Abstract
Rare diseases affect between 25 and 30 million people in the United States, and understanding their epidemiology is critical to focusing research efforts. However, little is known about the prevalence of many rare diseases. Given a lack of automated tools, current methods to identify and collect epidemiological data are managed through manual curation. To accelerate this process systematically, we developed a novel predictive model to programmatically identify epidemiologic studies on rare diseases from PubMed. A long short-term memory recurrent neural network was developed to predict whether a PubMed abstract represents an epidemiologic study. Our model performed well on our validation set (precision = 0.846, recall = 0.937, AUC = 0.967), and obtained satisfying results on the test set. This model thus shows promise to accelerate the pace of epidemiologic data curation in rare diseases and could be extended for use in other types of studies and in other disease domains.
Introduction
In the United States, a rare disease is defined as affecting fewer than 200,000 people.1 It is estimated that between 6,000 and 8,000 rare diseases exist,2 and that they affect between 25 and 30 million people in the United States.3 Among rare diseases, there is a significant range in prevalence. Some disorders with higher prevalence rates are well-documented in the population; for instance, sickle cell disease is estimated to affect 100,000 people in the United States.4 Other diseases are much rarer, affecting only a handful of patients. Fewer than twenty cases of Jansen's metaphyseal chondrodysplasia have been reported, for example.5 Still others are sporadically documented only in occasional case reports. Accurate estimates of prevalence and incidence rates are critical to developing an understanding of a disease's scope and population burden. Continued epidemiological data on a greater distribution of rare diseases can help in recognizing patterns in etiology and inform decisions on research funding by providing quantifiable indications of impact.6
Epidemiologic data can be discovered and presented in several ways. The most complete findings are provided through epidemiologic studies, which describe the frequency of a disease in a certain population group by both geographic and demographic distribution. Such studies are often found for rare diseases whose affected population sizes range closer to the upper margins of the US rare disease definition, as they are prevalent enough to warrant a large-scale study and for results to have sufficient statistical strength. For the majority of rare diseases, however, no epidemiology studies have been conducted and population estimates are often derived solely from expert opinions and published case reports.7 As such, remaining vigilant of newly published epidemiologic studies in these diseases is an important task in guiding research efforts focused on the broader field of rare diseases.
The Genetic and Rare Diseases (GARD) Information Center, a program managed by the National Center for Advancing Translational Sciences (NCATS) within the National Institutes of Health (NIH), aims to curate and disseminate freely accessible consumer health information on over 6,500 genetic and rare diseases.8 Currently, GARD curators search PubMed for relevant articles and manually review them for curation, which is a tedious and error-prone process. Curators noted that searching with keywords on PubMed returned relevant results, but they found less utility in the ranking of those results and were reliant on a manual process of reviewing and selecting evidence to pick as sources for curating knowledge. By leveraging natural language processing (NLP) techniques to automatically identify rare disease epidemiologic studies from a very large volume of PubMed articles, we aim to supplement this evidence selection process and reduce the need for strict manual review of publications.
Previously, traditional machine learning approaches have been applied to classify electronic health records for epidemiologic studies.9 Biomedical text classification has been performed using convolutional neural networks10 and support vector machines.11 In this work, we explored the use of a recurrent neural network (RNN) to predict the probability that a given scientific abstract on a rare disease is epidemiology related. In particular, we applied long short-term memory units,12 a type of recurrent neural network that is well-suited for NLP because their ability to store an internal state allows them to effectively process sequential data such as text.13 RNNs are considered state-of-the-art for sentiment analysis,14 machine translation,15 and speech recognition.16 RNNs have also shown to perform well for biomedicine-related NLP tasks, such as named entity recognition for biomedical related terms17 and chemical-protein interaction extraction from scientific papers.18
To our knowledge, this work represents the first attempt to automatically classify epidemiologic publications for rare diseases. Based on the performance of RNNs in related tasks, we hypothesize that this model will also be well-suited for epidemiology identification. We suspect that an RNN will be able to identify more sophisticated and semantically meaningful features than other machine learning approaches such as rule-based models or support vector machines, due to its mathematical complexity and broad success across NLP applications. This feature is important for this task because of the variation in the structure and content of epidemiologic studies, and the superficial similarities with other publication types, particularly in the limited dataset that is available. In addition, the flexibility of neural networks could allow for other types of publication classification tasks to benefit from this approach.
Methods
Dataset construction
We considered epidemiologic study identification as a binary classification task, which thus requires a positive set, containing PubMed abstracts that are rare disease-related epidemiologic studies, and a negative set, consisting of abstracts that are not rare diseases-related epidemiologic studies. As no such datasets already exist, and manually labeling articles would be labor-intensive, we utilized Medical Subject Headings (MeSH)19 and NLP techniques to create our own datasets.
Our positive dataset was constructed from a list of reference articles with epidemiologic data curated by Orphanet, which provides datasets relating to rare diseases.20 We selected only the references indexed by PubMed, which allowed us to retrieve their abstracts and MeSH terms through the EBI RESTful API.21 While many of these articles were epidemiologic studies, some focused on treatments or genetic causes, and instead contained references to data obtained in previous epidemiologic studies for the disease. Others were case reports, which were excluded from this study. To filter out these types of articles, we retrieved the MeSH terms tagged to each PubMed article through the EBI API. If the PubMed article is tagged with the epidemiology-related MeSH terms including "Epidemiology (MeSH:D004813)", "Prevalence(MeSH:D015995)," or "Incidence(MeSH:D015994)," then the article was retained; otherwise, it was excluded. Articles that are categorized as case reports based on their publication types were also removed. Abstracts were retrieved from the API if available based on their PMIDs.
To construct our negative dataset, we began with a list of 6,073 rare diseases included in GARD. For each of these diseases, we invoked the EBI API to retrieve the top five associated PubMed articles. From these results, we removed articles that fall into one of the following criteria: 1) the article is part of the reference list from the Orphanet epidemiologic data; 2) the article is associated with any of the aforementioned epidemiology related MeSH terms; 3) the abstract mentions any of the keywords of "epidemiology", "prevalence", or "incidence."
We combined the above two sets and used an 80:20 training/validation split. From the Orphanet dataset, we randomly selected one hundred articles to form a test set.
Text preprocessing
Text normalization.
Abstracts for epidemiologic studies often include the region in which the study was conducted and numerical statistics for prevalence data. The particular region and specific numerical values would add noise to the interpretation. Thus, we replaced all instances of percentage, geopolitical entities (countries, cities, and states), other locations, dates, times, quantities, ordinal values, and cardinal values with their entity types using the spaCy library.22 In addition, we applied the scispaCy package23 to normalize individual biomedical entities with their corresponding entity types, such as diseases, tissues, organs, and chemicals. We also removed stop words from the text. Figure 1 shows an example of the text normalization process for one abstract. All mentions of the specific disease, numeric values and geographic locations in this example have been normalized by their entity types.
Figure 1:
Text normalization example (the abstract is from24)
Tokenization.
We also fit a preprocessing tokenizer from the TensorFlow library on the training set. We limited our vocabulary size to 5,000, and the words that are not within the 5,000 most frequently used words in the training set are replaced with the <OOV> (out of vocabulary) token. In the example abstract shown in Figure 1, "seroprevalence," "immigrant," and "cruzi" were all replaced with "<OOV>," as these words do not occur frequently in rare disease texts. The tokenizer additionally vectorizes the set of abstracts and adds padding to standardize the length of the abstracts.
Recurrent neural network
We fit a shallow recurrent neural network on the training set. Figure 2 diagrams the model architecture. The network begins with an embedding layer, which converts the input into dense vectors representing the meaning of the abstract. The embedding layer is followed by two long short-term memory layers, the first with 64 units, and the second with 32 units. The output of the second LSTM layer feeds into a fully-connected (dense) layer with a ReLU activation function.25 The final output layer is followed by the softmax activation function, which adjusts the output to create probabilities.26 We used two LSTM layers as we found that this improved the model performance compared to one layer, and given the size of the dataset, we suspected that additional layers could cause overfitting. We begin with 64 units in the first LSTM layer to match the dimensionality of the embedding layer, and we decrease the dimensionality in the second LSTM layer to 32 to more densely represent the data. The model was compiled using the sparse categorical cross entropy loss function, and the Adam stochastic optimization function is applied.27 To reduce overfitting, we used early stopping28 with validation loss as the monitor. We set the maximum number of epochs to 10, as the preliminary results suggested that overfitting would compromise the performance with further epochs.
Figure 2:
The RNN model architecture. "Params" indicates the number of trainable parameters in the layer, and "units" indicates the number of basic computational nodes.
Evaluation
We conducted three steps to evaluate our model. 1) The model was evaluated on the hold-out validation set of 5,275 abstracts, of which 295 were epidemiologic studies. From this set, we calculated precision, recall, F1 score, and area under the ROC curve (AUC). 2) One GARD curator manually validated the predictive results on the test set consisting of 100 abstracts, none of which were included in the training or validation sets. 3) To further assess the performance of the model with practical cases, we performed five case studies with five rare diseases, namely Tay-Sachs disease, Turner syndrome, sickle cell disease, cystic fibrosis, and Ehlers-Danlos syndrome. Specifically, for each disease, we identified epidemiologic studies from their top fifty PubMed articles retrieved via EBI API. We sorted the articles in order of their predicted epidemiology probability and compared with our baseline results, which are the top five results by searching for the disease name and epidemiology related MeSH terms from PubMed.
Results
Dataset preparation
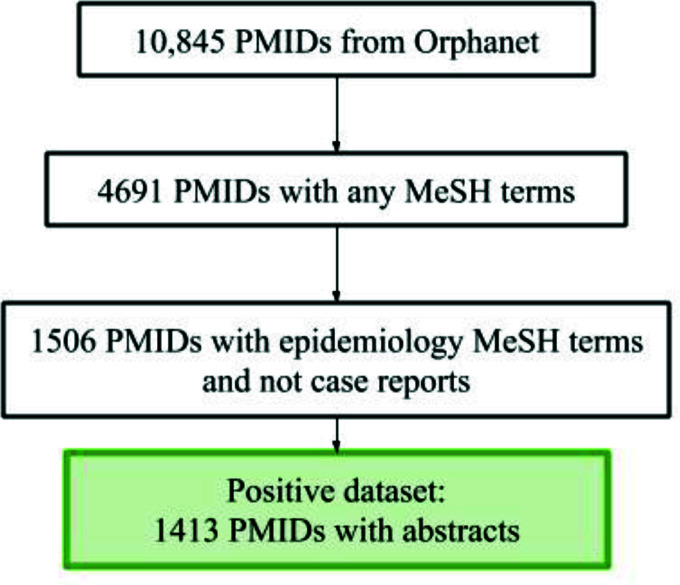
From the Orphanet epidemiology dataset, we extracted 10,845 articles with corresponding PMIDs. There are 4,691 PubMed articles associated with any MeSH terms. Of these, 1506 articles have been tagged with epidemiology-related MeSH terms ("Epidemiology," "Prevalence," or "Incidence") and were not categorized as case reports based on their publication types. After excluding 93 articles without abstracts, 1,413 articles comprised our positive set. Manual inspection on a sample set that confirmed that these articles represent epidemiologic studies. Figure 3 shows the results of creating the positive dataset.
Figure 3:
Stepwise results for the preparation of the positive dataset.
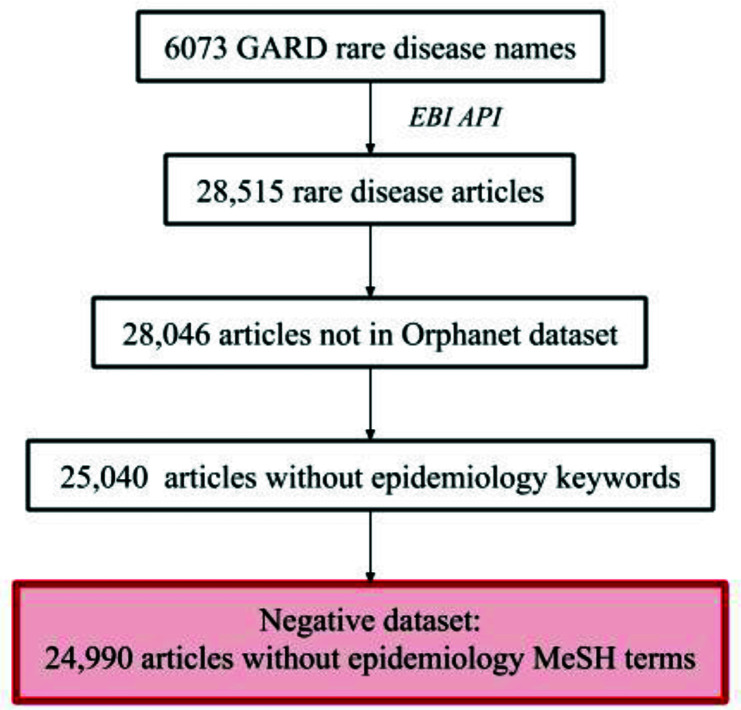
28,515 PubMed articles were retrieved for the 6,073 GARD rare diseases. Of these articles, we excluded 469 articles that are part of the Orphanet epidemiology dataset, and 3,056 articles with epidemiology related MeSH terms or keywords, leaving 24,990 articles in the negative dataset. Manual examination on randomly selected articles was performed and showed that they cover a wide spectrum of topics, including case reports, treatment explanations, genetic analyses, and general literature reviews of disorders. The results of the negative dataset preparation are shown in Figure 4.
Figure 4:
Stepwise results for the preparation of the negative dataset.
Table 1 provides the breakdown statistics of the dataset. In Discussion, we discuss the reason of having the imbalance in the training set and its influence on the model performance. Note that the total positive and negative dataset sizes were slightly reduced from the aforementioned numbers as articles in the test set were removed.
Table 1: The composition of the training and validation sets.
| Positive dataset (epidemiologic studies) | Negative dataset (not epidemiologic studies) | Total | |
| Training set | 1119 | 19,981 | 21,100 |
| Test set | 268 | 5007 | 5275 |
| Total | 1387 | 24,988 | 26,375 |
Holdout validation set evaluation
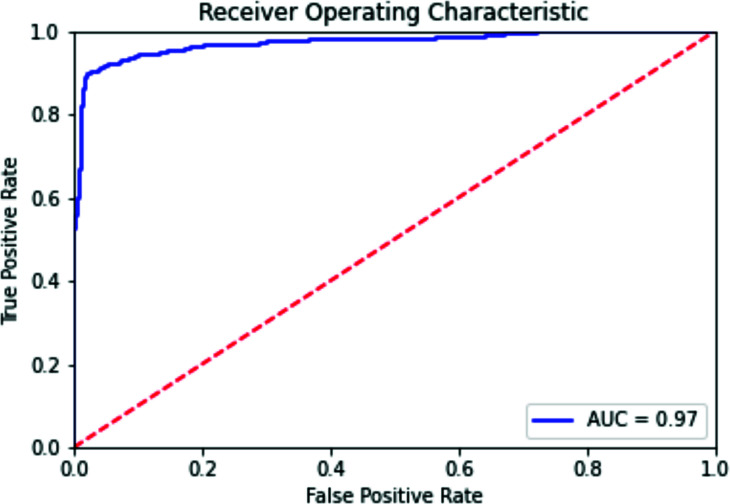
The recurrent neural network achieved promising results on the holdout validation set. Early stopping halted training after three epochs because of an increase in loss in the validation set. At this point, the precision on the validation set was 0.846, the recall was 0.937, the F1 score was 0.886, and the AUC was 0.967. The receiver operating characteristic (ROC) curve is given in Figure 5.
Figure 5:
ROC curve for the holdout validation set.
Overall, while the average epidemiology probability among the true positives was 0.966, the false positives received an average epidemiology probability of 0.892. Conversely, the epidemiology probability among the true negatives was 0.0229, while it was 0.0563 for false negatives. Of the 28 false negatives, only eight abstracts included epidemiologic information based on our manual review. Thus, given the focus of our study, the other twenty should be considered as true negatives, as our classification used only the abstract.
Manual evaluation
A GARD curator manually validated the predictive results on the test set consisting of 100 articles, and these results suffered slightly compared to results on the holdout validation set. The precision was 0.726, the recall was 0.700, the F1 score was 0.701, and the AUC was 0.751. We discuss the reasons behind this discrepancy in the Discussion section.
Of the twenty false negatives based on the test result, twelve articles described epidemiologic information only in the full text, instead of in the abstracts themselves, such as with the two article "Chromosome 1p36 deletions: the clinical phenotype and molecular characterization of a common newly delineated syndrome"29 and "Mutations in KANSL1 cause the 17q21.31 microdeletion syndrome phenotype".30 Thus, these errors were likely an artifact of the differing focus of the manual evaluation. The false positives included genealogy and genetics studies, a case report, and two epidemiologic studies in geographic regions that were too constricted for use by GARD.
Case studies
On the case studies we performed for five rare diseases, our model generally successfully identified epidemiologic studies from PubMed. We set the probability threshold for an epidemiology article to be 0.5, and additionally included the exact probability for analysis. In most cases, the results returned with our method were more relevant than those found via filtering by epidemiology related MeSH terms from PubMed. The PubMed search query was composed as "(((epidemiology[MeSH Terms]) OR (prevalence[MeSH Terms])) OR (incidence[MeSH Terms])) AND (Disease Name)", where "Disease Name" is replaced with the specific disease name.
Tay-Sachs disease.31
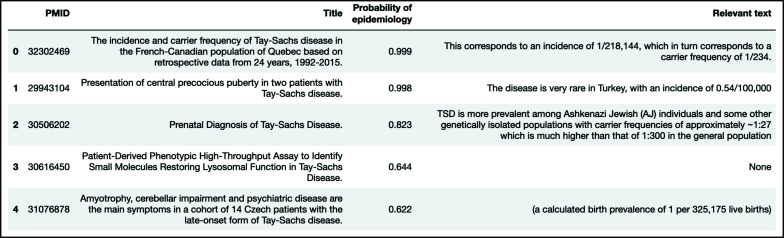
Four of the five articles that are predicted as epidemiologic studies by our model contain epidemiologic information, as shown in Figure 6. The article without epidemiologic information was ranked fourth of the five and had an epidemiology probability of 0.644. In contrast, out of the top five results from the manual PubMed search for Tay Sachs with epidemiology related MeSH terms, only one article titled "Insights into the genetic epidemiology of Crohn's and rare diseases in the Ashkenazi Jewish population"32 was epidemiology related and contained minimal information on Tay-Sachs disease. None of the four epidemiology articles discovered by our model appeared in the PubMed search results.
Figure 6:
Predictive results generated for the case study of Tay-Sachs disease.
Turner syndrome.33
Two articles were predicted as epidemiologic studies. The first article does in fact give the prevalence of the syndrome,34 while the second article described the risk of coronary artery disease, a known clinical complication amongst Turner syndrome patients.35 The manual PubMed search does not include any epidemiologic studies in the top five results; notably, two of them were not related to Turner syndrome at all, and another two articles detail bone fragility and autoimmune thyroid disease in Turner syndrome, but are not epidemiologic studies.
Sickle cell disease (SCD).36
Of the four articles predicted as epidemiologic studies for SCD, one stated a rough estimate for its prevalence in the United States,37 one referred to the millions of patients affected worldwide,38 one compared the prevalence of priapism in those with and without SCD,39 and one detailed an approach to treatment.40 None of the results from the manual PubMed search were epidemiologic studies or provided epidemiologic information in their abstracts.
Cystic fibrosis.41
One positive result from the model for cystic fibrosis described the prevalence of fungal disease within the disorder.42 None of the results from the manual PubMed search were epidemiologic studies, although one provided an estimate for the worldwide prevalence of the disease.43
Ehlers-Danlos syndrome.44
One of the two positive results generated from the model, detailed the prevalence of cardiovascular disorders in patients with this syndrome.45 The other was did not involve epidemiology.46 One result classified as negative did include a prevalence statistic, but the topic of the article was surgical outcomes.47 None of the manual search results were epidemiologic studies or included epidemiologic information for Ehlers-Danlos syndrome.
Discussion
Epidemiologic studies provide insights and directions for basic and clinical research to determine the causes and mechanisms of rare diseases and develop methodologies for prevention, diagnosis, and treatment. However, epidemiologic data curation in the rare disease field continues to rely heavily on human effort, from identification of epidemiologic studies from PubMed to data curation. In this study, we presented a computational model by applying recurrent neural networks and NLP techniques to programmatically identify epidemiologic studies from PubMed. This work can reduce the human effort required from the epidemiologic data curation process and holds promise for other applications beyond rare diseases and with other types of studies.
Quantitatively, our model performed very well on the holdout validation set, with a high AUC of 0.967. Our manual inspection of the results further proved that our model can consistently assign high epidemiology probabilities (above 0.98) for standard epidemiologic studies, and strong correlation is found between the predicted epidemiology probability and the amount of epidemiologic information mentioned in the abstract. For example, an article titled "Birth prevalence of Prader-Willi syndrome in Australia", whose abstract details an epidemiologic study,48 obtained an epidemiology probability of 0.999. However, the article titled "Th17 cytokine deficiency in patients with Aspergillus skull base osteomyelitis", which is a molecular study,49 is predicted to have an epidemiology probability of 0.00956. In addition, the five case studies demonstrated that this model was effective at surfacing epidemiologic studies for individual diseases. Compared to the baseline results with manual PubMed search, our model captured more epidemiologic studies, which were not part of the top five results, or were even not found in the entire list of PubMed search results. However, we observed the performance of the model on the test set was not as promising as the holdout validation set. Our analysis indicated this discrepancy was likely due to our focus on the content of the abstract, while the curators often examined the full text in addition to the abstract when labeling the dataset.
Notably, our model reached satisfying performance even with a dataset that is small and imbalanced: non-epidemiology articles outnumbered epidemiologic studies by roughly 20:1. Initially, we expanded our positive dataset by including articles tagged with epidemiology-related MeSH terms that were not referenced by Orphanet. However, this did not significantly improve the performance. This was likely because some of the MeSH terms may have been assigned incorrectly, whereas restricting the dataset to those also used by Orphanet added another layer of confirmation that the articles were likely related to epidemiology. The success of our model in light of this illustrates that the features of an epidemiologic study are easily identifiable and significantly distinct from those characteristic of case reports, clinical guidelines, genetic analyses, and other types of studies. For instance, of the 1702 case reports in the validation set, only 17 were predicted as epidemiologic studies. Since case reports rarely include epidemiology information about a disease, this result suggests that the model was able to identify features distinguishing case reports from epidemiologic studies.
Given the lack of available training data relating to epidemiology, we used a combination of Orphanet data, MeSH terms, and keyword searches to generate our dataset. This approach could introduce bias based on the types of sources selected by Orphanet and the process used to assign MeSH terms. The strategy of generating the negative set by excluding abstracts containing epidemiology keywords set might also bias the model toward over-relying on keywords to generate its predictions rather than more sophisticated linguistic features. We did not observe significant negative impact as a result, but a follow-up analysis could better characterize any bias. Relatedly, a more robust evaluation of the model from a larger and more consistently labeled dataset would assist in confirming our results.
The computational approach established in this study will be able to support the task of supplementing epidemiology curation for GARD and other applications in multiple ways. First, our model can identify and rank epidemiologic studies relating to rare diseases. This would allow curators to begin by reviewing the articles with the highest predicted epidemiology probability, rather than searching for relevant articles manually. Second, the model could be integrated into an alert system to notify curators about the publication of new epidemiologic studies. From a set of epidemiologic studies identified by the model, we could apply information extraction to their text following previous work50, which could lead to a process to fully automate the curation of epidemiology data.
Furthermore, there are several directions for expanding this work. A deeper analysis into the results of our model could reveal features or patterns in its predictions that would allow the model to be refined to achieve better performance, as the interpretability of the model at present is limited. The addition of more data, particularly epidemiology articles, could also improve performance. In this study, we limited the dataset to articles addressing rare diseases as this was the immediate use case of the model, and this approach accounts for any unique structural and content features of rare disease epidemiologic abstracts. In future work, epidemiologic studies addressing diseases that are not rare may also be included. Because the text processing steps remove the specific disease features, this change will likely improve the capacity of the model to identify rare disease epidemiologic studies, as the benefit of increasing the size of the dataset could outweigh any noise that is introduced. Furthermore, an expanded dataset could allow for more advanced approaches such as Bidirectional Encoder Representations from Transformers (BERT)51 or a deeper neural network architecture; these approaches were not used in this study due to concerns about overfitting on a limited dataset. In order to capture epidemiologic information beyond epidemiologic studies, our model framework could be applied to identify case reports, as these can be aggregated to determine case or family counts. When we combined case reports with epidemiologic studies in our dataset, the model performance suffered, likely because the structure and content of case reports differ significantly from epidemiologic studies. However, case reports could be considered independently in a separate model. Similarly, because of the generalizability of neural networks, our approach could also be used to develop classifiers for natural history studies or clinical trials, and in other domains beyond rare diseases.
Conclusion
In this paper, we demonstrated that a recurrent neural network with long short-term memory architecture achieved good performance in classifying epidemiologic studies of rare diseases. Our model can be leveraged to greatly shorten the manual curation process for evidence selection in curating epidemiologic information. We hypothesize that the success of our model suggests that our approach can be applied to other similar tasks such as classifying natural history studies and in other medical domains.
Acknowledgements
This research was supported in part by the Intramural/Extramural research program of NCATS, NIH. The authors thank Karen Hanson, from ICF International, Inc. for her help on manual evaluation; Dac-Trung Nguyen, from Division of Pre-Clinical Innovation, NCATS, participated in the valuable discussion. Dr. Anne Pariser, as Director of the Office of Rare Disease Research (ORDR), at NCATS, supported this work and also participated in the valuable discussion. Lastly, we thank the NIH Office of Data Science Strategy and the HHS Civic Digital Fellowship for supporting these efforts.
Figures & Table
References
- 1.Rare Diseases Act of 2002 Congress 107th Sess. 2002.
- 2.Dawkins HJ, Draghia-Akli R, Lasko P, et al. Progress in rare diseases research 2010–2016: an IRDiRC perspective. Clinical and Translational Science. 2018;11(1):11. doi: 10.1111/cts.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Griggs RC, Batshaw M, Dunkle M, et al. Clinical research for rare disease: opportunities, challenges, and solutions. Molecular Genetics and Metabolism. 2009;96(1):20–6. doi: 10.1016/j.ymgme.2008.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hassell KL. Population estimates of sickle cell disease in the US. American Journal of Preventive Medicine. 2010;38(4):S512–S21. doi: 10.1016/j.amepre.2009.12.022. [DOI] [PubMed] [Google Scholar]
- 5.Jansen Type Metaphyseal Chondrodysplasia: NORD - National Organization for Rare Disorders 2018. [Available from: https://rarediseases.org/rare-diseases/jansen-type-metaphyseal-chondrodysplasia/
- 6.Boat TF, Field MJ. Accelerating research and development. National Academies Press; 2011. Rare diseases and orphan products. [PubMed] [Google Scholar]
- 7.Wakap SN, Lambert DM, Olry A, et al. Estimating cumulative point prevalence of rare diseases: analysis of the Orphanet database. European Journal of Human Genetics. 2020 Feb;28(2):165–73. doi: 10.1038/s41431-019-0508-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.GARD Information Center [Available from: https://rarediseases.info.nih.gov/
- 9.Schuemie MJ, Sen E, 't Jong GW, van Soest EM, Sturkenboom MC, Kors JA. Automating classification of free-text electronic health records for epidemiological studies. Pharmacoepidemiology and Drug Safety. 2012;21(6):651–8. doi: 10.1002/pds.3205. [DOI] [PubMed] [Google Scholar]
- 10.Rios A, Kavuluru R, editors. Convolutional neural networks for biomedical text classification: application in indexing biomedical articles. Proceedings of the 6th ACM Conference on Bioinformatics, Computational Biology and Health Informatics; 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen AM. AMIA annual symposium proceedings. American Medical Informatics Association; 2006. An effective general purpose approach for automated biomedical document classification. [PMC free article] [PubMed] [Google Scholar]
- 12.Hochreiter S, Schmidhuber J. Long short-term memory. Neural Computation. 1997;9(8):1735–80. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- 13.Lipton ZC, Berkowitz J, Elkan C. A critical review of recurrent neural networks for sequence learning. arXiv preprint arXiv. 2015;150600019 [Google Scholar]
- 14.Tang D, Qin B, Liu T, editors. Document modeling with gated recurrent neural network for sentiment classification. Proceedings of the 2015 Conference on Empirical Methods in Natural Language Processing.2015. [Google Scholar]
- 15.Wu Y, Schuster M, Chen Z, et al. Google’s neural machine translation system: Bridging the gap between human and machine translation. arXiv preprint arXiv. 2016;160908144 [Google Scholar]
- 16.Graves A, Jaitly N, editors. Towards end-to-end speech recognition with recurrent neural networks. International Conference on Machine Learning; 2014. [Google Scholar]
- 17.Li L, Jin L, Jiang Z, Song D, Huang D, editors. Biomedical named entity recognition based on extended recurrent neural networks. 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM); IEEE; 2015. [Google Scholar]
- 18.Lu H, Li L, He X, Liu Y, Zhou A. Extracting chemical-protein interactions from biomedical literature via granular attention based recurrent neural networks. Computer Methods and Programs in Biomedicine. 2019;176:61–8. doi: 10.1016/j.cmpb.2019.04.020. [DOI] [PubMed] [Google Scholar]
- 19.Lipscomb CE. Medical subject headings (MeSH) Bulletin of the Medical Library Association. 2000;88(3):265. [PMC free article] [PubMed] [Google Scholar]
- 20.Epidemiological Data Orphanet, editor. orphadata.org2020.
- 21.Burke M, Armstrong D, Carvalho-Silva D, et al. EMBL-EBI, programmatically: take a REST from manual searches. European Bioinformatics Institute (EMBL-EBI); 2017. [Google Scholar]
- 22.spaCy: Explosion AI. 2020. [Available from: https://spacy.io/
- 23.Neumann M, King D, Beltagy I, Ammar W. Scispacy: Fast and robust models for biomedical natural language processing. arXiv preprint arXiv. 2019;190207669 [Google Scholar]
- 24.Bern C, Montgomery SP. An estimate of the burden of Chagas disease in the United States. Clinical Infectious Diseases. 2009;49(5):e52–e4. doi: 10.1086/605091. [DOI] [PubMed] [Google Scholar]
- 25.Nair V, Hinton GE, editors. Rectified linear units improve restricted boltzmann machines. ICML; 2010. [Google Scholar]
- 26.Bridle J. Training stochastic model recognition algorithms as networks can lead to maximum mutual information estimation of parameters. Advances in Neural Information Processing Systems. 1989;2:211–7. [Google Scholar]
- 27.Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv. 2014;14126980 [Google Scholar]
- 28.Caruana R, Lawrence S, Giles CL, editors. Overfitting in neural nets: Backpropagation, conjugate gradient, and early stopping. Advances in Neural Information Processing Systems. 2001.
- 29.Shapira SK, McCaskill C, Northrup H, et al. Chromosome 1p36 deletions: the clinical phenotype and molecular characterization of a common newly delineated syndrome. The American Journal of Human Genetics. 1997;61(3):642–50. doi: 10.1086/515520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zollino M, Orteschi D, Murdolo M, et al. Mutations in KANSL1 cause the 17q21. 31 microdeletion syndrome phenotype. Nature Genetics. 2012;44(6):636–8. doi: 10.1038/ng.2257. [DOI] [PubMed] [Google Scholar]
- 31.Tay-Sachs disease [Available from: https://rarediseases.info.nih.gov/diseases/7737/tay-sachs-disease .
- 32.Rivas MA, Avila BE, Koskela J, et al. Insights into the genetic epidemiology of Crohn’s and rare diseases in the Ashkenazi Jewish population. PLoS Genetics. 2018;14(5):e1007329. doi: 10.1371/journal.pgen.1007329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner syndrome [Available from: https://rarediseases.info.nih.gov/diseases/7831/turner-syndrome .
- 34.Abu-Halima M, Oberhoffer FS, El Rahman MA, et al. Insights from circulating microRNAs in cardiovascular entities in turner syndrome patients. PLoS One. 2020;15(4):e0231402. doi: 10.1371/journal.pone.0231402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Funck KL, Budde RPJ, Viuff MH, et al. Coronary plaque burden in Turner syndrome a coronary computed tomography angiography study. Heart Vessels. 2020. [DOI] [PubMed]
- 36.Sickle cell disease [Available from: https://www.genome.gov/genetics-glossary/Sickle-Cell-Disease .
- 37.Fantasia HC, Morse BL. Voxelotor for the treatment of sickle cell disease. Nurs Womens Health. 2020;24(3):233–7. doi: 10.1016/j.nwh.2020.03.003. [DOI] [PubMed] [Google Scholar]
- 38.Pavan AR, Dos Santos JL. Advances in sickle cell disease treatments. Curr Med Chem. 2020. [DOI] [PubMed]
- 39.Idris IM, Abba A, Galadanci JA, et al. Men with sickle cell disease experience greater sexual dysfunction when compared with men without sickle cell disease. Blood Adv. 2020;4(14):3277–83. doi: 10.1182/bloodadvances.2020002062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Herity LB, Vaughan DM, Rodriguez LR, Lowe DK. Voxelotor: a novel treatment for sickle cell disease. Ann Pharmacother. 2020:1060028020943059. doi: 10.1177/1060028020943059. [DOI] [PubMed] [Google Scholar]
- 41.Cystic fibrosis [Available from: https://rarediseases.info.nih.gov/diseases/6233/cystic-fibrosis .
- 42.Cuthbertson L, Felton I, James P, et al. The fungal airway microbiome in cystic fibrosis and non-cystic fibrosis bronchiectasis. J Cyst Fibros. 2020. [DOI] [PMC free article] [PubMed]
- 43.Baiardini I, Steinhilber G. DI Marco F, Braido F, Solidoro P. Anxiety and depression in cystic fibrosis. Minerva Med. 2015;106(5 Suppl 1):1–8. [PubMed] [Google Scholar]
- 44.Ehlers-Danlos syndrome [Available from: https://www.cedars-sinai.org/health-library/diseases-and-conditions/e/ehlers-danlos-syndrome-eds.html .
- 45.Paige SL, Lechich KM, Tierney ESS, Collins RT. Cardiac involvement in classical or hypermobile Ehlers-Danlos syndrome is uncommon. Genet Med. 2020. [DOI] [PubMed]
- 46.Miller AJ, Schubart JR, Sheehan T, Bascom R, Francomano CA. Arterial elasticity in Ehlers-Danlos syndromes. Genes (Basel) 2020;11(1) doi: 10.3390/genes11010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Louie A, Meyerle C, Francomano C, et al. Survey of Ehlers-Danlos patients’ ophthalmic surgery experiences. Mol Genet Genomic Med. 2020;8(4):e1155. doi: 10.1002/mgg3.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smith A, Egan J, Ridley G, et al. Birth prevalence of Prader-Willi syndrome in Australia. 2003;88(3):263–4. doi: 10.1136/adc.88.3.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Delsing CE, Becker KL, Simon A, et al. Th17 cytokine deficiency in patients with Aspergillus skull base osteomyelitis. BMC Infectious Diseases. 2015;15(1):140. doi: 10.1186/s12879-015-0891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karystianis G, Thayer K, Wolfe M, Tsafnat G. Evaluation of a rule-based method for epidemiological document classification towards the automation of systematic reviews. Journal of Biomedical Informatics. 2017;70:27–34. doi: 10.1016/j.jbi.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 51.Devlin J, Chang M-W, Lee K, Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv. 2018;181004805 [Google Scholar]