Abstract
COVID-19 is the cause of a pandemic associated with substantial morbidity and mortality. As yet, there is no available approved drug to eradicate the virus. In this review article, we present an alternative study area that may contribute to the development of therapeutic targets for COVID-19. Growing evidence is revealing further pathophysiological mechanisms of COVID-19 related to the disregulation of inflammation pathways that seem to play a critical role toward COVID-19 complications.
The NF-kB and JAK/STAT signaling pathways are highly activated in acute inflammation, and the excessive activity of these pathways in COVID-19 patients likely exacerbates the inflammatory responses of the host. A group of non-coding RNAs (miRNAs) manage certain features of the inflammatory process. In this study, we discuss recent advances in our understanding of miRNAs and their connection to inflammatory responses. Additionally, we consider the link between perturbations in miRNA levels and the onset of COVID-19 disease.
Furthermore, previous studies published in the online databases, namely web of science, MEDLINE (PubMed), and Scopus, were reviewed for the potential role of miRNAs in the inflammatory manifestations of COVID-19. Moreover, we disclosed the interactions of inflammatory genes using STRING DB and designed interactions between miRNAs and target genes using Cityscape software. Several miRNAs, particularly miR-9, miR-98, miR-223, and miR-214, play crucial roles in the regulation of NF-kB and JAK-STAT signaling pathways as inflammatory regulators. Therefore, this group of miRNAs that mitigate inflammatory pathways can be further regarded as potential targets for far-reaching-therapeutic strategies in COVID-19 diseases.
Keywords: COVID-19, miRNA, Inflammation, NF-kB, JAK/STAT
Abbreviations: COVID-19, Coronavirus disease 2019; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; NF-kB, Nuclear factor kappa B; JAK, Janus kinase; STAT, Signal transducers and activators of transcription; IL‐1, Interleukin-1; IL‐6, Interleukin-6; IL‐8, Interleukin-8; IL‐21, Interleukin-21; TNF‐β, Tumor necrosis factor-beta; MCP‐1, Monocyte chemoattractant protein-1; DPP4R, Dipeptidyl peptidase-4 receptor; TLR‐3, Toll-like receptor 3; IRFs, Interferon-regulatory factors; IFNs, Interferons; RAS, Renin–angiotensin–system; Ang I, Angiotensin I; Ang II, Angiotensin II; ACE, Angiotensin-converting enzyme; TNF-α, Tumor necrosis factor alpha; AT1R, Angiotensin II type I receptor; MCP-1, Monocyte chemoattractant protein-1*; TGF-β1, Transforming growth factor beta 1; CXCR2, Chemokine receptor 2; MCP-1/CCL2, Monocyte chemotactic protein; CCR2, Chemokine receptor type 2; IP-10, Interferon IFN-γ inducible protein-10; MIP1-α, Macrophage inflammatory protein 1-α; CRP, C-reactive protein; PCT, Procalcitonin; ESR, Erythrocyte sedimentation rate; B-cells inhibitor, Nuclear factor of kappa light polypeptide gene enhancer in; IκB-α, alpha; BALF, Bronchoalveolar lavage fluid; MIP-1A, Macrophage inflammatory protein-1α; IP-10, interferon gamma-induced protein 10; MIP1B, Macrophage inflammatory protein-1 beta; NK, Natural killer; FABP4, cells Fatty acid binding protein 4; CD8, Cluster of differentiation 8; CD4, Cluster of differentiation 4; DEGs, Differentially expressed genes; ISGs, Interferon stimulated genes; NLR, Neutrophil to lymphocyte ratio; IFITM3, Interferon-induced transmembrane protein 3; SOCS1, Suppressor of cytokine signaling 1; SOCS3, Suppressor of cytokine signaling 3; PCBP2, Poly (rC)-binding protein 2; MAPK, Mitogen‑activated protein kinase; VLDLR, Very low-density lipoprotein receptors; CNS, Central nervous system; EGF, Epidermal growth factor; PKC, protein kinase; ERK, Extracellular signal-regulated kinase; TLRs, Toll-like receptors; RhoB, Ras homolog family member B; GCS-F, Granulocyte colony-stimulating factor; GP130, Glycoprotein 130; MCPIP1, Monocyte chemotactic protein-1-induced protein-1; PBMCs, Peripheral blood mononuclear cells; SLE, Lupus erythematosus; IGF1R/PI3K, Insulin growth factor I receptor/phosphatidylinositol-3-kinase; KD, Kawasaki disease; IFNGR1, Interferon gamma receptor 1; NLRP3, NLR family pyrin domain containing 3; PIAS3, protein inhibitor of activated STAT 3; eMDSCs, early-stage myeloid-derived suppressor cells; PTEN, Phosphatase and tensin homolog; IgAN, Immunoglobulin a nephropathy; EDAR, Ectodysplasin A receptor; RIPK1, Receptor-interacting protein kinase 1; TRADD, TNFR1 associated death domain protein; TRAF2, Tumor necrosis factor receptor associated factor-2; TRAF5, Tumor necrosis factor receptor associated factor-5; NFKBIA, NFKB inhibitor alpha; TRIM25, Tripartite motif containing 25; ICAM1, Intercellular adhesion molecule 1
1. Introduction
The coronavirus disease 2019 (COVID-19) is the cause of morbidity and mortality, which is associated with severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection that is characterized by acute respiratory failure. Usually, COVID-19 is manifested as a mild respiratory disease, although it can occasionally lead to severe alveolar inflammation. Lung histopathological findings of patients with COVID-19 showed extensive alveolar damaging via widespread infiltration of immune cells, thrombosis of pulmonary vessels and fibrin deposits [8], [44].
In patients with COVID-19, an excessive or uncontrolled innate immune response leads to a cytokine storm and acute respiratory distress syndrome (ARDS). Infected macrophages with COVID-19 present COVID-19 antigens to T cells. Activated T cells produce cytokines that are associated with different T cell subtypes, and following the release of large quantities of cytokines, there is immune response amplification. In response to COVID-19 infection, the secretion of massive chemokines and cytokines like Interleukin-1 (IL‐1), Interleukin-6 (IL‐6), Interleukin-8 (IL‐8), Interleukin-21 (IL‐21), tumor necrosis factor-beta (TNF‐β), and monocyte chemoattractant protein-1 (MCP‐1) is promoted in infected cells. Afterward, these chemokines and cytokines, in turn, recruit lymphocytes and leukocytes to the infection site. CD8 T cells produce very effective mediators to destroy COVID-19, but the continued release of these mediators led to viral persistence and a negative effect on CD8 T cell activation [15].
S protein of COVID-19 attaches to dipeptidyl peptidase-4 receptor (DPP4R) on the host cell and leads to the appearance of genomic RNA (dsRNA) in the cytoplasm of the host cell. Toll-like receptor 3 (TLR‐3) sensitized by dsRNA and signaling cascades of interferon-regulatory factors (IRFs) and nuclear factor-kB (NF‐kB) are activated and then produced interferons (IFNs) (IFN-I, IFN-II, and IFN-III) and pro-inflammatory cytokines. The engagement of IFN (I and III) receptors induced different members of the Janus kinase (JAK) and signal transducer and activator of transcription (STAT) families and also helped form specific transcription factor complexes. In contrast, IFN-II activated JAK1 and JAK2, producing a phosphorylated STAT1 homodimer [88].
Small molecules (17–24nt) of non-coding RNAs known as miRNAs regulate the post-transcriptional expression thought to block the target genes by binding to the 3′UTR during transcription, repressing the messenger RNA, and promoting their degradation by cleavage in various cellular processes, such as cell proliferation, apoptosis, and differentiation. More than two thousand miRNAs have been identified in humans. One miRNA targets several genes, while one gene is targeted by multiple miRNAs. Some miRNAs are produced during viral infection, modulating the function of the immune response against respiratory viruses [2], [88]. This review will analyze the role of miRNAs in the regulation of immune response against COVID-19 especially inflammatory pathway based on bioinformatics prediction.
2. The role of JAK/STAT and NF-kB signaling pathways during viral infections particularly SARS-CoV-2
The renin–angiotensin–system (RAS) is a cascade, which results in the conversion of the inactive decapeptide angiotensin I (Ang I) into the active peptide angiotensin II (Ang II). The initial enzyme in the cascade is renin, an aspartic proteinase, which cleaves the amino terminus of the angiotensinogen, thereby releasing the decapeptide pro-hormone Ang I. Angiotensin-converting enzyme (ACE) removes two additional amino acids to yield the active octapeptide hormone Ang II [103]. Ang II exerts its actions via the (JAK-STAT) signaling pathway, which leads to vasoconstriction, hypertension, and chronic tissue injury [3], [75]. Renin-Angiotensin system mediates pro-inflammatory cytokine production. Indeed, Ang II increases the infiltration of immune cells, which, in turn, augments the local production of pro-inflammatory cytokines such as interferon (IFN)-γ, IL-1, IL-6, and tumor necrosis factor alpha (TNF-α) in target tissues [75].
Activation of the Ang II signaling by JAK STAT pathway can occur in the mesangial cells, hepatocytes, cardiovascular system, renal proximal tubular cells, and brainstem astrocytes [25]. In the pathogenesis of ARDS, Ang II increases through ACE and drives severe lung failure through the Angiotensin II type I receptor (AT1R) [29]. AT1R recruits and phosphorylates JAK2, and, based on the target cell, one of the STAT is phosphorylated by activated JAK2 (F. et al., 2020). In COVID-19, macrophage infiltration into the lung tissue occurs, which is accompanied by the apoptosis of epithelial cells and pneumocytes. Infiltration of macrophages into alveolar cavities may be mediated by monocyte chemoattractant protein-1 (MCP-1) through transforming growth factor beta 1 (TGF-β1) and TNF-α, which leads to cytokine production (He et al., 2008). Markers of infiltration include the neutrophil chemokine receptor 2 (CXCR2), along with monocyte chemotactic protein (MCP-1/CCL2) and its receptor chemokine receptor type 2 (CCR2). NF-kB signaling pathway genes (NFKBIA, NFKB1, RELA, NFKB2) were upregulated in COVID-19 patients [30], [77]. Cytokine storm is emerging as one of the mechanisms leading to ARDS, defined as the release of high concentrations of pro-inflammatory cytokines, including IL-1, IL-2, IL-6, IL-7, IL-10, TNF-α, granulocytecolony stimulating factor, interferon (IFN)-γ inducible protein-10 (IP-10), MCP-1, and macrophage inflammatory protein 1-α (MIP1-α) by immune cells. The cytokine storm initiates a fulminant response by the immune system, leading to multisystemic organ dysfunction and even death in severe COVID-19 [13]. Elevated concentrations of inflammatory markers including IL-6, c-reactive protein (CRP), procalcitonin (PCT), and erythrocyte sedimentation rate (ESR) are detected in fatal cases [96]. Cytokine storm on the IL-6/TNF-α axis appears to be mediated likely through the phosphorylation of the NF-kB subunit p65. In SARS-CoV infection, the viral spike protein induces the activation of NF-kB via the nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (IkB-α) degradation, resulting in the production of IL-6 and TNF-α [84]. The viral nucleocapsid protein of SARS-CoV can also bind to the NF-kB regulatory element on the IL-6 promoter, with the highest activity in the presence of the p65 subunit [99]. Regulatory elements in the ACE2 gene control the transcription of PIR (pirin), a negative regulator of NF-kB subunit RELA (p65). SARS-CoV-2 disruption of ACE2 has been proposed to decrease PIR expression [26]. PIR has been proposed to act as a reversible switch that provides the NF-kB response with changes in the level of redox (oxidative stress) in the cell nucleus [50]. Repression of PIR ablates the inhibition of IL-6 expression [90]. Studies in addressing bronchoalveolar lavage fluid (BALF) show promise since findings in the blood do not essentially describe the events occurring in tissues directly affected by the infection [28].
In a study conducted by Xiong et al [93], a comparison of the cytokine genes demonstrated that in BALFs of three COVID-19 patients, the genes for IL-10, CCL2/MCP-1 (together with its CCR2 receptor), macrophage inflammatory protein-1α (CCL3/MIP-1A) together with its CCR5 receptor, interferon gamma-induced protein 10 (CXCL10/IP-10), and macrophage inflammatory protein-1 beta (CCL4/MIP1β) increased. Interestingly, in PBMC, genes related to apoptosis, autophagy, and p53 pathways increased; a finding that could be related to the lymphopenia found in the COVID-19 patients. Moreover, IL-6 transcripts were not enhanced in PBMCs, although the patients had high plasma contents of such cytokine, suggesting that circulating IL-6 could have been produced in the lungs, either by alveolar epithelial cells or by recruited inflammatory cells.
One study showed that in BALFs of patients with severe COVID-19, macrophages were predominant, with a minor proportion of T and natural killer (NK) cells, compared with moderate COVID-19 patients. Fatty acid binding protein 4 (FABP4) produced by alveolar macrophages was preferentially expressed in healthy controls and in patients who had a moderate COVID-19 infection, while FCN1 (monocyte-derived) and SPP1 (pro-fibrotic) were expressed in patients with severe COVID-19. Besides, these results suggest that during COVID-19 infection, inflammatory monocytes (FCN1+) are recruited from the circulation into the lungs, where they differentiate into SPP1+ macrophages, constituents of the severe inflammatory reaction. Analysis of the BALF transcriptome displayed that T and NK cells increased in COVID-19 patients in comparison with healthy controls, which according to their gene expression can be classified in NK, cluster of differentiation 8 (CD8), (cluster of differentiation 4 (CD4), Tregs, and proliferating cells. Interestingly, genes related to activating molecules, migration, calcium signaling, and effector molecules were highly expressed by CD8 cells in patients who had a moderate COVID-19 infection compared with those with severe COVID-19 infection; this further supports the role of CD8 cells in the elimination of the virus and their subsequent protective immunity. In contrast, higher expression of genes related to proliferation, energy generation, and initiation of translation was seen in patients with severe disease, suggesting that in patients with moderate COVID-19 infection, CD8 cells are more differentiated and efficient, while in severe infection, T cells are in a proliferative stage. Furthermore, the finding that the TCR repertoire is higher in CD8 than CD4 cells suggests a larger clonal expansion of the CD8 cells taking part in the resolution of the infection [47]. The results of Zhou et al. [102] demonstrated that in the BALF of patients with COVID-19, the differentially expressed genes (DEGs) included more pro-inflammatory chemokines, such as CXCL2, CXCL8, and, CXCL17, as well as the CXCR2 receptor, essential to neutrophil recruitment, and CCL2 and CCL7, needed for monocyte recruitment. Besides, in patients with COVID-19, IL-1b, antiviral interferon stimulated genes (ISGs), and genes related to the TNF, IL-17, and NF-kB signaling pathways increased. Furthermore, the cellular analysis demonstrated an enhanced neutrophil to lymphocyte ratio (NLR) in COVID-19 patients compared with other pneumonias patients.
3. MiRNA biogenesis
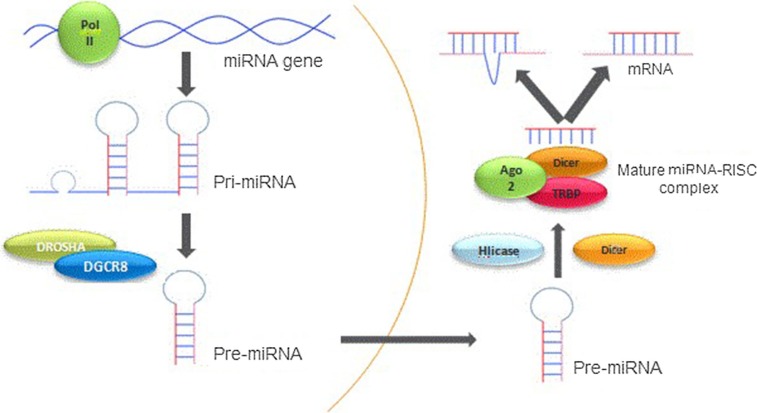
The miRNA is referred to a short RNA originated from a precursor with a stem-loop structure that interferes mainly with the regulation of gene expression by targeting mRNAs and dissecting or inhibiting their expression [87] (Fig. 1 ).
Fig. 1.
MiRNA biogenesis. The biogenesis of miRNAs begins with the production of the pri-miRNA transcript. The microprocessor complex which is consisted of Drosha and DiGeorge Syndrome Critical Region 8 (DGCR8), generates the pre-miRNA via cleaving the pri-miRNA. The pre-miRNA is exported to the cytoplasm in an Exportin5/RanGTP-dependent manner and processed to form the mature miRNA duplex. Eventually, either the 5p or 3p strands of the mature miRNA duplex is loaded into the Argonaute (AGO) family of proteins to form a miRNA-induced silencing complex (miRISC) and regulates gene expression [65].
Seven nucleotides situated at the 5′-end of mature miRNA molecules comprise what is known as the seed region, which binds to the non-coding end [3′-untranslated region (3′- UTR)] of its complementary mRNA. Thus, the miRNA identifies the target mRNA molecule. If the miRNA fully binds to the target mRNA, it will completely degrade the mRNA, while in case of an incomplete binding to the target mRNA, the miRNA will inhibit the translation of the mRNA [87].
The miRNA-coding genes make up around 3% of the human genome. Hence, around 30–92% of human genes are likely to be regulated by miRNAs. The regulatory role of the miRNA gene is now well documented [100]. The function of miRNAs is essential for the development of different physiological systems and the maintenance of their cell homeostasis and normal functioning. Numerous miRNA variants play roles in various biological processes such as cell signaling regulation, differentiation, cell cycle, growth, apoptosis, tumorigenesis, and neurogenesis [7], [6], [9], [34], [48]. MiRNAs comprise about 1–5% of the animal genomes, and it is estimated that there are approximately 800–1000 miRNAs in the human genome and that over one-third of the protein-coding genome is regulated by miRNAs. Each miRNA can target and regulate the mRNA transcription of hundreds of downstream genes. A miRNA can have several target sites in the mRNA of a downstream gene, while a mRNA can be targeted by multiple miRNAs. Therefore, miRNAs contribute to a recently known level of gene expression regulation and play an important role in biological processes, function of almost all tissues and organ systems in the body, and development of various diseases [87].
4. MiRNAs in SARS-CoV-2 infection
Coronaviruses are a large family of viruses that are known to cause illness ranging from the common cold to SARS [4]. MiR-155 augmented antiviral T cell responses including cytokine production, cytotoxic activity, and homing to the central nervous system (CNS) in coronavirus-induced neurologic disease model [22]. The human coronaviruse OC43 nucleocapsid protein potentiates NF-kB pathway activation. It has been demonstrated that miR-9 is capable of diminishing NF-kB translation by targeting its 3′ UTR region. SARS, a relatively rare disease, is caused by severe acute respiratory syndrome coronavirus (SARS-CoV or SARS-CoV-1). This is an acute infectious disease with a significant mortality rate and common clinical features such as pulmonary fibrosis and pulmonary insufficiency [78]. Induction of miR-574 and miR-214 expression was reported in bronchoalveolar stem cells infected by SARS-CoV [69]. The expression of miR-223 and miR-98 was downregulated in BASCs through some viral nucleocapsid proteins controling pro-inflammatory cytokine secretion [57].
A recent study showed that the response to tocilizumab which was tested on COVID-19-induced cytokine storm is associated with a significant reduction of plasma IL-6 and a significant elevation of miR-146a levels. However, COVID-19 patients who did not respond to this antibody had lower serum levels of miR-146a-5p after the treatment [73].
The other established effect of miRNAs on coronavirus is the regulation of ACE2 expression. This enzyme catalyzes the conversion of Ang I into Ang 1–9 and II into 1–7. Several studies have confirmed the potential role of ACE2 in COVID-19-related diseases [1], [32], [97]. MiR-125b acts as a negative regulator targeting 3′UTR of ACE2 mRNA. Similarly, miR-18 was disregulated in ACE-associated nephropathy, which offers antimiR-18 as a novel therapeutic candidate in COVID-19 related diseases [88].
Computational prediction analysis detected that 29 miRNA precursors in the SARS-CoV-2 genome can target host genes involved in vital signaling pathways such as Wnt and epidermal growth factor (EGF)-receptor signaling pathways. On the other hand, some SARS-Cov-2-encoded genes could be targeted by host miRNAs. The products of these genes are essential in the entrance and replication of the virus. For example, has-miR-203b-3p and has-miR-148-3p targeted ORF genes that play key roles in viral replication. Liu et al. recruited bioinformatics approaches and showed that miR-147-3p derived virus through serine 2 as a transmembrane protease, facilitating entrance into gastrointestinal cells. STAT1 is an essential element in IFN signaling and anti-virus defense. Panther analysis reported that some miRNAs derived from SARS-CoV-2 might suppress STAT1 protein [74]. Analyszing the transcriptomic data of human alveolar and bronchial epithelial cells demonstrated that the CSF1 gene, a known target of miR-1207-5p, is over-expressed during SARS-CoV-2 infection. CSF1 promotes macrophage migration and activation and may be associated with the acute inflammatory response in severe COVID-19 [12]. Yousefi et al. employed Viruses- STRING, a protein–protein interaction database, and illustrated that the signaling pathway through TGF-beta with the largest numbers of involved genes plays a crucial role during SARS-CoV-2 infection [94].
Several reports have confirmed that diabetes increases the severity and mortality of COVID-19 infection [42], [66]. It was indicated that the upregulation of the cardiac miR-133a levels in the diabetic heart diminished cardiac lipid accumulation. Thus, miR-133a is a promising candidate for investigating its role in heart failure in patients with diabetes and COVID-19 [40]. Upregulation of miRNA 197–5p was described in cardiovascular patients, and miRNA 197–5p was proposed as a prognostic and risk factor of such cardiac events. MiRNA 197–5p also has a role in defense against SARS-CoV-2 infection. Therefore, a higher COVID-19 mortality rate and susceptibility in patients with the cardiovascular disease may be connected to the loss of the miRNA 197–5p binding site across the SARS-CoV-2 genome [5]. On the other hand, Balme et al.’s prediction study found that an increased expression of hsa-miR-1307-3p might lead to a reduction in SARS-CoV-2 replication and had a high affinity to SARS-CoV-2 genome 3′UTR and high expression in lung tissue. The hsa-miR-1307-3p could promote glycogenesis and glucose uptake by inhibiting protein kinase c (PKC) and extracellular signal-regulated kinase (ERK), which are inhibitors of RS1 and indirectly reduce blood sugar [10].
In the field of miRNA-based antiviral therapies, Hum et al. introduced three microRNAs (miR-16, miR-200, and miR-24) that can serve as modulating approaches in SARS-CoV-2 infection [33]. It was demonstrated that miR-24 had a crucial role in furin regulation. It is a serine protease that activates proteolytic function and the host-cell entrance of some respiratory viruses and also increases TGF-β1secration that leads to lung damage [61]. MiR-200 binds to a highly conserved region of the ACE2 3′-UTR gene as a major regulator of heart function [33], and miR-16 induces apoptosis by downregulating BCL2 [21]. Suppression of miR-16 was also found to increase mitochondrial reactive oxygen species (ROS) generation and toll-like receptor 4 (TLR4) expression [63].
All in all, it seems that the identification of host and virus miRNAs may bring about perspective therapeutic strategies for the control of viral pathogenicity and its side effects in COVID-19 related diseases [24].
5. Further hypotheses about the role of miRNAs in regulating signaling pathways involved in COVID-19 pathogenesis
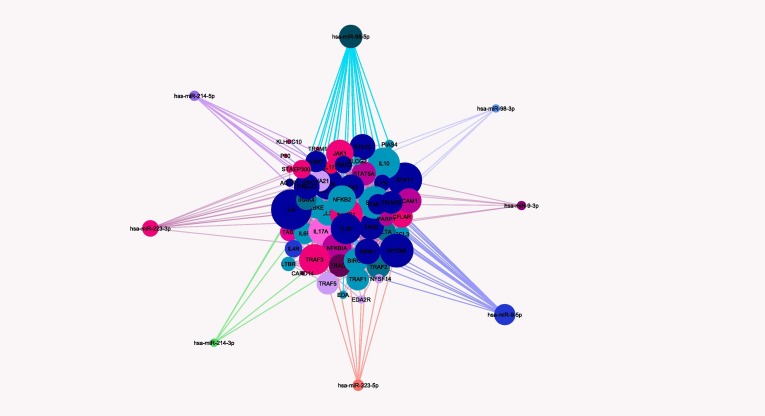
Potential targets of a number of most important miRNAs (miR-9, miR-98, miR-223, and miR-214), whose expression is predicted to change in host cells infected with COVID-19 [76], were specified using bioinformatics analysis and shown in Table 1 . Moreover, the interactions of protein targets with each other and with the mentioned miRNAs are shown in Fig. 2, Fig. 3, Fig. 4, Fig. 5 .
Table 1.
Potential targets of miR-9, miR-98, miR-223 and miR-214 involved in COVID19 pathogenesis.
| Gene | miRNA | Full name of the genes |
|---|---|---|
| IL6 | miR-98-5p,miR-223-3p, miR-9-5p | Interleukin 6 |
| IL6R | miR-98-5p | Interleukin 6 receptor |
| IL10 | miR-98-5p | Interleukin 10 |
| STAT2 | miR-98-5p,miR-214-3p, | Signal transducer and activator of transcription 2 |
| miR-223-3p | ||
| STAT3 | miR-98-5p | Signal transducer and activator of transcription 3 |
| IFNAR1 | miR-98-5p,miR-98-3p, miR-9-3p, miR-214-3p | Interferon alpha and beta receptor subunit 1 |
| IFNGR1 | miR-98-5p, miR-9-3p | Interferon gamma receptor 1 |
| PIAS4 | miR-98-5p | Protein inhibitor of activated STAT 4 |
| CCL3 | miR-98-5p | C-C motif chemokine ligand 3 |
| EDA | miR-98-5p | Ectodysplasin A |
| LYN | miR-98-5p, miR-223-3p | LYN proto-oncogene, Src family tyrosine kinase |
| PLCG2 | miR-98-5p | Phospholipase C gamma 2 |
| TAB2 | miR-98-5p,miR-98-3p, miR-9-3p, miR-214-5p | TGF-beta activated kinase 1/MAP3K7 binding protein 2 |
| BIRC3 | miR-98-5p | Baculoviral IAP repeat containing 3 |
| IKBKE | miR-98-5p | Inhibitor of nuclear factor kappa B kinase subunit epsilon |
| BCL2 | miR-98-5p, miR-9-5p | B-cell lymphoma 2 |
| BCL2L1 | miR-98-5p | BCL2 Like 1 |
| NFKB2 | miR-98-5p | Nuclear factor kappa B subunit 2 |
| LTBR | miR-98-5p | Lymphotoxin beta receptor |
| TLR8 | miR-98-5p | Toll like receptor 8 |
| TRAF1 | miR-98-5p | TNF receptor associated factor 1 |
| TRIM25 | miR-98-3p, miR-223-3p | Tripartite motif containing 25 |
| miR-214-5p, miR-9-5p | ||
| AGTR1 | miR-98-3p, miR-223-3p | Angiotensin II receptor type 1 |
| IL4R | miR-98-3p | Interleukin 4 receptor |
| IL6ST | miR-223-3p,miR-223-5p, miR-9-5p | Interleukin 6 signal transducer |
| STAT1 | miR-223-3p, miR-9-3p | Signal transducer and activator of transcription 1 |
| STAT5A | miR-223-3p | Signal transducer and activator of transcription 5A |
| ERC1 | miR-223-3P,miR-214-3p | ELKS/RAB6-interacting/CAST family member 1 |
| NFKBIA | miR-223-3p | NFKB inhibitor alpha |
| PARP1 | miR-223-3p | Poly(ADP-ribose) polymerase 1 |
| TLR8 | miR-223-3p, miR-98-5p | Toll like receptor 8 |
| ICAM1 | miR-223-3p | Intercellular adhesion molecule 1 |
| EDA2R | miR-223-5p | Ectodysplasin A2 receptor |
| IFNA21 | miR-223-5p | Interferon alpha 21 |
| MYD88 | miR-223-5p,miR-214-5p | Myeloid differentiation primary response 88 |
| TRAF5 | miR-223-5p | TNF receptor associated factor 5 |
| TNFSF14 | miR-223-5p | TNF superfamily member 14 |
| TLR7 | miR-223-5p,miR-9-5p | Toll like receptor 7 |
| RIPK1 | miR-223-5p, miR-9-5p | Receptor interacting serine/threonine kinase 1 |
| IL17D | miR-9-5p | Interleukin 17D |
| JAK1 | miR-9-5p | Janus kinase 1 |
| JAK3 | miR-9-5p, miR-214-5p, | Janus kinase 3 |
| miR-9-3p | ||
| STAM | miR-9-5p | Signal transducing adaptor molecule |
| PIAS3 | miR-9-5p, miR-214-5p | Protein inhibitor of activated STAT 3 |
| EP300 | miR-9-5p | E1A binding protein p300 |
| KLHDC10 | miR-9-5p | Kelch domain containing 10 |
| TRAF3 | miR-9-5p | TNF receptor associated factor 3 |
| TRAM1 | miR-9-5p | Translocation associated membrane protein 1 |
| NFKB1 | miR-9-5p | Nuclear factor kappa B subunit 1 |
| CFLAR | miR-9-5p | CASP8 and FADD like apoptosis regulator |
| TAB3 | miR-9-5p | TGF-beta activated kinase1 and MAP3K7 binding protein 3 |
| IL17A | miR-9-3p | Interleukin 17A |
| IRAK4 | miR-214-3p | Interleukin 1 receptor associated kinase 4 |
| TRAF2 | miR-214-5p | TNF receptor associated factor 2 |
| LTA | miR-214-5p | Lymphotoxin alpha |
| CARD14 | miR-214-3p | Caspase recruitment domain family member 14 |
| TRADD | miR-214-3p | TNFRSF1A associated via death domain |
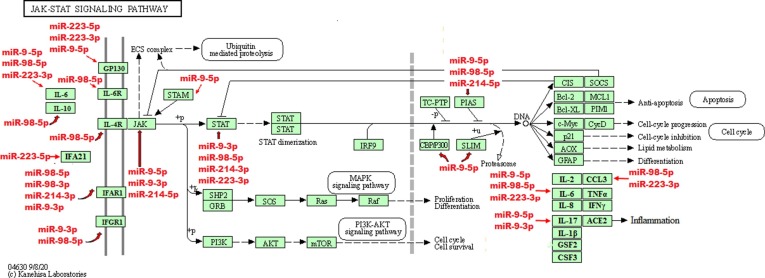
Fig. 2.
Potential targets of miR-223, miR-98, miR-9, and miR-214 in JAK/STAT pathway. The JAK/STAT signaling pathway is a transducer of a wide variety of signals for development and homeostasis in animals from humans to flies. In mammals, this pathway is the main mechanism of signal transduction from growth factors and cytokines. Following the binding of a cytokine to a receptor, the JAK tyrosine kinase activates STAT. Upon the activation of STAT, it is dimerized and transferred to the nucleus, leading to the regulation of the transcription of target genes. The targets of the miR-223, miR-98, miR-9, and miR-214 in these pathway are shown using red arrows (KEGG PATHWAY database). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
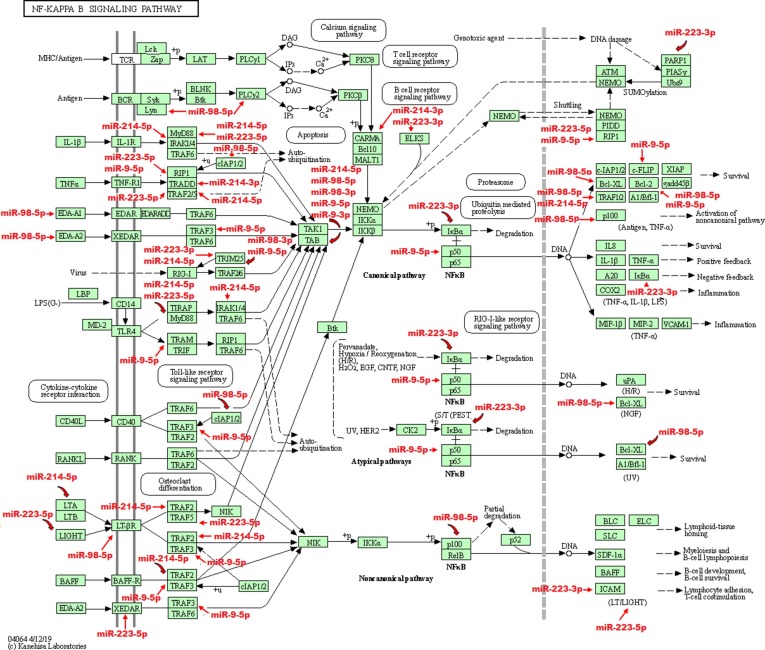
Fig. 3.
Potential targets of miR-223, miR-98, miR-9, and miR-214 in NFκB pathway. Nuclear factor-kappa B (NF-kappa B) regulates genes played important roles in immunity, inflammation and cell survival. There are several pathways resulting in NF-kappa B-activation. The canonical pathway is induced by TNF-α, IL-1 or byproducts of bacterial and viral infections. This pathway depends on IKK- mediated IkappaB-alpha phosphorylation on Ser32 and 36, leading to its degradation, which allows the p50/p65 NF-kappa B dimer to enter the nucleus and activate gene transcription. The targets of the miR-223, miR-98, miR-9, and miR-214 in these pathway are shown using red arrows (KEGG PATHWAY database). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
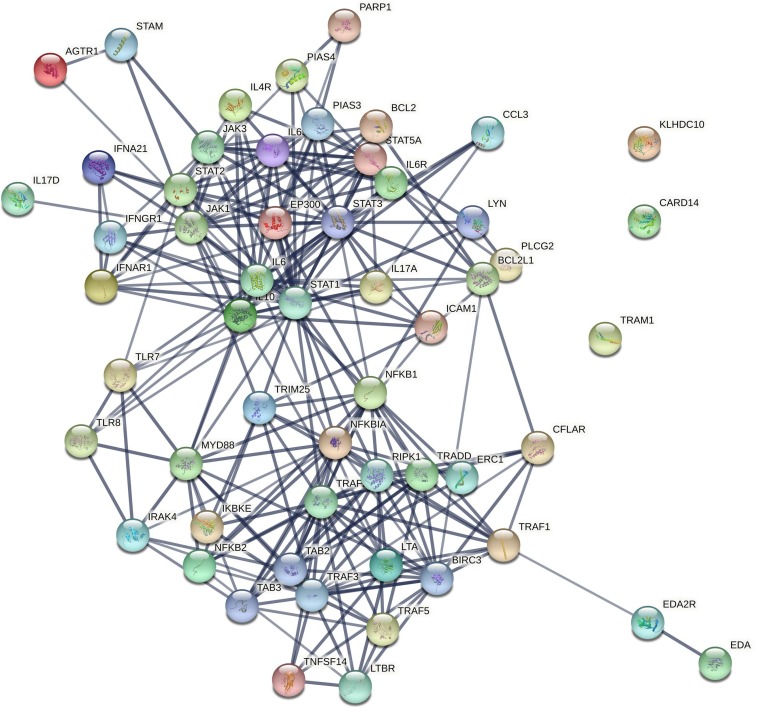
Fig. 4.
Potential interactions between genes involved in the JAK/STAT and NFκB pathways are depicted using the STRING database (string-db.org).
Fig. 5.
Potential interactions between genes involved in the JAK/STAT and NFκB pathways and miR-223, miR-98, miR-9, and miR-214 (extracted from databases of KEGG, WikiPathways, MiRTarBase, and Target scan). Potential target genes of miR-9-5p (pink), miR-9-3p (light pink), miR-98-5p (light blue), miR-98-3p (medium blue), miR-214-5p (green), miR-214-3p (dark purple), and miR-223-3p (medium purple), and miR-223-5p (light purple) are shown in the picture. MiRNAs with more interactions are illustrates with larger sizes. Potential interactions between genes with each other and miRNAs of interest were depicted via Cytoscape software. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
5.1. The role of miRNAs in regulating JAK-STAT signaling pathway
After the attachment of the spike protein of coronaviruses to the ACE2 receptor of host cells, the RNAs of the viral genome are detected by toll-like receptors (TLRs), especially TLR3 and TLR 7/8, as well as (RIG-I)/MDA5, which is a cytosolic RNA sensor, leading to the activation of antiviral responses through the activation of different signaling pathways including NF-kB and JAK-STAT [58]. The expression of TLRs is regulated by different miRNAs. The 3′untranslated region (3′ UTR) of TLR7 is directly targeted by miR-9-5p and miR-223-5p. TLR8 is also directly targeted by miR-98-5p and miR-223-3p. Interestingly, the activation of TLR2 and TLR7/8 in the MyD88-dependent pathway increased the expression of miR-9 in polymorphonuclear neutrophils (PMN) and monocyte cells [11].
Various cytokines including interleukins IL-10, IL-7, IL-6, IL-4, and IL-2 and interferons IFN-α, IFN-β, IFN-γ, and granulocyte colony-stimulating factor (GCS-F) induce different biologic effects through activating the JAK/STAT pathway [14]. IL-6 that severely increases in COVID-19 patients and plays a significant role in acute inflammatory response is a major potential target of miR-98-5p, miR-9-5p, and miR-223-3p. Moreover, it is predicted that the expression of IL6R, the membrane receptor of IL-6, is also partially regulated by miR-98-5p. Via activating various cells expressing glycoprotein 130 (GP130) and IL-6 receptors, IL-6 activated JAK/STAT pathway [19]. The IL-6/JAK/STAT pathway is highly activated in acute inflammation, and the excessive activity of this pathway in COVID-19 patients likely exacerbates the inflammatory responses of the host. Interestingly, the activation of this pathway in a positive feedback loop produced IL-6 [14]. Investigating samples of osteoarthritis (OA) patients indicated that miR-9-5p elevated the expression of IL-6 in human chondrocytes through inhibitng monocyte chemotactic protein-1-induced protein-1 (MCPIP1) [56].
The results of a study on the peripheral blood mononuclear cells (PBMCs) samples of systemic lupus erythematosus (SLE) patients showed that miR-98-5p directly targeted IL-6 and inhibited the production of inflammatory cytokines and cell proliferation through IL-6/STAT3 pathway [95]. Moreover, the results of a dual luciferase reporter assay in human adipose stem cells indicated that miR-223-3p regulated the expression of inflammatory cytokines IL-6 and STAT3 (J. [91]. Furthermore, the results of another study showed that miR-223-3p might be able to increase the expression of IL-6 in synoviocytes through the downregulation of IL-17 D receptor [64]. Note that miR-223-3p affects the secretion of IL-6 in mast cells via regulating insulin growth factor I receptor/phosphatidylinositol-3-kinase (IGF1R/PI3K) signaling pathway [83]. The GP130 (IL6ST) receptor is also targeted by miR-223-5p, miR-223-3p, and miR-9-5p. The results of an in vitro study on mouse NPCs demonstrated that miR-9 directly targeted LIFR beta, IL6ST (GP130), and JAK1 genes, lowered their expression, and thus inhibited astrogliogenesis in these cells [101]. A research on Kawasaki disease (KD) mouse found that the increased expression of miR-223-3p, which is an important regulatory factor in endothelial damage to the KD, lowered the expression of IL6ST [85]. On the other hand, angiotensin II that is produced locally in the inflamed vessels leads to the production of IL-6 after attachment to angiotensin II receptor through activating JAK/STAT pathway. AT1R is directly targeted by miR-98-3p and miR-223-3p.
IL-10, acting as a key mediator in immune response, and interleukin-4 receptor are also directly targeted by miR-98-5p. The results of a study on HEK-293 and RAW 264.7 cell lines demonstrated that miR-98-5p directly targeted the 3′ UTR of IL-10 and inhibited its expression at the post-transcriptional level [52]. IFNA21, a member of type 1 IFN family, is directly targeted by miR-223-3p. An in vitro functional study in macrophages demonstrated that miR-223-3p increased the production of type 1 IFN via directly targeting FOXO3. Moreover, a type 1 IFN raised the expression of miR-223-3p in a positive feedback loop [17]. Furthermore, miRNAs could also affect the expression of IFN receptors at the post-transcriptional level. IFNAR1 (interferon alpha receptor 1) is a direct target of miR-98-5p, miR-98-3p, miR-9-3p, and miR-214-3p. Interferon gamma receptor 1(IFNGR1) is also targeted by miR-9-3p and miR-98-5p. The expression of genes in the downstream signaling pathway of IFN receptors, JAK/STAT, is affected by many miRNAs [41], [89]. JAK1, a member of the Janus kinase (JAK), a family of non-receptor tyrosine kinases, is directly targeted by miR-9-5p. Moreover, JAK3, as another member of this family, is targeted by miR-9-5p, miR-9-3p, and miR-214-5p. The findings of an in vitro functional study indicated that the 3′ UTR region of JAK1 and MMP-13 genes was a direct target of miR-9 in human atherosclerosis models and miR-9 was able to inhibit downstream NLR family pyrin domain containing 3 (NLRP3) inflammasome through targeting JAK1 [86].
STAT transcription factors, which are phosphorylated and activated after the activation of JAK proteins, are the regulatory targets of miRNAs. The expression of STAT1 is regulated by miR-9-3p and miR-223-3p, and the expression of STAT2 is regulated by miR-98-5p, miR-223-3p, and miR-214-3p. In addition, STAT3 and STAT5A are also directly targeted by miR-98-5p and miR-223-3p, respectively. A luciferase reporter assay in human T cells revealed that miR-150 and miR-223 specifically targeted the 3′UTR of STAT1 and consequently reduced the expression of this gene and inhibited the downstream signaling pathway independent from or dependent on IFN [62]. Microarray analysis results in the nucleus pulposus tissues of patients with intervertebral disc degeneration showed that the miR-98 expression significantly declined. Furthermore, the results of an in vitro gain-of-function study indicated that miR-98 was able to target STAT3 and inhibit IL-6/STAT3 signaling pathway in IDD patients [36]. In addition, the knockdown of miR-98 in nasopharyngeal carcinoma cells reduced the expression of STAT3 protein and consequently inhibited the proliferation, migration, and invasion of these cells through miR-98/STAT3 signaling pathway [51]. In another study, it was shown that the induced underexpression of miR-223 in murine macrophages activated its direct target, STAT3, and thus significantly increased the production of pro-inflammatory cytokines IL-6 and IL-1 β [18]. The increased expression of miR-223 in mouse embryonic fibroblasts and HEK293 cells lowered the expression of different genes in the cell survival or death pathway including STAT5A, and more comprehensive analyses verified STAT5A as a direct target of miR-223 in this study [68].
MiRNAs are able to indirectly affect signaling pathways activated by cytokines and thus innate and adaptive immune responses through targeting the JAK/STAT pathway regulators [67]. PIAS3, an inhibitor of STAT3 transcription factor in cell nucleus, was identified as a direct target of miR-214 and miR-9-5p. Increasing the expression of miR-9-5p by reducing the expression of protein inhibitor of activated STAT 3 (PIAS3), an inhibitor of JAK/STAT pathway, increased the expression of STAT3, resulting in an increase in the population of Th17 cells in MS patients [55]. Furthermore, miR-9 and miR-181a led to the development of early-stage myeloid-derived suppressor cells (eMDSCs) in breast cancer through inhibiting the function of negative regulators of JAK/STAT pathway, SOCS3, and PIAS3 [37]. Histone acetyltransferase EP300 that functions in chromatin remodeling and transcription regulation of the target genes of JAK/STAT pathway is also a direct target of miR-9-5p. The overexpression of miR-9 increased the expression of miR-132 and indirectly affected the expression of the direct target genes of miR-132, namely phosphatase and tensin homolog (PTEN), FOXO3A, and EP300, and apoptosis in rheumatoid arthritis [46]. STAM, an adaptor molecule playing a role in facilitating the transcription of specific target genes of JAK/STAT signaling pathway [71], and SLIM, a nuclear E3 ubiquitin ligase that is a negative regulator of STAT pathway [80], are targets of miR-9-5p. Finally, JAK/STAT pathway activates the transcription of numerous genes in the inflammatory pathway, including ACE2, IL-8, IL-6, IL-2, IL-17, IL-1β, CSF3, CSF2, CCL3, TNF-α, and IFN-γ, which are regulated by different miRNAs [14], [53]. IL-7 was identified as a direct target of miR-9-5p and miR-9-3p, and also, CCL3 was identified as a direct target of miR-98-5p and miR-223-3p. The results of high-throughput sequencing demonstrated that the expression of miR-98-5p severely increased in immunoglobulin a nephropathy (IgAN) patients and this miRNA was able to directly target CCL3, and when its expression was inhibited, increase the expression of IL6 [49]. Through directly targeting CCL3 and IL-6, miR-223 controls the migration of inflammatory cells to the lung tissue and the formation of stable local inflammation in tuberculosis disease [23].
5.2. The role of miRNAs in regulating NF-kB signaling pathway
As mentioned earlier, RNAs of the viral genome are diagnosed via toll-like receptors (TLRs), inducing various antiviral responses via activating various signaling pathways such as NF-kB.
Investigating the performance of miR-223 in the regulation of the inflammatory responses activated through TLRs in macrophages indicated that the expression of miR-223 decreased in TRL ligand stimulation. The reduced expression of miR-223 via the increased expression of Ras homolog family member B (RhoB) led to the activation of the NF-kB pathway [98].
NF-kB transcription factor plays an important role in innate and adaptive immunity. In normal conditions, NF-kB is inhibited by IKB proteins in cytoplasm. Various cellular stimuli including pathogens lead to the induction of NF-kB and the displacement of its nucleus through the phosphorylation and ubiquitin-dependent proteasomal degradation of the inhibitor of IKB. NF-kB leads to the transcription of a wide variety of pro-inflammatory cytokines, chemokines, anti-apoptotic proteins, and stress-response proteins [14]. NF-kB pathway in lung inflammatory immunopathology, which is induced after infection with respiratory viruses such as SARS-CoV, is very active (DeDiego et al., 2014; Smits et al., 2010).
Genes regulating NF-kB signaling pathway are the regulatory targets of different miRNAs. EDA, a cytokine activating NF-kB pathway after attachment to ectodysplasin A receptor (EDAR) is a direct target of miR-98-5p. Lyn, a member of Src family of tyrosine kinases that activates NF-kB via activating PI3K signaling pathway, is a target of miR-98-5p [81]. PLCG2 is also directly targeted by miR-98-5p. Cytokines such as TNF and TGF- β1 can activate PLCG2 and lead to the activation of ERK and NF-kB signaling pathways [54]. On the other hand, miR-223 and miR-214 can directly target 3′ UTR of ERC1 gene. The miRNA microarray profiling results demonstrated that the expression of miR-223-3p increased by 13.01 times in patients with ulcerative colitis, and the results of in silico prediction tools determined ERC1 as one of the most important targets of this miRNA in NF-kB pathway [82]. MyD88, an adaptor protein in NF-κB signaling pathway, is also targeted by miR-214-5p and miR-223-5p. The increased expression of miR-214 by targeting MyD88 reduced the production of inflammatory cytokines in V. harveyi bacterial infection and prevented the exacerbation of inflammation [20]. Through the utilization and phosphorylation of IRAK and TRAF6 complexes, MyD88 activates MAPK and NF-kB pathways. IRAK4 enzyme, which is an important threonine/serine protein kinase in MAPK pathway and TLR-mediated immune responses [39], is directly targeted by miR-214. Receptor-interacting protein kinase 1 (RIPK1) that determines the cell fate, death or survival, through NF-kB signaling pathway after receiving a wide variety of pro-inflammatory or pro-death cytokines is a target of miR-223-5p and miR-9-5p [60]. The results of gene expression analysis and luciferase reporter assay showed that the expression of RIPK1 and RIPK3 in acute kidney injury induced in C57BL/6 mice increased, and miR-223 could directly target 3′ UTR of RIPK3 gene (Huang et al., 2018). TNFR1 associated death domain protein (TRADD), an adaptor protein mediating cell death and activating NF-kB pathway through interacting with TNFRSF1A/TNFR1, is a direct target of miR-214-3p. TRAF2 and TRAF5 were identified as targets of miR-214-5p and miR-223-5p, respectively. Tumor necrosis factor receptor associated factor-2 (TRAF2) and tumor necrosis factor receptor associated factor-5 (TRAF5) lead to the activation of TAK1 via the ubiquitination of IKKγ/NEMO. TAK1 finally activates IKK enzyme and leads to the phosphorylation and deactivation of IκBα and the activation NF-kB pathway [59]. MiR-98-5p is capable of directly targeting the epsilon subunit of this enzyme (IKBKE). BIRC3 (cIAP2), as a member of the family of proteins inhibiting apoptosis by NF-kB pathway, was determined as one target of miR-98-5p. TRAF3 was identified as a potential target of miR-214 and miR-9-5p. The expression of miR-214 increased in the tissues of osteosarcoma patients as well as osteosarcoma cell lines, and this miRNA was capable of directly targeting TRAF3 gene [72]. TAB2, an adaptor protein that forms a complex with TAK1 and is necessary for the activation of NF-kB pathway, is a target of many miRNAs including miR-9-5p, miR-9-3p, miR-98-5p, miR-98-3p, and miR-214-5p. The findings of qRT-PCR in ovariectomized rats (OVX-ASCs) suggested the increased expression of miR-214. In addition, two new targets of miR-214, CTNNB1 (β-catenin) and TAB2, were identified in this study via luciferase reporter assay [45]. MiRNAs can directly target the negative regulators of NF-kB pathway. In this regard, miR-223-3p is able to directly target NFKB inhibitor alpha (NFKBIA). It was shown that notch and NF-kB signaling pathways could raise the expression of miR-223, which in turn led to the reduced expression of onco-suppressor FBXW7 in T-cell acute lymphoblastic leukemia [43].
Tripartite motif containing 25 (TRIM25) that is a positive regulator of NF-kB signaling pathway is targeted by miR-98-3p, miR-9-5p, miR-223-3p, and miR-214-5p. PARP1, a DNA polymerase playing an important role in the identification of DNA damages and the utilization of DNA repair proteins, is a direct target of miR-223-3p. PARP1 leads to the activation of NF-kB pathway via a TNF-α-induced pathway. Microarray analysis results in JHesoAD1 and esophageal OE33 (adenocarcinoma) cell lines transfected with miR-223 mimics indicated the reduced expression of several target genes including PARP1, CYB5A, and SMARCD1. Afterwards, the luciferase assay established that the 3′ UTR of PARP1 gene is directly identified by miR-223 [79]. Note that transcription factors NF-kB1 and NF-kB2 are directly regulated by miR-9-5p and miR-98-5p. MiR-9 in human monocytes was induced by inflammatory cytokines TNF-α and IL-1β, and NF-kB1 was identified as an important target of miR-9 in both in silico and experimental investigations [11]. C-FLIP, Bcl-XL, and BCL2, as target genes of NF-kB pathway, are directly targeted by miR-9-5p, miR-98-5p, and both of these miRNAs, respectively. The elevated expression of miR-98 through targeting Bcl2 gene inhibits the growth and resistance tolerance in esophageal squamous cell carcinoma (ESCC) [38]. Moreover, miR-98 plays a critical role in the pathogenesis of osteoarthritis disease through targeting Bcl2 [83]. Furthermore, the expression of miR-98 in rat uterus declined in the receptive phase, and miR-98 inhibited apoptosis and increased proliferation in this phase by directly targeting Bcl-xl [92]. As other target genes of NF-kB pathway, TRAF1 and TRAF2 are directly targeted by miR-98-5p and miR-214-5p. EDA2R, which is a member of TNFR family and activates NF-kB signaling pathway, and intercellular adhesion molecule 1 (ICAM1), which is a target of this pathway, are also directly targeted by miR-223-5p and miR-223-3p, respectively. The targeting of other members of TNFR family by miR-223 has been established in the literature. The findings of luciferase reporter assay in HEK293 cells or rat cardiac H9c2 cells demonstrated that miR-223-5p was able to target 3′ UTR of TNFR1 and DR6 genes [70]. LTA and TNFSF14 ligands, activating NF-kB signaling pathway after attachment to LT-βR receptor, are targeted by miR-214-5p and miR-223-5p, respectively. LT-βR, a member of TNFR family, was also identified as a direct target of miR-98-5p. Finally, TRAM1, a member of steroid receptor coactivator (SRC) family inducing gene overexpression through NF-kB transcription factor, is a target of miR-9-5p.
Therefore, it can be concluded that the expression of genes in JAK/STAT and NF-kB signaling pathways is significantly affected by miR-9, miR-98, miR-223, and miR-214, and these miRNAs may play an important role in the pathogenesis of COVID-19.
6. Discussion
Multiple destructive inflammatory signaling pathways have been implicated in COVID-19 pathogenesis, which leads to multiple-organ damages, physiological deterioration, and eventually death. In the current study, the results obtained from targeting the aforementioned inflammatory pathways in COVID-19 patients were reviewed. It was found that NF-kB and JAK-STAT signaling pathways have key roles in inflammation involved in COVID-19 pathogenesis.
NF-kB signaling pathway genes (NFKBIA, NFKB1, RELA, NFKB2) were upregulated in patients with COVID-19 infection. JAK/STAT pathway induced various cytokines including interleukins IL-10, IL-7, IL-6, IL-4, and IL-2 and interferons IFN-α, IFN-β, IFN-γ, and GCS-F. Additionally, we reviewed research works in which miRNAs targeted inflammatory signaling pathways involved in the inflammatory diseases. Our study focused on miR-9, miR-98, miR-223, and miR-214, which make a unique contribution to the regulation of inflammatory pathways related to COVID-19 pathogenesis, and whose target genes are related to NF-kB and JAK-STAT pathways.
Based on the results, TRIM25, which is a positive regulator of NF-kB signaling pathway, is targeted by miR-98-3p, miR-9-5p, miR-223-3p, and miR-214-5p. PARP1, which leads to the activation of NF-kB pathway via a TNFα-induced pathway, is targeted by miR-223-3p. Transcription factors NFKB1 and NFKB2 are directly regulated by miR-9-5p and miR-98-5p. In addition, TNF-α, IL-1β, and NF-kB1 were targeted by miR-9. C-FLIP, Bcl-XL, and BCL2, as target genes of NF-kB pathway, are directly targeted by miR-9-5p, miR-98-5p, and both of these miRNAs, respectively. TRAF1 and TRAF2 are directly targeted by miR-98-5p and miR-214-5p. TNFR family and ICAM1 are also directly targeted by miR-223-5p and miR-223-3p, respectively.
Recently, several miRNA-based treatments have illustrated therapeutic promise in clinical trials. These treatments include the use of miRNA antagonists to increase the expression of specific proteins that are downregulated or miRNA mimics to decrease the expression of proteins that induce abnormally [31]. For example, miravirsen was the first miRNA-based drug that was successfully applied for hepatitis C virus (HCV) infection. This drug suprresses viral RNA levels by sequestering miR-122 away from the viral genome [35]. In regard to the delivery of miRNA targets, the poor half-life of miRNAs and the toxicity of delivery vehicles are obstacles to treatment. Nanoparticles, lipid-based carriers, and exosomes are highly recommended for direct delivery into the lungs in the clinical setting [16], [27].
In conclusion, miRNAs are associated with inflammation processes, and their target networks provide new knowledge that can pave the way for new therapeutic strategies for COVID-19 pathogenesis. In this regard, miR-9, miR-98, miR-223, and miR-214 can be considered as potential targets for new therapeutic strategies in COVID-19 diseases by targeting inflammatory pathways.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This review was supported by the Shahrekord University of Medical Sciences, Shahrekord, Iran, Semnan University of Medical Sciences, Semnan, Iran, and Mashhad University of Medical Sciences, Mashhad, Iran.
References
- 1.Abedi F., Rezaee R., Hayes A.W., Nasiripour S., Karimi G. MicroRNAs and SARS-CoV-2 life cycle, pathogenesis, and mutations: biomarkers or therapeutic agents? Cell Cycle. 2021;20(2):143–153. doi: 10.1080/15384101.2020.1867792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abu-Izneid T., AlHajri N., Ibrahim A.M., Javed M.N., Salem K.M., Pottoo F.H., Kamal M.A. Micro-RNAs in the regulation of immune response against SARS CoV-2 and other viral infections. J. Adv. Res. 2021;30:133–145. doi: 10.1016/j.jare.2020.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adrian Recinos I.I.I., LeJeune W.S., Sun H., Lee C.Y., Tieu B.C., Muping L.u., Hou T., Boldogh I., Tilton R.G., Brasiera A.R. Angiotensin II induces IL-6 expression and the Jak-STAT3 pathway in aortic adventitia of LDL receptor-deficient mice. Atherosclerosis. 2007;23(1):1–7. doi: 10.1016/j.atherosclerosis.2006.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Hazmi A. Challenges presented by MERS corona virus, and SARS corona virus to global health. Saudi J. Biol. Sci. 2016;23(4):507–511. doi: 10.1016/j.sjbs.2016.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ali H.R.S.M., McLellan A.D. Implications of SARS-CoV-2 mutations for genomic RNA structure and host microRNA targeting. BioRxiv. 2020 doi: 10.1101/2020.05.15.098947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Amini‐Farsani Z., Asgharzade S. The impact of miR-183/182/96 gene regulation on the maturation, survival, and function of photoreceptor cells in the retina. J. Comparative Neurol. 2020;528(9):1616–1625. doi: 10.1002/cne.v528.910.1002/cne.24833. [DOI] [PubMed] [Google Scholar]
- 7.Amini-Farsani Z., Sangtarash M.H., Shamsara M., Teimori H. MiR-221/222 promote chemoresistance to cisplatin in ovarian cancer cells by targeting PTEN/PI3K/AKT signaling pathway. Cytotechnology. 2018;70(1):203–213. doi: 10.1007/s10616-017-0134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Asgharzad S., Alizadeh A., Arab S. Regenerative Medicine Approaches in COVID -19 Pneumonia. Curr. Stem Cell Res. Ther. 2021;16 doi: 10.2174/1574888x16999210112205826. [DOI] [PubMed] [Google Scholar]
- 9.Asgharzade S., Sheikhshabani S.H., Ghasempour E., Heidari R., Rahmati S., Mohammadi M., Jazaeri A., Amini-Farsani Z. The effect of oleuropein on apoptotic pathway regulators in breast cancer cells. Eur. J. Pharmacol. 2020;886:173509. doi: 10.1016/j.ejphar.2020.173509. [DOI] [PubMed] [Google Scholar]
- 10.Balmeh N., Mahmoudi S., Mohammadi N., Karabedianhajiabadi A. Predicted therapeutic targets for COVID-19 disease by inhibiting SARS-CoV-2 and its related receptors. Inf. Med. Unlocked. 2020;20:100407. doi: 10.1016/j.imu.2020.100407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bazzoni F., Rossato M., Fabbri M., Gaudiosi D., Mirolo M., Mori L., Tamassia N., Mantovani A., Cassatella M.A., Locati M. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. PNAS. 2009;106(13):5282–5287. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertolazzi G., Cipollina C., Benos P.V., Tumminello M., Coronnello C. miR-1207-5p Can Contribute to Dysregulation of Inflammatory Response in COVID-19 via Targeting SARS-CoV-2 RNA. Front. Cell. Infect. Microbiol. 2020;10 doi: 10.3389/fcimb.2020.586592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y.i., Zhang L.i., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L.i., Xie J., Wang G., Jiang R., Gao Z., Jin Q.i., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan China. The Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catanzaro M., Fagiani F., Racchi M., Corsini E., Govoni S., Lanni C. Immune response in COVID-19: addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Sign. Trans. Targeted Ther. 2020;5(1) doi: 10.1038/s41392-020-0191-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chauhan A.J., Wiffen L.J., Brown T.P. COVID-19: A collision of complement, coagulation and inflammatory pathways. J. Thromb. Haemost. 2020;18(9):2110–2117. doi: 10.1111/jth.v18.910.1111/jth.14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chauhan N., Jaggi M., Chauhan S.C., Yallapu M.M. COVID-19: fighting the invisible enemy with microRNAs. Expert Review of Anti-Infective Therapy. 2021;19(2):137–145. doi: 10.1080/14787210.2020.1812385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L., Song Y., He L., Wan X., Lai L., Dai F., Liu Y., Wang Q. MicroRNA-223 promotes type i interferon production in antiviral innate immunity by targeting forkhead box protein O3 (FOXO3) J. Biol. Chem. 2016;291(28):14706–14716. doi: 10.1074/jbc.M115.700252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Q., Wang H., Liu Y., Song Y., Lai L., Han Q., Cao X., Wang Q., Viola J.P.B. Inducible microRNA-223 down-regulation promotes TLR-triggered IL-6 and IL-1β production in macrophages by targeting STAT3. PLoS ONE. 2012;7(8):e42971. doi: 10.1371/journal.pone.0042971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chi Z., Zhao W., Jia-Wen L., Hong Z., Gui-Qiang W. Cytokine Release Syndrome in Severe COVID-19: Interleukin-6 Receptor Antagonist Tocilizumab May Be the Key to Reduce Mortality. Int. J. Antimicrob. Agents. 2020;55(5) doi: 10.1016/j.ijantimicag.2020.105954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chu Q., Sun Y., Cui J., Xu T. Inducible microRNA-214 contributes to the suppression of NF-κB-mediated inflammatory response via targeting myd88 gene in fish. J. Biol. Chem. 2017;292(13):5282–5290. doi: 10.1074/jbc.M117.777078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cimmino, A., Calin, G. A., Fabbri, M., Iorio, M. V., Ferracin, M., Shimizu, M., Wojcik, S. E., Aqeilan, R. I., Zupo, S., Dono, M., Rassenti, L., Alder, H., Volinia, S., Liu, C. G., Kipps, T. J., Negrini, M., & Croce, C. M. (2005). miR-15 and miR-16 induce apoptosis by targeting BCL2. Proceedings of the National Academy of Sciences of the United States of America, 102(39), 13944–13949. DOI: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed]
- 22.Dickey L.L., Worne C.L., Glover J.L., Lane T.E., O’Connell R.M. MicroRNA-155 enhances T cell trafficking and antiviral effector function in a model of coronavirus-induced neurologic disease. J. Neuroinflammation. 2016;13(1) doi: 10.1186/s12974-016-0699-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorhoi A., Iannaccone M., Farinacci M., Faé K.C., Schreiber J., Moura-Alves P., Nouailles G., Mollenkopf H.-J., Oberbeck-Müller D., Jörg S., Heinemann E., Hahnke K., Löwe D., Del Nonno F., Goletti D., Capparelli R., Kaufmann S.H.E. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J. Clin. Invest. 2013;123(11):4836–4848. doi: 10.1172/JCI67604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Nabi S.H., Elhiti M., El-Sheekh M. A new approach for COVID-19 treatment by micro-RNA. Med. Hypotheses. 2020;143:110203. doi: 10.1016/j.mehy.2020.110203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.F., S., H., A., M., K., M., K., M., M., M., P., & D., M. (2020). JAK Inhibition as a New Treatment Strategy for Patients with COVID-19. International Archives of Allergy and Immunology. 10.1159/000508247. [DOI] [PMC free article] [PubMed]
- 26.Fadason, T., Gokuladhas, S., Golovina, E., Ho, D., Farrow, S., Nyaga, D., Pan, H., Karnani, N., Wong, C., Cooper, A., Schierding, W., & O’Sullivan, J. (2020). A transcription regulatory network within the ACE2 locus may promote a pro-viral environment for SARS-CoV-2 by modulating expression of host factors. DOI: 10.1101/2020.04.14.042002.
- 27.Fu Y., Chen J., Huang Z. Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA. 2019;1(1) doi: 10.1186/s41544-019-0024-y. [DOI] [Google Scholar]
- 28.García L.F. Immune Response, Inflammation, and the Clinical Spectrum of COVID-19. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gu H., Xie Z., Li T., Zhang S., Lai C., Zhu P., Wang K., Han L., Duan Y., Zhao Z., Yang X., Xing L.i., Zhang P., Wang Z., Li R., Yu J.J., Wang X., Yang P. Angiotensin-converting enzyme 2 inhibits lung injury induced by respiratory syncytial virus. Sci. Rep. 2016;6(1) doi: 10.1038/srep19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hadjadj J., Yatim N., Barnabei L., Corneau A., Boussier J., Pere H., Charbit B., Bondet V., Chenevier-Gobeaux C., Breillat P., Carlier N., Gauzit R., Morbieu C., Pene F., Marin N., Roche N., Szwebel T.-A., Smith N., Merkling S., Terrier B. Impaired type I interferon activity and exacerbated inflammatory responses in severe Covid-19 patients. Science. 2020 doi: 10.1101/2020.04.19.20068015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanna J., Hossain G.S., Kocerha J. The potential for microRNA therapeutics and clinical research. Front. Genet. 2019;10(MAY) doi: 10.3389/fgene.2019.00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hum C., Loiselle J., Ahmed N., Shaw T.A., Toudic C., Pezacki J.P. MicroRNA Mimics or Inhibitors as Antiviral Therapeutic Approaches Against COVID-19. Drugs. 2021;81(5):517–531. doi: 10.1007/s40265-021-01474-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Injinari N., Amini-Farsani Z., Yadollahi-Farsani M., Teimori H. Apoptotic effects of valproic acid on miR-34a, miR-520h and HDAC1 gene in breast cancer. Life Sci. 2021;269:119027. doi: 10.1016/j.lfs.2021.119027. [DOI] [PubMed] [Google Scholar]
- 35.Janssen H.L.A., Reesink H.W., Lawitz E.J., Zeuzem S., Rodriguez-Torres M., Patel K., van der Meer A.J., Patick A.K., Chen A., Zhou Y., Persson R., King B.D., Kauppinen S., Levin A.A., Hodges M.R. Treatment of HCV Infection by Targeting MicroRNA. N. Engl. J. Med. 2013;368(18):1685–1694. doi: 10.1056/nejmoa1209026. [DOI] [PubMed] [Google Scholar]
- 36.Ji M.L., Lu J., Shi P.L., Zhang X.J., Wang S.Z., Chang Q., Chen H., Wang C. Dysregulated miR-98 Contributes to Extracellular Matrix Degradation by Targeting IL-6/STAT3 Signaling Pathway in Human Intervertebral Disc Degeneration. J. Bone Miner. Res. 2016;31(4):900–909. doi: 10.1002/jbmr.2753. [DOI] [PubMed] [Google Scholar]
- 37.Jiang M., Zhang W., Zhang R., Liu P., Ye Y., Yu W., Guo X., Yu J. Cancer exosome-derived miR-9 and miR-181a promote the development of early-stage MDSCs via interfering with SOCS3 and PIAS3 respectively in breast cancer. Oncogene. 2020;39(24):4681–4694. doi: 10.1038/s41388-020-1322-4. [DOI] [PubMed] [Google Scholar]
- 38.Jin, Y. Y., Chen, Q. J., Wei, Y., Wang, Y. L., Wang, Z. W., Xu, K., He, Y., & Ma, H. B. (2016). Upregulation of microRNA-98 increases radiosensitivity in esophageal squamous cell carcinoma. Journal of Radiation Research, 57(5), 468–476. DOI: 10.1093/jrr/rrw068. [DOI] [PMC free article] [PubMed]
- 39.Kawagoe T., Sato S., Jung A., Yamamoto M., Matsui K., Kato H., Uematsu S., Takeuchi O., Akira S. Essential role of IRAK-4 protein and its kinase activity in Toll-like receptor-mediated immune responses but not in TCR signaling. J. Exp. Med. 2007;204(5):1013–1024. doi: 10.1084/jem.20061523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim G.H. MicroRNA regulation of cardiac conduction and arrhythmias. Translational Res. 2013;161(5):381–392. doi: 10.1016/j.trsl.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohanbash G., Okada H. MicroRNAs and STAT interplay. Semin. Cancer Biol. 2012;22(1):70–75. doi: 10.1016/j.semcancer.2011.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar A., Arora A., Sharma P., Anikhindi S.A., Bansal N., Singla V., Khare S., Srivastava A. Is diabetes mellitus associated with mortality and severity of COVID-19? A meta-analysis. Diabetes Metab. Synd. Clin. Res. Rev. 2020;14(4):535–545. doi: 10.1016/j.dsx.2020.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar V., Palermo R., Talora C., Campese A.F., Checquolo S., Bellavia D., Tottone L., Testa G., Miele E., Indraccolo S., Amadori A., Ferretti E., Gulino A., Vacca A., Screpanti I. Notch and NF-kB signaling pathways regulate miR-223/FBXW7 axis in T-cell acute lymphoblastic leukemia. Leukemia. 2014;28(12):2324–2335. doi: 10.1038/leu.2014.133. [DOI] [PubMed] [Google Scholar]
- 44.Li G., Fan Y., Lai Y., Han T., Li Z., Zhou P., Pan P., Wang W., Hu D., Liu X., Zhang Q., Wu J. Coronavirus infections and immune responses. J. Med. Virol. 2020;92(4):424–432. doi: 10.1002/jmv.v92.410.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li K.C., Chang Y.H., Hsu M.N., Lo S.C., Li W.H., Hu Y.C. Baculovirus-Mediated miR-214 Knockdown Shifts Osteoporotic ASCs Differentiation and Improves Osteoporotic Bone Defects Repair. Sci. Rep. 2017;7(1) doi: 10.1038/s41598-017-16547-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z., Li Y., Li Q., Zhang Z., Jiang L., Li X. Role of miR-9-5p in preventing peripheral neuropathy in patients with rheumatoid arthritis by targeting REST/miR-132 pathway. In Vitro Cell. Develop. Biol. Animal. 2019;55(1):52–61. doi: 10.1007/s11626-018-0310-2. [DOI] [PubMed] [Google Scholar]
- 47.Liao M., Liu Y., Yuan J., Wen Y., Xu G., Zhao J., Chen L., Li J., Wang X., Wang F., Liu L., Zhang S., Zhang Z. The landscape of lung bronchoalveolar immune cells in COVID-19 revealed by single-cell RNA sequencing. Nat. Med. 2020 doi: 10.1101/2020.02.23.20026690. [DOI] [PubMed] [Google Scholar]
- 48.Lim L.P., Glasner M.E., Yekta S., Burge C.B., Bartel D.P. Vertebrate microRNA genes. Science. 2003;299(5612):1540. doi: 10.1126/science.1080372. [DOI] [PubMed] [Google Scholar]
- 49.Liu D.i., Xia M., Liu Y., Tan X., He L., Liu Y.u., Chen G., Liu H. The upregulation of miR-98-5p affects the glycosylation of IgA1 through cytokines in IgA nephropathy. Int. Immunopharmacol. 2020;82:106362. doi: 10.1016/j.intimp.2020.106362. [DOI] [PubMed] [Google Scholar]
- 50.Liu F., Rehmani I., Esaki S., Fu R., Chen L., De Serrano V., Liu A. Pirin is an iron-dependent redox regulator of NF-κB. PNAS. 2013;110(24):9722–9727. doi: 10.1073/pnas.1221743110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu J., Chen W., Chen Z., Wen J., Yu H., Wang F., Wu L., He X. The effects of microRNA-98 inhibits cell proliferation and invasion by targeting STAT3 in nasopharyngeal carcinoma. Biomed. Pharmacother. 2017;93:869–878. doi: 10.1016/j.biopha.2017.06.094. [DOI] [PubMed] [Google Scholar]
- 52.Liu Y., Chen Q., Song Y., Lai L., Wang J., Yu H., Cao X., Wang Q. MicroRNA-98 negatively regulates IL-10 production and endotoxin tolerance in macrophages after LPS stimulation. FEBS Lett. 2011;585(12):1963–1968. doi: 10.1016/j.febslet.2011.05.029. [DOI] [PubMed] [Google Scholar]
- 53.Luo W., Li Y.X., Jiang L.J., Chen Q., Wang T., Ye D.W. Targeting JAK-STAT Signaling to Control Cytokine Release Syndrome in COVID-19. Trends Pharmacol. Sci. 2020;41(8):531–543. doi: 10.1016/j.tips.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma D., Lian F., Wang X. PLCG2 promotes hepatocyte proliferation in vitro via NF-κB and ERK pathway by targeting bcl2, myc and ccnd1. Artif. Cells Nanomed. Biotechnol. 2019;47(1):3786–3792. doi: 10.1080/21691401.2019.1669616. [DOI] [PubMed] [Google Scholar]
- 55.Majd M., Hosseini A., Ghaedi K., Kiani-Esfahani A., Tanhaei S., Shiralian-Esfahani H., Rahnamaee S.Y., Mowla S.J., Nasr-Esfahani M.H. MiR-9-5p and miR-106a-5p dysregulated in CD4+ T-cells of multiple sclerosis patients and targeted essential factors of T helper17/regulatory T-cells differentiation. Iranian J. Basic Med. Sci. 2018;21(3):277–283. doi: 10.22038/ijbms.2018.25382.6275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Makki M.S., Haseeb A., Haqqi T.M. MicroRNA-9 promotes IL-6 expression by inhibiting MCPIP1 expression in IL-1β-stimulated human chondrocytes HHS Public Access. Arthritis Rheumatol. 2015;67(8):2117–2128. doi: 10.1002/art.39173. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4519390/pdf/nihms-684964.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mallick B., Ghosh Z., Chakrabarti J., Jordan I.K. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PLoS ONE. 2009;4(11):e7837. doi: 10.1371/journal.pone.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Marcken M., Dhaliwal K., Danielsen A.C., Gautron A.S., Dominguez-Villar M. TLR7 and TLR8 activate distinct pathways in monocytes during RNA virus infection. Sci. Signaling. 2019;12(605):eaaw1347. doi: 10.1126/scisignal.aaw1347. [DOI] [PubMed] [Google Scholar]
- 59.Matsui S., Zhou L., Nakayama Y., Mezawa M., Kato A., Suzuki N., Tanabe N., Nakayama T., Suzuki Y., Kamio N., Takai H., Ogata Y. MiR-200b attenuates IL-6 production through IKKβ and ZEB1 in human gingival fibroblasts. Inflamm. Res. 2018;67(11–12):965–973. doi: 10.1007/s00011-018-1192-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mifflin L., Ofengeim D., Yuan J. Receptor-interacting protein kinase 1 (RIPK1) as a therapeutic target. Nat. Rev. Drug Discovery. 2020;19(8):553–571. doi: 10.1038/s41573-020-0071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mille J.K., Whittaker G.R. Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. PNAS. 2014;111(42):15214–15219. doi: 10.1073/pnas.1407087111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moles R., Bellon M., Nicot C. STAT1: A novel target of miR-150 and miR-223 is involved in the proliferation of HTLV-I-transformed and ATL cells. Neoplasia. 2015;17(5):449–462. doi: 10.1016/j.neo.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moon H.-G., Yang J., Zheng Y., Jin Y. miR-15a/16 Regulates Macrophage Phagocytosis after Bacterial Infection. J. Immunol. 2014;193(9):4558–4567. doi: 10.4049/jimmunol.1401372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Moriya N., Shibasaki S., Karasaki M., Iwasaki T., Roemer K. The impact of microRNA-223-3p on IL-17 receptor d expression in synovial cells. PLoS ONE. 2017;12(1):e0169702. doi: 10.1371/journal.pone.0169702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.O Brien, J., Hayder, H., Zayed, Y., & Peng, C. (2018). Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Frontiers in Endocrinology, 9. DOI: 10.3389/fendo.2018.00402/full. [DOI] [PMC free article] [PubMed]
- 66.Paul Zimmet, M. ., George Alberti, M. D., Stefan Bornstein, M. D., Robert H. Eckel, M. D., Geltrude Mingrone, M. D., Bernhard Boehm, M. D., Mark E. Cooper, Ph.D. Zhonglin Chai, P. D., Stefano Del Prato, M. D., & Eric Renard, M. D. (2020). New-Onset Diabetes in Covid-19. New England Journal of Medicine, 383(8), 787–789. Paul Zimmet, M. ., George Alberti, M. D., Stefan Bornstein, M. D., Robert H. Eckel, M. D., Geltrude Mingrone, M. D., Bernhard Boehm, M. D., Mark E. Cooper, Ph.D. Zhonglin Chai, P. D., Stefano Del Prato, M. D., & Eric Renard, M. D. (2020). New-Onset Diabetes in Covid-19. New England Journal of Medicine, 383(8), 787–789. [DOI] [PMC free article] [PubMed]
- 67.Piganis R.A.R., De Weerd N.A., Gould J.A., Schindler C.W., Mansell A., Nicholson S.E., Hertzog P.J. Suppressor of Cytokine Signaling (SOCS) 1 inhibits type I interferon (IFN) signaling via the interferon α receptor (IFNAR1)-associated tyrosine kinase tyk2. J. Biol. Chem. 2011;286(39):33811–33818. doi: 10.1074/jbc.M111.270207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pinatel E.M., Orso F., Penna E., Cimino D., Elia A.R., Circosta P., Dentelli P., Brizzi M.F., Provero P., Taverna D., Lafrenie R. miR-223 is a coordinator of breast cancer progression as revealed by bioinformatics predictions. PLoS ONE. 2014;9(1):e84859. doi: 10.1371/journal.pone.0084859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qian S., Ding J.-yong., Xie R., An J.-hong., Ao X.-jun., Zhao Z.-guo., Sun J.-guo., Duan Y.-zhong., Chen Z.-tang., Zhu B. MicroRNA expression profile of bronchioalveolar stem cells from mouse lung. Biochem. Biophys. Res. Commun. 2008;377(2):668–673. doi: 10.1016/j.bbrc.2008.10.052. [DOI] [PubMed] [Google Scholar]
- 70.Qin D., Wang X., Li Y., Yang L., Wang R., Peng J., Essandoh K., Mu X., Peng T., Han Q., Yu K.-J., Fan G.-C. MicroRNA-223-5p and -3p Cooperatively suppress necroptosis in ischemic/reperfused hearts. J. Biol. Chem. 2016;291(38):20247–20259. doi: 10.1074/jbc.M116.732735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rawlings J.S., Rosler K.M., Harrison D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004;117(8):1281–1283. doi: 10.1242/jcs.00963. [DOI] [PubMed] [Google Scholar]
- 72.Rehei A.L., Zhang L., Fu Y.X., Mu W.B., Yang D.S., Liu Y., Zhou S.J., Younusi A. MicroRNA-214 functions as an oncogene in human osteosarcoma by targeting TRAF3. Eur. Rev. Med. Pharmacol. Sci. 2018;22(16):5156–5164. doi: 10.26355/eurrev_201808_15711. [DOI] [PubMed] [Google Scholar]
- 73.Sabbatinelli J., Giuliani A., Matacchione G., Latini S., Laprovitera N., Pomponio G., Ferrarini A., Svegliati Baroni S., Pavani M., Moretti M., Gabrielli A., Procopio A.D., Ferracin M., Bonafè M., Olivieri F. Decreased serum levels of the inflammaging marker miR-146a are associated with non-clinical response to tocilizumab in COVID-19 patients. Mech. Ageing Dev. 2021;193 doi: 10.1016/j.mad.2020.111413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saçar Demirci M.D., Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020 doi: 10.1101/2020.03.15.992438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seif F., Khoshmirsafa M., Aazami H., Mohsenzadegan M., Sedighi G., Bahar M. The role of JAK-STAT signaling pathway and its regulators in the fate of T helper cells. Cell Commun. Signaling. 2017;15(1) doi: 10.1186/s12964-017-0177-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Soheilifar, M. H., Keshmiri Neghab, H., & Basiri, P. (2020). Biological Impacts of MicroRNAs in Covid-19: Implications for Anti-Viral miRNA-Based Therapies. Archives of Clinical Infectious Diseases, In Press(In Press). DOI: DOI: 10.5812/archcid.104140.
- 77.Sohn K.M., Lee S.G., Kim H.J., Cheon S., Jeong H., Lee J., Kim I.S., Silwal P., Kim Y.J., Paik S., Chung C., Park C., Kim Y.S., Jo E.K. COVID-19 patients upregulate toll-like receptor 4-mediated inflammatory signaling that mimics bacterial sepsis. J. Korean Med. Sci. 2020;35(38) doi: 10.3346/JKMS.2020.35.E343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Stark C.J., Atreya C.D. Molecular advances in the cell biology of SARS-CoV and current disease prevention strategies. Virology Journal. 2005;2 doi: 10.1186/1743-422X-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Streppel M.M., Pai S., Campbell N.R., Hu C., Yabuuchi S., Canto M.I., Wang J.S., Montgomery E.A., Maitra A. MicroRNA 223 is upregulated in the multistep progression of barrett’s esophagus and modulates sensitivity to chemotherapy by targeting PARP1. Clin. Cancer Res. 2013;19(15):4067–4078. doi: 10.1158/1078-0432.CCR-13-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tanaka T., Soriano M.A., Grusby M.J. SLIM is a nuclear ubiquitin E3 ligase that negatively regulates STAT signaling. Immunity. 2005;22(6):729–736. doi: 10.1016/j.immuni.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 81.Toubiana J., Rossi A.L., Belaidouni N., Grimaldi D., Pene F., Chafey P., Comba B., Camoin L., Bismuth G., Claessens Y.E., Mira J.P., Chiche J.D. Src-family-tyrosine kinase Lyn is critical for TLR2-mediated NF-κ activation through the PI 3-kinase signaling pathway. Innate Immunity. 2015;21(7):685–697. doi: 10.1177/1753425915586075. [DOI] [PubMed] [Google Scholar]
- 82.Valmiki S., Ahuja V., Paul J. MicroRNA exhibit altered expression in the inflamed colonic mucosa of ulcerative colitis patients. World J. Gastroenterol. 2017;23(29):5324–5332. doi: 10.3748/wjg.v23.i29.5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang J., Chen L., Jin S., Lin J., Zheng H., Zhang H., Fan H., He F., Ma S., Li Q. MiR-98 promotes chondrocyte apoptosis by decreasing Bcl-2 expression in a rat model of osteoarthritis. Acta Biochim. Biophy. Sin. 2016;48(10):923–929. doi: 10.1093/abbs/gmw084. [DOI] [PubMed] [Google Scholar]
- 84.Wang W., Ye L., Ye L., Li B., Gao B., Zeng Y., Kong L., Fang X., Zheng H., Wu Z., She Y. Up-regulation of IL-6 and TNF-α induced by SARS-coronavirus spike protein in murine macrophages via NF-κB pathway. Virus Res. 2007;128(1–2):1–8. doi: 10.1016/j.virusres.2007.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X., Ding Y.Y., Chen Y., Xu Q.Q., Qian H.G., Qian G.W., Cao L., Zhou P.W., Hou M., Lv T.H. MiR-223-3p alleviates vascular endothelial injury by targeting IL6ST in Kawasaki disease. Front. Pediatrics. 2019;7(JUN) doi: 10.3389/fped.2019.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Y., Han Z., Fan Y., Zhang J., Chen K., Gao L., Zeng H., Cao J., Wang C. MicroRNA-9 inhibits NLRP3 inflammasome activation in human atherosclerosis inflammation cell models through the JAK1/STAT signaling pathway. Cell. Physiol. Biochem. 2017;41(4):1555–1571. doi: 10.1159/000470822. [DOI] [PubMed] [Google Scholar]
- 87.Watanabe T., Keino H., Kudo A., Sato Y., Okada A.A. MicroRNAs in retina during development of experimental autoimmune uveoretinitis in rats. Br. J. Ophthalmol. 2016;100(3):425–431. doi: 10.1136/bjophthalmol-2015-306924. [DOI] [PubMed] [Google Scholar]
- 88.Widiasta A., Sribudiani Y., Nugrahapraja H., Hilmanto D., Sekarwana N., Rachmadi D. Potential role of ACE2-related microRNAs in COVID-19-associated nephropathy. Non-Coding RNA Research. 2020;5(4):153–166. doi: 10.1016/j.ncrna.2020.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Witte S., Muljo S.A. Integrating non-coding RNAs in JAK-STAT regulatory networks. Jak-Stat. 2014;3(1):e28055. doi: 10.4161/jkst.28055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu H., Zheng X., Dong L., Li C., Zhang M., Wang G., Wang K. Pir-B inhibits the DC function and disturbs the Th17/Treg balance in lung cancer murine model. Oncotarget. 2017;8(70):114710–114721. doi: 10.18632/oncotarget.21763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu J., Niu P., Zhao Y., Cheng Y., Chen W., Lin L., Lu J., Cheng X., Xu Z., Ahmad A. Impact of miR-223-3p and miR-2909 on inflammatory factors IL-6, IL-1ß, and TNF-α, and the TLR4/TLR2/NF-κB/STAT3 signaling pathway induced by lipopolysaccharide in human adipose stem cells. PLoS ONE. 2019;14(2):e0212063. doi: 10.1371/journal.pone.0212063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xia H.F., Jin X.H., Cao Z.F., Shi T., Ma X. MiR-98 is involved in rat embryo implantation by targeting Bcl-xl. FEBS Lett. 2014;588(4):574–583. doi: 10.1016/j.febslet.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 93.Xiong Y., Liu Y., Cao L., Wang D., Guo M., Guo D., Hu W., Yang J., Tang Z., Zhang Q., Shi M., Liu Y., Zhou Y., Lan K., Chen Y. Transcriptomic Characteristics of Bronchoalveolar Lavage Fluid and Peripheral Blood Mononuclear Cells in COVID-19 Patients. SSRN Electr. J. 2020 doi: 10.2139/ssrn.3549993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yousefi H., Poursheikhani A., Bahmanpour Z., Vatanmakanian M., Taheri M., Mashouri L., Alahari S.K. SARS-CoV infection crosstalk with human host cell noncoding-RNA machinery: An in-silico approach. Biomed. Pharmacother. 2020;130:110548. doi: 10.1016/j.biopha.2020.110548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Yuan S., Tang C., Chen D., Li F., Huang M., Ye J., He Z., Li W., Chen Y., Lin X., Wang X., Cai X. MiR-98 modulates cytokine production from human PBMCs in systemic lupus erythematosus by targeting IL-6 mRNA. J. Immunol. Res. 2019;2019:1–11. doi: 10.1155/2019/9827574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zeng F., Huang Y., Guo Y., Yin M., Chen X., Xiao L., Deng G. Association of inflammatory markers with the severity of COVID-19: A meta-analysis. Int. J. Infect. Dis. 2020;96:467–474. doi: 10.1016/j.ijid.2020.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhang H., Penninger J.M., Li Y., Zhong N., Slutsky A.S. Angiotensin-converting enzyme 2 (ACE2) as a SARS-CoV-2 receptor: molecular mechanisms and potential therapeutic target. Intensive Care Med. 2020;46(4):586–590. doi: 10.1007/s00134-020-05985-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang N., Fu L., Bu Y., Yao Y., Wang Y. Downregulated expression of miR-223 promotes Toll-like receptor-activated inflammatory responses in macrophages by targeting RhoB. Mol. Immunol. 2017;91:42–48. doi: 10.1016/j.molimm.2017.08.026. [DOI] [PubMed] [Google Scholar]
- 99.Zhang X., Wu K., Wang D., Yue X., Song D., Zhu Y., Wu J. Nucleocapsid protein of SARS-CoV activates interleukin-6 expression through cellular transcription factor NF-κB. Virology. 2007;365(2):324–335. doi: 10.1016/j.virol.2007.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang Z., Liu K., Chen Y., Li Z., Yan N., Zhang J. [The expression of miR-183 family in the pathogenesis and development of noise-induced deafness]. Lin Chuang Er Bi Yan Hou Tou Jing Wai Ke Za Zhi = J. Clin. Otorhinolaryngol. Head Neck Surg. 2014;28(7):468–472. [PubMed] [Google Scholar]
- 101.Zhao, J., Lin, Q., Kim, K. J., Dardashti, F. D., Kim, J., He, F., & Sun, Y. (2015). Ngn1 inhibits astrogliogenesis through induction of miR-9 during neuronal fate specification. ELife, 4(AUGUST2015). DOI: 10.7554/eLife.06885. [DOI] [PMC free article] [PubMed]
- 102.Zhou Z., Ren L., Zhang L., Zhong J., Xiao Y., Jia Z., Guo L., Yang J., Wang C., Jiang S., Yang D., Zhang G., Li H., Chen F., Xu Y., Chen M., Gao Z., Yang J., Dong J., Liu B., Zhang X., Wang W., He K., Jin Q., Li M., Wang J. Heightened Innate Immune Responses in the Respiratory Tract of COVID-19 Patients. Cell Host Microbe. 2020;27(6):883–890.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zisman L.S. ACE and ACE2: A tale of two enzymes. Eur. Heart J. 2005;26(4):322–324. doi: 10.1093/eurheartj/ehi043. [DOI] [PMC free article] [PubMed] [Google Scholar]