Abstract
When a cell runs out of nutrients, its growth rate substantially decreases. Here, we report that microorganisms speed up the return to a state of rapid growth by preventing the degradation of functional proteins while in the slow-growth state. The bacterium Salmonella enterica serovar Typhimurium reduced the proteolysis of functional proteins by adenosine triphosphate (ATP)-dependent proteases during stationary phase. This reduction resulted from a decrease in the intracellular concentration of ATP that, critically, was sufficient to allow the continued degradation of non-functional proteins by those very same proteases. Protein preservation was observed under limiting magnesium, carbon, or nitrogen conditions, indicating that it was not specific to limitation for a particular nutrient. Emergence from a slow-growth state was delayed by treatments that increased the amounts of intracellular ATP. Moreover, it required the transcriptional regulator PhoP and the alternative sigma factor RpoS when the slow growth state was caused by magnesium limitation. Reductions in intracellular ATP and ATP-dependent proteolysis also enabled the yeast Saccharomyces cerevisiae to recover faster from stationary phase. Our findings suggest that protein preservation during a slow-growth state is a conserved microbial strategy that facilitates the return to a growth state once nutrients become available.
Introduction
Proteins are the macromolecules that conduct the vast majority of cellular work. Protein homeostasis reflects the balance between protein synthesis and degradation as well as the activities of chaperones that aid protein folding. When an organism experiences slow-growth conditions, adenosine triphosphate (ATP) amounts decrease, which reduces the rate of protein synthesis (1, 2), chaperone activity (3) and growth (2, 4–6). Such a decrease in the rate of protein synthesis has been observed in a wide range of organisms from bacteria to humans (2, 7). We now report that protein preservation during slow growth is necessary for a swift return to a growth state.
Organisms from all domains of life harbor proteases of the ATPases associated with cellular activities (AAA+) family (8–10). These proteases are critical for the degradation of both non-functional (and potentially toxic) proteins and functional proteins that control a variety of biological processes (11–13). AAA+ proteases have two functional domains or subunits: an ATPase domain that unfolds and translocates substrates into a peptidase domain where proteolysis actually takes place (14). ATP is required not only for protein unfolding (14, 15), but also for the assembly of protease domains (16) and for substrate recognition by ubiquitylation in eukaryotes (17, 18).
Degradation of some substrates requires adaptor proteins that recognize the substrate and deliver it to the protease but are not themselves degraded by the protease (19, 20). For example, RssB is an adaptor that recognizes the alternative sigma factor RpoS and delivers it to the protease ClpXP in Escherichia coli (21). Degradation of substrates recognized by a given adaptor can be hindered by anti-adaptor proteins that bind to adaptors (19). For example, the anti-adaptors IraM and IraP bind to RssB, stabilizing RpoS when E. coli is starved for Mg2+ (22) and phosphate (23), respectively.
RpoS is also stabilized when E. coli experiences carbon starvation even though an anti-adaptor produced under this condition has not been described (24). RpoS stabilization under carbon starvation has been ascribed to a decrease in the ATP concentration, which is proposed to reduce degradation of RpoS, but not of SsrA-tagged green fluorescent protein (GFP), which is also a ClpXP substrate (15). When an mRNA is truncated within the coding region of a gene, the ribosome translating such a gene stalls at the end of the mRNA until rescued by ssrA, which specifies an RNA with both tRNA and mRNA properties (25, 26). ssrA encodes a short protein sequence followed by a stop codon that is translated and tags the protein specified by the truncated mRNA. This rescues the stalled ribosome and results in the recognition and degradation of the tagged protein by the proteases ClpXP and ClpAP. In this way, the ssrA gene helps maintain the quality of the proteome (27).
Here, we report that a decrease in ATP amounts taking place under nutrient-limiting conditions stabilized not only RpoS but also multiple substrates of several AAA+ proteases. The remaining ATP allowed proteolysis of non-functional proteins targeted for degradation. Stabilization of functional proteins during stationary phase helped microorganisms return to a growth state once nutrients became available. This behavior was displayed by both bacteria and yeast, suggesting that protein preservation is a conserved mechanism that governs escape from stationary phase into growth phase. Protein longevity likely plays a role in the germination of bacterial spores and in antibiotic persistence.
Results
A reduction in ATP abundance decreases proteolysis by ATP-dependent proteases
When the bacterium Salmonella enterica serovar Typhimurium (S. Typhimurium) experiences insufficient amounts of essential nutrients, it decreases the production of macromolecules and metabolites (2, 28, 29). This reduces ATP amounts, which lowers the rate of protein synthesis; thus, bacteria that are growing exponentially enter stationary phase (2). Because the activity of AAA+ proteases is dependent on the ATP concentration (14, 15, 30, 31), we wondered whether proteolysis by AAA+ proteases other than ClpXP also decreased during nutrient limitation. Thus, we examined both ATP amounts and protein stability in S. Typhimurium subjected to different starvation conditions.
Initially, we analysed the effects of starvation for Mg2+, the most abundant divalent cation in all living cells (32). We examined bacteria prior to and after a drop in the cytoplasmic Mg2+ concentration that triggers the production of proteins that reduce ATP amounts (2). When placed in media containing low (10 μM) Mg2+, S. Typhimurium grew exponentially for ~4 h (Fig. 1A). At this time, the Mg2+ concentration became limiting (2), the ATP concentration decreased (Fig. 1B and fig. S1A) and growth slowed down substantially (Fig. 1A). We collected samples at 2.5 and 5.5 h, times at which the ATP concentration was high and low, respectively (Fig. 1B), and examined substrates of the AAA+ proteases ClpXP, Lon, and FtsH.
Fig. 1. Nutritional starvation reduces ATP amounts, stabilizing substrates of ATP-dependent proteases.
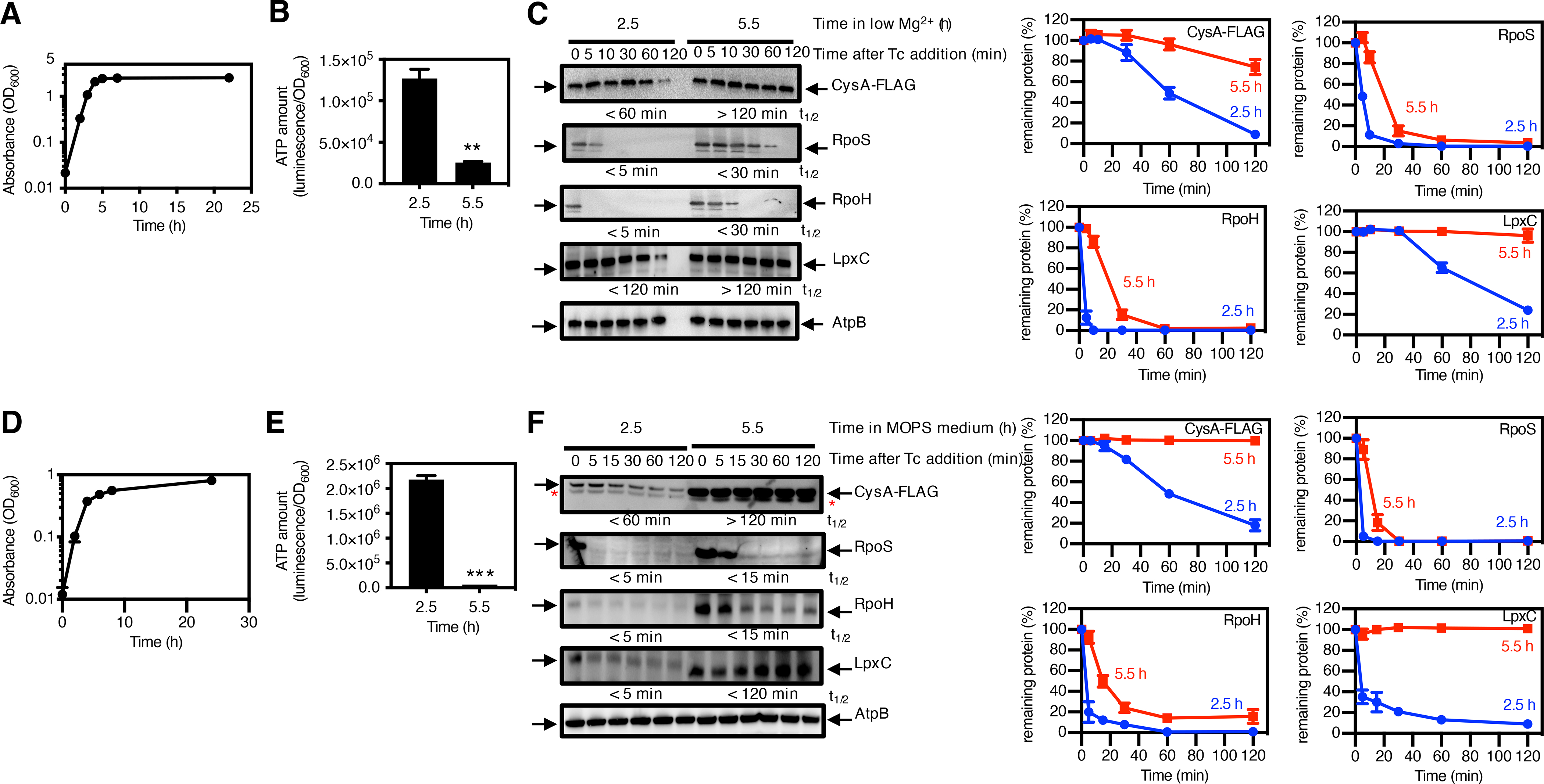
(A) Growth curve for wild-type S. Typhimurium expressing cysA-FLAG (strain JY767) under low-Mg2+ conditions. (B) Quantification of ATP amount and (C) immunoblotting and degradation curves for the indicated protease substrates in strain JY767 grown under low-Mg2+ conditions for 2.5 or 5.5 h and treated with tetracycline to inhibit protein synthesis. Numbers below blots correspond to the half-life (t1/2) of the indicated substrates. AtpB is a loading control. (D) Growth curve of strain JY767 in Mg2+-replete MOPS minimal medium and in the presence of tetracycline to inhibit protein synthesis. (E) Quantification of ATP amount and (F) immunoblotting, calculated half-lives (t1/2), and degradation curves of the indicated protease substrates in strain JY767 grown in Mg2+-replete MOPS medium for 2.5 or 5.5 h. Growth curves (A and D) show the average of 4 independent experiments, and error bars represent SDs. ATP amounts (B and E) were calculated with normalization luminescence by OD600 and represent the mean and SD from 4 independent experiments. Western blotting (C and F) was performed with antibodies directed to the FLAG peptide or to the AtpB, RpoS, RpoH, or LpxC proteins, and degradation curves show the mean and SD for protein amounts normalized to AtpB from 3 independent experiments. Half-lives of the protease substrates were calculated by regression analysis of the exponential decay of the proteins. Unpaired Student’s t tests were performed between 2.5 h with 5.5 h; ***P < 0.001, **P < 0.01.
The stability of the ClpXP substrates CysA (33) and RpoS (34, 35), the Lon and FtsH substrate RpoH (36, 37), and the FtsH substrate LpxC (38) was higher at 5.5 than at 2.5 h (Fig. 1C). Tetracycline, which was used to stop protein synthesis and thus, determine substrate stability, increased ATP concentrations slightly at the two time points (fig. S2A), but did not impact the fold ATP difference between the two time points. Therefore, the decrease in ATP concentration taking place during stationary phase correlated with reduced proteolysis of functional proteins by different AAA+ proteases.
Protein stability also increased upon starvation for nutrients other than Mg2+. That is, when bacteria reached stationary phase during growth in MOPS minimal medium with abundant Mg2+ (Fig. 1D), a condition that also lowers ATP amounts (Fig. 1E and fig. S1B), the four substrates (CysA, RpoS, RpoH, and LpxC) were similarly stabilized (Fig. 1F). Likewise, starvation for carbon or nitrogen reduced bacterial growth (fig. S3A), ATP amounts (fig. S3B), and the rate of proteolysis of CysA (fig. S3C). We conclude that stabilization of substrates of AAA+ proteases is a response to nutrient starvation regardless of the specific nutrient that becomes limiting.
A reduction in ATP amounts is necessary to stabilize substrates of ATP-dependent proteases
The PhoP protein is a transcriptional activator of the mgtA, mgtB, and mgtC genes (39), which encode the Mg2+ transporters MgtA (40) and MgtB (41) and the ATPase inhibitor MgtC (42), respectively. These proteins reduce ATP amounts when S. Typhimurium experiences Mg2+ starvation (2), a condition that activates the PhoP protein (43, 44) and stimulates expression of the mgtA, mgtB, and mgtC genes (39, 45). Therefore, the ATP concentration is higher in the phoP mutant than in wild-type S. Typhimurium following growth in low Mg2+ (2). If a reduction in ATP amounts was required to stabilize the substrates of AAA+ proteases, then a phoP mutant should have lower substrate amounts than the wild-type strain.
We determined that the steady-state amounts of the CysA, RpoS, RpoH, and LpxC proteins were lower in the phoP mutant than in wild-type S. Typhimurium following 5.5 h growth in low Mg2+ media (Fig. 2A). The ATP abundance at this time point was 40 times higher in the phoP mutant than in the wild-type strain (Fig. 2B). Therefore, there was an inverse correlation between substrate amounts and ATP abundance. By contrast, wild-type and phoP strains had similar amounts of both substrates (Fig. 2A) and ATP (Fig. 2B) following 2.5 h growth in the same media (Fig. 2A). These results reflect that expression of the ATP-reducing genes takes place at 5.5 h but not at 2.5 h in low Mg2+ media (2). These findings suggest that a PhoP-dependent reduction in ATP amounts stabilizes substrates of AAA+ proteases during Mg2+ starvation. As detailed below, this notion was supported by multiple lines of evidence.
Fig. 2. PhoP-mediated ATP reduction stabilizes protease substrates under Mg2+ starvation.
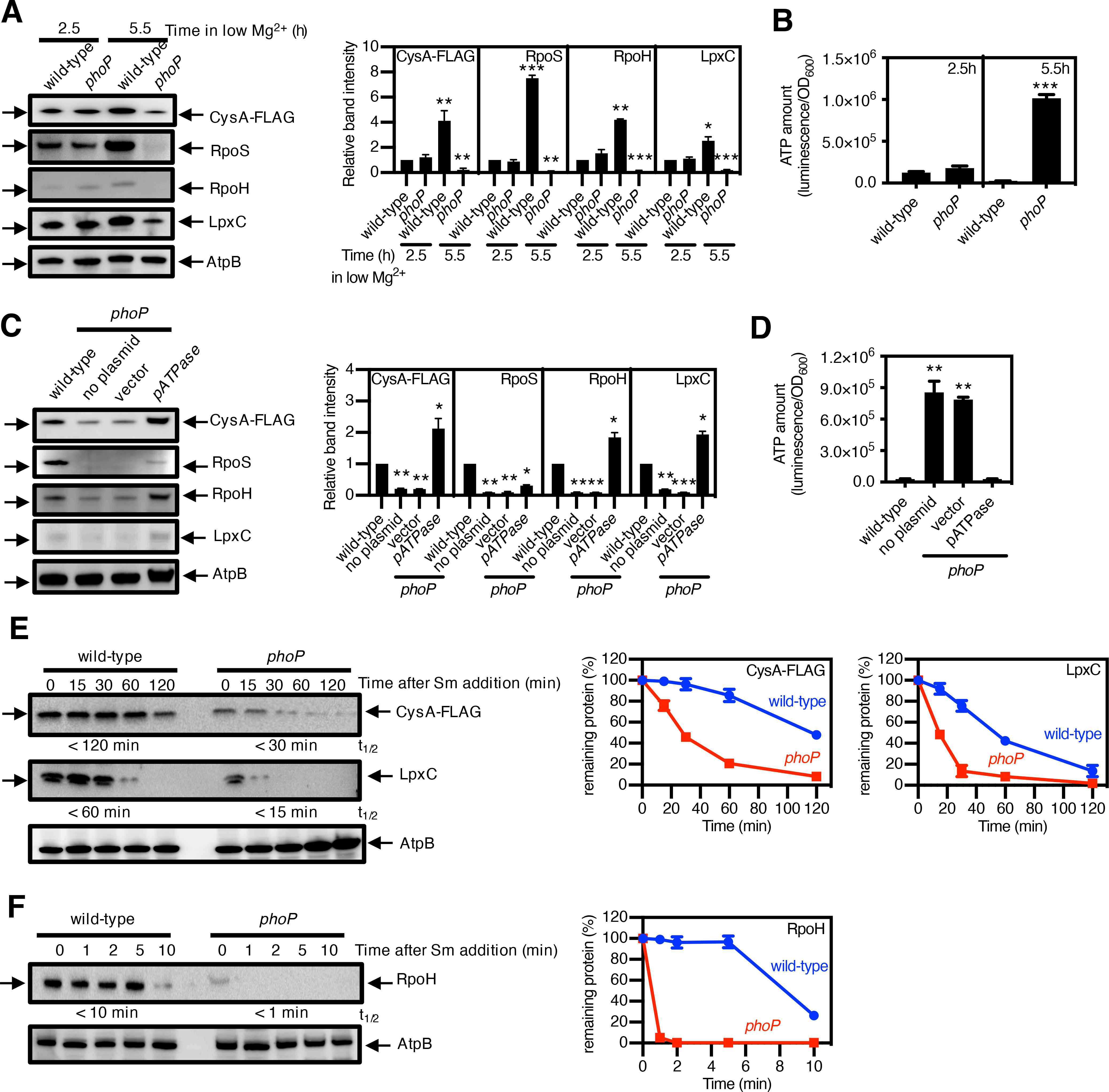
(A) Immunoblotting and quantification of the indicated protease substrates and (B) quantification of ATP amount in wild-type S. Typhimurium expressing cysA-FLAG (JY767, wild-type) and phoP S. Typhimurium expressing cysA-FLAG (JY901, phoP) grown under low-Mg2+ conditions for 2.5 or 5.5 h. AtpB is a loading control. (C) Immunoblotting and quantification of the indicated protease substrates and (D) quantification of ATP amount in strain JY767 (wild-type) and in strain JY901 with no plasmid, the plasmid vector alone (45), or expressing the soluble subunit of the F1Fo ATPase (pATPase) (54), grown under low-Mg2+ conditions for 5.5 h. (E and F) Immunoblotting and degradation curves of the indicated protease substrates in strains JY767 (wild-type) and JY901 (phoP) grown under low-Mg2+ conditions for 5.5 h with the protein synthesis inhibitor spectinomycin. Numbers below blots correspond to the half-lives (t1/2) of the indicated substrates, which were calculated by regression analysis of the exponential decay of the proteins. Western blotting (A, C, E, F) was performed with antibodies directed to the FLAG peptide or to the AtpB, RpoS, RpoH, or LpxC proteins, and degradation curves show the mean and SD for protein amounts normalized to AtpB from 3 independent experiments. ATP amounts (B and D) were calculated with normalization to luminescence by OD600 and represent the mean and SD from 3 independent experiments. Unpaired Student’s t tests were performed between wild-type and the mutant strains; ***P < 0.001, **P < 0.01.
First, a plasmid that encodes the soluble subunit of the F1Fo ATPase (pATPase) and thereby promotes ATP hydrolysis (2), corrected both ATP amounts and the abundance of CysA, LpxC, and RpoH (Fig. 2, C and D) in the phoP mutant. By contrast, the vector control had no effect (Fig. 2, C and D). Second, arsenate, a phosphate analogue that poisons ATP synthesis (46), reduced ATP amounts (fig. S4, A and B) and increased the abundance of RpoH, CysA, and LpxC (fig. S4, C and D) in both wild-type and phoP strains. And third, the three substrates displayed a shorter half-life in the phoP mutant than in the wild-type strain (Fig. 2, E and F), indicating that the absence of PhoP promoted substrate stability. Spectinomycin, which was used to stop protein synthesis and, thus, determine substrate stability, increased the ATP concentration 1.5-fold in the phoP mutant. By contrast, there was an 8-fold difference in the ATP concentration between wild-type and phoP Salmonella (fig. S2B).
Control experiments demonstrated that wild-type and phoP Salmonella exhibited similar abundances of the mRNAs encoding the substrates (fig. S5A), ruling out the possibility of differences in substrate abundance resulting from transcriptional effects of the regulatory protein PhoP on the corresponding genes. As expected, the mRNA abundance of the control PhoP-activated gene pagC was higher in the wild-type strain than in the phoP mutant (fig. S5A). In addition, the amounts of the protease FtsH and of the protease subunits ClpX and ClpP were similar in wild-type and phoP Salmonella at both 2.5 and 5.5 h in low Mg2+ media (fig. S5B), arguing against the observed differences in substrate abundance resulting from disparities in protease amounts. Furthermore, abundance of the Lon substrate RpoH was lower in the phoP mutant than in the wild-type strain (Fig. 2A) even though Lon amounts were ~50% lower in the phoP mutant than in the wild-type strain (fig. S5B).
A reduction in ATP amounts increases but does not fully restore wild-type RpoS abundance in the phoP mutant
Neither pATPase (Fig. 2D) nor arsenate (fig. S4C) fully restored RpoS abundance to the phoP mutant under Mg2+-starvation conditions. We ascribe these results to PhoP protecting RpoS both by decreasing ATP abundance and by promoting expression of the anti-adaptor protein IraP (35, 47). In agreement with this notion and previous results (35, 47), iraP mutant S. Typhimurium had low RpoS amounts (fig. S6B), albeit not as low as the phoP mutant (fig. S6B), but retained wild-type amounts of ATP (fig. S6A). As expected, the iraP phoP double mutant behaved like the phoP single mutant (fig. S6, A and B).
That PhoP stabilizes RpoS through two mechanisms is reminiscent of the MgtC protein protecting the PhoP protein from degradation by the AAA+ protease ClpAP through two mechanisms. MgtC reduces ATP amounts (42) and hinders ClpS access to PhoP (48). This is why plasmid pATPase does not fully restore PhoP abundance in a mgtC mutant (48) or RpoS abundance in a phoP mutant (Fig. 2D) despite normalizing ATP amounts (48) (Fig. 2C).
Degradation of non-functional proteins continues unabated during nutritional starvation
Because they arise from truncated mRNAs, SsrA-tagged proteins are generally not functional and thus targeted for degradation by the ATP-dependent proteases ClpXP and ClpAP (27). We investigated the amounts of GFP-LAA, a GFP variant with the complete ssrA-encoded degradation motif at its C terminus, as a model substrate for a non-functional protein. However, GFP-LAA amounts were similar at 2.5 and 5.5 h (Fig. 3A). By contrast, the abundance of the ClpXP substrate RpoS was higher at 5.5 than at 2.5 h (Fig. 3A). In agreement with the results presented above (Fig. 1B and 2B), ATP amounts were much higher in wild-type cells expressing GFP-LAA at 2.5 than at 5.5 h after a switch to low Mg2+ media (Fig. 3B).
Fig. 3. Degradation of SsrA-tagged proteins continues unimpeded despite the reduction in ATP amounts during Mg2+ starvation.
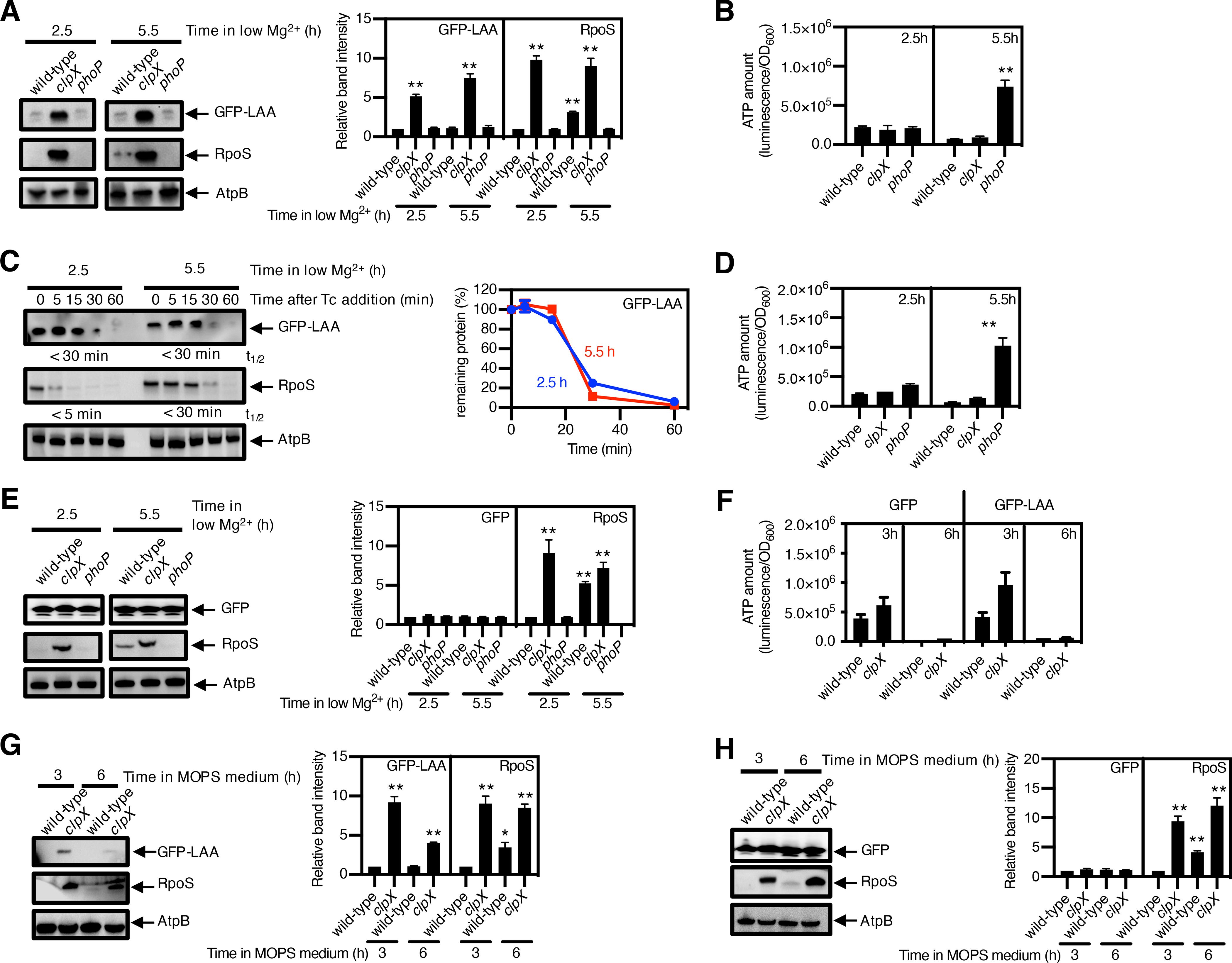
(A) immunoblotting and quantification of the indicated protease substrates; (B) quantification of ATP amount; and (C) stability of the protease substrates in cysA-FLAG (JY767, wild-type), cysA-FLAG clpX (JY1043, clpX) and cysA-FLAG phoP (JY901, phoP) S. Typhimurium expressing GFP-LAA. grown under low-Mg2+ conditions for 2.5 or 5.5 h. AtpB is a loading control. (D) Quantification of ATP and (E) immunoblotting and quantification of the indicated protease substrates in strains JY767 (wild-type), JY1043 (clpX), and JY901 (phoP) expressing GFP (100) and were grown under low-Mg2+ conditions for 2.5 h or 5.5 h. (F) Quantification of ATP and (G and H) the indicated protease substrates in strains JY767 (wild-type) and JY1043 (clpX) expressing GFP-LAA (G) or GFP (H) grown in Mg2+-replete MOPS medium for 3 or 6 h.
ATP amounts (A, D, F) were calculated with normalization to luminescence by OD600 and represent the mean and SD from 3 independent experiments. Unpaired Student’s t tests were performed between wild-type and the mutant strains; ***P < 0.001, **P < 0.01.
Densitometry graphs (B, E, G, H) show the average and SD of protein amounts from 3 independent experiments. Graphs correspond to the amounts in different strains relative to that of the wild-type, which was normalized to 1.0. Unpaired Student’s t tests were performed between wild-type samples at 2.5 h with the other combinations; **P < 0.01. Degradation curve and half-lives (t1/2) of protease substrates (C) were determined by band intensity from Western blots with protein amounts normalized to AtpB. The mean and SD from three 3 independent experiments are shown.
A similar situation is observed with pre-OmpF (49), a Lon substrate in secM mutant bacteria (50). That is, translocation-competent Sec proteins are required for maturation and export of outer membrane proteins (51); therefore, regulation of the SecA protein is compromised in a secM mutant (52), resulting in the aggregation of SecB substrates, such as pre-OmpF (49). Despite the higher ATP concentration present at 2.5 than at 5.5 h (fig. S7A), pre-OmpF amounts were similar at the two times (fig. S7B). By contrast, RpoH abundance was higher at 5.5 than at 2.5 h (fig. S7B). Control experiments demonstrated that the secM mutant exhibited wild-type ATP amounts, unlike the phoP mutant (fig. S7A). These results demonstrate that a reduction in ATP concentration reduces the degradation of functional proteins but not non-functional proteins, even though both classes of proteins are substrates of the same ATP-dependent proteases.
That the ATP amounts remaining in bacteria experiencing Mg2+ starvation are sufficient to proteolyze proteins targeted for degradation was supported by the behavior of wild-type and phoP Salmonella experiencing Mg2+ starvation. Both strains displayed similar amounts of GFP-LAA (Fig. 3B) and pre-OmpF (fig. S7B) even though wild-type Salmonella had much lower ATP amounts than the phoP mutant (Fig. 3A and fig. S7A). By contrast, RpoS and RpoH abundances were higher in the wild-type strain than in the phoP mutant, and both strains had similar abundances of the loading control protein AtpB (Fig. 3B and fig. S7B).
GFP-LAA stability was similar at 2.5 and 5.5 h (Fig. 3C), and the same was true for pre-OmpF (fig. S7C), again in contrast to lower stability of RpoS and RpoH at 2.5 than at 5.5 h (Fig. 3C and fig. S7C). The behaviors of GFP-LAA and RpoS reflected ClpXP-dependent proteolysis because the clpX mutant had much higher amounts of both proteins than the wild-type strain at both time points (Fig. 3B). Likewise, the behaviors of pre-OmpF and RpoH reflected Lon-dependent proteolysis because the lon secM double mutant had higher amounts of both proteins compared to the secM single mutant at both time points (fig. S7B). Although the ATP amounts in wild-type S. Typhimurium were higher at 2.5 than at 5.5 h after a switch to low Mg2+ media (Fig. 3D), a similar abundance was exhibited by untagged GFP (Fig. 3E), AtpB (Fig. 3E), and mature OmpF (fig. S7B) at the two time points. Taken together, these results indicate that bacteria continue to safeguard proteome quality even when experiencing low ATP amounts resulting from Mg2+ starvation.
The ATP remaining in bacteria in stationary phase was sufficient to degrade proteins targeted for proteolysis even when the limiting nutrient was not Mg2+. That is, ATP amounts were higher at 3 h than at 6 h of growth in MOPS media (Fig. 3F and fig. S7D), but the abundances of GFP-LAA (Fig. 3G) and pre-OmpF (fig. S7E) were the same at the two times. By contrast, RpoS and RpoH accumulated at 6 h but not at 3 h (Fig. 3G and fig. S7E). Controls showed the same abundance of untagged GFP and AtpB (Fig. 3H) and mature OmpF protein (fig. S7E) at the two times.
Preserving proteins during slow growth state favors return to growth state when nutrients become available
If proteins present during a slow- or no-growth state help organisms leave that state when nutrients become available (53), stimulating proteolysis during the slow- or no-growth state should hinder the return to the growth state. To test this hypothesis, we exposed bacteria to agents that arrest growth (fig. S8 A to E) and either increase or do not alter ATP abundance (fig. S9 A to F) (54), and then determined bacterial growth following a switch to fresh media (Fig. 4A) (55).
Fig. 4. Slow growth–induced protein longevity speeds bacterial entry into the growth state.
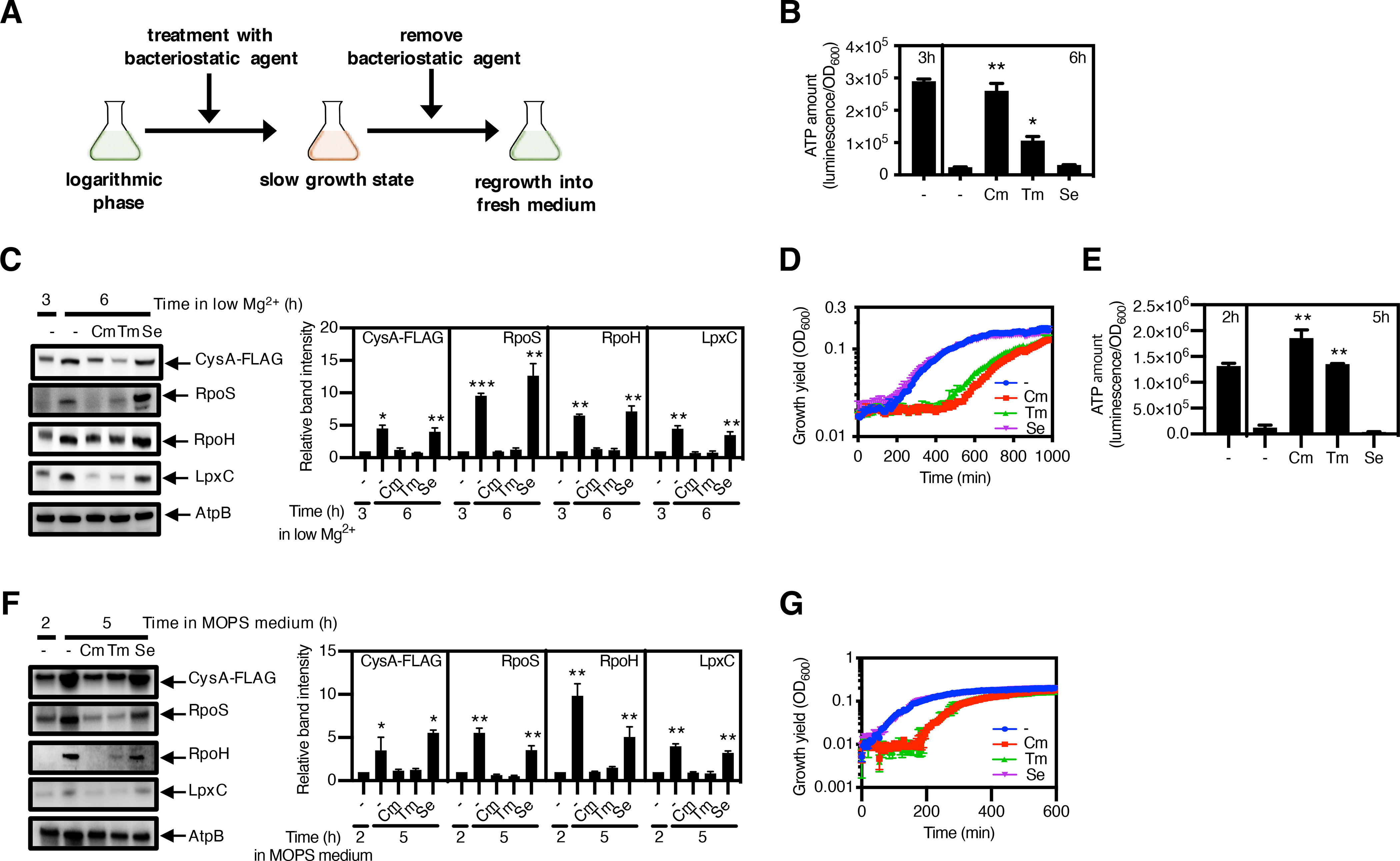
(A) Schematic of the strategy used to test the role that protein longevity plays in the entry into the growth state following starvation. Bacteria were grown for 2 h or 3 h, followed by 3 h or 21 h treatment with a bacteriostatic antibiotic to slow growth. Treated samples were washed with fresh medium and then inoculated into fresh medium to investigate escape from the slow growth state into the growth state. (B) Quantification of ATP amount and (C) immunoblotting and quantification of the indicated protease substrates in strain JY767 (wild-type) grown under low-Mg2+ conditions for 3 h, followed by 3 h treatment with chloramphenicol (Cm), trimethoprim (Tm) or sulfamethoxazole (Se). AtpB is a loading control. (D) Growth curves of strain JY767 under Mg2+-replete conditions after washing antibiotics from samples in (B) and (C). (E) Quantification of ATP amount and (F) immunoblotting and quantification of the indicated protease substrates in strain JY767 grown in Mg2+-replete MOPS medium for 2 h, followed by 3 h without glucose treatment in the presence of Cm, Tm, or Se. (G) Growth curves of strain JY767 in Mg2+-replete MOPS medium containing glucose after washing antibiotics from 5 h samples in (E) and (F). ATP amounts (B and E) were calculated with normalization by OD600, and are the average of 3 independent experiments. Samples were analysed by Western blotting (C and F) with antibodies directed to the FLAG peptide, or the RpoS, RpoH, LpxC or AtpB proteins. Data are representative of 3 independent experiments. Densitometry graphs show the average and SD of protein amounts relative to wild-type from 3 independent experiments. Growth curves (D and G) show the average of 4 independent experiments, and error bars represent SDs. Unpaired Student’s t tests were performed between untreated samples at 6 or 5 h with the other combinations; *P < 0.05 and **P < 0.01.
When the protein synthesis inhibitor chloramphenicol was added to wild-type S. Typhimurium experiencing Mg2+ starvation, the ATP concentration increased dramatically (Fig. 4B and fig. S9A), reflecting that protein synthesis was the activity that demanded most cellular energy (56, 57). The ATP increase was accompanied by a decrease in the abundance of protease substrates (Fig. 4C) and a growth delay upon a switch to Mg2+-rich media relative to organisms not exposed to chloramphenicol (Fig. 4D).
Plasmid pATPase (but not the vector control) reduced ATP amounts in chloramphenicol-treated bacteria experiencing Mg2+ starvation (fig. S10A), which restored both wild-type abundance of protease substrates (fig. S10B) and fast entry into the growth state upon addition of Mg2+ (fig. S10C). These results suggest that the decreased proteolysis resulting from a reduction in ATP concentration favored a speedy transition from the slow- or no-growth state to the growth state.
Protein synthesis inhibitors can exhibit a post-antibiotic effect whereby bacterial regrowth is initially inhibited upon a return to media lacking the drug. Therefore, we investigated the effects of trimethoprim, a bacteriostatic agent that inhibits the production of a DNA synthesis precursor (58), and sulfamethoxazole, a bacteriostatic agent that targets the same pathway as trimethoprim (59), because neither agent displays a post-antibiotic effect in Gram-negative bacteria (60). Like chloramphenicol treatment (fig. S9, A and B), trimethoprim addition increased ATP amounts (fig. S9C) and inhibited bacterial growth (fig. S9D). The increased ATP amounts (Fig. 4B) were accompanied by decreased substrate abundance (Fig. 4C), and prolonged the lag phase when Mg2+-starving bacteria were placed in Mg2+-rich media (Fig. 4D). The effects of chloramphenicol and trimethoprim are not due to bacterial stasis per se because ATP amounts (Fig. 4B), substrate abundance (Fig. 4C), and emergence from the slow/no-growth state (Fig. 4D) remained unaltered upon treatment with sulfamethoxazole, which inhibited bacterial growth without altering ATP amounts (fig. S9, E and F).
The association between protein preservation during a slow- or no-growth state and a speedy return to the growth state was not limited to insufficiency of a specific nutrient. Chloramphenicol and trimethoprim increased ATP amounts (Fig. 4E), decreased protease substrate abundance (Fig. 4F), and prolonged the lag phase upon the restoration of fresh media (Fig. 4G) in bacteria experiencing carbon limitation. By contrast, untreated and sulfamethoxazole-treated bacteria had similar ATP amounts (Fig. 4E), substrate abundance (Fig. 4F), and a speedy return to the growth state when carbon became plentiful (Fig. 4G). Moreover, chloramphenicol-treated and trimethoprim-treated bacteria media had high ATP amounts (fig. S11A), low substrate abundance (fig. S11B), and delayed entry into the growth state (fig. S11C) in MOPS media. Sulfamethoxazole-treated bacteria had similar ATP amounts (fig. S11A), substrate abundance (fig. S11B), and fast entry into the growth state as untreated bacteria (fig. S11C). We conclude that the reduction in ATP amounts that bacteria exhibit when nutrient limitation slows down or stops growth extends protein longevity and favors escape from a slow- or no-growth state regardless of the specific nutrient that was limited.
Preserving specific protease substrates during slow growth state helps return to growth state
Because the PhoP protein is the master regulator of the Mg2+ starvation response (32), PhoP abundance may determine the length of the lag phase displayed by Mg2+-starved S. Typhimurium once Mg2+ becomes available. To test this notion, we examined the behavior of four isogenic strains. One of these strains expresses the PhoP (L4P) variant instead of wild-type PhoP. The PhoP (L4P) variant exhibits decreased ClpS binding and reduced proteolysis by ClpAP (61). Another strain lacks PhoP because it is a phoP null mutant; a third strain fails to degrade PhoP because it lacks clpS; and the fourth strain is the wild-type parent.
The PhoP (L4P)-expressing strain returned to the growth state faster than the wild-type PhoP-expressing strain upon chloramphenicol treatment (Fig. 5A). Although the two strains had similar ATP amounts before chloramphenicol treatment (Fig. 5B) and reached similar ATP amounts following chloramphenicol treatment (Fig. 5B), wild-type PhoP amounts decreased upon chloramphenicol treatment but PhoP (L4P) amounts did not (Fig. 5C). The clpS mutant behaved liked the PhoP (L4P)-expressing strain, retaining high PhoP amounts and returning to the growth phase faster than the wild-type strain (Fig. 5A). By contrast, the phoP mutant displayed the longest lag phase of the four strains (Fig. 5A). The abundance of the chaperone GroEL, which was used as a loading control, was the same before and during starvation conditions (Fig. 5C). In sum, these data suggest that preserving the PhoP protein during Mg2+ starvation speeds the return to the growth state upon restoration of Mg2+.
Fig. 5. Preserving specific protease substrates during low Mg2–triggered slow growth state speeds the return to the growth state.
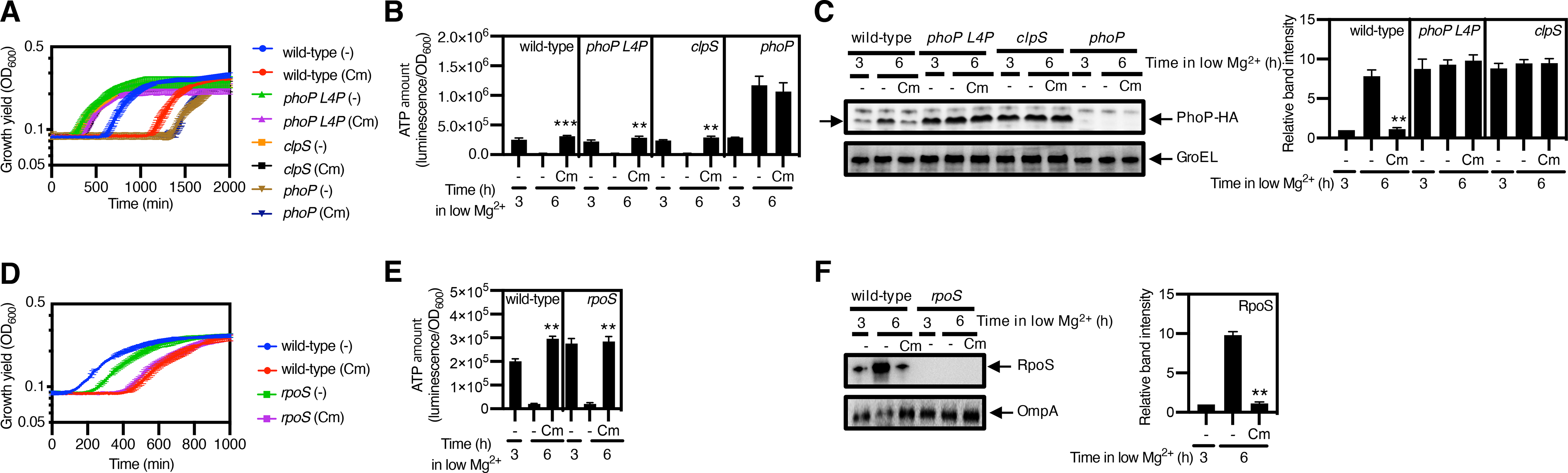
(A) Growth curves of wild-type (EG13918) clpS (JY881), and phoP (MS7953s) S. Typhimurium expressing PhoP-HA and wild-type S. Typhimurium expressing PhoP(L4P)-HA (JY992) under low-Mg2+ conditions after 3 h growth followed by 3 h treatment with chloramphenicol (Cm) to induce growth arrest or with no Cm treatment. (B) Quantification of ATP amount and (C) immunoblotting and quantification of protease substrate PhoP-HA in the indicated strains grown low-Mg2+ conditions for 3 h, followed by 3 h of treatment with or without Cm. GroEL is a loading control. (D) Growth curves of wild-type (14028s) and rpoS (EG18022) S. Typhimurium under Mg2+-replete conditions after washing antibiotics from 6 h samples in (A) and (B). (E) Quantification of ATP and (F) immunoblotting and quantification of the protease substrate RpoS in strains 14028s and EG18022 grown under low-Mg2+ conditions for 3 h, followed by 3 h treatment with or without Cm. OmpA is a loading control. Growth curves (A and D) show the average of 4 independent experiments, and error bars represent SDs. ATP amounts (B and E) were calculated with normalization to luminescence by OD600, and are the average of independent experiments. Western blotting (C and F) was performed with antibodies directed to the HA peptide, GroEL, or OmpA, and densitometry graphs show the average and SD for protein amounts in different strains relative to those in the untreated bacteria at 3 h from 3 independent experiments. Unpaired Student’s t tests were performed between untreated samples at 6 h with the other combinations; **P < 0.01 and ***P < 0.001.
PhoP stabilization helps S. Typhimurium emerge from Mg2+ starvation but not from carbon starvation, because the PhoP (L4P)-expressing strain and the phoP null mutant exhibited a wild-type lag phase following carbon restoration to carbon-starved bacteria (fig. S12A). The four strains had similar ATP amounts (fig. S12B), but differed in PhoP abundance (fig. S12C): PhoP (L4P) was the highest, reflecting resistance to degradation by ClpSAP (61), closely followed by the clpS mutant and then, by the wild-type strain (fig. S12C). The clpS mutant exhibited a longer lag phase than the other three strains (fig. S12A) even though its PhoP amounts were in between those of the PhoP (L4P)-expressing strain and the wild-type strain (fig. C12C). Thus, the longer lag displayed by the clpS mutant must have been due to a ClpS-regulated protein other than PhoP. The abundance of GroEL was the same before and after carbon starvation conditions (fig. S12C).
In addition to PhoP, S. Typhimurium requires RpoS to resume growth following Mg2+ starvation, because a rpoS null mutant took longer than the wild-type strain to grow when Mg2+ was restored (Fig. 5D). Chloramphenicol treatment increased ATP abundance (Fig. 5E) and decreased RpoS amounts >9 fold (Fig. 5F), in agreement with RpoS being a ClpXP substrate (21). Cumulatively, the data in this section argue that preserving specific protease substrates eases the return to the growth state once the limiting nutrient becomes available.
Protein longevity favors return to growth state in yeast
The ubiquitin-dependent 26S proteasome typically controls the abundance of proteins that require high amounts of ATP to be degraded in the yeast Saccharomyces cerevisiae (16). Despite obvious differences in the identity of the proteases and substrates involved, a decrease in ATP amounts during nutritional starvation should preserve proteins and favor exit from a slow- or no-growth state in yeast as in bacteria. To test this hypothesis, we exposed S. cerevisiae to the lowest concentration of the translation inhibitor cycloheximide (250 μg/ml) that increased ATP amounts (fig. S13A) and inhibited growth (fig. S13B).
The slowdown in the growth of wild-type S. cerevisiae as the cells reached stationary phase at about 8 h of culture in rich medium (Fig. 6A and fig. S1C) resulted in decreased ATP abundance (Fig. 6B) and stabilization of the 26S proteasome substrates H2B (62), Nap1 (63), and Rap1 (64) (Fig. 6C). The stability of β-actin, which was used as loading control, was the same before and during starvation conditions (Fig. 6C). Treatment with cycloheximide increased ATP amounts (Fig. 6D), decreased the abundance of 26S proteasome substrates (Fig. 6E), and substantially delayed entry into the growth state when nutrients were restored (Fig. 6F). The effect of cycloheximide on S. cerevisiae mimicked that of chloramphenicol on S. Typhimurium (Fig. 4, B to G). Therefore, yeast and bacteria respond to nutrient limitation by decreasing proteolysis by ATP-dependent proteases, which speeds the return to the growth state.
Fig. 6. Slow growth–induced protein longevity is critical for yeast entry into the growth state.
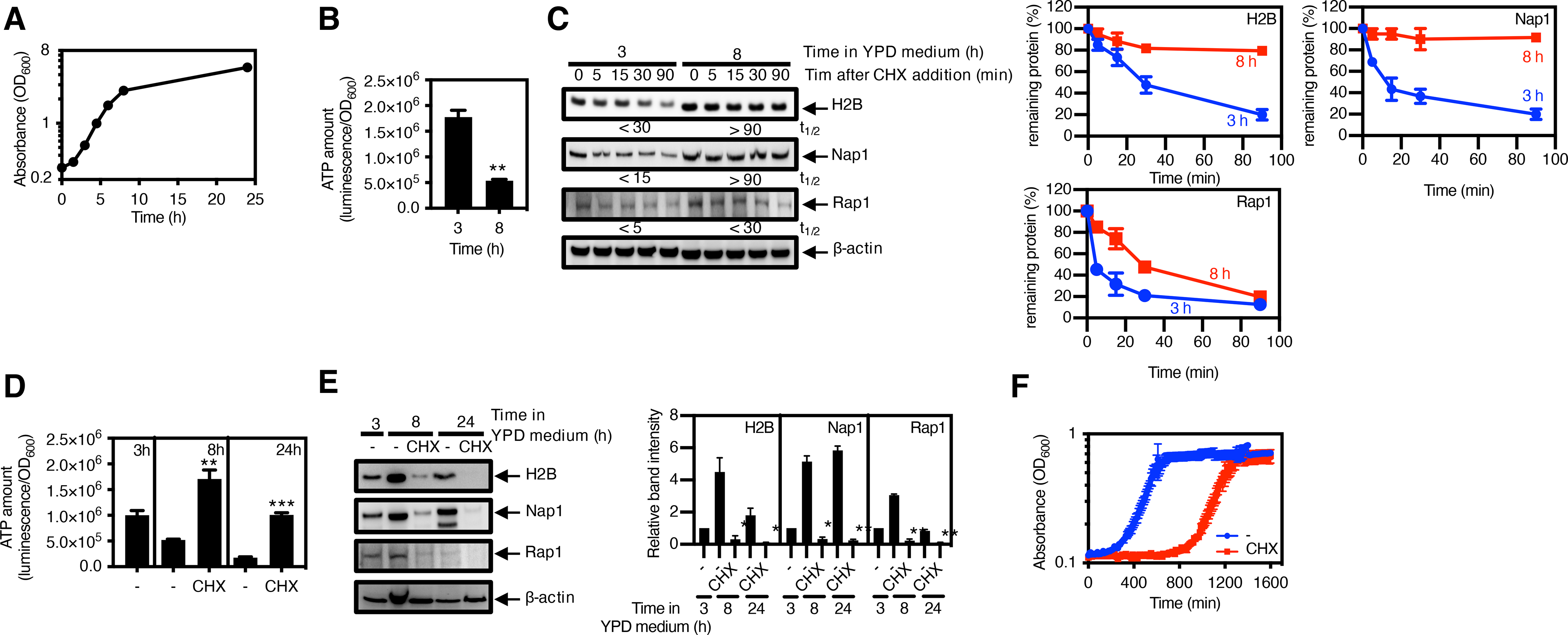
(A) Growth curve of wild-type S. cerevisiae (MP1361) in YPD medium. Data represents the average and SD from 4 independent experiments. (B) Quantification of ATP amount and (C) immunoblotting and quantification of the indicated protease substrates in wild-type S. cerevisiae (MP1361) grown in YPD medium for 3 or 8 h. Protein synthesis was inhibited by cycloheximide (CHX), and samples were removed at the indicated times for Western blotting and quantification. The half-lives (t1/2) of the substrates, indicated below the blots, were calculated by regression analysis of the exponential decay of proteins. Degradation curves and half-lives (t1/2) of protease substrates (C) were determined by band intensity from Western blots with protein amounts normalized to β-actin. The mean and SD from three 3 independent experiments are shown. (D) Quantification of ATP amount and (E) immunoblotting and quantification of the indicated protease substrates in wild-type S. cerevisiae (MP1361) grown in YPD medium for 3 h, followed by 5 h or 21 h treatment with CHX. Densitometry graphs show the average and SD for protein amounts relative to that in the untreated bacteria at 3 h from 3 independent experiments. (F) Growth curves of wild-type S. cerevisiae (MP1361) in fresh medium after washing cycloheximide from 8 h samples in (D) and (E). ATP amounts (B and D) were calculated with normalization to luminescence by OD600, and the mean and SD from 4 independent experiments are shown. Unpaired Student’s t tests were performed between 3 h with 8 h samples (B) or between untreated samples at 8 or 24 h (C) with the other combinations; **P < 0.01 and ***P < 0.001. Western blotting (C and E) was performed with antibodies specific for H2B, Nap1, Rap1, or β-actin.
Discussion
Nutrient limitation results in a slow- or no-growth state state that can be long-lasting in the case of bacterial spores. We have determined that, when nutrients became available to a slow- or no-growing cell, a speedy return to the growth state required proteins present during the slow- or no-growth state. We established that nutritional starvation decreased the ATP concentration from ~1–2 mM to ~200 μM in bacteria (fig. S1, A and B), and from ~3–4 mM to ~600 μM in yeast (fig. S1C), which reduced the degradation of functional proteins by ATP-dependent proteases (Fig. 7). The remaining ATP concentration was sufficient for proteolysis of proteins specifically targeted for degradation, such as SsrA-tagged GFP (Fig. 3, B and C) and pre-OmpF in a secM mutant (fig. S7, B and C). Therefore, bacteria, and possibly yeast, use the remaining ATP to maintain quality control over their proteomes in agreement with previous findings (15, 65). In other words, slow- or no-growing bacteria maintain both a healthy proteome and the longevity of functional proteins, which helps them emerge from arrested growth (66).
Fig. 7. A reduction in proteolysis during starvation preserves proteins and speeds the return to the growth state.
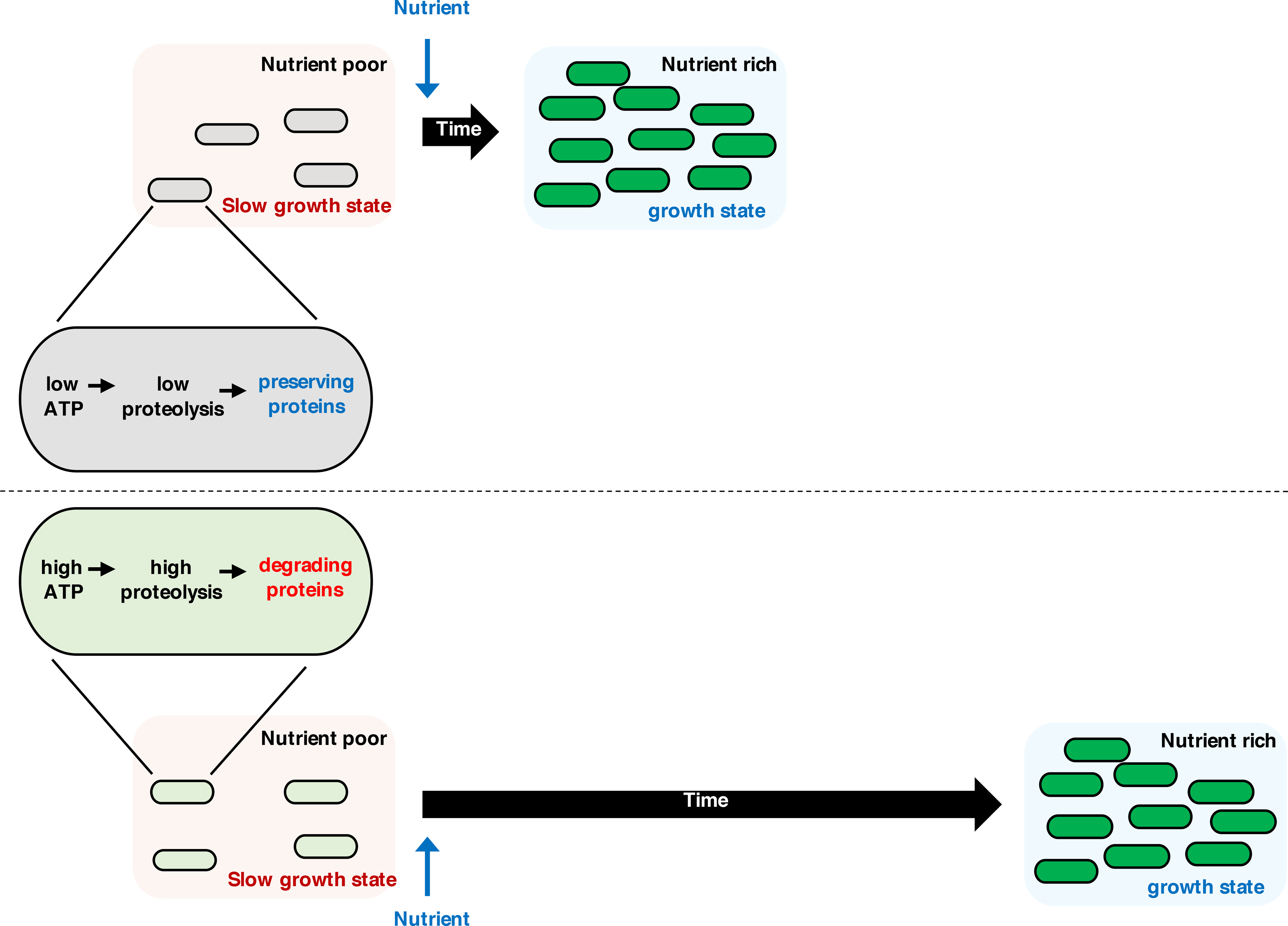
When bacteria or yeast experience nutrient starvation, they reduce intracellular ATP amounts, which decreases proteolysis by ATP-dependent proteases, thereby preserving the protein pool. Protein preservation speeds the return to the growth state when nutrients become available. By contrast, if the organisms fail to preserve proteins during starvation, they experience a delay in returning to the growth state.
A decrease in ATP concentration impacts all aspects of protein homeostasis including synthesis, solubility, folding, and degradation. For example, protein synthesis is readily compromised by a reduction in the ATP concentration (1, 2) from the millimolar to the micromolar range (56, 57). This is also the case for protein solubility because ATP functions as a hydrotrope only when present in millimolar concentrations (67). When the ATP concentration decreases below 670 μM, pre-existing proteins begin to aggregate (3). A further decrease in ATP concentration to the 200 μM range (fig. S1) hinders the degradation of multiple substrates of different AAA+ proteases (Figs. 1 to 5). In addition to nutritional starvation (1, 2, 5), a variety of conditions trigger a decrease in ATP concentration including acidic pH (68) and low oxygen (69), both of which pathogens often encounter during infection.
How do microbes return to the growth state once nutrients become available? On the one hand, they preserve proteins during the slow- or no-growth state by reducing the activity of ATP-dependent proteases (Fig. 7). On the other hand, the complex of the DnaK and ClpB chaperones disaggregates proteins (3). These two processes are physiologically connected because DnaK, the rate-limiting factor for disaggregation by DnaK-ClpB, is proteolyzed during prolonged starvation (70). That is, ClpB cannot disaggregate proteins in the absence of DnaK (71–73). and ATP amounts do not impact ClpB function in the absence of DnaK under physiological conditions (74).
Ribosome abundance can affect the kinetics with which bacteria emerge from a slow- or no-growth condition once a limiting nutrient becomes available. When bacteria transition from exponential to stationary growth phase, the composition of the ribosome changes with certain core ribosomal proteins being replaced by others (75). In addition, ribosomes are reversibly inactivated by hibernation factors (76) and degraded (77), with the extent of the decrease in ribosome content depending on the specific nutrient that becomes limiting (78). Moreover, bacteria that increase ribosome content during growth exit stationary phase more rapidly once nutrients become available (79).
We identified the ClpXP substrate RpoS and the ClpSAP substrate PhoP as being necessary for a normal emergence of wild-type S. Typhimurium from Mg2+ starvation (Fig. 5). Proteins helping bacteria leave a particular starvation condition may not help under a different starvation condition. For instance, PhoP protein amounts are not correlated with emergence from a slow growth state triggered by carbon starvation (fig. S12). Therefore, the return to a growth state requires both proteins that operate when bacteria experience limitation for particular nutrients, such as PhoP, and ribosomes that carry out protein synthesis regardless of the nutrient limitation that stops or slows growth.
Antibiotic persisters are metabolically-dormant bacteria that resist killing by bactericidal antibiotics (80, 81). Persisters arise from different stresses, including exposure to antibiotics (80) and nutritional starvation (66, 82). Persister pathogens can “wake up” spontaneously when the antibiotic concentration drops and return to their infection cycle (83), resulting in recurrent infections. The decrease in ATP concentration taking place during dormancy may impact the number of persisters for several reasons. First, SsrA tagging is required for awakening antibiotic tolerant bacteria from arrested growth (84), and proteolysis of SsrA-tagged substrates continues unabated during nutritional limitation (Fig. 3, B and C). Secondly, continuous degradation of misfolded proteins by the protease Lon is important to escape from dormancy because the formation of polypeptide aggregates during dormancy hinders the regrowth of antibiotic persisters (3). In addition, as discussed above for bacterial emergence from slow- or no-growth conditions, there is a positive correlation between ribosome content and the speed of persister resuscitation (85).
Our findings suggest that persisters may be eliminated by targeting the molecules responsible for protein preservation. In support of this notion, compounds that stimulate ATP production (86) or promote proteolysis by the ClpP protease (87) kill slow-growing bacteria exhibiting antibiotic tolerance (88).
Because it takes proteins to make proteins, the ability to preserve proteins under extreme nutrient-limiting conditions may be responsible for the initiation of bacterial spore germination, a process that does not require protein synthesis (89, 90). In addition, preserving proteins may be critical after spore germination because specific proteins such as ribosomal proteins and translation factors must be present in the spores for protein synthesis to take place (90). Given that tumor cells reduce protein synthesis to survive in nutrient-limited environments (7), it may be possible to sensitize dormant tumor cells using strategies analogous to those discussed above for bacteria.
Materials and Methods
Microbial strains, plasmids and growth conditions
Bacterial strains and plasmids used in this study are presented in Table S1. All S. enterica serovar Typhimurium strains are derived from strain wild-type strain 14028s (91) and following construction, the mutations were moved by phage P22-mediated transductions (92). DNA oligonucleotides used in this study are presented in Table S2. Bacteria were grown at 37°C in Luria-Bertani broth (LB), MOPS media (93) with 0.1% casamino acids and 0.4% glucose, and N-minimal media (pH 7.7) (41) supplemented with 0.1% casamino acids, 38 mM glycerol and the indicated concentrations of MgCl2. S. cerevisiae DY1457 (94) was grown in YPD medium supplemented with 0.4% glucose at 30°C. Ampicillin was used at 50 μg/ml, chloramphenicol at 25 μg/ml and tetracycline at 12.5 μg/ml for bacterial growth. For the protein stability assays, tetracycline was used at 50 μg/ml and spectinomycin at 1 mg/ml. To reduce ATP amounts, sodium arsenate was used at 1mM. For the bacterial growth assay with bacteriostatic antibiotics, chloramphenicol was used at 0, 5, 10, 20, 50 μg/ml, trimethoprim at 0, 50, 100, 200, 500 μg/ml, and sulfamethoxazole at 0, 0.1, 0.5, 1, 2 μg/ml. To induce expression from plasmid-borne genes, isopropyl thio-β-D-1 galactopyranoside (IPTG) was used at 1 mM.
Construction of bacterial mutants
Bacterial chromosomal mutants were constructed using the one-step disruption method (95) with minor modifications. To construct strains specifying a C-terminally FLAG-tagged CysA protein (JY766), a cat cassette was introduced at the 3′ end of the cysA gene: a cat gene fragment was amplified from plasmid pKD3 using primers 16490/16491 for cysA-FLAG, then introduced into wild-type S. Typhimurium (14028s) harboring plasmid pKD46 selecting for Cm resistance. The resulting strain (JY767) was kept at 30°C and transformed with pCP20 to remove the cat cassette. To construct the secM mutant (JYDN1), a kan cassette was introduced into the secM gene as follows: a kat gene fragment was amplified from pKD4 using primers 2E5/2E6, then introduced into wild-type S. Typhimurium (14028s) harbouring plasmid pKD46 selecting from Kan resistance.
Strains JY901 and JY747 were made by transducing the cysA-FLAG::cat insertion into strain MS7953s (96) using a P22 lysate generated in strain JY766. Strains EG16740 was made by transducing the mgtA::cat insertion into strain EL4 (42) using a P22 lysate generated in strain EG16734 (97). Strain EG17284 was made by transducing the phoP::Tn10 insertion into strain EG17133 (35) using a P22 lysate generated in strain MS7953s (96). Strain JY1043 was made by transducing the cysA-FLAG::cat insertion into strain JY649 (48) using a P22 lysate generated in strain JY766. Strains JYDN2 and JYDN3 were made by transducing the secM::kan insertion into strains EG16039 (98) and MS7953s (96), respectively, using a P22 lysate generated in strain JYDN1. Strain EG10822 was made by transforming with pCP20 to remove the cat cassette from rpoS::cat (35).
Western blot assay
Bacteria cells were grown in MOPS or N-minimal medium. Yeast cells were grown in YPD medium. Crude extracts were prepared in B-PER reagent (Pierce) with 100 μg/mL lysozyme and EDTA-free protease inhibitor (Roche). Samples were loaded on 4–12% NuPAGE gels (Life Technologies) and 4–15% TGX gels (Bio-rad), then transferred to nitrocellulose membrane using the iBot machine (Life Technologies) or the Trans-Blot Turbo machine (Bio-rad). Membranes were blocked with 3% skim milk solution at room temperature for 2 h. Then, samples were analysed using antibodies against the FLAG peptide or the RpoS, RpoH, LpxC, AtpB, GFP, OmpF, FtsH, ClpX, ClpP, Lon, H2B, Nap1, Rap1 or β-actin proteins. Mouse anti-RpoS (Neoclone, W0009) and anti-RpoH (Biolegend, 663402) antibodies were used at 1:2,000 dilution. Mouse anti-Rap1 (Santa Cruz Biotech, sc-166556) and anti-Nap1 (Santa Cruz Biotech, sc-25342) antibodies were used at 1:2,000 dilution. Rabbit anti-FtsH (Thermo Scientific, PA5-43806), anti-ClpX (a generous gift from Urs Jenal), anti-ClpP (LifeSpan BioSciences, LS-C675226), anti-Lon (LifeSpan BioSciences, LS-C488160), anti-H2B (Abcam, ab188291), anti-FLAG (Sigma-Aldrich, F7425), anti-GFP (Sigma-Aldrich, SAB4301138), anti-OmpF (Abcam, ab203223) and anti-LpxC (LifeSpan BioSciences, LS-C318823-50) antibodies were used at 1:2,000 dilution. Mouse anti-AtpB (Abcam, ab110280) and anti-β-actin (Abcam, ab8224) were used at 1:5,000 dilution. Secondary horseradish peroxidase-conjugated anti-rabbit or anti-mouse antisera (GE healthcare) were used at 1:5,000 dilution. Blots were developed with the Amersham ECL Western Blotting Detection Reagents (GE Healthcare) or SuperSignal West Femto Chemiluminescent system (Pierce).
Measurement of ATP amounts
ATP amounts were measured using a microplate reader (Tecan, Infinite M1000 PRO) as described (99) with a few modifications. Bacteria were grown overnight in MOPS media. One ml of the overnight culture was washed one time in MOPS media and resuspended in 1 ml of the same media. Diluted (1/50) bacteria were inoculated into 1 ml of media. Bacteria were grown overnight in N-minimal media containing 10 mM Mg2+. One ml of the overnight culture was washed three times in the N-minimal medium without Mg2+ and resuspended in 1 ml of the same media. Diluted (1/50) bacteria were inoculated into 1 ml of N-minimal media containing 10 μM Mg2+. Yeast cells were grown overnight in YPD media. One ml of the overnight culture was washed one time in the YPD media and resuspended in 1 ml of the same media. Diluted (1/50) yeast cells were inoculated into 1 ml of media. Cells were normalized by OD600 and heated at 70°C for 10 min. Intracellular ATP was measured using the BacTiter-Glo Microbial Cell Viability Assay Kit (Promega) according to the manufacturer’s instructions.
In vivo protein degradation assay
To measure protein stability, bacteria were treated with tetracycline (50 μg/ml) or spectinomycin (1mg/ml), and 1.5 mL samples were removed at the indicated times and harvested at 4°C. Yeast cells were treated with cycloheximide (1 mg/ml), and 1.5 mL samples were removed at the indicated times and harvested at 4°C. Pelleted cells were kept on dry ice for 30 min. Samples were then resuspended in B-PER reagent (Pierce) with 100 μg/mL lysozyme and EDTA-free protease inhibitor (Roche). After addition of the same volume of SDS sample buffer, samples were separated on 4–12% SDS-polyacrylamide gel and analysed by Western blotting.
Return to growth state assay
Cells were grown in MOPS medium, N-minimal medium or YPD medium for 2 or 3 h, followed by 3 h for N-minimal medium, 3 or 21 h for MOPS medium, and 5 h for YPD medium treatment with chloramphenicol (10 μg/ml), trimethoprim (100 μg/ml), sulfamethoxazole (1 mg/ml) or cycloheximide (250 μg/ml). Treated samples were washed two times in fresh medium, then inoculated into fresh nutrient rich medium to investigate escape from the slow growth state into the growth state. Microbial growth was measured using the Victor3 (Bio-rad) or Infinite M1000 PRO (Tecan) at 37°C or 30°C for 16 h.
Quantitative RT-PCR
To measure mRNA abundance, cells were grown in N-minimal medium containing 10 μM MgCl2 and 38 mM glycerol at 37°C for 6 h. Total RNA was purified using RNeasy Kit (Qiagen) with on-column DNase treatment, and cDNA was synthesized by using VILO Super Mix (Life Technologies). Quantification of transcripts was carried out by quantitative RT-PCR using SYBR Green PCR Master Mix (Applied Biosystems) in a QuantStudio™ 6 Flex Real-Time PCR System (Applied Biosystems). mRNA abundance was determined by using a standard curve obtained from PCR products generated with serially diluted genomic DNA, and results were normalized to the levels of the ompA gene. Data shown are an average from at least three independent experiments. Primers used in quantitative RT-PCR assay are presented in Table S2.
Data analysis and statistics
To quantify protein amounts, each blot was analysed by the ImageJ (NIH, ver. 1.49u) program by selecting a rectangular area around the target band for quantitative analysis. Calculated intensity of target bands was normalized to the levels of the loading protein control. Numbers are the average corresponding to protein abundance relative to the leftmost lane from independent experiments. For measurement of protein stability, protein half-lives (t1/2) were calculated by regression analysis of the exponential decay of proteins. ATP amounts were calculated with normalization by OD600. The mean and SD from independent experiments are shown. Unpaired Student’s t tests were performed between samples, and P < 0.05 was defined as statistically significant.
Supplementary Material
Acknowledgments:
We thank Jennifer Aronson for comments on the manuscript; Dr. Andrea Lay-Hoon Kwa for useful advice on bacteriostatic antibiotics; Dr. Kwangsoo Kim for useful advice on statistical analysis. Dr. Urs Jenal for his generous gift of anti-ClpX antibody.
Funding: This research was supported by NIH grant AI49561 to EAG and new faculty startup funds from the Duke-NUS Medical School and the Seoul National University College of Medicine to JY.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: All data needed to evaluate the conclusions of the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Manning BD, Adaptation to starvation: translating a matter of life or death. Cancer Cell 23, 713–715 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Pontes MH, Yeom J, Groisman EA, Reducing ribosome biosynthesis promotes translation during low Mg(2+) stress. Mol. Cell 64, 480–492 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pu Y, Li Y, Jin X, Tian T, Ma Q, Zhao Z, Lin SY, Chen Z, Li B, Yao G, Leake MC, Lo CJ, Bai F, ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Mol. Cell 73, 143–156 e144 (2019). [DOI] [PubMed] [Google Scholar]
- 4.Gengenbacher M, Kaufmann SH, Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 36, 514–532 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Watson SP, Clements MO, Foster SJ, Characterization of the starvation-survival response of Staphylococcus aureus. J. Bacteriol. 180, 1750–1758 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saldanha AJ, Brauer MJ, Botstein D, Nutritional homeostasis in batch and steady-state culture of yeast. Mol. Biol. Cell 15, 4089–4104 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leprivier G, Remke M, Rotblat B, Dubuc A, Mateo AR, Kool M, Agnihotri S, El-Naggar A, Yu B, Somasekharan SP, Faubert B, Bridon G, Tognon CE, Mathers J, Thomas R, Li A, Barokas A, Kwok B, Bowden M, Smith S, Wu X, Korshunov A, Hielscher T, Northcott PA, Galpin JD, Ahern CA, Wang Y, McCabe MG, Collins VP, Jones RG, Pollak M, Delattre O, Gleave ME, Jan E, Pfister SM, Proud CG, Derry WB, Taylor MD, Sorensen PH, The eEF2 kinase confers resistance to nutrient deprivation by blocking translation elongation. Cell 153, 1064–1079 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mogk A, Schmidt R, Bukau B, The N-end rule pathway for regulated proteolysis: prokaryotic and eukaryotic strategies. Trends Cell Biol. 17, 165–172 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Nishimura K, Kato Y, Sakamoto W, Essentials of proteolytic machineries in chloroplasts. Mol. Plant 10, 4–19 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Wang N, Maurizi MR, Emmert-Buck L, Gottesman MM, Synthesis, processing, and localization of human Lon protease. J. Biol. Chem. 269, 29308–29313 (1994). [PubMed] [Google Scholar]
- 11.Gur E, Biran D, Ron EZ, Regulated proteolysis in Gram-negative bacteria — how and when? Nat. Rev. Microbiol. 9, 839–848 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Konovalova A, Sogaard-Andersen L, Kroos L, Regulated proteolysis in bacterial development. FEMS Microbiol. Rev. 38, 493–522 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varshavsky A, The ubiquitin system, autophagy, and regulated protein degradation. Annu. Rev. Biochem. 86, 123–128 (2017). [DOI] [PubMed] [Google Scholar]
- 14.Sauer RT, Baker TA, AAA+ proteases: ATP-fueled machines of protein destruction. Annu. Rev. Biochem. 80, 587–612 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Peterson CN, Levchenko I, Rabinowitz JD, Baker TA, Silhavy TJ, RpoS proteolysis is controlled directly by ATP levels in Escherichia coli. Genes Dev. 26, 548–553 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eytan E, Ganoth D, Armon T, Hershko A, ATP-dependent incorporation of 20S protease into the 26S complex that degrades proteins conjugated to ubiquitin. Proc. Natl. Acad. Sci. U.S.A. 86, 7751–7755 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ciechanover A, Finley D, Varshavsky A, The ubiquitin-mediated proteolytic pathway and mechanisms of energy-dependent intracellular protein degradation. J. Cell. Biochem. 24, 27–53 (1984). [DOI] [PubMed] [Google Scholar]
- 18.Ciechanover A, Heller H, Katz-Etzion R, Hershko A, Activation of the heat-stable polypeptide of the ATP-dependent proteolytic system. Proc. Natl. Acad. Sci. U.S.A. 78, 761–765 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battesti A, Gottesman S, Roles of adaptor proteins in regulation of bacterial proteolysis. Curr. Opin. Microbiol. 16, 140–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kirstein J, Molière N, Dougan DA, Turgay K, Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. Microbiol. 7, 589–599 (2009). [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y, Gottesman S, Hoskins JR, Maurizi MR, Wickner S, The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev. 15, 627–637 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bougdour A, Cunning C, Baptiste PJ, Elliott T, Gottesman S, Multiple pathways for regulation of σS (RpoS) stability in Escherichia coli via the action of multiple anti-adaptors. Mol. Microbiol. 68, 298–313 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Bougdour A, Wickner S, Gottesman S, Modulating RssB activity: IraP, a novel regulator of sigma(S) stability in Escherichia coli. Genes Dev. 20, 884–897 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zgurskaya HI, Keyhan M, Matin A, The sigma S level in starving Escherichia coli cells increases solely as a result of its increased stability, despite decreased synthesis. Mol. Microbiol. 24, 643–651 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Roche ED, Sauer RT, SsrA-mediated peptide tagging caused by rare codons and tRNA scarcity. EMBO J. 18, 4579–4589 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keiler KC, Waller PR, Sauer RT, Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993 (1996). [DOI] [PubMed] [Google Scholar]
- 27.Farrell CM, Grossman AD, Sauer RT, Cytoplasmic degradation of ssrA-tagged proteins. Mol. Microbio. 57, 1750–1761 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Li SH, Li Z, Park JO, King CG, Rabinowitz JD, Wingreen NS, Gitai Z, Escherichia coli translation strategies differ across carbon, nitrogen and phosphorus limitation conditions. Nat. Microbiol. 3, 939–947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Albers E, Larsson C, Andlid T, Walsh MC, Gustafsson L, Effect of nutrient starvation on the cellular composition and metabolic capacity of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 73, 4839–4848 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans AC Jr., Wilkinson KD, Ubiquitin-dependent proteolysis of native and alkylated bovine serum albumin: effects of protein structure and ATP concentration on selectivity. Biochemistry 24, 2915–2923 (1985). [DOI] [PubMed] [Google Scholar]
- 31.Charette MF, Henderson GW, Markovitz A, ATP hydrolysis-dependent protease activity of the lon (capR) protein of Escherichia coli K-12. Proc. Natl. Acad. Sci. U.S.A. 78, 4728–4732 (1981). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Groisman EA, Hollands K, Kriner MA, Lee E-J, Park S-Y, Pontes MH, Bacterial Mg2+homeostasis, transport, and virulence. Annu. Rev. Genet. 47, 625–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Flynn JM, Neher SB, Kim YI, Sauer RT, Baker TA, Proteomic discovery of cellular substrates of the ClpXP protease reveals five classes of ClpX-recognition signals. Mol. Cell 11, 671–683 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Zhou Y, Gottesman S, Regulation of proteolysis of the stationary-phase sigma factor RpoS. J. Bacteriol. 180, 1154–1158 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tu X, Latifi T, Bougdour A, Gottesman S, Groisman EA, The PhoP/PhoQ two-component system stabilizes the alternative sigma factor RpoS in Salmonella enterica. Proc. Natl. Acad. Sci. U.S.A. 103, 13503–13508 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Arsene F, Tomoyasu T, Bukau B, The heat shock response of Escherichia coli. Int. J. Food Microbiol. 55, 3–9 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Kanemori M, Nishihara K, Yanagi H, Yura T, Synergistic roles of HslVU and other ATP-dependent proteases in controlling in vivo turnover of sigma32 and abnormal proteins in Escherichia coli. J. Bacteriol. 179, 7219–7225 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fuhrer F, Langklotz S, Narberhaus F, The C-terminal end of LpxC is required for degradation by the FtsH protease. Mol. Microbiol. 59, 1025–1036 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Soncini FC, Garcia Vescovi E, Solomon F, Groisman EA, Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178, 5092–5099 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Snavely MD, Florer JB, Miller CG, Maguire ME, Magnesium transport in Salmonella typhimurium: 28Mg2+ transport by the CorA, MgtA, and MgtB systems. J. Bacteriol. 171, 4761–4766 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snavely MD, Miller CG, Maguire ME, The mgtB Mg2+ transport locus of Salmonella typhimurium encodes a P-type ATPase. J. Biol. Chem. 266, 815–823 (1991). [PubMed] [Google Scholar]
- 42.Lee E-J, Pontes MH, Groisman EA, A bacterial virulence protein promotes pathogenicity by inhibiting the bacterium’s own F1Fo ATP synthase. Cell 154, 146–156 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chamnongpol S, Cromie M, Groisman EA, Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J. Mol. Biol. 325, 795–807 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Garcia Vescovi E, Soncini FC, Groisman EA, Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84, 165–174 (1996). [DOI] [PubMed] [Google Scholar]
- 45.Soncini FC, Vescovi EG, Groisman EA, Transcriptional autoregulation of the Salmonella typhimurium phoPQ operon. J. Bacteriol. 177, 4364–4371 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Conlon BP, Rowe SE, Gandt AB, Nuxoll AS, Donegan NP, Zalis EA, Clair G, Adkins JN, Cheung AL, Lewis K, Persister formation in Staphylococcus aureus is associated with ATP depletion. Nat. Microbiol. 1, 16051 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Park M, Nam D, Kweon DH, Shin D, ATP reduction by MgtC and Mg(2+) homeostasis by MgtA and MgtB enables Salmonella to accumulate RpoS upon low cytoplasmic Mg(2+) stress. Mol. Microbiol. 110, 283–295 (2018). [DOI] [PubMed] [Google Scholar]
- 48.Yeom J, Wayne KJ, Groisman EA, Sequestration from protease adaptor confers differential stability to protease substrate. Mol. Cell 66, 234–246.e235 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crowlesmith I, Gamon K, Henning U, Precursor proteins are intermediates in vivo in the synthesis of two major outer membrane proteins, the OmpA and OmpF proteins, of Escherichia coli K12. Eur. J. Biochem. 113, 375–380 (1981). [DOI] [PubMed] [Google Scholar]
- 50.Sakr S, Cirinesi AM, Ullers RS, Schwager F, Georgopoulos C, Genevaux P, Lon protease quality control of presecretory proteins in Escherichia coli and its dependence on the SecB and DnaJ (Hsp40) chaperones. J. Biol. Chem 285, 23506–23514 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Driessen AJ, Fekkes P, van der Wolk JP, The Sec system. Curr. Opin. Microbiol 1, 216–222 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Tian H, Beckwith J, Genetic screen yields mutations in genes encoding all known components of the Escherichia coli signal recognition particle pathway. J. Bacteriol. 184, 111–118 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuang Z, Ji H, Boeke JD, Stress response factors drive regrowth of quiescent cells. Curr. Genet. 64, 807–810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pontes MH, Groisman EA, Protein synthesis controls phosphate homeostasis. Genes Dev. 32, 79–92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pontes MH, Sevostyanova A, Groisman EA, When too much ATP is bad for protein synthesis. J. Mol. Biol. 427, 2586–2594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rodnina MV, Serebryanik AI, Ovcharenko GV, El’Skaya AV, ATPase strongly bound to higher eukaryotic ribosomes. Eur. J. Biochem. 225, 305–310 (1994). [DOI] [PubMed] [Google Scholar]
- 57.Gaal T, Bartlett MS, Ross W, Turnbough CL Jr., Gourse RL, Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278, 2092–2097 (1997). [DOI] [PubMed] [Google Scholar]
- 58.Gleckman R, Blagg N, Joubert DW, Trimethoprim: mechanisms of action, antimicrobial activity, bacterial resistance, pharmacokinetics, adverse reactions, and therapeutic indications. Pharmacotherapy 1, 14–20 (1981). [DOI] [PubMed] [Google Scholar]
- 59.Hong YL, Hossler PA, Calhoun DH, Meshnick SR, Inhibition of recombinant Pneumocystis carinii dihydropteroate synthetase by sulfa drugs. Antimicrob. Agents Chemother. 39, 1756–1763 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katzung BG, Trevor AJ, Basic and clinical pharmacology. (McGraw-Hill, New York, ed. 13th, 2015), pp. 1203. [Google Scholar]
- 61.Gao X, Yeom J, Groisman EA, The expanded specificity and physiological role of a widespread N-degron recognin. Proc. Natl. Acad. Sci. U.S.A. 116, 18629–18637 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Clift D, McEwan WA, Labzin LI, Konieczny V, Mogessie B, James LC, Schuh M, A method for the acute and rapid degradation of endogenous proteins. Cell 171, 1692–1706 e1618 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao W, Wang L, Zhang M, Wang P, Yuan C, Qi J, Meng H, Gao C, Tripartite motif-containing protein 38 negatively regulates TLR3/4- and RIG-I-mediated IFN-beta production and antiviral response by targeting NAP1. J. Immunol. 188, 5311–5318 (2012). [DOI] [PubMed] [Google Scholar]
- 64.Jordan JD, He JC, Eungdamrong NJ, Gomes I, Ali W, Nguyen T, Bivona TG, Philips MR, Devi LA, Iyengar R, Cannabinoid receptor-induced neurite outgrowth is mediated by Rap1 activation through G(alpha)o/i-triggered proteasomal degradation of Rap1GAPII. J. Biol. Chem. 280, 11413–11421 (2005). [DOI] [PubMed] [Google Scholar]
- 65.St John AC, Goldberg AL, Effects of reduced energy production on protein degradation, guanosine tetraphosphate, and RNA synthesis in Escherichia coli. J. Biol. Chem. 253, 2705–2711 (1978). [PubMed] [Google Scholar]
- 66.Wilmaerts D, Windels EM, Verstraeten N, Michiels J, General mechanisms leading to persister formation and awakening. Trends Genet. 35, 401–411 (2019). [DOI] [PubMed] [Google Scholar]
- 67.Patel A, Malinovska L, Saha S, Wang J, Alberti S, Krishnan Y, Hyman AA, ATP as a biological hydrotrope. Science 356, 753–756 (2017). [DOI] [PubMed] [Google Scholar]
- 68.O’Sullivan E, Condon S, Relationship between acid tolerance, cytoplasmic pH, and ATP and H+-ATPase levels in chemostat cultures of Lactococcus lactis. Appl. Environ. Microbiol. 65, 2287–2293 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmidt RR, Fulda M, Paul MV, Anders M, Plum F, Weits DA, Kosmacz M, Larson TR, Graham IA, Beemster GTS, Licausi F, Geigenberger P, Schippers JH, van Dongen JT, Low-oxygen response is triggered by an ATP-dependent shift in oleoyl-CoA in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 115, E12101–E12110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rockabrand D, Arthur T, Korinek G, Livers K, Blum P, An essential role for the Escherichia coli DnaK protein in starvation-induced thermotolerance, H2O2 resistance, and reductive division. J. Bacteriol. 177, 3695–3703 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Doyle SM, Hoskins JR, Wickner S, Collaboration between the ClpB AAA+ remodeling protein and the DnaK chaperone system. Proc. Natl. Acad. Sci. U.S.A. 104, 11138–11144 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Martin I, Celaya G, Alfonso C, Moro F, Rivas G, Muga A, Crowding activates ClpB and enhances its association with DnaK for efficient protein aggregate reactivation. Biophys. J 106, 2017–2027 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosenzweig R, Moradi S, Zarrine-Afsar A, Glover JR, Kay LE, Unraveling the mechanism of protein disaggregation through a ClpB-DnaK interaction. Science 339, 1080–1083 (2013). [DOI] [PubMed] [Google Scholar]
- 74.Klosowska A, Chamera T, Liberek K, Adenosine diphosphate restricts the protein remodeling activity of the Hsp104 chaperone to Hsp70 assisted disaggregation. Elife 5, e15159 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lilleorg S, Reier K, Pulk A, Liiv A, Tammsalu T, Peil L, Cate JHD, Remme J, Bacterial ribosome heterogeneity: Changes in ribosomal protein composition during transition into stationary growth phase. Biochimie 156, 169–180 (2019). [DOI] [PubMed] [Google Scholar]
- 76.Prossliner T, Skovbo Winther K, Sorensen MA, Gerdes K, Ribosome Hibernation. Annu. Rev. Genet. 52, 321–348 (2018). [DOI] [PubMed] [Google Scholar]
- 77.Piir K, Paier A, Liiv A, Tenson T, Maivali U, Ribosome degradation in growing bacteria. EMBO Rep 12, 458–462 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fessler M, Gummesson B, Charbon G, Svenningsen SL, Sorensen MA, Short-term kinetics of rRNA degradation in Escherichia coli upon starvation for carbon, amino acid or phosphate. Mol. Microbiol. 113, 951–963 (2020). [DOI] [PubMed] [Google Scholar]
- 79.Remigi P, Ferguson GC, McConnell E, De Monte S, Rogers DW, Rainey PB, Ribosome provisioning activates a bistable switch coupled to fast exit from stationary phase. Mol. Biol. Evol. 36, 1056–1070 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lewis K, Persister cells. Annu. Rev. Microbiol. 64, 357–372 (2010). [DOI] [PubMed] [Google Scholar]
- 81.Pontes MH, Groisman EA, A physiological basis for nonheritable antibiotic resistance. mBio 11, e00817–00820 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nguyen D, Joshi-Datar A, Lepine F, Bauerle E, Olakanmi O, Beer K, McKay G, Siehnel R, Schafhauser J, Wang Y, Britigan BE, Singh PK, Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science 334, 982–986 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cohen NR, Lobritz MA, Collins JJ, Microbial persistence and the road to drug resistance. Cell Host Microbe 13, 632–642 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li J, Ji L, Shi W, Xie J, Zhang Y, Trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J. Antimicrob. Chemother. 68, 2477–2481 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim JS, Yamasaki R, Song S, Zhang W, Wood TK, Single cell observations show persister cells wake based on ribosome content. Environ. Microbiol 20, 2085–2098 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Lobritz MA, Belenky P, Porter CB, Gutierrez A, Yang JH, Schwarz EG, Dwyer DJ, Khalil AS, Collins JJ, Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. U.S.A 112, 8173–8180 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Conlon BP, Nakayasu ES, Fleck LE, LaFleur MD, Isabella VM, Coleman K, Leonard SN, Smith RD, Adkins JN, Lewis K, Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature 503, 365–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pontes MH, Groisman EA, Slow growth determines nonheritable antibiotic resistance in Salmonella enterica . Sci. Signal 12, pii: eaax3938 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Korza G, Setlow B, Rao L, Li Q, Setlow P, Changes in Bacillus spore small molecules, rrna, germination, and outgrowth after extended sublethal exposure to various temperatures: evidence that protein synthesis is not essential for spore germination. J. Bacteriol. 198, 3254–3264 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boone T, Driks A, Protein Synthesis during Germination: Shedding New Light on a Classical Question. J. Bacteriol. 198, 3251–3253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fields PI, Swanson RV, Haidaris CG, Heffron F, Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. U.S.A. 83, 5189–5193 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Davis RW, Botstein D, Roth JR, Advanced bacterial genetics. (Cold Spring Harbor Laboratory, 1980). [Google Scholar]
- 93.Neidhardt FC, Bloch PL, Smith DF, Culture medium for enterobacteria. J. Bacteriol. 119, 736–747 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Askwith C, Eide D, Van Ho A, Bernard PS, Li L, Davis-Kaplan S, Sipe DM, Kaplan J, The FET3 gene of S. cerevisiae encodes a multicopper oxidase required for ferrous iron uptake. Cell 76, 403–410 (1994). [DOI] [PubMed] [Google Scholar]
- 95.Datsenko KA, Wanner BL, One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fields PI, Groisman EA, Heffron F, A Salmonella locus that controls resistance to microbicidal proteins from phagocytic cells. Science 243, 1059–1062 (1989). [DOI] [PubMed] [Google Scholar]
- 97.Park SY, Groisman EA, Signal-specific temporal response by the Salmonella PhoP/PhoQ regulatory system. Mol. Microbiol. 91, 135–144 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yeom J, Groisman EA, Activator of one protease transforms into inhibitor of another in response to nutritional signals. Genes Dev. 33, 1280–1292 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pontes MH, Lee E-J, Choi J, Groisman EA, Salmonella promotes virulence by repressing cellulose production. Proc. Natl. Acad. Sci. U. S. A. 112, 5183–5188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Valdivia RH, Falkow S, Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction. Mol. Microbiol. 22, 367–378 (1996) [DOI] [PubMed] [Google Scholar]
- 101.Shin D, Groisman EA, Signal-dependent binding of the response regulators PhoP and PmrA to their target promoters in vivo. J. Biol. Chem. 280, 4089–4094 (2005). [DOI] [PubMed] [Google Scholar]
- 102.Yeom J, Gao X, Groisman EA, Reduction in adaptor amounts establishes degradation hierarchy among protease substrates. Proc. Natl. Acad. Sci. U. S. A 115, E4483–e4492 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


