Abstract
Porcine reproductive and respiratory syndrome (PRRS) is a serious swine disease causing great economic impact worldwide. The emergence of highly pathogenic strains in Asian countries is associated with large scale mortality in all age groups of pigs besides the classical presentation of severe respiratory distress, pneumonia, and a series of reproductive disorders in sows, like late-term abortion, premature farrowing, and an increased number of stillborn piglets. The present study was designed with the aim of isolation and characterization of the Betaarterivirus suid 2 from outbreaks in Mizoram in primary porcine alveolar macrophage and subsequently characterized the GP5 gene sequence of the isolate in terms of phylogenetic analysis and deduce amino acid sequence comparison. Virus propagation was performed in the porcine alveolar macrophage (PAM) primary cell culture and confirmed by immunoperoxidase test, FAT, and nested RT-PCR. The full-length GP5 gene (603nt) was amplified from the isolate and subsequently cloned and sequenced (MN928985). Phylogenetic analysis and sequence comparison of the present isolate was found to have similarity 98.7–98.8% with Myanmar HP-PRRS strains, 98–98.5% with Vietnam strains, 98.2–98.3% with China strains, indicating a close lineage with highly pathogenic PRRS strains. In deduced amino acid sequence analysis, one mutation was found in the primary neutralizing epitope (PNE) at position 39L → I39 and one more mutation was also found in the decoy epitope (DCE) at position 30 N → D30. The amino acid at this position is an N-linked glycosylation site, and mutation of the N-linked glycosylation is an immune escaped strategy adopted by this virus causing a persistent infection in the natural host.
Keywords: Porcine reproductive and respiratory syndrome, Betaarterivirus suid 2, Isolatio, GP5 glycoprotein, Molecular characterization
Introduction
Porcine reproductive and respiratory syndrome (PRRS) is a major systemic viral infection of pigs and is recognized as an important viral disease of swine causing severe economic losses of pig husbandry worldwide. Since its first occurrence in 1987 in North America and 1991 in Europe, the disease has spread to Southeast Asian countries [16] and was introduced to India in 2013 in Mizoram [29]. The emergence of highly pathogenic strain and its rapid spread in Asian countries become a major concern in pig husbandry practice in this region as diseased animals exhibit severe respiratory distress, pneumonia, and a series of reproductive disorders in sows, like late-term abortion, premature farrowing, an increased number of stillborn piglets including high mortality by highly pathogenic strains [1–3, 32–34]. The disease is caused by Betaarterivirus suid 1 (European type) and Betaarterivirus suid 2 (North American type) of the subgenus Eurpobartevirus and Ampobartevirus respectively under the genus Betaarterivirus of the subfamily Variarterivirinae of the family Arteriviridae in the order Nidovirales. However, both species are genetically diverse (60% homology) [23] and the North American type is mostly circulating in this region. The virus is having a selective tropism for porcine alveolar macrophages (PAM) expressing CD163 and sialoadhesin virus receptors and can be best isolated in primary PAM culture [1, 33, 34] as well as MARC-145 cells expressing the receptors [18, 33, 34]. The virus genome is around 15.1–15.5 kb having ten overlapping open reading frames (ORF) [7]. Immunogenic glycoprotein GP5 coded by ORF5 is around 603 nt length (MK315208) has been targeted for the development of recombinant vaccines [10, 25]. ORF5 sequence is used for phylogenetic study and virus characterization [5]. Since its first outbreaks in 2013 in Mizoram, subsequent years in 2015, 2016, and 2018 outbreaks of the disease have been reported [29, 30, 42]. This study was designed to isolate the virus from an outbreak in Mizoram, India, and to study the GP5 gene sequence.
Materials and methods
Virus isolation
Isolation of the virus was performed using archival tissue samples preserved in the Department of Veterinary Microbiology from the previous outbreaks of PRRS in Mizoram, India reported by Zohlimpuia et al. [42]. The archival samples used are lung and spleen tissues of dead piglets which were collected in the 2016 PRRS outbreak from Lawngtlai and Saiha districts of Mizoram. One isolate (MZ/5/2016) was used for molecular characterization in our present study. Porcine alveolar macrophage (PAM) primary cell culture was prepared from bronchoalveolar lavage from a 3 months old pig as per the method described by Mayer and Lam, 1984 [20]. These terminally differentiated PAM cells were used for virus isolation.
Virus detection
After 72 h of incubation at 37˚C in a CO2 incubator, the infected PAM cells were processed for virus growth detection. Growth of the virus was detected by immunoperoxidase test (IPT) using rabbit anti GP5 antibody (Cat. No. bs-4504R, Bioss Antibodies) and DAB nickel substrate (Ref. No. SK-4100, Vector Laboratories). Further fluorescent antibody test (FAT) using a rabbit anti GP5 antibody (Cat. No. bs-4504R, Bioss Antibodies) and anti-rabbit FITC conjugate (Cat. No. FTC2, Genei) was performed following the method described by OIE, 2018 [24]. A nested reverse transcriptase-polymerase chain reaction (RT-PCR) was carried out to detect the virus by molecular method [9].
Virus titration
One of the positive isolates was passaged in PAM cells twice and then virus titer was determined. A five-fold dilution of the virus isolate was prepared in the RPMI-1640 media without added serum and inoculated in a 96 well cell culture plate (Cat. No. 260836, Thermo Scientific) keeping 5 replica. Then the plate was incubated for 3 days and processed for IPT staining as described by OIE, 2018 [24]. TCID50 value was determined by the Spearman-Karber method.
GP5 gene sequence analysis
RNA extraction was carried out from the virus isolate using the QIAamp Viral RNA extraction kit (Cat. No. 52904 QIAGEN) as per the manufacturer’s protocol. The cDNA was synthesized using the Maxima H Minus First Strand kit (Cat. No. K1651, Thermo Scientific) as per the manufacturer’s protocol. The full-length GP5 gene was amplified using published primers [40] (Table 1) and was cloned in a TA cloning vector and sequenced by outsourcing. A reaction mixture of 25 µl containing 2.5 µl of 10X Taq buffer with 20 mM MgCl2, 2 µl of 10 mM dNTPs, 1 µl of each of 10 pM forward and reverse primers, 0.2 µl of 5 U/µl Taq polymerase, and 2 µl of cDNA as template was prepared. Cycling condition was carried out as an initial denaturation at 94ºC for 2 min, and 30 cycles of denaturation at 94 °C for 30 s, annealing at 56.1 °C for 45 s and extension at 72ºC for 30 s. A final extension was set at 72 °C for 10 min. The amplified product was verified on 1.5% agarose gel electrophoresis and was gel eluted using GeneJET Gel Extraction Kit (Cat. No. K0691, Thermo Scientific) as per the manufacturer’s protocol. The purified product was used for cloning using InsTAclone PCR Cloning Kit (Cat. No. K1213, Thermo Scientific) as per the manufacturer’s protocol. The gel eluted PCR product was ligated with pTZ57R/T cloning vector and transformation was performed in competent DH5α (mutant Escherichia coli). The transformed cells were plated in LB ampicillin and incubated for 18 h at 37 °C. The clone was confirmed by colony PCR and subsequently sequenced by outsourcing. Phylogenetic analysis was carried out based on the full-length GP5 sequence using MEGA9 software. The phylogenetic tree was generated by using the Neighbor-Joining method, keeping bootstrap consensus from 1000 replicates. Deduced amino acid sequence of GP5 was compared using BioEdit software, with sequences of previous outbreaks and prototype and vaccine virus sequences retrieved from GenBank.
Table 1.
Primer sequences used in the present study
| Primer name | Sequence | Amplicon size | Use | References |
|---|---|---|---|---|
| US OUT F | TCGTGTTGGGTGGCAGAAAAGC | 484 bp | Detection of Betaarterivirus suid 2 by nested RT-PCR | Christopher-Hennings et al. [30] |
| US OUT R | GCCATTCACCACACATTCTTCC | |||
| US IN F | CCAGATGCTGGGTAAGATCATC | 235 bp | ||
| US IN R | CAGTGTAACTTATCCTCCCTGA | |||
| GP5 Forward | CGGGAATTCATGTTGGGGAAATGC | 603 bp | Amplification of full-length GP5 gene | Xia et al. [42] |
| GP5 Reverse | ATACTCGAGCTAGAGACGACCCCATTG |
Results and discussion
Virus isolation
After 24 h incubation, terminally differentiated healthy PAM cells attaching to the surface could be observed (Fig. 1a). Isolation of the virus was detected by immunoperoxidase test, fluorescent antibody test as well as by nested RT-PCR. Primary porcine alveolar macrophage (PAM) cell culture is a standard system for the isolation of Betaarterivirus suid 2. This virus can be isolated in very few cell types of porcine and monkey origin like MARC-145. In our present study, we could isolate this virus in PAM cells and after two passages virus titer of 105.55 TCID50/ml could be achieved. PAM cells have been used extensively for the isolation of Betaarterivirus suid 2 in different outbreaks [1, 33]. However, a cell line derived from MA-104 cells expressing high and stable porcine CD163 named MARC-145 is a very suitable cell line for isolation of the North American genotype of the virus and is also used for vaccine production [18, 33, 34, 39]. A newly derived cell line SJPL from monkey origin was found to be permissive for this virus and reported to be an additional tool to study in vitro pathogenesis [27].
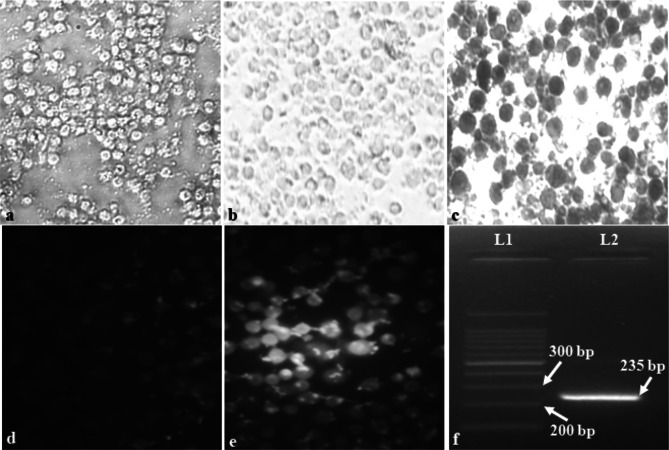
Fig. 1.
Isolation and detection of Betaarterivirus suid 2. a Harvested Healthy PAM cells (400X). b Control PAM cells after IPT staining (400X). c Infected PAM cells after IPT staining showing the color of the substrate (gray–black) (400X). d Control PAM cells after FAT staining (400X). e Infected PAM cells after FAT staining showing apple-green fluorescence (400X). f Positive amplicon of 235 bp of target product observed after nested PCR
Detection of Betaarterivirus suid 2
Virus isolation was detected by immunoperoxidase test and infected cells appeared gray-black color indicating virus infected cells (Fig. 1c) than compared with no color development in uninfected control cells (Fig. 1b). Virus growth was also confirmed by indirect fluorescent antibody test and a distinct apple-green fluorescence of infected cells (Fig. 1e) could be seen under a fluorescent microscope (Olympus, Model no. IX51) whereas uninfected control cells did not produce any fluorescence (Fig. 1d). Further virus growth was detected by molecular detection by reverse transcriptase nested polymerase chain reaction (RT-nPCR) test and targeted amplification of 235 bp product could be detected from the sample (Fig. 1f). Detection of this virus in a cell culture system can be done by indirect immunoperoxidase test as well as by fluorescent antibody test, which are standard tests for the detection of viral infection in cell culture. Viral antigen expressed on the surface of infected cells by non-cytopathic or mild cytopathic viruses can be detected by these tests as well as can be used for virus titration assay [11, 35, 37]. In viral pathogenicity studies, an immunoperoxidase test is a powerful tool for in situ detection of the virus in infected tissues [13]. FAT is a highly sensitive assay for the accurate detection of virus-laden cells. The test can detect even when a fewer number of infected cells are present in the monolayer [1, 6, 19, 36]. Molecular detection can be done by OIE recommended RT-PCR protocol which is considered to be a standard procedure for confirming the virus nucleic acid in the sample. This assay can detect both Betaarterivirus suid 1 and 2 viruses and has been reported in many literatures [9, 8, 17]. Nested PCR reactions are very sensitive and specific and can detect even from the sample with a low virus load [21]. However, the test cannot differentiate the genotypes of the virus.
Molecular characterization of GP5 gene of Betaarterivirus suid 2
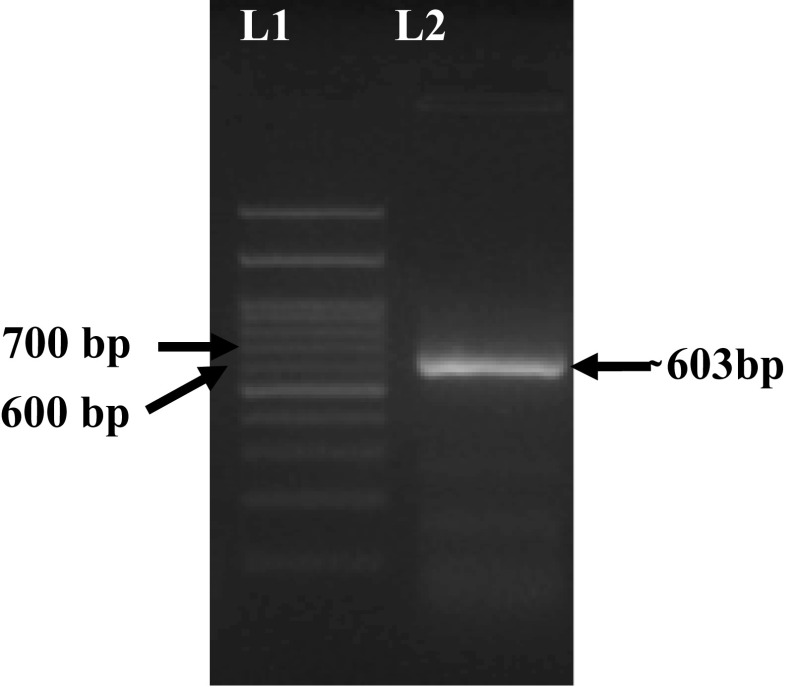
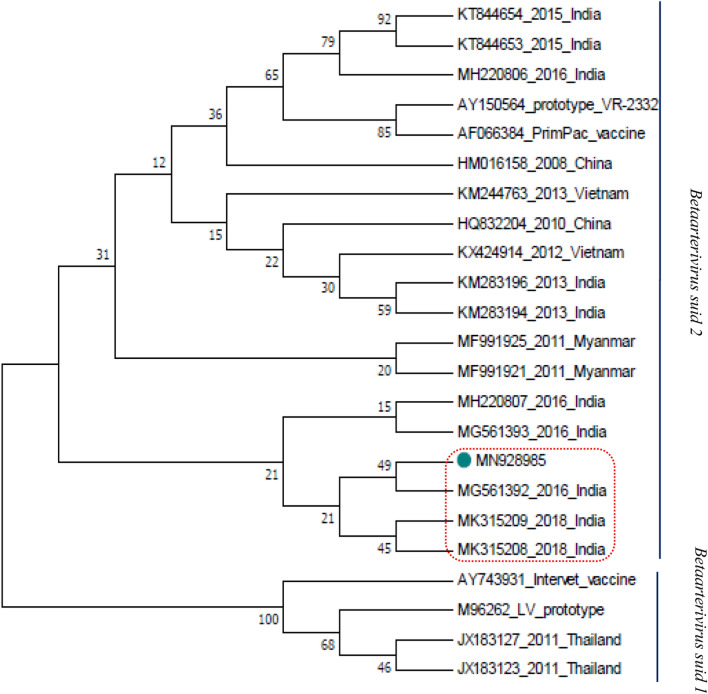
Positive amplification of full-length GP5 of 603 bp was observed after electrophoresis (Fig. 2) and the present isolate was cloned and sequenced and also annotated and submitted to the GenBank and accession number received is MN928985 for the present sequence. Phylogenetic analysis was carried out by comparing with GP5 sequences of previous outbreaks in India as well as different outbreaks of south-east Asian countries including China, prototype VR-2332, and Lelystad Virus (LV) strains and vaccine strains (Table 2). The present isolate was placed within a cluster of Indian isolates of 2016 and 2018 recorded outbreaks of Mizoram, India (Fig. 3). Sequence similarity within this cluster was ranged between 96.6 and 100%. However, the sequence of the first outbreak of PRRS in India during 2013 was found phylogenetically placed in a different clade within the same cluster. Sequence similarity with 2013 isolates was found to be 97.7–98.2%. When compared with the highly pathogenic strains from other south-east Asian countries present sequence showed a phylogenetic similarity with all those reported sequences. Sequence similarity of the present isolate was found 98.7–98.8% with Myanmar HP-PRRS strains, 98–98.5% with Vietnam strain, 98.2–98.3% with China strains (Table 3). A phylogenetic cluster of present isolate within the NA strains and a close similarity with the highly pathogenic strains indicates the present isolate is a highly pathogenic strain of Betaarterivirus suid 2 (North American genotype). All the strains of Betaarterivirus suid 1 were forming a separate branch in the phylogenetic tree and are also phylogenetically diverse from NA strains with a homology percent of 62.3%. The deduced amino acid sequence of GP5 protein of the present isolate was compared with other sequences of genotype 2 from the GenBank. The deduced amino acid sequence showed a total of 26 mutations in the signal peptide domain including decoy epitope (DCE), ectodomain including primary neutralizing epitope (PNE) (Table 4), transmembrane domain, and endodomain region. There are five N-linked (Asparagine-linked) glycosylation sites in the amino acid position viz. N30, N32/ N33/ N34, N35, N44, N51 of the GP5 glycoprotein of Betaarterivirus suid 2 [22, 38]. The first potential N-linked glycosylation site of this virus is N32/N33/N34, the second and third potential N-linked glycosylation sites are N44 and N51 respectively reported by Music and Gagnon, 2010 [22]. They also mentioned a fourth potential N-linked glycosylation site at position N30 among few NA strains such as VR-2332 and Quebec IAF Klop. N30 comes under the decoy epitope (DCE) which comprises four amino acids at position 27VLAN30 in the NA prototype VR-2332. DCE delays the neutralizing antibody response induced by primary neutralizing epitope (PNE)20. In our study we could observe the present isolate does not have N-linked glycosylation at position N30, rather the asparagine (N) has been substituted by aspartic acid (D) (30 N → D30). This change was not observed in the viruses of the 2013, 2015, and 2018 outbreaks in Mizoram. The substitution of amino acid of the present isolate was observed within the PNE in position 39 where leucine (L) has been substituted by isoleucine (I) (39L → I39) when compared with prototype strain. The same change has also been observed from Mizoram and Meghalaya isolates. In the present study, some random amino acid substitutions in the signal peptide, ectodomain, transmembrane domain, and endodomain were also observed when compared with prototype VR-2332. Amino acid substitutions observed in the position 3E → G3, 9G → C9, 10C → S10, 16S → F16, 23F → S23, 24C → Y24, 25F → L25 within the signal peptide; 34D → S34, 35S → N35, 58 N → Q58 within the ectodomain; 66S → T66, 92A → G92, 94 V → A94, 101F → Y101, 102 V → Y102, 121 T → I121, 127F → L127 within the transmembrane domain; 137A → S137, 161I → V161, 164R → G164, 170E → G170, 185 V → A185, 189I → L189, 200P → L200 within the endodomain. The similar amino acid substitutions of other isolates of 2013, 2015, and 2016 outbreaks of Mizoram were also reported by Rajkhowa et al. [30] at the position 3E → G3, 9G → C9, 10C → S10, 16S → F16, 23F → S23, 24C → Y24, 25F → L25, 34D → S34, 35S → N35, 58 N → Q58, 66S → T66, 92A → G92, 94 V → A94, 101F → Y101, 102 V → Y102, 121 T → I121, 127F → L127, 137A → S137, 161I → V161, 164R → G164, 170E → G170, 185 V → A185, 189I → L189, 200P → L200. However, some changes were not observed in our present isolate at the position 6L → L/S6, 11C → C/Y11, 34D → N34, 58 N → K58. GP5 gene (ORF5) is the most commonly targeted gene for the phylogenetic analysis of this virus strain. Since ORF5 is the most variable region containing both neutralizing and non-neutralizing epitopes, N-linked glycosylation sites, apoptotic cell death induced region; so phylogenetic differences are more prominent in this region [12, 14, 26]. The close similarity of Indian strains and their relation with other highly pathogenic strains were also reported by Rajkhowa et al. [28]; Zohlimpuia et al. [42]; Chaithra et al. [4]. If we observe the timeline of the highly pathogenic strain of Betaarterivirus suid 2 outbreaks in the south-east Asian region then it can be assumed that the virus has entered Mizoram from neighboring Myanmar. Close sequence identity within the Mizoram outbreaks was also recorded by Rajkhowa et al. [28] where a sequence similarity of 99.4–100% was observed within 2015 outbreaks and in comparison to the 2013 outbreak, it was reported 96.15–97.35%. Similarly compared with highly pathogenic 10 HEB-3 strain and 2013 outbreak was 98.33–99.17%. In another observation by Zohlimpuia et al. [42] reported 98.3–100% similarity among the four isolates of the 2016 outbreak in Mizoram, 98.2–99.7% similarity with Meghalaya isolates. He also observed 98–99% sequence similarity with the highly pathogenic strains of the 2011 outbreak in Myanmar. Amino acid substitution at DCE was also reported by Yu et al. [41] and Hong et al. [15] from China. N-linked glycan of GP5 ectodomain is important for the biological activity of the protein. Modification of this glycosylation site is one way of altering the receptor binding site, membrane fusion, and entry and exit process from the infected cells. Studies on enveloped viruses revealed that glycosylation of viral envelope protein is a major mechanism of immune escape as well as viral persistence or minimize the virus-neutralizing antibody response [2, 31]. Ansari et al. [2] reported that the loss of glycan residues in positions N34, N44, N51 enhances the immunogenicity of the nearby neutralizing epitope. The PNE within GP5 of NA prototype VR-2332 comprises 9 amino acids ranging from position 37 to 45 (37SHLQLIYNL45). Substitution within PNE was also observed among HP-PRRS strains in China (JXA1, HUN4, JXwn06) which was assumed to be due to large scale vaccination with modified live vaccines (MLV) in China. This type of alteration in neutralizing epitope maybe because of the immune escape strategy adapted by the virus as mentioned by Hong et al. [15].
Fig. 2.
Full-length GP5 amplification of Betaarterivirussuid 2
Table 2.
Sequence used for analysis of the present isolate
| Accession number | Country | Year | Strain/isolate | Genotype |
|---|---|---|---|---|
| MN928985 | India | 2016 | MZ/5/2016 | Betaarterivirus suid 2 |
| AY150564 | North America | 2002 | VR-2332 | Prototype of Betaarterivirus suid 2 |
| M96262 | Netherlands | 2000 | Lelystad virus | Prototype of Betaarterivirus suid 1 |
| KM283195 | India | 2013 | PRRS/MZ/AZ/50/13 | Betaarterivirus suid 2 |
| KM283196 | India | 2013 | PRRS/MZ/AZ/49/13 | Betaarterivirus suid 2 |
| KT844653 | India | 2015 | PRRS/MZ/IND/5/15 | Betaarterivirus suid 2 |
| KT844654 | India | 2015 | PRRS/MZ/IND/2/15 | Betaarterivirus suid 2 |
| MG561392 | India | 2016 | MZ/1/2016 | Betaarterivirus suid 2 |
| MG561393 | India | 2016 | MZ/2/2016 | Betaarterivirus suid 2 |
| MK315208 | India | 2018 | PRRSV/MZ/IND/399A/18 | Betaarterivirus suid 2 |
| MK315209 | India | 2018 | PRRS/MZ/IND/DBT1/18 | Betaarterivirus suid 2 |
| MH220806 | India | 2016 | ML-8 | Betaarterivirus suid 2 |
| MH220807 | India | 2016 | ML-9 | Betaarterivirus suid 2 |
| MF991921 | Myanmar | 2011 | HP/MYANMAR/1908AM/2011 | Betaarterivirus suid 2 |
| MF991925 | Myanmar | 2011 | HP/MYANMAR0411AM/2011 | Betaarterivirus suid 2 |
| KX424914 | Vietnam | 2012 | HUA/HP5 | Betaarterivirus suid 2 |
| KM244763 | Vietnam | 2013 | MN1 | Betaarterivirus suid 2 |
| HQ832204 | China | 2010 | HUBWH10 | Betaarterivirus suid 2 |
| JQ663553 | China | 2010 | 10-10HEB-3 | Betaarterivirus suid 2 |
| JX183123 | Thailand | 2011 | SCP0311EU1/1 | Betaarterivirus suid 1 |
| JX183127 | Thailand | 2011 | FDT0111EU2/1 (clone) | Betaarterivirus suid 1 |
| AY743931 | Italy | 2004 | Intervet vaccine strain | Betaarterivirus suid 1 |
| AF066384 | USA | 1998 | PrimePac PRRS vaccine acronym: PRRS virus | Betaarterivirus suid 2 |
Fig. 3.
Phylogenetic tree generated on the basis of full-length GP5 (603nt)of present isolate when compared with sequences of previous outbreaks in India and other countries(both type1 and type 2 strains) including vaccine strains using MEGA9 software
Table 3.
Sequence homology study of present isolate with other published isolates
| Percent identity with published sequences | GenBank published sequences |
|---|---|
| 88.3 | AY150564 prototype VR-2332 |
| 97.7 | KM283194 2013 Mizoram, Pig |
| 98.2 | KM283196 2013 Mizoram, Pig |
| 96.2 | KT844653 2015 Mizoram, Pig |
| 95.9 | KT844654 2015 Mizoram, Pig |
| 100 | MG561392 Mizoram 2016, Pig lung Lwangtlai |
| 99.2 | MG561393 Mizoram 2016, Pig lung Saiha |
| 98.8 | MK315208 Mizoram 2018 |
| 98.3 | MK315209 Mizoram 2018 |
| 96.6 | MH220806 Meghalaya 2016, Pig serum |
| 99 | MH220807 Meghalaya 2016, Pig serum |
| 98.8 | MF991921 Myanmar 2011, highly pathogenic strain, serum, wild boar |
| 98.7 | MF991925 Myanmar 2011, highly pathogenic strain, serum, wild boar |
| 98.5 | KX424914 Vietnam 2012, NA, wild boar |
| 98 | KM244763 Vietnam 2013, highly pathogenic strain, Pig |
| 98.2 | HM016158 China 2008, highly pathogenic strain, Pig |
| 98.3 | HQ832204 China 2010, NA, Pig |
| 89.8 | AF066384 USA, PrimePac PRRS Vaccine |
| 63.3 | M96262 Lelystad virus, Protoype Betaarterivirus suid 1 |
| 63.3 | AY743931 Italy, Intervet Vaccine |
| 62.5 | JX183123 Thailand 2011, Betaarterivirus suid 1, Pig |
| 62.4 | JX183127 Thailand 2011, Betaarterivirus suid 1, Pig |
Table 4.
Amino acid sequence of decoy epitope (DCE) and primary neutralizing epitope (PNE) of GP5
| Accession no | Decoy epitope (DCE) (27VLAN30) | Primary Neutralizing Epitope (PNE) (37SHLQLIYNL45) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 27 | 28 | 29 | 30 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | |
| AY150564 VR-2332 | V | L | A | N | S | H | L | Q | L | I | Y | N | L |
| MN928985 (Present study) MZ/5/2016 Mizoram 2016 | V | L | A | D | S | H | I | Q | L | I | Y | N | L |
| KM283196 Mizoram 2013 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| KT844653 Mizoram 2015 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| KT844654 Mizoram 2015 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| MG561392 Mizoram 2016 | V | L | A | D | S | H | I | Q | L | I | Y | N | L |
| MG561393 Mizoram 2016 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| MH220806 Meghalaya 2016 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| MH220807 Meghalaya 2016 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| MK315208 Mizoram 2018 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| MK315209 Mizoram 2018 | V | L | V | N | S | H | I | Q | L | I | Y | N | L |
| MF991921 Myanmar 2011 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| MF991925 Myanmar 2011 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| KX424914 Vietnam 2012 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| KM244763 Vietnam 2013 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| HM016158 China 2008 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| HQ832204 China 2010 | V | L | A | N | S | H | I | Q | L | I | Y | N | L |
| AF066384 PrimPac Vaccine | V | L | V | N | S | H | L | Q | L | I | Y | N | L |
Conclusion
In conclusion, based on the complete gene sequence of Betaarterivirus suid 2 GP5 gene analysis of one isolate of 2016 outbreak in Mizoram revealed that the isolate belonged to the highly pathogenic strain which is circulating in South East Asian countries. Our phylogenetic analysis indicates that 95.9–100% sequence similarity with previous outbreaks in Mizoram and 98.7–98.8% with neighboring Myanmar isolates. Furthermore, deduced amino acid sequence analysis reveals a mutation in the primary neutralizing epitope (PNE) as well as an amino acid substitution in decoy epitope which has not been reported earlier from India. Periodical sequence analysis of the virus associated with the outbreak is necessary to understand the disease status in this region.
Acknowledgements
Authors are thankful to the Dean, College of Veterinary Sciences and Animal Husbandry, and Vice-Chancellor, Central Agricultural University for providing the financial support and necessary facility to complete the research studies.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.De Abin MF, Spronk G, Wagner M, Fitzsimmons M, Abrahante JE, Murtaugh MP. Comparative infection efficiency of porcine reproductive and respiratory syndrome virus field isolates on MA104 cells and porcine alveolar macrophages. Can J Vet Res. 2009;73:200–204. [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari IH, Kwon B, Osorio FA, Pattnaik AK. Influence of N-linked glycosylation of porcine reproductive and respiratory syndrome virus GP5 on virus infectivity, antigenicity and ability to induce neutralizing antibodies. J Virol. 2006;80:3994–4004. doi: 10.1128/JVI.80.8.3994-4004.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai J, Ma Y, Li J, Yan C, Hu R, Zhang J. Construction and characterization of a recombinant CanineAdenovirus expressing GP5 and M proteins of porcine reproductive and respiratory syndrome virus. J Vet Med Sci. 2010;72:1035–1040. doi: 10.1292/jvms.10-0061. [DOI] [PubMed] [Google Scholar]
- 4.Chaithra G, Ravishankar C, Sebastian SR, Rajasekhar R, Anoopraj R, Mani BK, Sumod K, Deorao CV, Logeshwaran G, Nandhakumar D, John K. Molecular characterisation of porcine reproductive and respiratory syndrome virus from pigs in Kerala. Virus Dis. 2020;31:560–564. doi: 10.1007/s13337-020-00634-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen C, Fan W, Jia X, Li J, Bi Y, Liu W. Development of a recombinant N-GP5c fusion protein-based ELISA for detection of antibodies porcine reproductive and respiratory syndrome virus. J Virol Methods. 2013;189:213–220. doi: 10.1016/j.jviromet.2013.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Chiou MT, Jeng CR, Chueh LL, Cheng CH, Pang VF. Effects of porcine reproductive and respiratory syndrome virus (isolate tw91) on porcine alveolar macrophages in vitro. Vet Microbiol. 2000;71:9–25. doi: 10.1016/S0378-1135(99)00159-5. [DOI] [PubMed] [Google Scholar]
- 7.Choi HW, Nam E, Lee YJ, Noh YH, Lee SC, Yoon IJ, Kim HS, Kang SY, Choi YK, Lee C. Genomic analysis and pathogenic characteristics of type 2 porcine reproductive and respiratory syndrome virus nsp2 deletion strains isolated in Korea. Vet Mic. 2014;170:232–245. doi: 10.1016/j.vetmic.2014.02.027. [DOI] [PubMed] [Google Scholar]
- 8.Christopher-Hennings J, Holler LD, Benfield DA, Nelson EA. Detection and duration of porcine reproductive and respiratory syndrome virus in semen, serum, peripheral blood mononuclear cells, and tissues from Yorkshire, Hampshire, and Landrace boars. J Vet Diagn Invest. 2001;13:133–142. doi: 10.1177/104063870101300207. [DOI] [PubMed] [Google Scholar]
- 9.Christopher-Hennings J, Nelson EA, Nelson JK, Hines RJ, Swenson SL, Hill HT, Zimmerman JJ, Katz JB, Yaeger MJ, Chase CCL, Benfield DA. Detection of porcine reproductive and respiratory syndrome in boar semen by PCR. J Clin Microbiol. 1995;33:1730–1734. doi: 10.1128/jcm.33.7.1730-1734.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cui J, O’Connell CM, Costa A, Pan Y, Smyth JA, Verardi PH, Burgess DJ, Kruininjen HJV, Germandia AE. A PRRSV GP5-Mosaic vaccine: protection of pigs from challenge and ex vivo detection of IFN-γ responses against several genotypes 2 strains. PLoS ONE. 2019;14:e0208801–e208815. doi: 10.1371/journal.pone.0208801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Decorte I, Van Breedam W, Van der Stede Y, Nauwynck HJ, De Regge N, Cay AB. Detection of total and PRRSV-specific antibodies in oral fluids collected with different rope types from PRRSV-infected and experimentally infected pigs. BMC Vet Res. 2014;10:134–144. doi: 10.1186/1746-6148-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez A, Suarez P, Castro JM, Tabares E, Guerra MD. Characterization of regions in the GP5 protein of porcine reproductive and respiratory syndrome virus required to induce apoptotic cell death. Virus Res. 2002;83:103–118. doi: 10.1016/S0168-1702(01)00426-9. [DOI] [PubMed] [Google Scholar]
- 13.Halbur PG, Andrews JJ, Huffman EL, Paul PS, Meng XJ, Niyo Y. Development of a streptavidin-biotin immunoperoxidase procedure for the detection of porcine reproductive and respiratory syndrome virus antigen in porcine lung. J Vet Diagn Invest. 1994;6:254–257. doi: 10.1177/104063879400600219. [DOI] [PubMed] [Google Scholar]
- 14.Han J, Wang Y, Faaberg KS. Complete genome analysis of RFLP 184 isolates of porcine reproductive and respiratory syndrome virus. Virus Res. 2006;122:175–182. doi: 10.1016/j.virusres.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 15.Hong S, Wei Y, Lin S, Huang J, He W, Yao J, Chen Y, Kang O, Huang W, Wei Z. Genetic analysis of porcine productive and respiratory syndrome virus between 2013 and 2014 in southern parts of China: identification of several novel strains with amino acid deletions or insertions in nsp2. BMC Vet Res. 2019;15:171–181. doi: 10.1186/s12917-019-1906-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kedkovid R, Sirisereewan C, Thanawongnuwech R. Major swine viral diseases: an Asian perspective after the African swine fever introduction. Porc Health Manag. 2020;6:20–30. doi: 10.1186/s40813-020-00159-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kleiboeker SB, Schommer SK, Lee SM, Watkins S, Chittick W, Polson D. Simultaneous detection of North American and European porcine reproductive and respiratory syndrome virus using real-time quantitative reverse transcriptase–PCR. J Vet Diagn Invest. 2005;17:165–170. doi: 10.1177/104063870501700211. [DOI] [PubMed] [Google Scholar]
- 18.Kwon T, Yoo SJ, Park JW, Kang SC, Park CK, Lyoo YS. Genomic characteristics and pathogenicity of natural recombinant porcine reproductive and respiratory syndrome virus 2 harboring genes of a Korean field strain and VR-2332-like strain. Virol. 2019;530:89–98. doi: 10.1016/j.virol.2019.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee YJ, Park CK, Nam E, Kim SH, Lee OS, Lee DS, Lee C. Generation of a porcine alveolar macrophage cell line for the growth of porcine reproductive and respiratory syndrome virus. J Virol Methods. 2010;163:410–415. doi: 10.1016/j.jviromet.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Mayer P, Lam C. Porcine alveolar macrophages, isolation, morphological and functional characteristics. Zbl Vet Med A. 1984;31:59–71. doi: 10.1111/j.1439-0442.1984.tb01262.x. [DOI] [PubMed] [Google Scholar]
- 21.Mijatovic-Rustempasic S, Esona MD, Williams AL, Bowen MD. Sensitive and specific nested PCR assay for detection of Rotavirus A in samples with low virus load. J Virol Methods. 2016;236:41–46. doi: 10.1016/j.jviromet.2016.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Music N, Gagnon CA. The role of porcine reproductive and respiratory syndrome (PRRS) virus structural and non-structural proteins in virus pathogenesis. Anim Health Res Rev. 2010;11:135–163. doi: 10.1017/S1466252310000034. [DOI] [PubMed] [Google Scholar]
- 23.Nelsen CJ, Murtaugh MP, Faaberg KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. 1999;73:270–280. doi: 10.1128/JVI.73.1.270-280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.OIE. Porcine reproductive and respiratory syndrome. In: OIE Terrestrial Manual. 2018;1579–1593. Accessed 16 March 2021.
- 25.Park SI, Seo JY, Kim TJ. Heterologous expression of porcine reproductive and respiratory syndrome virus glycoprotein 5 in Bordetella bronchiseptica aroA mutant. J Vet Med Sci. 2016;78:1625–1629. doi: 10.1292/jvms.15-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirzadeh B, Gagnon CA, Dea S. Genomic and antigenic variations of porcine reproductive and respiratory syndrome virus major envelope GP5 glycoprotein. Can J Vet Res. 1998;62:170–177. [PMC free article] [PubMed] [Google Scholar]
- 27.Provost C, Jia JJ, Music N, Levesque C, Lebel ME, Castillo JRE, Jacques M, Gagnon CA. Identification of a new cell line permissive to porcine reproductive and respiratory syndrome virus infection and replication which is phenotypically distinct from MARC-145 cell line. Virol J. 2012;9:267–280. doi: 10.1186/1743-422X-9-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajkhowa TK, Mohanarao GJ, Gogoi A, Hauhnar L. Indian porcine reproductive and respiratory syndrome virus bears discontinuous deletion of 30 amino acids in nonstructural protein 2. Virus Dis. 2016;27:287–293. doi: 10.1007/s13337-016-0341-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajkhowa TK, Mohanarao GJ, Gogoi A, Hauhnar L, Isaac L. Porcine reproductive and respiratory syndrome virus (PRRSV) from the first outbreak of India shows close relationship with the highly pathogenic variant of China. Vet Q. 2015;4:186–193. doi: 10.1080/01652176.2015.1066043. [DOI] [PubMed] [Google Scholar]
- 30.Rajkhowa TK, Vanlalruati C, Singh YD, Ravindran R, Arya RS. Genetic variation of highly pathogenic Indian porcine reproductive and respiratory syndrome viruses after introduction in 2013. Indian J Anim Sci. 2018;88:1118–1126. [Google Scholar]
- 31.Reitter JN, Means RE, Desrosiers RC. A role for carbohydrates in immune evasions in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 32.Roques E, Girard A, Louis MCS, Massie B, Gagnon CA, Lessard M, Archambault D. Immunogenic and protective properties of GP5 and M structural proteins of porcine reproductive and respiratory syndrome virus expressed from replicating but non-disseminating adenovectors. Vet Res. 2013;44:17–29. doi: 10.1186/1297-9716-44-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Senthilkumar D, Rajukumar K, Kumar M, Kalaiyarasu S, Shrivastava D, Katare M, Kulkarni DD, Singh VP. Porcine reproductive and respiratory syndrome virus induces concurrent elevation of High Mobility Group Box-1 protein and pro-inflammatory cytokines in experimentally infected piglets. Cytokine. 2018;113:21–30. doi: 10.1016/j.cyto.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 34.Senthilkumar D, Rajukumar K, Sen A, Kumar M, Shrivastava D, Kalaiyarasu S, Gautam S, Singh F, Kulkarni DD, Singh VP. Pathogenic characterization of porcine reproductive and respiratory syndrome virus of Indian origin in experimentally infected piglets. Transbound Emerg Dis. 2018:1–15. [DOI] [PubMed]
- 35.Takashima H, Takai K, Goto S. A modified immunoperoxidase assay for detection of antibody porcine reproductive and respiratory syndrome (PRRS) virus in swine sera. J Vet Med Sci. 1999;61:195–196. doi: 10.1292/jvms.61.195. [DOI] [PubMed] [Google Scholar]
- 36.Tong GZ, Zhou YJ, Hao XF, Tian ZJ, An TQ, Qiu HJ. Highly pathogenic porcine reproductive and respiratory syndrome. China Emerg Infect Dis. 2007;13:1434–1435. doi: 10.3201/eid1309.070399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vanhee M, Delputte PL, Delrue I, Geldhof MF, Nauwynck HJ. Development of an inactivated PRRSV vaccine that induces virus-neutralizing antibodies. Vet Res. 2009;40:63. doi: 10.1051/vetres/2009046. [DOI] [PubMed] [Google Scholar]
- 38.Wei Z, Lin T, Sun L, Li Y, Wang X, Gao F, Liu R, Chen C, Tong G, Yuan S. N-linked glycosylation of GP5 of porcine reproductive and respiratory syndrome virus is critically important for virus replication in vivo. J Virol. 2012;86:9941–9951. doi: 10.1128/JVI.07067-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu X, Qi J, Cong X, Chen L, Hu Y, Yoo D, Wang G, Tian F, Li F, Sun W, Chen Z, Guo L, Wu J, Li J, Wang J, Zhao X, Du Y. Establishment and characterization of a high and stable porcine CD163-expressing MARC-145 cell line. Biomed Res Int. 2018;2018:4315861. doi: 10.1155/2018/4315861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xia P, Dang Z, Qiu H, Fan X, Zhou B, Cui B, Chen P. Function of PRRSV GP5 envelope protein by using pseudotyped virus. Vet Mic. 2009;138:297–303. doi: 10.1016/j.vetmic.2009.04.013. [DOI] [PubMed] [Google Scholar]
- 41.Yu L, Zhao P, Dong J, Liu Y, Zhang L, Liang P, Wang L, Song C. Genetic characterization of 11 porcine reproductive and respiratory syndrome virus isolates in South China from 2014 to 2015. Virol J. 2017;14:139–148. doi: 10.1186/s12985-017-0807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zohlimpuia RP, Subudhi PK, Dutta TK, Behera PS, Kalita G. Molecular detection and characterization of Orf5 gene of Porcine reproductive and respiratory syndrome virus from outbreaks in Mizoram. Indian J Comp Microbiol Immunol Infect Dis. 2018;39:101–104. [Google Scholar]