Abstract
Chemical investigation of an organic extract of a fungus isolated from submerged wood collected from fresh water (strain G173), identified as a Talaromyces amestolkiae (Eurotiales; Trichocomaceae), led to the isolation of three coumarins, three dihydroisocoumarins, a dibenzo-α-pyrone, a meroterpenoid, and a merodrimane. Three of the isolated compounds, namely 7-chloropestalasin A (3), 4-hydroxyaspergillumarin (6), and ent-thailandolide B (9) were new. The structures were elucidated using a combination of spectroscopic and spectrometric techniques. The absolute configurations of 2, 3, 5, and 6 were established via a modified Mosher’s ester method, whereas for 9 a combination of TDDFT ECD and ORD calculations were employed. Compounds 1–9 were evaluated for antimicrobial activity against a group of bacteria and fungi.
Keywords: Freshwater Fungi, Coumarins, Dihydroisocoumarins, Dibenzo-α-pyrones, Meroterpenoids, Merodrimanes
As part of an ongoing project to uncover new chemistry from nature,1–5 our group has been investigating freshwater fungi.6–11 Lignicolous freshwater fungi represent a viable resource for discovering new secondary metabolites with a broad range of biological activities.12–14
A fungal strain accessioned as G173 and identified as Talaromyces amestolkiae (Eurotiales; Trichocomaceae) was isolated from submerged wood in a small pond near Bur-Mil Park, Guilford County, North Carolina. From an ecological point of view, strain G173 is not a true indweller of freshwater but can be defined as an immigrant species.15, 16 Fractionation of the organic extract of G173 using flash chromatography, followed by preparative RP-HPLC, resulted in the isolation of three coumarins (1–3), three dihydroisocoumarins (4–6), a dibenzo-α-pyrone (7), a meroterpenoid (8), and a merodrimane (9), with >97% purity according to UPLC-PDA (Fig. S1).
Compounds 1 (12.2 mg) and 2 (1.0 mg) were isolated as colorless amorphous solids with molecular formulas of C12H12O5 and C14H16O5, respectively, as determined by HRESIMS. The NMR (Fig. S2) and HRMS data identified 1 as the known compound, 3-hydroxymethyl-6,8-dimethoxycoumarin (Fig. 1), which was previously isolated from the soil fungus Talaromyces flavus.17 In addition, 2 was identified as pestalasin A (Fig. S3), a coumarin that was reported from the endophytic fungus Pestalotiopsis sp., which was isolated from the leaves of the Chinese mangrove Rhizophora mucronata.18 The absolute configuration of 2 was not previously reported; therefore it was assigned via a modified Mosher’s ester method,19 establishing the configuration as 2′S (Fig. 2 and S4).
Fig. 1.
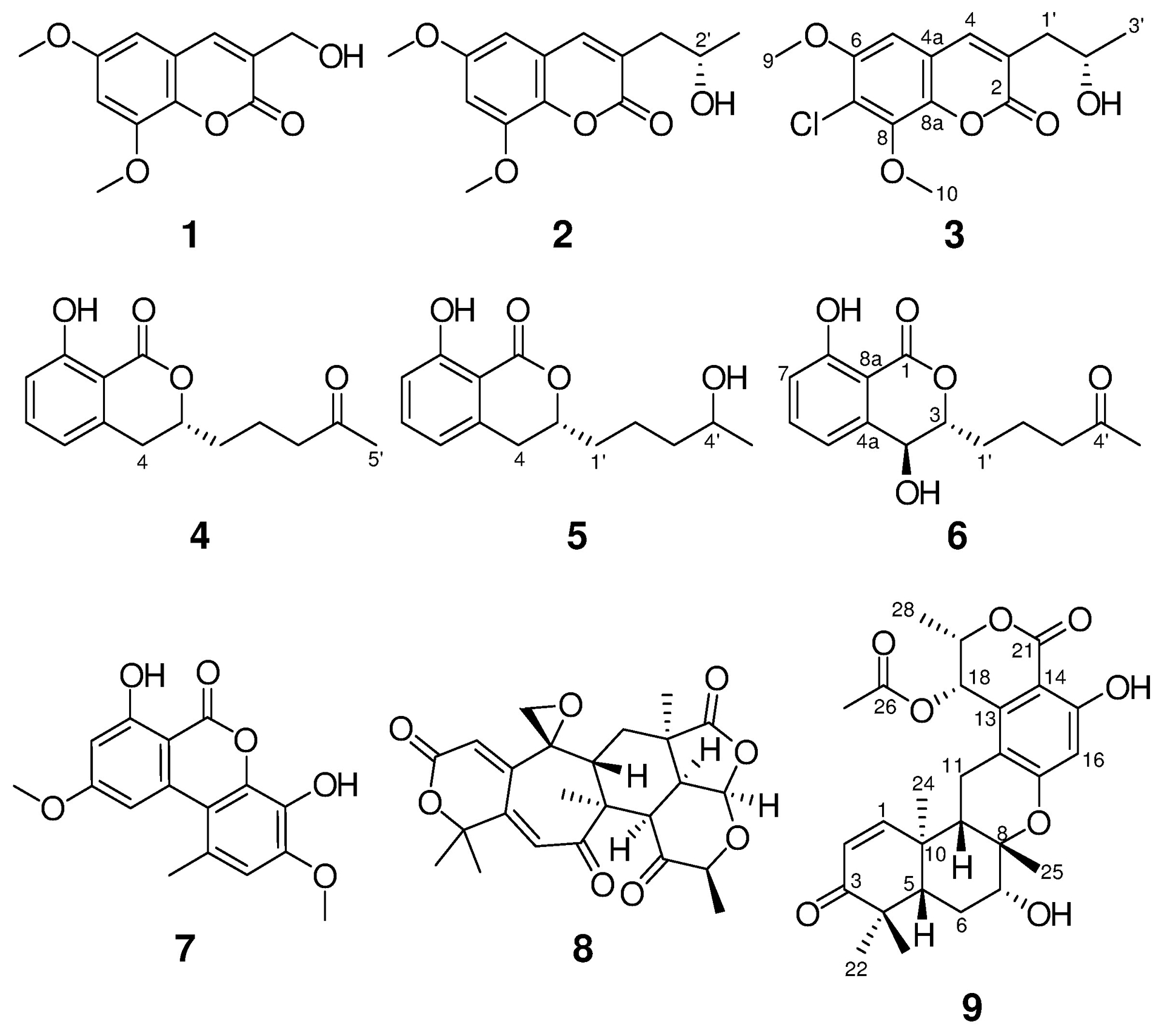
Structures of compounds 1-9.
Fig. 2.
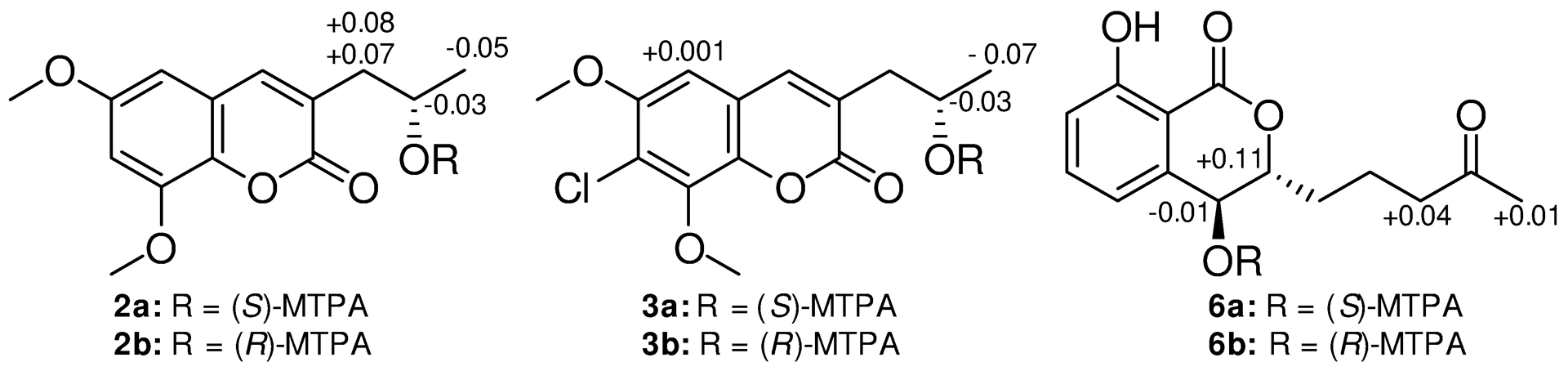
ΔδH values [Δδ (in ppm) = δS − δR] obtained for (S)- and (R)-MTPA esters 2a and 2b for pestalasin A (2), 3a and 3b for 7-chloro-pestalasin A (3), and 6a and 6b for 4-hydroxy-aspergillumarin A (6), in pyridine-d5.
Compound 3 (0.5 mg) was obtained as a white solid.20 The molecular formula was determined as C14H15ClO5 by HRESIMS and analysis of 1H, HMBC, and edited-HSQC NMR data (Table 1 and Fig. S5–S7). The HRMS and NMR data indicated 3 as a chlorinated analogue of 2, which was supported by both the presence of the characteristic isotopic pattern of chlorine in the HRMS data of 3, and the replacement of the meta-coupled aromatic protons (δH 6.45 and 6.65 for H-5 and H-6, respectively, JH-5/H-7 = 2.65 Hz) in 2 (Fig. S3), by a singlet aromatic proton (δH 6.70 for H-5) in 3 (Fig. S5). Analyses of the 2D NMR data (Fig. 3) gave the structure of 3, which was ascribed the trivial name 7-chloropestalasin A. The absolute configuration of 3 was assigned via a modified Mosher’s ester method,19 establishing the configuration as 2′S (Fig. 2 and S8).
Table 1.
1H and 13C NMR data of 3 (400 MHz for 1H; 100 MHz for 13C, CDCl3) and 6 (700 MHz for 1H; 175 MHz for 13C, CDCl3)
| Position | 3 | 6 | ||
|---|---|---|---|---|
| δ C * | δH mult (J in Hz) | δ C | δH mult (J in Hz) | |
| 1 | 168.8 | |||
| 2 | 161.4 | |||
| 3 | 128.4 | 83.3 | 4.43, ddd (8.6, 3.4, 2.9) | |
| 4 | 137.6 | 7.95, s | 67.4 | 4.78, dd (8.6, 2.3) |
| 4a | 109.6 | 141.9 | ||
| 5 | 100.0 | 6.70, s | 116.1 | 7.07, d (7.5) |
| 6 | 151.8 | 137.1 | 7.53, dd (8.0, 7.5) | |
| 7 | 118.1 | 117.8 | 6.98, d (8.0) | |
| 8 | 146.1 | 162.2 | ||
| 8a | 137.9 | 106.8 | ||
| 9 | 57.4 | 3.92, s | ||
| 10 | 56.9 | 3.95, s | ||
| 1’ | 41.1 | 2.64, dd (13.7, 8.2) | 30.7 | 1.76, m |
| 2.83, dd (13.7, 3.7) | 1.92, m | |||
| 2’ | 66.7 | 4.16, m | 18.4 | 1.75, m |
| 1.90, m | ||||
| 3’ | 23.7 | 1.28, d (6.4) | 42.9 | 2.56, ddd (9.2, 6.3, 2.9) |
| 4’ | 209.2 | |||
| 5’ | 30.3 | 2.16, s | ||
| 4-OH | 2.76, br. s. | |||
| 8-OH | 10.91, s | |||
13C NMR data for 3 were obtained from HMBC and edited-HSQC spectra.
Fig. 3.
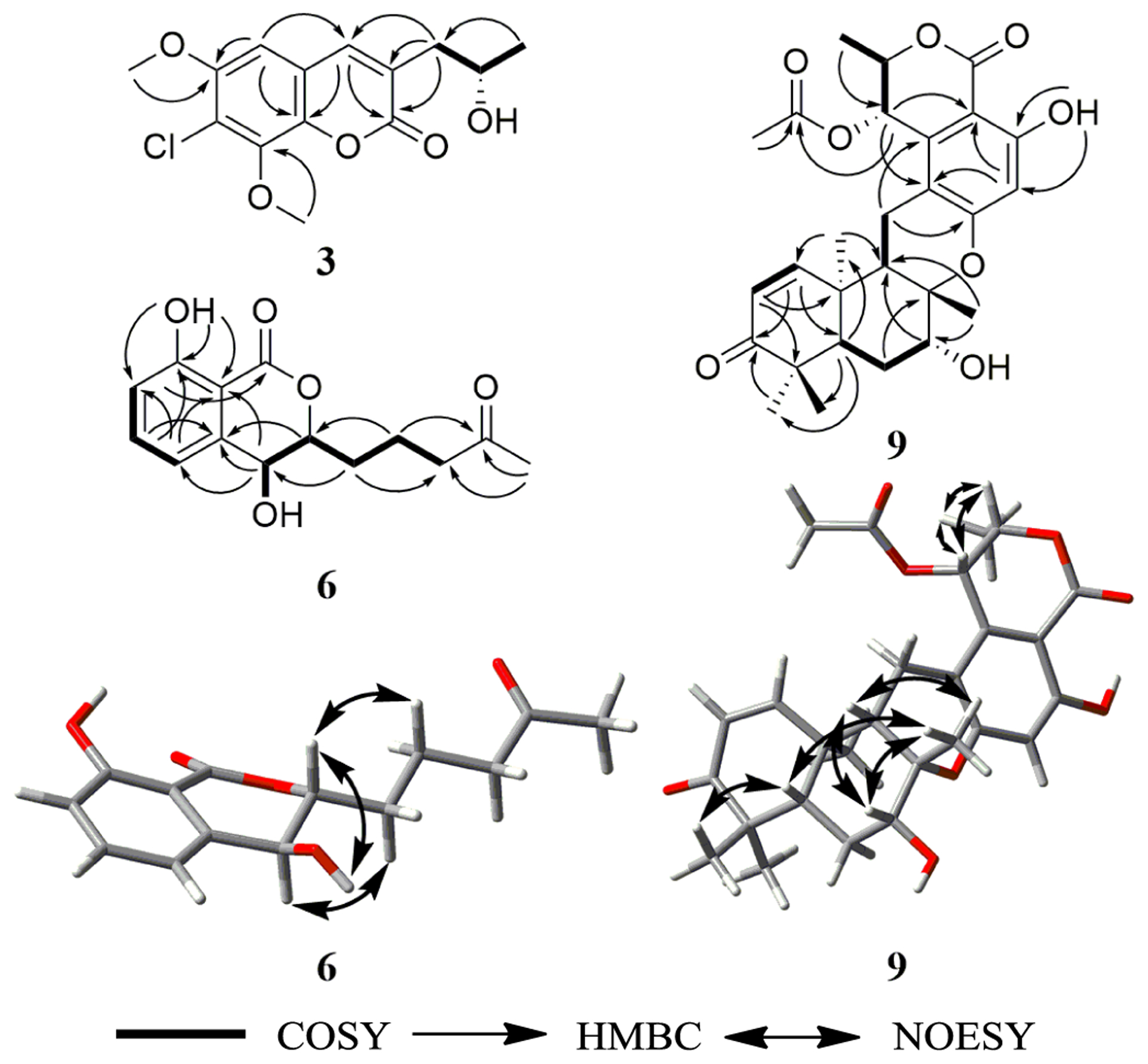
Key HMBC, COSY, and NOESY correlations of 3, 6 and 9.
Compounds 4 (10.5 mg; colorless oil) and 5 (2.0 mg; colorless crystal) were isolated with molecular formulas of C14H16O4 and C14H18O4, respectively, as determined by HRESIMS. The NMR (Fig. S9 and S10) and HRMS data identified 4 and 5 as the known dihydroisocoumarins, aspergillumarins A and B, respectively (Fig. 1), which were previously reported from the culture broth of a marine-derived fungus Aspergillus sp. isolated from the fresh leaf of the mangrove tree Bruguiera gymnorrhiza collected from the South China Sea.21 The NMR data of 5 matched those reported by Li and co-workers, except for the chemical shift of the 5′-methyl group (δH 2.34, d, J = 6 Hz),21 which was observed at δH 1.22, d, J = 6 Hz (Fig. S10). The absolute configuration of 5 at C-4′ was not determined by Li and co-workers.21 Therefore, we attempted to assign the absolute configuration via a modified Mosher’s ester method;19 however, these results indicated that 5 was a racemic mixture. Indeed, four products were observed, a major and a minor product from each reaction in a 3:1 ratio (Fig. S11).
Compound 6 (0.6 mg) was obtained as a white solid,22 with a molecular formula of C14H16O5 as determined by HRESIMS along with 1H, 13C, and edited-HSQC NMR data (Table 1, Fig. S12 and S13), establishing an index of hydrogen deficiency of 7. The NMR data suggested 6 as a dihydroisocoumarin analogue of 4. A key difference was replacement of the allylic methylene moiety (δH/δC 2.93/34.1, m, for H2-4/C-4) in 4 by an oxymethine in 6 (δH/δC 4.78/67.4, dd, J = 8.6, 2.3 for H-4/C-4). These data, along with a 16 amu difference in the HRMS data between 4 and 6, indicated hydroxylation at the C-4 position in 6. The coupling constant (JH-4/H-3 = 8.6 Hz) implied a pseudoaxial/pseudoequatorial trans orientation in 6 (Table 1, Fig. S12). A NOESY correlation observed between 4-OH and H-3 indicated that these two protons were on the same face (Fig. 3 and S15). Analyses of the COSY and HMBC NMR data (Fig. 3 and S14), established the structure of 6, which was given the trivial name 4-hydroxyaspergillumarin A. The absolute configuration of 6 was assigned via a modified Mosher’s ester method19 as 4S (Fig. 2 and S16).
Compounds 7 (5.8 mg) and 8 (6.3 mg) were isolated as colorless crystalline solids and identified using HRMS and NMR data as graphislactone A (a dibenzo-α-pyrone)23 and berkeleyacetal C (a meroterpenoid)24 (Fig. S17 and S18), respectively. Graphislactone A was first isolated from the lichen Graphis scripta var. pulverulenta,25 while berkeleyacetal C was isolated from extracts of a Penicillium sp.24
Compound 9 (2.9 mg) was obtained as a white solid,26 with a molecular formula of C27H32O8 as determined by HRESIMS and NMR data (Table S3 and Fig. 3 and S19–S22), establishing an index of hydrogen deficiency of 12. The HRMS and NMR data of 9, including the NOESY spectrum, were identical to that of thailandolide B, a merodrimane isolated from Talaromyces thailandiasis.27 However, the specific rotation of 9 ([α]D20 −47, CHCl3, c 0.05) was found to be opposite to that of thailandolide B ([α]D24 +134, CHCl3, c 0.1), suggesting that 9 could be an enantiomer of thailandolide B.27 Thus, the absolute configuration of 9 was determined using electronic circular dichroism (ECD) and optical rotatory dispersion (ORD) spectroscopy combined with time-dependent density functional theory (TDDFT) and quantum chemical calculations. The calculated TDDFT-ECD spectrum of 9 matched the measured data, displaying two positive (~230 and ~310 nm) and two negative (~270 and ~350 nm) Cotton effects, respectively (Fig. 4). The calculated spectra for thailandolide B was, as expected, opposite to 9 (Fig. 4). Unfortunately, no experimental data were published for thailandolide B for comparison purposes. However, the calculated ORD value for 9 ([α]D20 −88.5 in CHCl3) agreed with the experimental data. Thus, the absolute configuration of 9 was established as 5S,7R,8S,9S,10R,18S,19S and given the trivial name ent-thailandolide B.
Fig. 4.
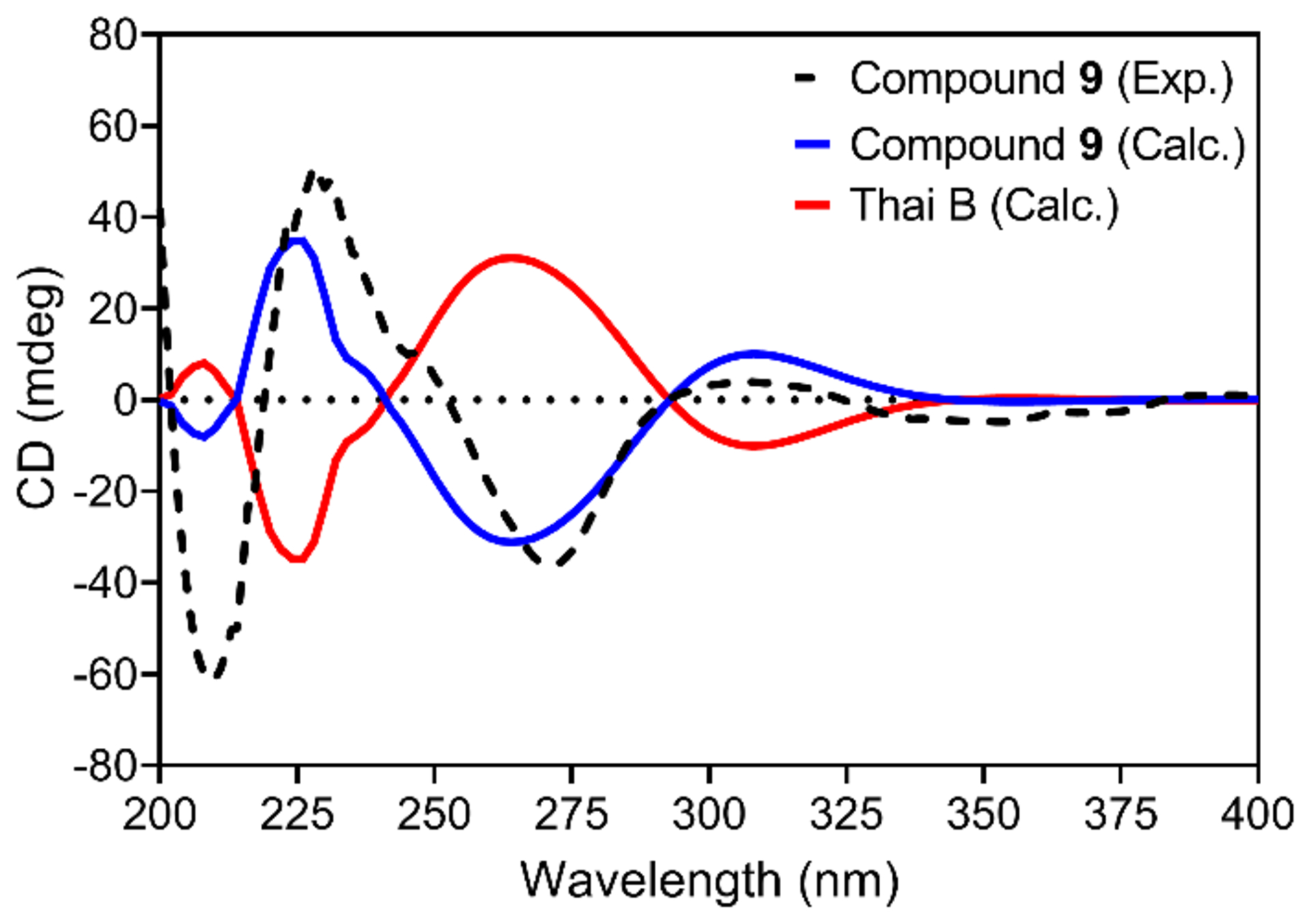
Comparison of experimental and calculated ECD spectra of 9 and thailandolide B in MeOH.
Compounds 1–9 were tested for antimicrobial activity against a group of bacteria and fungi28 and found to be inactive.
Supplementary Material
Acknowledgments
This research was supported in part by the National Institutes of Health/National Cancer Institute via P01 CA125066. TE acknowledges the partial financial support by Deanship of Research, Jordan University of Science and Technology, Irbid, Jordan (Grant No. 38/2018). MF acknowledges partial financial support via grants from UNAM-DGAPA IN222220 and FQ-PAIP 5000-9145. High-resolution mass spectrometry data were acquired at the Triad Mass Spectrometry Laboratory at the University of North Carolina at Greensboro. This work was performed in part at the Joint School of Nanoscience and Nanoengineering, a member of the Southeastern Nanotechnology Infrastructure Corridor (SENIC) and National Nanotechnology Coordinated Infrastructure (NNCI), which is supported by the National Science Foundation (Grant ECCS-1542174).
References and notes
- 1.Al Subeh ZY; Raja HA; Monro S; Flores-Bocanegra L; El-Elimat T; Pearce CJ; McFarland SA; Oberlies NH J. Nat. Prod 2020, 83, 2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.El-Elimat T; Alhawarri MB; Rivera-Chávez J; Burdette JE; Czarnecki A; Al-Gharaibeh M; Al Sharie AH; Alhusban A; Alali FQ; Oberlies NH Fitoterapia 2020, 146, 104706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El-Elimat T; Figueroa M; Raja HA; Adcock AF; Kroll DJ; Swanson SM; Wani MC; Pearce CJ; Oberlies NH Tetrahedron Lett 2013, 54, 4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Elimat T; Raja HA; Ayers S; Kurina SJ; Burdette JE; Mattes Z; Sabatelle R; Bacon JW; Colby AH; Grinstaff MW; Pearce CJ; Oberlies NH Org. Lett 2019, 21, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.El-Elimat T; Raja HA; Figueroa M; Swanson SM; Falkinham Iii JO; Lucas DM; Grever MR; Wani MC; Pearce CJ; Oberlies NH J. Antibiot 2015, 68, 191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Elimat T; Raja HA; Day CS; Chen W-L; Swanson SM; Oberlies NH J. Nat. Prod 2014, 77, 2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.El-Elimat T; Raja HA; Day CS; McFeeters H; McFeeters RL; Oberlies NH Bioorg. Med. Chem 2017, 25, 795. [DOI] [PubMed] [Google Scholar]
- 8.El-Elimat T; Raja HA; Figueroa M; Falkinham JO; Oberlies NH Phytochemistry 2014, 104, 114. [DOI] [PubMed] [Google Scholar]
- 9.Paguigan ND; Raja HA; Day CS; Oberlies NH Phytochemistry 2016, 126, 59. [DOI] [PubMed] [Google Scholar]
- 10.Paguigan ND; Rivera-Chávez J; Stempin JJ; Augustinović M; Noras AI; Raja HA; Todd DA; Triplett KD; Day C; Figueroa M; Hall PR; Cech NB; Oberlies NH J. Nat. Prod 2019, 82, 550. [DOI] [PubMed] [Google Scholar]
- 11.Rivera-Chávez J; El-Elimat T; Gallagher JM; Graf TN; Fournier J; Panigrahi GK; Deep G; Bunch RL; Raja HA; Oberlies NH Planta Med 2019, 85, 62. [DOI] [PubMed] [Google Scholar]
- 12.Gloer J In Applications of Fungal Ecology in the Search for New Bioactive Natural Products; Springer, Berlin Heidelberg New York, 1997; Vol. IV. pp 249. [Google Scholar]
- 13.Gloer JB In Applications of Fungal Ecology in the Search for New Bioactive Natural Products; Springer, Berlin, Heidelberg, 2007; Vol. 4. [Google Scholar]
- 14.Hernández-Carlos B; Gamboa-Angulo MM Phytochem. Rev 2011, 10, 261. [Google Scholar]
- 15.Park D Transactions of the British Mycological Society 1972, 58, 291. [Google Scholar]
- 16.El-Elimat T; Raja HA; Figueroa M; Sharie AHA; Bunch RL; Oberlies NH J. Nat. Prod 2021, 84, 898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ayer WA; Racok JS Can. J. Chem 1990, 68, 2085. [Google Scholar]
- 18.Xu J; Kjer J; Sendker J; Wray V; Guan H; Edrada R; Müller WEG; Bayer M; Lin W; Wu J; Proksch P Bioorg. Med. Chem 2009, 17, 7362. [DOI] [PubMed] [Google Scholar]
- 19.Hoye TR; Jeffrey CS; Shao F Nat. Protoc 2007, 2, 2451. [DOI] [PubMed] [Google Scholar]
- 20. 7-Chloropestalasin A (3): white solid; [α]D20 = +23 (c = 0.05, Chloroform); 1H NMR (CDCl3, 400 MHz); see Table S1 and Fig. S5–S7; HRESIMS m/z 299.0670 [M+H]+ (calcd for C14H16ClO5 299.0681); UV (MeOH) λmax (log ε) 295 (3.02), 258 (2.97), 214 (3.14) nm.
- 21.Li S; Wei M; Chen G; Lin Y Chem. Nat. Compd 2012, 48, 371. [Google Scholar]
- 22. 4-Hydroxyaspergillumarin A (6): white solid; [α]D20 = +1 (c = 0.04, Chloroform); 1H NMR (CDCl3, 700 MHz) and 13C NMR (CDCl3, 175 MHz); see Table S2 and Fig. S12–S15; HRESIMS m/z 265.1069 [M+H]+ (calcd for C14H17O5 265.1071); UV (MeOH) λmax (log ε) 315 (3.16), 245 (3.18), 220 (3.10) nm.
- 23.Altemöller M; Gehring T; Cudaj J; Podlech J; Goesmann H; Feldmann C; Rothenberger A Eur. J. Org. Chem 2009, 2009, 2130. [Google Scholar]
- 24.Stierle DB; Stierle AA; Patacini BJ Nat. Prod 2007, 70, 1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanahashi T; Kuroishi M; Kuwahara A; Nagakura N; Hamada N Chem. Pharm. Bull 1997, 45, 1183. [Google Scholar]
- 26. ent-thailandolide B (9): white solid; [α]D20 = −47 (c = 0.05, Chloroform); 1H NMR (CDCl3, 500 MHz) and 13C NMR (CDCl3, 125 MHz); see Table S3 and Fig. S19–S22; HRESIMS m/z 485.2153 [M+H]+ (calcd for C27H33O8 485.2170); UV (MeOH) lmax (log e) 314 (3.42), 271 (3.57), 237 (3.61) nm.
- 27.Dethoup T; Manoch L; Kijjoa A; Pinto M; Gales L; Damas AM; Silva AMS; Eaton G; Herz WJ Nat. Prod 2007, 70, 1200. [DOI] [PubMed] [Google Scholar]
- 28.Alali F; El-Elimat T; Albataineh H; Al-Balas Q; Al-Gharaibeh M; Falkinham JO; Chen W-L; Swanson SM; Oberlies NH J. Nat. Prod 2015, 78, 1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


