Abstract
HL7 Fast Healthcare Interoperability Resources (FHIR) is one of the current data standards for enabling electronic healthcare information exchange. Previous studies have shown that FHIR is capable of modeling both structured and unstructured data from electronic health records (EHRs). However, the capability of FHIR in enabling clinical data analytics has not been well investigated. The objective of the study is to demonstrate how FHIR-based representation of unstructured EHR data can be ported to deep learning models for text classification in clinical phenotyping. We leverage and extend the NLP2FHIR clinical data normalization pipeline and conduct a case study with two obesity datasets. We tested several deep learning-based text classifiers such as convolutional neural networks, gated recurrent unit, and text graph convolutional networks on both raw text and NLP2FHIR inputs. We found that the combination of NLP2FHIR input and text graph convolutional networks has the highest F1 score. Therefore, FHIR-based deep learning methods has the potential to be leveraged in supporting EHR phenotyping, making the phenotyping algorithms more portable across EHR systems and institutions.
Introduction
Electronic health record (EHR) data is being increasingly used for conducting clinical and translational research. Large scale research networks such as the electronic Medical Records and Genomics (eMERGE) network1, Pharmacogenomics Research Network (PGRN)2, The National Patient-Centered Clinical Research Network (PCORnet)3, and the UK BioBank4 have enabled multi-institutional studies using EHR data5-8.
However, the lack of interoperability of EHR systems is a challenge for healthcare institutions and clinical research centers. Health Level 7 (HL7) Fast Healthcare Interoperability Resources (FHIR)9 is one of the current data standards for representation of EHR data, and has been adopted by major EHR vendors to enhance data and system interoperability among different EHR implementations. The overall goal of FHIR is to facilitate available, discoverable and interpretable data sharing across institutions. Many research communities and medical centers are supporting the advancement and development of FHIR standards, including i2b210, SMART on FHIR11 and eMERGE12.
Due to its advantages on implementation readiness and interoperability among different EHR systems, FHIR is increasingly being used for exchanging EHR data. On top of representing normalized structured data, NLP2FHIR13 has been developed as a data normalization pipeline, which provides a reference implementation of the FHIR standard for modeling unstructured data. A follow-up study was done on computational phenotyping with FHIR- based EHR representation, which demonstrated that NLP2FHIR-based representation of EHR data can effectively identify phenotypes using the case study on patients with obesity and multiple comorbidities from discharge summaries14, 15. Machine learning models such as Decision Tree, Support Vector Machine and Random Forest were also tested for effectively identification of obesity and multiple comorbidities using semi-structured information from discharge summaries.
However, little work has been done in standards and clinical research informatics communities on adopting FHIR for deep learning models. In this study, we use existing deep learning methods including convolutional neural networks (CNN)16, Gated Recurrent Unit (GRU)17 and Text Graph Convolutional Network (GCN)18 to demonstrate how FHIR-based data representation can be integrated into deep learning models. We leveraged the NLP2FHIR pipeline and deep learning models on a case study to predict obesity and its comorbidities in two different datasets. We found that the combination of NLP2FHIR input, which is a graph-based input format, and the text graph convolutional networks has the highest F1 score. It shows promises to effectively use NLP2FHIR outputs as an input standard for deep learning methods in supporting EHR phenotyping, making the phenotyping algorithms more portable across data systems and institutions.
Related Work
Standard-based phenotype algorithms and execution workflow have been studied in the clinical research informatics community to allow implementations of clinical logic and value sets in a modular software architecture19. Various machine learning algorithms have been leveraged by the Phenotype Execution and Modeling Architecture (PhEMA) project to identify phenotypes and sub-phenotypes for a number of conditions including acute kidney injury, heart failure, major depression and Alzheimer’s disease20-22. Rasmussen et al23 also proposed a framework using a common data model (CDM), standardized representation of the phenotype algorithms logic, and technical solutions to facilitate federated execution of queries. It is envisioned to help guide future research in operationalizing phenotype algorithm portability at scale. Hripcsak et al. described the process of transferring the phenotypes of type 2 diabetes mellitus (T2DM) and attention deficit and hyperactivity disorder (ADHD) to the Observational Medical Outcomes Partnership (OMOP) CDM within the eMERGE network24.
Standardized preprocessing pipelines for machine learning25, 26 can enable fair comparisons among machine learning models on publicly available datasets such as MIMIC III27. To further standardize these datasets into the clinical interoperable standard of FHIR, one of the representative work is done by Rajkomar et al28. They represented EHR data using FHIR and demonstrated that FHIR is capable of medical event prediction when tested on de-identified structured and unstructured EHR data from two US academic medical centers. Sharma et al. have studied a phenotyping system to integrate both rule-based and statistical machine learning methods29. The system has leveraged OHDSI’s OMOP CDM with Unified Medical Language System (UMLS) Concept Unique Identifiers (CUIs) to represent clinical NLP concepts as input features for machine learning based classifiers in phenotype identification systems. Hong et al. has demonstrated FHIR-based EHR phenotyping can be applied to semi-structured discharge summaries for multiple comorbidities identification15. The NLP2FHIR implementation contains several different NLP components leveraging existing information extraction systems including cTAKES30, MedTagger31, MedXN32 and UMLS VTS33. On the same task, Yao et al. used word embeddings and entity embeddings on CNN adapted from rule-based systems34, but the system did not leverage any standards or CDMs and hence have limited interoperability.
Materials and Methods Materials
In this work, we selected two datasets for our analysis: the i2b2 2008 obesity dataset35 and MIMIC III dataset27.
The i2b2 2008 obesity dataset is a fully de-identified dataset consisting of discharge summaries. The dataset contains human-curated obesity status explicitly mentioned in the texts as well as 15 comorbidities consisting of asthma, atherosclerotic cardiovascular disease (CAD), congestive heart failure (CHF), depression, diabetes mellitus, hypertension, gastroesophageal reflux disease (GERD), gallstones, hypercholesterolemia, hypertriglyceridemia, obstructive sleep apnea (OSA), osteoarthritis (OA), peripheral vascular disease (PVD), and venous insufficiency35. For each comorbidity, there are 4 labels as the prediction target: present, absent, questionable or unmentioned. Both textual and intuitive judgments are provided in the dataset for each patient. While the judgment of textual is based on explicit mentions, the intuitive judgments are based on the annotators’ judgment, and may lead to additional inference (e.g. the statement of weights to infer obesity).
The MIMIC III (Medical Information Mart for Intensive Care) is a publicly available dataset containing vital signs, medications, lab test results, observations and clinical notes of 53,423 adult admissions of critical care units. To build an obesity related comorbidity prediction dataset which is similar to the i2b2 dataset, we follow the experiment settings of Hong et al. to validate the design of portability15. The obesity and non-obesity groups are selected based on body mass index (BMI) for adult patients. Adults with a BMI value larger than 30 at admission with discharge summaries are categorized in the obesity group, while adults with a BMI value between 18.5 to 24.9 at admission are categorized as the control group. A total of 2000 discharge summaries are randomly selected among all available discharge summary notes, with 1000 each for case and control. 70% of the notes are selected as the training set (n=1400), and 30% of the notes are selected as the test set (n=600).
We present dataset characteristics in Table 1.
Table 1.
Statistics of the i2b2 and MIMIC IIIs datasets
| Dataset | Train | Test | Average # of words/doc | Vocabulary size | Average # concepts/doc | Extracted concepts |
|---|---|---|---|---|---|---|
| I2b2 2008 | 719 | 496 | 833.4 | 8633 | 106.6 | 1919 |
| MIMIC III | 1400 | 600 | 1491.1 | 13129 | 158.6 | 3220 |
Methods
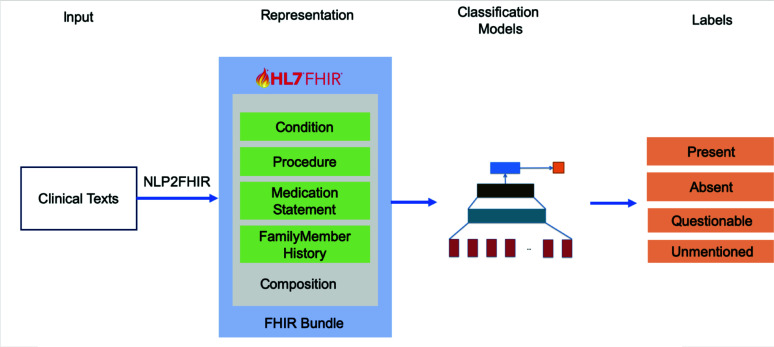
The proposed workflow of FHIR-enabled text classification application for clinical phenotyping is illustrated in Figure 1. Given the original texts from the two datasets, a document is first tokenized into to a list of tokens as the input of the deep learning models. During the preprocessing, stop words and words appearing less than three times are removed for the purpose of better performance in the embedding training phase. Then, FHIR resources in JSON format produced by the NLP2FHIR pipeline are concatenated into token-like representations, which are categorized into different resources (Condition, Procedure, MedicationStatement, and FamilyMemberHistory) and grouped into FHIR Bundles. The NLP2FHIR representation is based on an existing system primarily validated on various data types36. Figure 2 shows an example of NLP2FHIR output of the sentence “Ms. [Name] is a 64-year-old female with nonischemic cardiomyopathy and class II-III symptoms who presented with worsening volume overload”. There are 2 concepts (“cardiomyopathy” and “worsening volume overload”) identified by cTAKES to be normalized to SNOMED CT codes. To make the extracted concept objects compatible with word-based input formats, the coding system URL (i.e. http://snomed.info/sct), the code, and the polarity from the “abatementString” field are concatenated into one “word” to represent their uniqueness. As an important factor for the learning and prediction for many machine learning models, although word orders are not supposed to be preserved by FHIR, it is preserved sequentially in the NLP2FHIR output naturally as the dictionary lookup generates sequential concepts as outputs.
Figure 1.
Workflow of text classification tasks for clinical phenotyping using NLP2FHIR
Figure 2.
NLP2FHIR JSON representation of a sample clinical text to concept-based representation for deep learning models
After both the texts and NLP2FHIR representations are ready, the data are fed into machine learning/deep learning-based classifiers to classify the documents. We tested three different deep learning models in this study: CNN, GRU and Text GCN. The details of the models in this study are described as follows.
CNN
Convolutional neural networks are one of the earliest and most commonly used deep learning models in text classification tasks16, 37. Experiments in the biomedical domain have shown that CNN can achieve good performance without extensive model tuning. In a typical 1-dimensional CNN for text classification tasks, it can capture local contexts by leveraging a convolutional kernel (or filter) acting as a sliding window among tokens.
In this study, we use a fixed length CNN where the length is a hyperparameter. If the input document is shorter than the expected length, the end of the sequence was padded with zeros. If the input document was longer than the expected length, the input document was truncated.
RNN/GRU
One challenge for CNN is that it does not capture long-term information when the contexts are not close. RNNs are capable to handle long-term patterns in sequential inputs, because the state of previous RNN units can be passed to the units behind them until the end of the sequence. There are multiple variations of RNN38 which usually have better performance than vanilla RNN, including Long short-term memory (LSTM)39 or Gated Recurrent Unit (GRU)17. Experiments showed that there is no consistency on which model would perform better in general. In our experiments, we selected GRU due to its faster convergence time, and it should not impact our conclusion as the performances of these two models are usually comparable with each other17.
Text GCN
Kipf et al. proposed GCN40, a graph neural network architecture for node classification. GCN is one of the methods to generalize neural networks to structured datasets. While CNNs or RNNs are typically good at modeling “array-like” input data, they will face challenges to model graphs as the data connections are more challenging to capture. Common characteristics like depths, degrees, density, or node connectivity cannot be easily modeled without adapting to graph specific models.
To make GCN work better for text classification tasks, Text GCN is proposed by Yao et al. as an extension of GCN18. Text GCN uses words and documents as nodes, and uses the trained embedding to classify document nodes into categories. The major differences between GCN and Text GCN is how the edges in the graph are represented.
There are 2 types of nodes in Text GCN: document nodes and token nodes. When a word appears in a document, an edge between the document node and the token node will be generated. Each element of adjacent matrix A which keeps the edge weights between two nodes (m and n) are defined as follows:

The PMI (point-wise mutual information) given a word pair m, n can be calculated by:
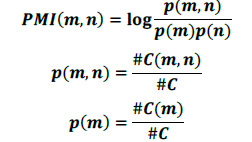
where #C(m) is the count of the sliding windows with the token m from the whole corpus, #C(m, n) is the count of sliding windows containing both m and n, and #C is the count of windows in the corpus. Only the positive PMI edges are added, since a negative PMI means there is little to no correlation of the words within the corpus. The constructed graph is then fed into a 2-layer GCN for training as proposed in Yao el al.18 and Kipf and Welling40, which allows messages passing among different nodes and layers.
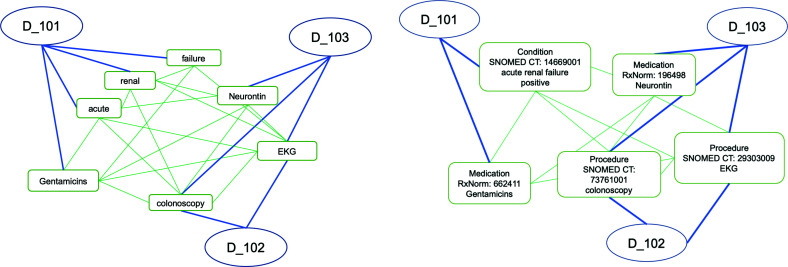
The illustration of how the Text GCN graph can be adapted to the token and NLP2FHIR representations is shown in Figure 3. As a comparison, the phrase “acute renal failure” is normalized to a Condition concept with a SNOMED CT code 14669001.
Figure 3.
Token-based (left) and FHIR-concept-based (right) graphs for Text GCN text classification. The nodes denoted by circles are document nodes with document IDs, and the nodes denoted by round rectangles are token (left) or concept (right).
Results
In this section, we compare the performances of original tokens with the NLP2FHIR representations, used in different deep learning methods (CNN, GRU and Text GCN) in clinical text classification tasks. In the i2b2 obesity dataset, we use the official training and test set for evaluation, while the training and testing set of the MIMIC III dataset in this study is split by a ratio of 70% and 30% from the randomly selected notes described in the Materials section. All discharge summaries are flattened as lists of lower-case tokens with all the line-breaks removed before entering into the deep learning models.
The Text GCN implementation is adopted from Yao et al41 and is implemented by scikit-learn42 and TensorFlow43. The CNN and GRU implementations are based on Keras44 using a TensorFlow backend. All the embedding layers are trained on the training set, and no pre-trained word embedding models are used in the experiments. The hyperparameters are tuned for different datasets separately. For the CNN model, we used 1 convolutional layer before 1 fully connected layer with the number of filters as 200, maximum input length as 600, the embedding dimension as 50, and the convolution kernel size as 3. For GRU, the hidden dimension is set to 128. The number of epochs for both the CNN and GRU are 40, with an early stopping patience of 5 monitoring the validation loss. The validation set consists of 10% of the training data. The source code of the implementation can be found at https://github.com/BD2KOnFHIR/nlp2fhir-deep-learning.
For Text GCN, only text data is used, and for FHIR, the concepts consist of code and polarity (positive, negative). It is transformed to a multi-class document classification problem. The graph statistics of the i2b2 and MIMIC datasets are shown in Table 2. We used the default settings of 2 GCN layers as it shows better performance than the 1-layer model, and it is more likely to converge compared to 3 or more layer models in our early experiments.
Table 2.
Statistics of the Text GCN graph for i2b2 and MIMIC datasets
| Dataset | # of nodes | # of edges | Density |
|---|---|---|---|
| I2b2 2008 | 6134 | 79598 | 4.23 * 10e-3 |
| MIMIC III | 9204 | 168143 | 3.97 * 10e-3 |
The reported performances are the accuracy (equivalent to micro-precision, recall and F1-score), macro-averaged precision, recall and F1-score of the obesity and its 15 comorbidities and its 95% confidence intervals (95% CI) among different comorbidities. Table 3 and 4 show the mean macro-averaged precision, recall and F1-scores among obesity and different comorbidities. For the i2b2 dataset, we used textual gold labels instead of intuitive, which is more relevant to show how models understand the contexts without additional inferences by human experts.
Table 3.
Performances of different experiment settings on the i2b2 dataset by accuracy, macro averaged precision, recall and F1-score with the 95% CI. The highest scores are on bold
| Dataset | CNN | CNN-FHIR | LSTM | LSTM-FHIR | Text GCN | Text GCN - FHIR |
|---|---|---|---|---|---|---|
| Accuracy | 0.737 ± 0.068 | 0.748 ± 0.068 | 0.652 ± 0.075 | 0.697 ± 0.075 | 0.707 ± 0.055 | 0.795 ± 0.044 |
| Precision | 0.489 ± 0.080 | 0.493 ± 0.058 | 0.400 ± 0.074 | 0.520 ± 0.064 | 0.347 ± 0.057 | 0.525 ± 0.049 |
| Recall | 0.504 ± 0.055 | 0.519 ± 0.055 | 0.457 ± 0.055 | 0.478 ± 0.056 | 0.500 ± 0.056 | 0.523 ± 0.045 |
| F1-score | 0.495 ± 0.060 | 0.505 ± 0.056 | 0.415 ± 0.059 | 0.495 ± 0.061 | 0.410 ± 0.057 | 0.524 ± 0.048 |
Table 4.
Performances of different experiment settings the MIMIC III dataset by accuracy, macro averaged precision, recall and F1-score with the 95% CI. The highest scores are on bold
| Dataset | CNN | CNN-FHIR | LSTM | LSTM-FHIR | Text GCN | Text GCN - FHIR |
|---|---|---|---|---|---|---|
| Accuracy | 0.859 ± 0.080 | 0.857 ± 0.083 | 0.824 ± 0.083 | 0.873 ± 0.079 | 0.746 ± 0.077 | 0.914 ± 0.070 |
| Precision | 0.628 ± 0.052 | 0.607 ± 0.036 | 0.587 ± 0.041 | 0.614 ± 0.069 | 0.388 ± 0.076 | 0.616 ± 0.044 |
| Recall | 0.645 ± 0.053 | 0.638 ± 0.021 | 0.596 ± 0.001 | 0.643 ± 0.057 | 0.623 ± 0.037 | 0.721 ± 0.058 |
| F1-score | 0.625 ± 0.052 | 0.616 ± 0.036 | 0.590 ± 0.025 | 0.622 ± 0.057 | 0.478 ± 0.047 | 0.664 ± 0.050 |
From the experiments, we observe that NLP2FHIR representations provide better performances when used as input compared to the original texts. In most cases the use of FHIR representation have positive impacts on classification performance, with the CNN vs CNN-FHIR on MIMIC III dataset the exception in our experiments. CNN models are one of the strong baseline models for text classification on raw texts, with little to none preprocessing needed, in many studies. Therefore, applying information extraction pipeline (dictionary lookup) on texts may not lead to favorable performances on CNN models.
Text GCN, which is a graph-based algorithm, also outperforms other deep learning models. The main reason is that the data we tested are very sparse. Unlike other text classification tasks presented in the Text GCN experiments, such as movie reviews or abstracts, only a few tokens are related to the classification results. That results in difference in density of the graphs.
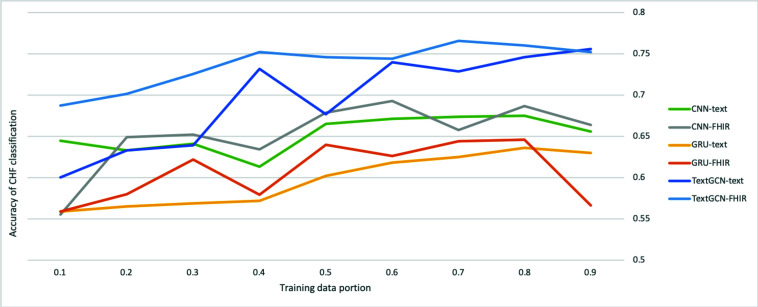
We also experimented with multiple settings with different portions of training data. The impacts of accuracy in one of the comorbidities (CHF) on the deep learning models are shown in Figure 4. We can observe increasing trends in general from left (fewer training samples) to right (more training samples), meaning increasing the amount of data into training while reducing the amount of data into testing. However, the trend is not obvious when the proportion is larger than 0.5, indicating the amount of training data can be considered sufficient to learn the hidden patterns in a fully annotated dataset. After the amount of data reaching the threshold, the model may at risk of overfitting that may have negative impact on the generalizability of the trained models due to the lack of generalizable test samples.
Figure 4.
The impact of proportions of data into training on one of the comorbidity classifications (Congestive Heart Failure, CHF). The x-axis is the ratio of training data used from the all labeled data (training + testing), and the y-axis is the accuracy of CHF as an example of the comorbidities.
Discussion
In this paper, we designed and experimented with token-based and NLP2FHIR representations for text classification models. The tested models represent three different type of information for classification: CNN primarily classifies texts based on collections (max pooling) of local contexts, RNN on the actual sequences and Text GCN on graph structures of the tokens or concepts. The experimental results show that the sequence of normalized concept models from FHIR representation is better than the input data from the raw texts, or sequence of tokens, when applied to vanilla deep learning models without feature extraction and feature engineering. One potential reason for that is the contribution of the normalized representations that may be more informative comparing with sequences of tokens. With normalized concepts, the input sequence of the model is more condensed and standardized with potentially more edges in the graph. Another advantage of migrating data into the FHIR representation is the implementations and toolkits available through open-source FHIR development efforts. For the standardized implementation with improved portability and interoperability, deep learning applications can be deployable with minimal efforts across different datasets and systems.
One usage of the proposed standard-based design is to allow de-identified data sharing regarding protected health information (PHI). The FHIR elements will only contain higher-level concepts from clinical ontologies and knowledge bases. As general concepts (separated from any specific patient), they are intrinsically free of PHI as defined by Health Insurance Portability and Accountability Act (HIPAA)45 that may make the data identifiable. The NLP2FHIR representation includes the annotated clinical mentions with normalized entities that are expected to be PHI-free. This can further inspire more pilot studies on distributing NLP2FHIR representations without the original texts as a standard format to facilitate standard-based phenotype identification algorithms without sharing PHI or de-identification efforts.
There are also several limitations in this study. First, for the text classification problems, we only demonstrate a few models as a case study, and it is not an exhaustive evaluation to determine the best performing ML methods. As comparisons, our overall macro F1 score of 0.524 in the i2b2 dataset is higher than the decision tree on CUI performances (0.5121)29 but lower than 0.6578 when a decision tree classifier is used including section information in Hong et al15. Likewise, other studies have demonstrated better performance than that in our experiments, although we note they were conducted based on different pre-processing steps and experimental settings. For instance, many top systems from the i2b2 challenge filtered the discharge summaries that are not relevant to the patients and developed keyword-based approaches to identify comorbidities46-48, which contain hand-crafted rules and regular expressions that are not portable. However, our major goal in this study was to demonstrate the portability of deep learning models when applied to NLP2FHIR representations. Therefore, we did not work towards building corpus-specific dictionaries or rules, as such efforts and models tend to overfit to a specific task or corpus.
Second, the implementation and evaluation did not utilize any document or textual structures. The current structure represents the document structure such as sections or sentences, but in the experiments, we did not weigh in the structure due to the lack of sentence-level and section-level gold standard labels.
Third, the semantic based representations may not fully utilize syntax-based features that may be helpful for phenotype classification49-51, because the sentence structural information is omitted by the concepts and thus are not retained in the FHIR based representations. This makes it challenging to apply NLP2FHIR outputs for contextual pre-trained language models such as BERT (Bidirectional Encoder Representations from Transformers)52 and RoBERTa53, which are intended to handle natural languages as neural language models rather than coded phenotypic representations. This can cause some contextual information eliminated before feeding into the neural models for the classification task.
Conclusion
NLP2FHIR outputs can be ported and integrated into deep learning methods. We found that the classification results of NLP2FHIR based methods outperformed the methods with original texts. We demonstrated that FHIR-based deep learning methods could be leveraged in supporting EHR phenotyping, making the phenotyping algorithms more portable across data systems and institutions. In the future, we will work on improving the performance by adding document structure such as sentences and sections into the document modeling.
Acknowledgment
Research reported in this publication was supported by National Institutes of Health under the awards FHIRCat (R56EB028101), BD2K (U01 HG009450), and PhEMA (R01 GM105688). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Reference
- 1.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genetics in Medicine. 2013;15(10):761–71. doi: 10.1038/gim.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pharmacogenomics Research Network [Google Scholar]
- 3.PCORnet: the National Patient-Centered Clinical Research Network [Google Scholar]
- 4.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562(7726):203–9. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin-Sanchez FJ, Aguiar-Pulido V, Lopez-Campos GH, Peek N, Sacchi L. Secondary Use and Analysis of Big Data Collected for Patient Care. Yearb Med Inform. 2017;26(1):28–37. doi: 10.15265/IY-2017-008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danciu I, Cowan JD, Basford M, Wang X, Saip A, Osgood S, et al. Secondary use of clinical data: the Vanderbilt approach. Journal of biomedical informatics. 2014;52:28–35. doi: 10.1016/j.jbi.2014.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy SN, Mendis ME, Berkowitz DA, Kohane I, Chueh HC. Integration of clinical and genetic data in the i2b2 architecture. AMIA Annual Symposium proceedings AMIA Symposium. 2006;2006:1040. [PMC free article] [PubMed] [Google Scholar]
- 8.Benincasa G, Marfella R, Della Mura N, Schiano C, Napoli C. Strengths and Opportunities of Network Medicine in Cardiovascular Diseases. Circulation Journal. 2020;84(2):144–52. doi: 10.1253/circj.CJ-19-0879. [DOI] [PubMed] [Google Scholar]
- 9.FHIR Overview. [Available from: https://www.hl7.org/fhir/overview.html. [Google Scholar]
- 10.i2b2: Informatics for Integrating Biology & the Bedside. [Available from: https://www.i2b2.org/ [Google Scholar]
- 11.Mandel JC, Kreda DA, Mandl KD, Kohane IS, Ramoni RB. SMART on FHIR: a standards-based, interoperable apps platform for electronic health records. Journal of the American Medical Informatics Association. 2016;23(5):899–908. doi: 10.1093/jamia/ocv189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Taylor CO, Lemke KW, Richards TM, Roe KD, He T, Arruda-Olson A, et al. Comorbidity Characterization Among eMERGE Institutions: A Pilot Evaluation with the Johns Hopkins Adjusted Clinical Groups® System. AMIA Jt Summits Transl Sci Proc. 2019;2019:145–52. [PMC free article] [PubMed] [Google Scholar]
- 13.NLP2FHIR: A FHIR-based Clinical Data Normalization Pipeline and Its Applications. [Available from: https://github.com/BD2KOnFHIR/NLP2FHIR. [Google Scholar]
- 14.Hong N, Wen A, Shen F, Sohn S, Wang C, Liu H, et al. Developing a scalable FHIR-based clinical data normalization pipeline for standardizing and integrating unstructured and structured electronic health record data. JAMIA Open. 2019;2(4):570–9. doi: 10.1093/jamiaopen/ooz056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hong N, Wen A, Stone DJ, Tsuji S, Kingsbury PR, Rasmussen LV, et al. Developing a FHIR-based EHR phenotyping framework: A case study for identification of patients with obesity and multiple comorbidities from discharge summaries. J Biomed Inform. 2019;99:103310. doi: 10.1016/j.jbi.2019.103310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim Y, editor. Proceedings of the 2014 Conference on Empirical Methods in Natural Language Processing (EMNLP) Convolutional Neural Networks for Sentence Classification. 2014 oct; Doha, Qatar: Association for Computational Linguistics. [Google Scholar]
- 17.Chung J, Gulcehre C, Cho K, Bengio Y. arXiv preprint arXiv:14123555. 2014. Empirical evaluation of gated recurrent neural networks on sequence modeling. [Google Scholar]
- 18.Yao L, Mao C, Luo Y, editors. Proceedings of the AAAI Conference on Artificial Intelligence. 2019. Graph convolutional networks for text classification. [Google Scholar]
- 19.Rasmussen LV, Kiefer RC, Mo H, Speltz P, Thompson WK, Jiang G, et al. A Modular Architecture for Electronic Health Record-Driven Phenotyping. AMIA Joint Summits on Translational Science proceedings. AMIA Joint Summits on Translational Science. 2015;2015:147–51. [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Z, Feng Y, Li Y, Srivastava A, Adekkanattu P, Ancker JS, et al. Predictive Modeling of the Risk of Acute Kidney Injury in Critical Care: A Systematic Investigation of The Class Imbalance Problem. AMIA Jt Summits Transl Sci Proc. 2019;2019:809–18. [PMC free article] [PubMed] [Google Scholar]
- 21.Xu Z, Luo Y, Adekkanattu P, Ancker JS, Jiang G, Kiefer RC, et al. Stratified Mortality Prediction of Patients with Acute Kidney Injury in Critical Care. Stud Health Technol Inform. 2019;264:462–6. doi: 10.3233/SHTI190264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Z, Chou J, Zhang XS, Luo Y, Isakova T, Adekkanattu P, et al. Identifying sub-phenotypes of acute kidney injury using structured and unstructured electronic health record data with memory networks. J Biomed Inform. 2020;102:103361. doi: 10.1016/j.jbi.2019.103361. [DOI] [PubMed] [Google Scholar]
- 23.Rasmussen LV MS, Brandt PS MSc, Jiang G MD PhD, Kiefer RC, Pacheco JA MS, Adekkanattu P PhD, et al., editors. Considerations for Improving the Portability of Electronic Health Record-Based Phenotype Algorithms. AMIA Annual Symposium. 2019;2019 Washington DC, USA. [PMC free article] [PubMed] [Google Scholar]
- 24.Hripcsak G, Shang N, Peissig PL, Rasmussen LV, Liu C, Benoit B, et al. Facilitating phenotype transfer using a common data model. Journal of Biomedical Informatics. 2019;96:103253. doi: 10.1016/j.jbi.2019.103253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang S, Davarmanesh P, Song Y, Koutra D, Sjoding MW, Wiens J. Democratizing EHR analyses with FIDDLE: a flexible data-driven preprocessing pipeline for structured clinical data. J Am Med Inform Assoc. 2020;27(12):1921–34. doi: 10.1093/jamia/ocaa139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang S, McDermott MBA, Chauhan G, Ghassemi M, Hughes MC, Naumann T. Proceedings of the ACM Conference on Health, Inference, and Learning. MIMIC-Extract: a data extraction, preprocessing, and representation pipeline for MIMIC-III. Toronto, Ontario, Canada: Association for Computing Machinery; 2020. p. 222– 35. [Google Scholar]
- 27.Johnson AE, Pollard TJ, Shen L, Lehman LW, Feng M, Ghassemi M, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016;3:160035. doi: 10.1038/sdata.2016.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajkomar A, Oren E, Chen K, Dai AM, Hajaj N, Hardt M, et al. Scalable and accurate deep learning with electronic health records. npj Digital Medicine. 2018;1(1):18. doi: 10.1038/s41746-018-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sharma H, Mao C, Zhang Y, Vatani H, Yao L, Zhong Y, et al. Developing a portable natural language processing based phenotyping system. BMC Medical Informatics and Decision Making. 2019;19(3):78. doi: 10.1186/s12911-019-0786-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savova GK, Masanz JJ, Ogren PV, Zheng J, Sohn S, Kipper-Schuler KC, et al. Mayo clinical Text Analysis and Knowledge Extraction System (cTAKES): architecture, component evaluation and applications. Journal of the American Medical Informatics Association. 2010;17(5):507–13. doi: 10.1136/jamia.2009.001560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torii M, Wagholikar K, Liu H. Using machine learning for concept extraction on clinical documents from multiple data sources. Journal of the American Medical Informatics Association : JAMIA. 2011;18(5):580–7. doi: 10.1136/amiajnl-2011-000155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sohn S, Clark C, Halgrim SR, Murphy SP, Chute CG, Liu H. MedXN: an open source medication extraction and normalization tool for clinical text. Journal of the American Medical Informatics Association : JAMIA. 2014;21(5):858–65. doi: 10.1136/amiajnl-2013-002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.UMLS Vocabulary and Terminology Service. 2018 [Google Scholar]
- 34.Yao L, Mao C, Luo Y. Clinical text classification with rule-based features and knowledge-guided convolutional neural networks. BMC Medical Informatics and Decision Making. 2019;19(3):71. doi: 10.1186/s12911-019-0781-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Uzuner O. Recognizing obesity and comorbidities in sparse data. J Am Med Inform Assoc. 2009;16(4):561–70. doi: 10.1197/jamia.M3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hong N, Wen A, Shen F, Sohn S, Liu S, Liu H, et al. Integrating Structured and Unstructured EHR Data Using an FHIR-based Type System: A Case Study with Medication Data. AMIA Jt Summits Transl Sci Proc. 2018;2017:74–83. [PMC free article] [PubMed] [Google Scholar]
- 37.Rios A, Kavuluru R. Convolutional Neural Networks for Biomedical Text Classification: Application in Indexing Biomedical Articles. ACM BCB. 2015;2015:258–67. doi: 10.1145/2808719.2808746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rumelhart DE, Hinton GE, Williams RJ. Learning representations by back-propagating errors. Nature. 1986;323(6088):533–6. [Google Scholar]
- 39.Hochreiter S, Schmidhuber J. Long Short-Term Memory. Neural Computation. 1997;9(8):1735–80. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 40.Kipf TN, Welling M. arXiv preprint arXiv:160902907. 2016. Semi-supervised classification with graph convolutional networks. [Google Scholar]
- 41.Graph Convolutional Networks for Text Classification. AAAI 2019. [Available from: https://github.com/yao8839836/text_gcn. [Google Scholar]
- 42.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. Scikit-learn: Machine learning in Python. Journal of machine learning research. 2011;12(Oct):2825–30. [Google Scholar]
- 43.Abadi M, Agarwal A, Barham P, Brevdo E, Chen Z, Citro C, et al. arXiv preprint arXiv:160304467. 2016. Tensorflow: Large-scale machine learning on heterogeneous distributed systems. [Google Scholar]
- 44.Keras 2015. [Available from: https://keras.io/ [Google Scholar]
- 45.(OCR) OfCR. Summary of the HIPAA Security Rule 2013. [Available from: https://www.hhs.gov/hipaa/for-professionals/security/laws-regulations/index.html. [Google Scholar]
- 46.Yang H, Spasic I, Keane JA, Nenadic G. A Text Mining Approach to the Prediction of Disease Status from Clinical Discharge Summaries. Journal of the American Medical Informatics Association. 2009;16(4):596–600. doi: 10.1197/jamia.M3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Childs LC, Enelow R, Simonsen L, Heintzelman NH, Kowalski KM, Taylor RJ. Description of a rule-based system for the i2b2 challenge in natural language processing for clinical data. Journal of the American Medical Informatics Association : JAMIA. 2009;16(4):571–5. doi: 10.1197/jamia.M3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mishra NK, Cummo DM, Arnzen JJ, Bonander J. A rule-based approach for identifying obesity and its comorbidities in medical discharge summaries. Journal of the American Medical Informatics Association : JAMIA. 2009;16(4):576–9. doi: 10.1197/jamia.M3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komninos A, Manandhar S, editors. Proceedings of the 2016 conference of the North American chapter of the association for computational linguistics: human language technologies. 2016. Dependency based embeddings for sentence classification tasks. [Google Scholar]
- 50.Luo Y, Sohani AR, Hochberg EP, Szolovits P. Automatic lymphoma classification with sentence subgraph mining from pathology reports. Journal of the American Medical Informatics Association. 2014;21(5):824–32. doi: 10.1136/amiajnl-2013-002443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Banarescu L, Bonial C, Cai S, Georgescu M, Griffitt K, Hermjakob U, et al., editors. Proceedings of the 7th linguistic annotation workshop and interoperability with discourse. 2013. Abstract meaning representation for sembanking. [Google Scholar]
- 52.Devlin J, Chang M-W, Lee K, Toutanova K, editors. Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. Volume 1 (Long and Short Papers); 2019 jun; Minneapolis, Minnesota: Association for Computational Linguistics. [Google Scholar]
- 53.Liu Y, Ott M, Goyal N, Du J, Joshi M, Chen D, et al. ArXiv. 2019;abs/1907.11692. RoBERTa: A Robustly Optimized BERT Pretraining Approach. [Google Scholar]