Abstract
A well-accepted hallmark of cancer is genomic instability, which drives tumorigenesis. Therefore, understanding the molecular and cellular defects that destabilize chromosomal integrity is paramount to cancer diagnosis, treatment and cure. DNA repair and the replication stress response are overarching paradigms for maintenance of genomic stability, but the devil is in the details. ATP-dependent helicases serve to unwind DNA so it is replicated, transcribed, recombined and repaired efficiently through coordination with other nucleic acid binding and metabolizing proteins. Alternatively folded DNA structures deviating from the conventional anti-parallel double helix pose serious challenges to normal genomic transactions. Accumulating evidence suggests that G-quadruplex (G4) DNA is problematic for replication. Although there are multiple human DNA helicases that can resolve G4 in vitro, it is debated which helicases are truly important to resolve such structures in vivo. Recent advances have begun to elucidate the principal helicase actors, particularly in cellular DNA replication. FANCJ, a DNA helicase implicated in cancer and the chromosomal instability disorder Fanconi Anemia, takes center stage in G4 resolution to allow smooth DNA replication. We will discuss FANCJ’s role with its protein partner RPA to remove G4 obstacles during DNA synthesis, highlighting very recent advances and implications for cancer therapy.
INTRODUCTION
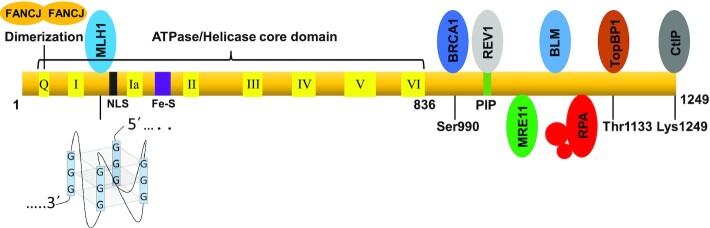
Biallelic mutations in the gene encoding the FANCJ/BRIP1/BACH1 DNA helicase are linked to Fanconi Anemia (FA) (1–3), a disease characterized by hematopoietic stem cell defects, progressive bone marrow failure and cancer, particularly blood cancers at an early age. Somatic mutations in FANCJ are associated with various cancers, especially those of the breast and ovary, classifying it as a tumor suppressor (4). Like the 21 other FA genes identified so far (5), FANCJ is implicated in interstrand cross-link (ICL) repair (1–3). Specifically, FANCJ is believed to function at the homologous recombination (HR) step of repairing double-strand breaks (DSBs) generated after unhooking of ICLs (3,6,7). FANCJ, originally discovered by its interaction with the tumor suppressor BRCA1 (8) (Figure 1), also plays a role in DSB repair beyond the correction of ICL-induced lesions (3,8,9). Many of FANCJ’s protein partners are implicated in DNA damage sensing, and DSB repair including the mismatch repair proteins MLH1/MutLα (10) and MSH5 (11), Bloom's syndrome helicase (BLM) (12), and the DNA end-processing nuclease MRE11 (9) and facilitator CtIP (13) (Figure 1). Recent work suggests that FANCJ must balance its function in DNA strand transactions during HR repair with DNA end-processing events orchestrated by factors such as the DNA end binding protein RAP80 to ensure timely and faithful repair (14). A Fancj mutant mouse model displayed a defect in spermatogenesis that is accompanied by defective regulation of DSB repair (15).
Figure 1.
G4 binding pocket, catalytic domain, mapped protein interaction sites, and sites for post-translational modifications of FANCJ. Certain key residues are shown, but not all for simplicity. Lys 141 and Lys142 are implicated in G4 binding. The nuclear localization sequence (NLS) and Fe-S domain are comprised by residues 159–174 and residues 270–263, respectively. The conserved PCNA interaction motif (PIP) corresponds to residues 1001–1017. FANCJ directly binds to the RPA70 subunit of the RPA heterotrimer. The FANCJ Q25A substitution disables the ability of FANCJ to effectively dimerize. The binding site on FANCJ for MSH5 (not shown) has not yet been mapped. See text for additional details and references.
In addition to FANCJ’s well accepted importance for ICL repair and DSB repair, the Shiekhattar group provided early evidence for a role of FANCJ in replication progression (16). Indeed, FANCJ protects from replication stress and FANCJ-deficient cells are sensitive to the replication stress-inducing agent hydroxyurea (12). Research from the Cantor laboratory demonstrated that FANCJ helicase coordinates with the replication fork remodeling enzyme helicase-like transcription factor (HLTF) to promote fork integrity (17). FANCJ interacts with Replication Protein A (RPA) (18) (Figure 1), a eukaryotic single-stranded DNA-binding protein originally identified as an essential player in SV40 DNA replication (19,20) and found to be implicated in mammalian DNA replication as well (21,22). RPA is also implicated in DNA damage recognition (23–26) and facilitates the initiation of checkpoint signaling by binding to exposed single-stranded DNA at sites of DNA damage and recruiting the protein kinase complex ATR-ATRIP necessary for ATR-mediated Chk1 activation (27). Indeed, FANCJ and RPA strongly co-localize in human cells exposed to agents that impose replication stress or DNA damage (18). FANCJ also interacts with Topoisomerase Binding Protein 1 (TopBP1) (28) (Figure 1) and helps to ensure replication fork integrity and restart by mechanisms that still remain to be fully characterized (17,29). FANCJ deficiency in both mice and human cells results in microsatellite instability (30,31), suggesting a difficulty in replicating these repetitive elements of the genome.
Aside from its ability to catalytically unwind duplex DNA structures associated with replication forks (32) and branch-migrate mobile D-loop structures that represent strand invasion intermediates of HR (14), FANCJ is a potent resolvase of G-quadruplex (G4) DNA structures derived from guanine-rich DNA sequences located in various regions of the genome including oncogene promoters, ribosomal DNA and telomeres (33,34). Missense mutations in FANCJ’s Fe-S cluster domain (Figure 1) that are linked to FA (35) or associated with cancer (36) result in the inactivation of its helicase activity on G4 or duplex DNA substrates. There is much interest in G4 DNA, given its unique structure and potential to interfere with nucleic acid transactions such as DNA synthesis, transcription or recombination (37). FANCJ suppresses DNA damage and genomic instability caused by G4 in human cells (33,34). However, until only very recently did we begin to appreciate more mechanistic details of FANCJ’s involvement in G4 resolution as it pertains to DNA synthesis in vivo. The body of experimental work, and most recently an important advance by Lee et al. (38), provide molecular details for an emerging picture of FANCJ’s unique role in cellular DNA replication; moreover, FANCJ closely collaborates with its protein partner RPA allowing the single-stranded DNA binding protein to bind the unwound single-strand to facilitate smooth DNA synthesis past guanine-rich sequences prone to form G4 and trigger the ATR-mediated replication stress response.
FANCJ’s UNIQUE ROLE IN G4 RESOLUTION TO FACILITATE DNA REPLICATION
In two new publications, Odermatt et al. (36) and Lee et al. (38) provide elegant data from single-molecule localization microscopy (SMLM) imaging studies conducted in human cancer cells and in vitro experiments with purified recombinant proteins and defined G4 DNA substrates which together demonstrate that FANCJ plays a uniquely important role in G4 unwinding to allow smooth DNA replication. By coordination with its interacting partner RPA, FANCJ was shown to efficiently resolve stable G4 DNA structures to allow binding by RPA, which serves two purposes: (i) efficient replicative DNA synthesis past a formidable endogenous obstacle and (ii) RPA-mediated signaling to suppress DSBs. Collectively, these findings represent a significant advance in understanding a key mechanistic event whereby persistent fork stalling is avoided and the replication stress response is preserved. At this juncture, we contend that placing these findings in the context of recent work will help to inform both mechanistic aspects of DNA replication in cancer cells and therapeutic opportunities.
G4-INDUCED REPLICATION STRESS RESPONSE IS DETERMINED BY ROBUSTNESS OF FANCJ CATALYTIC ACTIVITY
A minimal threshold of FANCJ catalytic activity is required in cancer cells to overcome replication stress induced by the G4 ligand telomestatin (TMS) or DNA polymerase inhibitor aphidicolin (APH), as well as DNA breakage caused by bleomycin (39). In contrast, a greater level of FANCJ helicase activity is required for repair of pharmacologically induced ICLs. An additional distinction between the two responses is that FANCJ rescues TMS or APH sensitivity independent of the FA pathway, whereas ICL repair is dependent on an intact FA pathway (39). These findings suggest that optimal FANCJ helicase activity and an intact FA pathway are required for cross-link resistance. Furthermore, FANCJ’s role in G4 DNA metabolism may be more relevant to replication stress observed in cancer cells as opposed to the molecular pathology of FA, but this remains to be substantiated. Although a threshold of FANCJ catalysis was not assessed in the SM imaging studies (36,38) to determine a quantitative level of FANCJ ATPase/helicase activity required for RPA loading onto thermodynamically stable G4 structures, it would be of interest to ascertain the relationships of FANCJ catalytic efficiency, G4 stability and replication stress response. Rapidly dividing cancer cells are likely to be characterized by an elevated frequency of replication fork encounters with G4 and other alternatively folded DNA structures. Presumably, interactions of the replication machinery with auxiliary replisome components (e.g. DNA helicases) and modulation of replication proteins by post-translational modifications are key determinants of cancer cell proliferation, in addition to checkpoint mechanisms. How these factors and events precisely interplay with the robustness of FANCJ-catalyzed resolution of G4 structures with varying degrees of stability dictated by nucleotide sequence or architecture remains to be seen. Consistent with the most recent advance by Lee et al. (38), FANCJ plays a conserved role to promote DNA synthesis past G4 structures as elegantly demonstrated in a reconstituted Xenopus egg extract replication system (40).
DISTINCTIVE IMPORTANCE OF FANCJ IN G4 METABOLISM DISTINGUISHES IT FROM OTHER G4-RESOLVING DNA HELICASES
The employment of multi-color SMLM provided researchers the opportunity to visualize and quantitatively assess the spatial arrangement of nascently synthesized DNA with replication factors and G-quadruplexes in living cells (38). The authors delivered strong evidence that FANCJ’s ability to resolve stable G4 structures enables RPA to load on to the unwound single-stranded genomic DNA that otherwise would be refractory to RPA binding; consequently, a cellular response to G4-induced replication stress is enacted and smooth replication past G4 obstacles takes place. The paramount role of FANCJ in this process cannot be overstated. While it has been reported that many purified recombinant helicase proteins can bind and/or resolve various G4 DNA structures in vitro (41), it is apparent from the Lee et al. study and previously published work from several laboratories (34,40,42) that human FANCJ is a prominent player during replication to resolve G4 DNA structures in vivo. Thus, the presence of other DNA helicases capable of unwinding G4 DNA substrates in vitro such as BLM (43), WRN (44), RTEL1 (45), PIF1 (46) and ChlR1/DDX11 (47) in FANCJ-deficient human cells does not compensate for the specific loss of FANCJ to deal with G4-induced replication stress. Interestingly, none of these G4-resolving helicases possess the G4 recognition site mapped in FANCJ (48) (Figure 1), suggesting FANCJ has a distinct mechanism of G4 unwinding. Nonetheless, many of these aforementioned helicases are also important for fork progression and maintaining genomic integrity, leaving it unexplained how FANCJ could have a uniquely critical role compared to the other G4 unwinding helicases. BLM was found to directly interact with FANCJ (12) (Figure 1), which may help to regulate its G4 unwinding function and response to replication stress. The ChlR1/DDX11 helicase is also implicated in G4 DNA metabolism to support sister chromatid cohesion (49), consistent with its importance in chromosomal stability in yeast (50), as well as chicken (51) and human (52) cells. Perhaps properties relating to its mechanism of action [e.g. oligomerization (53) (Figure 1)] in addition to substrate specificity (54) is distinct for FANCJ.
The reported observation that G4 DNA metabolism is unaffected in Fancj(-/-) mice (31) suggests FANCJ’s importance in G4 DNA metabolism in human cells may be unique. It is tempting to speculate that the much longer G-rich telomeres in mice may contribute to the observed difference (55,56), but the underlying basis for FANCJ’s apparent importance in human versus mouse is unclear and additional factors may come into play. The FANCJ homolog in C. elegans known as dog-1 has been implicated in the stability of regions upstream of G-rich tracts during DNA synthesis of the lagging strand (57). A recent report from the Tijsterman group (58) highlights the importance of dog-1 in preventing deletions at G4 sites from polymerase alpha generated repriming of the lagging strand, building off their previous identification of pol theta mediated end-joining as a key repair pathway for G4 (59). This most recent study provides a striking example of how the FANCJ homolog controls pathway choice and genome stability near G4 structures, but further studies of FANCJ’s role(s) in human cells are warranted. G4 accumulates in FANCJ-deficient human cells (60,61), suggesting that there are likely to be consequences for transcription, recombination and DNA repair as well.
FANCJ’s ABILITY TO RESOLVE ENTROPICALLY FAVORED INTRAMOLECULAR G4 DNA AND COLLABORATE WITH RPA
From our earlier biochemical studies, we found that purified recombinant human FANCJ protein is distinct from other Fe-S cluster DNA helicases including human DDX11 in its ability to efficiently resolve entropically favored unimolecular G4 DNA (54). Aside from the Fe-S helicases, PIF1 helicase was reported to bind parallel intramolecular G4 DNA with high affinity but unfold it with very slow kinetics, suggesting that auxiliary factors are required for efficient G4 unwinding (62). It is tempting to speculate that PIF1 has a specialized role in G4 DNA metabolism (e.g. at telomeres (63)) whereas FANCJ has a more universal role in general genome G4 DNA resolution during cellular replication (40,42). However, yeast strains deficient for the Pif1 helicase exhibit replication defects around sequences across the genome predicted to form G4 structures (64,65). It is plausible that Pif1 (or other G4-resolving helicases) respond locally to replication stress induced by an unusual DNA structure such as G4 at telomeres (or some other specific chromosomal region) and elicit global fork slowing in a checkpoint-dependent manner, as recently demonstrated for DNA damage induced by cellular exposure to a DNA cross-linking agent (66). In chicken DT40 cells, FANCJ may coordinate its G4 resolvase activity with the RECQ helicases WRN and BLM or the translesion polymerase REV1 to maintain epigenetic stability (67). Interestingly, FANCJ biochemically interacts with BLM (12) and REV1 (68) (Figure 1), but their cooperative molecular mechanism in cellular G4 metabolism has not yet been revealed.
FANCJ physically binds RPA (18) (Figure 1), which stimulates its helicase activity on both duplex (18) and G-quadruplex (34) DNA substrates, and the two proteins co-localize with each other in human cells exposed to agents that induce replication stress (18). In this latest work, Rothenberg et al. undertook a mechanistic study of the FANCJ–RPA interaction using a SM FRET-based assay to show that FANCJ (by virtue of its ATPase-dependent helicase activity) operates with RPA to efficiently destabilize stable intramolecular G4 DNA structures, allowing RPA to load on to the resolved secondary structure which is otherwise refractory to RPA binding (38). RPA loading on to the unwound G4-forming sequence allows the ATR-mediated replication stress response. Based on in vitro experimental observations using a SM system, Wu and Spies proposed that FANCJ repeatedly unfolds and refolds unimolecular G4 DNA (48). Despite FANCJ’s G4-specific recognition site (48) and its catalytic motor comprised by the conserved ATPase/helicase motifs (Figure 1), FANCJ helicase enzyme requires RPA to stabilize the unfolded G4 (38), consistent with RPA’s stimulation of FANCJ G4 resolvase activity under multi-turnover conditions (34). On the other hand, RPA itself can bind to and destabilize certain G4 DNA structures in vitro (69–71), suggesting that its interaction with G4 may independently regulate G4 stability of certain DNA sequences (e.g. those found at telomeres). It is plausible that RPA acting on its own may resolve less stable G4 structures, whereas FANCJ coordinates with RPA to unwind more stable G4. Thus, there may be two effects of RPA on G4 unwinding: (i) direct interaction with FANCJ to stimulate FANCJ-catalyzed G4 resolution; (ii) thermodynamic shift in the equilibrium of G4 versus single-stranded DNA, favoring single-stranded DNA. FANCJ is found at active replication forks in human cells (17), supporting its specialized role during replication stress. The FANCJ–RPA interaction provides an excellent modern-day example of molecular matchmakers, originally coined by Aziz Sancar in a different context (nucleotide excision repair) (72). In addition to its stimulation of FANCJ helicase activity, RPA binds the G4 unwound single-strand to trigger the ATR-mediated replication stress response.
Although the model proposed by Lee et al. provides new insight to the interactive mechanism of FANCJ and RPA to resolve G4 enabling smooth fork progression, there remain some unresolved questions. Cellular G4 formation induced by the G4 ligand pyridostatin (PDS) did not decrease overall DNA synthesis, as assessed by nuclear staining intensity of the thymidine analog 5-Ethynyl-2’-deoxyruridine (EdU) (38). It is plausible that firing of new G4-associated origins induced by pharmacological G4 stabilization, as recently reported in an in vitro Xenopus laevis replication system (73), may be at play. In this scenario, global EdU signal remains robust irrespective of G4 ligand stabilization. A more sensitive and specific readout for in vivo DNA synthesis at specific G4 loci and origins of DNA synthesis employing DNA fiber analysis would be informative. The data reported by Lee et al. (38) also raise some questions about the effect of FANCJ depletion on RPA binding to all replisomes or just those associated with G4 as a function of PDS exposure. Surprisingly, they found that RPA levels at G4-replisomes remained constant irrespective of PDS treatment. It might have been expected that FANCJ-deficient cells would have shown less RPA at G4-replisomes because the G4 structure would have not been resolved to create single-stranded DNA for RPA to bind. It is possible that because a base level of RPA readily binds to single-stranded DNA created in the wake of the advancing MCM replicative helicase complex as it opens parental double-stranded DNA at the fork (74), a similar amount of RPA was detected irrespective of FANCJ status or G4 ligand stabilization. Given the importance of FANCJ G4 resolution and its coordination with RPA (and possibly other factors) to shift the equilibrium from G4 to single-stranded state to promote DNA synthesis, alternative approaches such as proximity ligation assay to examine juxtaposition of a replisome protein with an antibody that specifically binds G4 as a function of FANCJ status will likely provide further insight.
IMPLICATIONS
Replication stress occurs due to various reasons including collisions of replisome-transcription machinery, replication slow zones, fragile sites, template damage and DNA bound proteins. Although G4 DNA blocks DNA replication and cause replication arrest, only a small percent of replication sites in the human cells coincide with G-rich motifs, as measured by the fraction of PCNA foci that non-randomly colocalize with G4s detected by quantitative SMLM clustering (38). This estimate may seem low, given evidence from G4-seq analysis that >700 000 G4s exist in single-stranded, purified human genomic DNA (75). However, the determination was made under G4-promoting conditions that favored DNA polymerase stalling at G4s during the Illumina next-generation sequencing procedure. Moreover, effects of chromatin on genomic G4 DNA formation are likely to come into play. Nonetheless, the SM localization data demonstrate that association between G4 and replisomes is likely to exist in human cells (38). The highly stable nature of G4 DNA (76) may lead to its persistence in cells, requiring specialized DNA helicases to resolve such structures in an efficient manner.
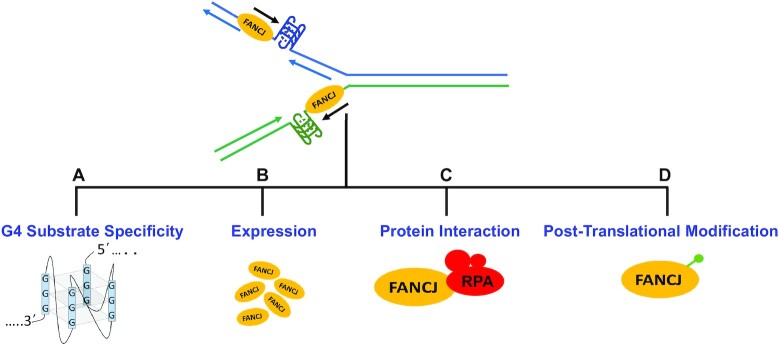
The collective evidence demonstrates that FANCJ is a true and prominent G4 resolvase that allows smooth DNA replication in human cancer cells. Moreover, FANCJ collaborates with RPA to not only efficiently unwind intramolecular G4 DNA but also elicit a timely and robust replication stress response. There are likely multiple factors that contribute to FANCJ’s unique involvement in G4 resolution to facilitate DNA replication (Figure 2). FANCJ’s nucleic acid substrate specificity may come into play. For example, unlike the sequence-related DDX11 helicase, FANCJ efficiently resolves unimolecular G-quadruplexes (54). During replication, FANCJ may act to resolve G4 structures embedded in the template single-strand for lagging strand synthesis between Okazaki fragments (Figure 2). Alternatively, FANCJ may load on to the leading strand template generated by the replicative MCM helicase ahead of DNA synthesis (Figure 2). In either capacity, FANCJ through its coordination with RPA serves to resolve G4 formed by transient single-stranded DNA to aid in template-directed DNA synthesis. In the simplest model, the 5’ to 3’ directionality FANCJ uses to resolve G4 (34) necessitates coordination with the converging DNA polymerase using the same template strand for replicative synthesis; however, the details of this synchronization remain a mystery. It was recently reported that G4 structures on the lagging strand template are more disruptive to replication than G4 structures in the leading strand template (77), suggesting FANCJ’s role to resolve G4 is prominent on the lagging strand template characterized by greater single-stranded DNA character, even if transient. Moreover, cytosine-rich DNA sequences known as i-motifs residing in the strand opposite to the G-rich strand may form a non-canonical DNA structure of two intercalated parallel-stranded duplexes stabilized by hemi-protonated cytosine-cytosine base pairs (78,79); however, it is debatable if G4 and i-motif quadruplexes can form simultaneously or if they are mutually exclusive due to steric hindrance (80). To our knowledge, there are no published studies on the activity of FANCJ or any other DNA helicases to resolve i-motif DNA structures.
Figure 2.
Factors that may contribute to FANCJ’s unique requirement to resolve G4 DNA structures to enable smooth cellular DNA replication of the leading and lagging strands. (A) FANCJ unique ability to efficiently resolve intramolecular G4 DNA. (B) FANCJ expression abundance in a cell-type or tissue-specific manner. (C) FANCJ’s protein partnerships with factors directly involved in replication (e.g. RPA). (D) FANCJ’s post-translational modifications affect catalytic function or replisome association. Factors are not mutually exclusive and may influence one another.
Because of the growing evidence that FANCJ plays a specialized role during replication, it will be important to assess its expression in tissues enriched for rapidly dividing cells in which there is a high turnover and compare it to other G4-resolving helicases (Figure 2). Further studies are required to characterize FANCJ’s regulation mediated by its protein interactions and post-translational modifications (Figure 2). FANCJ phosphorylation at Ser-990 and Thr-1133 mediate the BRCA1 (81) and TopBP1 (28) interactions, respectively (Figure 1). FANCJ acetylation at Lys-1249 (Figure 1) facilitates DNA end-processing (82), by recruiting the DNA damage response protein CtIP to the sites of DSBs as recently elucidated (13). Despite these advances, it remains undetermined if these or other post-translation modifications of FANCJ directly affect its involvement in G4 metabolism. It also remains to be seen if FANCJ localizes by itself to the sites of G4 at stalled forks, is aided by other factors, or exists as a constitutive component of replisomes (or a sub-population of replisomes). Conversely, FANCJ may regulate the recruitment of other proteins to G4 sites, as suggested by the stimulatory effect of replication-coupled G4 formation on FANCJ-mediated RPA loading (38).
Further efforts will help to guide researchers who are interested in G4 stabilization to achieve synthetic lethality in defined mutant backgrounds deficient in DNA repair or the replication stress response (83,84). Indeed, BRCA1- or BRCA2-deficient cancer cells are hypersensitive to ligands that bind to and stabilize G4 DNA structures (85). Helicases represent a provocative target for synthetic lethality (83,84,86). One would suspect that FANCJ-deficient tumors would be highly susceptible to G4-specific drugs. Furthermore, tumors selectively deficient in other helicases that act specifically on a subset of alternatively folded DNA structures may also be useful targets in anticancer therapy. Alternatively, small molecule inhibitors that target FANCJ or other G4-resolving helicases [e.g. WRN (87–89)] may be employed to combat cancer with defined genetic deficiencies and/or in concert with chemotherapy drugs that stabilize G4 structures.
Recently, a comprehensive mutational analysis of FANCJ was performed to assess functionality in the cell-based response to ICL-inducing agents (4). The loss-of-function (LOF) mutations for ICL resistance were largely localized to the helicase domain of FANCJ and not so much in the C-terminal region where protein interactions with such factors as BRCA1, BLM and TOPBP1 are located, suggesting that catalytic unwinding function is the critical function of FANCJ for ICL resistance whereas FANCJ–protein interactions modulate its other duties such as DNA repair or checkpoint activities. Interestingly, only 12% of the clinically relevant FANCJ mutations were associated with LOF for ICL resistance, suggesting that the remaining (majority) of cancer-associated FANCJ mutations disrupt other functions, one possibly being G4 resolution. Given the growing number of tumor suppressor genes implicated in G4 metabolism [e.g. BRCA1 (90), BRCA2 (90), RTEL1 (91), BLM (92), WRN (93)], it is of high priority to assess how this information can be utilized for cancer diagnostics and therapeutics.
Contributor Information
Robert M Brosh, Jr, Translational Gerontology Branch, National Institute on Aging, NIH, Baltimore, MD 21224, USA.
Yuliang Wu, Department of Biochemistry, Microbiology and Immunology, University of Saskatchewan, Saskatoon, Saskatchewan S7N 5E5, Canada.
FUNDING
Natural Sciences and Engineering Research Council of Canada (NSERC) [RGPIN-2019-05487 to Y.W.]; Cancer Research Society [CRS-24139 to Y.W.]; National Institute on Aging, NIH (1ZIAAG000753-12 to R.M.B.).
Conflict of interest statement. None declared.
REFERENCES
- 1. Levitus M., Waisfisz Q., Godthelp B.C., de Vries Y., Hussain S., Wiegant W.W., Elghalbzouri-Maghrani E., Steltenpool J., Rooimans M.A., Pals G. et al. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet. 2005; 37:934–935. [DOI] [PubMed] [Google Scholar]
- 2. Levran O., Attwooll C., Henry R.T., Milton K.L., Neveling K., Rio P., Batish S.D., Kalb R., Velleuer E., Barral S. et al. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 2005; 37:931–933. [DOI] [PubMed] [Google Scholar]
- 3. Litman R., Peng M., Jin Z., Zhang F., Zhang J., Powell S., Andreassen P.R., Cantor S.B. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell. 2005; 8:255–265. [DOI] [PubMed] [Google Scholar]
- 4. Calvo J.A., Fritchman B., Hernandez D., Persky N.S., Johannessen C.M., Piccioni F., Kelch B.A., Cantor S.B. Comprehensive mutational analysis of the BRCA1-associated DNA helicase and tumor-suppressor FANCJ/BACH1/BRIP1. Mol. Cancer Res. 2021; 19:1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Niraj J., Farkkila A., D’Andrea A.D. The Fanconi anemia pathway in cancer. Ann. Rev. Cancer Biol. 2019; 3:457–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cantor S., Drapkin R., Zhang F., Lin Y., Han J., Pamidi S., Livingston D.M. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl. Acad. Sci. 2004; 101:2357–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nath S., Somyajit K., Mishra A., Scully R., Nagaraju G. FANCJ helicase controls the balance between short- and long-tract gene conversions between sister chromatids. Nucleic Acids Res. 2017; 45:8886–8900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cantor S.B., Bell D.W., Ganesan S., Kass E.M., Drapkin R., Grossman S., Wahrer D.C., Sgroi D.C., Lane W.S., Haber D.A. et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001; 105:149–160. [DOI] [PubMed] [Google Scholar]
- 9. Suhasini A.N., Sommers J.A., Muniandy P.A., Coulombe Y., Cantor S.B., Masson J.Y., Seidman M.M., Brosh R.M. Jr Fanconi anemia group J helicase and MRE11 nuclease interact to facilitate the DNA damage response. Mol. Cell. Biol. 2013; 33:2212–2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Peng M., Litman R., Xie J., Sharma S., Brosh R.M. Jr, Cantor S.B. The FANCJ/MutLalpha interaction is required for correction of the cross-link response in FA-J cells. EMBO J. 2007; 26:3238–3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu Y., Wu X., Her C. hMSH5 facilitates the repair of Camptothecin-induced double-strand breaks through an interaction with FANCJ. J. Biol. Chem. 2015; 290:18545–18558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Suhasini A.N., Rawtani N.A., Wu Y., Sommers J.A., Sharma S., Mosedale G., North P.S., Cantor S.B., Hickson I.D., Brosh R.M. Jr Interaction between the helicases genetically linked to Fanconi anemia group J and Bloom's syndrome. EMBO J. 2011; 30:692–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Nath S., Nagaraju G. FANCJ helicase promotes DNA end resection by facilitating CtIP recruitment to DNA double-strand breaks. PLoS Genet. 2020; 16:e1008701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Awate S., Sommers J.A., Datta A., Nayak S., Bellani M.A., Yang O., Dunn C.A., Nicolae C.M., Moldovan G.L., Seidman M.M. et al. FANCJ compensates for RAP80 deficiency and suppresses genomic instability induced by interstrand cross-links. Nucleic Acids Res. 2020; 48:9161–9180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun X., Brieño-Enríquez M.A., Cornelius A., Modzelewski A.J., Maley T.T., Campbell-Peterson K.M., Holloway J.K., Cohen P.E. FancJ (Brip1) loss-of-function allele results in spermatogonial cell depletion during embryogenesis and altered processing of crossover sites during meiotic prophase I in mice. Chromosoma. 2016; 125:237–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kumaraswamy E., Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol. Cell. Biol. 2007; 27:6733–6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peng M., Cong K., Panzarino N.J., Nayak S., Calvo J., Deng B., Zhu L.J., Morocz M., Hegedus L., Haracska L. et al. Opposing Roles of FANCJ and HLTF Protect Forks and Restrain Replication during Stress. Cell Rep. 2018; 24:3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gupta R., Sharma S., Sommers J.A., Kenny M.K., Cantor S.B., Brosh R.M. Jr FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007; 110:2390–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fairman M.P., Stillman B. Cellular factors required for multiple stages of SV40 DNA replication in vitro. EMBO J. 1988; 7:1211–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wold M.S., Kelly T. Purification and characterization of replication protein A, a cellular protein required for in vitro replication of simian virus 40 DNA. Proc. Natl. Acad. Sci. 1988; 85:2523–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ishimi Y., Matsumoto K., Ohba R. DNA replication from initiation zones of mammalian cells in a model system. Mol. Cell. Biol. 1994; 14:6489–6496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yuzhakov A., Kelman Z., Hurwitz J., O’Donnell M Multiple competition reactions for RPA order the assembly of the DNA polymerase delta holoenzyme. EMBO J. 1999; 18:6189–6199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Burns J.L., Guzder S.N., Sung P., Prakash S., Prakash L. An affinity of human replication protein A for ultraviolet-damaged DNA. J. Biol. Chem. 1996; 271:11607–11610. [DOI] [PubMed] [Google Scholar]
- 24. Clugston C.K., McLaughlin K., Kenny M.K., Brown R. Binding of human single-stranded DNA binding protein to DNA damaged by the anticancer drug cis-diamminedichloroplatinum (II). Cancer Res. 1992; 52:6375–6379. [PubMed] [Google Scholar]
- 25. Patrick S.M., Turchi J.J. Human replication protein A preferentially binds cisplatin-damaged duplex DNA in vitro. Biochemistry. 1998; 37:8808–8815. [DOI] [PubMed] [Google Scholar]
- 26. Suhasini A.N., Sommers J.A., Mason A.C., Voloshin O.N., Camerini-Otero R.D., Wold M.S., Brosh R.M. Jr FANCJ helicase uniquely senses oxidative base damage in either strand of duplex DNA and is stimulated by replication protein A to unwind the damaged DNA substrate in a strand-specific manner. J. Biol. Chem. 2009; 284:18458–18470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zou L., Elledge S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science (New York, N.Y.). 2003; 300:1542–1548. [DOI] [PubMed] [Google Scholar]
- 28. Gong Z., Kim J.E., Leung C.C., Glover J.N., Chen J. BACH1/FANCJ acts with TopBP1 and participates early in DNA replication checkpoint control. Mol. Cell. 2010; 37:438–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Raghunandan M., Chaudhury I., Kelich S.L., Hanenberg H., Sobeck A. FANCD2, FANCJ and BRCA2 cooperate to promote replication fork recovery independently of the Fanconi Anemia core complex. Cell Cycle. 2015; 14:342–353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barthelemy J., Hanenberg H., Leffak M. FANCJ is essential to maintain microsatellite structure genome-wide during replication stress. Nucleic Acids Res. 2016; 44:6803–6816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Matsuzaki K., Borel V., Adelman C.A., Schindler D., Boulton S.J. FANCJ suppresses microsatellite instability and lymphomagenesis independent of the Fanconi anemia pathway. Genes Dev. 2015; 29:2532–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gupta R., Sharma S., Sommers J.A., Jin Z., Cantor S.B., Brosh R.M. Jr Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 2005; 280:25450–25460. [DOI] [PubMed] [Google Scholar]
- 33. London T.B., Barber L.J., Mosedale G., Kelly G.P., Balasubramanian S., Hickson I.D., Boulton S.J., Hiom K. FANCJ is a structure-specific DNA helicase associated with the maintenance of genomic G/C tracts. J. Biol. Chem. 2008; 283:36132–36139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu Y., Shin-ya K., Brosh R.M. Jr FANCJ helicase defective in Fanconia anemia and breast cancer unwinds G-quadruplex DNA to defend genomic stability. Mol. Cell. Biol. 2008; 28:4116–4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu Y., Sommers J.A., Suhasini A.N., Leonard T., Deakyne J.S., Mazin A.V., Shin-Ya K., Kitao H., Brosh R.M. Jr Fanconi anemia group J mutation abolishes its DNA repair function by uncoupling DNA translocation from helicase activity or disruption of protein-DNA complexes. Blood. 2010; 116:3780–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Odermatt D.C., Lee W.T.C., Wild S., Jozwiakowski S.K., Rothenberg E., Gari K. Cancer-associated mutations in the iron-sulfur domain of FANCJ affect G-quadruplex metabolism. PLos Genet. 2020; 16:e1008740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Varshney D., Spiegel J., Zyner K., Tannahill D., Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020; 21:459–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee W.T.C., Yin Y., Morten M.J., Tonzi P., Gwo P.P., Odermatt D.C., Modesti M., Cantor S.B., Gari K., Huang T.T. et al. Single-molecule imaging reveals replication fork coupled formation of G-quadruplex structures hinders local replication stress signaling. Nat. Commun. 2021; 12:2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bharti S.K., Sommers J.A., Awate S., Bellani M.A., Khan I., Bradley L., King G.A., Seol Y., Vidhyasagar V., Wu Y. et al. A minimal threshold of FANCJ helicase activity is required for its response to replication stress or double-strand break repair. Nucleic Acids Res. 2018; 46:6238–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Castillo Bosch P., Segura-Bayona S., Koole W., van Heteren J.T., Dewar J.M., Tijsterman M., Knipscheer P. FANCJ promotes DNA synthesis through G-quadruplex structures. EMBO J. 2014; 33:2521–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012; 13:770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schwab R.A., Nieminuszczy J., Shin-ya K., Niedzwiedz W. FANCJ couples replication past natural fork barriers with maintenance of chromatin structure. J. Cell Biol. 2013; 201:33–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sun H., Karow J.K., Hickson I.D., Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998; 273:27587–27592. [DOI] [PubMed] [Google Scholar]
- 44. Fry M., Loeb L.A. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999; 274:12797–12802. [DOI] [PubMed] [Google Scholar]
- 45. Vannier J.B., Sandhu S., Petalcorin M.I., Wu X., Nabi Z., Ding H., Boulton S.J. RTEL1 is a replisome-associated helicase that promotes telomere and genome-wide replication. Science (New York, N.Y.). 2013; 342:239–242. [DOI] [PubMed] [Google Scholar]
- 46. Sanders C.M. Human Pif1 helicase is a G-quadruplex DNA-binding protein with G-quadruplex DNA-unwinding activity. Biochem. J. 2010; 430:119–128. [DOI] [PubMed] [Google Scholar]
- 47. Wu Y., Sommers J.A., Khan I., de Winter J.P., Brosh R.M. Jr Biochemical characterization of Warsaw breakage syndrome helicase. J. Biol. Chem. 2012; 287:1007–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu C.G., Spies M. G-quadruplex recognition and remodeling by the FANCJ helicase. Nucleic Acids Res. 2016; 44:8742–8753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. van Schie J.J.M., Faramarz A., Balk J.A., Stewart G.S., Cantelli E., Oostra A.B., Rooimans M.A., Parish J.L., de Almeida Estéves C., Dumic K. et al. Warsaw Breakage Syndrome associated DDX11 helicase resolves G-quadruplex structures to support sister chromatid cohesion. Nat. Commun. 2020; 11:4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Skibbens R.V. Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics. 2004; 166:33–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lerner L.K., Holzer S., Kilkenny M.L., Šviković S., Murat P., Schiavone D., Eldridge C.B., Bittleston A., Maman J.D., Branzei D. et al. Timeless couples G-quadruplex detection with processing by DDX11 helicase during DNA replication. EMBO J. 2020; 39:e104185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Parish J.L., Rosa J., Wang X., Lahti J.M., Doxsey S.J., Androphy E.J. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. J. Cell Sci. 2006; 119:4857–4865. [DOI] [PubMed] [Google Scholar]
- 53. Wu Y., Sommers J.A., Loiland J.A., Kitao H., Kuper J., Kisker C., Brosh R.M. Jr The Q motif of Fanconi anemia group J protein (FANCJ) DNA helicase regulates its dimerization, DNA binding, and DNA repair function. J. Biol. Chem. 2012; 287:21699–21716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bharti S.K., Sommers J.A., George F., Kuper J., Hamon F., Shin-ya K., Teulade-Fichou M.P., Kisker C., Brosh R.M. Jr Specialization among iron-sulfur cluster helicases to resolve G-quadruplex DNA structures that threaten genomic stability. J. Biol. Chem. 2013; 288:28217–28229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Blasco M.A., Lee H.W., Hande M.P., Samper E., Lansdorp P.M., DePinho R.A., Greider C.W. Telomere shortening and tumor formation by mouse cells lacking telomerase RNA. Cell. 1997; 91:25–34. [DOI] [PubMed] [Google Scholar]
- 56. Zijlmans J.M., Martens U.M., Poon S.S., Raap A.K., Tanke H.J., Ward R.K., Lansdorp P.M. Telomeres in the mouse have large inter-chromosomal variations in the number of T2AG3 repeats. Proc. Natl. Acad. Sci. 1997; 94:7423–7428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Cheung I., Schertzer M., Rose A., Lansdorp P.M. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 2002; 31:405–409. [DOI] [PubMed] [Google Scholar]
- 58. van Schendel R., Romeijn R., Buijs H., Tijsterman M. Preservation of lagging strand integrity at sites of stalled replication by Pol a-primase and 9-1-1 complex. Sci. Adv. 2021; 7:eabf2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Koole W., van Schendel R., Karambelas A.E., van Heteren J.T., Okihara K.L., Tijsterman M. A Polymerase Theta-dependent repair pathway suppresses extensive genomic instability at endogenous G4 DNA sites. Nat. Commun. 2014; 5:3216. [DOI] [PubMed] [Google Scholar]
- 60. Henderson A., Wu Y., Huang Y.C., Chavez E.A., Platt J., Johnson F.B., Brosh R.M. Jr, Sen D., Lansdorp P.M Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014; 42:860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Summers P.A., Lewis B.W., Gonzalez-Garcia J., Porreca R.M., Lim A.H.M., Cadinu P., Martin-Pintado N., Mann D.J., Edel J.B., Vannier J.B. et al. Visualising G-quadruplex DNA dynamics in live cells by fluorescence lifetime imaging microscopy. Nat. Commun. 2021; 12:162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Byrd A.K., Raney K.D. A parallel quadruplex DNA is bound tightly but unfolded slowly by pif1 helicase. J. Biol. Chem. 2015; 290:6482–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Geronimo C.L., Zakian V.A. Getting it done at the ends: Pif1 family DNA helicases and telomeres. DNA Repair (Amst.). 2016; 44:151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Paeschke K., Bochman M.L., Garcia P.D., Cejka P., Friedman K.L., Kowalczykowski S.C., Zakian V.A. Pif1 family helicases suppress genome instability at G-quadruplex motifs. Nature. 2013; 497:458–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Sabouri N., Capra J.A., Zakian V.A. The essential Schizosaccharomyces pombe Pfh1 DNA helicase promotes fork movement past G-quadruplex motifs to prevent DNA damage. BMC Biol. 2014; 12:101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mutreja K., Krietsch J., Hess J., Ursich S., Berti M., Roessler F.K., Zellweger R., Patra M., Gasser G., Lopes M. ATR-Mediated Global Fork Slowing and Reversal Assist Fork Traverse and Prevent Chromosomal Breakage at DNA Interstrand Cross-Links. Cell Rep. 2018; 24:2629–2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sarkies P., Murat P., Phillips L.G., Patel K.J., Balasubramanian S., Sale J.E. FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res. 2012; 40:1485–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Lowran K., Campbell L., Popp P., Wu C.G. Assembly of a G-quadruplex repair complex by the FANCJ DNA helicase and the REV1 polymerase. Genes. 2019; 11:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Safa L., Delagoutte E., Petruseva I., Alberti P., Lavrik O., Riou J.F., Saintome C. Binding polarity of RPA to telomeric sequences and influence of G-quadruplex stability. Biochimie. 2014; 103:80–88. [DOI] [PubMed] [Google Scholar]
- 70. Safa L., Gueddouda N.M., Thiebaut F., Delagoutte E., Petruseva I., Lavrik O., Mendoza O., Bourdoncle A., Alberti P., Riou J.F. et al. 5′ to 3′ unfolding directionality of DNA secondary structures by replication protein A: G-quadruplexes and duplexes. J. Biol. Chem. 2016; 291:21246–21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Salas T.R., Petruseva I., Lavrik O., Bourdoncle A., Mergny J.L., Favre A., Saintome C. Human replication protein A unfolds telomeric G-quadruplexes. Nucleic Acids Res. 2006; 34:4857–4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sancar A., Hearst J.E. Molecular matchmakers. Science (New York, N.Y.). 1993; 259:1415–1420. [DOI] [PubMed] [Google Scholar]
- 73. Prorok P., Artufel M., Aze A., Coulombe P., Peiffer I., Lacroix L., Guédin A., Mergny J.L., Damaschke J., Schepers A. et al. Involvement of G-quadruplex regions in mammalian replication origin activity. Nat. Commun. 2019; 10:3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tanaka T., Nasmyth K. Association of RPA with chromosomal replication origins requires an Mcm protein, and is regulated by Rad53, and cyclin- and Dbf4-dependent kinases. EMBO J. 1998; 17:5182–5191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chambers V.S., Marsico G., Boutell J.M., Di Antonio M., Smith G.P., Balasubramanian S. High-throughput sequencing of DNA G-quadruplex structures in the human genome. Nat. Biotechnol. 2015; 33:877–881. [DOI] [PubMed] [Google Scholar]
- 76. Sen D., Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988; 334:364–366. [DOI] [PubMed] [Google Scholar]
- 77. Gadgil R.Y., Romer E.J., Goodman C.C., Rider S.D. Jr, Damewood F.J., Barthelemy J.R., Shin-Ya K., Hanenberg H., Leffak M Replication stress at microsatellites causes DNA double-strand breaks and break-induced replication. J. Biol. Chem. 2020; 295:15378–15397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Abou Assi H., Garavís M., González C., Damha M.J. i-Motif DNA: structural features and significance to cell biology. Nucleic Acids Res. 2018; 46:8038–8056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gehring K., Leroy J.L., Guéron M. A tetrameric DNA structure with protonated cytosine.cytosine base pairs. Nature. 1993; 363:561–565. [DOI] [PubMed] [Google Scholar]
- 80. Sengupta P., Bose D., Chatterjee S. The Molecular Tête-à-Tête between G-Quadruplexes and the i-motif in the Human Genome. ChemBioChem. 2021; 22:1517–1537. [DOI] [PubMed] [Google Scholar]
- 81. Yu X., Chini C.C., He M., Mer G., Chen J. The BRCT domain is a phospho-protein binding domain. Science (New York, N.Y.). 2003; 302:639–642. [DOI] [PubMed] [Google Scholar]
- 82. Xie J., Peng M., Guillemette S., Quan S., Maniatis S., Wu Y., Venkatesh A., Shaffer S.A., Brosh R.M. Jr, Cantor S.B. FANCJ/BACH1 acetylation at lysine 1249 regulates the DNA damage response. PLoS Genet. 2012; 8:e1002786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. McLuckie K.I., Di Antonio M., Zecchini H., Xian J., Caldas C., Krippendorff B.F., Tannahill D., Lowe C., Balasubramanian S. G-quadruplex DNA as a molecular target for induced synthetic lethality in cancer cells. J. Am. Chem. Soc. 2013; 135:9640–9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Nakanishi C., Seimiya H. G-quadruplex in cancer biology and drug discovery. Biochem. Biophys. Res. Commun. 2020; 531:45–50. [DOI] [PubMed] [Google Scholar]
- 85. Zimmer J., Tacconi E.M.C., Folio C., Badie S., Porru M., Klare K., Tumiati M., Markkanen E., Halder S., Ryan A. et al. Targeting BRCA1 and BRCA2 deficiencies with G-quadruplex-interacting compounds. Mol. Cell. 2016; 61:449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Datta A., Dhar S., Awate S., Brosh R.M. Jr Synthetic lethal interactions of RECQ helicases. Trends Cancer. 2021; 7:146–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Aggarwal M., Banerjee T., Sommers J.A., Brosh R.M. Jr Targeting an Achilles’ heel of cancer with a WRN helicase inhibitor. Cell Cycle. 2013; 12:3329–3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Aggarwal M., Banerjee T., Sommers J.A., Iannascoli C., Pichierri P., Shoemaker R.H., Brosh R.M. Jr Werner syndrome helicase has a critical role in DNA damage responses in the absence of a functional fanconi anemia pathway. Cancer Res. 2013; 73:5497–5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Aggarwal M., Sommers J.A., Shoemaker R.H., Brosh R.M. Jr Inhibition of helicase activity by a small molecule impairs Werner syndrome helicase (WRN) function in the cellular response to DNA damage or replication stress. Proc. Natl. Acad. Sci. 2011; 108:1525–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Xu H., Di Antonio M., McKinney S., Mathew V., Ho B., O’Neil N.J., Santos N.D., Silvester J., Wei V., Garcia J. et al. CX-5461 is a DNA G-quadruplex stabilizer with selective lethality in BRCA1/2 deficient tumours. Nat. Commun. 2017; 8:14432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Vannier J.B., Pavicic-Kaltenbrunner V., Petalcorin M.I., Ding H., Boulton S.J. RTEL1 dismantles T loops and counteracts telomeric G4-DNA to maintain telomere integrity. Cell. 2012; 149:795–806. [DOI] [PubMed] [Google Scholar]
- 92. van Wietmarschen N., Merzouk S., Halsema N., Spierings D.C.J., Guryev V., Lansdorp P.M. BLM helicase suppresses recombination at G-quadruplex motifs in transcribed genes. Nat. Commun. 2018; 9:271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tang W., Robles A.I., Beyer R.P., Gray L.T., Nguyen G.H., Oshima J., Maizels N., Harris C.C., Monnat R.J. Jr The Werner syndrome RECQ helicase targets G4 DNA in human cells to modulate transcription. Hum. Mol. Genet. 2016; 25:2060–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]