Abstract
The gut microbiota of insects usually plays an important role in the development and reproduction of their hosts. The fecundity of Henosepilachna vigintioctopunctata (Fabricius) varies greatly when they develop on different host plants. Whether and how the gut microbiota regulates the fecundity of H. vigintioctopunctata was unknown. To address this question, we used 16S rRNA sequencing to analyze the gut microbiomes of H. vigintioctopunctata adults fed on two host plant species (Solanum nigrum and Solanum melongena) and one artificial diet. The development of the ovaries and testes was also examined. Our results revealed that the diversity and abundance of gut microorganisms varied significantly in insects reared on different diets. The gut microbiota of H. vigintioctopunctata raised on the two host plants was similar, with Proteobacteria being the dominant phylum in both groups, whereas Firmicutes was the dominant phylum in the group reared on the artificial diet. The predominant microbiota in the S. nigrum group were Acinetobacter soli and Acinetobacter ursingii (Acinetobacter, Moraxellaceae); Moraxella osloensis (Enhydrobacter, Moraxellaceae); and Empedobacter brevis (Empedobacter, Weeksellaceae). The microbiota in this group are associated with high lipid metabolism. In addition, the beetles’ ovaries and testes were more highly developed in the S. nigrum group than in the other two groups. These findings provide valuable information for elucidating the complex roles the gut microbiota play in the fecundity of H. vigintioctopunctata, and may also contribute to developing future novel control strategies involving this economically important pest.
Keywords: Henosepilachna vigintioctopunctata, diet, gut microorganism, reproduction
The insect gut, which is a vital organ for digestion and nutrient absorption, is inhabited by a large complex of microorganisms. Insects and their gut microorganisms share symbiotic relationships that have proven crucial in adaptation and evolution (Janson et al. 2008). The gut microorganisms in insects directly or indirectly influence the adaptability and foraging behavior of their host (Akami et al. 2019b), while also providing a vital role in their metabolism and reproduction (Behar et al. 2008; Sharon et al. 2011a, b). They play an important role in growth, development, resistance to pathogens, and the maintenance of homoeostasis in insects (Shin et al. 2011, Shuai et al. 2020). In recent years, the gut microorganisms in insects have attracted increased attention owing to their significance from an ecological and economic perspective. As a result, research on the relationship between the gut microbiome and physiological and developmental events in the host has become vital for understanding this aspect of insect biology.
Microorganisms found in insect guts often have metabolic characteristics that the insect lacks and thereby contribute to vitamin (Salem et al. 2014) and other nutrient provisions for the host (Scully et al. 2014, Hu et al. 2018). These microbes also aid in digesting recalcitrant food (Kaufman and Klug 1991, Salem et al. 2017) and plant allelochemicals (Mason et al. 2014, Ceja-Navarro et al. 2015, Welte et al. 2016). As a result of prolonged periods of co-evolution, gut microorganisms have formed symbiotic relationships with their insect hosts, enabling the insects to adapt to different plant species (Medina et al. 2011). Considering how susceptible the insect gut microbiota is to change, variations in the host plant can cause transformations in gut microbial diversity (Lee et al. 2017), as observed in studies on Hylobius abietis (Berasategui et al. 2016), Thaumetopoea pityocampa (Strano et al. 2018), Spodoptera frugiperda and Helicoverpa zea (Jones et al. 2019). Changes in host plants are also known to cause differences in reproduction in several insects (Zhou et al. 2006, Liang et al. 2008, Lu et al. 2016). Therefore, clarifying the ‘host plant–insect gut microbiota–insect fecundity’ relationships can lead to a more thorough understanding of the variations that occur in insect host fecundity.
Henosepilachna vigintioctopunctata (Fabricius) is a widely distributed vegetable pest from Asia that feeds on a variety of solanaceous plants, including Solanum nigrum, Solanum melongena, Solanum tuberosum, Solanum lycopersicum, Capsicum annuum, Cucumis sativus, Benincasa hispida, and Nicotiana tabacum (Zhou et al. 2015). Due to the rapid growth of greenhouse vegetable cultivation in Asia and the year-round presence of its natural host (S. nigrum), H. vigintioctopunctata are now known to actively feed on plants year-round, increasing their economic impact throughout the region. Accordingly, there is an urgent need for effective management and control of this pest. Previous studies have shown that H. vigintioctopunctata adults reared on different host plants have different fecundities (Zhou et al. 2015, Wang et al. 2017). In addition, using 16S rRNA gene sequencing to identify the gut microorganisms in fourth-instar larvae, the composition of the gut microbiota was shown to differ in larvae fed on different host plants (Lü et al. 2019a). However, the composition of the gut microbial fauna in H. vigintioctopunctata adults fed on different host plants was unknown. It is possible that elucidation of these food source-dependent variations can provide new strategies for use in their control.
In the present study, we used high-throughput sequencing (16S rRNA gene sequencing) to identify the composition and diversity of the gut microbiota of H. vigintioctopunctata adults reared on different food sources (S. nigrum, S. melongena, and an artificial diet [based on our previous study]). The objective was to determine whether diets can influence gut microorganisms in H. vigintioctopunctata adults in order to provide a baseline for identifying and studying the functions of the gut microorganisms in this insect and to aid in developing effective and environmentally friendly techniques for controlling this important pest.
Materials and Methods
Insect Food and Sample Preparation
Henosepilachna vigintioctopunctata adults were collected from Solanum nigrum at Yangtze University, Jingzhou, Hubei province, China in July 2020, and were reared on S. nigrum at 25 ± 1°C, 75% ± 5% RH, and a photoperiod of 16:8 (L:D) for three generations (Lü et al. 2019b). Adults that emerged on the same day were divided into three groups. One group was transferred to S. melongena leaves (Sm group), one to S. nigrum leaves (Sn group), and one reared on artificial diet (Ad group).
Solanum melongena (cv. Heilongchangfeng F1) and S. nigrum (transplanted) were cultivated in planting pots (diameter, 20 cm; height, 11 cm) under a natural temperature regime (24–30°C). During the experimental period, fresh leaves were obtained daily, washed with ddH2O, and fed to H. vigintioctopunctata. The artificial diet used in the study consisted of the following: Leptinotarsa decemlineata artificial diet (Frontier Corporation, CT) 14%, agar powder 1.5%, yeast extract 3%, sorbic acid 0.1%, methyl para-hydroxybenzoate 0.1%, locust bean gum 0.2%, α-cellulose 3%, and 75% KOH 1%. The S. melongena leaves, S. nigrum leaves, and the artificial diet were replaced daily.
After H. vigintioctopunctata adults were allowed to feed for 10 d, the midguts of the unsexed beetles were extracted by placing the adults on ice (freezing anesthesia), disinfecting them with 75% ethanol for 90 s, washing 3× with ddH2O, and then dissecting the abdomens using a microscope under sterile conditions. The midguts were then collected in a 1.5-ml centrifuge tube and cryopreserved at −80°C for subsequent DNA extraction. Each complete sample consisted of 30 midguts, with three replicates per group.
Extraction of Gut Microbial DNA and 16S rRNA Sequencing
The CTAB method was used to extract total genomic DNA, with the solution diluted to 1 ng/µl using sterile water. Using the diluted DNA as a template, universal primers (515F: 5′-GTGCCAGCMGCCGCGGTAA-3′; 806R: 5′-GGACTACHVGGG TWTCTAAT-3′; Sampson et al. 2016, Snijders et al. 2016) were selected to target the V4 hypervariable region of 16S rRNA. Each sample was subjected to PCR in 30 µl of the reaction mixture, including 15 µl of Master Mix (New England Biolabs, MA), 0.2 µM of forward and reverse primers, and ca. 10 ng template DNA. The PCR conditions were as follows: initial denaturation at 98°C for 1 min followed by 30 cycles of 98°C (10 s), 50°C (30 s), and 72°C (30 s), and a post-PCR incubation at 72°C for 5 min. Samples containing bright main bands between 400 and 450 bp were retained for further experiments. The sequencing library was generated using the Truseq DNA PCR-Free kit (Illumina, CA), with an index code added. Library quality was evaluated using a Qubit 2.0 Fluorometer (Thermo Scientific, MA). Finally, the library was sequenced on the Illumina NovaSeq platform and 250-bp paired-end read segments were generated.
Bioinformatics and Data Analysis
If the similarity between sequences was more than 97%, they were assigned to the same operational taxonomic units (OTUs). The alpha diversity (within samples) of OTUs was analyzed through abundance-based coverage estimator (ACE) and Chao1, Shannon, and Simpson indices to obtain the species richness and uniformity information in the samples, as well as to identify common and unique OTUs among different samples. In the beta diversity analysis, weighted UniFrac-based Principal Coordinates Analysis (PCoA) was used to analyze the clustering of microbial communities in different samples through complex and multidimensional data. We also used unweighted pair-group method with arithmetic mean (UPGMA), which is based on average links, to interpret the distance matrix. The linear discriminant analysis effect size (LefSe) function was used to explore the differences in clusters among samples in a group, and the significance of differences in species composition and clustering was examined. Subsequently, the function of gut microorganisms was demonstrated using Tax4Fun.
Comparison of Reproductive System Development According to Diet
To observe the developmental progress in H. vigintioctopunctata adults reared on different diets, 20 4-d-old and 20 7-d-old female and male adults were randomly selected for dissection. Phenotypes were judged visually, and images were obtained using a Leica M205A stereomicroscope with motorized zoom and focus control. The length (mm) and width (mm) of the ovary and testis were measured, and the index (length × width) was calculated using LAS (Version 4.3) software. SPSS software (Version 22) was used to analyze the different development parameters. The parameters were expressed as mean ± standard error. The paired Tukey test was used for multiple comparisons of mean values, with P ≤ 0.05 considered to be statistically significant.
Results
Diversity and Abundance of the Gut Microbiota in H. vigintioctopunctata Adults Reared on Different Diets
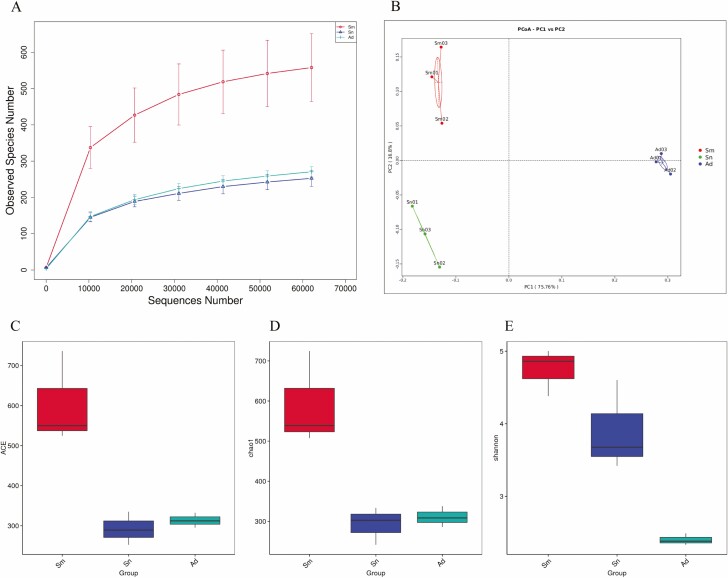
We examined the gut microbiota of H. vigintioctopunctata adults reared on different diets and identified the V4 hypervariable region of the 16S rRNA gene using high-throughput sequencing. The coverage was over 99%, indicating that the number of clones was sufficient to adequately sample the microbial diversity in the adult beetles (Table 1). Rarefaction analysis showed that the number of species increased rapidly before plateauing (Fig. 1A), indicating that the data covered most of the microbial information in the sample and was therefore valid and had research value. UniFrac-based PCoA showed that the Sm, Sn, and Ad groups were separated along the first two axes (a measure of the phylogenetic differences among gut microbial communities of the three groups), accounting for 75.76% and 16.8% of the data changes, respectively. There was some distance among the three groups, but the within-group distance was relatively short, indicating that H. vigintioctopunctata reared on different food sources showed differences in their gut microbial communities (Fig. 1B).
Table 1.
Diversity and abundance of gut microbial communities in H. vigintioctopunctata
| Sample | Good’s coverage | Observed species | Shannon | Simpson | Chao1 | Ace | PD whole tree index |
|---|---|---|---|---|---|---|---|
| Sm01 | 0.999 | 507 | 5.002 | 0.930 | 538.897 | 549.840 | 43.763 |
| Sm02 | 0.999 | 478 | 4.382 | 0.842 | 507.583 | 524.701 | 33.197 |
| Sm03 | 0.999 | 689 | 4.861 | 0.905 | 724.442 | 736.163 | 52.058 |
| Sn01 | 0.999 | 284 | 4.603 | 0.925 | 333.763 | 334.947 | 26.100 |
| Sn02 | 1.000 | 235 | 3.420 | 0.793 | 241.591 | 252.358 | 22.353 |
| Sn03 | 0.999 | 239 | 3.676 | 0.828 | 302.370 | 289.106 | 19.486 |
| Ad01 | 0.999 | 265 | 2.489 | 0.733 | 308.571 | 312.190 | 25.922 |
| Ad02 | 0.999 | 257 | 2.332 | 0.700 | 286.057 | 295.366 | 32.528 |
| Ad03 | 0.999 | 290 | 2.381 | 0.650 | 337.838 | 332.425 | 32.286 |
Fig. 1.
Diversity and abundance of the gut microbial communities in H. vigintioctopunctata examined via Rarefaction analysis (A), PcoA (B), ACE analysis (C), the Chao1 index (D), and the Shannon index (E).
When the effects of the diet on species diversity were examined, significant differences in gut microbial diversity were observed in the Sm group, as shown by ACE and Chao1 indices (Fig. 1C and D, Table 1). According to the Shannon index, the microbial diversity was higher in the Sm and Sn groups than in the Ad group, and the species distribution was more uniform (Fig. 1E; Table 1).
Composition and Structure of Gut Microbial Communities in H. vigintioctopunctata Reared on Different Food Sources
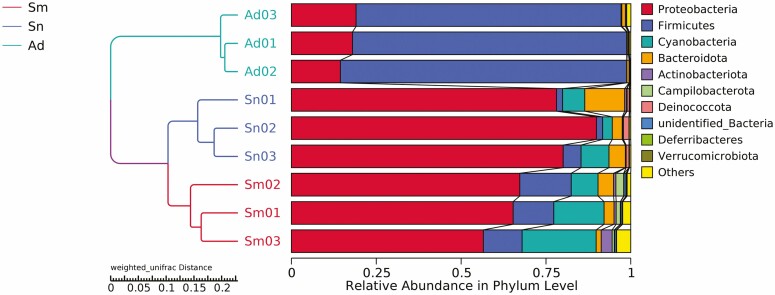
The diet of H. vigintioctopunctata affected the composition and structure of the gut microbial community at six taxonomic levels (Supp Fig. S1 [online only]). We observed similarity between different samples, and a cluster tree could be constructed through UPGMA. The composition of the Sm group was similar to that of the Sn group but differed from the Ad group (Fig. 2).
Fig. 2.
Weighted UniFrac UPGMA tree of the gut microorganisms in H. vigintioctopunctata.
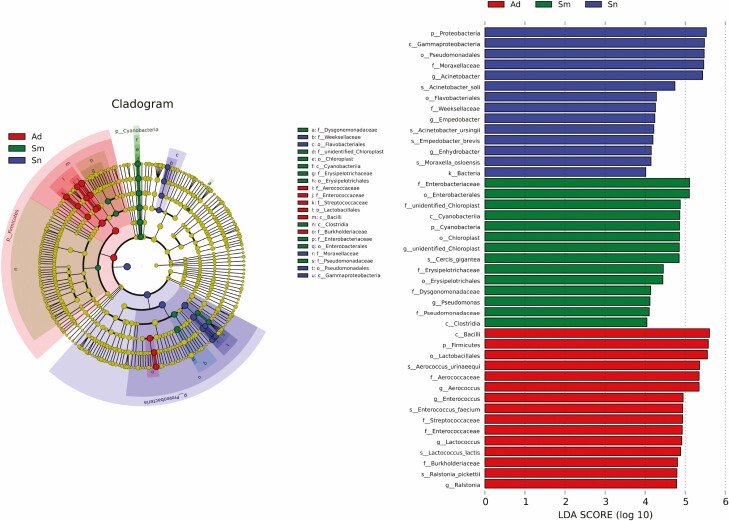
LefSe was used to recognize key phylotypes responsible for diet-related microbial differences. The Sn group had a preponderance of Acinetobacter soli and Acinetobacter ursingii (Acinetobacter, Moraxellaceae, Pseudomonadales, Gammaproteobacteria, Proteobacteria, Bacteria); Moraxella osloensis (Enhydrobacter, Moraxellaceae); and Empedobacter brevis (Empedobacter, Weeksellaceae, Flavobacteriales). The Sm group showed Enterobacteriaceae (Enterobacterales); Cercis gigantea (unidentified Chloroplast, unidentified Chloropla, Chloroplast, Cyanobacteria, Cyanobacteria); Erysipelotrichaceae (Erysipelotrichales); Dysgonomonadaceae; Pseudomonas (Pseudomonadaceae); and Clostridia. However, Aerococcus urinaeequi (Aerococcus, Aerococcaceae, Lactobacillales, Bacilli, Firmicutes); Enterococcus faecium (Enterococcus, Enterococcaceae); Lactococcus lactis (Lactococcus, Streptococcaceae); and Ralstonia pickettii (Ralstonia, Burkholderiaceae) were more consistently present in the Ad group than in the other groups (Fig. 3).
Fig. 3.
Abundance of different gut microbial taxa observed in the three groups using linear discriminant analysis effect size (LEfSe analysis).
Effects of Different Diets on Gut Microbial Function in H. vigintioctopunctata
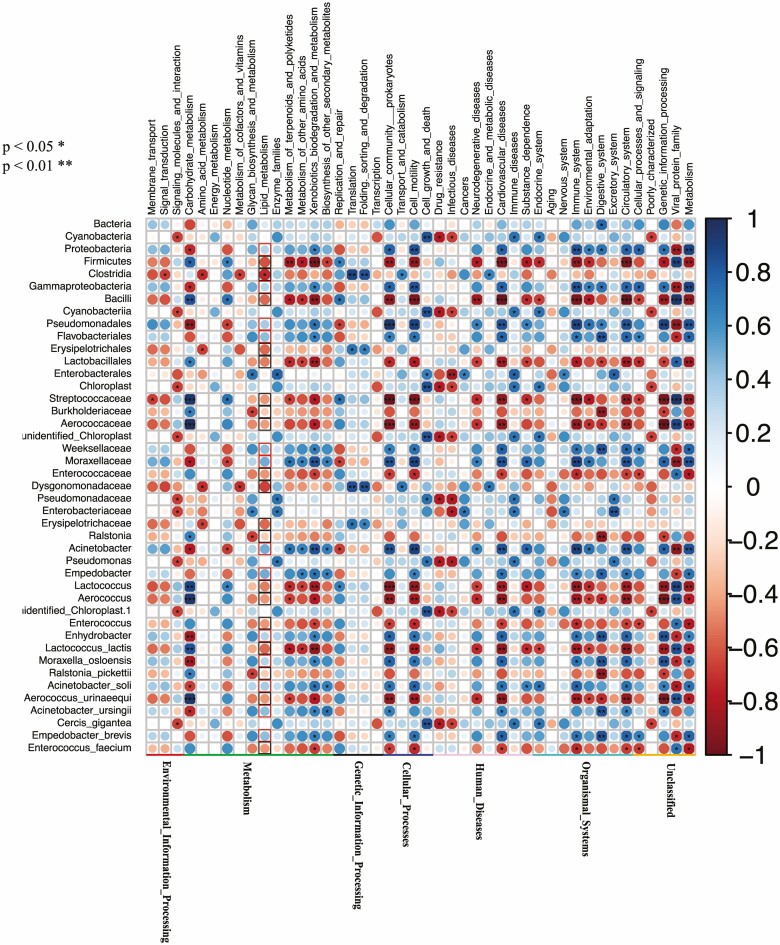
The Tax4Fun function predicted that at the KEGG level 2, the Sm group was similar to the Ad group, but different from the Sn group (Supp Fig. S2 [online only]). Using Tax4Fun and correlating the identified microbial communities from the LEfSe analysis with biological pathway analysis (KEGG) to examine the key phylotypes causing changes in gut microbial function (Fig. 4), we did observe relationships between gut microbes and functions, although there were certain differences in the functions of different gut microorganisms. Of note, the gut microbes in the Sn group were associated with high lipid metabolism (red circles, Fig. 4). In contrast, the gut microbes in the Ad group showed a negative correlation with lipid metabolism levels (Fig. 4, black circles). The Erysipelotrichaceae, Erysipelotrichales, Dysgonomonadaceae, and Clostridia observed in the Sm group had a similar negative effect on lipid metabolism (Fig. 4, black circles). However, the relationship of the other gut microorganisms in the Sm group with lipid metabolism was not obvious.
Fig. 4.
Correlation between the microbes identified in the LEfSe analyses with Tax4Fun functional predictions.
Effect of Different Diets on Reproductive Development
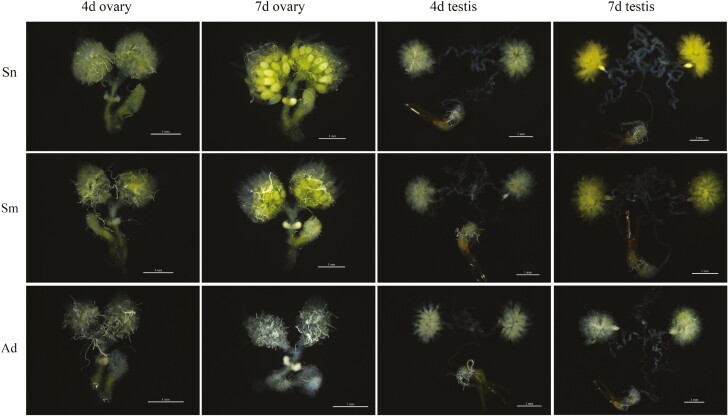
As shown in Table 2, there were significant differences in the developmental parameters in H. vigintioctopunctata fed on different diets. Ovary and testis development were the most rapid in the Sn group, followed by the Sm group, with the Ad group having the slowest rate of development. On day 7 postemergence, the ovaries of female adults were enlarged in both the Sn and Sm groups, with yellowish egg cells appearing in the middle ovarioles, single ovarioles were plainly visible, and the entire ovary was obviously yellow. The ovaries were slightly larger in females from the Sn group than they were in the Sm group, while in the Ad group, the individual ovarioles did not separate and were difficult to distinguish. On day 7 postemergence, males from the Sn group had completely developed testes that were distinctly yellow in color, whereas the testes from the Sm group were yellowish and those from the Ad group white (Fig. 5).
Table 2.
Comparison of developmental parameters (mean ± SE, n = 20) in H. vigintioctopunctata fed on different diets
| Treatment | Sn | Sm | Ad | Sn vs Sm vs Ad | |||
|---|---|---|---|---|---|---|---|
| df | F | P | |||||
| 4d Ovary | Ovary length (mm) | 1.64 ± 0.00a | 1.44 ± 0.00b | 1.22 ± 0.00c | 2, 57 | 11,723.890 | <0.001 |
| Ovary width (mm) | 1.47 ± 0.00a | 1.23 ± 0.00b | 1.00 ± 0.00c | 2, 57 | 16,029.252 | <0.001 | |
| Ovarian index (mm2) | 2.40 ± 0.01a | 1.77 ± 0.00b | 1.22 ± 0.00c | 2, 57 | 19,403.858 | <0.001 | |
| 7d Ovary | Ovary length (mm) | 2.36 ± 0.00a | 1.90 ± 0.00b | 1.28 ± 0.00c | 2, 57 | 128,473.303 | <0.001 |
| Ovary width (mm) | 1.91 ± 0.00a | 1.89 ± 0.00b | 1.03 ± 0.00c | 2, 57 | 196,517.716 | <0.001 | |
| Ovarian index (mm2) | 4.50 ± 0.00a | 3.59 ± 0.00b | 1.32 ± 0.00c | 2, 57 | 153,176.065 | <0.001 | |
| 4d Testis | Testis length (mm) | 1.85 ± 0.01a | 1.75 ± 0.01b | 1.74 ± 0.01b | 2, 57 | 119.842 | <0.001 |
| Testis width (mm) | 1.70 ± 0.00a | 1.44 ± 0.00b | 1.40 ± 0.01c | 2, 57 | 1,391.714 | <0.001 | |
| Testis index (mm2) | 3.15 ± 0.01a | 2.53 ± 0.01b | 2.44 ± 0.00c | 2, 57 | 3,794.193 | <0.001 | |
| 7d Testis | Testis length (mm) | 2.50 ± 0.00a | 1.87 ± 0.00b | 1.77 ± 0.00c | 2, 57 | 11,553.412 | <0.001 |
| Testis width (mm) | 1.92 ± 0.00a | 1.59 ± 0.00b | 1.48 ± 0.00c | 2, 57 | 4,244.647 | <0.001 | |
| Testis index (mm2) | 4.80 ± 0.00a | 2.98 ± 0.01b | 2.62 ± 0.01c | 2, 57 | 24,177.694 | <0.001 |
Data indicated by the different letters in the same line showed significant differences at a level of 5% (P ≤ 0.05).
Fig. 5.
Ovary and testis development in H. vigintioctopunctata adults reared on different diets (white bars indicate a scale of 1 mm).
Discussion
Diets play an important role in shaping the composition and functional contribution of the gut microbiota. It has been reported that Leptinotarsa decemlineata, which has a host range similar to that of H. vigintioctopunctata, developed a different gut microbiota when reared on different plants. It has also been shown that symbiotic microorganisms can decrease plant defenses in all Solanum hosts (Chung et al. 2017). Previous study on gut microorganisms in H. vigintioctopunctata, which focused on fourth instar larvae, reported that Proteobacteria and Gammaproteobacteria were the most abundant and dominant microorganisms in the guts of larvae reared on S. nigrum and S. melongena (Lü et al. 2019a). Proteobacteria are the predominant gut microorganisms found in insects when fed on natural foods or plants (e.g., Bactrocera minax [Wang et al. 2014], Ectropis obliqua [Jin et al. 2013], Musca domestica [Gupta et al. 2011], and Nilaparvata lugens [Wang et al. 2019]), indicating that Proteobacteria are widely present in the gut in insects that feed on natural food sources. Several differences were noted between our current results and previous findings. For example, Cyanobacteria was found in both the Sn and Sm groups in the present study, but were only found in H. vigintioctopunctata reared on S. nigrum in the previous study. Moreover, the differences in microbial clusters were mainly concentrated at three taxonomic levels: family, genus, and species. In the previous study involving H. vigintioctopunctata fourth instar larvae, the predominant microorganisms were unclassified Enterobacteriaceae (Enterobacteriaceae), whereas in this study, Acinetobacter (Moraxellaceae) was predominant in both groups. These differences may reflect differences in geographic populations (Wang et al. 2020), life stages (Yang et al. 2020), growth environments, or nutrient accumulation in the host plant. Studies have shown that although host plants are the primary driving factors of gut microbial composition (Jones et al. 2019), other factors, including habitat (Paniagua et al. 2018) and developmental stage (Yun et al. 2014), can also affect gut microbial composition in insects.
Artificial diets are a poorer microbial source for insects than natural plant diets (Priya et al. 2012). The gut microbial composition of H. vigintioctopunctata fed on artificial diets (Ad group) was significantly different from that of H. vigintioctopunctata fed on host plants (Sm and Sn groups). The dominant phylum was Proteobacteria in the Sm and Sn groups, and Firmicutes in the Ad group. Firmicutes are involved in energy absorption, which is associated with high fiber diets (Jandhyala et al. 2015), and thereby modulate the ability of insects to obtain energy from their diet (Chen et al. 2016). Previous studies, including those on Ostrinia nubilalis (Eugeni et al. 2011) and Helicoverpa armigera (Xiang et al. 2006), found that Firmicutes become the dominant gut microorganism after rearing on artificial diets. These results indicate that Firmicutes play an important role in the gut of insects reared on artificial diets, and their roles in carbohydrate and nucleotide metabolism may facilitate the digestion and absorption of artificial foods and nutrients. It is worth noting that the number of Cyanobacteria was extremely low in the Ad group. This has also been observed in a variety of animals, including Bombyx mori (Dong et al. 2018), chickens (Ding et al. 2016), pigs (Niu et al. 2015), and horses (Zhao et al. 2016) reared on artificial diets. This phenomenon may be related to the fact that there are abundant components for nutrition in the leaves of the host plant that are lacking in artificial diets.
It is important to note that the Enterobacteriaceae were found in the Sm and Sn groups, whereas Enterococcaceae were found in the Ad group. Enterobacteriaceae can help their hosts fix nitrogen from plants and hydrolyze pectin (Behar et al. 2005, 2008). They also affect pheromone synthesis (Wada-Katsumata et al. 2015), reduce the effect of trichlorfon, increase insecticide resistance (Cheng et al. 2017), and play an important functional role in host fitness. A study showed that a reduction in Enterobacteriaceae, which were the most abundant microbes in the gut of Bactrocera dorsalis, may cause a decrease in nitrogen fixation, obstruct pheromone synthesis, and contribute to a decrease in fertility, reducing the ecological fitness of sterile males (Cai et al. 2018). Studies on B. dorsalis and Zeugodacus tau have also confirmed that Enterobacter have a positive effect on reproduction, while Enterococcus have a negative effect (Akami et al. 2019a, Noman et al. 2020). This is consistent with our finding that ovipositing did not occur in the Ad group when Enterococcaceae were observed in the gut, whereas ovipositing was successful in the Sn and Sm groups, when Enterobacteriaceae were present in the gut.
Lipid biosynthesis pathways and lipophorin are linked to reproductive fitness in insects (Ziegler and Antwerpen 2006, Fruttero et al. 2011). Lipids are needed for the synthesis of vitellogenin (Vg; Tufail and Takeda 2008), which affects the number and quality of eggs (Liu et al. 2015), provides nutrition for embryo development, and serves as an energy reserve (Tiu et al. 2009), directly or indirectly affecting insect fecundity (Tian et al. 2004, Xu et al. 2010, Zhai et al. 2015). Decreasing lipid accumulation may then result in an inhibition of Vg protein synthesis leading to a reduction of fecundity in insects. In addition, lipids are an important component of insect oocytes, not only accounting for a significant proportion of oocyte dry weight (30–40%; Kawooya and Law 1988, Hans 1990) but also providing energy to the embryos (Beenakkers et al. 1981). It can be concluded from the above that lipids do play an important role in insect reproduction. In the present study, LEfSe and Tax4Fun analyses demonstrated a corresponding relationship between the gut microbes present in H. vigintioctopunctata reared on different food sources and their functions. We found that the major microorganisms involved in lipid metabolism (which is likely related to insect fecundity) were associated with the high fecundity of H. vigintioctopunctata reared on S. nigrum (Fig. 4). Furthermore, the advanced development of the testes and ovaries seen in adults that fed on S. nigrum (Table 2; Fig. 5) further supports this conclusion. Therefore, gut microorganisms involved in lipid metabolism may affect insect fecundity through the regulation of lipid metabolism. The diversity of insect gut microorganisms varies with variations in food intake; hence, gut microbial diversity may be a major factor contributing to diet-related insect fecundity.
Supplementary Data
Supplementary data are available at Journal of Insect Science online.
Supp Fig. S1. Relative abundance of the gut microorganism in H. vigintioctopunctata at the phylum (A), class (B), order (C), family (D), genus (E), and species (F) levels
Supp Fig. S1. Relative abundance of predicted gut microbial functions in three groups, as predicted by Tax4fun using KEGG Orthologs
Acknowledgments
We are grateful to the reviewers and the editor for their valuable comments and suggestions, all of which have greatly helped us in improving this manuscript. This study was supported by the National Natural Science Foundation of China (31572010).
Author Contributions
H. W. Li and C. R. Li conceived the original idea. H. W. Li carried out the experiment and wrote the manuscript with support from C. R. Li. C. W. Zhao assisted in the conduct of experiment and data collection. Y. Yang and J. W. Qi reared the insects required for experiments. Z. X. Zhou revised the manuscript.
References Cited
- Akami, M., Ren X. M., Qi X., Mansour A., Gao B., Cao S., and Niu C. Y.. . 2019a. Symbiotic bacteria motivate the foraging decision and promote fecundity and survival of Bactrocera dorsalis (Diptera: Tephritidae). BMC Microbiol. 19: 293–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akami, M., Andongma A. A., Zhengzhong C., Nan J., Khaeso K., Jurkevitch E., Niu C. Y., and Yuval B.. . 2019b. Intestinal bacteria modulate the foraging behavior of the oriental fruit fly Bactrocera dorsalis (Diptera: Tephritidae). PLoS One 14: e0210109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenakkers, A. M. T., Horst D. J. V. D., and Marrewijk W. J. A. V.. . 1981. Role of lipids in energy metabolism, 53–100. In Roger G. H. Downer, Energy Metabolism in Insects. Springer, Boston, MA, America. [Google Scholar]
- Behar, A., Yuval B., and Jurkevitch E.. . 2005. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol. Ecol. 14: 2637–2643. [DOI] [PubMed] [Google Scholar]
- Behar, A., Yuval B., and Jurkevitch E.. . 2008. Gut bacterial communities in the Mediterranean fruit fly (Ceratitis capitata) and their impact on host longevity. J. Insect Physiol. 54: 1377–1383. [DOI] [PubMed] [Google Scholar]
- Berasategui, A., Axelsson K., Nordlander G., Schmidt A., Borg-Karlson A. K., Gershenzon J., Terenius O., and Kaltenpoth M.. . 2016. The gut microbiota of the pine weevil is similar across Europe and resembles that of other conifer-feeding beetles. Mol. Ecol. 25: 4014–4031. [DOI] [PubMed] [Google Scholar]
- Cai, Z., Yao Z., Li Y., Xi Z., Bourtzis K., Zhao Z., Bai S., and Zhang H.. . 2018. Intestinal probiotics restore the ecological fitness decline of Bactrocera dorsalis by irradiation. Evol. Appl. 11: 1946–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceja-Navarro, J. A., Vega F. E., Karaoz U., Hao Z., Jenkins S., Lim H. C., Kosina P., Infante F., Northen T. R., and Brodie E. L.. . 2015. Gut microbiota mediate caffeine detoxification in the primary insect pest of coffee. Nat Commun. 6: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, B., Teh B. S., Sun C., Hu S., Lu X., Boland W., and Shao Y.. . 2016. Biodiversity and activity of the gut microbiota across the life history of the insect herbivore Spodoptera littoralis. Sci. Rep. 6: 1120–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, D., Guo Z., Riegler M., Xi Z., Liang G., and Xu Y.. . 2017. Gut symbiont enhances insecticide resistance in a significant pest, the oriental fruit fly Bactrocera dorsalis (Hendel). Microbiome 5: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung, S. H., Scully E. D., Peiffer M., Geib S. M., Rosa C., Hoover K., and Felton G. W.. . 2017. Host plant species determines symbiotic bacterial community mediating suppression of plant defenses. Sci. Rep. 7: 39690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, J., Zhao L., Wang L., Zhao W., Zhai Z., Leng L., Wang Y., He C., Zhang Y., Zhang H., . et al. 2016. Divergent selection-induced obesity alters the composition and functional pathways of chicken gut microbiota. Genet. Sel. Evol. 48: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, H. L., Zhang S. X., Chen Z. H., Tao H., and Xu S. Q.. . 2018. Differences in gut microbiota between silkworms (Bombyx mori) reared on fresh mulberry (Morus alba var. multicaulis) leaves or an artificial diet. RSC Adv. 8: 26188–26200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugeni, B., Laia P., Juli P., Martínez-Blanch J. F., Arnau M., Emilio N., Javier U., Daniel R., Andrés M., and Manuel P.. . 2011. Microbial diversity in the midguts of field and lab-reared populations of the European corn borer Ostrinia nubilalis. PLoS One 6: e21751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fruttero, L. L., Frede S., Rubiolo E. R., and Canavoso L. E.. . 2011. The storage of nutritional resources during vitellogenesis of Panstrongylus megistus (Hemiptera: Reduviidae): the pathways of lipophorin in lipid delivery to developing oocytes. J. Insect Physiol. 57: 475–486. [DOI] [PubMed] [Google Scholar]
- Gupta, A. K., Dana N., Pankaj V., Bhavin S., Ghate H. V., Patole M. S., and Shouche Y. S.. . 2011. Phylogenetic characterization of bacteria in the gut of house flies (Musca domestica L.). FEMS Microbiol. Ecol. 79: 581–593. [DOI] [PubMed] [Google Scholar]
- Hans, B. 1990. Fecundity, metabolism, and body size in Anopheles (Diptera: Culicidae),vectors of malaria. J. Med. Entomol. 27: 839–850. [DOI] [PubMed] [Google Scholar]
- Hu, Y., Sanders J. G., Lukasik P., D’Amelio C. L., Millar J. S., Vann D. R., Lan Y., Newton J. A., Schotanus M., and Kronauer D. J. C.. . 2018. Herbivorous turtle ants obtain essential nutrients from a conserved nitrogen-recycling gut microbiome. Nat. Commun. 9: 1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandhyala, S. M., Talukdar R., Subramanyam C., Vuyyuru H., Sasikala M., and Nageshwar Reddy D.. . 2015. Role of the normal gut microbiota. World J. Gastroenterol. 21: 8787–8803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson, E. M., J. O.Stireman, 3rd, Singer M. S., and Abbot P.. . 2008. Phytophagous insect-microbe mutualisms and adaptive evolutionary diversification. Evolution 62: 997–1012. [DOI] [PubMed] [Google Scholar]
- Jin, L., Wang J. C., Wang H. X., Zhang J. G., Yang G., and Jin H. Y.. . 2013. Bacterial community in midguts of Ectropic oblique larvae by PCR-DGGE and 16S rRNA gene library analysis. Jiangxi Sci. 31: 759–763,829. (in Chinese) [Google Scholar]
- Jones, A. G., Mason C. J., Felton G. W., and Hoover K.. . 2019. Host plant and population source drive diversity of microbial gut communities in two polyphagous insects. Sci. Rep. 9: 17–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman, M. G., and Klug M. J.. . 1991. The contribution of hindgut bacteria to dietary carbohydrate utilization by crickets (Orthoptera: Gryllidae). Comp. Biochem. Physiol. 98: 117–123. [Google Scholar]
- Kawooya, J. K., and Law J. H.. . 1988. Role of lipophorin in lipid transport to the insect egg. J. Biol. Chem. 263: 8748–8753. [PubMed] [Google Scholar]
- Lee, J. H., Lee K. A., and Lee W. J.. . 2017. Microbiota, gut physiology, and insect immunity. Adv. Insect Physiol. 52: 111–138. [Google Scholar]
- Liang, Y. Y., Li H. G., He Z., and Ge F.. . 2008. Effect of alternative hosts on the growth and development of massion pine caterpillar, Dendrolimus punctatus. Chin. Bull. Entomol. 45: 739–743. (in Chinese) [Google Scholar]
- Liu, C., Mao J., and Zeng F.. . 2015. Chrysopa septempunctata (Neuroptera: Chrysopidae) Vitellogenin functions through effects on egg production and hatching. J. Econ. Entomol. 108: 2779–2788. [DOI] [PubMed] [Google Scholar]
- Lu, H., Yang P., Xu Y., Luo L., Zhu J., Cui N., Kang L., and Cui F.. . 2016. Performances of survival, feeding behavior, and gene expression in aphids reveal their different fitness to host alteration. Sci. Rep. 6: 810–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü, J., Guo W., Chen S., Guo M., Qiu B., Yang C., Lian T., and Pan H.. . 2019a. Host plants influence the composition of the gut bacteria in Henosepilachna vigintioctopunctata. PLoS One 14: e0224213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lü, J., Chen S., Guo M., Ye C., Qiu B., Wu J., Yang C., and Pan H.. . 2019b. Corrigendum: selection and validation of reference genes for RT-qPCR analysis of the Ladybird Beetle Henosepilachna vigintioctopunctata. Front. Physiol. 10: 981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason, C. J., Couture J. J., and Raffa K. F.. . 2014. Plant-associated bacteria degrade defense chemicals and reduce their adverse effects on an insect defoliator. Oecologia 175: 901–910. [DOI] [PubMed] [Google Scholar]
- Medina, R. F., Nachappa P., and Tamborindeguy C.. . 2011. Differences in bacterial diversity of host-associated populations of Phylloxera notabilis Pergande (Hemiptera: Phylloxeridae) in pecan and water hickory. J. Evol. Biol. 24: 761–771. [DOI] [PubMed] [Google Scholar]
- Niu, Q., Li P., Hao S., Zhang Y., and Huang R.. . 2015. Dynamic distribution of the gut microbiota and the relationship with apparent crude fiber digestibility and growth stages in pigs. Sci. Rep. 5: 15485–15490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noman, M. S., Shi G., Liu L. J., and Li Z. H.. . 2020. The diversity of bacteria in different life stages and their impact on the development and reproduction of Zeugodacus tau (Diptera: Tephritidae). Insect Sci. 28: 363–376. [DOI] [PubMed] [Google Scholar]
- Paniagua, V. L. R., Enric F., Martin K., Monika H., and Fatouros N. E.. . 2018. Bacterial symbionts in Lepidoptera: their diversity, transmission, and impact on the host. Front. Microbiol. 9: 556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priya, N. G., Ojha A., Kajla M. K., Raj A., and Rajagopal R.. . 2012. Host plant induced variation in gut bacteria of Helicoverpa armigera. PLoS One 7: e30768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem, H., Bauer E., Strauss A. S., Vogel H., Marz M., and Kaltenpoth M.. . 2014. Vitamin supplementation by gut symbionts ensures metabolic homeostasis in an insect host. Proc. Biol. Sci. 281: 20141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem, H., Bauer E., Kirsch R., Berasategui A., Cripps M., Weiss B., Koga R., Fukumori K., Vogel H., Fukatsu T., . et al. 2017. Drastic genome reduction in an herbivore’s pectinolytic symbiont. Cell 171: 1520–1531.e13. [DOI] [PubMed] [Google Scholar]
- Sampson, T. R., Debelius J. W., Thron T., Janssen S., Shastri G. G., Ilhan Z. E., Challis C., Schretter C. E., Rocha S., Gradinaru V., . et al. 2016. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell. 167: 1469–1480.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully, E. D., Geib S. M., Carlson J. E., Tien M., McKenna D., and Hoover K.. . 2014. Functional genomics and microbiome profiling of the Asian longhorned beetle (Anoplophora glabripennis) reveal insights into the digestive physiology and nutritional ecology of wood feeding beetles. BMC Genomics 15: 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon, G., Segal D., Zilber-Rosenberg I., and Rosenberg E.. . 2011a. Symbiotic bacteria are responsible for diet-induced mating preference in Drosophila melanogaster, providing support for the hologenome concept of evolution. Gut Microbes 2: 190–192. [DOI] [PubMed] [Google Scholar]
- Sharon, G., Segal D., Ringo J. M., Hefetz A., Zilber-Rosenberg I., and Rosenberg E.. . 2011b. Commensal bacteria play a role in mating preference of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 107: 20051–20056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, S. C., Kim S. H., You H., Kim B., Kim A. C., Lee K. A., Yoon J. H., Ryu J. H., and Lee W. J.. . 2011. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334: 670–674. [DOI] [PubMed] [Google Scholar]
- Shuai, B., Zhichao Y., Fahim R. M., Zhaohui C., and Hongyu Z.. . 2020. Regulatory mechanisms of microbial homeostasis in insect gut. Insect Sci. 28: 286–301. [DOI] [PubMed] [Google Scholar]
- Snijders, A. M., Langley S. A., Kim Y. M., Brislawn C. J., Noecker C., Zink E. M., Fansler S. J., Casey C. P., Miller D. R., Huang Y., . et al. 2016. Influence of early life exposure, host genetics and diet on the mouse gut microbiome and metabolome. Nat. Microbiol. 2: 16221. [DOI] [PubMed] [Google Scholar]
- Strano, C. P., Malacrinò A., Campolo O., and Palmeri V.. . 2018. Influence of host plant on Thaumetopoea pityocampa gut bacterial community. Microb. Ecol. 75: 487–494. [DOI] [PubMed] [Google Scholar]
- Tian, H., Vinson S. B., and Coates C. J.. . 2004. Differential gene expression between alate and dealate queens in the red imported fire ant, Solenopsis invicta Buren (Hymenoptera: Formicidae). Insect Biochem. Mol. Biol. 34: 937–949. [DOI] [PubMed] [Google Scholar]
- Tiu, S. H., Hui H. L., Tsukimura B., Tobe S. S., He J. G., and Chan S. M.. . 2009. Cloning and expression study of the lobster (Homarus americanus) vitellogenin: conservation in gene structure among decapods. Gen. Comp. Endocrinol. 160: 36–46. [DOI] [PubMed] [Google Scholar]
- Tufail, M., and Takeda M.. . 2008. Molecular characteristics of insect vitellogenins. J. Insect Physiol. 54: 1447–1458. [DOI] [PubMed] [Google Scholar]
- Wada-Katsumata, A., Zurek L., Nalyanya G., Roelofs W. L., Zhang A., and Schal C.. . 2015. Gut bacteria mediate aggregation in the German cockroach. Proc. Natl. Acad. Sci. USA 112: 15678–15683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, A., Yao Z., Zheng W., and Zhang H.. . 2014. Bacterial communities in the gut and reproductive organs of Bactrocera minax (Diptera: Tephritidae) based on 454 pyrosequencing. PLoS One 9: e106988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. L., Li C. R., Yuan J. J., Li S. X., Wang X. P., and Chi H.. . 2017. Demographic comparison of Henosepilachna vigintioctopunctata (F.) (Coleoptera: Coccinellidae) reared on three cultivars of Solanum melongena L. and a wild hostplant Solanum nigrum L. J. Econ. Entomol. 110: 2084–2091. [DOI] [PubMed] [Google Scholar]
- Wang, T. Z., Wang Z. L., Zhu H. F., Wang Z. Y., and Yu X. P.. . 2019. Analysis of the gut microbial diversity of the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) by high-throughput sequencing. Acta Entomol. Sin. 62: 323–333. (in Chinese) [Google Scholar]
- Wang, Z. Y., Wang Y., He M. T., and Lu Y. J.. . 2020. Diversity of intestinal bacteria in different geographic populations of Tribolium castaneum (Coleoptera: Tenebrionidae). Chin. J. Appl. Entomol. 57: 617–622. (in Chinese) [Google Scholar]
- Welte, C. U., Graaf R. M. D., Bosch T. J. M. V. D., Camp H. J. M. O. D., Dam N. M. V., and Jetten M. S. M.. . 2016. Plasmids from the gut microbiome of cabbage root fly larvae encode SaxA that catalyses the conversion of the plant toxin 2-phenylethyl isothiocyanate. Environ. Microbiol. 18: 1379–1390. [DOI] [PubMed] [Google Scholar]
- Xiang, H., Wei G. F., Jia S., Huang J., Miao X. X., Zhou Z., Zhao L. P., and Huang Y. P.. . 2006. Microbial communities in the larval midgut of laboratory and field populations of cotton bollworm (Helicoverpa armigera). Can. J. Microbiol. 52: 1085–1092. [DOI] [PubMed] [Google Scholar]
- Xu, J., Tan A., and Palli S. R.. . 2010. The function of nuclear receptors in regulation of female reproduction and embryogenesis in the red flour beetle, Tribolium castaneum. J. Insect Physiol. 56: 1471–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y. J., Liu X. G., Xu H. X., Liu Y. H., Ali P., Bodlah M. A., and Lu Z. X.. . 2020. The abundance and diversity of gut bacteria of rice leaffolder Cnaphalocrocis medinalis (Guenée) across life stages. J. Asia-Pacif. Entomol. 23: 430–438. [Google Scholar]
- Yun, J. H., Roh S. W., Whon T. W., Jung M. J., and Bae J. W.. . 2014. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 80: 5254–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai, Y., Sun Z., Zhang J., Kang K., Chen J., and Zhang W.. . 2015. Activation of the TOR signalling pathway by glutamine regulates insect fecundity. Sci. Rep. 5: 10694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Li B., Bai D., Huang J., and Dugarjaviin M.. . 2016. Comparison of fecal microbiota of mongolian and thoroughbred horses by high-throughput sequencing of the V4 region of the 16S rRNA gene. Asian-Australas. J. Anim. Sci. 29: 1345–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, F. C., Ren S. X., Du Y. Z., and Wang Y.. . 2006. Effects of host plant exchange on survivorship, development and reproduction of B biotype Bemisia tabaci. Chin. J. Appl. Entomol. 43: 524–526. (in Chinese) [Google Scholar]
- Zhou, L., Xie B. G., Wang X. P., Li C. R., and Wang W. K.. . 2015. Population dynamic of Henosepilachna vigintioctopunctata on different host plant in Jianghan plain. Northern Horticult. 39: 103–105. (in Chinese) [Google Scholar]
- Ziegler, R., and Van Antwerpen R.. . 2006. Lipid uptake by insect oocytes. Insect Biochem. Mol. Biol. 36: 264–272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.