SUMMARY
There is a lack of appropriate melanoma models that can be used to evaluate the efficacy of novel therapeutic modalities. Here, we discuss the current state of the art of melanoma models including genetically engineered mouse, patient-derived xenograft, zebrafish, and ex vivo and in vitro models. We also identify five major challenges that can be addressed using such models, including metastasis and tumor dormancy, drug resistance, the melanoma immune response, and the impact of aging and environmental exposures on melanoma progression and drug resistance. Additionally, we discuss the opportunity for building models for rare subtypes of melanomas, which represent an unmet critical need. Finally, we identify key recommendations for melanoma models that may improve accuracy of preclinical testing and predict efficacy in clinical trials, to help usher in the next generation of melanoma therapies.
INTRODUCTION
Melanoma constitutes a prime example of how basic and translational research has improved the prognosis for cancer. We have made remarkable progress in the last 10 years, with 13 new melanoma therapies approved in the United States, including both targeted and immune-based therapies (Figure 1) (Luke et al., 2017). Standard of care includes treatment with immune checkpoint inhibitors (ICI), either alone or in combination, as well as targeted inhibitors of BRAFV600E and MEK kinases in the mitogen-activated protein kinase (MAPK) pathway. Inter-leukin-2 (IL-2), oncolytic viral, and interferon therapy remain options for a subset of patients.
Figure 1. Timeline of treatment approvals by the US Food and Drug Administration for advanced melanoma patients.
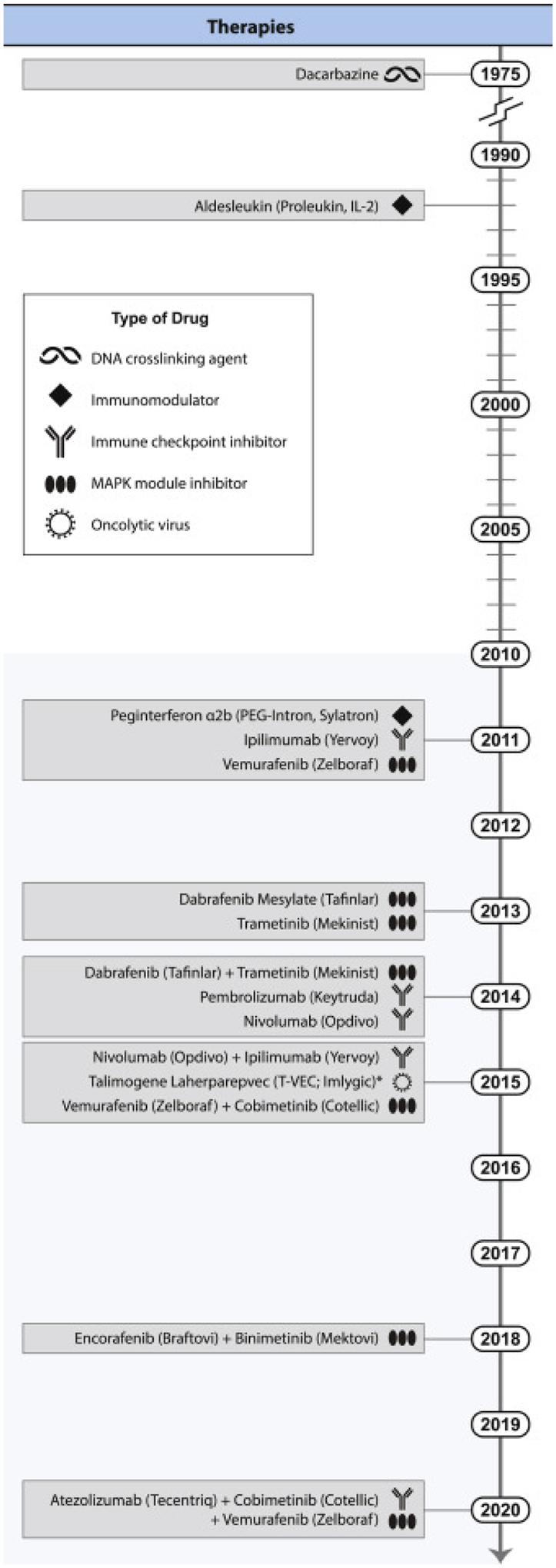
Thirteen new treatments, including both single agents and combination therapies, have been approved since 2011. 2011 also marked the first approval of an immune checkpoint inhibitor, ipilimumab, which ushered in the modern era of cancer immunotherapy. Several of these new treatments have also been approved in the adjuvant setting (after surgery). Credit: Heather McDonald, Nancy R. Gough, BioSerendipity.
These treatments have led to tremendous improvements in the survival and quality of life for advanced patients, who before the modern era could expect a 5-year survival rate of ~10% (Balch et al., 2001). For instance, an objective response rate of 61% was seen in patients treated with combination ipilimumab (anti-CTLA-4) and nivolumab (anti-PD-1) (Postow et al., 2015). In a separate study, 52% of patients receiving this combination were alive at 5 years compared with 26% treated with ipilimumab alone (Larkin et al., 2019). Approvals of targeted and immunotherapies for patients with stage III operable melanomas (adjuvant therapy), have further extended the number of patients who can benefit from these therapeutic advances (Eggermont et al., 2018; Long et al., 2017; Robert et al., 2019; Weber et al., 2017). Likely reflecting these advances, deaths from melanoma dropped by an unprecedented 18% from 2013 to 2016 among people of European ancestry, the group most affected by melanoma, which is the largest drop seen over such a short period for any type of cancer (Berk-Krauss et al., 2020).
Despite this progress, current therapies are not a cure for most patients with metastatic melanoma, which is a genetically heterogeneous disease, particularly in ~50% of patients whose tumors lack a BRAF mutation, and in patients with rare melanoma subtypes (e.g., uveal, acral, mucosal). Many patients are inherently resistant to these therapies (primary resistance), or melanomas recur and become resistant following an initial response (acquired resistance). Moreover, no targeted treatment options are available for rare forms of melanoma, and immunotherapies are less effective in these patient populations compared with patients with cutaneous melanoma (Klemen et al., 2020).
We advocate for melanoma models as critical drivers of discovery, innovation and translation research for next-generation therapies. Melanoma models capture the disease processes in an experimental platform and are necessary for (1) fundamental discoveries in melanoma biology, leading to new therapeutic target and (bio)marker identification, (2) preclinical discoveries and insights into therapeutic modalities, and (3) understanding and predicting therapy mechanisms of action and response (Figure 2). No single melanoma model can capture the complex landscape of melanoma progression representing all patients, yet a multitude of model systems have together become a cornerstone to the understanding, diagnosis, and treatment of melanoma (Figure 3). That animal studies are consequential to melanoma therapy is exemplified by the efficacy of anti-PD-1 and anti-CTLA-4 antibodies to inhibit immune checkpoints in mouse models, leading to autoimmune disease and rejection of transplanted tumors (Iwai et al., 2002, 2005; Leach et al., 1996; van Elsas et al., 1999, 2001). This work transformed the care of patients with melanoma and other cancers, and led to the award of a Nobel Prize for Honjo and Allison (Hodi et al., 2010; Robert et al., 2015).
Figure 2. Timeline of melanoma models and opportunities for addressing key challenges.
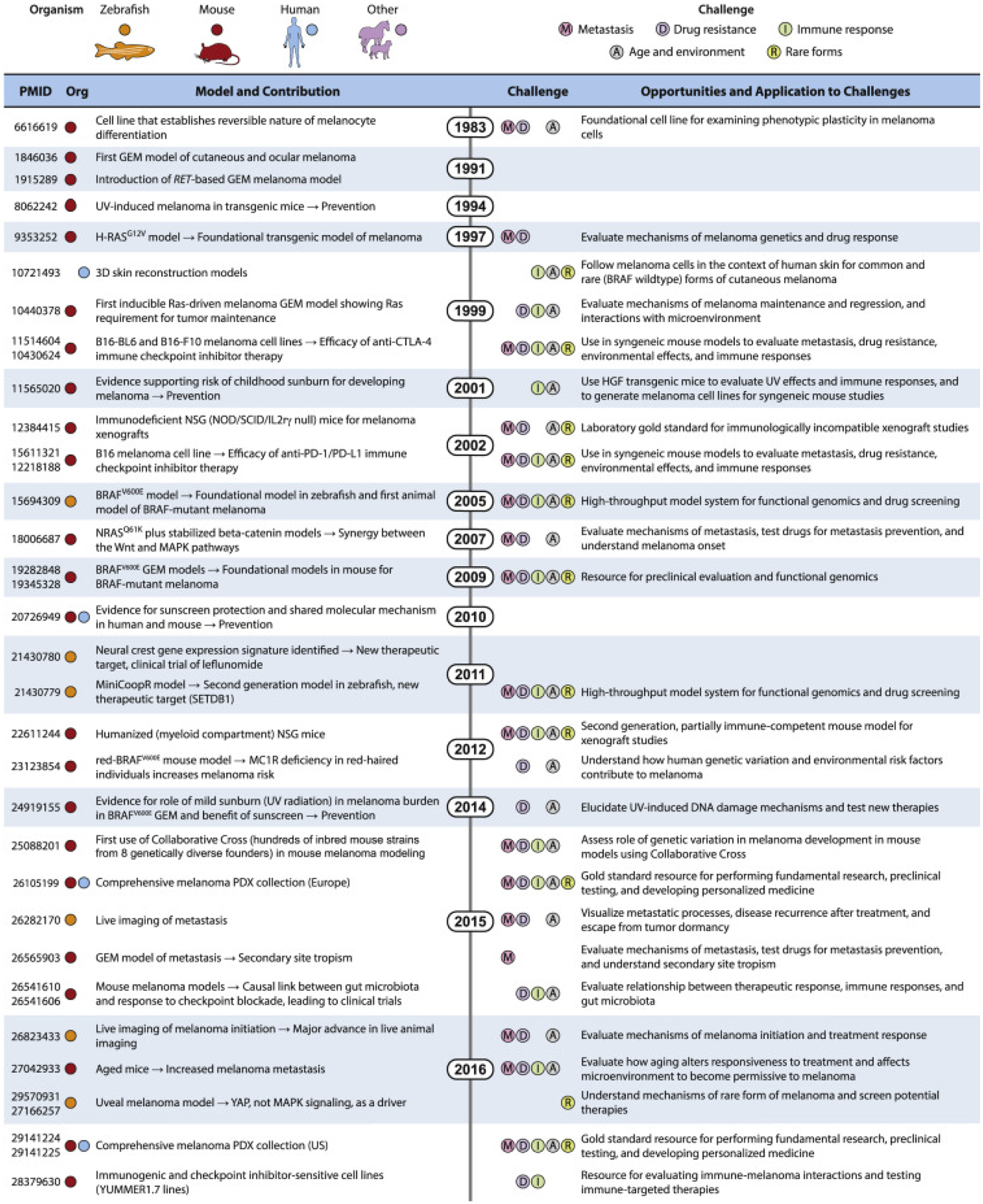
A brief selection of models from mouse, zebrafish, human, and other species that have made major contributions to our understanding of melanoma biology and in the discovery of new therapies. Such models are critical to address the most pressing challenges in melanoma, including metastasis, drug resistance, immune response, aging and the microenvironment, and rare forms of melanoma. Due to space constrictions, unfortunately only a subset of available melanoma models is presented. Species are indicated by colored circles, and challenges are indicated by letters in colored circles. Credit: Heather McDonald, Nancy R. Gough, BioSerendipity.
Figure 3. Schematic representation of current melanoma models and the many ways they can be employed to develop new therapies.
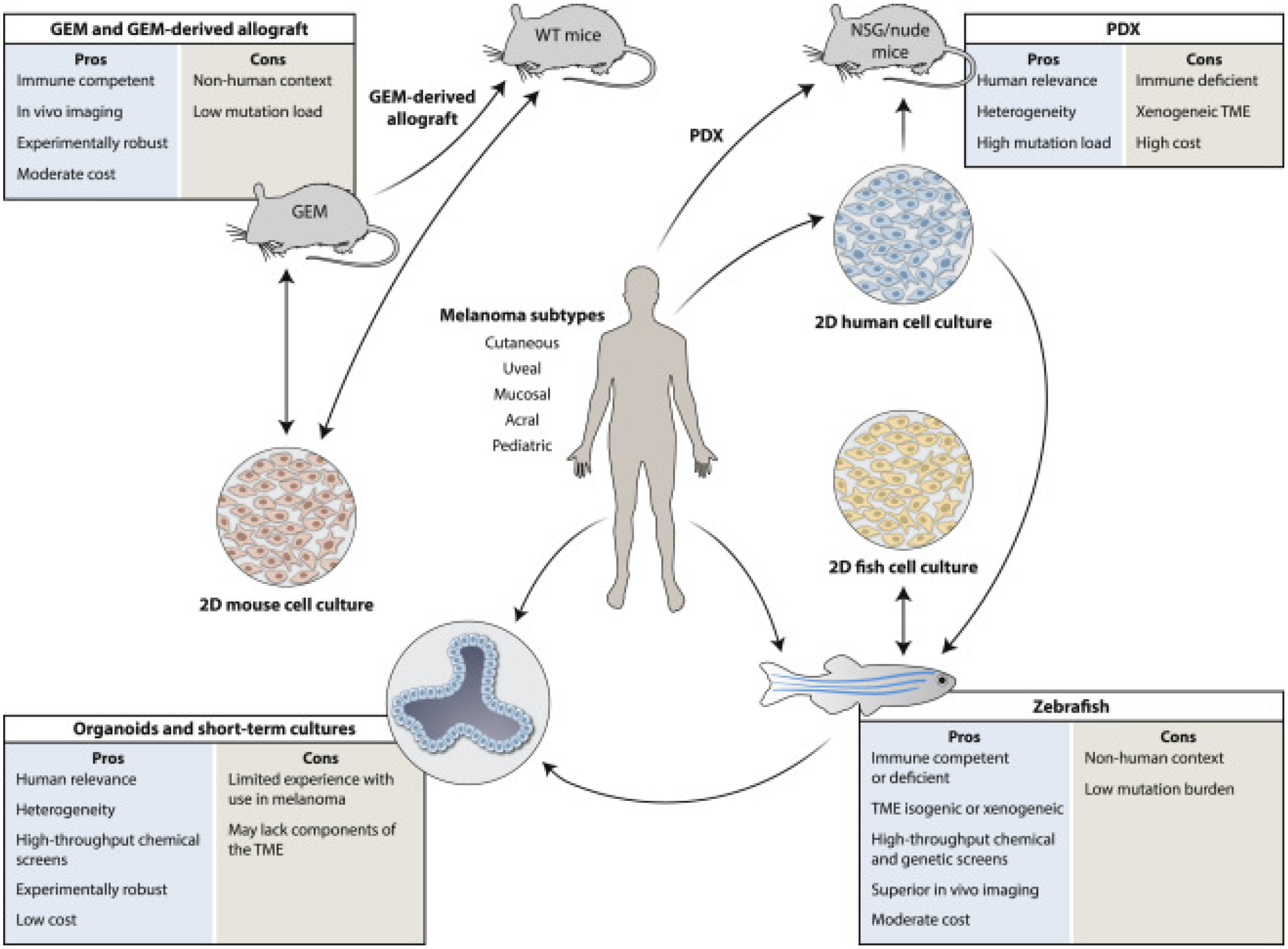
Animal models are first generated to resemble their human counterparts as closely as possible, but are also constantly being improved through genetic engineering, environmental intrusions, and/or technical advances. In general, in vitro models are more amenable to mechanistic analysis, while in vivo models help capture the complex microenvironment and immune responses that contribute to melanoma outcomes. A critical evolving aspect to all animal models is that “omics” data generated from their study can now be compared directly with comparable human datasets, providing powerful validation of relevance. The establishment of in vitro cultures generated from the models and directly from patients (such as organoids) provides additional layers of experimental possibilities. 2D, two-dimensional; GEM, genetically engineered mouse; GDA, GEM-derived allograft; NSG, NOD/SCID/IL-2rγnull immunodeficient mouse; PDX, patient-derived xenograft; TME, tumor microenvironment; WT, wild-type.
In this Perspective, we discuss state-of-the-art melanoma models, identify five of the most significant challenges for new melanoma therapies, and discuss how models are necessary to help overcome them. Furthermore, we identify necessary advances to improve models, now and in the future, to make breakthroughs in these critical areas.
MELANOMA MODELS
Genetically engineered mouse models
Mouse models of melanoma genetically engineered to harbor one or more known drivers of subtypes of human melanoma have greatly enhanced our understanding of the functional genetics underlying melanoma formation and progression (Figure 2) (Bardeesy et al., 2001; Bonet et al., 2017; Marsh Durban et al., 2013; Olvedy et al., 2017; Perez-Guijarro et al., 2017; You et al., 2002). Genetically engineered mouse (GEM) melanoma models have more physiological growth rates relative to other models and are useful for evaluating therapies and sunscreens. Accordingly, numerous studies have evaluated small-molecule drugs including BRAFV600E and MEK inhibitors with response characteristics resembling human melanoma. The emergence of ICIs as standard melanoma therapy has raised the possibility that GEM melanoma models could be used to evaluate immune-oncology therapies (Galvani et al., 2020; Perez-Guijarro et al., 2020). GEM models capture the intricacies of the tumor immune response and microenvironment for spontaneous melanomas, biological responses that are otherwise difficult to study due to the complexity and variation within human patient cohorts. Genotype-specified GEM models will undoubtedly continue to be useful for uncovering mechanistic aspects of melanoma response and resistance to targeted therapy (Dankort et al., 2009; Das Thakur et al., 2013; Deuker et al., 2015; Dhomen et al., 2009; Marsh Durban et al., 2013; Petit et al., 2019; Vredeveld et al., 2012).
Given that melanomas arise from the melanocyte lineage, GEM models designed to study melanocyte stem cells and development have been crucial for providing modeling tools and for demonstrating the dependence of melanomas on developmental lineages (Conde-Perez and Larue, 2014; Delmas et al., 2007; Dorard et al., 2017; Nishimura et al., 2005; Shakhova et al., 2012; Sun et al., 2019; Zurkirchen and Sommer, 2017). Emerging gene-editing technologies will improve GEM models, since they can replace current models that inactivate genes simultaneously with inducible, temporally controlled changes, recapitulating the accumulation of genetic changes over time commonly seen in human disease. Unlike humans, mice have fur, and mouse melanocytes are mainly located in hair follicles; however, this limitation can be overcome. Hepatocyte growth factor-based GEMs demonstrate “humanized” skin in which mouse cutaneous melanocytes are ectopically localized at the epidermal/dermal junction as well as hair follicles (Noonan et al., 2001). Alternatively, melanomagenesis can be induced on the tail or ear, which are inhabited by inter-follicular cutaneous melanocytes and are useful for studying tanning and melanoma development (Cui et al., 2007; Glover et al., 2015; Kohler et al., 2017).
UV radiation (UVR) exposure is a major risk factor for human melanoma, and the cause of genome-wide mutations (Cancer Genome Atlas Network, 2015). UVR has been incorporated into GEMs to address tanning and burns and genetic risk factors, and to test sunscreens and tanning chemoprevention (Cui et al., 2007; D’Orazio et al., 2006; Day et al., 2017; Hennessey et al., 2017; Mujahid et al., 2017; Nguyen and Fisher, 2019; Noonan et al., 2001; Viros et al., 2014). Mouse models provided evidence to support the concept that UVR burns in childhood increase the risk of melanoma in adulthood (Klug et al., 2010; Noonan et al., 2001). UVR models also demonstrated the molecular mechanisms linking DNA damage to tanning, science which underpins public health advice on indoor tanning (Fisher and James, 2010). Notably, despite the importance of the BRAFV600E mutation in nevi and melanoma initiation, it is not itself a UVR signature mutation. However, BRAFV600E mutant mice were critical in demonstrating that even mild sunburn is sufficient to cause melanocyte clonal expansion and that repeated mild UVR exposure causes melanoma (Dhomen et al., 2009; Viros et al., 2014). These models recapitulate the UVR-induced genetic signature and recurrent mutations prevalent in human melanoma (Trucco et al., 2019). Together, GEM UVR models have directly demonstrated the efficacy of sunscreen and have provided the molecular evidence to support public health melanoma prevention campaigns.
Syngeneic models
Syngeneic transplantation models have long been a workhorse for preclinical testing and offer the advantages of relative ease of use, low cost, and fast turnaround time. However, model-specific features should be considered. The B16 mouse cell line has been widely used for decades to model anticancer immune responses in mice with intact immune systems, but typically curative responses are not observed unless a vaccine approach is incorporated. In recent years, several new genetically defined syngeneic transplantation models have been developed from GEM melanoma models, termed GEM-derived allografts (GDAs) (Figure 3). Melanomas used to derive cell lines typically arose on mice backcrossed to C57BL/6, and the lines are typically transplantable into C57BL/6 host mice without rejection (Jenkins et al., 2014; Meeth et al., 2016). With the advent and success of immune therapies in melanoma and other cancers, models with enhanced immunogenicity that exhibit curative responses to ICIs have been developed (Wang et al., 2017; Zelenay et al., 2015). Some of these have involved UVR or other forms of mutagenesis followed by isolation and characterization of single-cell-derived clones (Wang et al., 2017; Wolf et al., 2019). Other approaches have utilized expression of model antigens such as ovalbumin or lymphocytic choriomeningitis virus antigens to enhance immunogenicity and enable analysis of antigen-specific T cell responses using reagents that were previously defined for these antigens (Lelliott et al., 2019). A comparison of the immunogenic features of a variety of syngeneic melanoma models was recently completed (Perez-Guijarro et al., 2020).
Patient-derived xenograft models
Patient-derived xenograft (PDX) models, in which patient cancers are grown in an immunocompromised mouse, have revealed important aspects of melanoma evolution and drug-resistance mechanisms. PDX models can accurately predict response to targeted therapies (Fiebig et al., 1984; Sanmamed et al., 2016; Einarsdottir et al., 2014) and can be used to explore the role of metabolism (Tasdogan et al., 2019) and of primate-specific genes, such as long non-coding RNAs in melanoma that are enriched for genetic alterations related to cancer (Leucci, 2018; Leucci et al., 2016).
A sufficiently large collection of PDX models can capture much of the clinical, histologic, and genetic diversity of human melanoma compared with GEM models, which recapitulate only a limited number of genetic alterations. Krepler et al. (2017) described a collection of 459 PDX models derived from 384 patients. Most of these samples were derived from metastatic lesions, representing all major genetic drivers (e.g., BRAF, NRAS, NF1) and subtypes of melanoma (e.g., cutaneous, mucosal, acral, uveal). Moreover, PDXs obtained from patients who had progressed on ICI and targeted therapies were represented, and a subset of these models was subjected to in-depth molecular analyses (Garman et al., 2017), thereby offering an invaluable resource for drug development and translational research. Importantly, parallel sequencing analysis of primary patient tumor samples and patient-derived cell lines showed that the PDXs generally faithfully preserve genetic lesions present in the original tumor (Kemper et al., 2015; Woo et al., 2019). These models were used to establish preclinical efficacy of multiple interventions in the context of targeted therapy-resistant melanoma, such as PI3K pathway, MDM2, and BET inhibitors (Echevarria-Vargas et al., 2018; Krepler et al., 2016, 2017).
PDX models also have limitations. Following the initial in vivo passage, the tumor-stromal component becomes that of the mouse host, rather than the original human patient, potentially complicating studies of tumor-stroma interactions. Also lost are species-specific signaling interactions between mouse and human ligands and receptors. Moreover, mouse genomes encode retroviruses capable of infecting human cells, potentially serving as a driver of insertional mutagenesis in the engrafted tumor cells (Naseer et al., 2015). Conversely, the immunocompromised mice used as PDX hosts are highly susceptible to infection by human pathogens that may be present in human tumor samples (Manuel et al., 2017). PDX models of uveal melanoma, notoriously resistant to targeted and ICI therapies, have proved to be challenging, highlighting the need for further technical advancements in PDX approaches for this melanoma subtype (Chua and Aplin, 2018). Notably, current PDX models are limited to analysis in immunocompromised host mice, preventing the testing of immune-based therapies. Many of these issues could be overcome when more robust and tractable humanized mouse models are developed, which is currently a daunting challenge (see below).
Organoids as melanoma models
To complement in vivo studies in mice, a variety of novel ex vivo approaches have been developed. Upon in vitro culture in defined conditions, cancer cells derived from primary patient material organize into organoids that morphologically and functionally mimic significant aspects of the original tumor (Tuveson and Clevers, 2019). Organoids can be expanded, passaged, and banked for further use if maintained under appropriate conditions, and can be genetically and phenotypically profiled to identify outstanding candidates for in vitro drug screening. Unlike for other diseases such as colon cancer, limited experience so far exists in utilizing melanoma organoids for drug screens and preclinical studies (Tuveson and Clevers, 2019; van de Wetering et al., 2015). However, emerging evidence suggests that organoids can be readily generated from melanoma biopsies and might even be supplemented with autologous immune cells for evaluation of immunotherapeutic responses (Votanopoulos et al., 2019). In fact, the National Cancer Institute is building repositories of well-annotated organoids from melanomas and other diseases that will become available to the community (https://pdmr.cancer.gov).
Other advanced in vitro melanoma models
Alternatives for advanced non-animal, in vitro melanoma modeling and experimental therapeutics include skin reconstructs, tissue explants, and three-dimensional (3D) spheroids. Skin reconstructs co-culture various primary and/or established cell types embedded in collagen such as fibroblasts, endothelial cells, and keratinocytes in addition to melanoma cells (Li et al., 2011; Satyamoorthy et al., 1999). Advantages include the 3D-and multi-cell type nature of the reconstruct that more closely mimics in vivo features. However, skin reconstructs have limited scalability for multi-cellular processes, can be difficult to generate using allogeneic material, and lack immune or other key cells of the tumor microenvironment (TME).
Tissue explants are small pieces of surgical specimens resected from patients that can be cultivated for a limited time (Powley et al., 2020). Such explants contain all components of a patient’s cancer in situ at the time of culture initiation. Different protocols exist for culture, but over time this composition will likely change significantly. A clear advantage of this system is the native cellular composition, with autologous material and cells of all kinds present on site for analysis and drug testing. In colon cancer, for example, drug candidates that later were found to have activity in patients could be successfully identified using this approach (Halama et al., 2016). Tissue explants have been shown to work for melanoma (Berezhnaya et al., 2006; Hegerfeldt et al., 2002) but with substantial heterogeneity in yield, indicating that additional studies will be required to expand their utility.
Zebrafish melanoma models
Melanocyte development and the genetic basis of melanoma is shared between fish and human (Figure 2) (Mort et al., 2015). Zebrafish have epithelium-associated melanocytes, and in zebrafish models of cutaneous melanoma, tumors develop near the epithelium, as is typical of human disease. Rapid and scalable melanocyte-specific transgenesis can overexpress or knock out genes in tumor-prone strains (Ablain et al., 2018; Ceol et al., 2011; Patton et al., 2005), enabling modeling of human melanoma genetics. More recently, an electroporation method called TEAZ (transgene electroporation in adult zebrafish) has been developed to introduce transgenes directly into the skin of living zebrafish, making temporally defined manipulations of melanocytes and melanomas possible (Callahan et al., 2018).
The small size of zebrafish embryos enables phenotype-based screening of drug libraries and drug-led therapeutic development (MacRae and Peterson, 2015). Drug administration in zebrafish is simple and inexpensive because animals can be soaked in low volumes of drug-laden solutions, making screens and combinatorial tests of drugs straightforward. Notably, zebrafish studies have highlighted transcriptional elongation as a novel therapeutic target in melanoma, leading to clinical trials (Johansson et al., 2020a; Santoriello et al., 2020; Tan et al., 2016; White et al., 2011). Drug-discovery approaches can now encompass patient-derived tumors engrafted in zebrafish (Yan et al., 2019).
One of the strengths of the zebrafish system is that melanomas can be directly imaged using confocal and other high-resolution modalities in real time (Heilmann et al., 2015; Kaufman et al., 2016; Travnickova et al., 2019; White et al., 2008, 2011). By incorporating cell-type-specific labels onto melanoma backgrounds, interactions between melanoma cells and other cells can be monitored, including innate and adaptive immune cells (zebrafish have a full representation of all key components of innate and adaptive immunity), other skin-resident cell types, and stromal cells (Feng et al., 2012; Roh-Johnson et al., 2017). The ability to edit genomic loci with precision and in a temporal-specific fashion remains challenging, although recently precise genome editing has been performed in zebrafish (Prykhozhij and Berman, 2018). Experiments in zebrafish have pioneered the development of lineage-tracing technologies such as zebrabow and gestalt (McKenna et al., 2016; Pan et al., 2013) which, when combined with single-cell analyses, constitutes a powerful approach to the study of tumor clonality and heterogeneity.
MELANOMA CHALLENGES
Challenge 1: metastasis
Metastasis is the spread of tumor cells from a primary lesion to a distant site within the body and is responsible for most cancer-related deaths (Nguyen et al., 2009). In addition to the intrinsic properties of the cancer cell that enable this process, extrinsic cues from the TME of the metastatic site also provide support for the growth of disseminated tumor cells (Fares et al., 2020). To identify new drug targets for melanoma metastases, it is critical that we understand intrinsic and extrinsic factors that (1) drive metastasis, (2) dictate tumor cell affinity for specific distant sites, and (3) enable tumor cells to remain dormant at distant sites for long periods of time and resist immune and therapeutic destruction. Extensive analyses of patient samples have been invaluable for addressing these questions by providing a plethora of data and hypotheses for further validation and mechanistic study in experimental model systems (Cancer Genome Atlas Network, 2015; Lauss et al., 2016).
Time to metastatic progression
GEM models can mimic human melanoma metastatic progression and have provided great insights into factors that promote metastasis to distant sites such as the brain, including CCR4 (Klein et al., 2017) and AKT (Cho et al., 2015). A key issue with current animal models is that in order to be tractable, primary melanomas require relatively short latency, which does not provide time to develop metastases. Developing models with longer latency for melanoma development is one option but makes the practicality of the experiments challenging. Resection of a rapidly growing primary melanoma will buy some time for metastatic progression, but this approach is not always sufficient (Day et al., 2015). A potentially faster alternative would be to create mouse models that permit manipulation of gene expression in somatic cells, such as those based on the viral RCAS/TVA system with TVA (tumor virus A) expression controlled by a melanocyte-specific promoter (Kircher et al., 2019). The RCAS/TVA system enables more rapid, high-throughput analyses of specific genes than can be achieved with GEM models developed by germline mutagenesis. Evolving CRISPR/Cas9 technology will also make it possible to genetically manipulate somatic cells in juvenile and adult inbred mice. Although tracking tumor heterogeneity and rare cell subpopulations is challenging, the novel methodology CaTCH (CRISPRa tracing of clones in heterogeneous cell populations) provides new opportunities to study clonal evolution in cancers (Umkehrer et al., 2020).
Metastatic initiation and dormancy
Modeling metastasis is a major challenge because in patients it can occur rapidly and be detected early, or it can take decades following long periods of dormancy (Aguirre-Ghiso, 2018). Recent long-term studies that show while stage I melanoma patients have good outcomes within the first 5 years, 15%–29% of patients with stage I melanomas die from metastasis up to 20 years following initial treatment (Lo et al., 2018). Unlike humans, animal models have short lifespans, and models need to be redesigned to capture the biology of dormancy in vivo. Incorporation of creative intravital imaging approaches would enhance our ability to employ animals as models for dormancy.
Capturing the live dynamics of disease spread or response to therapy requires detailed spatiotemporal observation of the interactions between cancer cells and their microenvironment, an approach that is particularly powerful when high-resolution intravital imaging of fluorescently labeled melanoma cells is performed in semi-transparent zebrafish (van Rooijen et al., 2017). Combined with microenvironment cell-specific knockdown, this allowed the identification of keratinocyte-derived endothelin 3 as a metastasis-promoting factor that regulates phenotype switching of melanoma cells between invasive and proliferative states (Kim et al., 2017). Furthermore, upon invasion into adipocyte-rich environments, lipid uptake is dependent on the melanoma-expressed fatty acid transporter proteins (FATPs), and blocking lipid transfer inhibits tumor growth and dissemination in zebrafish (Zhang et al., 2018).
A novel advance in mouse modeling is the “MET Alert” mouse, engineered with a VEGFR3-luciferase reporter expressed specifically in lymphatic vessels and that reveals metastatic dispersion through lymph vessels (Martinez-Corral et al., 2012; Olmeda et al., 2017). Live imaging of the lymphatic system in these mice reveals the lymphangiogenic preconditioning of distant sites to melanoma metastatic niches by the cytokine midkine (Olmeda et al., 2017). This induction at distal premetastatic niches is uncoupled from lymphangiogenesis at the primary melanoma, enabling visualization of a whole-body systemic response to metastatic lesions. Capturing the live dynamics of metastatic “seeds” and the homing to preconditioned metastatic niches are critical steps for understanding how metastatic cells survive and may help identify biomarkers to assist in disease management in patients.
Melanoma organotropism
Melanomas metastasize to specific organ sites, including the skin, lung, liver, brain, bone, and intestines. PDXs can model the specific metastatic spread observed in an individual patient (Quintana et al., 2008), and metastases from GEM melanomas can also retain the ability to target specific sites upon transplantation. PDX models harbor the inter- and intra-tumor heterogeneity that is frequently found in patient tumors, which likely plays a role in site-specific metastasis. Moreover, while GEM-derived melanoma cells have the same engineered genetic aberrations, heterogeneity can be incorporated by exposing mice to a relevant, mutagenic dose of UVR (Noonan et al., 2001; Viros et al., 2014). Current PDX models require immunocompromised mouse hosts, limiting their usefulness in analyzing extrinsic metastatic factors that could be critical for organotropism. In contrast, GEM models have a fully functional immune system and appropriate tumor-host interactions.
Tumors derived from GEM and other models of human melanoma can also be “trained” to home to selected organs through a series of transplantations. Josh Fidler discovered that by resecting a metastatic lesion from a specific organ, he could enhance the frequency of metastasis to that organ in a second round of subcutaneous transplantation (Fidler, 1973). After several cycles of this transplantation strategy, melanomas could be trained to target specific organs with high efficiency. Specific unmet needs in melanoma metastasis include the creation of additional models for melanoma dissemination to the brain, which tends to be more resistant to currently approved therapies and a site where patients often relapse, and models that accurately mimic tumor cell dormancy at metastatic sites. Overcoming the challenges of understanding and eventually treating or preventing metastatic melanoma will benefit from novel melanoma models that permit live imaging of specific steps in the metastatic process at the whole-body level (Olmeda et al., 2017) and at single-cell resolution (Benjamin and Hynes, 2017).
Challenge 2: drug resistance
Most patients with BRAF mutations initially show response to BRAF inhibitors, yet the melanoma often recurs only a few months after starting treatment. In 58% of cases the putative resistance mechanism is linked to MAPK-pathway mutations that overwhelm the BRAF inhibitor therapy (Johnson et al., 2015). Drug resistance is also seen in patients receiving ICI, an important point that deserves attention in future studies. It is therefore essential to gain a deeper understanding of the mechanisms of intrinsic and acquired resistance to current therapies.
Primary resistance
Most GEM melanoma models include dominant BRAF or RAS mutations, and respond to MAPK-pathway inhibition (Dankort et al., 2009; Perez-Guijarro et al., 2017; Xiao et al., 2019). However, it is now clear that many infrequent spontaneous genetic alterations confer conditional adaptive growth or survival advantages. Furthermore, in 39% of cases resistance was not accounted for by any validated mutations but displayed transcriptomic alterations (Cancer Genome Atlas Network, 2015). Thus, research may be well served by focusing on identifying the pathways, such as changes in autophagy, mitochondrial metabolism, or epigenetic modifications, that drive resistance, rather than on studying single mutated genes. For example, therapy that confers sensitivity to MAPK-pathway inhibition has recently shown efficacy in conjunction with the autophagy inhibitor, chloroquine, in pancreatic ductal adenocarcinoma and in other cancers including NRAS-driven and uveal melanoma (Kinsey et al., 2019; Truong et al., 2020). Furthermore, targeting the NAMPT → NAD+ metabolic pathway restores sensitivity of resistant cells to BRAF inhibitors (Audrito et al., 2018; Ballotti et al., 2019; Ohanna et al., 2018).
Addressing innate resistance to current targeted therapies requires models that are driven by non-BRAF/RAS mutations (Perez-Guijarro et al., 2020; Scahill et al., 2017; Travnickova et al., 2019). Moreover, models are needed that reflect specific transcriptional subtypes that are not linked to driver mutations and are predictive of patient outcomes (Kawakami and Fisher, 2016). For example, zebrafish models of the MITF-low transcriptional signature, which is associated with early resistance to targeted therapy, are independent of BRAF mutations (Muller et al., 2014; Travnickova et al., 2019).
Adaptive resistance
Resistance can arise from pre-existing rare tumor cells that are selected by the therapy or from resistant cells as an adaptive response to therapy, through newly occurring mutations and/or the activation of adaptive responses (Boumahdi and de Sauvage, 2020). Unbiased genome-wide screens in cells have been instrumental in the identification of acquired drug-resistance mechanisms to targeted and immune checkpoint therapies (Kong et al., 2017a; Vredevoogd et al., 2019; Strub et al., 2018). Exemplifying the importance of metabolism in drug-resistance mechanisms, Bernards and colleagues demonstrated that MAPK-pathway resistance is associated with drug-induced increased levels of reactive oxygen species (ROS), making resistant cells selectively vulnerable to further increases in ROS (Wang et al., 2018).
Adaptive resistance mechanisms are also problematic for immunotherapy, with >60% of melanoma patients progressing on combination immunotherapy. Transcriptional signatures that characterize immunotherapy resistance and predict patient outcomes have been identified in human samples and using syngeneic mouse models (Hugo et al., 2016; Peng et al., 2016; Perez-Guijarro et al., 2020). Modeling resistance mechanisms in vivo is highlighted by the rationale for combining targeted therapy with immune-based therapies. For instance, upon MAPK inhibition macrophages secrete tumor necrosis factor α (TNF α) to promote melanoma growth; inhibiting TNFα signaling rescues this effect by targeting not only the melanoma cells but also the microenvironment (Smith et al., 2014).
Models are also required to capture the dynamic cellular heterogeneity that contributes to therapy resistance (Arozarena and Wellbrock, 2019). This is directed in part by microphthalmia-associated transcription factor (MITF), a critical regulator of melanoma cell-intrinsic phenotypic plasticity that facilitates adaptive cellular reprogramming and intra-tumor heterogeneity (Goding and Arnheiter, 2019; Rambow et al., 2019). In particular, a melanoma subpopulation, featured by low MITF/high AXL, is considered a drug-resistance phenotype (Muller et al., 2014; Ohanna et al., 2013); AXL-targeted antibody drug conjugates are currently in clinical trials for different types of solid tumors including melanoma (Boshuizen et al., 2018). The concept of melanoma cell phenotype switching has been proposed to push invasive melanoma cells toward a differentiated cell state where they are sensitive to a tyrosinase-dependent prodrug (Saez-Ayala et al., 2013). Other cell states are distinguished by differences in cell-cycle and stem cell characteristics, including a slow-cycling JARID1B-high subpopulation that is enriched upon drug treatment and can be targeted by inhibition of mitochondrial respiration (Roesch et al., 2013).
PDX models may be superior for modeling resistance to targeted agents (Krepler et al., 2017), in that they faithfully recapitulate acquired resistance and melanoma patient responses to targeted therapy (Rambow et al., 2018) and have also been used to show increased dependency of drug-tolerant lesions on mitochondrial translational machinery (Vendramin et al., 2020). Crucially, PDXs generated from therapy-refractory patients can be leveraged to elucidate mechanisms of resistance and define alternative or second-line treatments. Using PDX models, ER translocation of the MAPK pathway was shown to lead to ERK reactivation and protective autophagy, new druggable targets for therapy resistance (Ojha et al., 2019).
Targeting residual disease
As part of modeling adaptive resistance, there is a need to model the cell types and states that emerge at the residual disease site, including the “persister” cells and a preconditioned tumor site (Marine et al., 2020). Persister cells represent a small subpopulation of cancer cells that may not necessarily have acquired genetic mutations that drive resistance, and their plasticity can be characterized by metabolic, transcriptional, and translational states. For example, following exposure to BRAF and MEK inhibitors, melanoma persister cells reversibly remodel their translational program to survive in a quiescent state, and targeting the eIF4A RNA helicase counteracts this reprogramming to the detriment of persister cell survival (Shen et al., 2019). Single-cell RNA sequencing (RNA-seq) of BRAF-mutant PDXs was used to identify four BRAF/MEK inhibitor-tolerant residual subpopulations, with one subpopulation contributing to disease recurrence featuring MITF-low-to-no activity and regulated by RXR-γ (Rambow et al., 2018). RXR-γ antagonist in combination with BRAF/MEK inhibitor treatments was shown to both improve bulk tumor regression and target the adaptive resistant state to prevent recurrence. Understanding the complex milieu of the heterogeneous states of persister cells with their microenvironmental surroundings will be bolstered by models enabling live imaging coupled with single-cell technologies of the residual disease state, such as in zebrafish (Travnickova and Patton, 2020; Travnickova et al., 2019). The concept of targeting the bulk of the tumor in addition to persister cell states, including adaptive states and those that pre-exist in the primary tumor, will be crucial in delaying or even eliminating recurrent disease.
Challenge 3: the immune response
Most patients do not respond to immune-based therapy and/or are affected by adverse toxicities. Immunocompetent animal models allow for in-depth analyses of pending questions about the immune microenvironment of both primary and metastatic melanomas, as well as local and systemic responses at central and peripheral sites. Models should be tailored to specific questions and their physiological relevance defined. Key unmet needs include improved understanding of how melanomas evade immune recognition during tumorigenesis, especially at anatomical sites such as the eye, acral surfaces (palm and sole), and the mucosa. Aspects of metastatic tropism to different organs, particularly the brain, are unclear. Models need to address the complex role of the immune system as conditioned by exosomes, immune responses in the tissue, and the role of local immunity and systemic inflammation on the premetastatic niche. Furthermore, immune-related adverse events (irAEs) that now affect up to 30%–40% of patients have yet to be addressed in model systems.
Immunogenicity
Reflecting patient responses, BRAFV600E GEM models mount a mixed response to anti-PD-1 checkpoint blockade, some with significant responses (Galvani et al., 2020). However, as in patients, many mice fail to respond. It is thought that low mutation rate and associated neoepitope burden in most GEM melanoma models account for the observed lack of response, and indeed BRAFV600E/UVR models with higher mutation burdens are associated with better immunotherapy responses. However, increased mutations alone cannot fully explain ICI responses, as BRAFV600E/UVR melanomas are associated with stromal remodeling and reduced tumor cell proliferation, a response shown in patients who benefit from sustained immunotherapy response (Galvani et al., 2020).
Progress has also been reported in harnessing the predictive value of GEM models to ICIs across human cutaneous melanoma subtypes (Perez-Guijarro et al., 2020). The use of a preclinical platform consisting of GEM models representing major human cutaneous melanoma genetic subtypes, in conjunction with patient datasets and computation data analytics, has uncovered key determinants of ICI efficacy and a melanocytic plasticity signature able to predict ICI responses (Perez-Guijarro et al., 2020). This approach demonstrates the potential of GEM models, especially when used in conjunction with sophisticated computational methodology, and their ability to elucidate mechanisms associated with drug response and resistance as well as to inform clinical melanoma trials (Perez-Guijarro et al., 2020).
New models are being developed to help understand why many patients are unresponsive to ICI, while others suffer side effects associated with an autoimmune disease yet benefit from sustained long-term cancer suppression (Chamoto et al., 2017; Kumar et al., 2020). New mouse models may avoid foreign or viral antigens, such as ovalbumin and viral antigens, and instead stimulate an immune response using more relevant endogenous neoepitopes (e.g., through UVR) (Galvani et al., 2020; Perez-Guijarro et al., 2020; Wang et al., 2017) or through models that create inducible neoantigens de novo (Damo et al., 2020). Such models more accurately reflect the antigenic profile of human melanoma, enabling the generation of reagents for characterizing epitope-specific responses.
Humanizing immune systems in models
PDX models have limited utility for preclinical testing of ICIs or evaluating irAEs because human tumors must be grown in immune-compromised mice to prevent rejection. The most popular host is the NSG mouse strain (NOD/SCID/IL-2rγnull), with defects in both adaptive and innate immunity, including an absence of natural killer (NK) cells (Ito et al., 2002). Additionally, the mouse immune system is substantively different from its human counterpart, making direct translational interpretation challenging. Mice with humanized immune systems can help to overcome these limitations. Two mouse strains, MITRG and MISTRG, have been engineered to express several human cytokines from the corresponding endogenous mouse loci (Rongvaux et al., 2014). Once engrafted with human CD34+ hematopoietic stem cells, these models are permissive for development of NK cells, macrophages, and monocytes, which have proved challenging to reconstitute in other systems (Rongvaux et al., 2014). Using this approach, a human melanoma xenograft was found to be infiltrated by immunosuppressive myeloid cells, promoting tumor growth. However, this model does not reconstitute a humanized adaptive immune system. Developing a mouse models with functional mature human lymphocytes represents a major technical challenge but remains a high priority (Forsberg et al., 2019; Jespersen et al., 2017). Very recently, humanized mice challenged with HLA-matched melanoma cells demonstrated a critical role for mast cells in mediating resistance to anti-PD-1 antibody therapy (Somasundaram et al., 2021). Because T lymphocytes require thymic education, generation of a fully humanized immune system in mice will likely require implantation of thymic tissue using stem cells from the melanoma patient that produced the PDX.
Modeling biological heterogeneity
Understanding the plasticity of melanoma cells and its relationship with microenvironment heterogeneity is critical to understanding immunotherapy resistance (HöIzel and Tüting, 2016). These include, but are not limited to, the (lymph)angiogenic vasculature, stroma (e.g., fibroblasts, adipocytes), tumor-associated myeloid cells, and various immune cells. Other variables that may regulate the host immune response include aging, diet, exercise, sex, and the microbiome. Models are needed that recapitulate known mechanisms of ICI resistance, including loss of major histocompatibility complex I (MHC I) presentation and perturbed interferon-γ signaling. In addition, unbiased models and approaches, including CRISPR-based screening, should be developed to effectively mimic resistance to immunotherapy (Vredevoogd et al., 2019).
Modeling the heterogeneity commonly seen in patient melanoma cells themselves represents a tremendous challenge. Within any one patient’s melanoma exists a myriad of tumor cells with varying mutations, transcription patterns, and phenotypes, each potentially responding differently to immunotherapy. Beyond that, the human population constitutes the most heterogeneous genetic background imaginable. In contrast, animals used as models are typically largely genetically identical, and their melanomas tend to be much more homogeneous, arising from the engineering of dominant oncogenic mutations and harboring fewer spontaneous mutations. The collaborative cross, a large panel of newly developed inbred strains of mice, has been created to add genetic diversity to melanoma models (Ferguson et al., 2015). Moreover, tumor-bearing animals are bred and raised under strictly controlled conditions, deviating dramatically from the multitude of environmental factors to which humans are exposed daily. UVR is being used in modeling to help compensate for this, and genetically defined mice can now be imbued with a natural, diversified microbiota, through the creation of so-called “wildling mice” (Rosshart et al., 2019); future GEM models will be adapted to harbor such a natural microbiome as well.
Characterization and standardized reporting
Maximizing the utility of any newly developed model requires adequate characterization and standardized reporting of the immune response and widely available access to such information. This includes standardization on reporting time of tumor growth, aiming to generate tumor regression rather than tumor growth inhibition, and long-term or sustained responses. This standardization should include Kaplan-Meier plots and survival curves as well as granularity in reporting individual mouse responses using spider and spaghetti plots. Newly developed animal melanoma models should be fully characterized with regard to immune profiles, including cytotoxic CD8 T cells, regulatory T cells, B cells, macrophages, and dendritic cells. These profiles may vary across genetic backgrounds. Additionally, the immunogenicity of the models should be characterized, including the relative antigenicity of the melanomas, the frequency of spontaneous regression, the affinity and avidity of the immune response to the tumor, the repertoire of the T cell receptors that recognize the melanoma cells, and the repertoire of MHC class I and II epitopes presented by the melanoma cells. These steps will elucidate the baselines of immune responses in models and can be directly compared with human disease. Importantly, these analyses may also help us to understand the irAEs associated with current immunotherapies.
Challenge 4: the impact of age and environmental exposures
Age remains the greatest risk factor for cancer development overall (Fane and Weeraratna, 2020). Aging provides time for premalignant cells to accumulate mutations, such as those from UVR in cutaneous melanoma, and epigenetic changes that favor cancerous progression and immune escape. The malignant cells and their TME must adapt to support tumor survival and growth at primary and metastatic sites. The adaptations arise over time from pressures in the TME and the physiological macroenvironment of the organism. Similarly, the TME itself ages, and this may place pressures on initiated cancer cells, driving them to enhanced malignancy (Fane and Weeraratna, 2020).
A notable feature of the aged TME is the senescence-associated secretory phenotype (Faget et al., 2019). Senescence, the loss over time of a cell’s ability to divide, and the cornucopia of molecules secreted by a senescent cell, usually a fibroblast, has been used as a measure of aging. Currently, many models of aging and cancer are based on acceleration of senescence and have led to important insights into the role of senescence and cancer (Demaria et al., 2014; Luo et al., 2016). Overall, such models allow for the examination of pressures placed upon a developing, dormant, or progressing cancer as the TME and organism age (Fane and Weeraratna, 2020).
Natural aging and age-appropriate models
Addressing questions related to the dynamic adaptation of both the cancer cells and TME over time in response to natural aging or environmental factors requires inducible, site-specific models that capture the long latency of human melanoma and can be examined at different ages. Evidence points to older patients with melanoma responding better to ICIs compared with younger patients (Kugel et al., 2018), but in vivo studies of melanoma tend to use 6- to 8-week-old mice, which corresponds to ~20-year-old humans, and developing/young-adult zebrafish. However, subcutaneous implantation of melanoma tumors into 12- to 18-month-old mice can increase melanoma metastasis and affect response to therapy. For example, lipid uptake from aged but not young fibroblasts by FATP2 can drive resistance to BRAF/MEK inhibitors in aged mouse melanoma models, and inhibiting FATP2 overcame resistance in aged mice (Alicea et al., 2020).
Particularly important are age-appropriate models that enable (1) quantitative dissection of adaptive relationships between nascent or dormant tumors and their aging hosts, (2) direct measurement of the progression-promoting crosstalk between tumor and host and between cancer cells and the TME, and (3) analysis of the response to therapy and age-associated toxicities. One such GEM model for melanoma is MT-RET, whereby mice express the human RET transgene in melanocytes and develop skin melanosis, benign melanocytic tumors, and metastatic melanoma in a stepwise manner (Kato et al., 2000). Tumor cells disseminate as early as 6 weeks to the lung; however, they remain slow cycling and do not develop overt metastases until about 18 months of age. Depletion of CD8+ cells leads to rapid development of metastases (Lengagne et al., 2008), suggesting that the immune microenvironment plays a role in containing the metastatic process. Implanting melanoma cells into syngeneic immunocompetent mice of different ages can also provide important information about the TME during aging. Evaluating these issues within an appropriate age context, given the known epidemiology of melanoma, is critical to accurate modeling of human disease, understanding underlying drivers, and identifying unique age-related tumor vulnerabilities that may be targeted for patient benefit.
Environmental exposures
Age-associated changes also reflect effects of chronic environmental exposures, such as UVR, diet, smoking, infection, and pollution. Existing models generally fail to consistently incorporate such chronic environmental exposures, which exert direct and indirect effects on tumor growth. Other than UVR (Galvani et al., 2020; Noonan et al., 2001; Shain et al., 2018; Trucco et al., 2019), contributions of environmental exposures to melanoma are not well characterized. A significant challenge in the near future is assessing and accounting for the influence of the microbiota in the gut and other organs in melanoma development and phenotype. Notably, mouse melanoma models were used to demonstrate a causal link between microbiome and response to ICIs, which has since been extended to melanoma patients (Gopalakrishnan et al., 2018; Matson et al., 2018; Routy et al., 2018; Sivan et al., 2015; Vetizou et al., 2015). A clear focus is now on the mechanisms by which the microbiome influences melanoma behavior, which should provide clues to its influence in clinical care (Li et al., 2019). Ultimately, the superimposition of aging together with chronic environmental exposures in melanoma models will be required to dissect the full complexity of the human disease (Lotz et al., 2020).
Experienced immune systems
In addition to directly contributing to the mutational burden in cancer cells, environmental stressors can also affect the microenvironment and systemic immune response, which can in turn influence melanoma development and progression. Chronic infections acquired during the lifetime, especially common viruses such as human cytomegalovirus, Epstein-Barr virus, influenza viruses (Kugel et al., 2018), and possibly even COVID-19, shape the immune repertoire and thus can affect immune system-targeted responses to developing or re-emerging tumors and contribute to therapy-induced toxicities. Pathogen-free animals lack this immunological experience and exhibit profoundly altered responses to challenge, such as with a pathogen or the presence of tumor cells, indicating the need for melanoma models with more experienced immune systems (Masopust et al., 2017).
Challenge 5: rare forms of melanoma
In countries with populations of predominantly European descent, most melanoma diagnoses are of the UVR-induced cutaneous subtypes. However, most cases in many countries in Latin America, Africa, and Asia are acral (occurring on the palms, soles, and in the nail bed) and mucosal melanomas (occurring within sinuses, nasal passages, mouth, vagina, and anus) (Ossio et al., 2017). Less common forms of melanoma also include those that occur in the uvea and conjunctiva of the eye, a subset of pediatric melanomas, and a form that invades the nervous system called leptomeningeal melanocytosis. Due to their low incidence, these non-cutaneous forms are often referred to as rare forms of melanoma. Critically, there are no approved therapies specific for rare forms of melanoma.
Rare melanoma genetic drivers
Unlike cutaneous forms of melanoma, UVR plays only a minorrole if any, in the etiology of acral, mucosal, and uveal melanoma at sun-protected sites (Hayward et al., 2017; Kong et al., 2017b; Liang et al., 2017; Robertson et al., 2017; Yeh et al., 2019). However, sun exposure is a risk factor for a subset of ocular melanomas (Johansson et al., 2020b; Karlsson et al., 2020; Mundra et al., 2021). In contrast to cutaneous melanoma, acral and mucosal melanomas are characterized by chromosomal alterations that result in large-scale genomic amplifications and deletions (Ablain et al., 2018; Yeh et al., 2019). The exception is uveal melanoma, most of which have activating mutations in either GNAQ or GNA11, encoding alpha subunits of G proteins activated by G protein-coupled receptors (Perez et al., 2018; Van Raamsdonk et al., 2009; Van Raamsdonk et al., 2010), and in ~40% of tumors BAP1, encoding a deubiquitinase (Harbour et al., 2010).
Mutations in genes characterizing The Cancer Genome Atlas (TCGA) cutaneous melanoma subtypes, namely BRAF, RAS, and NF1, are less common in mucosal, acral, and uveal melanoma than in cutaneous and conjunctival melanoma, although NRAS is fairly common in mucosal and acral melanoma (Wong et al., 2019). Hence, model generation approaches need to be amenable to rapidly testing potential driver genes both individually and in combinations. One possible approach is the use of embryonic stem cells harboring a melanocyte-specific Cre recombinase allele, Tyr-CreERt2 (Bok et al., 2020). Similarly, transposon-mediated forward genetic screens have been performed to identify drivers of melanoma development, for example genes whose mutation cooperates with BRAFV600E, as well as mediators of resistance to BRAF inhibitor therapy and new regulators of metastasis (Mann et al., 2015; Perna et al., 2015). In vitro modification followed by the generation of experimental chimeras enables the rapid assessment of driver genes alone and in combination. A parallel approach through genomic alteration analysis of human mucosal melanoma and tissue-specific CRISPR implementation in zebrafish identified SPRED1 as a tumor suppressor in mucosal melanoma in the context of KIT mutation (Ablain et al., 2018). Further increased use of CRISPR technology to knock out multiple genes should enhance progress, and genetic approaches may be supplemented with localized application (e.g., viruses in mice) to induce melanoma at different sites such as the mucosa or uveal tract. Unbiased whole-genome CRISPR/Cas9 screens are being used to identify rare melanoma dependencies (Hayward, personal communication).
Disease progression
In vivo and cell-culture-based strategies are useful for rapidly modeling rare forms of melanoma and enable high-throughput drug screening. A few transplantation models of rare melanomas from patients are available (Krepler et al., 2017), and the value of these models is demonstrated by their use in preclinical testing of palbociclib, a Food and Drug Administration-approved CDK4/6 inhibitor for mucosal melanoma (Zhou et al., 2019). Within each rare melanoma disease, we need to capture cell lines that represent the distinct stages of disease progression; for instance, although some uveal melanoma cell lines are available, few are from metastatic tumors. Increased numbers of cell-line models representing both early and advanced lesions are still required.
For optimal representation of rare melanomas, cell-based models should incorporate the appropriate aspects of the TME through transplantation into the correct orthotopic site (Stei et al., 2016). This is especially important for the ~50% of uveal melanomas that eventually metastasize to the liver, a more selective metastatic site tropism than observed in other melanoma subtypes (Ozaki et al., 2016; Richards et al., 2020). An imperative is the cataloging and characterization of available rare melanoma cell lines representing the repertoire of genomic alterations. Models of rare forms of melanoma should be optimized to enable in vivo imaging for quantitative and spatiotemporal assessment of tumor tropism and growth.
Genetic and ethnic diversity
Patient-derived cell lines of rare forms of melanoma exist and are growing in number but have been difficult to derive. For all melanoma research, but especially for rare melanomas, cell-based models need to include representation from different ethnicities, sites on the body, and genomic profiles to capture the extensive diversity of human melanomas (Ossio et al., 2017). Since rare forms of melanoma are proportionally more common in individuals of Asian, Latin American, and African descent, it is important to foster international collaborations to understand the germline contribution to disease presentation and predisposition. Furthermore, the poorer prognosis of patients with rare forms of melanoma is a clear example of health disparity (Klemen et al., 2020). These collaborations will also help improve outcomes through knowledge exchange between researchers and clinicians at major global centers, including those in low- and middle-income countries, who have considerable experience and expertise in these forms of the disease, and where high volumes of patients are seen. At present a resource of acral melanoma PDX and cell lines is currently being developed by researchers in Mexico and Brazil with assistance from a United Kingdom-based team.
Veterinary oncology
Pet dogs develop several forms of melanoma, including cutaneous, mucosal, acral, and uveal. Of these canine models, oral mucosal melanoma shows similar histology and has some of the genomic features common to oral mucosal melanoma in humans (Prouteau and Andre, 2019; Wong et al., 2019). In fact, trials in dogs with oral melanoma led to the first federal approval of a therapeutic cancer vaccine for commercial use (Bergman et al., 2003; Grosenbaugh et al., 2011). Pigs and horses may be other animals that could serve as models of rare forms of melanoma (van der Weyden et al., 2020).
MELANOMA MODELS: RISING TO THE CHALLENGE
Across models and challenges, common themes emerge as key opportunities to advance melanoma therapy discovery.
Integrating multiple models
There is tremendous value in conducting a cross-species approach for the development of novel melanoma therapies. For example, the ability to perform high-throughput screens to identify new targets or pathways in resistant cells using CRISPR/Cas9 technology can be performed with organoids or patient-derived cell lines co-cultured with immune, endothelial, and/or fibroblast cells. By screening libraries of potential therapeutics in the CRISPR-modified cells, drugs or drug combinations targeting those vulnerabilities can be rapidly tested. Candidate therapies can then be tested in vivo in preclinical fish and mouse models, and the impact on melanoma biology studied in the context of a whole animal system. Predictive preclinical modeling will also greatly benefit from the integration of multiple models. The advantages of an intensive cross-species (e.g., zebrafish, mouse, human) approach can also be applied to the examination of the TME over time, in various environmental conditions and in response to therapy. Tumor initiation can be evaluated for common mechanisms, such as the induction of the neural crest state, to determine whether age and environmental exposures influence melanogenesis pathways. A simplified melanoma genome in models compared with human melanoma can further be exploited to search for new melanoma genes (Ablain et al., 2018; Olvedy et al., 2017; Venkatesan et al., 2018; Yen et al., 2013).
Computational approaches
The tools with which we capture large “omics” data are becoming ever more sophisticated, allowing the capture of transcriptomic, exome, proteomic, chromatin immunoprecipitation-sequencing, and ATAC-sequencing data, opening up avenues to analyze potentially rare cell types, subpopulations, or limited patient tissue. Innovative analyses will enable direct comparisons between models and human disease. Cross-referencing with normal datasets, including melanocytic or neural crest lineage and the immune compartment, can be used to parse out meaning and patterns (Cao et al., 2019; Marie et al., 2020; Rambow et al., 2015, 2018; Soldatov et al., 2019; Varum et al., 2019; Zilionis et al., 2019). Evolutionarily conserved signatures/pathways identified in models have proved useful for establishing which melanoma subtype or process the model represents and for translation to the clinic (Galvani et al., 2020; Johansson et al., 2020a; Kaufman et al., 2016; Marie et al., 2020; Perez-Guijarro et al., 2020; Travnickova et al., 2019; Trucco et al., 2019).
A unified, bespoke approach between biologists and data scientists as to how and which data are generated and analyzed will help answer novel questions. For example, single-cell RNA-seq atlases of patient cohorts or melanoma models, generated by collating disparate experiments representing multiple biological contexts, are beneficial for comprehensive analyses. Heterogeneous multi-sample data can then be aligned and analyzed using applications such as Conos and cellAlign (Alpert et al., 2018; Barkas et al., 2019).
Melanoma patient datasets should be used to explore translational benefit with patient characteristics, such as patient treatment, mutational subtype, sex, pathological stage, and ethnic origin, taken into account (Lotz et al., 2020). TCGA was revolutionary for its amount of available melanoma patient transcriptomic data (Cancer Genome Atlas Network, 2015), and now computational approaches that infer the TME component based on bulk sequencing are fully exploiting these data (Jimenez-Sanchez et al., 2019). However, some annotation is incomplete and there is often a lag between the date of tumor staging and collection (Cancer Genome Atlas Network, 2015; Liu et al., 2018; Marie et al., 2020). Other patient datasets are also available that have their own unique setup and data collection, including patient-matched whole-exome and transcriptome sequencing data (Van Allen et al., 2015), pretreatment samples (Hugo et al., 2016; Van Allen et al., 2015), and matched histopathological analysis (Nsengimana et al., 2018).
With the advent of single-cell sequencing technology, we are able to obtain more information from limited samples from patients and models to better understand intra-tumor heterogeneity (de Andrade et al., 2019; Jerby-Arnon et al., 2018; Li et al., 2019; Rambow et al., 2018; Sade-Feldman et al., 2018; Tirosh et al., 2016; Travnickova et al., 2019). Models can be used to explore sparse cells and samples, such as micro-metastases, dormant cells, and residual disease, which would be difficult to obtain in the clinic. It is also possible to collect sufficient cells for analysis from one biopsy, thereby allowing monitoring of tumor progression on-therapy in real time. This allows melanoma modeling to generate dynamic signatures of tumor progression and therapy response, which can be used in the clinic for personalized medicine applications.
Quantitative dynamic in vivo information
Addressing questions related to in vivo tumor-stromal crosstalk and dynamic adaptation ideally requires inducible, site-specific models. While this remains a challenge, the careful design of models to ask specific questions and incorporate novel technologies to facilitate quantitative analyses will provide a path forward. Intravital imaging is a mainstay approach by which to examine the dynamic TME. Models that permit live imaging of specific steps in melanoma development, as well as melanoma/stromal interactions, immune responses, and metastatic events, will address each of the challenges. Ideally, these should enable imaging at the level of the whole body and entire tumor, and at the single-cell level (Hirata et al., 2015). Zebrafish are particularly well suited for intravital imaging; however, there are limitations to its application at some tissue sites, and it remains technologically challenging and resource intensive.
New techniques that provide quantitative dynamic information in vivo (e.g., signaling reporters, lineage tracing, barcoding, inducible labeling), paired with computational approaches to predict evolutionary trajectories and receptor/ligand interactions across diverse cell types, will increase throughput and capture spatiotemporal regulation of the tumor niche at primary and metastatic sites. These efforts will be supported by continued adoption of multiplexed image analysis and spatial omics platforms that are capable of quantitatively localizing relationships within intact tissues and simultaneously capturing tumor intrinsic and extrinsic heterogeneity. Such assays can then provide a link to human samples for future validation and data integration across modalities, paramount to developing a complex understanding of the dynamic networks that feed tumor growth.
Improved preclinical modeling
Future development of mice bearing a humanized immune system to host PDX transplants has been discussed, as have the advantages of integrating diverse models and of computational analytics in preclinical approaches. However, to fully exploit preclinical models to accurately predict patient responses and inform clinical trial decisions, rigorous conduct and reporting, including appropriate design and sample size, blinding and randomization, and statistical analyses, must be strictly employed to ensure reproducibility (Percie du Sert et al., 2020). Truly informative preclinical trials often require extensive resources and development time, items that are not always readily available to individual academic laboratories. We recommend that grants with ample resources be established to specifically support the development of preclinical models for drug development, including drug efficacy, biomarkers, and ADME-tox properties. Moreover, dedicated regional centers of preclinical modeling excellence should be instituted to support and educate researchers who wish to faithfully develop a drug lead from the discovery phase into the drug-development pipeline, and provide critical links with clinical centers and the pharmaceutical industry.
Repositories
A key requirement for realizing the full potential of models is ensuring their widespread availability and dissemination to the research community. Several representative PDX models from the collection described by Krepler et al. (2017) have been provided to a commercial vendor for distribution (Horizon/Envigo), and there are plans to enhance the availability of the collection still further via the NCI PDXNet (www.pdxnetwork.org). There is a conceptually similar initiative to make genetically characterized melanoma and other tumor PDXs available in Europe (www.europdx.eu). Melanoma models characterized for copy-number alteration and RNA-seq are hosted by Trace (https://www.uzleuven-kuleuven.be/lki/trace/trace-leuven-pdx-platform) and are available through the europdx research infrastructure (https://www.europdx.eu/europdxri-ta). All these models are annotated in the PDX finder (http://www.pdxfinder.org/).
A central repository is needed to provide methodological guidelines for the reporting and analysis of cell lines and animal models. Rigorous standardization, quality control, and genetic characterization when generating models should be emphasized. This should include how to use the models for the implantation of cell lines or for the testing of endogenous genetic modifications. Information regarding the validity of each model for studying the responses of primary and metastatic lesions to targeted therapies and ICIs, as well as irAEs, will be important. Caveats to each model (e.g., limited metastatic potential) should also be addressed. This goal includes sharing knowledge on how to use these models for immune assessments, including the window of opportunity for treatment, (neo)adjuvant regimes, vaccination, or treatment of established metastases. Details regarding early versus late therapeutic escape will also be informative to the research community.
Repositories will also facilitate progress toward effective therapies for rare melanoma subtypes, which would benefit greatly from generating a catalog of cell lines derived from rare melanomas coupled with various animal models that capture the range of rare melanoma subtypes, the genetic lesions, and the diversity of the patient population. These models can significantly advance our understanding of rare melanoma etiology, disease progression, and drug treatment. Importantly, we need to understand the unique drug targets in each subtype within the context of the diversity of the human population in order to develop the best therapeutic strategies.
CONCLUSIONS
The use of animal models derived from varied species combined with cell-based models will provide a mechanistic understanding of melanoma occurrence and outcome, generating critically needed insights into new targets and biomarkers to guide novel clinical paradigms. Furthermore, it is increasingly apparent that tissue context regulates immune surveillance, activation, localization, and function, and targets within the TME could provide orthogonal strategies for synergistic therapeutic combinations with ICIs or mitigation of off-target toxicities. Ultimately, the appropriate use of animal models provides a rapid route, relative to observational human studies, to fully understand and elucidate in vivo crosstalk, to differentiate the normal from the pathological, and to characterize the dynamic maladapted processes that drive disease. Finally, we propose that conducting cross-species studies of melanoma biology is advantageous and may provide synergic outcomes. Where models mimic or diverge from human disease, this will focus our use of these systems to probe specific mechanisms relevant to the clinic. In this way, we can identify the unique vulnerabilities that can be targeted and the specific challenges that must be overcome to eradicate this disease.
ACKNOWLEDGMENTS
We are grateful to the Melanoma Research Alliance for funding the Melanoma Models Workshop 2020 (Washington, DC, USA) that enabled the authors to discuss and prepare the manuscript. The authors thank Nancy R. Gough (BioSerendipity) for assistance with drafting the manuscript and Ray Gustin (MRA Patient Advocate) for discussions and note preparations. This work was supported by funding from the Intramural Research Program, National Institutes of Health, the National Cancer Institute (R13CA250294), Pigment Cell and Melanoma Research (Wiley) and Disease Models and Mechanisms (Company of Biologists). D.J.A. is supported by Cancer Research UK and the Wellcome Trust. N.A. is supported by the National Institute of Arthritis and Musculoskeletal and Skin Disease R01 AR070234. A.E.A. is supported by grants R01 CA182635, R01 CA196278, R01 CA253977, and P01 CA114046 from the National Institutes of Health/National Cancer Institute and by The Helman Family-Melanoma Research Alliance Team Science award (#559058). C.B. is supported by INCa (grant INCa_10573) and Le Fonds de dotation de La Société Française de Dermatologie. M.B. is supported by NIH/NCI grants (P50 CA121974, U01 CA238728, U01 CA233096, P30 CA016359, P01 CA128814, P01 CA206980, R01 CA244660, R01 CA216101, R01 CA212376, R01 CA204002, R01 CA216846, R01 CA196566, and R01 CA196660). C.E.B. is supported by National Institutes of Health R01CA237213. C.J.C. is supported by DOD CDMRP: W81XWH2010288. P.C. is supported by NIH/NCI grants (R01 CA228216, DP2 CA174499, P50 CA217694), Cycle for Survival Fund, and Geoffrey Beene Cancer Research Fund. M.H. is supported by NIH grants R01 CA238237, U54 CA224070, P01 CA114046, and the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation. S.L.H. is supported by grants from the National Institutes of Health (R01 CA121118), the Leveraged Finance Fights Melanoma-Melanoma Research Alliance Established Investigator award, and the Huntsman Cancer Foundation. F.A.K. is supported by NIH/NCI K22CA197058, NIH/NCI R03CA227349, Melanoma Research Alliance Young Investigator award, MRF Mucosal Melanoma Career Development award, and Harry J. Lloyd Charitable Trust Career Development award. C.K.K. is supported by NIH R01CA240633 and Melanoma Research Alliance Young Investigator award. S. Khan is supported by Melanoma Research Alliance-Society for Immunotherapy of Cancer Young Investigator award in Immune-Related Adverse Events. S. Kobold is supported by the Marie-Sklodowska-Curie Program Training Network for Optimizing Adoptive T Cell Therapy of Cancer funded by the H2020 Program of the European Union (grant 955575), the Heritage Foundation, the International Doctoral Program i-Target: Immunotargeting of Cancer funded by the Elite Network of Bavaria, the Bristol Myers Squibb-Melanoma Research Alliance Young Investigator award, the Ernst-Jung-Stiftung, the Bundesministerium für Bildung und Forschung Project Oncoattract, the European Research Council grant 756017, ARMOR-T, the German Research Foundation, the Fritz Bender Foundation, and the José-Carreras Foundation. E.L. is supported by the Amanda and Jonatan Eilian-Melanoma Research Alliance Young Investigator award, Belgian Federation for Cancer, Kom op tegen Kanker, and is a member of the EurOPDX Consortium and receives funding from the European Union’s Horizon 2020 research and innovation programme (EDIReX grant, no.731105, www.europdx.eu). C.L. is supported by Saban Family Foundation-Melanoma Research Alliance Team Science award (402792). D.B.L. is supported by NIH: R01GM101171 and R21ES032305: DOD: CA190267, CA170628, and NF170044. A.W.L. is funded by NIH NCI R01CA238163, Cancer Research Institute Lloyd J. Old STAR award, The Mark Foundation for Cancer Research Emerging Leader award, and American Cancer Society RSG-18-169-01-LIB. K.L.M. is supported by the NCI Intramural Research Program of the NIH, and in part by NCI Director’s Innovation award. J.-C.M. is supported by FWO (#G.0929.16N), Stichting Tegen Kanker, Melanoma Research Alliance-Established Investigator award (#623591), and KU Leuven (C1 grant). R.M. is supported by Cancer Research UK (A27412, A22902, and C5759/A29061), Wellcome Trust (100282/Z/12/Z), European Research Council Advanced grant (ERC-ADG-2014 671262), and the European Commission (Horizon 2020 program: UM Cure; project no. 667787). M.M. is supported by the National Cancer Institute, CA176839 & CA042014. E.E.P. is supported by MRC HGU Program (MC_UU_00007/9), ERC (ZF-MEL-CHEMBIO-648489), and the Anna-Maria and Stephen Kellen Foundation-Melanoma Research Alliance Team Science award (#687306). C.D.R.-E. is supported by MRC (MR/S01473X/1), Consejo Nacional de Ciencia y Tecnología of Mexico (Projects A3-S-31603 and A1-S-30165), Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT UNAM) (IA202020), Academy of Medical Sciences Newton Advanced Fellowship (NAF/R2/180782), and by a Wellcome Sanger Institute International Fellowship. Z.A.R. is supported by NCI CA197465 and NCI CA128814. M.S.S. is supported by grants from the Spanish Ministry of Economy and Innovation (SAF2017-89533-R), the Sokoloff Family-Melanoma Research Alliance-Established Investigator award, and by grants from the and Fundación “La Caixa and the Asociación Española Contra el Cáncer” (AECC). Y.S. is supported by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. 770854), and the Melanoma Research Alliance-Established Investigator award (#622106). J.V. is supported by P01 CA114046, R01 CA215733, R01 CA226888, P30 CA010815, DoD W81XWH2010356, and PA Department of Health. A.T.W. is supported by R01CA207935, P01 CA114046, R01 CA2322456, a Melanoma Research Alliance Team Science award, a Bloomberg Distinguished Professorship, and the E.V. McCollum Endowed Chair. R.M.W. is supported by the Anna-Maria and Stephen Kellen Foundation-Melanoma Research Alliance Team Science award, NIH Research Program grant R01CA229215, NIH Director’s New Innovator award DP2CA186572, NIH R01CA238317, NIH R01CA229215, The Pershing Square Sohn Foundation, American Cancer Society, The Alan and Sandra Gerry Metastasis Research Initiative at the Memorial Sloan Kettering Cancer Center, The Harry J. Lloyd Foundation, Consano, and the Starr Cancer Consortium. I.Y. is supported by NCI: R37CA240914, The Black Family-Melanoma Research Alliance Team Science award in Acral Melanoma, and Dermatology Foundation Stiefel Scholar award in Skin Cancer. J.Z. is supported by Melanoma Research Alliance Acral Melanoma Team Science award #579152 and NIH grant R01GM071725. L.I.Z. is supported by Cancer Biology R01 CA103846, NIH Melanoma PPG, P01CA63222, Melanoma Research Alliance-Established Investigator award, and Starr Cancer Consortium grant. G.M. is funded by the NIH Intramural Research Program. The content of this article is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. All authors read and contributed to the manuscript. We wish to apologize to the authors of substantive melanoma model studies that could not be cited or discussed here due to limitations in size and scope.
DECLARATION OF INTERESTS
D.J.A. is a paid consultant for Microbiotica and receives grant support from AstraZeneca and Open Targets. N.A. is a consultant for Viela Bio. A.E.A. reports receiving a commercial research grant from Pfizer. (2013–2017) and has ownership interest in patent number 9880150. M.B. is a consultant for Eli Lilly and Company and Bristol-Myers Squibb. P.C. received grants and personal fees from Deciphera, personal fees from Exelixis, grants from Array/Pfizer, and personal fees from Zai lab. S. Khan has a US patent pending (62/654,025). S. Kobold received research support from TCR2 Inc and Arcus Bioscience for work unrelated to this manuscript, has licensed IP to TCR2 Inc, has received consultancy fees from TCR2 Inc and Novartis, and is an inventor of several patents and patent applications in the field of cancer immunotherapy. R.M. consults for Pfizer, and as a former employee of the Institute of Cancer Research he may benefit financially from drug-discovery programs that are commercialized. M.M. receives research funding from Pfizer and Deciphera Pharma; serves as a scientific advisor to Pfizer, Deciphera Pharma, Revolution Medicine, and ARO; and receives royalty income from the University of California for Braf(CA) mice, which are the basis of several GEM models of BRAF(V600E)-driven melanoma described in this review. Z.A.R. is founder and scientific advisor of Pangea Therapeutics. Y.S. is a consultant of Achilles Therapeutics Limited. A.T.W. sits on the advisory board for Melanoma Research Foundation, Phoremost Technologies, and Healthe Scientific. R.M.W. is a paid consultant to N-of-One Therapeutics, a subsidiary of Qiagen; is on the Scientific Advisory Board of Consano but receives no income for this; and receives royalty payments for the use of the casper line from Carolina Biologicals. L.I.Z. is a founder and stockholder of Fate Therapeutics, CAMP4 Therapeutics, and Scholar Rock, and a consultant for Celularity.
REFERENCES
- Ablain J, Xu M, Rothschild H, Jordan RC, Mito JK, Daniels BH, Bell CF, Joseph NM, Wu H, Bastian BC, et al. (2018). Human tumor genomics and zebrafish modeling identify SPRED1 loss as a driver of mucosal melanoma. Science 362, 1055–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguirre-Ghiso JA (2018). How dormant cancer persists and reawakens. Science 361, 1314–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alicea GM, Rebecca VW, Goldman AR, Fane ME, Douglass SM, Behera R, Webster MR, Kugel CH 3rd, Ecker BL, Caino MC, et al. (2020). Changes in aged fibroblast lipid metabolism induce age-dependent melanoma cell resistance to targeted therapy via the fatty acid transporter FATP2. Cancer Discov. 10, 1282–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, et al. (2015). Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert A, Moore LS, Dubovik T, and Shen-Orr SS (2018). Alignment of single-cell trajectories to compare cellular expression dynamics. Nat. Methods 15, 267–270. [DOI] [PubMed] [Google Scholar]
- de Andrade LF, Lu Y, Luoma A, Ito Y, Pan D, Pyrdol JW, Yoon CH, Yuan GC, and Wucherpfennig KW (2019). Discovery of specialized NK cell populations infiltrating human melanoma metastases. JCI Insight 4, 10.1172/jci.insight.133103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arozarena I, and Wellbrock C (2019). Phenotype plasticity as enabler of melanoma progression and therapy resistance. Nat. Rev. Cancer 19, 377–391. [DOI] [PubMed] [Google Scholar]
- Audrito V, Manago A, La Vecchia S, Zamporlini F, Vitale N, Baroni G, Cignetto S, Serra S, Bologna C, Stingi A, et al. (2018). Nicotinamide phosphoribosyltransferase (NAMPT) as a therapeutic target in BRAF-mutated metastatic melanoma. J. Natl. Cancer Inst 110, 10.1093/jnci/djx198. [DOI] [PubMed] [Google Scholar]
- Balch CM, Buzaid AC, Soong SJ, Atkins MB, Cascinelli N, Coit DG, Fleming ID, Gershenwald JE, Houghton A Jr., Kirkwood JM, et al. (2001). Final version of the American Joint Committee on Cancer staging system for cutaneous melanoma. J. Clin. Oncol 19, 3635–3648. [DOI] [PubMed] [Google Scholar]
- Ballotti R, Healy E, and Bertolotto C (2019). Nicotinamide as a chemopreventive therapy of skin cancers. Too much of good thing? Pigment Cell Melanoma Res. 32, 601–602. [DOI] [PubMed] [Google Scholar]
- Bardeesy N, Bastian BC, Hezel A, Pinkel D, DePinho RA, and Chin L (2001). Dual inactivation of RB and p53 pathways in RAS-induced melanomas. Mol. Cell. Biol 21, 2144–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkas N, Petukhov V, Nikolaeva D, Lozinsky Y, Demharter S, Khodosevich K, and Kharchenko PV (2019). Joint analysis of heterogeneous single-cell RNA-seq dataset collections. Nat. Methods 16, 695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin DC, and Hynes RO (2017). Intravital imaging of metastasis in adult Zebrafish. BMC Cancer 17, 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezhnaya NM, Vinnichuk UD, Belova OB, and Baranovich VV (2006). Antitumor action of lymphokin-activated cells of patients with soft tissue sarcomas and melanomas in dependence on expression of MHC classes I and II antigenes. Exp. Oncol 28, 231–234. [PubMed] [Google Scholar]
- Bergman PJ, McKnight J, Novosad A, Charney S, Farrelly J, Craft D, Wulderk M, Jeffers Y, Sadelain M, Hohenhaus AE, et al. (2003). Long-term survival of dogs with advanced malignant melanoma after DNA vaccination with xenogeneic human tyrosinase: a phase I trial. Clin. Cancer Res 9, 1284–1290. [PubMed] [Google Scholar]
- Berk-Krauss J, Stein JA, Weber J, Polsky D, and Geller AC (2020). New systematic therapies and trends in cutaneous melanoma deaths among US Whites, 1986–2016. Am. J. Public Health 110, 731–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok I, Vera O, Xu X, Jasani N, Nakamura K, Reff J, Nenci A, Gonzalez JG, and Karreth FA (2020). A versatile ES cell-based melanoma mouse modeling platform. Cancer Res. 80, 912–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonet C, Luciani F, Ottavi JF, Leclerc J, Jouenne FM, Boncompagni M, Bille K, Hofman V, Bossis G, Marco de Donatis G, et al. (2017). Deciphering the role of oncogenic MITFE318K in senescence delay and melanoma progression. J. Natl. Cancer Inst 109, 10.1093/jnci/djw340. [DOI] [PubMed] [Google Scholar]
- Boshuizen J, Koopman LA, Krijgsman O, Shahrabi A, van den Heuvel EG, Ligtenberg MA, Vredevoogd DW, Kemper K, Kuilman T, Song JY, et al. (2018). Cooperative targeting of melanoma heterogeneity with an AXL antibody-drug conjugate and BRAF/MEK inhibitors. Nat. Med 24, 203–212. [DOI] [PubMed] [Google Scholar]
- Boumahdi S, and de Sauvage FJ (2020). The great escape: tumour cell plasticity in resistance to targeted therapy. Nat. Rev. Drug Discov 19, 39–56. [DOI] [PubMed] [Google Scholar]
- Callahan SJ, Tepan S, Zhang YM, Lindsay H, Burger A, Campbell NR, Kim IS, Hollmann TJ, Studer L, Mosimann C, and White RM (2018). Cancer modeling by transgene electroporation in adult zebrafish (TEAZ). Dis. Models Mech 11, dmm034561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2015). Genomic classification of cutaneous melanoma. Cell 161, 1681–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, et al. (2011). The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471, 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamoto K, Chowdhury PS, Kumar A, Sonomura K, Matsuda F, Fagarasan S, and Honjo T (2017). Mitochondrial activation chemicals synergize with surface receptor PD-1 blockade for T cell-dependent antitumor activity. Proc. Natl. Acad. Sci. U S A 114, E761–E770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A, Fernandez del Arna L, Ferguson J, Kamarashev J, Wellbrock C, and Hurlstone A (2014). Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep 8, 688–695, 10.1016/j.celrep.2014.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JH, Robinson JP, Arave RA, Burnett WJ, Kircher DA, Chen G, Davies MA, Grossmann AH, VanBrocklin MW, McMahon M, and Holmen SL (2015). AKT1 activation promotes development of melanoma metastases. Cell Rep. 13, 898–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua V, and Aplin AE (2018). Novel therapeutic strategies and targets in advanced uveal melanoma. Curr. Opin. Oncol 30, 134–141. [DOI] [PubMed] [Google Scholar]
- Conde-Perez A, and Larue L (2014). Human relevance of NRAS/BRAF mouse melanoma models. Eur. J. Cell Biol 93, 82–86. [DOI] [PubMed] [Google Scholar]
- Cui R, Widlund HR, Feige E, Lin JY, Wilensky DL, Igras VE, D’Orazio J, Fung CY, Schanbacher CF, Granter SR, and Fisher DE (2007). Central role of p53 in the suntan response and pathologic hyperpigmentation. Cell 128, 853–864. [DOI] [PubMed] [Google Scholar]
- D’Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, et al. (2006). Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature 443, 340–344. [DOI] [PubMed] [Google Scholar]
- Damo M, Fitzgerald B, Lu Y, Nader M, William I, Cheung JF, Connolly KA, Foster GG, Akama-Garren E, Lee DY, et al. (2020). Inducible de novo expression of neoantigens in tumor cells and mice. Nat. Biotechnol 39, 64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE Jr., You MJ, DePinho RA, McMahon M, and Bosenberg M (2009). Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat. Genet 41, 544–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CP, Merlino G, and Van Dyke T (2015). Preclinical mouse cancer models: a maze of opportunities and challenges. Cell 163, 39–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day CP, Marchalik R, Merlino G, and Michael H (2017). Mouse models of UV-induced melanoma: genetics, pathology, and clinical relevance. Lab. Invest 97, 698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmas V, Beermann F, Martinozzi S, Carreira S, Ackermann J, Kumasaka M, Denat L, Goodall J, Luciani F, Viros A, et al. (2007). Beta-catenin induces immortalization of melanocytes by suppressing p16INK4a expression and cooperates with N-Ras in melanoma development. Genes Dev. 21, 2923–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria M, Ohtani N, Youssef SA, Rodier F, Toussaint W, Mitchell JR, Laberge RM, Vijg J, Van Steeg H, Dolle ME, et al. (2014). An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell 31, 722–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuker MM, Marsh Durban V, Phillips WA, and McMahon M (2015). PI3’-kinase inhibition forestalls the onset of MEK1/2 inhibitor resistance in BRAF-mutated melanoma. Cancer Discov. 5, 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhomen N, Reis-Filho JS, da Rocha Dias S, Hayward R, Savage K, Delmas V, Larue L, Pritchard C, and Marais R (2009). Oncogenic Braf induces melanocyte senescence and melanoma in mice. Cancer Cell 15, 294–303. [DOI] [PubMed] [Google Scholar]
- Dorard C, Estrada C, Barbotin C, Larcher M, Garancher A, Leloup J, Beermann F, Baccarini M, Pouponnot C, Larue L, et al. (2017). RAF proteins exert both specific and compensatory functions during tumour progression of NRAS-driven melanoma. Nat. Commun 8, 15262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echevarria-Vargas IM, Reyes-Uribe PI, Guterres AN, Yin X, Kossenkov AV, Liu Q, Zhang G, Krepler C, Cheng C, Wei Z, et al. (2018). Co-targeting BET and MEK as salvage therapy for MAPK and checkpoint inhibitor-resistant melanoma. EMBO Mol. Med 10, e8446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggermont AMM, Blank CU, Mandala M, Long GV, Atkinson V, Dalle S, Haydon A, Lichinitser M, Khattak A, Carlino MS, et al. (2018). Adjuvant pembrolizumab versus placebo in resected stage III melanoma. N. Engl. J. Med 378, 1789–1801. [DOI] [PubMed] [Google Scholar]
- Einarsdottir BO, Bagge RO, Bhadury J, Jespersen H, Mattsson J, Nilsson LM, Truvé K, López MD, Naredi P, Nilsson O, et al. (2014). Melanoma patient-derived xenografts accurately model the disease and develop fast enough to guide treatment decisions. Oncotarget 5, 9609–9618, 10.18632/oncotarget.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elsas A, Hurwitz AA, and Allison JP (1999). Combination immunotherapy of B16 melanoma using anti-cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) and granulocyte/macrophage colony-stimulating factor (GMCSF)-producing vaccines induces rejection of subcutaneous and metastatic tumors accompanied by autoimmune depigmentation. J. Exp. Med 190, 355–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Elsas A, Sutmuller RP, Hurwitz AA, Ziskin J, Villasenor J, Medema JP, Overwijk WW, Restifo NP, Melief CJ, Offringa R, and Allison JP (2001). Elucidating the autoimmune and antitumor effector mechanisms of a treatment based on cytotoxic T lymphocyte antigen-4 blockade in combination with a B16 melanoma vaccine: comparison of prophylaxis and therapy. J. Exp. Med 194, 481–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faget DV, Ren Q, and Stewart SA (2019). Unmasking senescence: context-dependent effects of SASP in cancer. Nat. Rev. Cancer 19, 439–453. [DOI] [PubMed] [Google Scholar]
- Fane M, and Weeraratna AT (2020). How the ageing microenvironment influences tumour progression. Nat. Rev. Cancer 20, 89–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fares J, Fares MY, Khachfe HH, Salhab HA, and Fares Y (2020). Molecular principles of metastasis: a hallmark of cancer revisited. Signal Transduct. Target Ther 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y, Renshaw S, and Martin P (2012). Live imaging of tumor initiation in zebrafish larvae reveals a trophic role for leukocyte-derived PGE(2). Curr. Biol 22, 1253–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B, Ram R, Handoko HY, Mukhopadhyay P, Muller HK, Soyer HP, Morahan G, and Walker GJ (2015). Melanoma susceptibility as a complex trait: genetic variation controls all stages of tumor progression. Oncogene 34, 2879–2886. [DOI] [PubMed] [Google Scholar]
- Fidler IJ (1973). Selection of successive tumour lines for metastasis. Nat. New Biol 242, 148–149. [DOI] [PubMed] [Google Scholar]
- Fiebig HH, Schuchhardt C, Henss H, Fiedler L, and Lohr GW (1984). Comparison of tumor response in nude mice and in the patients. Behring Inst. Mitt 343–352. [PubMed] [Google Scholar]
- Fisher DE, and James WD (2010). Indoor tanning—science, behavior, and policy. N. Engl. J. Med 363, 901–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg EMV, Lindberg MF, Jespersen H, Alsén S, Bagge RO, Donia M, Svane IM, Nilsson O, Ny L, Nilsson LM, and Nilsson JA (2019). HER2 CAR-T cells eradicate uveal melanoma and T-cell therapy-resistant human melanoma in IL2 transgenic NOD/SCIO IL2 receptor knockout mice. Cancer Res. 79, 899–904, 10.1158/0008-5472.CAN-18-3158. [DOI] [PubMed] [Google Scholar]
- Galvani E, Mundra PA, Valpione S, Garcia-Martinez P, Smith M, Green-all J, Thakur R, Helmink B, Andrews MC, Boon L, et al. (2020). Stroma remodeling and reduced cell division define durable response to PD-1 blockade in melanoma. Nat. Commun 11, 853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garman B, Anastopoulos IN, Krepler C, Brafford P, Sproesser K, Jiang Y, Wubbenhorst B, Amaravadi R, Bennett J, Beqiri M, et al. (2017). Genetic and genomic characterization of 462 melanoma patient-derived xenografts, tumor biopsies, and cell lines. Cell Rep. 21, 1936–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JD, Knolle S, Wells KL, Liu D, Jackson IJ, Mort RL, and Headon DJ (2015). Maintenance of distinct melanocyte populations in the interfollicular epidermis. Pigment Cell Melanoma Res. 28, 476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goding CR, and Arnheiter H (2019). MITF—the first 25 years. Genes Dev. 33, 983–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan V, Spencer CN, Nezi L, Reuben A, Andrews MC, Karpinets TV, Prieto PA, Vicente D, Hoffman K, Wei SC, et al. (2018). Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill C, Bergsteinsdóttir K, Ogmundsdóttir MH, Pogenberg V, Schepsky A, Wilmanns M, Pingault V, and Steingrímsson E (2013). MITF mutations associated with pigment deficiency syndromes and melanoma have different effects on protein function. Hum. Mel. Genet 22, 4357–4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosenbaugh DA, Leard AT, Bergman PJ, Klein MK, Meleo K, Susaneck S, Hess PR, Jankowski MK, Jones PD, Leibman NF, et al. (2011). Safety and efficacy of a xenogeneic DNA vaccine encoding for human tyrosinase as adjunctive treatment for oral malignant melanoma in dogs following surgical excision of the primary tumor. Am. J. Vet. Res 72, 1631–1638. [DOI] [PubMed] [Google Scholar]
- Halama N, Zoernig I, Berthel A, Kahlert C, Klupp F, Suarez-Carmona M, Suetterlin T, Brand K, Krauss J, Lasitschka F, et al. (2016). Tumoral immune cell exploitation in colorectal cancer metastases can be targeted effectively by anti-CCR5 therapy in cancer patients. Cancer Cell 29, 587–601. [DOI] [PubMed] [Google Scholar]
- Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, Council ML, Matatall KA, Helms C, and Bowcock AM (2010). Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330, 1410–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch AM, Kakavand H, Alexandrov LB, Burke H, et al. (2017). Whole-genome landscapes of major melanoma subtypes. Nature 545, 175–180. [DOI] [PubMed] [Google Scholar]
- Hegerfeldt Y, Tusch M, Brocker EB, and Friedl P (2002). Collective cell movement in primary melanoma explants: plasticity of cell-cell interaction, beta1-integrin function, and migration strategies. Cancer Res. 62, 2125–2130. [PubMed] [Google Scholar]
- Heilmann S, Ratnakumar K, Langdon E, Kansler E, Kim I, Campbell NR, Perry E, McMahon A, Kaufman C, van Rooijen E, et al. (2015). A quantitative system for studying metastasis using transparent zebrafish. Cancer Res. 75, 4272–4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennessey RC, Holderbaum AM, Bonilla A, Delaney C, Gillahan JE, Tober KL, Oberyszyn TM, Zippin JH, and Burd CE (2017). Ultraviolet radiation accelerates NRas-mutant melanomagenesis: a cooperative effect blocked by sunscreen. Pigment Cell Melanoma Res. 30, 477–487. [DOI] [PubMed] [Google Scholar]
- Hirata E, Girotti MR, Viros A, Hooper S, Spencer-Dene B, Matsuda M, Larkin J, Marais R, and Sahai E (2015). Intravital imaging reveals how BRAF inhibition generates drug-tolerant microenvironments with high integrin beta1/FAK signaling. Cancer Cell 27, 574–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, et al. (2010). Improved survival with ipilimumab in patients with metastatic melanoma. N. Engl. J. Med 363, 711–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höizel M, and Tüting T (2016). Inflammation-induced plasticity in melanoma therapy and metastasis. Trends lmmunol. 37, 364–374, 10.1016/j.it.2016.03.009. [DOI] [PubMed] [Google Scholar]
- Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, et al. (2016). Genomic and transcriptomic features of response to anti-PD-1 therapy in metastatic melanoma. Cell 165, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, Hiramatsu H, Kobayashi K, Suzue K, Kawahata M, Hioki K, Ueyama Y, Koyanagi Y, Sugamura K, Tsuji K, et al. (2002). NOD/SCID/gamma(c)(null) mouse: an excellent recipient mouse model for engraftment of human cells. Blood 100, 3175–3182. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Ishida M, Tanaka Y, Okazaki T, Honjo T, and Minato N (2002). Involvement of PD-L1 on tumor cells in the escape from host immune system and tumor immunotherapy by PD-L1 blockade. Proc. Natl. Acad. Sci. U S A 99, 12293–12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwai Y, Terawaki S, and Honjo T (2005). PD-1 blockade inhibits hematogenous spread of poorly immunogenic tumor cells by enhanced recruitment of effector T cells. Int. Immunol 17, 133–144. [DOI] [PubMed] [Google Scholar]
- Jenkins MH, Steinberg SM, Alexander MP, Fisher JL, Ernstoff MS, Turk MJ, Mullins DW, and Brinckerhoff CE (2014). Multiple murine BRaf(V600E) melanoma cell lines with sensitivity to PLX4032. Pigment Cell Melanoma Res. 27, 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su MJ, Melms JC, Leeson R, Kanodia A, Mei S, Lin JR, et al. (2018). A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell 175, 984–997.e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen H, Lindberg MF, Donia M, Söderberg EMV, Andersen R, Keller U, Ny L, Svane IM, Nilsson LM, and Nilsson JA (2017). Clinical responses to adoptive T-cell transfer can be modeled in an autologous immune-humanized mouse model. Nat. Comm 8, 707, 10.1038/s41467-017-00786-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Sanchez A, Cast O, and Miller ML (2019). Comprehensive benchmarking and integration of tumor microenvironment cell estimation methods. Cancer Res. 79, 6238–6246. [DOI] [PubMed] [Google Scholar]
- Johansson JA, Marie KL, Lu Y, Brombin A, Santoriello C, Zeng Z, Zich J, Gautier P, von Kriegsheim A, Brunsdon H, et al. (2020a). PRL3-DDX21 transcriptional control of endolysosomal genes restricts melanocyte stem cell differentiation. Dev. Cell 54, 317–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson PA, Brooks K, Newell F, Palmer JM, Wilmott JS, Pritchard AL, Broit N, Wood S, Carlino MS, Leonard C, et al. (2020b). Whole genome landscapes of uveal melanoma show an ultraviolet radiation signature in iris tumours. Nat. Commun 11, 2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DB, Menzies AM, Zimmer L, Eroglu Z, Ye F, Zhao S, Rizos H, Sucker A, Scolyer RA, Gutzmer R, et al. (2015). Acquired BRAF inhibitor resistance: a multicenter meta-analysis of the spectrum and frequencies, clinical behaviour, and phenotypic associations of resistance mechanisms. Eur. J. Cancer 51, 2792–2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson J, Nilsson LM, Mitra S, Alsén S, Shelke GV, Sah VR, Forsberg EMV, Stierner U,AII-Eriksson C, Einarsdottir B, et al. (2020). Molecular profiling of driver events in metastatic uveal melanoma. Nat. Commun 11, 1894, 10.1038/s41467-020-15606-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Liu W, Akhand AA, Hossain K, Takeda K, Takahashi M, and Nakashima I (2000). Ultraviolet radiation induces both full activation of ret kinase and malignant melanocytic tumor promotion in RFP-RET-transgenic mice. J. Invest. Dermatol 115, 1157–1158. [DOI] [PubMed] [Google Scholar]
- Kaufman CK, Mosimann C, Fan ZP, Yang S, Thomas AJ, Ablain J, Tan JL, Fogley RD, van Rooijen E, Hagedorn EJ, et al. (2016). A zebrafish melanoma model reveals emergence of neural crest identity during melanoma initiation. Science 351, aad2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami A, and Fisher DE (2016). Bioinformatic analysis of gene expression for melanoma treatment. J. Invest. Dermatol 136, 2342–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper K, Krijgsman O, Cornelissen-Steijger P, Shahrabi A, Weeber F, Song JY, Kuilman T, Vis DJ, Wessels LF, Voest EE, et al. (2015). Intra- and inter-tumor heterogeneity in a vemurafenib-resistant melanoma patient and derived xenografts. EMBO Mol. Med 7, 1104–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IS, Heilmann S, Kansler ER, Zhang Y, Zimmer M, Ratnakumar K, Bowman RL, Simon-Vermot T, Fennell M, Garippa R, et al. (2017). Micro-environment-derived factors driving metastatic plasticity in melanoma. Nat Commun. 8, 14343, 10.1038/ncomms14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinsey CG, Camolotto SA, Boespflug AM, Guillen KP, Foth M, Truong A, Schuman SS, Shea JE, Seipp MT, Yap JT, et al. (2019). Protective autophagy elicited by RAF–>MEK–>ERK inhibition suggests a treatment strategy for RAS-driven cancers. Nat. Med 25, 620–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Xiong C, Martin B, Schubach M, Inoue F, Bell RJA, Costello JF, Shendure J, and Ahituv N (2019). Saturation mutagenesis of twenty disease-associated regulatory elements at single base-pair resolution. Nat. Commun 10, 3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein A, Sagi-Assif O, Meshel T, Telerman A, Izraely S, Ben-Menachem S, Bayry J, Marzese DM, Ohe S, Hoon DSB, et al. (2017). CCR4 is a determinant of melanoma brain metastasis. Oncotarget 8, 31079–31091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemen ND, Wang M, Rubinstein JC, Olino K, Clune J, Ariyan S, Cha C, Weiss SA, Kluger HM, and Sznol M (2020). Survival after checkpoint inhibitors for metastatic acral, mucosal and uveal melanoma. J. Immunother. Cancer 8, e000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug HL, Tooze JA, Graff-Cherry C, Anver MR, Noonan FP, Fears TR, Tucker MA, De Fabo EC, and Merlino G (2010). Sunscreen prevention of melanoma in man and mouse. Pigment Cell Melanoma Res. 23, 835–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler C, Nittner D, Rambow F, Radaelli E, Stanchi F, Vandamme N, Baggiolini A, Sommer L, Berx G, van den Oord JJ, et al. (2017). Mouse cutaneous melanoma induced by mutant BRaf arises from expansion and dedifferentiation of mature pigmented melanocytes. Cell Stem Cell 21, 679–693.e76. [DOI] [PubMed] [Google Scholar]
- Kong X, Kuilman T, Shahrabi A, Boshuizen J, Kemper K, Song JY, Niessen HWM, Rozeman EA, Geukes Foppen MH, Blank CU, et al. (2017a). Cancer drug addiction is relayed by an ERK2-dependent phenotype switch. Nature 550, 270–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Sheng X, Wu X, Yan J, Ma M, Yu J, Si L, Chi Z, Cui C, Dai J, et al. (2017b). Frequent genetic aberrations in the CDK4 pathway in acral melanoma indicate the potential for CDK4/6 inhibitors in targeted therapy. Clin. Cancer Res 23, 6946–6957. [DOI] [PubMed] [Google Scholar]
- Krepler C, Xiao M, Sproesser K, Brafford PA, Shannan B, Beqiri M, Liu Q, Xu W, Garman B, Nathanson KL, et al. (2016). Personalized preclinical trials in BRAF inhibitor-resistant patient-derived xenograft models identify second-line combination therapies. Clin. Cancer Res 22, 1592–1602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepler C, Sproesser K, Brafford P, Beqiri M, Garman B, Xiao M, Shannan B, Watters A, Perego M, Zhang G, et al. (2017). A comprehensive patient-derived xenograft collection representing the heterogeneity of melanoma. Cell Rep. 21, 1953–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugel CH 3rd, Douglass SM, Webster MR, Kaur A, Liu Q, Yin X, Weiss SA, Darvishian F, Al-Rohil RN, Ndoye A, et al. (2018). Age correlates with response to anti-PD1, reflecting age-related differences in intratumoral effector and regulatory T-cell populations. Clin. Cancer Res 24, 5347–5356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Chamoto K, Chowdhury PS, and Honjo T (2020). Tumors attenuating the mitochondrial activity in T cells escape from PD-1 blockade therapy. Elife 9, e52330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin J, Chiarion-Sileni V, Gonzalez R, Grob JJ, Rutkowski P, Lao CD, Cowey CL, Schadendorf D, Wagstaff J, Dummer R, et al. (2019). Five-year survival with combined nivolumab and ipilimumab in advanced melanoma. N. Engl. J. Med 381, 1535–1546. [DOI] [PubMed] [Google Scholar]
- Lauss M, Nsengimana J, Staaf J, Newton-Bishop J, and Jonsson G (2016). Consensus of melanoma gene expression subtypes converges on biological entities. J. Invest. Dermatol 136, 2502–2505. [DOI] [PubMed] [Google Scholar]
- Leach DR, Krummel MF, and Allison JP (1996). Enhancement of antitumor immunity by CTLA-4 blockade. Science 271, 1734–1736. [DOI] [PubMed] [Google Scholar]
- Lelliott EJ, Cullinane C, Martin CA, Walker R, Ramsbottom KM, Souza-Fonseca-Guimaraes F, Abuhammad S, Michie J, Kirby L, Young RJ, et al. (2019). A novel immunogenic mouse model of melanoma for the preclinical assessment of combination targeted and immune-based therapy. Sci. Rep 9, 1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengagne R, Graff-Dubois S, Garcette M, Renia L, Kato M, Guillet JG, Engelhard VH, Avril MF, Abastado JP, and Prevost-Blondel A (2008). Distinct role for CD8 T cells toward cutaneous tumors and visceral metastases. J. Immunol 180, 130–137. [DOI] [PubMed] [Google Scholar]
- Leucci E (2018). Cancer development and therapy resistance: spotlights on the dark side of the genome. Pharmacol. Ther 189, 22–30. [DOI] [PubMed] [Google Scholar]
- Leucci E, Vendramin R, Spinazzi M, Laurette P, Fiers M, Wouters J, Radaelli E, Eyckerman S, Leonelli C, Vanderheyden K, et al. (2016). Melanoma addiction to the long non-coding RNA SAMMSON. Nature 531, 518–522. [DOI] [PubMed] [Google Scholar]
- Li L, Fukunaga-Kalabis M, and Herlyn M (2011). The three-dimensional human skin reconstruct model: a tool to study normal skin and melanoma progression. J. Vis. Exp 10.3791/2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ, van den Braber M, Rozeman EA, Haanen J, Blank CU, et al. (2019). Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell 176, 775–789.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang WS, Hendricks W, Kiefer J, Schmidt J, Sekar S, Carpten J, Craig DW, Adkins J, Cuyugan L, Manojlovic Z, et al. (2017). Integrated genomic analyses reveal frequent TERT aberrations in acral melanoma. Genome Res. 27, 524–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lichtenberg T, Hoadley KA, Poisson LM, Lazar AJ, Cherniack AD, Kovatich AJ, Benz CC, Levine DA, Lee AV, et al. (2018). An integrated TCGA pan-cancer clinical data resource to drive high-quality survival outcome analytics. Cell 173, 400–416.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo SN, Scolyer RA, and Thompson JF (2018). Long-term survival of patients with thin (T1) cutaneous melanomas: a Breslow thickness cut point of 0.8 mm separates higher-risk and lower-risk tumors. Ann. Surg. Oncol 25, 894–902. [DOI] [PubMed] [Google Scholar]
- Long GV, Hauschild A, Santinami M, Atkinson V, Mandala M, Chiarion-Sileni V, Larkin J, Nyakas M, Dutriaux C, Haydon A, et al. (2017). Adjuvant dabrafenib plus trametinib in stage III BRAF-mutated melanoma. N. Engl. J. Med 377, 1813–1823. [DOI] [PubMed] [Google Scholar]
- Lotz M, Budden T, Furney SJ, and Viros A (2020). Molecular subtype, biological sex and age shape melanoma tumour evolution. Br. J. Dermatol 10.1111/bjd.19128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke JJ, Flaherty KT, Ribas A, and Long GV (2017). Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat. Rev. Clin. Oncol 14, 463–482. [DOI] [PubMed] [Google Scholar]
- Luo X, Fu Y, Loza AJ, Murali B, Leahy KM, Ruhland MK, Gang M, Su X, Zamani A, Shi Y, et al. (2016). Stromal-initiated changes in the bone promote metastatic niche development. Cell Rep. 14, 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacRae CA, and Peterson RT (2015). Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov 14, 721–731. [DOI] [PubMed] [Google Scholar]
- Mann MB, Black MA, Jones DJ, Ward JM, Yew CC, Newberg JY, Dupuy AJ, Rust AG, Bosenberg MW, McMahon M, et al. (2015). Transposon mutagenesis identifies genetic drivers of Braf(V600E) melanoma. Nat. Genet 47, 486–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manuel CA, Bagby SM, Reisinger JA, Pugazhenthi U, Pitts TM, Keysar SB, Arcaroli JJ, and Leszczynski JK (2017). Procedure for horizontal transfer of patient-derived xenograft tumors to eliminate Corynebacterium bovis. J. Am. Assoc. Lab. Anim. Sci 56, 166–172. [PMC free article] [PubMed] [Google Scholar]
- Marie KL, Sassano A, Yang HH, Michalowski AM, Michael HT, Guo T, Tsai YC, Weissman AM, Lee MP, Jenkins LM, et al. (2020). Melanoblast transcriptome analysis reveals pathways promoting melanoma metastasis. Nat. Commun 11, 333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marine JC, Dawson SJ, and Dawson MA (2020). Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 20, 743–756. [DOI] [PubMed] [Google Scholar]
- Marsh Durban V, Deuker MM, Bosenberg MW, Phillips W, and McMahon M (2013). Differential AKT dependency displayed by mouse models of BRAFV600E-initiated melanoma. J. Clin. Invest 123, 5104–5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Corral I, Olmeda D, Dieguez-Hurtado R, Tammela T, Alitalo K, and Ortega S (2012). In vivo imaging of lymphatic vessels in development, wound healing, inflammation, and tumor metastasis. Proc. Natl. Acad. Sci. U S A 109, 6223–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D, Sivula CP, and Jameson SC (2017). Of mice, dirty mice, and men: using mice to understand human immunology. J. Immunol 199, 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matson V, Fessler J, Bao R, Chongsuwat T, Zha Y, Alegre ML, Luke JJ, and Gajewski TF (2018). The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Findlay GM, Gagnon JA, Horwitz MS, Schier AF, and Shendure J (2016). Whole-organism lineage tracing by combinatorial and cumulative genome editing. Science 353, aaf7907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meeth K, Wang JX, Micevic G, Damsky W, and Bosenberg MW (2016). The YUMM lines: a series of congenic mouse melanoma cell lines with defined genetic alterations. Pigment Cell Melanoma Res. 29, 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort RL, Jackson IJ, and Patton EE (2015). The melanocyte lineage in development and disease. Development 142, 620–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujahid N, Liang Y, Murakami R, Choi HG, Dobry AS, Wang J, Suita Y, Weng QY, Allouche J, Kemeny LV, et al. (2017). A UV-independent topical small-molecule approach for melanin production in human skin. Cell Rep. 19, 2177–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller J, Krijgsman O, Tsoi J, Robert L, Hugo W, Song C, Kong X, Possik PA, Cornelissen-Steijger PD, Geukes Foppen MH, et al. (2014). Low MITF/AXL ratio predicts early resistance to multiple targeted drugs in melanoma. Nat. Commun 5, 5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundra PA, Dhomen N, Rodrigues M, Mikkelsen LH, Cassoux N, Brooks K, Valpione S, Reis-Filho JS, Heegaard S, Stern MH, et al. (2021). Ultraviolet radiation drives mutations in a subset of mucosal melanomas. Nat. Commun 12, 259, 10.1038/s41467-020-20432-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naseer A, Terry A, Gilroy K, Kilbey A, Watts C, Mackay N, Bell M, Mason S, Blyth K, Cameron E, and Neil JC (2015). Frequent infection of human cancer xenografts with murine endogenous retroviruses in vivo. Viruses 7, 2014–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen NT, and Fisher DE (2019). MITF and UV responses in skin: from pigmentation to addiction. Pigment Cell Melanoma Res. 32, 224–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen DX, Bos PD, and Massague J (2009). Metastasis: from dissemination to organ-specific colonization. Nat. Rev. Cancer 9, 274–284. [DOI] [PubMed] [Google Scholar]
- Nishimura EK, Granter SR, and Fisher DE (2005). Mechanisms of hair graying: incomplete melanocyte stem cell maintenance in the niche. Science 307, 720–724. [DOI] [PubMed] [Google Scholar]
- Noonan FP, Recio JA, Takayama H, Duray P, Anver MR, Rush WL, De Fabo EC, and Merlino G (2001). Neonatal sunburn and melanoma in mice. Nature 413, 271–272. [DOI] [PubMed] [Google Scholar]
- Nsengimana J, Laye J, Filia A, O’Shea S, Muralidhar S, Pozniak J, Droop A, Chan M, Walker C, Parkinson L, et al. (2018). beta-Catenin-mediated immune evasion pathway frequently operates in primary cutaneous melanomas. J. Clin. Invest 128, 2048–2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohanna M, Cheli Y, Bonet C, Bonazzi VF, Allegra M, Giuliano S, Bille K, Bahadoran P, Giacchero D, Lacour JP, et al. (2013). Secretome from senescent melanoma engages the STAT3 pathway to favor reprogramming of naive melanoma towards a tumor-initiating cell phenotype. Oncotarget 4, 2212–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohanna M, Cerezo M, Nottet N, Bille K, Didier R, Beranger G, Mograbi B, Rocchi S, Yvan-Charvet L, Ballotti R, and Bertolotto C (2018). Pivotal role of NAMPT in the switch of melanoma cells toward an invasive and drug-resistant phenotype. Genes Dev. 32, 448–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha R, Leli NM, Onorati A, Piao S, Verginadis II, Tameire F, Rebecca VW, Chude CI, Murugan S, Fennelly C, et al. (2019). ER translocation of the MAPK pathway drives therapy resistance in BRAF-mutant melanoma. Cancer Discov. 9, 396–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olmeda D, Cerezo-Wallis D, Riveiro-Falkenbach E, Pennacchi PC, Contreras-Alcalde M, Ibarz N, Cifdaloz M, Catena X, Calvo TG, Canon E, et al. (2017). Whole-body imaging of lymphovascular niches identifies premetastatic roles of midkine. Nature 546, 676–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olvedy M, Tisserand JC, Luciani F, Boeckx B, Wouters J, Lopez S, Rambow F, Aibar S, Thienpont B, Barra J, et al. (2017). Comparative oncogenomics identifies tyrosine kinase FES as a tumor suppressor in melanoma. J. Clin. Invest 127, 2310–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossio R, Roldan-Marin R, Martinez-Said H, Adams DJ, and Robles-Espinoza CD (2017). Melanoma: a global perspective. Nat. Rev. Cancer 17, 393–394. [DOI] [PubMed] [Google Scholar]
- Ozaki S, Vuyyuru R, Kageyama K, Terai M, Ohara M, Cheng H, Manser T, Mastrangelo MJ, Aplin AE, and Sato T (2016). Establishment and characterization of orthotopic mouse models for human uveal melanoma hepatic colonization. Am. J. Pathol 186, 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan YA, Freundlich T, Weissman TA, Schoppik D, Wang XC, Zimmerman S, Ciruna B, Sanes JR, Lichtman JW, and Schier AF (2013). Zebrabow: multispectral cell labeling for cell tracing and lineage analysis in zebrafish. Development 140, 2835–2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patton EE, Widlund HR, Kutok JL, Kopani KR, Amatruda JF, Murphey RD, Berghmans S, Mayhall EA, Traver D, Fletcher CD, et al. (2005). BRAF mutations are sufficient to promote nevi formation and cooperate with p53 in the genesis of melanoma. Curr. Biol 15, 249–254. [DOI] [PubMed] [Google Scholar]
- Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, et al. (2016). Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov. 6, 202–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, Browne WJ, Clark A, Cuthill IC, Dirnagl U, et al. (2020). The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. PLoS Biol. 18, e3000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez DE, Henle AM, Amsterdam A, Hagen HR, and Lees JA (2018). Uveal melanoma driver mutations in GNAQ/11 yield numerous changes in melanocyte biology. Pigment Cell Melanoma Res. 31, 604–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Guijarro E, Day CP, Merlino G, and Zaidi MR (2017). Genetically engineered mouse models of melanoma. Cancer 123, 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Guijarro E, Yang HH, Araya RE, El Meskini R, Michael HT, Vodnala SK, Marie KL, Smith C, Chin S, Lam KC, et al. (2020). Multimodel preclinical platform predicts clinical response of melanoma to immunotherapy. Nat. Med 26, 781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perna D, Karreth FA, Rust AG, Perez-Mancera PA, Rashid M, Iorio F, Alifrangis C, Arends MJ, Bosenberg MW, Bollag G, et al. (2015). BRAF inhibitor resistance mediated by the AKT pathway in an oncogenic BRAF mouse melanoma model. Proc. Natl. Acad. Sci. U S A 112, E536–E545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit V, Raymond J, Alberti C, Pouteaux M, Gallagher SJ, Nguyen MQ, Aplin AE, Delmas V, and Larue L (2019). C57BL/6 congenic mouse NRAS(Q61K) melanoma cell lines are highly sensitive to the combination of Mek and Akt inhibitors in vitro and in vivo. Pigment Cell Melanoma Res. 32, 829–841. [DOI] [PubMed] [Google Scholar]
- Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, Linette GP, Meyer N, Giguere JK, Agarwala SS, et al. (2015). Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. N. Engl. J. Med 372, 2006–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powley IR, Patel M, Miles G, Pringle H, Howells L, Thomas A, Kettle-borough C, Bryans J, Hammonds T, MacFarlane M, and Pritchard C (2020). Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery. Br. J. Cancer 122, 735–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prouteau A, and Andre C (2019). Canine melanomas as models for human melanomas: clinical, histological, and genetic comparison. Genes (Basel) 10, 501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prykhozhij SV, and Berman JN (2018). Zebrafish knock-ins swim into the mainstream. Dis. Models Mech 11, dmm037515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, and Morrison SJ (2008). Efficient tumour formation by single human melanoma cells. Nature 456, 593–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O’Brien JM, Simpson EM, Barsh GS, and Bastian BC (2009). Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature 457, 599–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, et al. (2010). Mutations in GNA11 in uveal melanoma. N. Engl. J. Med 363, 2191–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooijen E, Fazio M, and Zon LI (2017). From fish bowl to bedside: the power of zebrafish to unravel melanoma pathogenesis and discover new therapeutics. Pigment Cell Melanoma Res. 30, 402–412, 10.1111/pcmr.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambow F, Job B, Petit V, Gesbert F, Delmas V, Seberg H, Meurice G, Van Otterloo E, Dessen P, Robert C, et al. (2015). New functional signatures for understanding melanoma biology from tumor cell lineage-specific analysis. Cell Rep. 13, 840–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambow F, Rogiers A, Marin-Bejar O, Aibar S, Femel J, Dewaele M, Karras P, Brown D, Chang YH, Debiec-Rychter M, et al. (2018). Toward minimal residual disease-directed therapy in melanoma. Cell 174, 843–855.e19. [DOI] [PubMed] [Google Scholar]
- Rambow F, Marine JC, and Goding CR (2019). Melanoma plasticity and phenotypic diversity: therapeutic barriers and opportunities. Genes Dev. 33, 1295–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards JR, Yoo JH, Shin D, and Odelberg SJ (2020). Mouse models of uveal melanoma: strengths, weaknesses, and future directions. Pigment Cell Melanoma Res. 33, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E, et al. (2015). Nivolumab in previously untreated melanoma without BRAF mutation. N. Engl. J. Med 372, 320–330. [DOI] [PubMed] [Google Scholar]
- Robert C, Grob JJ, Stroyakovskiy D, Karaszewska B, Hauschild A, Levchenko E, Chiarion Sileni V, Schachter J, Garbe C, Bondarenko I, et al. (2019). Five-year outcomes with dabrafenib plus trametinib in metastatic melanoma. N. Engl. J. Med 381, 626–636. [DOI] [PubMed] [Google Scholar]
- Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, Hess JM, Uzunangelov V, Walter V, Danilova L, et al. (2017). Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 32, 204–220.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, Korbel C, Laschke MW, Gimotty PA, Philipp SE, et al. (2013). Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell 23, 811–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh-Johnson M, Shah AN, Stonick JA, Poudel KR, Kargl J, Yang GH, di Martino J, Hernandez RE, Gast CE, Zarour LR, et al. (2017). Macrophage-dependent cytoplasmic transfer during melanoma invasion in vivo. Dev. Cell 43, 549–562.e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A, Willinger T, Martinek J, Strowig T, Gearty SV, Teichmann LL, Saito Y, Marches F, Halene S, Palucka AK, et al. (2014). Development and function of human innate immune cells in a humanized mouse model. Nat. Biotechnol 32, 364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart SP, Herz J, Vassallo BG, Hunter A, Wall MK, Badger JH, McCulloch JA, Anastasakis DG, Sarshad AA, Leonardi I, et al. (2019). Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science 365, eaaw4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Routy B, Le Chatelier E, Derosa L, Duong CPM, Alou MT, Daillere R, Fluckiger A, Messaoudene M, Rauber C, Roberti MP, et al. (2018). Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97. [DOI] [PubMed] [Google Scholar]
- Sade-Feldman M, Yizhak K, Bjorgaard SL, Ray JP, de Boer CG, Jenkins RW, Lieb DJ, Chen JH, Frederick DT, Barzily-Rokni M, et al. (2018). Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell 175, 998–1013.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Ayala M, Montenegro MF, Sanchez-Del-Campo L, Fernandez-Perez MP, Chazarra S, Freter R, Middleton M, Pinero-Madrona A, Cabezas-Herrera J, Goding CR, and Rodriguez-Lopez JN (2013). Directed phenotype switching as an effective antimelanoma strategy. Cancer Cell 24, 105–119. [DOI] [PubMed] [Google Scholar]
- Sanmamed MF, Chester C, Melero I, and Kohrt H (2016). Defining the optimal murine models to investigate immune checkpoint blockers and their combination with other immunotherapies. Ann. Oncol 27, 1190–1198. [DOI] [PubMed] [Google Scholar]
- Santoriello C, Sporrij A, Yang S, Flynn RA, Henriques T, Dorjsuren B, Custo Greig E, McCall W, Stanhope ME, Fazio M, et al. (2020). RNA helicase DDX21 mediates nucleotide stress responses in neural crest and melanoma cells. Nat. Cell Biol 22, 372–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyamoorthy K, Meier F, Hsu MY, Berking C, and Herlyn M (1999). Human xenografts, human skin and skin reconstructs for studies in melanoma development and progression. Cancer Metastasis Rev. 18, 401–405. [DOI] [PubMed] [Google Scholar]
- Scahill CM, Digby Z, Sealy IM, Wojciechowska S, White RJ, Collins JE, Stemple DL, Bartke T, Mathers ME, Patton EE, and Busch-Nentwich EM (2017). Loss of the chromatin modifier Kdm2aa causes BrafV600E-independent spontaneous melanoma in zebrafish. PLoS Genet. 13, e1006959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Joseph NM, Yu R, Benhamida J, Liu S, Prow T, Ruben B, North J, Pincus L, Yeh I, et al. (2018). Genomic and transcriptomic analysis reveals incremental disruption of key signaling pathways during melanoma evolution. Cancer Cell 34, 45–55.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakhova O, Zingg D, Schaefer SM, Hari L, Civenni G, Blunschi J, Claudinot S, Okoniewski M, Beermann F, Mihic-Probst D, et al. (2012). Sox10 promotes the formation and maintenance of giant congenital naevi and melanoma. Nat. Cell Biol 14, 882–890. [DOI] [PubMed] [Google Scholar]
- Shen S, Faouzi S, Bastide A, Martineau S, Malka-Mahieu H, Fu Y, Sun X, Mateus C, Routier E, Roy S, et al. (2019). An epitranscriptomic mechanism underlies selective mRNA translation remodelling in melanoma persister cells. Nat. Commun 10, 5713, 10.1038/s41467-019-13360-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MP, Sanchez-Laorden B, O’Brien K, Brunton H, Ferguson J, Young H, Dhomen N, Flaherty KT, Frederick DT, Cooper ZA, et al. (2014). The immune microenvironment confers resistance to MAPK pathway inhibitors through macrophage-derived TNFalpha. Cancer Discov. 4, 1214–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldatov R, Kaucka M, Kastriti ME, Petersen J, Chontorotzea T, Englmaier L, Akkuratova N, Yang Y, Haring M, Dyachuk V, et al. (2019). Spatiotemporal structure of cell fate decisions in murine neural crest. Science 364, eaas9536. [DOI] [PubMed] [Google Scholar]
- Somasundaram R, Connelly T, Choi R, Choi H, Samarkina A, Li L, Gregario E, Chen Y, Thakur R, Abdel-Moshen M, et al. (2021). Tumor-infiltrating mast cells are associated with therapy resistance to anti-PD-1. Nat. Commun 12, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stei MM, Loeffler KU, Holz FG, and Herwig MC (2016). Animal models of uveal melanoma: methods, applicability, and limitations. Biomed. Res. Int 2016, 4521807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strub T, Ghiraldini FG, Carcamo S, Li M, Wroblewska A, Singh R, Goldberg MS, Hasson D, Wang Z, Gallagher SJ, et al. (2018). SIRT6 haploinsufficiency induces BRAF V600E melanoma cell resistance to MAPK inhibitors via IGF signaling. Nat. Commun 9, 3440, 10.1038/s41467-018-05966-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Lee W, Mohri Y, Takeo M, Lim CH, Xu X, Myung P, Atit RP, Taketo MM, Moubarak RS, et al. (2019). A novel mouse model demonstrates that oncogenic melanocyte stem cells engender melanoma resembling human disease. Nat. Commun 10, 5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan JL, Fogley RD, Flynn RA, Ablain J, Yang S, Saint-Andre V, Fan ZP, Do BT, Laga AC, Fujinaga K, et al. (2016). Stress from nucleotide depletion activates the transcriptional regulator HEXIM1 to suppress melanoma. Mol. Cell 62, 34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdogan A, Faubert B, Ramesh V, Ubellacker JM, Shen B, Solmonson A, Murphy MM, Gu Z, Gu W, Martin M, et al. (2020). Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115–120, 10.1038/s41586-019-1847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das Thakur M, Salangsang F, Landman AS, Sellers WR, Pryer NK, Levesque MP, Dummer R, McMahon M, and Stuart DD (2013). Modelling vemurafenib resistance in melanoma reveals a strategy to forestall drug resistance. Nature 494, 251–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, et al. (2016). Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 352, 189–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travnickova J, and Patton EE (2020). Deciphering melanoma cell states and plasticity with zebrafish models. J. Invest. Dermatol 10.1016/j.jid.2020.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travnickova J, Wojciechowska S, Khamseh A, Gautier P, Brown DV, Lefevre T, Brombin A, Ewing A, Capper A, Spitzer M, et al. (2019). Zebrafish MITF-low melanoma subtype models reveal transcriptional subclusters and MITF-independent residual disease. Cancer Res. 79, 5769–5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trucco LD, Mundra PA, Hogan K, Garcia-Martinez P, Viros A, Mandal AK, Macagno N, Gaudy-Marqueste C, Allan D, Baenke F, et al. (2019). Ultraviolet radiation-induced DNA damage is prognostic for outcome in melanoma. Nat. Med 25, 221–224. [DOI] [PubMed] [Google Scholar]
- Truong A, Yoo JH, Scherzer MT, Sanchez JMS, Dale KJ, Kinsey CG, Richards JR, Shin D, Ghazi PC, Onken MD, et al. (2020). Chloroquine sensitizes GNAQ/11-mutated melanoma to MEK1/2 inhibition. Clin. Cancer Res 26, 6374–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson D, and Clevers H (2019). Cancer modeling meets human organoid technology. Science 364, 952–955. [DOI] [PubMed] [Google Scholar]
- Umkehrer C, Holstein F, Formenti L, Jude J, Froussios K, Neumann T, Cronin SM, Haas L, Lipp JJ, Burkard TR, et al. (2020). Isolating live cell clones from barcoded populations using CRISPRa-inducible reporters. Nat. Biotechnol 10.1038/s41587-020-0614-0. [DOI] [PubMed] [Google Scholar]
- Varum S, Baggiolini A, Zurkirchen L, Atak ZK, Cantu C, Marzorati E, Bossart R, Wouters J, Hausel J, Tuncer E, et al. (2019). Yin yang 1 orchestrates a metabolic program required for both neural crest development and melanoma formation. Cell Stem Cell 24, 637–653.e39. [DOI] [PubMed] [Google Scholar]
- Vendramin R, Konnova A, Adnane S, Cinque S, Katopodi V, Knezevic Z, Karras P, Demesmaeker E, Bosisio FM, Rizzotto L, et al. (2020). Activation of the integrated stress response in drug-tolerant melanoma cells confers vulnerability to mitoribosome-targeting antibiotics. bioRxiv. 10.1101/2020.06.26.173492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesan AM, Vyas R, Gramann AK, Dresser K, Gujja S, Bhatnagar S, Chhangawala S, Gomes CBF, Xi HS, Lian CG, et al. (2018). Ligand-activated BMP signaling inhibits cell differentiation and death to promote melanoma. J. Clin. Invest 128, 294–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viros A, Sanchez-Laorden B, Pedersen M, Furney SJ, Rae J, Hogan K, Ejiama S, Girotti MR, Cook M, Dhomen N, and Marais R (2014). Ultraviolet radiation accelerates BRAF-driven melanomagenesis by targeting TP53. Nature 511, 478–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votanopoulos KI, Forsythe S, Sivakumar H, Mazzocchi A, Aleman J, Miller L, Levine E, Triozzi P, and Skardal A (2019). Model of patient-specific immune-enhanced organoids for immunotherapy screening: feasibility study. Ann. Surg. Oncol 27, 1956–1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vredeveld LC, Possik PA, Smit MA, Meissl K, Michaloglou C, Horlings HM, Ajouaou A, Kortman PC, Dankort D, McMahon M, et al. (2012). Abrogation of BRAFV600E-induced senescence by PI3K pathway activation contributes to melanomagenesis. Genes Dev. 26, 1055–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vredevoogd DW, Kuilman T, Ligtenberg MA, Boshuizen J, Stecker KE, de Bruijn B, Krijgsman O, Huang X, Kenski JCN, Lacroix R, et al. (2019). Augmenting immunotherapy impact by lowering tumor TNF cytotoxicity threshold. Cell 178, 585–599.e15. [DOI] [PubMed] [Google Scholar]
- Wang J, Perry CJ, Meeth K, Thakral D, Damsky W, Micevic G, Kaech S, Blenman K, and Bosenberg M (2017). UV-induced somatic mutations elicit a functional T cell response in the YUMMER1.7 mouse melanoma model. Pigment Cell Melanoma Res. 30, 428–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Leite de Oliveira R, Huijberts S, Bosdriesz E, Pencheva N, Brunen D, Bosma A, Song JY, Zevenhoven J, Los-de Vries GT, et al. (2018). An acquired vulnerability of drug-resistant melanoma with therapeutic potential. Cell 173, 1413–1425.e14. [DOI] [PubMed] [Google Scholar]
- Weber J, Mandala M, Del Vecchio M, Gogas HJ, Arance AM, Cowey CL, Dalle S, Schenker M, Chiarion-Sileni V, Marquez-Rodas I, et al. (2017). Adjuvant nivolumab versus ipilimumab in resected stage III or IV melanoma. N. Engl. J. Med 377, 1824–1835. [DOI] [PubMed] [Google Scholar]
- van de Wetering M, Francies HE, Francis JM, Bounova G, Iorio F, Pronk A, van Houdt W, van Gorp J, Taylor-Weiner A, Kester L, et al. (2015). Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Weyden L, Brenn T, Patton EE, Wood GA, and Adams DJ (2020). Spontaneously occurring melanoma in animals and their relevance to human melanoma. J. Pathol 252, 4–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, Bourque C, Dovey M, Goessling W, Burns CE, and Zon LI (2008). Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2, 183–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RM, Cech J, Ratanasirintrawoot S, Lin CY, Rahl PB, Burke CJ, Langdon E, Tomlinson ML, Mosher J, Kaufman C, et al. (2011). DHODH modulates transcriptional elongation in the neural crest and melanoma. Nature 471, 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Y, Bartok O, Patkar S, Eli GB, Cohen S, Litchfield K, Levy R, Jimenez-Sanchez A, Trabish S, Lee JS, et al. (2019). UVB-induced tumor heterogeneity Diminishes immune response in melanoma. Cell 179, 219–235.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong K, van der Weyden L, Schott CR, Foote A, Constantino-Casas F, Smith S, Dobson JM, Murchison EP, Wu H, Yeh I, et al. (2019). Cross-species genomic landscape comparison of human mucosal melanoma with canine oral and equine melanoma. Nat. Commun 10, 353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo XY, Giordano J, Srivastava A, Zhao Z-M, Lloyd MW, de Bruijn R, Suh Y-S, Patidar R, Chen L, Scherer S, et al. (2019). Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts. Nat. Genet 10.1038/s41588-020-00750-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Rebecca VW, and Herlyn M (2019). A melanoma patient-derived xenograft model. J. Vis. Exp 10.3791/59508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Brunson DC, Tang Q, Do D, Iftimia NA, Moore JC, Hayes MN, Welker AM, Garcia EG, Dubash TD, et al. (2019). Visualizing engrafted human cancer and therapy responses in immunodeficient zebrafish. Cell 177, 1903–1914.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh I, Jorgenson E, Shen L, Xu M, North JP, Shain AH, Reuss D, Wu H, Robinson WA, Olshen A, et al. (2019). Targeted genomic profiling of acral melanoma. J. Natl. Cancer Inst 111, 1068–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen J, White RM, Wedge DC, Van Loo P, de Ridder J, Capper A, Richardson J, Jones D, Raine K, Watson IR, et al. (2013). The genetic heterogeneity and mutational burden of engineered melanomas in zebrafish models. Genome Biol. 14, R113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You MJ, Castrillon DH, Bastian BC, O’Hagan RC, Bosenberg MW, Parsons R, Chin L, and DePinho RA (2002). Genetic analysis of Pten and Ink4a/Arf interactions in the suppression of tumorigenesis in mice. Proc. Natl. Acad. Sci. U S A 99, 1455–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelenay S, van der Veen AG, Bottcher JP, Snelgrove KJ, Rogers N, Acton SE, Chakravarty P, Girotti MR, Marais R, Quezada SA, et al. (2015). Cyclooxygenase-dependent tumor growth through evasion of immunity. Cell 162, 1257–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Di Martino JS, Bowman RL, Campbell NR, Baksh SC, Simon-Vermot T, Kim IS, Haldeman P, Mondal C, Yong-Gonzales V, et al. (2018). Adipocyte-derived lipids mediate melanoma progression via FATP proteins. Cancer Discov. 8, 1006–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Shi C, Tao W, Li J, Wu J, Han Y, Yang G, Gu Z, Xu S, Wang Y, et al. (2019). Analysis of mucosal melanoma whole-genome landscapes reveals clinically relevant genomic aberrations. Clin Cancer Res. 25, 3548–3560, 10.1158/1078-0432.CCR-18-3442. [DOI] [PubMed] [Google Scholar]
- Zilionis R, Engblom C, Pfirschke C, Savova V, Zemmour D, Saatcioglu HD, Krishnan I, Maroni G, Meyerovitz CV, Kerwin CM, et al. (2019). Single-cell transcriptomics of human and mouse lung cancers reveals conserved myeloid populations across individuals and species. Immunity 50, 1317–1334.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurkirchen L, and Sommer L (2017). Quo vadis: tracing the fate of neural crest cells. Curr. Opin. Neurobiol 47, 16–23. [DOI] [PubMed] [Google Scholar]


