARS-CoV-2 RNA triggered targeted clinical testing of almost all residents within the specific dorm. Consequently, fecal shedding rates were calculable since virus wastewater concentrations from dorm-specific manholes and the corresponding numbers of infected dorm residents were both known. These shedding rate estimations are reasonable since each dorm was a theoretically closed population: nonresidents were prohibited from entering dorms, and previously identified COVID-19 positive individuals were isolated within other facilities. This also prevented recounting of positive individuals and further shedding into monitored wastwater (Cevik et al., 2020; Gupta et al., 2020). Therefore, the shedding rates calculated from the dorms represent incident infections only.
For estimating the number of individuals shedding and contributing to the viral load in wastewater samples, a 6-day range of clinical data was considered (see Section 2.5). Then, fecal shedding rates of SARS-CoV-2 RNA were extrapolated from wastewater viral loads in samples specific to UArizona dorms in which the exact number of infected individuals was known. This estimation is based on a calculation that accounts for the number of infected individuals within a defined community; the amount of fecal material excreted per person; the flow rate of wastewater in the sewer pipeline; and the viral load within the wastewater (see Section 2.5).
The aggregation of wastewater and clinical data enabled quantification of SARS-CoV-2 fecal shedding rates per infected person. Mean shedding rates averaged over all dorms with positive wastewaters was 7.30 ± 0.67 log10 gc/g-feces based on the N1 gene (Table 3). Statistical parameters calculated include number of samples (N), average values (Avg), standard deviation (Std dev), median (Med), minimum (Min), and maximum (Max) (Table 3). These statistics are important to establish the precision of the calculated shedding rates since several factors may have influenced final estimates. Sensitivity analyses suggest that detected RNA concentrations in wastewater samples is the most influential factor for estimating viral shedding rates in infected individuals. Other factors such as demographics of a specific community (i.e., median age) and disease severity may also influence viral shedding rates in feces. Further, the duration of shedding appears to be affected by the severity of the COVID-19 disease: asymptomatic (6 days) < mild symptoms (10 days) < moderate symptoms (12 days) < serious (14 days) < critical (32 days) (Chen et al., 2020). Recent reports also suggest that asymptomatic individuals had a shorter duration of viral shedding than pre-symptomatic individuals (Hu et al., 2020). Further investigations are needed to determine observed virus concentrations in wastewater samples across various demographics and the viral shedding rate variance among specific demographics.
Our shedding rate figure is an assumed early infection shedding rate estimate because students were identified within the 6-day period and transferred from the dorm into an isolated living quarters. However, it is possible that some individuals may have started and/or continued shedding viral RNA in feces outside of the assumed 6-day period. Although fecal shedding rates may vary within this 6-day period, literature has not defined fluctuations in shedding in initial stages of infection. Therefore, individuals were assumed to be at peak shedding for the first 6 days in which infected individuals could be identified from UArizona WBE protocols. The shedding rate may also be overestimated based on case capture definitions, as although the vast majority of individuals in each dorm were tested after positive wastewater sample results, a minority of residents were not tested. Shedding rates in this study are based on young university students (18–20 years old) who reside in dormitories and do not necessarily approximate the general population. Nevertheless, a recent study which estimated the total number of active shedders from SARS-CoV-2 RNA levels in wastewater from a large metropolitan area similarly demonstrated that virus shedding occurred in a high proportion of asymptomatic individuals (Chavarria-Miró et al., 2021).
Some studies have reported similar initial shedding rates from symptomatic and asymptomatic infections (Lavezzo et al., 2020; Van Vinh Chau et al., 2020); whereas, other studies have found lower viral loads in asymptomatic cases (Han et al., 2020; Zhou et al., 2020). Also, some reports indicate that only approximately 50% of COVID-19 patients shed viral RNA in stool samples (Gupta et al., 2020; Medema et al., 2020a), suggesting the possibility that not all infected individuals shed viral RNA in feces. Regardless of these considerations, viral load in wastewater in this study were based on all individuals (symptomatic and asymptomatic) contributing to the sewage system. Therefore, fecal shedding estimates are general for defined communities of mixed clinical cases, while considering the high proportion of asymptomatic to symptomatic reported cases.
Lastly, it is important to note that no fecal samples were collected from individual COVID-19 patients directly. Therefore, results in this study are specific to wastewater from defined communities. Data and results from this study should be compared with clinical research and evaluations.
4.3. Concluding remarks
This study may have significant implications for public health. The fecal shedding rate of SARS-CoV-2 RNA derived in this study may be utilized to estimate the total number of infections in communities with similar demographics based on wastewater viral concentrations. However, direct estimates may require clinical fecal shedding rates from COVID-19 positive patients as a reliable comparison to this study, and more generalizable shedding rate estimates may require a community with more diverse demographics. Knowledge of disease prevalence, especially as a leading indicator, can be used to assist communities in efficient resource allocation to prevent and contain COVID-19 outbreaks. This study also provides further understanding for the total number of cases that are symptomatic versus asymptomatic. Moving forward, the number of reported cases can provide context for estimating the number of cases that were asymptomatic and/or unreported. This, in turn, could have implications for understanding the proportion of individuals that has been exposed to COVID-19 and for understanding progress towards immunity within a community.
CRediT authorship contribution statement
Bradley W. Schmitz: Formal Analysis, Project Administration, Visualization, Investigation, Writing – original draft.
Gabriel K. Innes: Formal Analysis, Visualization, Investigation, Writing – original draft.
Sarah M. Prasek: Formal Analysis, Resource, Writing – original draft.
Walter W. Betancourt: Methodology, Supervision, Resource.
Erika R. Stark: Resource.
Aidan R. Foster: Resource.
Alison G. Abraham: Resource, Writing – reviewing & editing.
Charles P. Gerba: Resource.
Ian L. Pepper: Formal analysis, Funding acquisition, Project administration, Visualization, Supervision, Investigation, Writing – original draft.
Bradley W. Schmitz and Gabriel K. Innes are co-first authors and contributed equally.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors thank the UArizona Task Force, Amy Glicken, and Jeff Bliznick for their contributions. Financial support for the study was provided by the University of Arizona Campus Re-Entry Task Force. The IRB at the University of Arizona reviewed the study and verified that all data was de-identified and complied with the Human Subjects Protection.
Editor: Adrian Covaci
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2021.149794.
Appendix A. Supplementary data
Supplementary tables
References
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W.J.M., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akgiray Ö. Explicit solutions of the manning equation for partially filled circular pipes. Can. J. Civ. Eng. 2005;32:490–499. doi: 10.1139/L05-001. [DOI] [Google Scholar]
- Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., Gerba C.P., Pepper I.L. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779 doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brönimann S., Rebhan K., Lemberger U., Misrai V., Shariat S.F., Pradere B. Secretion of severe acute respiratory syndrome coronavirus 2 in urine. Curr. Opin. Urol. 2020;30:735–739. doi: 10.1097/MOU.0000000000000808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., Vandesompele J., Wittwer C.T. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Cevik M., Tate M., Lloyd O., Maraolo A.E., Schafers J., Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2020 doi: 10.1016/S2666-5247(20)30172-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavarria-Miró G., Anfruns-Estrada E., Martínez-Velázquez A., Vázquez-Portero M., Guix S., Paraira M., Galofré B., Sánchez G., Pintó R.M., Bosch A. Time evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in wastewater during the first pandemic wave of COVID-19 in the metropolitan area of Barcelona, Spain. Appl. Environ. Microbiol. 2021;87 doi: 10.1128/AEM.02750-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhu B., Hong W., Zeng J., He X., Chen J., Zheng H., Qiu S., Deng Y., Chan J.C.N., Wang J., Zhang Y. Associations of clinical characteristics and treatment regimens with the duration of viral RNA shedding in patients with COVID-19. Int. J. Infect. Dis. 2020;98:252–260. doi: 10.1016/j.ijid.2020.06.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis K., Keeling D., Yetka K., Larson A., Gonzalez R. medRxiv; 2020. Wastewater SARS-CoV-2 Concentration and Loading Variability from Grab and 24-Hour Composite Samples. [DOI] [Google Scholar]
- Gupta S., Parker J., Smits S., Underwood J., Dolwani S. Persistent viral shedding of SARS-CoV-2 in faeces - a rapid review. Color. Dis. Off. J. Assoc. Coloproctol. G. B. Irel. 2020;22:611–620. doi: 10.1111/codi.15138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han M.S., Seong M.-W., Kim N., Shin S., Cho S.I., Park H., Kim T.S., Park S.S., Choi E.H. Viral RNA load in mildly symptomatic and asymptomatic children with COVID-19, Seoul, South Korea. Emerg. Infect. Dis. 2020 doi: 10.3201/eid2610.202449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Song C, Xu C, Jin G, Chen Y, Xu X, Ma H, Chen W, Lin Y, Zheng Y, Wang J, Hu Z, Yi Y, Shen H. Clinical characteristics of 24 asymptomatic infections with COVID-19 screened among close contacts in Nanjing, China. China. Sci. China Life Sci. 2020;63:706–711. doi: 10.2139/ssrn.3543598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauer S.A., Grantz K.H., Bi Q., Jones F.K., Zheng Q., Meredith H.R., Azman A.S., Reich N.G., Lessler J. The incubation period of coronavirus disease 2019 (COVID-19) from publicly reported confirmed cases: estimation and application. Ann. Intern. Med. 2020;172:577–582. doi: 10.7326/M20-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavezzo E., Franchin E., Ciavarella C., Cuomo-Dannenburg G., Barzon L., Del Vecchio C., Rossi L., Manganelli R., Loregian A., Navarin N., Abate D., Sciro M., Merigliano S., De Canale E., Vanuzzo M.C., Besutti V., Saluzzo F., Onelia F., Pacenti M., Parisi S.G., Carretta G., Donato D., Flor L., Cocchio S., Masi G., Sperduti A., Cattarino L., Salvador R., Nicoletti M., Caldart F., Castelli G., Nieddu E., Labella B., Fava L., Drigo M., Gaythorpe K.A.M., Brazzale A.R., Toppo S., Trevisan M., Baldo V., Donnelly C.A., Ferguson N.M., Dorigatti I., Crisanti A., Ainslie K.E.C., Baguelin M., Bhatt S., Boonyasiri A., Boyd O., Cattarino L., Ciavarella C., Coupland H.L., Cucunubá Z., Cuomo-Dannenburg G., Djafaara B.A., Donnelly C.A., Dorigatti I., van Elsland S.L., FitzJohn R., Flaxman S., Gaythorpe K.A.M., Green W.D., Hallett T., Hamlet A., Haw D., Imai N., Jeffrey B., Knock E., Laydon D.J., Mellan T., Mishra S., Nedjati-Gilani G., Nouvellet P., Okell L.C., Parag K.V., Riley S., Thompson H.A., Unwin H.J.T., Verity R., Vollmer M.A.C., Walker P.G.T., Walters C.E., Wang H., Wang Y., Watson O.J., Whittaker C., Whittles L.K., Xi X., Ferguson N.M., Team, I.C.C.-19 R Suppression of a SARS-CoV-2 outbreak in the Italian municipality of Vo’. Nature. 2020;584:425–429. doi: 10.1038/s41586-020-2488-1. [DOI] [PubMed] [Google Scholar]
- Lundy L., Fatta-Kassinos D., Slobodnik J., Karaolia P., Cirka L., Kreuzinger N., Castiglioni S., Bijlsma L., Dulio V., Deviller G., Lai F.Y., Alygizakis N., Barneo M., Baz-Lomba J.A., Béen F., Cíchová M., Conde-Pérez K., Covaci A., Donner E., Ficek A., Hassard F., Hedström A., Hernandez F., Janská V., Jellison K., Hofman J., Hill K., Hong P.-Y., Kasprzyk-Hordern B., Kolarevic S., Krahulec J., Lambropoulou D., de Llanos R., Mackulak T., Martinez-García L., Martínez F., Medema G., Micsinai A., Myrmel M., Nasser M., Niederstätter H., Nozal L., Oberacher H., Ocenášková V., Ogorzaly L., Papadopoulos D., Peinado B., Pitkänen T., Poza M., Rumbo-Feal S., Sánchez M.B., Székely A.J., Soltysova A., Thomaidis N.S., Vallejo J., van Nuijs A., Ware V., Viklander M. Making waves: collaboration in the time of SARS-CoV-2 - rapid development of an international co-operation and wastewater surveillance database to support public health decision-making. Water Res. 2021;199 doi: 10.1016/j.watres.2021.117167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Curr. Opin. Environ. Sci. Health. 2020 doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-Coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in the Netherlands. Environ. Sci. Technol. Lett. 2020 doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- Morone G., Palomba A., Iosa M., Caporaso T., De Angelis D., Venturiero V., Savo A., Coiro P., Carbone D., Gimigliano F., Iolascon G., Paolucci S. Incidence and persistence of viral shedding in COVID-19 post-acute patients with negativized pharyngeal swab: a systematic review. Front. Med. 2020;7:562. doi: 10.3389/fmed.2020.00562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose C., Parker A., Jefferson B., Cartmell E. The characterization of feces and urine: a review of the literature to inform advanced treatment technology. Crit. Rev. Environ. Sci. Technol. 2015;45:1827–1879. doi: 10.1080/10643389.2014.1000761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RStudio . 2020. RStudio: Integrated Development for R. RStudio, PBC, Boston, MA.http://www.rstudio.com/ [Google Scholar]
- Sun J., Zhu A., Li H., Zheng K., Zhuang Z., Chen Z., Shi Y., Zhang Z., Chen S.-B., Liu X., Dai J., Li X., Huang S., Huang X., Luo L., Wen L., Zhuo J., Li Y., Wang Y., Zhang L., Zhang Y., Li F., Feng L., Chen X., Zhong N., Yang Z., Huang J., Zhao J., Li Y.-M. Isolation of infectious SARS-CoV-2 from urine of a COVID-19 patient. Emerg. Microbes Infect. 2020 doi: 10.1080/22221751.2020.1760144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vinh Chau N., Lam V.T., Dung Nguyen Thanh, Yen L.M., Minh N.N.Q., Hung L.M., Ngoc N.M., Dung Nguyen Tri, Man D.N.H., Nguyet L.A., Nhat L.T.H., Nhu L.N.T., Ny N.T.H., Hong N.T.T., Kestelyn E., Dung N.T.P., Xuan T.C., Hien T.T., Phong N.T., Tu T.N.H., Geskus R.B., Thanh T.T., Truong N.T., Binh N.T., Thuong T.C., Thwaites G., Van Tan L. The natural history and transmission potential of asymptomatic severe acute respiratory syndrome coronavirus 2 infection. Clin. Infect. Dis. 2020;71:2679–2687. doi: 10.1093/cid/ciaa711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., Tan W. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., Hoelscher M., Bleicker T., Brünink S., Schneider J., Ehmann R., Zwirglmaier K., Drosten C., Wendtner C. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., Zhao Jingxian, Huang J., Zhao Jincun. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg. Infect. Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen C., Zhu S., Shu C., Wang D., Song J., Song Y., Zhen W., Feng Z., Wu G., Xu J., Xu W. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Wkly. 2020;2:123–124. doi: 10.46234/ccdcw2020.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R., Li F., Chen F., Liu H., Zheng J., Lei C., Wu X. Viral dynamics in asymptomatic patients with COVID-19. Int. J. Infect. Dis. 2020;96:288–290. doi: 10.1016/j.ijid.2020.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary tables