Graphical abstract
Keywords: SARS-CoV-2, Coronaviruses, Proteases, Peptides, Antivirals
Abstract
Specific anti-coronaviral drugs complementing available vaccines are urgently needed to fight the COVID-19 pandemic. Given its high conservation across the betacoronavirus genus and dissimilarity to human proteases, the SARS-CoV-2 main protease (Mpro) is an attractive drug target. SARS-CoV-2 Mpro inhibitors have been developed at unprecedented speed, most of them being substrate-derived peptidomimetics with cysteine-modifying warheads. In this study, Mpro has proven resistant towards the identification of high-affinity short substrate-derived peptides and peptidomimetics without warheads. 20 cyclic and linear substrate analogues bearing natural and unnatural residues, which were predicted by computational modelling to bind with high affinity and designed to establish structure–activity relationships, displayed no inhibitory activity at concentrations as high as 100 μM. Only a long linear peptide covering residues P6 to P5′ displayed moderate inhibition (Ki = 57 µM). Our detailed findings will inform current and future drug discovery campaigns targeting Mpro.
With over 200 million reported cases and 4 million deaths,1 the ongoing COVID-19 pandemic is among the most devastating pandemics in human history.2 Specific antiviral drug candidates targeting SARS-CoV-2 are urgently needed to complement available vaccines and prepare for future coronavirus outbreaks.3 Inspired by the successful discovery of HIV and HCV protease inhibitors and their development into drugs,4 coronaviral proteases are currently among the most promising targets.5, 6, 7
The betacoronavirus RNA genome encodes two proteases, the papain-like protease (PLpro) and the main protease (Mpro or 3CLpro), which process the viral polyproteins pp1a and pp1ab into smaller non-structural proteins that assemble the replisome.8 Mpro is structurally conserved across SARS-CoV-1, MERS-CoV and SARS-CoV-2, which may allow the development of pan-coronaviral drugs.7, 9 The majority of polyprotein cleavage events are performed by Mpro, making it an attractive drug target. Mpro forms a homodimer and is a cysteine protease with distinct substrate specificity ranging from P4 to P1′ (using the nomenclature of Schechter and Berger),10 with a particularly strong preference for glutamine in P1.7, 11 No human host proteases with similar substrate recognition are known, rendering Mpro an ideal drug target with respect to off-target effects.5, 6, 7
Before the emergence of SARS-CoV-2, Mpro had already attracted attention as a potential drug target against the related coronaviruses SARS-CoV-1 and MERS-CoV,12 which emerged in 2002 and 2012, respectively. None of the small molecules and peptidomimetics reported to inhibit Mpro of SARS-CoV-1 and MERS-CoV were developed further into antiviral drugs.13, 14 Substrate-derived peptidomimetics relied on electrophilic reactive groups modifying the catalytic cysteine residue (commonly known as cysteine warheads) to achieve sufficient affinity. Inhibitors included Michael acceptors,15, 16, 17, 18, 19, 20 aldehydes,20, 21, 22, 23, 24 aldehyde prodrugs,25, 26 α-ketoamides,27 epoxides and aziridines,28, 29 and α-halomethyl ketones.30, 31
Since the emergence of SARS-CoV-2 in late 2019, several more Mpro inhibitors have been discovered at unprecedented speed. The same dependence on reactive warheads prevails for substrate-derived peptides and peptidomimetics. Warheads employed include Michael acceptors,32 aldehydes,33, 34 aldehyde prodrugs,35 α-ketoamides,36 vinylsulfones,11 azanitriles,37 and ketones.38 In April 2021, Pfizer revealed the orally available Mpro inhibitor PF-07321332, which is a short substrate analogue featuring a C-terminal nitrile warhead.39 Very recently, a cyclic peptide has been reported, which binds to SARS-CoV-2 Mpro without forming a covalent bond.40 With an IC50 value of about 160 µM, however, the activity of this compound is many orders of magnitudes below those of substrate-based analogues with warheads.
Reactive warheads pose the risk of pronounced off-target effects, potentially compromising the advancement of lead compounds into clinical drugs.41 Therefore, we set out to investigate the possibility of high-affinity short substrate-based SARS-CoV-2 Mpro inhibitors without warheads. We were particularly interested in exploring short cyclic substrate analogues with higher proteolytic stability than linear peptides.
Inspired by previous successes with generating nanomolar cyclic inhibitors of the Zika virus protease NS2B-NS3,42, 43, 44, 45 we designed various cyclic and linear analogues of the substrate amino acid sequence of SARS-CoV-2 Mpro (Table 1 ). We applied our in-house peptide-cyclization technique, which is based on the unique reactivity of 2-cyanopyridine and N-terminal cysteine or analogues.42, 43 The peptide sequences explored cover the entire substrate range from P6 to P5′ (most being short peptides comprising only 4–6 residues of the substrate recognition sequence), and included unnatural amino acids where those have been reported as suitable replacement for canonical amino acids in covalent Mpro inhibitors (Table 1).7, 11
Table 1.
Cyclic and linear Mpro substrate analogues assessed in this study.
 |
[a] Structural formulas of 1–21 are shown in Fig. S1.
[b] List of three letter codes of unnatural amino acids and stapling reagent: Abu: l-2-aminobutanoic acid; Cpa: l-2-amino-3-(2-cyanopyridin-4-yl)-propanoic acid; Cyl: l-2-amino-3-cyclopropylpropanoic acid; Dab: l-2,4-diaminobutanoic acid; DCP: 2,6-dicyanopyridine; Thz: l-thiazolidine-4-carboxylic acid; Tle: l-2-amino-3,3-dimethylbutanoic acid.
[c] Activity determined in FRET activity assays with 25 µM substrate and 25 nM enzyme for IC50 determination or with 10 µM, 20 µM, 35 µM and 50 µM substrate and 12.5 nM enzyme for Ki determination.
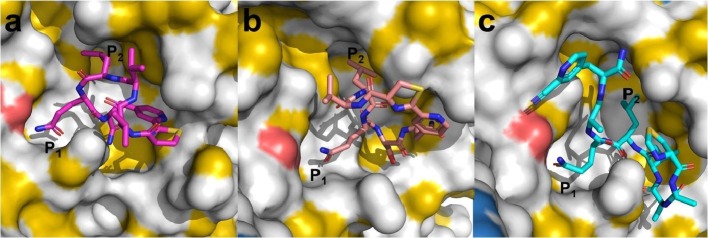
Initially, we chose two short cyclic peptides (1, 6) and one short linear peptide (20) for molecular modelling and docking experiments. All three peptides cover major recognition motifs of Mpro. Compound 1 features a non-prime site substrate recognition sequence (VVLQ, P4 – P1), while compound 6 also includes a prime site residue (VLQS, P3 – P1′). Peptide 20 has a P5 – P1′ recognition sequence and serves as a linear control, as the 2-cyanopyridine cannot react with the N-terminal thiazolidine. Molecular docking of 1, 6 and 20 with the dimer structure of SARS-CoV-2 Mpro (PDB: 6XQT) predicted promising binding orientations and interactions, especially with the S1 and S2 sub-cavity of the active site (Fig. 1 ), similar to previously co-crystallized peptidomimetics. The three compounds also showed almost similar predicted binding energies from computational docking experiments (Table 2 ) and were predicted to interact with critical active-site residues via hydrogen bonds (Fig. S3).46
Fig. 1.
Docking poses of compounds (a) 1, (b) 6, and (c) 20 against a dimeric X-ray crystal structure of SARS-CoV-2 Mpro (PDB: 6XQT). Protein shading was realized with the YRB highlighting script by Hagemans et al.55
Table 2.
Glide GScores and binding free energies of compounds 1, 6 and 20 docked with Mpro (PDB: 6XQT).
| Compound | Glide GScore [kcal/mol] | Binding free energy[a] [kcal/mol] |
|---|---|---|
| 1 | −7.373 | −60.47 |
| 6 | −6.695 | −58.85 |
| 20 | −10.094 | −58.50 |
Prime/MM-GBSA calculation performed in Maestro 2019–1, Schrödinger.
Encouraged by the computational predictions, we designed, synthesized, and purified 21 substrate-derived peptides (Table 1). Compounds 1–16 were prepared using standard Fmoc solid-phase peptide synthesis (SPPS) and the aforementioned side-chain-to-tail cyclization strategy.42 The method was successfully applied to more hydrophobic peptides and was compatible with sequences as long as 13 amino acids without major impact on the reaction yield. Compound 17 was synthesized following our in-house peptide stapling approach post Fmoc SPPS.43 All of these chemical transformations have proven to be biocompatible and deliver high-affinity ligands of viral proteases.42, 43 Peptides 18–21 were designed as linear analogues for comparison.
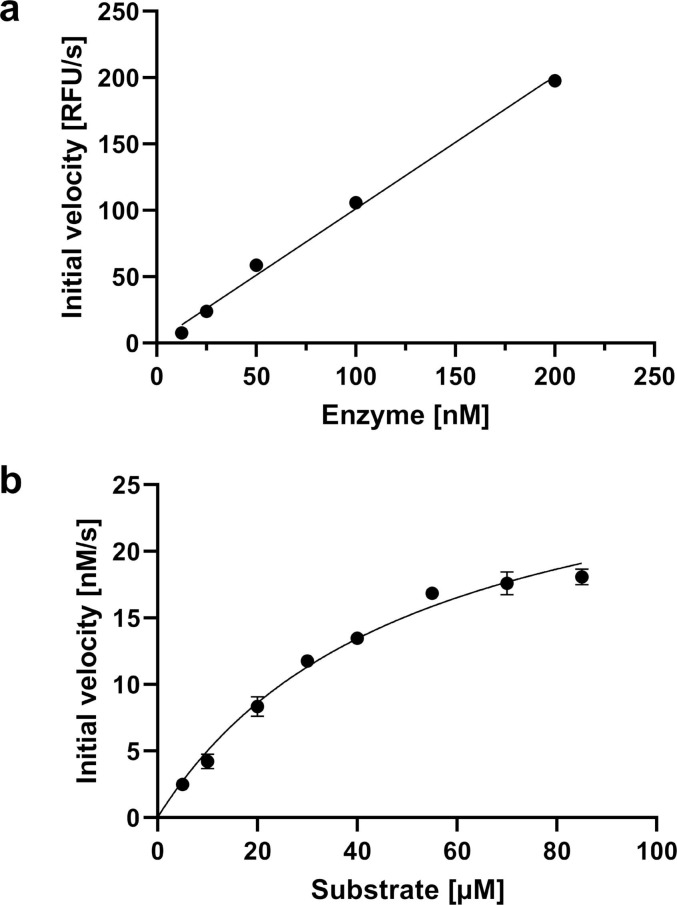
To assess Mpro inhibition by the substrate analogues 1–21, we employed an established Mpro inhibition assay using the FRET-based substrate DABCYL-KTSAVLQ↓SGFRKM-E(EDANS)–NH2 and recombinant SARS-CoV-2 Mpro.21, 36 Mpro dimer formation was confirmed by size exclusion chromatography and NMR spectroscopy (not shown). EDTA and DTT were added to the assay buffer to exclude any interferences from metal ions, oxidation, or cysteine modification. The use of reducing agent has proven to be particularly crucial to avoid nonspecific inhibition as, for example, observed for the covalent Mpro modifier ebselen.47 We also confirmed that the enzymatic activity depends linearly on the Mpro concentration (Fig. 2 a) and determined Michaelis Menten kinetics (Fig. 2 b), which yielded a k cat/K M ratio (23,500 M−1s−1) that is consistent with other studies.35, 36, 48
Fig. 2.
Activity assay of Mpro. (a) Linear dependence of Mpro activity (expressed by initial velocity) from Mpro concentration using 25 µM FRET substrate. (b) Michaelis-Menten kinetics using 25 nM Mpro. KM = 51 μM, kcat = 1.2 s−1.
In stark contrast to the promising computational results, compound 1 showed no inhibition of Mpro, even at the highest assayed concentration of 100 µM. We did not pursue tests at higher concentrations due to potential solubility problems, and because acceptable drug candidates are expected to show nanomolar or even picomolar affinities in biochemical assays.49 In order to investigate whether the lack of inhibition at 100 µM was a compound-specific result caused by peptide length or cyclization constraints, we explored additional cyclic and linear analogues (Table 1). Compounds 2–4 and 17–19 are short analogues of 1 encompassing four natural and unnatural amino acids from P4 to P1. Compound 5 was designed as a retro-peptide analogue of compound 1. We further expanded the substrate recognition sequence towards the non-prime site residue P5 and prime site residues P1′ and P2′ in compounds 6–11, 16 and 20–21. We also tested compounds 12–14, where the P1 glutamine was replaced by glutamate or asparagine. Compound 15 was inspired by the low-affinity inhibitor reported by Kreutzer et al.40 Remarkably, none of the peptides 1–20 displayed IC50 values below 100 µM in our well-validated Mpro activity assay (Table 1).
To further explore this finding, we performed NMR studies of uniformly 15N/2H-labeled Mpro with compounds 1 and 7. Neither compound induced any significant chemical shift perturbations in 15N-HSQC spectra at 100 µM, confirming that these cyclic substrate analogues do not bind to Mpro at concentrations relevant to drug design (Fig. S7).
Additionally, we carried out molecular dynamics simulations (MDS) in triplicate with 900 ns total simulation time for compound 1 docked to the SARS-CoV-2 Mpro dimer. One of the three simulations revealed large fluctuations in the root-mean-square deviation (RMSD) between 1 and the Mpro dimer during a short interval between 248 ns and 256 ns (Fig. S4), corresponding to 1 diffusing out of the active site briefly before returning to occupy it until the end of the simulation (Fig. S5). In the remaining two simulations, 1 remained bound within the active site for the duration, albeit undergoing conformational fluctuations, particularly in the orientation of the Gln (P1) and Leu (P2) sidechains of 1. Representative structures generated from the first cluster for all replicates revealed major differences in the positioning of these two sidechains compared to the molecular docking (Fig. S6). In contrast to the docked structure, in which the P1 Gln of 1 interacted with the S1 subpocket residues Leu141 and Glu166, the same sidechain does not interact to a similar degree with the S1 subpocket in the MDS. The position of the Leu sidechain in 1 (P2) also deviates from the pose observed in the docked structure. The results of this MDS study suggest that cyclic peptide 1 binds weakly and reversibly to the active site of Mpro, which may contribute to its poor in vitro activity.
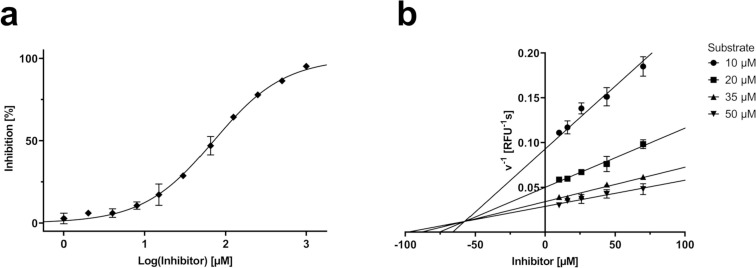
It is clear from this study that short peptides without a warhead cannot establish high affinity interactions to the Mpro dimer. Our study also suggests that cyclic substrate analogues are not a suitable alternative to address the insufficient affinity of linear peptides, a strategy that has previously been successful with other viral proteases.42, 43, 44 It is possible that our specific cyclization linkers are the cause of this observation; however, Kreutzer et al., who used an unrelated cyclization chemistry, equally failed to produce cyclic substrates with sufficient affinity.40 It is notable that peptide 16, which is a long (13 amino acids) cyclic analogue of the assay substrate, did not display inhibition at 100 µM. Only its linear analogue 21 showed moderate inhibition with an IC50 value of 71 ± 5 µM (Fig. 3 a). Compound 21, which is an acetylated analogue of the FRET substrate, is a competitive inhibitor with a K i value of 57 ± 10 µM as confirmed by a Dixon plot (Fig. 3 b). Its similarity to the natural Mpro substrate sequences and inhibition mode suggest that it acts as a competitive substrate. Thus, since peptide 16, which is a cyclic version of 21, did not show inhibition at 100 µM, it is reasonable to conclude that the macrocyclic peptide may be too constrained to bind the active site of the Mpro dimer in a high affinity conformation.
Fig. 3.
(a) Dose-response curve of Mpro FRET assay and compound 21. IC50 = 71 µM. (b) Mpro inhibition of compound 21 at multiple FRET substrate concentrations visualized in a Dixon plot. Ki = 57 µM.
Our results reveal major challenges associated with the discovery of short substrate-based Mpro inhibitors without electrophilic warheads. Substrate analogues of shorter lengths did not show any significant activity, while a longer linear analogue displayed moderate affinity. Previously successful strategies, including cyclization and the use of unnatural amino acids, did not help to overcome these challenges. The lack of affinity of cyclic substrate analogues described here and previously by Kreutzer et al.40 is particularly noteworthy as computational work in both studies predict binding to the active site with poses very similar to ligands observed in crystal structures. Given that our non-covalent inhibitors appear to be unusually ineffective against Mpro, it is not surprising that the first generation of SARS-CoV-2 Mpro inhibitors discovered at the beginning of the COVID-19 pandemic in 2020 were substrate derived covalent inhibitors bearing α-ketoamide, aldehyde, and Michael acceptor reactive groups.32, 33, 36 It should be noted that the substrate specificity of Mpro may be overruled by the electrophilicity of a warhead, as previously demonstrated.21 It should also be noted that the first generation of drugs targeting the HCV protease NS3-4A, such as telaprevir and boceprevir, required covalent warheads (α-ketoamides) as well,50, 51 while subsequent generations of drug candidates, such as faldaprevir or danoprevir, no longer require warheads.52, 53 It is thus not inconceivable that substrate-inspired inhibitors of Mpro without warheads may eventually become available, although present inhibitors still require warheads to boost affinity. Perhaps the larger diversity of peptide libraries available from phage or mRNA displays might help to overcome these challenges.54
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
C.N. thanks the Australian Research Council (ARC) for a Discovery Early Career Research Award (DE190100015) and Discovery Project funding (DP200100348). This study was supported by a RAMR (MAWA) grant awarded to S.U. and C.N. G.O. thanks the ARC for a Laureate Fellowship (FL170100019). C.J. and G.O. acknowledge support by the ARC Centre of Excellence for Innovations in Peptide & Protein Science (CE200100012).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bmcl.2021.128333.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fontanet A., Autran B., Lina B., Kieny M.P., Karim S.S.A., Sridhar D. SARS-CoV-2 variants and ending the COVID-19 pandemic. Lancet. 2021;397(10278):952–954. doi: 10.1016/s0140-6736(21)00370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asai A., Konno M., Ozaki M., et al. COVID-19 drug discovery using intensive approaches. Int J Mol Sci. 2020;21(8):2839. doi: 10.3390/ijms21082839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agbowuro A.A., Huston W.M., Gamble A.B., Tyndall J.D.A. Proteases and protease inhibitors in infectious diseases. Med Res Rev. 2018;38(4):1295–1331. doi: 10.1002/med.2018.38.issue-410.1002/med.21475. [DOI] [PubMed] [Google Scholar]
- 5.Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antiviral Res. 2013;100(1):286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hilgenfeld R. From SARS to MERS: crystallographic studies on coronaviral proteases enable antiviral drug design. FEBS J. 2014;281(18):4085–4096. doi: 10.1111/febs.2014.281.issue-1810.1111/febs.12936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg Med Chem Lett. 2020;30(17):127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thiel V., Ivanov K.A., Putics Á., et al. Mechanisms and enzymes involved in SARS coronavirus genome expression. J Gen Virol. 2003;84(9):2305–2315. doi: 10.1099/vir.0.19424-0. [DOI] [PubMed] [Google Scholar]
- 9.Stoermer M. Homology models of coronavirus 2019-nCoV 3CLpro protease. ChemRxiv. 2020 doi: 10.26434/chemrxiv.11637294.v3. [DOI] [Google Scholar]
- 10.Schechter I., Berger A. On the size of the active site in proteases. I. Papain. Biochem Biophys Res Commun. 1967;27(2):157–162. doi: 10.1016/s0006-291x(67)80055-x. [DOI] [PubMed] [Google Scholar]
- 11.Rut W., Groborz K., Zhang L., et al. SARS-CoV-2 Mpro inhibitors and activity-based probes for patient-sample imaging. Nat Chem Biol. 2021;17(2):222–228. doi: 10.1038/s41589-020-00689-z. [DOI] [PubMed] [Google Scholar]
- 12.Wu A., Peng Y., Huang B., et al. Genome composition and divergence of the novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe. 2020;27(3):325–328. doi: 10.1016/j.chom.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pillaiyar T., Manickam M., Namasivayam V., Hayashi Y., Jung S.-H. An overview of severe acute respiratory syndrome-coronavirus (SARS-CoV) 3CL protease inhibitors: peptidomimetics and small molecule chemotherapy. J Med Chem. 2016;59(14):6595–6628. doi: 10.1021/acs.jmedchem.5b0146110.1021/acs.jmedchem.5b01461.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liang R., Wang L., Zhang N., et al. Development of small-molecule MERS-CoV inhibitors. Viruses. 2018;10(12):721. doi: 10.3390/v10120721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang H., Xie W., Xue X., et al. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3(10):e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghosh A.K., Xi K., Grum-Tokars V., et al. Structure-based design, synthesis, and biological evaluation of peptidomimetic SARS-CoV 3CLpro inhibitors. Bioorg Med Chem Lett. 2007;17(21):5876–5880. doi: 10.1016/j.bmcl.2007.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghosh A.K., Xi K., Ratia K., et al. Design and synthesis of peptidomimetic severe acute respiratory syndrome chymotrypsin-like protease Inhibitors. J Med Chem. 2005;48(22):6767–6771. doi: 10.1021/jm050548m10.1021/jm050548m.s001. [DOI] [PubMed] [Google Scholar]
- 18.Ren Z., Yan L., Zhang N., et al. The newly emerged SARS-like coronavirus HCoV-EMC also has an 'Achilles’ heel': current effective inhibitor targeting a 3C-like protease. Protein Cell. 2013;4(4):248–250. doi: 10.1007/s13238-013-2841-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shie J.-J., Fang J.-M., Kuo T.-H., et al. Inhibition of the severe acute respiratory syndrome 3CL protease by peptidomimetic α, β-unsaturated esters. Bioorg Med Chem. 2005;13(17):5240–5252. doi: 10.1016/j.bmc.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang S., Chen S.-J., Hsu M.-F., et al. Synthesis, crystal structure, structure−activity relationships, and antiviral activity of a potent SARS coronavirus 3CL protease inhibitor. J Med Chem. 2006;49(16):4971–4980. doi: 10.1021/jm060392610.1021/jm0603926.s001. [DOI] [PubMed] [Google Scholar]
- 21.Zhu L., George S., Schmidt M.F., Al-Gharabli S.I., Rademann J., Hilgenfeld R. Peptide aldehyde inhibitors challenge the substrate specificity of the SARS-coronavirus main protease. Antiviral Res. 2011;92(2):204–212. doi: 10.1016/j.antiviral.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar V., Shin J.S., Shie J.-J., et al. Identification and evaluation of potent Middle East respiratory syndrome coronavirus (MERS-CoV) 3CLpro inhibitors. Antiviral Res. 2017;141:101–106. doi: 10.1016/j.antiviral.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akaji K., Konno H., Onozuka M., Makino A., Saito H., Nosaka K. Evaluation of peptide-aldehyde inhibitors using R188I mutant of SARS 3CL protease as a proteolysis-resistant mutant. Bioorg Med Chem. 2008;16(21):9400–9408. doi: 10.1016/j.bmc.2008.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akaji K., Konno H., Mitsui H., et al. Structure-based design, synthesis, and evaluation of peptide-mimetic SARS 3CL protease inhibitors. J Med Chem. 2011;54(23):7962–7973. doi: 10.1021/jm200870n. [DOI] [PubMed] [Google Scholar]
- 25.Kim Y., Liu H., Galasiti Kankanamalage A.C., et al. Reversal of the progression of fatal coronavirus infection in cats by a broad-spectrum coronavirus protease inhibitor. PLoS Pathog. 2016;12(3):e1005531. doi: 10.1371/journal.ppat.1005531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Galasiti Kankanamalage A.C., Kim Y., Damalanka V.C., et al. Structure-guided design of potent and permeable inhibitors of MERS coronavirus 3CL protease that utilize a piperidine moiety as a novel design element. Eur J Med Chem. 2018;150:334–346. doi: 10.1016/j.ejmech.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang L., Lin D., Kusov Y., et al. α-Ketoamides as broad-spectrum inhibitors of coronavirus and enterovirus replication: structure-based design, synthesis, and activity assessment. J Med Chem. 2020;63(9):4562–4578. doi: 10.1021/acs.jmedchem.9b0182810.1021/acs.jmedchem.9b01828.s00110.1021/acs.jmedchem.9b01828.s002. [DOI] [PubMed] [Google Scholar]
- 28.Martina E., Stiefl N., Degel B., et al. Screening of electrophilic compounds yields an aziridinyl peptide as new active-site directed SARS-CoV main protease inhibitor. Bioorg Med Chem Lett. 2005;15(24):5365–5369. doi: 10.1016/j.bmcl.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee T.-W., Cherney M.M., Liu J., et al. Crystal structures reveal an induced-fit binding of a substrate-like aza-peptide epoxide to SARS coronavirus main peptidase. J Mol Biol. 2007;366(3):916–932. doi: 10.1016/j.jmb.2006.11.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H.-Z., Zhang H., Kemnitzer W., et al. Design and synthesis of dipeptidyl glutaminyl fluoromethyl ketones as potent severe acute respiratory syndrome coronovirus (SARS-CoV) inhibitors. J Med Chem. 2006;49(3):1198–1201. doi: 10.1021/jm050767810.1021/jm0507678.s001. [DOI] [PubMed] [Google Scholar]
- 31.Sydnes M.O., Hayashi Y., Sharma V.K., et al. Synthesis of glutamic acid and glutamine peptides possessing a trifluoromethyl ketone group as SARS-CoV 3CL protease inhibitors. Tetrahedron. 2006;62(36):8601–8609. doi: 10.1016/j.tet.2006.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jin Z., Du X., Xu Y., et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582(7811):289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 33.Dai W., Zhang B., Jiang X.-M., et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science. 2020;368(6497):1331–1335. doi: 10.1126/science:abb4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qiao J., Li Y.-S., Zeng R., et al. SARS-CoV-2 Mpro inhibitors with antiviral activity in a transgenic mouse model. Science. 2021;371(6536):1374–1378. doi: 10.1126/science:abf1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma C., Sacco M.D., Hurst B., et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30(8):678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang L., Lin D., Sun X., et al. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368(6489):409–412. doi: 10.1126/science:abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Breidenbach J., Lemke C., Pillaiyar T., et al. Targeting the main protease of SARS-CoV-2: from the establishment of high throughput screening to the design of tailored inhibitors. Angew Chem Int Ed. 2021;60(18):10423–10429. doi: 10.1002/anie.v60.1810.1002/anie.202016961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Steuten K., Kim H., Widen J.C., et al. Challenges for targeting SARS-CoV-2 proteases as a therapeutic strategy for COVID-19. ACS Infect Dis. 2021;7(6):1457–1468. doi: 10.1021/acsinfecdis.0c0081510.1021/acsinfecdis.0c00815.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vandyck K., Deval J. Considerations for the discovery and development of 3-chymotrypsin-like cysteine protease inhibitors targeting SARS-CoV-2 infection. Curr Opin Virol. 2021;49:36–40. doi: 10.1016/j.coviro.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kreutzer A.G., Krumberger M., Diessner E.M., et al. A cyclic peptide inhibitor of the SARS-CoV-2 main protease. Eur J Med Chem. 2021;221:113530. doi: 10.1016/j.ejmech.2021.113530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flanagan M.E., Abramite J.A., Anderson D.P., et al. Chemical and computational methods for the characterization of covalent reactive groups for the prospective design of irreversible inhibitors. J Med Chem. 2014;57(23):10072–10079. doi: 10.1021/jm501412a. [DOI] [PubMed] [Google Scholar]
- 42.Nitsche C., Onagi H., Quek J.-P., Otting G., Luo D., Huber T. Biocompatible macrocyclization between cysteine and 2-cyanopyridine generates stable peptide inhibitors. Org Lett. 2019;21(12):4709–4712. doi: 10.1021/acs.orglett.9b0154510.1021/acs.orglett.9b01545.s001. [DOI] [PubMed] [Google Scholar]
- 43.Morewood R., Nitsche C. A biocompatible stapling reaction for in situ generation of constrained peptides. Chem Sci. 2021;12(2):669–674. doi: 10.1039/d0sc05125j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Braun N.J., Quek J.P., Huber S., et al. Structure-based macrocyclization of substrate analogue NS2B-NS3 protease inhibitors of Zika, West Nile and dengue viruses. ChemMedChem. 2020;15(15):1439–1452. doi: 10.1002/cmdc.v15.1510.1002/cmdc.202000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Patil N.A., Quek J.-P., Schroeder B., et al. 2-Cyanoisonicotinamide conjugation: a facile approach to generate potent peptide inhibitors of the Zika virus protease. ACS Med Chem Lett. 2021;12(5):732–737. doi: 10.1021/acsmedchemlett.0c0065710.1021/acsmedchemlett.0c00657.s001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li J., Abel R., Zhu K., Cao Y., Zhao S., Friesner R.A. The VSGB 2.0 model: a next generation energy model for high resolution protein structure modeling. Proteins. 2011;79(10):2794–2812. doi: 10.1002/prot.23106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ma C., Hu Y., Townsend J.A., et al. Ebselen, disulfiram, carmofur, PX-12, tideglusib, and shikonin are nonspecific promiscuous SARS-CoV-2 main protease inhibitors. ACS Pharmacol Transl Sci. 2020;3(6):1265–1277. doi: 10.1021/acsptsci.0c00130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuo C.-J., Chao T.-L., Kao H.-C., et al. Kinetic characterization and inhibitor screening for the proteases leading to identification of drugs against SARS-CoV-2. Antimicrob Agents Chemother. 2021;65(4) doi: 10.1128/AAC.02577-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hefti F.F. Requirements for a lead compound to become a clinical candidate. BMC Neurosci. 2008;9(3):S7. doi: 10.1186/1471-2202-9-s3-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Njoroge F.G., Chen K.X., Shih N.-Y., Piwinski J.J. Challenges in modern drug discovery: a case study of boceprevir, an HCV protease inhibitor for the treatment of hepatitis C virus infection. Acc Chem Res. 2008;41(1):50–59. doi: 10.1021/ar700109k. [DOI] [PubMed] [Google Scholar]
- 51.Kwong A.D., Kauffman R.S., Hurter P., Mueller P. Discovery and development of telaprevir: an NS3-4A protease inhibitor for treating genotype 1 chronic hepatitis C virus. Nat Biotechnol. 2011;29(11):993–1003. doi: 10.1038/nbt.2020. [DOI] [PubMed] [Google Scholar]
- 52.White P.W., Llinàs-Brunet M., Amad M., et al. Preclinical characterization of BI 201335, a C-terminal carboxylic acid inhibitor of the hepatitis C virus NS3-NS4A protease. Antimicrob Agents Chemother. 2010;54(11):4611–4618. doi: 10.1128/AAC.00787-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang Y., Andrews S.W., Condroski K.R., et al. Discovery of danoprevir (ITMN-191/R7227), a highly selective and potent inhibitor of hepatitis C virus (HCV) NS3/4A protease. J Med Chem. 2014;57(5):1753–1769. doi: 10.1021/jm400164c. [DOI] [PubMed] [Google Scholar]
- 54.Passioura T., Suga H. A RaPID way to discover nonstandard macrocyclic peptide modulators of drug targets. Chem Commun. 2017;53(12):1931–1940. doi: 10.1039/c6cc06951g. [DOI] [PubMed] [Google Scholar]
- 55.Hagemans D., van Belzen I.A.E.M., Morán Luengo T., Rüdiger S.G.D. A script to highlight hydrophobicity and charge on protein surfaces. Front Mol Biosci. 2015;2:56. doi: 10.3389/fmolb.2015.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.