Abstract
Background and aims
Vaccines, to limit SARS-CoV-2 infection, were produced and reliable assays are needed for their evaluation. The WHO produced an International-Standard (WHO-IS) to facilitate the standardization/comparison of serological methods. The WHO-IS, produced in limited amount, was never tested for reproducibility. This study aims at developing a reproducible and accessible working standard (WS) to complement the WHO-IS.
Materials and methods
Sera from vaccinated individuals were used to produce the WSs. The WHO-IS, the WSs and single serum samples (n = 48) were tested on 6 quantitative serological devices. Neutralization assays were performed for the 48 samples and compared with their antibody titers.
Results
The WS carry an antibody titer 20-fold higher than the WHO-IS. It was reproducible, showed both good linearity and insignificant intra- and inter-laboratory variability. However, the WSs behave differently from the WHO-IS. Analysis of the 48 samples showed that single correlation factors are not sufficient to harmonize results from different assays. Yet, all the devices predict neutralization activity based on the antibody titer.
Conclusions
A reproducible and highly concentrated WS, specific for IgG against SARS-CoV-2 Spike-glycoprotein was produced. Such characteristics make it particularly suited for the harmonization of commercially available assays and the consequent evaluation of post-vaccinated individuals.
Keywords: Serological standard, WHO, SARS-CoV-2, COVID-19, Comirnaty vaccine, Neutralization
1. Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), the aetiological agent of the Coronavirus Disease 2019 (COVID-19), has threatened the health of the world’s population, leading to an unprecedented social and economic burden since the end of 2019. On January 30th, 2020, the World Health Organization (WHO) declared COVID-19 a Public Health Emergency of International Concern, and a Pandemic on March 11th. As of July 10th nearly 190 million people have been infected and less than 4 million have died as a result [1]. Exceptional research efforts led to the rapid development of several vaccines [2], [3], some of which have already been distributed to the general population [4], [5], [6], [7], [8]. The response to vaccination can be monitored by means of quantitative serological assays detecting serum antibodies recognizing the viral Spike glycoprotein (S-protein). However, most of the commercially available devices are based on different technologies, evaluate different types of immunoglobulins and use a plethora of different S-protein targets: monomeric soluble form, trimeric form, receptor binding domain (RBD) motif [9], [10], [11]. To date, 55 serological assays have received emergency use authorization from the Food and Drug Administration [12] and their different readouts make all possible comparisons challenging; even when comparing analogue immunoglobulin isotypes or when using the same S-protein domain as target [10], [11], [13], [14]. Thus, development and harmonization of quantitative serological assays for COVID-19 antibodies are pivotal in evaluating the vaccine response/efficacy and potentially assessing the acquired immunity.
In this regard, the National Institute for Biological Standards and Control (NIBSC), and WHO adopted on December 10th 2020 an International Standard (IS) to allow the accurate calibration of assays into an arbitrary unit, in order to reduce inter-laboratory variation [15], [16]. The WHO-IS is based on a pool of human plasma from eleven convalescent patients [15]. It was lyophilized in 3500 ampoules and each aliquot, after reconstitution with 0.25 mL of distilled water, was arbitrary assigned 250 international units (IU) for neutralizing activity and 1000 binding antibody units (BAU) per mL for binding assays.
As mentioned above, the aim of the WHO-IS is to provide a benchmark for the serological assays detecting an immunoglobulin class specific for the same molecular target. Because of the limited amount of IS ampoules, the intended purpose of the WHO-IS is to provide a standard reference for further calibration of secondary reference standards to be used worldwide [17]. Unfortunately, not all of the serological standards described so far fit the attribute of commutability [17]. In this specific case, commutability between secondary reference reagents and the WHO-IS might be hindered by the antibody variability observed in subjects recovered from SARS-CoV-2 [18] as well as by the presence of different viral variants undergoing adaptive selection, described to date [19]. In our opinion, such a problem can be partly mitigated by developing novel reference standards from sera of vaccinated subjects.
Taking advantage of our ongoing multicenter longitudinal study Covidiagnostix, “health technology assessment in COVID-19 serological diagnostics” (funded by the Italian Ministry of Health), which investigates the antibody responses of thousands of healthcare professionals [8], [20], we explored the possibility of developing an inexpensive, reproducible and easily available working standard (WS), specific for post-vaccination evaluation and following the same criteria used for the WHO-IS. The WS utilizes serum collected 21 days after the first dose of the Comirnaty vaccine, from subjects showing high antibody titers. The WS was calibrated against the WHO-IS and then analyzed by six quantitative serological testing devices located in different laboratories.
2. Material and methods
2.1. Laboratories involved in the study and devices used
The centers involved in the study were all based in Italy: the IRCCS San Raffaele Hospital (OSR), Milan; the IRCCS Galeazzi Orthopedic Institute (IOG), Milan; the IRCCS Casa Sollievo della Sofferenza Hospital (CSS), San Giovanni Rotondo and the IRCCS Bambino Gesù Children's Hospital (BG), Rome. The centers were all part of the Covidiagnostix study. The study was approved by the OSR Institutional Ethical Review Board (which has jurisdiction also on IOG), the CSS Ethical Review Board and the BG Ethical Review Board.
The six quantitative serological tests and devices used in the study were:
1) The Roche Anti-SARS-CoV-2-S test, run on COBAS 601 platform (Roche, Basel, Switzerland): an electrochemiluminescence immunoassay (ECLIA) detecting total Immunoglobulins (IgTot: IgA, IgG and IgM) against the receptor binding domain (RBD) of the viral S-protein. The quantification range is between 0.4 and 250.0 U/mL which is further extended to 2500.0 U/mL by a 1:10 dilution of the sample automatically performed by the instrument. Specificity and sensitivity (≥14 days after diagnosis) are, 99.98% and 98.8%, respectively, when the manufacturer’s suggested cutoff of 0.8 U/mL is used. The instrument was available at both OSR and BG.
2) The Diasorin LIAISON SARS-CoV-2 S1/S2 IgG (Diasorin, Saluggia, Italy), a chemiluminescence immunoassay (CLIA) detecting IgG specific for the S1/S2 viral S-protein. The quantification range is between 3.8 and 400.0 AU/mL. Specificity and sensitivity (>15 days after diagnosis) are, 98.5% and 97.4%, respectively, when the manufacturer’s suggested cutoff > 15 U/mL is used. Results between 12.0 and 15.0 U/mL are considered borderline. The instrument was available at OSR.
3) The Diasorin LIAISON SARS-CoV-2 Trimerics IgG (Diasorin, Saluggia, Italy), a CLIA detecting IgG specific for the Trimeric Spike Glycoprotein. The quantification range is between 1.85 and 800 U/mL. Specificity and sensitivity (>15 days after diagnosis) are 99.5% and 98.7%, respectively, when the manufacturer’s suggested cutoff ≥ 13 U/mL is used. The instrument was available at BG.
4) The Atellica IM SARS-CoV-2 IgG (COV2G) assay, run on the Atellica IM Analyzer (Siemens Healthineers, Erlangen, Germany), a CLIA detecting IgG specific for the S-protein RBD. The quantification range is between 0.05 and 150 U/mL which is further extended to 750.0 U/mL by a 1:5 dilution of the sample automatically performed by the instrument. Specificity and sensitivity (>14 days after diagnosis) are 100% and 99.8%, respectively, when the manufacturer’s suggested cutoff ≥ 1 U/mL is used. The instrument was available at IOG.
5) The Diesse CHORUS SARS-CoV-2 “NEUTRALIZING” Ab (Diesse-Diagnostica Senese, Siena, Italy), a competition assay which detects IgTot specific for the S1 subunit of the viral S-protein. The instrument was available at IOG. The quantification range is between 20 and 1500 U/mL (or BAU/mL). The manufacturer’s suggested cutoff is 20 U/mL (or BAU/mL).
6) The Euroimmun Anti-SARS-CoV-2 (Euroimmun, Lüebeck, Germany), an enzyme-linked immunosorbent assay which detects IgG specific for the S1 subunit of the viral S-protein. The quantification range is between 3.2 and 348 U/mL (or BAU/mL). Specificity and sensitivity (>10 days after diagnosis) are 99.8% and 90.3%, respectively, when the manufacturer’s suggested cutoff ≥ 35.2 U/mL (or BAU/mL) is used. Results between 25.6 and 35.2 U/mL (or BAU/mL) are considered borderline. A solution used for diluting samples above 348 U/mL was included in the measurement kits. The instrument was available at CSS.
A seventh device, the CLIA Diasorin LIAISON SARS-CoV-2 S1/S2 IgM (Diasorin, Saluggia, Italy) detecting IgM specific for the S1/S2 viral S-protein, was used exclusively to test for the presence of IgM and was not compared with the above listed six instrumentations. No quantification range was defined by the manufacturer. Results < 1.1 AU are considered as negative (no presence of IgM), whereas results ≥ 1.1 AU are considered as positive (presence of IgM).
2.1.1. U/mL to BAU/mL conversion factors
Binding antibody units per milliliter (BAU/mL) proposed by the WHO to standardize any device to the WHO-IS were calculated by applying the conversion factors suggested by the manufacturers, whenever it was possible. Roche, Euroimmun and Diesse all claim a conversion factor of 1. Therefore, their results in U/mL correspond to BAU/mL (BAU/mL = U/mL*1). In this regards, Diesse and Euroimmun manufacturer’s datasheets claim readouts in BAU/mL for both assays. Siemens provides a conversion factor of 21.8 (BAU/mL = U/mL*21.8), whereas Diasorin Trimerics specifies a conversion factor of 2.6 (BAU/mL = U/mL*2.6). No conversion factor was available for Diasorin S1/S2.
2.2. WS preparation and samples
At OSR>5000 healthcare professionals were offered the Comirnaty mRNA BNT162b2 vaccine within the first two months of 2021 and were included in the Covidiagnostix study. Blood samples were withdrawn at different time points, as previously described [21]; time 0 (T0), 1–2 min before receiving the first vaccination dose, to discriminate subjects with or without natural presence of anti-SARS-CoV-2 antibodies, and at time 1 (T1), 21 days after T0, 1–2 min before the injection of the second dose.
Discrimination between naturally seropositive and seronegative subjects at T0 was carried out using the Elecsys Anti-SARS-CoV-2 (ECLIA) run on a COBAS 601 platform (Roche, Basel, Switzerland) which detects IgTot specific for the viral nucleocapsid-protein. Thanks to an instrument query upon a positive result, the samples were further tested on the same platform with the Roche anti-SARS-CoV-2-S test (Spike protein). The latter assay was also used to evaluate vaccine response at T1. For a limited number of subjects (n = 1100) a third blood sample was collected 21 days after the second vaccination dose (T2) and evaluated with the Roche Anti-SARS-CoV-2-S test.
2.2.1. WS1 preparation
Ten randomly chosen T1 serum samples (1 mL each) fulfilling the single inclusion criteria of having antibody titers (Roche Anti-SARS-CoV-2-S) above the 2500 U/mL upper instrument limit were pooled together in order to obtain the WS1 (“solution 0”). A 1:10 dilution was performed using a pool of pre-pandemic sera to obtain instrument readouts within the quantification range of the majority of the utilized devices (solution 1). Further 1:2 dilutions (from 1:20 to 1:10240; corresponding to solution 2 to 11) were performed using a pool of pre-pandemic sera for the purpose of testing the linearity of the various assays. The ten samples were all from naturally seropositive subjects.
2.2.2. WS2 preparation
Fifty-two randomly chosen T1 samples (1–2 mL each), fulfilling the single inclusion criteria of having antibody titers (Roche Anti-SARS-CoV-2-S) above the 2500 U/mL upper instrumental limit, were the starting material to produce the WS2. The 52 samples were pooled together (in different proportions) in order to obtain the second working standard solution (WS2: “solution 0”) matching the antibody titer of WS1 “solution 0” (according to the Roche Anti-SARS-CoV-2-S assay). As for WS1, a 1:10 dilution with a pool of pre-pandemic sera was performed to make solution 1. Further 1:2 dilutions with pre-pandemic serum were done to prepare solutions 2 to 11. Of the 52 subjects, 46 were from naturally seropositive subjects whereas 6 were seronegative at T0 thus showing no evidence of previous SARS-CoV-2 infection.
2.2.3. WHO International standard
An aliquot (0.25 mL after reconstitution with distilled water) of the WHO-IS for anti-SARS-CoV-2 (code: 20/136) was obtained from NIBSC (Hertfordshire, UK). As stated by the WHO [16], the assigned potency of the WHO-IS was 250 IU/ampoule for neutralizing antibody activity. After reconstitution, the final concentration is 1000 IU/mL. For binding antibody assays, an arbitrary measure of 1000 binding antibody units (BAU/mL) can be used to assist in the comparison of assays detecting the same class of immunoglobulins with the same specificity. Due to the scarcity of material, the reconstituted WHO-IS was diluted 1:2 (500 BAU/mL), using a pool of pre-pandemic sera, to reach the minimal volume needed for measurements on the Roche and Siemens assays. Further 1:4 dilutions (pre-pandemic sera) were performed five times to test the WHO-IS linearity (125, 31.25, 7.81, 1.95 and 0.49 BAU/mL).
For the Diasorin Trimerics and Euroimmun devices the reconstituted WHO-IS was diluted 1:4.4 (228.6 BAU/mL) using a pool of pre-pandemic sera to reach the minimal volume needed for the measurements. Further 1:4 dilutions (pre-pandemic sera) were performed five times to test the WHO-IS linearity (82.14, 22.32, 5.58, 1.39 and 0.22 BAU/mL).
2.3. Individual samples and neutralization assay
Forty-eight T2 samples (1–2 mL) from both naturally seropositive and seronegative subjects, chosen to cover a broad antibody titer range (Roche Anti-SARS-CoV-2-S), were serologically tested with the six devices used in this study as well as for neutralization activity. Samples showing antibody titers exceeding the upper instrument limits were diluted with a pool of pre-pandemic sera in order to obtain test results within the quantification range.
2.4. Micro-neutralization experiments
Vero E6 cells were seeded into 96-well plates 24 h before the experiment was performed at 95% cell confluency for each well. Serum samples were decomplemented by incubation at 56 °C for 30 min, diluted 1:80 and incubated with SARS-CoV-2 G614 strain at 0.001 multiplicity of infection (MOI) for 1 h at 37 °C. Virus-serum mixtures and positive infection control were applied to Vero E6 monolayers after washing cells with PBS 1×, and virus adsorption was carried out at 37 °C for 1 h. Cells were then washed with PBS 1 × to remove cell-free virus particles and virus-containing mixtures, while controls were replaced with complete DMEM supplemented with 2% FBS. The plates were incubated at 37 °C in the presence of CO2 for 72 h. The experiments were performed in triplicate. Neutralization activity was evaluated by comparing the percentage of cytopathic effect (CPE) presence detected in the virus-serum mixtures with the positive infection control. Neutralization activity was ranked as follow: 100%, 66.7%, 33.3% and 0% if all, two, one and none of the triplicate experiments showed neutralization, respectively.
2.5. Statistical analysis
WS and WHO-IS measurements were performed in triplicate. Values are expressed as arithmetic average ± standard deviation (SD). Commutability between WS1 and WS2 was assessed by Passing and Bablok regression [22] using the software Excel (Microsoft, Redmond, WA, USA). A significant (p < 0.05) bias was deemed to be present if the 95% confidence interval (CI) of the bias did not include zero (intercept) or one (slope). The linearity of the WS and the WHO-IS as well as the single samples instrument-to-instrument correlations were evaluated by statistical linear regression using the Sigmaplot software (Systat-Software, San Jose, CA, USA). The receiver operating characteristic (ROC) curves analyses were performed using R software v4.1.0 (R Core Team, Wien, Austria). Thresholds were calculated to minimize the optimality criterion ((1-sensitivities)^2+(1-specificities)^2). ROC curves were compared using Venkatraman statistic [23] with 500 permutations. P values < 0.05 were considered statistically significant.
3. Results
3.1. WS analysis and comparison with WHO-IS
WS1 and WS2 were prepared by mixing serum samples from 10 and 52 subjects, respectively (solution 0). Such subjects mounted an exceptionally high immune response after the first vaccination dose; thus, a pool of their sera represents a source of very concentrated antibodies specific for the viral S-protein. For the WS1, all the 10 subjects had previously been infected by SARS-CoV-2 whereas for WS2, 6 subjects (11.5%) had never experienced COVID-19. After a 1:10 dilution with pre-pandemic serum, the back-calculated WS1 and WS2 “solution 0” titers (Roche Anti-SARS-CoV-2-S) were between 18810 ± 970 and 19230 ± 476 U/mL (Table 1 ). Similar to what was observed in previous studies (4,6,8), the corresponding antibody titers at T0 (before vaccination) of the seropositive subjects were, on average, 35.6 ± 32.7 U/mL (Roche Anti-SARS-CoV-2-S), representing 0.19% of the “solution 0” antibody titers. Thus, vaccination increased their S-protein specific antibody titers by approximately 500-fold. No S-protein specific IgM were present in both WS1 and WS2 (Diasorin LIAISON SARS-CoV-2 S1/S2 IgM, data not shown).
Table 1.
WS1 and WS2 average measurements ± SD performed with the six different assays used in the study. The manufacturer’s suggested cutoff levels are highlighted in yellow. “Negative” results (i.e., below the cutoff level) are highlighted in light blue.
 |
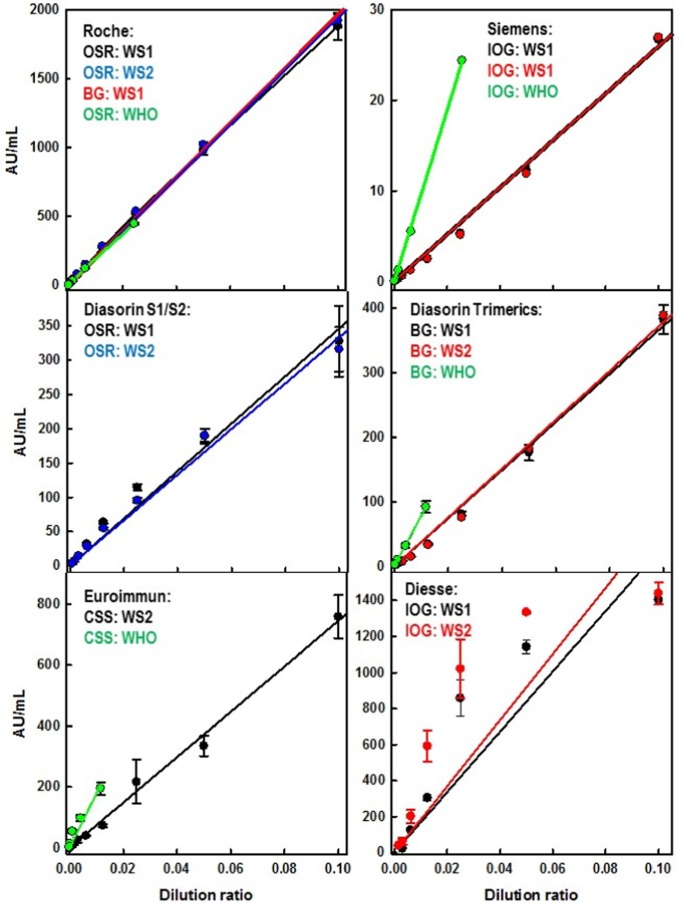
Solutions 1 to 11 were used to evaluate the linearity of the different assays in different laboratories for both WS1 and WS2. Fig. 1 and Table 1 show that all the devices displayed a good linearity (R2 > 0.977) except for the Diesse assay which exhibited a poor linearity (Fig. 1) consistent with an R2 = 0.776, in the selected range. Both WS1 and WS2 have been tested on the Roche, Diasorin (both S1/S2 and Trimerics), Siemens and Diesse devices. In all cases, except for Diesse, the two WSs were statistically identical as demonstrated by Passing Bablock regression. WS1 was not tested on the Euroimmun device because we have run out of material. It must be noted that, for the Roche device, the equivalence between WS1 and WS2 was maintained even when measurements were performed on devices located at different centers (OSR and BG) (Fig. 1, Table 1).
Fig. 1.
Graphical representation of the WS1 and WS2 measurements showed in Table 1. Dilution ratio of 0.1 corresponds to solution 1. Further 1:2 dilutions (from 0.05 to 9.7•10-5) correspond to solutions 2 to 11. Measurements were performed in triplicate. Error bars represent the corresponding SD. The higher WHO-IS concentration corresponds to (500 BAU/mL) for Roche and Siemens measurements and to 228.6 BAU/mL for the Diasorin Trimerics and Euroimmun devices (for details see Materials and Methods, section 2.2.3).
Table 1 shows that the cutoff levels suggested by different manufacturers impact the discrimination between positive and negative samples. For the Roche device, all the 11 solutions used in this study were considered as positive whereas for the remaining 5 assays, solutions 6 to 11 were considered as negative except for WS2 “solution 6” measured on the Diesse device (Table 1).
The Roche device, which shows a manufacturer’s suggested U/mL to BAU conversion factor for the WHO-IS equal to 1 (Table 2 ), was used to calibrate the WS against the WHO-IS. Fig. 1 shows that, after calibration, the other assays revealed some discrepancy. Because of the limited amount sent by NIBSC, the WHO-IS was not tested on the Diesse device. Such differences have been summarized in Table 2 which displays the comparison between the WHO-IS U/mL to BAU conversion factors (both suggested by the manufacturers and experimentally calculated in our study) and the WS. Experimentally calculated conversion factors, either for WS or WHO-IS, were obtained as ratios between the slopes of the linear regressions showed in Fig. 1. The experimentally calculated WHO-IS conversion factors were remarkably similar to those suggested by the manufacturers (Table 2). In contrast, the Siemens assay displayed a large discrepancy between the WHO-IS and the WS conversion factors, with a ratio equal to 3.4 (Table 2). Similar, albeit lower, discrepancies were observed in the other devices (Table 2), except for the Diesse assay which showed similar conversion factors (1 and 1.1 for the WHO-IS and WS, respectively (Table 2)).
Table 2.
U/mL to BAU conversion factors. “WHO-IS” column: manufacturers’ suggested WHO-IS conversion factors (the same conversion factors experimentally evaluated in our study are displayed in brackets); “WS” column: conversion factors experimentally evaluated in our study using the WS; “WS/WHO” column: ratio between the manufacturers’ suggested WHO-IS conversion factors and the corresponding WS ones.
| U/mL to BAU conversion factors |
|||
|---|---|---|---|
| WHO-IS | WS | WS/WHO | |
| Roche | 1 (1a) | 1a | 1 a |
| Siemens | 21.8 (20.8) | 74.7 | 3.4 |
| Diasorin Trimerics | 2.6 (2.5) | 5.2 | 2.0 |
| Diasorin S1/S2b | 5.7 | ||
| Diesse | 1 | 1.1 | 1.1 |
| Euroimmun | 1 (1.1) | 2.6 | 2.6 |
the perfect agreement between the WHO-IS and the WS for the Roche device is the consequence of having used the latter assay as the calibration reference.
no manufacturer’s suggested conversion factors were proposed for the WHO-IS.
Through the analysis of the WS slopes (Table 1) and the manufacturers’ suggested U/mL to BAU conversion factors (Table 2), we calculated the instrument-to-instrument U/mL to U/mL conversion factors for every type of conversion between the six assays (Table 3 ).
Table 3.
WS conversion factors (U/mL to U/mL) between different devices. In brackets the conversion factors between different devices (U/mL to U/mL) obtained from the corresponding manufacturer’s suggested WHO-IS U/mL to BAU/mL conversion factors.
| To |
||||||
|---|---|---|---|---|---|---|
| From | Roche | Diasorin S1/S2 | Diasorin Trimerics | Siemens | Diesse | Euroimmun |
| Roche | 1 | 0.18 | 0.19 (0.38) | 0.013 (0.046) | 0.86 (1) | 0.38 (1) |
| Diasorin S1/S2a | 5.60 | 1 | 1.08 | 0.075 | 4.8 | 2.15 |
| Diasorin Trimerics | 5.2 (2.6) | 0.92 | 1 | 0.069 (0.12) | 4.4 (2.6) | 1.98 (2.6) |
| Siemens | 74.7 (21.8) | 13.3 | 14.47 (8.4) | 1 | 64.6 (21.8) | 28.71 (21.8) |
| Diesse | 1.1 (1) | 0.21 | 0.23 (0.38) | 0.016 (0.046) | 1 | 0.45 (1) |
| Euroimmun | 2.6 (1) | 0.46 | 0.50 (0.38) | 0.03 (0.046) | 2.2 (1) | 1 |
No U/mL to BAU/mL conversion factor was available for Diasorin S1/S2
3.2. Single samples analysis
In parallel to the WS analysis, 48 T2 serum samples were analyzed with the six devices. Samples were chosen to cover a wide range of antibody titers (Table 1S). For instance, we chose 24 samples showing antibody titers within the Roche device range (0.4–2500 U/mL) and 24 above the 2500 U/mL instrument limit. Samples exceeding the instrument limits of any of the six devices used in this study were diluted 1:50 with a pool of pre-pandemic sera (Table 1S) in order to get readouts within the instrument quantification range.
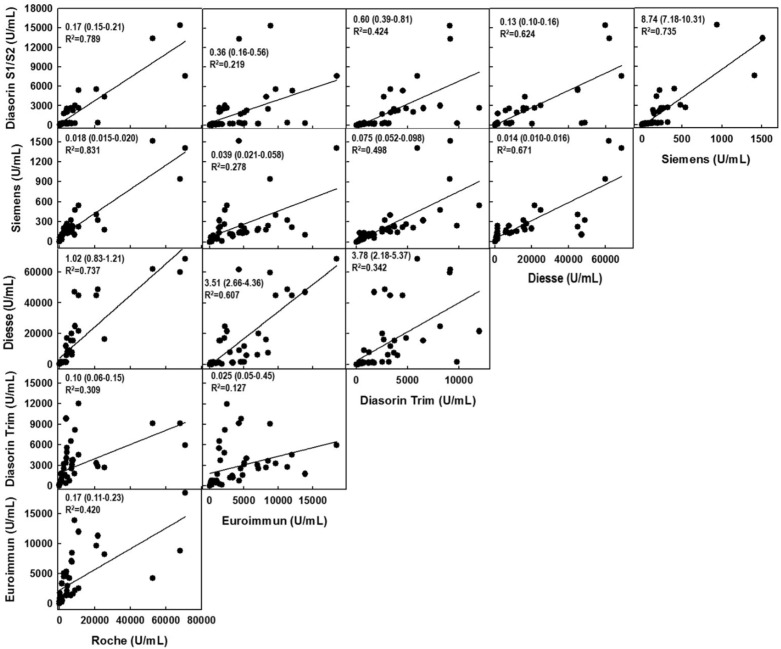
Fig. 2 shows the correlations between the serological results obtained with the different assays. The linear regressions showed that the best correlation was observed when the Roche and the Siemens devices were compared (R2 = 0.831, Fig. 2), whereas the weakest was observed when the Diasorin Trimerics assay was compared to the Euroimmun (R2 = 0.127, Fig. 2).
Fig. 2.
Linear regression of the “instrument-to-instrument” correlations for the 48 individual samples analyzed in the study. The slopes and the corresponding 95% CI in brackets are shown together with the corresponding coefficient of determinations (R2).
Fig. 2 also shows the slopes (and their corresponding 95% CI) of the linear regression calculated for each instrument-to-instrument correlation. These slopes correspond also to the “instrument-to-instrument” (U/mL to U/mL) conversion factors. When compared with either the WHO-IS or the WS conversion factors (Table 3) 5 out of 10 of the WS conversion factors (Diasorin S1/S2 was not considered because of the missing WHO-IS conversion factor) fell within the 95% CIs (Siemens vs Euroimmun; Siemens vs Diasorin Trimerics; Siemens vs DS; Diesse vs Roche; Diesse vs Diasorin Trimerics). In contrast, only 4 out of 10 WHO-IS conversion factors fell within the same 95% CIs (Siemens vs Euroimmun; Diesse vs Roche; Diesse vs Diasorin Trimerics.; Diasorin Trimerics vs Euroimmun). Furthermore, the 5 WS conversion factors not included within the 95% CIs showed values which were different from the single samples calculated conversion factors by: 27.8% (Siemens vs Roche), 37.3% (DS vs Euroimmun), 29.0% (Diasorin Trimerics vs Roche), 100.0% (Diasorin Trimerics vs Euroimmun) and 123.5% (Euroimmun vs Roche), respectively. In contrast, the 6 WHO-IS conversion factors not included within the 95% CI showed higher differences: 155.0% (Siemens vs Roche), 60.0% (Siemens vs Diasorin Trimerics), 228.6% (Siemens vs DS), 71.5% (DS vs Euroimmun), 280% (Diasorin Trimerics vs Roche), and 488.2% (Euroimmun vs Roche), respectively.
3.3. Neutralization assays
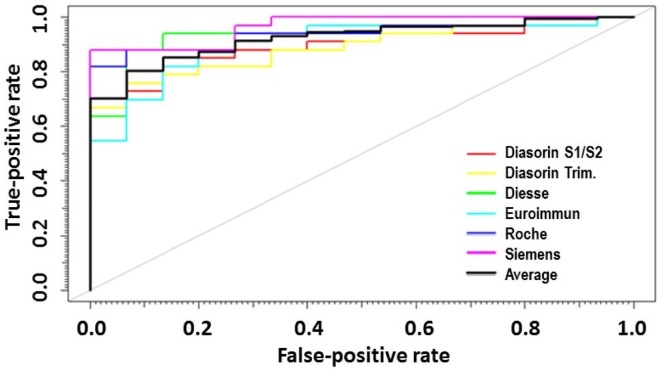
The 48 T2 samples described above in section 3.2 were also tested for neutralizing activity against the SARS-CoV-2 G614 strain (supplementary Table 1S). The ROC curves (Fig. 3 ) showed that neutralization activity correlates well with the antibody titers for all the six assays. Table 4 shows that the highest AUC was observed in the Siemens assay (93.8%), whereas the lowest in the Diasorin Trimerics (88.9%). Nevertheless, no statistically significant differences were observed between the ROC curves of the six considered devices. Table 4 additionally displays thresholds representing the antibody titers which maximize both specificity and sensitivity for each assay, expressed in U/mL.
Fig. 3.
ROC curves obtained by considering the antibody titers of the 48 individual samples measured with the six assays analyzed in the study and their corresponding neutralization activity. A sample was considered “neutralizing” if it showed at least 30% of neutralizing activity.
Table 4.
ROC parameters obtained by analyzing the 48 individual samples. “Threshold” represents the antibody titers which maximize both specificity and sensitivity.
| Threshold (U/mL) | Specificity(%) | Sensitivity(%) | AUC (CI95%) (%) |
|
|---|---|---|---|---|
| Siemens | 86.5 | 100.0 | 87.9 | 96.6 (92.3–100.0) |
| Roche | 1392.0 | 93.3 | 84.8 | 93.8 (87.3–100.0) |
| Diesse | 1266.7 | 93.3 | 87.5 | 93.8 (86.6–100.0) |
| Euroimmun | 1268.4 | 86.7 | 81.8 | 90.5 (81.6–99.4) |
| Diasorin S1/S2 | 206.0 | 86.7 | 81.8 | 89.3 (80.4–98.2) |
| Diasorin Trimerics | 789.0 | 86.7 | 78.8 | 88.9 (80.7 – 97.8) |
4. Discussion
In the current COVID-19 vaccination era, quantitative serological assays play a crucial role in determining the individual’s immunization status which may predict immunity from future infection. Nevertheless, there is currently a lack of harmonization/standardization among the devices produced by different manufacturers. Most assays report their outcomes in arbitrary-units which often differ substantially from one assay to another. The only currently available standard was produced by the WHO before the vaccines became accessible. It was prepared in limited amount using serum from post-convalescent subjects and, to the best of our knowledge, its reproducibility has never been demonstrated.
Using the serum from vaccinated subjects we produced a WS specific for serological assays targeting the S-protein. The WS showed a titer approximately 20-fold higher than the 250 µL solution obtained after reconstitution of the WHO-IS. The WS also exhibited a good linearity with all the tested devices, except for the Diesse assay whose limitations were a short analytical range and an early signal saturation. The greater than 500-fold increase in antibody titers showed, upon vaccination, by the samples used to prepare the WS, suggests a very high specificity for SARS-CoV-2 excluding concerns about possible cross-reactivity with other human coronaviruses. The purpose of the WS was to be used as a secondary reagent after calibration against the WHO-IS using the Roche assay. However, we observed that the WS and the WHO-IS behaved differently when tested on the remaining five quantitative serological devices. Such discrepancy was as high as 3.4-fold (Siemens) and was different from one assay to another. We might speculate that such differences arise from the different sources of material used for the two standards: recovered patients for the WHO-IS, and vaccinated subjects for the WS. In other words, it seems that the range of S-protein epitopes recognized by the two standards is different because their antibodies were generated against two different targets: the S-protein embedded in the virus for the WHO-IS and the isolated S-protein for the WS. Nevertheless, we demonstrated that our WS can be easily reproduced by using a pool of sera from the same type of subjects (i.e. vaccinated individuals showing a high antibody titers 21 days after the first vaccine dose). In this context, reproducibility of the WHO-IS might be hindered by the high variability observed in post-infection SARS-CoV-2 antibody levels as a consequence of variable antigen exposure [18], and also, in view of the different SARS-CoV-2 variants [19]. In contrast, such differences are mitigated in the context of vaccination as demonstrated by the WS reproducibility.
When testing serum from single samples, the linearity observed with both the WHO-IS and the WS was lost, demonstrating that a single correlation factor is not sufficient to harmonize the results from the different quantitative serological tests examined in this study. The best correlation was observed between the Roche and the Siemens assays and might be due to the same protein target (S-protein RBD) shared by the two assays. On the other hand, the Diasorin Trimerics, the only one using the trimeric S-protein as target, showed poor correlations with all other assays.
The fact that the linear regression slopes of the 48 individual samples (Fig. 2) are more similar to the WS conversion factors rather than to the manufacturers’ provided WHO-IS conversion factors is likely the consequence of the use of different materials for the preparation of the two standards. Indeed, the WS as well as the 48 analyzed samples were taken from vaccinated subjects, whereas the WHO-IS was prepared from COVID-19 recovered patients. These data further confirm that the range of S-protein epitopes recognized by the IgG produced after SARS-CoV-2 infection is different from the one exhibited by vaccinated subjects (regardless of previous SARS-CoV-2 infection).
Despite the poor instrument-to-instrument correlations observed when analyzing individual samples, all of the six assays showed a good agreement between antibody titers and neutralization activity. Thus, quantitative serological assays could play a pivotal role in prognosticating post-vaccination immunity. The ROC calculated thresholds may be used by clinicians to associate each device with an antibody titer reference allowing to differentiate between subjects who acquired post-vaccination immunity against SARS-CoV-2 infection and those who might still be at risk of an infection. A further element confirming the lack of harmonization/standardization among SARS-CoV-2 serological quantitative assays came from the positivity/negativity analysis. The Roche device showed a very low cutoff level when compared to the other five assays. As a consequence, individuals might be listed as “positive” or “negative” depending on the device available in the clinical laboratory where they underwent the serological test, generating confusion for both final users and medical personnel.
5. Conclusions
We developed an easy to prepare, reproducible and highly concentrated WS specific for IgG against SARS-CoV-2 S-protein. Analysis of both the WS and single samples showed that the WS behaves differently from the WHO-IS, and that the instrument-to-instrument conversion factors calculated with both the WS and the WHO-IS are not sufficient to obtain interchangeable instruments’ readouts from single samples.
Nevertheless, the six assays performed well in predicting neutralization activity based on the antibody titers. The reproducibility and high antibody titers of the WS (up to almost 20,000 BAU/mL, according to the Roche device) make it an excellent standard for the evaluation of post-vaccinated individuals who often show high antibody titers [4], [6]. In contrast, to the best of our knowledge, no reproducibility has yet been demonstrated for the WHO-IS. Furthermore, the latter is available only in limited amount and its reconstitution to a volume suitable for quantitative serological tests (0.5 mL) leads to a solution with a relatively low antibody titer (approximately 500 BAU/mL).
Funding
This project was supported by Ministry of Health of Italy, “Bando Ricerca COVID-19”; project number: COVID-2020–12371619; project title: COVIDIAGNOSTIX - Health Technology Assessment in Covid serological diagnostics.
Author Contributions
Davide Ferrari: Conceptualization, Formal analysis, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing. Nicola Clementi: Conceptualization, Investigation, Methodology, Writing – review & editing. Sestina Maria Spanò: Conceptualization, Investigation, Methodology. Sami Albitar-Nehme: Conceptualization, Investigation, Methodology, Writing – review & editing. Stefania Ranno: Conceptualization, Investigation, Methodology, Writing – review & editing. Alessandra Colombini: Conceptualization, Investigation, Methodology, Writing – review & editing. Elena Criscuolo: Conceptualization, Investigation, Methodology. Chiara Di Resta: Conceptualization, Project administration. Rossella Tomaiuolo: Conceptualization, Project administration. Marco Viganò: Formal analysis, Software. Nicasio Mancini: Conceptualization. Elena De Vecchi: Conceptualization, Investigation, Methodology, Writing – review & editing. Massimo Locatelli: Conceptualization, Supervision, Funding acquisition. Alessandra Mangia: Conceptualization, Supervision, Funding acquisition, Investigation, Methodology. Carlo Federico Perno: Conceptualization, Supervision, Funding acquisition. Giuseppe Banfi: Conceptualization, Supervision, Funding acquisition
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cca.2021.08.024.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Worldometers, COVID-19 statistics. https://www.worldometers.info/coronavirus/, 2008 (accessed 10 July 2021).
- 2.Izda V., Jeffries M.A., Sawalha A.H. COVID-19: A review of therapeutic strategies and vaccine candidates. Clin. Immunol. 2021;222:108634. doi: 10.1016/j.clim.2020.108634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le T.T., Cramer J.P., Chen R., Mayhew S. Evolution of the COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19(10):667–668. doi: 10.1038/d41573-020-00151-8. [DOI] [PubMed] [Google Scholar]
- 4.F. Krammer, K. Srivastava, P. Team, V. Simon, H. Alshammary, A. Amoako, M. Awawda, K. Beach, C.M. 1 3 Bermúdez-González, D. Bialak, J.M. Carreño, R. Chernet, L. Eaker, E. Ferreri, D. Floda, C. Gleason, J. Hamburger, K. Jiang, G.D. Kleiner, J. Jurczyszak, W. Matthews, L. Mendez, I. Mulder, A. Nabeel, K. Raskin, A. Russo, M. Salimbangon, A. Saksena, G. Shin, L. Singh, D. Sominsky, Antibody responses in seropositive persons after a single dose of SARS-CoV-2 mRNA vaccine, N. Engl. J. Med. 384 (2021) 1372-1374. doi: 10.1056/NEJMc2101667. [DOI] [PMC free article] [PubMed]
- 5.M. Prendecki, C. Clarke, J. Brown, A. Cox, S. Gleeson, M. Guckian, P. Randell, A.D. Pria, L. Lightstone, X.-N. Xu, W. Barclay, S.P. McAdoo, P. Kelleher, M. Willicombe, Effect of previous SARS-CoV-2 infection on humoral and T-cell responses to single-dose BNT162b2 vaccine., Lancet (London, England). 6736 (2021) 10–12. doi:10.1016/S0140-6736(21)00502-X. [DOI] [PMC free article] [PubMed]
- 6.Manisty C., Otter A.D., Treibel T.A., McKnight Á., Altmann D.M., Brooks T., Noursadeghi M., Boyton R.J., Semper A., Moon J.C. Antibody response to first BNT162b2 dose in previously SARS-CoV-2-infected individuals. Lancet. 2021;397(10279):1057–1058. doi: 10.1016/S0140-6736(21)00501-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.S. Saadat, Z.T. Rikhtegaran, L. James, N. Michelle, F. Matthew B., A.D. Harris, M.M. Sajadi, Binding and Neutralization Antibody Titers After a Single Vaccine Dose in Health CareWorkers Previously InfectedWith SARS-CoV-2, JAMA - J. Am. Med. Assoc. Mar 1;e213 (2021). doi:10.1001/jama.2021.3341. [DOI] [PMC free article] [PubMed]
- 8.C. Di Resta, D. Ferrari, M. Viganò, M. Moro, E. Sabetta, M. Minerva, A. Ambrosio, M. Locatelli, R. Tomaiuolo, The Gender Impact Assessment among Healthcare Workers in the SARS-CoV-2 Vaccination-An Analysis of Serological Response and Side Effects, Vaccines. May 18 (2021) 522. doi:10.3390/vaccines9050522. [DOI] [PMC free article] [PubMed]
- 9.C. Therrien, B. Serhir, M. Bélanger-Collard, J. Skrzypczak, D.K. Shank, C. Renaud, J. Girouard, V. Loungnarath, M. Carrier, G. Brochu, F. Tourangeau, B. Gilfix, A. Piche, R. Bazin, R. Guérin, M. Lavoie, V. Martel-Laferrière, C. Fortin, A. Benoit, D. Marcoux, N. Gauthier, A.M. Laumaea, R. Gasser, A. Finzi, M. Roger, Multicenter evaluation of the clinical performance and the neutralizing antibody activity prediction properties of 10 high-throughput serological assays used in clinical laboratories, J. Clin. Microbiol. 59 (2021). doi:10.1128/JCM.02511-20. [DOI] [PMC free article] [PubMed]
- 10.Kittel M., Muth M.C., Zahn I., Roth H.J., Thiaucourt M., Gerhards C., Haselmann V., Neumaier M., Findeisen P. Clinical evaluation of commercial automated SARS-CoV-2 immunoassays. Int. J. Infect. Dis. 2021;103:590–596. doi: 10.1016/j.ijid.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonelli F., Blocki F.A., Bunnell T., Chu E., Arriel De La O., Grenache D.G., Marzucchi G., Montomoli E., Okoye L., Pallavicini L., Streva V.A., Torelli A., Wagner A., Zanin D., Zierold C., Wassenberg J.J. Evaluation of the automated LIAISON® SARS-CoV-2 TrimericS IgG assay for the detection of circulating antibodies. Clin. Chem. Lab. Med. 2021;49:1463–1467. doi: 10.1515/cclm-2021-0023. [DOI] [PubMed] [Google Scholar]
- 12.Tang M.S., Farnsworth C.W. Associating SARS-CoV-2 Serological Assays with Protection: Where the Field Stands. Clin. Chem. 2021;67:707–709. doi: 10.1093/clinchem/hvab039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnurra C., Reiners N., Biemann R., Kaiser T., Trawinski H., Jassoy C. Comparison of the diagnostic sensitivity of SARS-CoV-2 nucleoprotein and glycoprotein-based antibody tests. J. Clin. Virol. 2020;129:104544. doi: 10.1016/j.jcv.2020.104544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Padoan A., Bonfante F., Pagliari M., Bortolami A., Negrini D., Zuin S., Bozzato D., Cosma C., Sciacovelli L., Plebani M. Analytical and clinical performances of five immunoassays for the detection of SARS-CoV-2 antibodies in comparison with neutralization activity. EBioMedicine. 2020;62:103101. doi: 10.1016/j.ebiom.2020.103101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.WHO, Establishment of the WHO International Standard and Reference Panel for anti-SARS-CoV-2 antibody, 2020. https://www.who.int/publications/m/item/WHO-BS-2020.2403, 2020 (accessed 10 February 2021).
- 16.World Health Organization, WHO TRS: Recommendations for the preparation, characterization and establishment of international and other biological reference standards, http://www.who.int/immunization_standards/vaccine_reference_preparations/TRS932Annex 2_Inter _biol ef standards rev2004.pdf?ua=1, 2006 (accessed 12 March 2021).
- 17.Page M., Wilkinson D.E., Mattiuzzo G., Efstathiou S., Minor P. Developing biological standards for vaccine evaluation. Future Virol. 2017;12(8):431–437. doi: 10.2217/fvl-2017-0003. [DOI] [Google Scholar]
- 18.F. Muecksch, H. Wise, B. Batchelor, M. Squires, E. Semple, C. Richardson, J. McGuire, S. Clearly, E. Furrie, N. Greig, G. Hay, K. Templeton, J.C.C. Lorenzi, T. Hatziioannou, S. Jenks, P.D. Bieniasz, Longitudinal Serological Analysis and Neutralizing Antibody Levels in Coronavirus Disease 2019 Convalescent Patients, J. Infect. Dis. 223 (2020) 389–398. doi:10.1093/infdis/jiaa659. [DOI] [PMC free article] [PubMed]
- 19.Centers for Disease Control and Prevention (US), SARS-CoV-2 Variants of Concern, Centers Dis. Control Prev. (2021). https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html#Concern%0Ahttps://www.cdc.gov/coronavirus/2019-ncov/cases-updates/variant-surveillance/variant-info.html. (accessed 12 June 2021).
- 20.Ferrari Davide, Di Resta Chiara, Tomaiuolo Rossella, Sabetta Eleonora, Pontillo Marina, Motta Andrea, Locatelli Massimo. Long-term antibody persistence and exceptional vaccination response on previously SARS-CoV-2 infected subjects. Vaccine. 2021;39(31):4256–4260. doi: 10.1016/j.vaccine.2021.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrari D., Manca M., Premaschi S., Banfi G., Locatelli M. Toxicological investigation in blood samples from suspected impaired driving cases in the Milan area : Possible loss of evidence due to late blood sampling. Forensic Sci Int. 2018;288:211–217. doi: 10.1016/j.forsciint.2018.04.038. [DOI] [PubMed] [Google Scholar]
- 22.Payne R B. Method comparison: Evaluation of least squares, Deming and Passing/Bablok regression procedures using computer simulation. Ann. Clin. Biochem. 1997;34(3):319–320. doi: 10.1177/000456329703400317. [DOI] [PubMed] [Google Scholar]
- 23.Venkatraman E.S. A permutation test to compare receiver operating characteristic curves. Biometrics. 2000;56:1134–1138. doi: 10.1111/j.0006-341X.2000.01134.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.