Abstract
Human annotations are the established gold standard for evaluating natural language processing (NLP) methods. The goals of this study are to quantify and qualify the disagreement between human and NLP. We developed an NLP system for annotating clinical trial eligibility criteria text and constructed a manually annotated corpus, both following the OMOP Common Data Model (CDM). We analyzed the discrepancies between the human and NLP annotations and their causes (e.g., ambiguities in concept categorization and tacit decisions on inclusion of qualifiers and temporal attributes during concept annotation). This study initially reported complexities in clinical trial eligibility criteria text that complicate NLP and the limitations of the OMOP CDM. The disagreement between and human and NLP annotations may be generalizable. We discuss implications for NLP evaluation.
Introduction
Named entity recognition (NER)-the process of automatically recognizing named entities and assigning appropriate semantic categories-is a fundamental task of natural language processing (NLP) [1] and has spurred the development of biomedical NLP systems [2,3]. Because of its importance, the evaluation of NER has been an active field of research [4]. In literature, most of the evaluation of NER research focused on comparative evaluation of performance of commonly used NER systems [5-7], proposing new evaluation metrics [8-11]. However, the inconsistencies in NER evaluations, preventing objective cross-system comparisons, are underexplored.
Biomedical terminologies, also referred as ontologies, are rich sources of biomedical domain knowledge. Therefore, an ontology can be employed to validate whether a predicted entity is correct or not. For an entity, if its exact term or its synonym(s) exist in a reference ontology, the probability of recognizing them correctly is high. Thus, the involvement of ontologies to evaluate biomedical NER tools promise to facilitate error analysis. A possible drawback of evaluating clinical NER using a single ontology is that the same concept can be phrased or categorized differently across ontologies and cause discrepancies in concept normalization. For example, the concept Breast cancer can be represented by a post-coordinated term Neoplasm with "body location" being "breast" in SNOMED [12] but by a pre-coordinated term Breast Neoplasms in MeSH [13]. In CLEF [14], the annotations were mapped to concepts in the Unified Medical Language System (UMLS) [15]. CliCR [16] — a dataset of annotated clinical case reports — also used UMLS to obtain alternative phrase forms (synonyms, abbreviations and acronyms) for any recognized entity.
A common data model harmonizes concepts across various biomedical ontologies. The Observational Medical Outcomes Partnership (OMOP) Common Data Model (CDM) [17] is such a standard common data model that unifies 81 frequently used vocabularies in biomedical domain and is adopted by the OHDSI network [18]. We hypothesize that a corpus annotated based on the OMOP CDM can minimize discrepancies in human annotation and NLP-based concept normalization during NER evaluation. With the availability of a large-scale manually annotated medical corpus conforming to the OMOP CDM, this study compared the human and NLP-based annotations. We experimented with entity boundary relaxation and categorical relaxation. This in-depth analysis identifies challenges originating from the complexities in the eligibility criteria text and the limitations in the OMOP CDM, and makes recommendations regarding leveraging ontologies or common data models to facilitate clinical NER evaluations.
Background and Related Work
Only a handful of studies have conducted comparative evaluation of the recognition and classification performance of commonly used NER systems [5-7]. A few studies proposed new evaluation metrics to more precisely appraise NER performance [8-11], or compared the existent evaluation strategies [2]. Although most of the evaluations adopt the standard quantitative metrics such as precision and recall, the processes of computing such metrics vary significantly and deep understanding of NER errors and their root causes is still lacking [6]. Some measure precision and recall at the word or token level, while others calculate these metrics at the concept level. Some use "exact match", which requires that a candidate entity can only be counted as a correct recognition if both its text spans and its class label fully agrees with an annotated entity [2], while others count partial match. Besides, we believe a proper evaluation of a clinical NER system should also leverage domain expertise [19] due to the complexity and imparity across entity categories in clinical corpora. For example, "HIV positive" can be categorized as a measurement entity ("HIV") with a value entity ("positive") or a condition entity ("HIV positive"). In clinical NER, classification of an entity's category, such as condition, measurement, or drug, can vary depending on the language context [2]. Therefore, during evaluation, effective leverage of domain knowledge is indispensable to recognize the right concept.
The Annotated Corpus for clinical NER
In a newly published corpus [20], we manually annotated the clinical trial eligibility criteria text extracted from 1,000 randomly selected clinical trials from Clinicaltrials.gov, and named the corpus Chia. The Chia dataset reached an 81% Kappa of inter-annotator agreement, which was calculated on annotations from randomly selected 50 clinical trials.
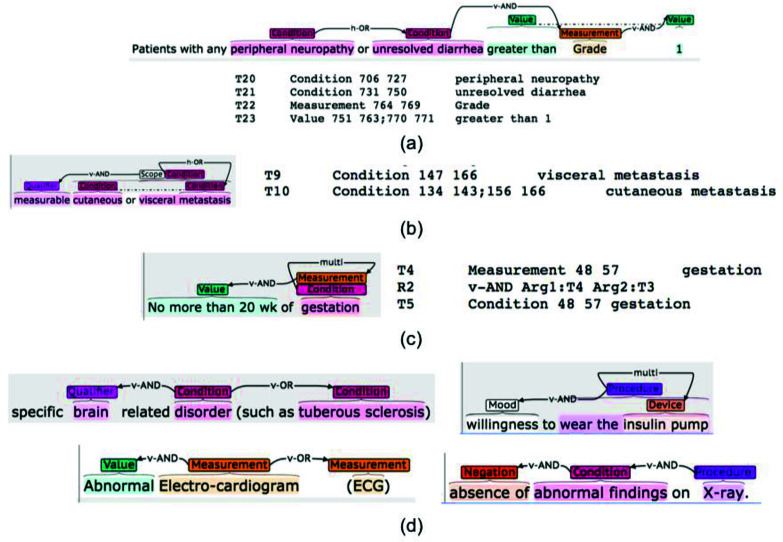
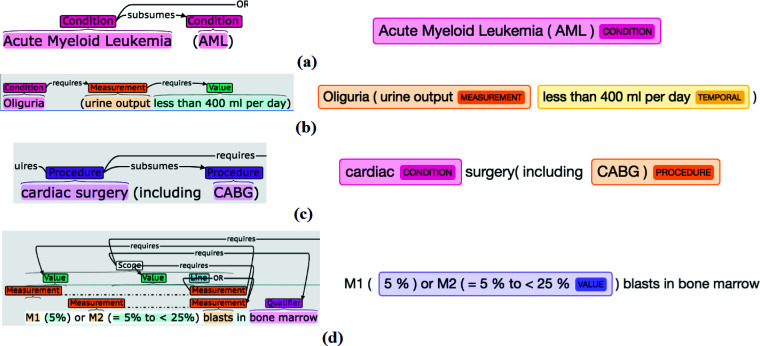
Alex et al. [21] categorized overlapping entities into three types: (1) entities containing one or more shorter embedded entities (e.g., "wear the insulin pump" and "insulin pump" shown in Figure 1(d)); (2) entities with more than one entity category (e.g., "gestation" can be categorized as Condition and Measurement as shown in Figure 1(c)); (3) coordination ellipsis ("cutaneous metastasis" and "visceral metastasis" in Figure 1(b)). Unlike most other flat corpora that exclude nested or overlapping entities [21], Chia uses a non-flat annotation scheme to accommodate these. The GENIA corpus supports overlapping entities [22] but focuses on biological entities, such as DNA, RNA, and protein.
Figure 1.
Various annotation examples from Chia and example C2Q predictions for some of them (highlighted by the green rectangles). (a). Top: visualized annotation interface suitable for human review. Bottom: text file storing the annotation data suitable for machine processing. (b). An example of a coordinated entity. (c). An example where one same piece of text corresponds to two distinct annotated entities. (d) Examples of relationships annotated in Chia.
The NER system: Criteria2Query
The NER system used in this study is Criteria2Query (C2Q) [23], which translates free-text eligibility criteria to OMOP CDM-based cohort queries. Its online demo is available at http://www.ohdsi.org/web/criteria2query/. Its output are computable queries in JSON format that can be directly fed into ATLAS [24] to define a patient cohort. In this study, we focused the comparison on the NER module, which recognizes eight entity types defined in the OMOP CDM, including CONDITION, DEMOGRAPHIC, DRUG, MEASUREMENT, OBSERVATION, PROCEDURE, VALUE, and TEMPORAL. An entity's type, indicating which category an entity is classified to, are denoted in uppercase (e.g., CONDITION). Noted that as C2Q is constantly being updated, the latest online version of C2Q may cover more entity types that were not be available for inclusion in the evaluation at the time of this study.
Material and Methods
In this study, we refer to a recognized entity as a prediction and a manually annotated entity a reference (as ground truth). If the prediction and the reference are exactly same, we call it an exact match. Besides exact match, there are partial matches. For example, in text "intercostal post-herpetic neuralgia", if the NER system recognizes "neuralgia" as opposed to "post-herpetic neuralgia," there is a partial match. Researchers have developed a variety of rules that relax boundary matching criteria to different degrees, including Left match, Right match, Partial match, Approximate match, Name part/fragment match, Core-term match, etc. [2]. When there is no exact match, a discrepancy can occur at the syntactic level, where the human annotation and NLP annotation disagrees on the entity's boundaries, or at the semantic level, where the human annotation and NLP annotation disagrees on the semantics (e.g., concepts or categories). The latter requires adjudication by domain experts. We compared the output of Criteria2Query with Chia's annotations in terms of entity span and type (category). An entity's span is composed of one or more words or phrases.
We defined the following matches for analysis of the disagreement between human and NLP-assisted annotations:
- “relaxed match”—the prediction's span overlaps with the reference's span, which include
- “exact match”—the prediction's span exactly overlaps with the reference's span.
- “extra match”—the prediction's span strictly contains the reference's span.
- “partial match”—the prediction's span is strictly contained by the reference's span.
“spurious match”—the prediction's span has no overlaps with any reference's span.
“missing match”—no prediction's span overlaps with the reference's span.
If there is a match, we further compared the agreement on concept categories as one of the following:
“correct”—the prediction's category agrees with the reference's category.
“incorrect”—otherwise.
“N/A”—not applicable.
We counted the frequency of each combination of disagreement scenarios. We also grouped the entities by its annotated categories and check the disagreement type distributions in each. These can be conducted completely systematically without human inspection of the texts. The questions of our main interest include:
Which type of disagreement between human and NLP-based annotations are frequent?
What is the distribution of the disagreement types?
Between which categories are mis-categorization frequent?
The disagreement type depends on how a reference aligns with a prediction. During the NER evaluation, a prediction and a reference align if their spans overlap. It is possible that a prediction aligns with multiple references, or vice versa. Hence, disagreement can occur for one prediction if different references are selected as alignments. This multi-matching issue is commonly observed in many NER evaluation and are particularly frequent for evaluations against non-flat annotations [4]. To simplify our error analysis, we designed a rule-based method to align the predictions and references with the highest overlapping ratio in their spans. After the alignment, each prediction was matched to at most one reference (if aligned with nothing, the case is a "missing"), and was assigned a unique disagreement type.
We designed the following 3-step workflow for our disagreement analysis: 1) we first manually inspected the disagreement instances and examples; 2) we abstracted a disagreement pattern from similar instances and formulated the definition for the pattern; and 3) according to the definition, we programmatically fetched all the incidents belonging to this pattern and determined if it represents substantial portion of all the disagreement.
Results
Data Statistics
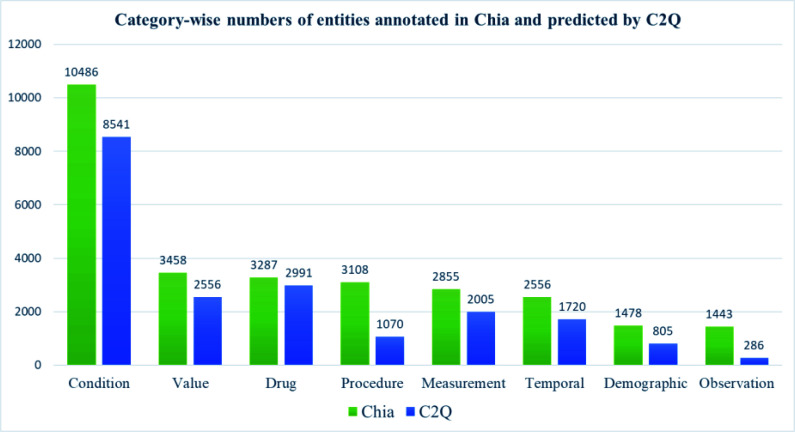
A total of 1000 trials in Chia were used for the evaluation after excluding 66 trials due to errors in Chia annotations or technical barriers in running C2Q due to server instability issues. From the eligibility criteria text of those 934 trials, there were 28,671 distinct references from Chia, while C2Q generated 19,974 distinct predictions, which fell into the aforementioned eight entity categories. Figure 2 shows entities annotated in Chia and predicted by C2Q for each category. We saw CONDITION was the dominant category in Chia with the highest frequency, while the frequencies of the other five categories (DEMOGRAPHIC, DRUG, MEASUREMENT, VALUE, TEMPORAL) shared the same order of magnitude. The number of entities predicted by C2Q for each category were roughly proportional to the corresponding category of Chia's annotations except categories of OBSERVATION and PROCEDURE.
Figure 2.
Category-wise numbers of entities annotated in Chia and predicted by Criteria2Query, respectively.
In-depth human- and NLP-based annotation disagreement analysis
We identified six disagreement patterns and reported the number of incidents for each pattern in Table 1. These patterns are indictors of the inconsistences between human annotations and machine learning-based annotations, not necessarily NER errors due to the imperfection of human annotations. Among these patterns, the most prevalent pattern includes 1532 disagreement instances, where C2Q predicted an entity together with its descriptive qualifier, temporal or value attributes. The pattern with the least number of cases (120) is mis-categorization between CONDITION and OBSERVATION. Each disagreement pattern along with its example(s) and error analysis are elaborated later.
Table 1. Prevalence of the disagreement pattern.
| No. | Type of Match | Disagreement patterns | Frequency |
| 1 | Extra Match (N=3300) | Recognizing qualifier, temporal and value attributes together with an entity | 1532 (46.4%) |
| 2 | Recognizing a coordinated elliptical expression as an entity | 591 (17.9%) | |
| 3 | Recognizing parentheses together with an entity | 338 (10.2%) | |
| 4 | Missing (N=10433) | Missing predictions around multi-labeled or nested entities | 601 (5.8%) |
| 5 | Partial Match (N=1366) | Omitting the reference point of a TEMPORAL entity | 392 (28.7%) |
| 6 | Exact Match (N=5217) | Mis-categorization between CONDITION and OBSERVATION | 120 (2.3%) |
Table 2 shows the entity count for each boundary matching criteria and correct predictions. C2Q predicted 12,584 "exact" matches with Chia's entities with 11,501 correct predictions. In addition, 3,801 "extra" matches and 1,853 "partial" matches were identified with boundary relaxing. With our relaxed matching criteria, we found additional 4,666 correct predictions (28.9% of total correct predications) to obtain 16,167 correct predictions.
Table 2. Comparison between human and Criteria2Query annotations.
| Type of Match | Match w Chia | No Match w Chia | <NA> | Total predictions by Criteri2Qery |
| exact | 11501 (91.4%) | 1083 (8.6%) | 0 | 12584 |
| extra | 3300 (86.8%) | 501 (13.2%) | 0 | 3801 |
| partial | 1366 (73.7%) | 487 (26.3%) | 0 | 1853 |
| spurious | 0 | 0 | 1736 | 1736 |
| missing | 0 | 0 | 10433 | 10433 |
| Total | 16167 | 2071 |
For entities in the "exact" matching, we further computed their categorization contingency table (Table 3). From the two tables we can see C2Q achieved a good overall prediction accuracy in categorization. We observed that 74 (42.5% =74/174) observation entities are mis-categorized into CONDITION, and 214 (22.1%=214/969) procedure entities are mis-categorized into DRUG. Possible reasons for this observation are discussed later.
Table 3. The contingency table of categorization over "exact" predictions.
| Criteria2Query Annotations | |||||||||
| Condition | Demographic | Drug | Measurement | Observation | Procedure | Value | Temporal | ||
| Chia | Condition | 4956 (96.2%) | 0 | 35 (0.7%) | 80 (1.6%) | 46 (0.9%) | 33 (0.6%) | 1 (0.02%) | 0 |
| Demographic | 9 (1.2%) | 722 (96.4%) | 3 (0.4%) | 7 (0.9%) | 6 (0.8%) | 0 | 0 | 2 (0.3%) | |
| Drug | 64 (3.3%) | 0 | 1819 (94.8%) | 28 (1.5%) | 0 | 8 (0.4%) | 0 | 0 | |
| Measurement | 67 (5.8%) | 1 (0.09%) | 36 (3.1%) | 1033 (89.9%) | 7 (0.6%) | 4 (0.3%) | 1 (0.09%) | 0 | |
| Observation | 74 (42.5%) | 0 | 8 (4.6%) | 7 (4.0%) | 81 (46.6%) | 4 (2.3%) | 0 | 0 | |
| Procedure | 125 (12.9%) | 0 | 214 (22.1%) | 70 (7.2%) | 9 (0.9%) | 550 (56.8%) | 0 | 1 (0.1%) | |
| Value | 8 (0.4%) | 3 (0.2%) | 0 | 3 (0.2%) | 0 | 0 | 1693 (94.4%) | 87 (4.8%) | |
| Temporal | 0 | 0 | 0 | 0 | 0 | 1 (0.15%) | 31 (4.6%) | 647 (95.3%) | |
We elaborate on the six disagreement patterns. For each pattern, one or more examples are analyzed to delineate it. Then a more systematic definition of the pattern was used to programmatically fetch all the instances following such pattern. Then the representation of this patterns among all the disagreement cases are discussed. Note that these disagreement patterns are not necessarily mutually exclusive, many disagreement instances being the compound of two or more patterns.
1). Recognizing the qualifier together with an entity
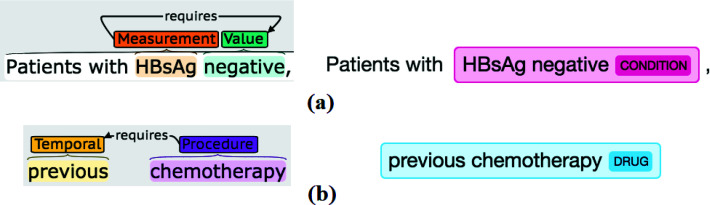
We observed that entities classified into "extra" was much more frequent than "partial." This is due to C2Q frequently included an entity's qualifiers as part of the real entity. Figure 3 (b) shows an example where C2Q predicted the "severe respiratory disease" as a whole entity while in Chia (Figure 3(a)) the "severe" is annotated as the qualifier of the "respiratory disease". We introduced a disagreement type called extra_qualifier to indicate cases where the prediction is "exact" or "partial" to the reference if dropping the "extra" qualifier(s). We found 1049 instances could be classified into this pattern, making this the most frequent pattern among "extra" recognitions.
Figure 3.
Example of recognizing the qualifier together with an entity (Left: Chia; Right: C2Q).
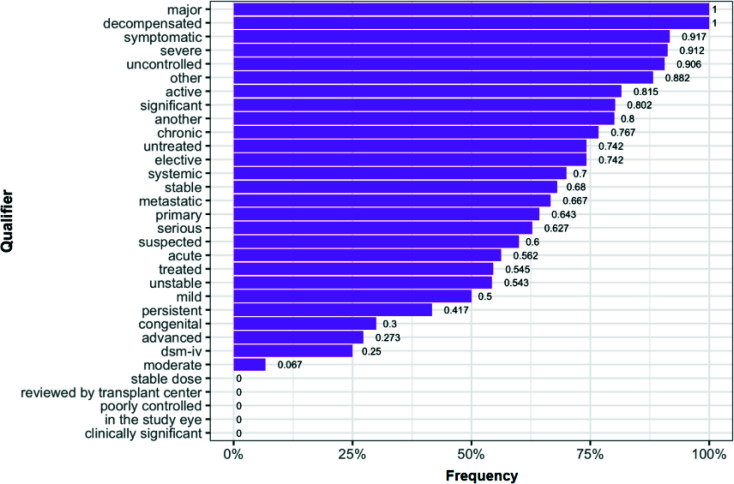
Two follow-up questions we then had were: (1) What qualifier terms were prone to be recognized as part of the target entity being qualified? (2) Was the probability of a qualifier to be recognized as part of its target term dependent on the target term itself? To address the first questions, we computed each qualifier term's relative frequency if a qualifier was parsed together with its target entity by C2Q for at least 10 times. Figure 4 shows the top portion of the results. Four qualifiers (i.e., major, decompensated, symptomatic, severe) were recognized together with their target entity over 90% of occurrences while some other qualifiers (clinically significant, in the study eye, etc.) rarely were. To answer the second question, we checked that for a qualifier that was recognized together with entities for at least 80% of cases, whether it is dominant by certain target entities. Interestingly, none of them had a dominant target entity, implying that they were considered part of the entity regardless of the real target entity.
Figure 4.
Frequency of qualifiers being parsed together with the target entity.
Recognizing an entity's value (Figure 5(a)) or temporal (Figure 5(b)) as part of an entity also caused a substantial number of "extra" disagreement instances. Systematically, we defined the disagreement type extra_value and extra_temporal to be the cases where the prediction is "extra" to the reference but can be updated to "exact" or "partial" if its value(s) or temporal(s) are dropped, respectively. A total of 329 (9.9%) "extra" cases were assigned to extra_value. Similarly, 154 (4.7%) "extra" cases were classified to extra_temporal.
Figure 5.
Example of recognizing the value or temporal together with an entity (Left: Chia; Right: C2Q).
2). Recognizing a coordinated elliptical expression as an entity
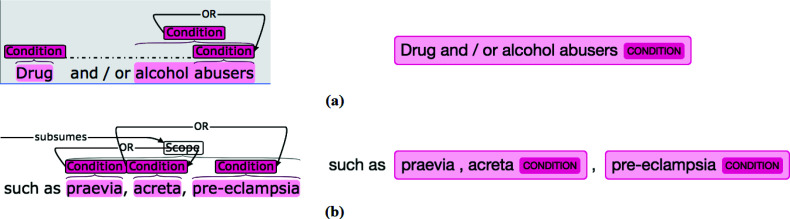
Another major cause of "extra" was the recognition of a coordination ellipsis as a single entity. For example, in Figure 6 (a), C2Q recognized "Drug and / or alcohol abusers" as a single entity, which was annotated as two separate entities "Drug abusers" and "alcohol abuser" connected by "or" in Chia. In Figure 6 (b), C2Q recognized "praevia, acreta" as a single entity while the "praevia" and "acreta" were two separate entities in Chia. Systematically, we defined this disagreement type as extra_logic to represent cases where: 1) a prediction is matched to more than one references; 2) the prediction is "extra" to each of the reference; 3) the prediction's text contains " and ", " or ", or ", " (note the spaces around the keywords) or "/" (excluding the cases where "/" is used for unit or mathematical formula). This pattern can be further subdivided into two types: the complete recognition of a simple logic group where the entities in the group are mutually independent (Figure 6 (b)); the complete recognition of a coordinated ellipsis group where the entities in the group share a piece of text (Figure 6 (a)). We found Criteria2Query mistaken 591 coordination ellipsis expressions for one semantic unit (17.9% of all "extra match"), implying the significance of this phenomenon in criteria text and the need for dedicated technology to address it. Yuan et al. contributed a related method [25].
Figure 6.
Example of recognizing a logic group in its entirety as an entity (Left: Chia; Right: C2Q).
3). Recognizing parentheses together with an entity
Figure 7 shows four examples of inconsistency between Chia and Criteria2Query around parentheses. Systematically, we defined the disagreement type extra_parenthesis to be the case where a prediction is "extra" to the "reference" and the prediction text contains "(" or ")". We found 338 (10.2%) "extra match" disagreement could be assigned to extra_parenthesis, and they can be further divided into four scenario: 1) recognition combining the entity and its acronym in parentheses (N=153 instances, e.g. Figure 7(a)); 2) recognition combining an entity followed by an open parenthesis plus the first term (N=119 instances, e.g. Figure 7(b)); 3) recognition combining an ending parenthesis in addition to the last entity inside the parentheses (N=63 instances, e.g. Figure 7(c)); 4) recognition combining multiple close and open parentheses were also observed (N=3 instances, e.g. Figure 7(d)).
Figure 7.
Example of recognizing parentheses together with an entity (Left: Chia; Right: C2Q).
4). Missing predictions around multi-labeled or nested entities
We have stressed the existent of multi-labeled or nested entities in Chia. However, C2Q made flat predictions, i.e. it made no predictions for overlapping spans. Hence it inevitably missed some entities that are part of non-flatness annotations. For example, the "anti-inflammatory" in Figure 8 is a DRUG entity nested in the PROCEDURE entity "anti-inflammatory therapy", while C2Q could only make one prediction for such text. Systematically, we defined an disagreement pattern missing_multi/nested to be cases where: a prediction aligns with multiple references and its span overlaps with each of their spans. We found this pattern caused 601 (5.8%) of "missing" disagreement.
Figure 8.
Example of missing predictions around multi-labeled or nested entities (Left: Chia; Right: C2Q).
5). Omitting the reference point of a TEMPORAL entity
In general, disagreement was less frequent when using "partial match" than "extra match". However, for TEMPORAL entities, "partial" disagreement manifested more frequently than the "extra" disagreement. The reason is that C2Q omitted the reference point of recognitions for TEMPORAL entities. An example is shown in Figure 9. The most often used keywords for the reference time points were "after", "before", "of", "from", "since", "past", "preceding", "pre" and "post". Thereby, systematically, we defined this disagreement type as partial_reference_point, to represent prediction of TEMPORAL entities with "partial" disagreement due to omitting the reference time points indicated by the keywords listed above. We found this pattern was responsible for 392 (28.7%) disagreed "partial matches".
Figure 9.
Example of omitting the reference point of a TEMPORAL entity (Left: Chia; Right: C2Q).
6). Mis-categorization between CONDITION and OBSERVATION
From Table 3 we observe that 74 of the observation entities were mis-categorized as CONDITION and 46 condition entities were annotated as OBSERVATION. For example, in Figure 10(a) the "sexually active" was annotated as OBSERVATION in Chia but was predicted as CONDITION by C2Q in Figure 10(b). However, we noticed Chia contains inconsistent annotations of the same term. For example, in Chia "sexually active" was annotated as CONDITION in Figure 10(c). We found that 45 terms were miscategorized from OBSERVATION to CONDITION at least once, and 15 terms were miscategorized from CONDITION to OBSERVATION at least once.
Figure 10.
Example of mis-categorization between CONDITION and OBSERVATION.
Discussion
In this study, we compared human and NLP annotations of clinical trial eligibility criteria text that both followed the OMOP CDM, an increasingly popular and widely adopted clinical data standard by the clinical research informatics community as a rich clinical data model. Therefore, the disagreement patterns identified in this study may offer generalizability implications for many NER tasks of interest with similar setup. The disagreement patterns help unveil the complex semantic expressions and sophisticated logic used in clinical trial eligibility criteria text, such as the frequently combined conditions in coordination ellipsis (pattern 2) and the importance of reserving temporal reference point for TEMPORAL entities (pattern 5). Patterns 1 and 6 provide insights to the ambiguity of the OMOP CDM, while other patterns (pattern 2, 4, and 5) manifested themselves as the semantic/logic complexity in eligibility criteria text that may require tailoring the ML-based NER system accordingly. Meanwhile, explicit definition of principles for annotating descriptive modifiers and temporal or value attributes for named entities is important for achieving the comparative effectiveness of NER evaluations.
Standard NER systems evaluated using corpora from challenges such as MUC, CONLL or ACEU employed "exact match". However, strict exact-boundary match may not always reflect the true performance of an NER system. For example, a human annotator annotated "SARS-CoV-2 infection" from an eligibility criterion "progressive disease suggestive of ongoing SARS-CoV-2 infection." A clinical NER system makes the prediction of "progressive disease suggestive of ongoing <CONDITION>SARS-CoV-2</CONDITION> infection." We noticed a boundary mismatch disagreement where "infection" was not included in the NER system's extraction. However, this disagreement may not be an error of NLP systems if "SARS-CoV-2" as a condition is adequate for downstream tasks such as concept normalization, document indexing, or relationship extraction. The extra_qualifier, extra_value and extra_temporal patterns, refer to disagreement where descriptive adjectives were annotated as parts of following entities. In reality, even annotators are oftentimes confused and make subjective decisions on whether descriptive adjectives such as "severe" or "secondary" should be considered part of entity names based on the clinical context. Therefore, these three patterns should not be judged as errors bluntly because some cases may be acceptable in the context of applications. From the perspective of OMOP CDM vocabulary, some pre-coordinated terms are included. For example, "severe cytopenia" is in the terminology while "severe arrhythmia" is not. Pre-coordination vs. post-coordination of the terms has been actively discussed and poses challenge to NER systems for predicting descriptive adjectives. Of course, specific rules and examples in annotation guidelines may help alleviate this issue; but ambiguity seems inevitable.
The open-ended definition of Observations in OMOP CDM leads to some concepts existing in both the CONDITION and OBSERVATION domains. This is due to the catch-all nature of the ambiguous OBSERVATION domain (https://www.ohdsi.org/web/wiki/doku.php?id=documentation:cdm:observation) in the OMOP CDM: "The OBSERVATION table captures clinical facts about a Person obtained in the context of examination, questioning or a procedure. Any data that cannot be represented by any other domains, such as social and lifestyle facts, medical history, family history, etc. are recorded here." Fan and Friedman previously reported the abundant ambiguity in the "finding" semantic type in the UMLS also cause similar problems for semantic classification of named entities [26]. Moreover, the human annotations are not perfect. Chia has an inter-rater agreement of 81%, which imposes an accuracy ceiling for the machine learning algorithm for NER. Reasons for human errors could be complex, e.g., the ambiguities in the OMOP CDM, the lack of details in annotation guidelines, subjective decisions by annotators, or even human mistakes due to fatigue. Similarly, Xu et al. found that nearly half of the discrepancy between the system and gold standard were due to errors in gold standard annotation [27]. Therefore, imperfections in human annotations is a common problem; yet its impact on machine learning-based NLP needs more careful evaluation.
For categories that are not clearly defined or not consistently exclusive, one can employ categorical relaxation to merge the two categories to reduce the ambiguity of NER evaluations [2]. This technique is often used to merge protein, DNA and RNA when no distinctions are required. In our study, 174 disagreement will be eliminated if categorical relaxation is employed to merge the evaluation of CONDITION with OBSERVATION. The aforementioned ambiguous representations in the OMOP CDM and inconsistent annotations in the evaluation corpus can diminished an NER's real performance. With Chia's inter-annotator agreement being around 81%, even if an NER system is trained with Chia corpus, the annotation inconsistencies and ambiguity exist in Chia will inevitably propagate to the machine learning based NER system. If the strict exact-boundary match is used as gold standard, the trained machine learning NER model could be penalized for overfitting the human annotations in Chia. It is another reason that for certain applications, exact match can weaken the reliability of an NER system's performance and customized relaxed matching criteria should be leveraged.
Limitations: One limitation of this study is that Criteria2Query was not able to resolve multiple or nested annotated entities at the time of the study. This limitation is due to current Criteria2Query's conditional random field (CRF) implementation cannot handle nested NER, where an entity can be contained in other entities [21]. Alex et al. [21] attempted to recognize the nested entities in GENIA corpus by specially pre-processing the annotation and saw an improvement over the baseline flat system. Byrne [28] used a multi-word token method and obtained a promising result in recognizing nested named entities in historical archive texts. Nevertheless, the work on nested NER has almost been entirely ignored until recently [29], and the technology is not so mature as that of regular flat NER. Another limitation of this study is the lack of integration of our method for recognizing coordination ellipsis-another case that the non-flatness concerns. The coordinated ellipses is particular hard [30] in NER. The coordinated ellipses, along with the simple logic group and the parenthesized insertion, are called composite mention in some research [31]. Buyko et al. [30] utilized CRF to resolve the coordinated ellipses in GENIA corpus and attained a good performance. Wei et al. [31] integrated machine learning and pattern identification to handle all types of the composite mentions. Yuan et al. [25] proposed a graph-based representation model to reconstruct concepts from coordinated elliptical expressions. This model is being incorporated into Criteria2Query to further improve the NER performance.
Conclusions
This study describes an in-depth analysis of NER annotation disagreement between human and NLP for clinical trial eligibility criteria by comparing human annotations with NLP annotations. We identified six types of disagreement between human and NLP annotations following the OMOP CDM, and highlighted the complexities in logic and temporal information, all requiring further improvement of NER methods. Our study also shows that relaxed match increased accuracy reporting by about 28.9% over exact match. We recommend reporting prevalent disagreement patterns in the context of application in addition to quantitative metrics to formulate a comprehensive NER assessment.
Acknowledgements
This research was sponsored by the National Library of Medicine grant R01LM009886 (PI: Weng) and the National Center for Advancing Translational Science grant U24TR001579 (PI: Harris).
Conflicts of Interest
Dr. Xu and The University of Texas Health Science Center at Houston have research-related financial interests in Melax Technologies, Inc.
Figures & Table
References
- [1].Marrero M, Urbano J, Sánchez-Cuadrado S, Morato J, Gómez-Berbís JM. Named entity recognition: fallacies, challenges and opportunities. Computer Standards & Interfaces. 2013;35(5):482–9. [Google Scholar]
- [2].Tsai RT-H, Wu S-H, Chou W-C, Lin Y-C, He D, Hsiang J, et al. Various criteria in the evaluation of biomedical named entity recognition. BMC bioinformatics. 2006;7(1):92. doi: 10.1186/1471-2105-7-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Chen Y, Lasko TA, Mei Q, Denny JC, Xu H. A study of active learning methods for named entity recognition in clinical text. Journal of biomedical informatics. 2015;58:11–8. doi: 10.1016/j.jbi.2015.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Galibert O, Rosset S, Grouin C, Zweigenbaum P, Quintard L. Structured and extended named entity evaluation in automatic speech transcriptions. Proceedings of 5th International Joint Conference on Natural Language Processing. 2011.
- [5].Atdağ S, Labatut V. 2nd International conference on systems and computer science. IEEE; 2013. A comparison of named entity recognition tools applied to biographical texts. [Google Scholar]
- [6].Marrero M, Sánchez-Cuadrado S, Lara JM, Andreadakis G. Evaluation of named entity extraction systems. Advances in Computational Linguistics, Research in Computing Science. 2009;41:47–58. [Google Scholar]
- [7].Jiang R, Banchs RE, Li H. Evaluating and combining name entity recognition systems. Proceedings of the Sixth Named Entity Workshop. 2016 [Google Scholar]
- [8].Chinchor N. MUC-4 evaluation metrics. Proceedings of the 4th conference on Message understanding; McLean, Virginia. Association for Computational Linguistics; 1992. pp. 22–9. [Google Scholar]
- [9].Chinchor N, Sundheim BM. MUC-5 evaluation metrics. Fifth Message Understanding Conference (MUC-5): Proceedings of a Conference Held in Baltimore; Maryland, August 25-27. 1993 1993. [Google Scholar]
- [10].Jannet MB, Adda-Decker M, Galibert O, Kahn J, Rosset S. Eter: a new metric for the evaluation of hierarchical named entity recognition. 2014.
- [11].Makhoul J, Kubala F, Schwartz R, Weischedel R. Proceedings of DARPA broadcast news workshop. VA: Herndon; 1999. Performance measures for information extraction. [Google Scholar]
- [12].SNOMED Available from: http://www.snomed.org/
- [13].Medical Subject Headings Available from: https://www.nlm.nih.gov/mesh/meshhome.html .
- [14].Roberts A, Gaizauskas R, Hepple M, Demetriou G, Guo Y, Setzer A, et al. Semantic annotation of clinical text: The CLEF corpus. Proceedings of the LREC 2008 workshop on building and evaluating resources for biomedical text mining. 2008.
- [15].Bodenreider O. The unified medical language system (UMLS): integrating biomedical terminology. Nucleic acids research. 2004;32(suppl_1):D267–D70. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Šuster S, Daelemans W. CliCR: a dataset of clinical case reports for machine reading comprehension. arXiv preprint arXiv. 2018;180309720 [Google Scholar]
- [17].OMOP Common Data Model – OHDSI Available from: https://www.ohdsi.org/data-standardization/the-common-data-model/
- [18].Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, et al. Observational Health Data Sciences and Informatics (OHDSI): opportunities for observational researchers. Studies in health technology and informatics. 2015;216:574. [PMC free article] [PubMed] [Google Scholar]
- [19].Cimino JJ, Shortliffe EH. Biomedical Informatics: Computer Applications in Health Care and Biomedicine (Health Informatics) Springer-Verlag; 2006. [Google Scholar]
- [20].Kury F, Butler A, Yuan C, Fu L-h, Sun Y, Liu H, et al. Chia, a large annotated corpus of clinical trial eligibility criteria. Scientific data; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Alex B, Haddow B, Grover C. Recognising nested named entities in biomedical text. Biological, translational, and clinical language processing. 2007.
- [22].Kim J-D, Ohta T, Tateisi Y, Tsujii Ji. GENIA corpus—a semantically annotated corpus for bio-textmining. Bioinformatics. 2003;19(suppl_1):i180–i2. doi: 10.1093/bioinformatics/btg1023. [DOI] [PubMed] [Google Scholar]
- [23].Yuan C, Ryan PB, Ta C, Guo Y, Li Z, Hardin J, et al. Criteria2Query: a natural language interface to clinical databases for cohort definition. Journal of the American Medical Informatics Association. 2019;26(4):294–305. doi: 10.1093/jamia/ocy178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].ATLAS Available from: http://www.ohdsi.org/web/atlas/#/home .
- [25].Yuan C, Wang Y, Shang N, Li Z, Zhao R, Weng C. A graph-based method for reconstructing entities from coordination ellipsis in medical text. Journal of the American Medical Informatics Association. 2020. [DOI] [PMC free article] [PubMed]
- [26].Fan J-W, Friedman C. Semantic classification of biomedical concepts using distributional similarity. Journal of the American Medical Informatics Association. 2007;14(4):467–77. doi: 10.1197/jamia.M2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Xu H, Anderson K, Grann VR, Friedman C. Facilitating cancer research using natural language processing of pathology reports. Studies in health technology and informatics. 2004;107(Pt 1):565. [PubMed] [Google Scholar]
- [28].Byrne K. International Conference on Semantic Computing (ICSC 2007) IEEE; 2007. Nested named entity recognition in historical archive text. [Google Scholar]
- [29].Finkel JR, Manning CD. Nested named entity recognition. Proceedings of the 2009 conference on empirical methods in natural language processing. 2009.
- [30].Buyko E, Tomanek K, Hahn U. PACLING 2007-Proceedings of the 10th Conference of the Pacific Association for Computational Linguistics. Citeseer; 2007. Resolution of coordination ellipses in biological named entities using Conditional Random Fields. [Google Scholar]
- [31].Wei C-H, Leaman R, Lu Z. SimConcept: a hybrid approach for simplifying composite named entities in biomedical text. IEEE journal of biomedical and health informatics. 2015;19(4):1385–91. doi: 10.1109/JBHI.2015.2422651. [DOI] [PMC free article] [PubMed] [Google Scholar]