Abstract
Several studies have shown that COVID-19 patients with prior comorbidities have a higher risk for adverse outcomes, resulting in a disproportionate impact on older adults and minorities that fit that profile. However, although there is considerable heterogeneity in the comorbidity profiles of these populations, not much is known about how prior comorbidities co-occur to form COVID-19 patient subgroups, and their implications for targeted care. Here we used bipartite networks to quantitatively and visually analyze heterogeneity in the comorbidity profiles of COVID-19 inpatients, based on electronic health records from 12 hospitals and 60 clinics in the greater Minneapolis region. This approach enabled the analysis and interpretation of heterogeneity at three levels of granularity (cohort, subgroup, and patient), each of which enabled clinicians to rapidly translate the results into the design of clinical interventions. We discuss future extensions of the multigranular heterogeneity framework, and conclude by exploring how the framework could be used to analyze other biomedical phenomena including symptom clusters and molecular phenotypes, with the goal of accelerating translation to targeted clinical care.
Introduction
Despite extreme measures to contain the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the resulting corona virus disease 2019 (COVID-19) continues to have a devastating impact on the physical, social, cultural, and economic health of humans around the world. Although this novel corona virus is serologically close to the known SARS-CoV, it has spread more widely primarily due to virus shedding from asymptomatic patients. As of December 23rd 2020 more than 78.6 million people were infected worldwide, with more than 1.73 million dead due to fatal complications. Because many countries have yet to peak in their cases and fatalities and prepare for subsequent waves and potential reinfections, there is an urgent need to analyze and treat the causes for fatal complications in COVID-19 patients.
A key trait of COVID-19 is the high fatality rate in older adults and minorities1–3 resulting from the following molecular and socio-demographic factors: (1) Molecular Mechanisms Precipitating Fatal Complications. SARS-CoV-2 has the characteristic spiked protein 3-D structure, with a strong binding affinity to the human cell receptor angiotensin-converting enzyme 2 (ACE2).4 Because ACE2 is expressed in many organs including the heart, lungs, and kidneys, in addition to the nervous system and skeletal muscle, SARS-CoV-2 can infect multiple sites by traversing the hematogenous or the retrograde neuronal routes.5,6 Furthermore, laboratory tests of critical patients have shown abnormal levels for key markers including cardiac troponin (myocardial infarction), D-dimer (compromised blood clotting), lymphocytes (lymphopenia), lactate dehydrogenase (multiple organ failure), and liver enzymes (damage to liver cells).7 These results suggest that SARS-CoV-2 can exacerbate already compromised organs in older patients with multiple chronic conditions, resulting in a high rate of complications, multiple organ failures, and fatalities;4,8 Furthermore, the higher expression of ACE2 in males9 is a critical factor in putting them at a higher risk for severe complications with COVID-19; (2) Prevalence of Multiple Chronic Conditions in Older Adults and Minorities. Due to a wide range of factors including improved treatments and increased life-expectancy, a growing number of older adults live with and manage multiple chronic conditions (MCCs) defined as <2 concomitant chronic conditions (also referred to as multimorbidities, or comorbidities when used in the context of an index condition).10 This trend has resulted in almost 75% of Americans aged 65 years and older having more than one chronic condition, 20% having five or more comorbidities, and 50% receiving five or more medications.11 Furthermore, due to systemic health inequities, MCCs is also growing in subgroups including women, African Americans, and non-Hispanic Whites.
The above two factors have resulted in a disproportionate number of infected older adults and minorities having adverse outcomes, including respiratory failure requiring ventilators, and multiple organ failure needing management in intensive care units (ICUs), and of mortality.1–3 However, despite the high prevalence of MCCs in these populations, the emerging clinical practice guidelines to treat COVID-19 patients have focused on treating single conditions. For example, recommendations to treat COVID-19 patients with diabetes have little to no cross-referencing to other comorbidities if they co-occur in the same patient.12 This single-condition focus is not unique to COVID-19; few existing clinical practice guidelines (CPGs) to treat conditions such as congestive heart failure are designed to treat multiple co-occurring conditions.13–17 Such condition-specific guidelines can substantially increase the burden of treatment on older adults to manage complex treatment regimes, and require constant monitoring by primary care physicians to change a treatment plan if it leads to adverse or null outcomes.15–17
We therefore attempted to address the above gap in understanding of how comorbidities co-occur in COVID-19 patients with the goal of designing guidelines for treating patients with multiple chronic conditions. We begin with describing the current methods used to analyze heterogeneity in the comorbidity profiles of patients, and the advantages of using bipartite networks to automatically identify patient subgroups and their most frequently co-occurring comorbidities at different levels of granularity. Next, we discuss how we used that approach to analyze COVID-19 patients at three levels of granularity, each providing direct clinical implications related to frequency, risk, and similarity of comorbidity profiles. We conclude with how the multigranular heterogeneity approach could be used to analyze other biomedical phenomena, and how it could be extended to include other analytical methods, with the goal of accelerating translation of results into clinical care.
Current Methods Used to Analyze Co-Occurrence of Multimorbidities
Because having MCCs is associated with several adverse outcomes including poor quality of life, physical disabilities, high healthcare use, drug-drug and drug-disease interactions, and mortality, several studies have attempted to analyze MCCs in older adults and minorities, with the goal of optimizing care.13,14 These studies have used a wide range of methods to analyze multimorbidities, each with critical trade-offs. For example, many studies have attempted to identify frequently co-occurring multimorbidities using combinatorial approaches18 (identify all pairs, all triples etc.). However, while such approaches are intuitive to understand, they lead to a combinatorial explosion (e.g., finding all combinations of the 31 Elixhauser comorbidities would lead to 231 or 2147483648 combinations), but with no simple way of addressing the overlap of patients between the combinations to identify patient subgroups.
Several studies have used unipartite clustering methods19,20 (clustering patients or comorbidities, but not both simultaneously) such as k-means, and hierarchical clustering to help identify either clusters of frequently co-occurring multimorbidities, or patients that have a high similarity in their multimorbidity profiles. Other studies have used dimensionality-reduction methods such as principal component analysis (PCA)19 combined with k-means to identify clusters of either MCCs or patients. However, because such methods produce unipartite outputs, there is no agreed upon method to identify the patient subgroups defined by a cluster of MCCs because patients can belong to more than one MCC cluster, and vice-versa. Furthermore, such methods have well-known limitations including the requirement of user-selected parameters such as similarity measures and the number of expected clusters, in addition to the absence of a quantitative measure to describe the quality of the clustering, critical for measuring its statistical significance.
Researchers have used the above methods to analyze multimorbidities at different levels of granularities in the data. Several studies have focused on analyzing multimorbidities at the cohort-level to identify co-occurring multimorbidities in an entire dataset.21 Other studies have focused on analyzing pre-defined patient subgroups within specific diseases such as COPD to determine their risk for adverse outcomes.22 Finally, a few studies have analyzed multimorbidities at an individual patient-level to determine through a case study approach, their burden of treatment resulting from clinical practice guidelines focused on treating single conditions.13
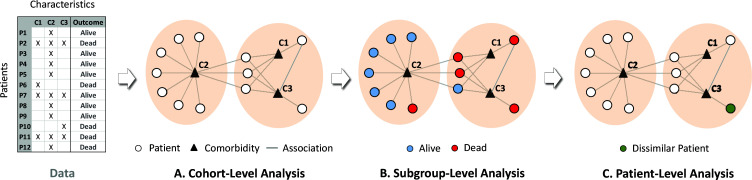
More recently, bipartite approaches have attempted to address the limitations of unipartite methods by identifying biclusters20,23 of patients and comorbidities simultaneously. For example, as shown in Fig. 1A, bipartite networks24 can be used to represent patients as well as their comorbidities as nodes (circles and triangles respectively), and the pair-wise associations between patients and comorbidities can be represented as edges (lines). Furthermore, algorithms such as modularity maximization24 can be used to automatically (1) identify the number and members of biclusters consisting of patient subgroups and their most frequently co-occurring comorbidities, (2) measure the quality of the biclustering through a quantity called modularity, used to measure its significance compared to random permutations of the data;20,25 and (3) visualize the results using force-directed algorithms such as Kamada Kawai26 and ExplodeLayout.27
Fig. 1.
Heterogeneity analysis at three levels of granularity: (A) cohort-level analysis enables the identification of frequently co-occurring comorbidities; (B) subgroup-level analysis enables the measurement of comparative risk of patient subgroups and how they differ in their characteristics; and (C) patient-level analysis helps to determine whether a specific patient is similar or dissimilar to other patients in their respective subgroup.
Need to Analyze Heterogeneity at Multiple Levels of Granularity
Although bipartite networks have been effective in automatically identifying statistically significant and clinically meaningful patient subgroups, recent results have revealed that heterogeneity in patient profiles could exist at other levels of granularity. For example, while a bipartite network analysis of comorbidities in hip-fracture patients helped to reveal heterogeneities at the cohort-level, there were additional heterogeneities within patient subgroups such as a significantly different proportion of patients that had one or more comorbidities across the subgroups.28 Such heterogeneities could impact how patients within each subgroup receive treatment. These results suggest that bipartite networks could be used to analyze heterogeneity at multiple levels of granularity in the data, each level providing different insights for designing clinical interventions.
For example, at the cohort-level (Fig. 1A) a bipartite network analysis can automatically identify biclusters consisting of patient subgroups defined by frequently co-occurring comorbidities. This level of analysis enables clinicians to determine which combinations of comorbidities need to be addressed in clinical practice guidelines, and identify potential underlying disease mechanisms. Furthermore, the bipartite network can be used to analyze heterogeneity at the subgroup-level by ranking each subgroup based on their risk for an outcome such as mortality (shown as red and blue nodes in Fig. 1B) enabling clinicians to design patient triage strategies in resource-constrained situations such as in COVID-19 hotspots. Finally, analysis at the patient-level (Fig. 1C) could help identify the degree of similarity of a specific patient to the rest in a subgroup, enabling clinicians to determine whether that patient should be treated using a generalized intervention designed for the entire subgroup, or requires individualized treatment.
The bipartite representation therefore provides a unified approach to quantitatively analyze and visually interpret heterogeneity at all three levels of granularity. This granularity approach enables the inspection of whether and how each level of granularity contributes to the translation of the analytical results into clinic interventions. Given the many unknowns in how multiple comorbidities impact outcomes in COVID-19 patients, and the urgent need for clinical interventions, we used the above multigranular heterogeneity analytical framework to guide our analysis of COVID-19 patients, with the explicit goal of enabling a more systematic approach for designing clinical interventions targeted to patients with multiple comorbidities.
Method
Research Questions. To analyze heterogeneity in the comorbidity profiles of COVID-19 inpatients at multiple levels of granularity, we posed the following three research questions: (1) Cohort-Level: How do comorbidities frequently co-occur to form subgroups of COVID-19 inpatients? (2) Subgroup-Level: What is the risk of inpatient subgroups for adverse outcomes (ICU No-Vent, ICU With-Vent, and mortality)? (3) Patient-Level: What is the degree of similarity in the profile of a specific inpatient compared to the rest of the inpatients in the respective subgroup?
Data. Using IRB (#STUDY00009771) from the University of Minnesota, we analyzed electronic health records (EHR) data from the University of Minnesota M Health Fairview COVID-19 patient registry. This registry includes patient data (spanning 135 unique zip codes) from 12 hospitals and 60 clinics in the Minneapolis-St. Paul twin-city area, and currently accounts for over 20% of inpatients in Minnesota. On August 2nd 2020, the registry contained health records of COVID-19 inpatients (n=858) with complete data for: (1) 31 comorbidities (Elixhauser comorbidities in 2019-2020). A subset of the COVID-19 inpatients had no comorbidities (n=69), which were considered as the control group; (2) 7 complications (acute respiratory distress (ARDS), acute kidney injury (AKI), hypotension, bleeding, delirium, and VTE/CVA/MI). Of these, 5 had <2% prevalence in the cases and controls and therefore were dropped from the analysis; (3) 55 laboratory test results (e.g., platelet count, creatinine, hemoglobin, D-dimer, troponin, IL6) that were dichotomized into normal vs. abnormal. Of these, 13 laboratory tests had <2% prevalence in the cases and controls and were therefore dropped from the analysis; and (4) markers of adverse outcomes including use of an intensive care unit without a ventilator (ICU No-Vent, n=273), use of an intensive care unit with a ventilator (ICU With-Vent, n=672), and mortality (n=103). As all our data relates to COVID-19 positive inpatients, henceforth we refer to them simply as patients.
Analysis. We used bipartite networks to quantitatively and visually analyze the above COVID-19 patient data with the goal of enabling domain experts design targeted interventions, by using the following steps:
Feature Selection. We used the chi-squared test to measure the univariable significance (corrected for multiple testing using false discovery rate) of each comorbidity to each of three outcomes (ICU No-Vent, ICU With-Vent, and mortality), compared to the control group (patients with no comorbidities), and selected those comorbidities that were significant at the .05 level for at least one of the three adverse outcomes (ICU No-Vent, ICU With-Vent, and mortality).
- Bipartite Network Analysis. Similar to Fig. 1A, we represented patients and comorbidities as nodes (circles and triangles respectively), and the pair-wise association between them as edges (lines). Patients (n=789) with at least one of the significant comorbidity were used in the bipartite network analysis, and patients with no comorbidities (n=69) were used as controls. The resulting network was analyzed at the following three levels of granularity:
- Cohort-Level. The quantitative analysis consisted of the following: (1) used a bicluster modularity maximization24 algorithm to identify the number and boundaries of patient-comorbidity biclusters and the degree of biclustering (Q); and (2) measured the significance of Q by comparing it to a distribution of Q generated from 1000 random permutations of the network by preserving the size (number of nodes and edges in the network), and the distribution of edges for each comorbidity. The biclustering was tested for stability by measuring the Adjusted Rand Index (ARI)19 between the comorbidity clustering in the real data, to 1000 random bootstrap resamples of patients in the data. The visual analysis of the above results consisted of the following: (1) used Kamada-Kawai24 to layout the network; and (2) applied ExplodeLayout27 to separate the identified biclusters for improving their interpretability.
- Subgroup-Level. The quantitative analysis consisted of the following steps applied to each patient subgroup: (1) used logistic regression to measure the odds ratio (OR) and tested its significance (corrected for multiple testing using FDR) for each of the three outcomes, in comparison to the control group; and (2) used logistic regression to measure the OR and tested its significance (corrected for multiple testing using FDR) for each demographic, complication, and laboratory test variable, compared to the control group. The visual analysis consisted of coloring the patient nodes based on mortality (as shown in Fig. 1B), and displaying in text the outcomes for each bicluster. Additionally, to increase interpretability of the results by the domain experts, a profile of each bicluster was compiled in a table (Table 4) showing the respective comorbidities, outcomes, demographics, complications, and laboratory test results.
- Patient-Level. The quantitative analysis consisted of the following: (1) defined an augmented bicluster as all patients within a bicluster (identified through modularity maximization in Step-2A), and all comorbidities within and outside that bicluster that were connected to at least one patient within the bicluster; (2) used the Jaccard Distance (JD),19 defined as: 1 – (number of shared comorbidities between a patient pair / union of comorbidities in that pair), to measure the similarity in comorbidity profile of each patient to the other patients within each of the augmented biclusters, and calculated the mean JD for each patient; and (3) generated a distribution of the mean JDs for each augmented bicluster. This distribution was used to test whether the mean JD of a specific patient was significantly higher than the mean of the distribution in its augmented bicluster. A patient which had a significantly higher mean JD compared to the mean of the above distribution was considered dissimilar to other patients in the respective bicluster. Outlier patients with the minimum and the maximum number of comorbidities were selected for interpretation by the domain experts to examine whether they would require treatment that was different from their respective bicluster.
Clinical Interpretation. The results of the above quantitative and visual analysis at each level of granularity were presented to two domain experts with experience in treating COVID-19 inpatients. To guard against confirmation bias,29 we used the following steps with each domain expert: (1) presented the network visualization generated from Step-2A (which did not contain the associations of each bicluster to any of the variables); (2) asked them to use their clinical judgement to rank the biclusters based on the risk for mortality; (3) revealed the quantitative profile of each bicluster, and asked them to discuss discrepancies between their prediction and the results, with the goal of determining which of the bicluster profiles were clinically meaningful. To translate the results into clinical interventions, we asked the two domain experts to (1) independently use their clinical judgement to design interventions to treat COVID-19 patients at each level of granularity, and (2) together arrive at a consensus.
Results
The analysis revealed statistically and clinically significant heterogeneity at all three levels of granularity. Below we present the quantitative, visual, and clinical interpretive results from each of the three levels.
1. Cohort-Level
Quantitative and Visual Results. The feature selection method identified 18 comorbidities that were significant after FDR correction for at least one of the three outcomes (13 were significant for all 3 outcomes, 1 was significant for 2 outcomes, and 4 were significant for one of the outcomes). All subsequent analyses were conducted on COVID-19 patients with these 18 comorbidities (n=789), compared to COVID-19 patients with none of these comorbidities (n=69).
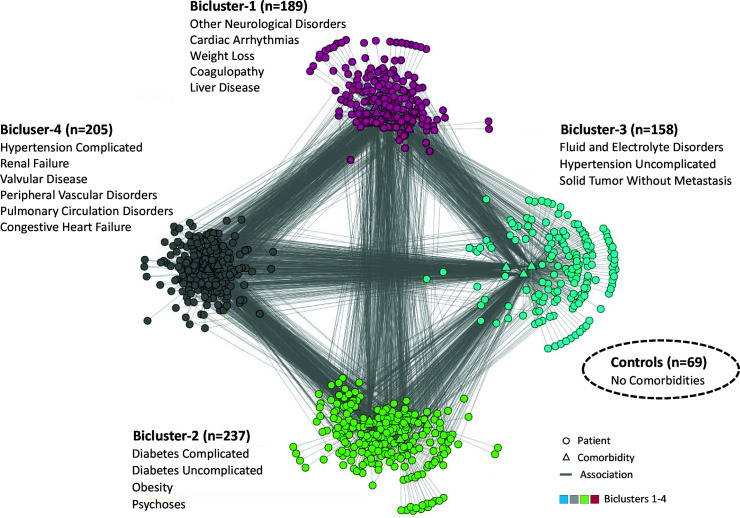
The modularity maximization algorithm identified 4 biclusters consisting of patient subgroups and their most frequently co-occurring comorbidities. The biclustering was statistically significant compared to 1000 random permutations of the data (COVID-19 Q=0.22, Random Median Q=0.19, P<.001), with clusters that were stable based on 1000 random bootstrap selections of the data (Median ARI=0.76, P<.001). As shown in Fig. 2, the visualization revealed biclusters consisting of different numbers of patients (ranging from 237-158) and different numbers of comorbidities (6-3). The comorbidities within each bicluster shown in Fig. 2 are ranked by their univariable significance. Not included in the above network analysis was the control group with no comorbidities, but shown in the lower right hand-side of the figure as a dotted oval.
Fig. 2.
Bipartite network visualization showing four biclusters each consisting of patient subgroups and their most frequently co-occurring comorbidities. Controls with no comorbidities are shown as a dotted oval.
Qualitative Results. The following was the consensus ranking of the four biclusters, with explanations and recommended treatments for each:
Bicluster-4 (hypertension complicated, renal failure, valvular disease, peripheral vascular disorders, pulmonary circulation disorders, and congestive heart failure). The domain experts stated that this bicluster had the highest risk for mortality. They concurred that hypertension often leads to renal failure and peripheral vascular disease. Furthermore, long-standing hypertension and pulmonary circulation disorders are a common cause for congestive heart failure. Given the high risk of cardiac involvement in such patients, they recommended that treatment for COVID-19 patients in this subgroup be focused on monitoring cardiac function through an echocardiogram and cardiac telemetry (requiring ICU use), in addition to monitoring renal function through judicious use of IV fluids.
Bicluster-1 (other neurological disorders, cardiac arrhythmias, weight loss, coagulopathy, liver disease). This bicluster was considered the next highest risk for mortality. They concurred that liver disease and coagulopathy tend to co-occur, often accompanied by weight loss. Furthermore, several studies have shown the strong association of liver disease to neurological disorders (e.g., encephalopathy), and to cardiac arrhythmias. Given the risk of excessive bleeding through coagulopathy, they recommended that treatment for COVID-19 patients in this subgroup be focused on monitoring coagulation results (e.g., platelet count, fibrinogen, and partial thromboplastin time), and treatment through replacement therapy (e.g., transfusion through blood plasma, and fibrinogen with tranexamic acid). Furthermore, given the risk for arrhythmias, they recommended monitoring cardiac function through cardiac telemetry (requiring ICU use).
Bicluster-2 (diabetes complicated, diabetes uncomplicated, obesity, psychoses). This bicluster was ranked third for risk of mortality. Initially, one domain expert ranked this bicluster second for risk of mortality due to the presence of obesity, but noted that Bicluster-1 was also of high risk. They concurred that because obesity is the main cause of diabetes, the co-occurrence with uncomplicated or complicated diabetes was expected. Furthermore, medications from psychosis are a known risk for high energy consumption, leading to obesity. Given the metabolic complications arising from diabetes, they recommended that treatment for COVID-19 patients in this subgroup be focused on monitoring glucose levels, which could be done at the inpatient floor, or at home.
Bicluster-3 (fluid and electrolyte disorders, hypertension uncomplicated, solid tumor without metastasis). This cluster was ranked by both as the lowest risk for mortality. The co-occurrence for these comorbidities in a subgroup was probably related to medications because hypertension medication and chemotherapy are both known to cause electrolyte imbalance. Given the low risk of dying, they recommended that treatment for this COVID-19 patient subgroup should be focused on monitoring fluid and electrolytes either on the inpatient floor, or at home if their laboratory test results were normal.
2. Subgroup-Level
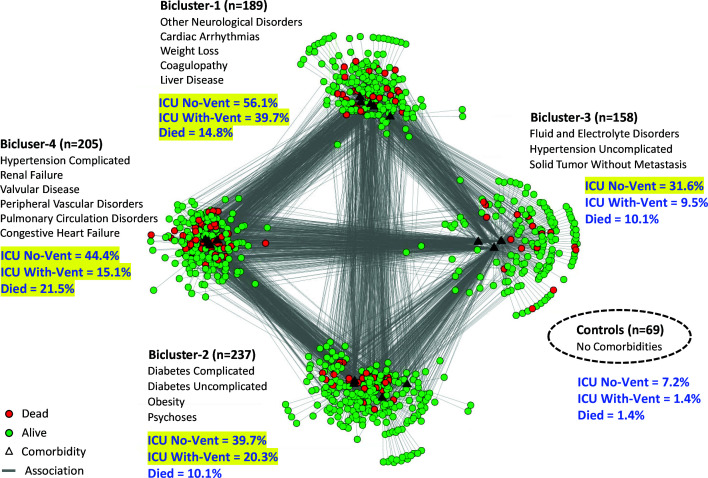
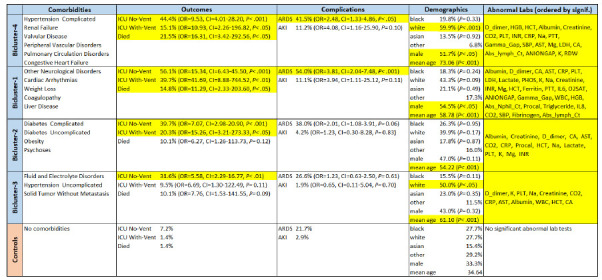
Quantitative and Visual Results. The analysis of risk at the subgroup-level entailed comparing the outcomes of the four patient subgroups in each of the biclusters, to the control group. As shown in Table 1, the results showed that Bicluster-4 and Bicluster-1 had significant ORs for all three adverse outcomes (ICU No-Vent, ICU With-Vent, and mortality), whereas Bicluster-2 had a significant OR for only ICU No-Vent and ICU With-Vent, and Bicluster-3 had a significant OR for only ICU No-Vent, compared to the controls. Furthermore, Bicluster-4 and Bicluster-1 had significantly high ORs for ARDS, but neither of the other two biclusters had significant risks for any complications, compared to the controls. Finally, Bicluster-4 and Bicluster-1 both had significant ORs for males, Bicluster-4 had significantly OR for white race, and Bicluster3 had a significantly higher OR for white race, compared to the controls. Fig. 3 shows the same network as in Fig. 2, but the patient nodes were colored based on their mortality status. Furthermore, the blue text shows the percentage of patients for each of the three outcomes, and which of them had significant ORs (highlighted in yellow) compared to the controls.
Table 1.
Subgroup-level analysis showing for each of the four biclusters, their outcomes, complications, demographics, and abnormal laboratory tests (only significant laboratory test results after multiple testing correction are shown; OR and CI values are not shown due to space constraints). Cells highlighted in yellow are significant compared to the control group after multiple testing correction.
|
|---|
Fig. 3.
Bipartite network visualization with the same 4 biclusters shown in Fig. 2, but showing patients that died (colored red) in each bicluster, in addition to percentages of all three outcomes (shown in blue text) and which of them were significant (highlighted in yellow) compared to the control group.
Qualitative Results. The domain experts used both the visualization in Fig. 2, and Table-1 to interpret the results, and to propose triage strategies for each of the biclusters. COVID-19 patients can arrive either at (1) the clinic, from where they can be triaged to home or to the ER, or (2) the emergency room (ER), from where they can be triaged to go to the ICU, inpatient floor, or home. Using this patient flow model, the domain experts recommended why and how they would triage patients from each bicluster if they arrived at the clinic, or at the ER.
Bicluster-4. The patients in this bicluster had a significant OR for dying from ARDS, were older (mean age 73.4), and had abnormal labs related to cardiac (systolic blood pressure), renal (creatinine), and pulmonary (CO2 and HGB) dysfunction. Given the comorbidities, it was not surprising that this bicluster had a high mean age, and a significantly higher percentage of whites and males known to have a high-risk for COVID-19 severity. The domain experts therefore recommended the following triage strategies: if such patients arrived at the clinic, they should be triaged to the ER, and if they arrived at the ER they should be triaged to the ICU.
Bicluster-1. Despite having differences in comorbidities, this bicluster had a similar risk profile for adverse outcomes compared to Bicluster-4. However, the difference in comorbidity profile (liver disease, weight loss, and coagulopathy) was reflected in significant ORs for albumin, gamma gap, fibrinogen, and platelets with significantly higher AST, INR, PTT compared to the controls. In addition, indirect evidence of cardiac arrythmias is demonstrated by significant ORs for Na, K, Phos, Mg, and Ca, compared to controls. This bicluster also had a higher proportion of males known to have a high risk for COVID-19 severity. Despite these differences, the domain experts recommended the same triage strategy as for Bicluster-4: patients arriving at the clinic should be triaged to the ER, and patients arriving at the ER should be triaged to the ICU.
Bicluster-2. Patients in this bicluster had a high risk for ventilator use and ICU, but not for ARDS or for dying. Furthermore, this bicluster had evidence of sequelae/complications of diabetes including renal insufficiency and electrolyte abnormalities (ketoacidosis) demonstrated in significant ORs for Cr, CO2, lactate, Na, K, and Mg, compared to the controls. Given the lower risk of dying, the domain experts recommended the following triage strategies: patients arriving at the clinic with respiratory difficulties, should be triaged to the ER, else sent home with instructions to monitor glucose levels; patients arriving at the ER, should be triaged to the inpatient floor and monitored closely for the need of a ventilator.
Bicluster-3. Although the patients in this bicluster had no significant risk for ARDS or for dying, they did have a high risk for ICU use. The electrolyte imbalances in their profile is reflected in the significant ORs for Na, K, and Ca, compared to controls. However, the domain experts believed that the use of ICU may not be warranted for such patients during resource-constrained situations, as it should be reserved for more critical patients. Therefore, they recommended that patients in this bicluster be sent home if the lab values were normal, with recommendations to monitor changes in electrolytes.
3. Patient-Level
Quantitative and Visual Results. As shown in Fig. 3, the network layout revealed that the biclusters appeared to have different internal topologies. While Bicluster 1-3 had many patients on their periphery that had only one comorbidity (shown by the single edge that connects them to cluster), Bicluster-4 had very few of such patients. Furthermore, patients on the inner part of each bicluster had many comorbidities outside their bicluster (shown by the many inter-bicluster edges). These differences in topologies strongly suggested that patients in each bicluster varied in their degree of similarity to each other, and therefore warranted examination at the patient-level of granularity.
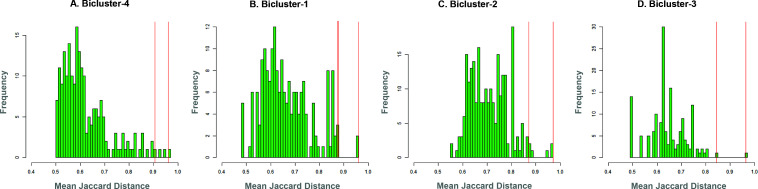
Fig. 4 shows the distributions of patient similarity in each bicluster. Each plot shows the distribution of the mean Jaccard Distance (JD) of each patient to the other patients in their bicluster (patients with smaller mean JD share more comorbidities with the rest of the patient in their bicluster, and are therefore are more similar). For example, Bicluster-4 (Fig. 4A) had a majority of the patients that were more similar to each other (median JD=0.59), compared to Bicluster-3 (Fig. 4C) which had a majority of the patients that were less similar (median JD=0.63) to each other in their comorbidity profiles (the pairs of biclusters had significantly different medians except for Bicluster-1 and Bicluster 3). Patients that were significantly more dissimilar compared to the rest of the patients in their bicluster would therefore tend to be in the right tail of the respective mean JD distribution, and were considered outliers to their respective biclusters. To examine a representative sample of such patient outliers in each bicluster, we selected all patients that had a significantly higher mean JD (patients to the right of the each distribution) compared to the rest of the patients, and from those selected for examination those that had the minimum (capturing patients in the outer periphery of their bicluster), and the maximum (capturing patients in the inner periphery of their bicluster) number of comorbidities. The vertical red lines in Fig. 4 A-D shows the resulting two patients within each of the four bicluster that were selected for examination by the domain experts.
Fig. 4.
Distributions of mean Jaccard Distance (JD) showing how the similarity among patients differed across the biclusters. The vertical red lines denote patients who were significantly dissimilar in their comorbidity profile compared to the rest in their bicluster, with the maximum and minimum comorbidities.
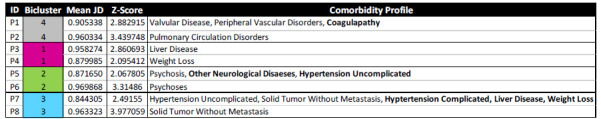
Qualitative Results. Table-2 shows the above eight outlier patients, the biclusters to which they belonged, and their comorbidity profiles. The domain experts examined these patients and their comorbidity profiles to recommend appropriate clinical interventions (to reduce the risk of reidentification, we were unable to examine and present the full profile of the patients including outcomes, complications, lab values, and demographics).
Table 2.
Eight outlier patients, two from each cluster that were significantly different from the rest in their bicluster, which were inspected by the domain expert to determine whether and how their treatments would be different from the bicluster to which they belonged. Colors denote biclusters shown in Fig. 2, and bolded comorbidities denote those that were outside the biclusters.
|
|---|
The results showed that a majority of the outlier patients selected for examination had comorbidities within their biclusters (P2, P3, P4, P6, P8), whereas a few (P1, P5, P7) had comorbidities outside their clusters (shown bolded in Table 1). However, despite having comorbidities within their bicluster, the generic treatment plans for the entire subgroup were often not appropriate. For example, while the comorbidities in Bicluster-1 overall suggest cardiac monitoring with pharmacologic control of the heart rate and correction of coagulopathy (with anticoagulation), its outliers P3 and P4 would require only replacement of clotting factors and nutritional support respectively. Similarly, while patients in Bicluster-2 overall would require glycemic control with insulin or other medications, the same treatment would cause harm to its outliers P5 and P6. Additionally, while Bicluster-3 as a whole suggests hypertension control using treatments such as diuretics, beta blockers, angiotensin converting enzyme inhibitors, its outlier P8 might be harmed with utilization of such anti-hypertensive medication. In contrast, the outliers P1 and P2 in Bicluster-4 would benefit from the same cardiac support therapy as the full bicluster. The analysis of heterogeneity at the patient-level of granularity therefore revealed that while subgroups provide efficiency in the design of treatment plans, outliers that need different treatments from their biclusters need to be flagged, underscoring the complexity of treating patients with multimorbidities.
Discussion
Several studies have shown that COVID-19 patients with prior MCCs, being older, male, and a minority are all high risk factors for having adverse outcomes. However, little is known about how prior comorbidities co-occur to form COVID-19 patient subgroups, their risks for adverse outcomes, and their implications for clinical interventions. This is particularly important because multimorbidities are common and well-studied in older adults and minorities. However, while these studies have analyzed how multimorbidities co-occur in different populations such as patients that have been readmitted to the hospital after a hip fracture,28 they have been done using a wide range of different methods, and at different levels of granularities.
Given the critical importance of designing interventions to reduce the risk of adverse outcomes in COVID-19 patients, here we explored a unified approach to (1) automate and therefore accelerate the quantitative analysis of heterogeneity at different levels of granularity, and (2) visualize the results using the same representation to increase interpretability of the results with the explicit goal of designing clinical interventions. We used the bipartite network representation because it explicitly represented both patients and comorbidities simultaneously using a computable graph representation consisting of nodes and edges, which enabled their quantitative and visual analysis at different levels of granularity. The application of this approach to COVID-19 EHR patient data led to the following two insights:
Explicit and Suggestive Clinical Insights at Each Level of Granularity. Analysis of heterogeneity at each level of granularity led to explicit clinical insights that were enabled by the respective analytical methods used. At the cohort-level, modularity maximization identified biclusters which provided insights on which comorbidities frequently co-occurred. This led to inferences about the disease mechanisms that potentially connected them (e.g., hypertension → pulmonary circulation disorders → congestive heart failure), enabling a focus on monitoring the organ or system (e.g., using telemetry to monitor the heart) that could precipitate an adverse outcome. Other applications could include the design of clinical trials. At the subgroup-level, comparative analysis between subgroups enabled ranking biclusters based on their risks for specific outcomes, resulting in the design of triage strategies critical during resource-constrained situations such as COVID-19 hotspots. Furthermore, this analysis also revealed how age and gender were stratified among the high-risk biclusters. Finally, at the patient-level, analysis of similarity helped to identify which patients were outliers to their biclusters, and whether they would require different treatments compared to their subgroups. Examining each level separately therefore helped to elucidate the contribution (frequency, risk, and similarity) each played in the design of clinical intervention for COVID-19 patients.
However, each level of granularity also provided suggestive insights for the next level. For example, at the cohort-level, the use of telemetry suggested that the patients needed to be triaged to the ICU, an insight which was more fully realized when analyzing subgroups at the next level of granularity. Similarly, analysis at the subgroup-level was suggestive that patients within each bicluster ranged in the degree to which they were similar to the rest in their bicluster, but more fully realized when analyzing individual patients at the next level of granularity. Such connections between levels could be the result of using a uniform bipartite representation to analyze heterogeneity across all levels of granularity, which needs to be further explored.
Extensions and Limitations. While using the multigranular heterogeneity framework, we realized that although the analysis at the cohort-level was within the cohort, the analysis at the subgroup and patient-levels were between subgroups and patients respectively. The framework could therefore be elaborated to include inter-and intra-level analysis. For example, at the cohort-level intra-cohort analysis would be the current modularity maximization to identify biclusters within the cohort, but inter-cohort analysis could include analyzing if the co-occurrence patterns of comorbidities in one dataset, replicate in another using methods such as the Rand Index.19 Furthermore, the framework could include other analytical approaches such as causal modeling and association rule mining for conducting intra-subgroup analysis. Additionally, the domain experts noted that as the comorbidities were defined by disease categories such as liver disease, the data lacked details about subtypes resulting in their inability to incorporate etiology, severity, and chronicity into their design of clinical interventions. However, despite this limitation, the current dataset was sufficient to enable a provider to triage and make initial treatment decisions quickly and efficiently. Finally, the outlier detection at the patient-level analysis currently does not take into consideration the shape and size of the distributions, and our current research is exploring other statistical methods to more precisely identify those outliers.
Conclusions and Future Research
Heterogeneity and granularity are well-known and critical concepts in biomedical research. Heterogeneity embraces the notion that patients are similar and different depending on the characteristics used to describe them. This notion has led to an understanding of phenomena such as phenotypes and symptom clusters, and is a corner stone of precision medicine. Granularity embraces the notion that patients can be analyzed at different levels of detail ranging from the molecular to the environmental. This notion has led to approaches such as molecular medicine, and is a corner stone of translational science. Here we attempted to merge both concepts to analyze the co-occurrence of comorbidities in COVID-19 patients, with the explicit goal of translating heterogeneity results from each level of granularity into clinical interventions. Such an analysis was possible through the use of bipartite networks as they provided a unified quantitative and visual representation, which enabled (1) automation for the quantitative analysis of heterogeneity at each level, and (2) the rapid interpretation of that heterogeneity by domain experts through the visualization, leading to the design of clinical interventions.
The results suggest that each level of granularity can provide distinct insights into the co-occurrence of comorbidities: (1) cohort-level analysis can be used to provide insights into the frequency of co-occurrence patterns enabling recommendations on which co-occurrences should be included in clinical practice guidelines to address multimorbidities; (2) subgroup-level analysis can be used to provide insights into the risk of each subgroup, enabling the design of triage strategies critical in resource-constrained situations such as COVID-19 hotspots; (3) patient-level analysis can be used to provide insights into the similarity of patients in each subgroup useful to determine which patients can use interventions designed for the subgroup, and which require individualized interventions. While ultimately each patient is an individual and requires personalized care, the goal of such analysis is to enable the design of evidence-based proactive strategies, which are adaptable to specific situations with the goal of improving the quality and efficiency of care.
A critical limitation of the current research is that we analyzed only one dataset, and our current and future research will test the replicability of these results in another COVID-19 dataset. Furthermore, we will explore the use of the multigranular heterogeneity framework to analyze other phenomena such as symptom clusters and clinical phenotypes with the explicit goal of translating the analytical results from each level of granularity, to the design of clinical interventions and their evaluation.
Acknowledgements
This research was supported in part by the UTMB Clinical and Translational Science Award (UL1 TR000071) from NCATS, and the UTMB Claude D. Pepper Older Americans Independence Center Award #P30-AG024832 from NIA, and the UTMB Cancer Center.
References
- 1.Yang J, Zheng Y, Gou X, et al. Prevalence of comorbidities in the novel Wuhan coronavirus (COVID-19) infection: a systematic review and meta-analysis. Int J Infect Dis. 2020 doi: 10.1016/j.ijid.2020.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Centers for Disease Control and Prevention. Interim Clinical Guidance for Management of Patients with Confirmed Coronavirus Disease (COVID-19) https://www.cdc.gov/coronavirus/2019-ncov/hcp/clinical-guidance-management-patients.html. Accessed April, 6, 2020. [Google Scholar]
- 3.Shi S, Qin M, Shen B, et al. Association of Cardiac Injury With Mortality in Hospitalized Patients With COVID-19 in Wuhan, China. JAMA cardiology. 2020 doi: 10.1001/jamacardio.2020.0950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y-C, Bai W-Z, Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol. 2020 doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zou X, Chen K, Zou J, Han P, Hao J, Han Z. Single-cell RNA-seq data analysis on the receptor ACE2 expression reveals the potential risk of different human organs vulnerable to 2019-nCoV infection. Front Med. 2020 doi: 10.1007/s11684-020-0754-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mao L, Wang M, Chen S, et al. Neurological Manifestations of Hospitalized Patients with COVID-19 in Wuhan, China: a retrospective case series study. medRxiv. 2020:2020.2002.2022.20026500 [Google Scholar]
- 7.McIntosh K, Hirsch M S, Bloom A. Coronavirus disease 2019 (COVID-19) 2020 https://www.uptodate.com/contents/coronavirus-disease-2019-covid-19, 2020. [Google Scholar]
- 8.World Health Organization. Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19) https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf. Accessed April, 6, 2020. [Google Scholar]
- 9.Griffith DM, Sharma G, Holliday CS, et al. Men and COVID-19: A Biopsychosocial Approach to Understanding Sex Differences in Mortality and Recommendations for Practice and Policy Interventions. Preventing chronic disease. 2020;17:E63. doi: 10.5888/pcd17.200247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hajat C, Stein E. The global burden of multiple chronic conditions: A narrative review. Prev Med Rep. 2018;12:284–293. doi: 10.1016/j.pmedr.2018.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.HHS. About the Multiple Chronic Conditions Initiative. 2020. https://www.hhs.gov/ash/about-ash/multiple-chronic-conditions/about-mcc/index.html#:~:text=MCC%20are%20concurrent%20chronic%20conditions,both%20have%20multiple%20chronic%20conditions. Accessed 8/14, 2020. [Google Scholar]
- 12.Gupta A, Madhavan MV, Sehgal K, et al. Extrapulmonary manifestations of COVID-19. Nat Med. 2020;26(7):1017–1032. doi: 10.1038/s41591-020-0968-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyd CM, Darer J, Boult C, Fried LP, Boult L, Wu AW. Clinical practice guidelines and quality of care for older patients with multiple comorbid diseases: implications for pay for performance. Jama. 2005;294(6):716–724. doi: 10.1001/jama.294.6.716. [DOI] [PubMed] [Google Scholar]
- 14.Boyd CM, Wolff JL, Giovannetti E, et al. Healthcare task difficulty among older adults with multimorbidity. Medical care. 2014;52 Suppl 3(0 3):S118–S125. doi: 10.1097/MLR.0b013e3182a977da. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tinetti ME, Bogardus ST, Jr., Agostini JV. Potential pitfalls of disease-specific guidelines for patients with multiple conditions. The New England journal of medicine. 2004;351(27):2870–2874. doi: 10.1056/NEJMsb042458. [DOI] [PubMed] [Google Scholar]
- 16.Muth C, Blom JW, Smith SM, et al. Evidence supporting the best clinical management of patients with multimorbidity and polypharmacy: a systematic guideline review and expert consensus. Journal of internal medicine. 2019;285(3):272–288. doi: 10.1111/joim.12842. [DOI] [PubMed] [Google Scholar]
- 17.Guthrie B, Payne K, Alderson P, McMurdo MET, Mercer SW. Adapting clinical guidelines to take account of multimorbidity. BMJ : British Medical Journal. 2012;345:e6341. doi: 10.1136/bmj.e6341. [DOI] [PubMed] [Google Scholar]
- 18.Lochner KA, Cox CS. Prevalence of multiple chronic conditions among Medicare beneficiaries, United States, 2010. Preventing chronic disease. 2013;10:E61. doi: 10.5888/pcd10.120137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. New York, NY, USA: Springer New York Inc.; 2001. [Google Scholar]
- 20.Abu-jamous B, Fa R, Nandi AK. Integrative Cluster Analysis in Bioinformatics. Chichester, West Sussex, United Kingdom: John Wiley & Sons, Ltd.; 2015. [Google Scholar]
- 21.Violán C, Roso-Llorach A, Foguet-Boreu Q, et al. Multimorbidity patterns with K-means nonhierarchical cluster analysis. BMC Family Practice. 2018;19(1):108. doi: 10.1186/s12875-018-0790-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Triest FJJ, Franssen FME, Reynaert N, et al. Disease-Specific Comorbidity Clusters in COPD and Accelerated Aging. Journal of clinical medicine. 2019;8(4) doi: 10.3390/jcm8040511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Padilha VA, Campello RJGB. A systematic comparative evaluation of biclustering techniques. BMC bioinformatics. 2017;18(1):55. doi: 10.1186/s12859-017-1487-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Newman MEJ. Networks: An Introduction. Oxford, United Kingdom: Oxford University Press; 2010. [Google Scholar]
- 25.Chauhan R, Ravi J, Datta P, et al. Reconstruction and topological features of the sigma factor regulatory network of Mycobacterium tuberculosis. doi: 10.1038/ncomms11062. In Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kamada T, Kawai S. An algorithm for drawing general undirected graphs. Information Processing Letters. 1989;31:7–15. [Google Scholar]
- 27.Bhavnani SK, Chen T, Ayyaswamy A, et al. Enabling Comprehension of Patient Subgroups and Characteristics in Large Bipartite Networks: Implications for Precision Medicine. Proceedings of AMIA Joint Summits on Translational Science. 2017:21–29. [PMC free article] [PubMed] [Google Scholar]
- 28.Bhavnani SK, Dang B, Penton R, et al. How High-Risk Comorbidities Co-Occur in Readmitted Patients With Hip Fracture: Big Data Visual Analytical Approach. JMIR Med Inform. 2020;8(10):e13567. doi: 10.2196/13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nickerson RS. Confirmation Bias: A Ubiquitous Phenomenon in Many Guises. Review of General Psychology. 1998;2(2):175–220. [Google Scholar]