Abstract
Artemisia annua and its phytocompounds have a rich history in the research and treatment of malaria, rheumatoid arthritis, systemic lupus erythematosus, and other diseases. Currently, the World Health Organization recommends artemisinin-based combination therapy as the first-line treatment for multi-drug-resistant malaria. Due to the various research articles on the use of antimalarial drugs to treat coronaviruses, a question is raised: would A. annua and its compounds provide anti-severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) properties? PubMed/MEDLINE, Scopus, and Google Scholar were searched for peer-reviewed articles that investigated the antiviral effects and mechanisms of A. annua and its phytochemicals against SARS-CoVs. Particularly, articles that evidenced the herb’s role in inhibiting the coronavirus-host proteins were favored. Nineteen studies were retrieved. From these, fourteen in silico molecular docking studies demonstrated potential inhibitory properties of artemisinins against coronavirus-host proteins including 3CLPRO, S protein, N protein, E protein, cathepsin-L, helicase protein, nonstructural protein 3 (nsp3), nsp10, nsp14, nsp15, and glucose-regulated protein 78 receptor. Collectively, A. annua constituents may impede the SARS-CoV-2 attachment, membrane fusion, internalization into the host cells, and hinder the viral replication and transcription process. This is the first comprehensive overview of the application of compounds from A. annua against SARS-CoV-2/coronavirus disease 2019 (COVID-19) describing all target proteins. A. annua’s biological properties, the signaling pathways implicated in the COVID-19, and the advantages and disadvantages for repurposing A. annua compounds are discussed. The combination of A. annua’s biological properties, action on different signaling pathways and target proteins, and a multi-drug combined-therapy approach may synergistically inhibit SARS-CoV-2 and assist in the COVID-19 treatment. Also, A. annua may modulate the host immune response to better fight the infection.
Keywords: COVID-19, SARS-CoV-2, Artemisia annua, Artemisinin, Main protease, Spike protein
1. Introduction
Artemisia annua has more than 2000 years of history in the treatment of diseases. Since its first record in the Mawangdui texts (Wu Shi Er Bing Fang) from 168 BCE, it has been mentioned in different ancient medical treatises [1]. Traditionally, this herb was used for respiratory infections, wound healing, longevity, fevers, and notably, “intermittent fevers” [1]. Since then, A. annua and its phytocompounds, such as artemisinin and artesunate, have found their way into modern research and the treatment of malaria, rheumatoid arthritis (RA), systemic lupus erythematosus (SLE), and allergic contact dermatitis [2], [3]. Today, the World Health Organization (WHO) recommends artemisinin-based combination therapy (ACT) as the first-line treatment for uncomplicated multi-drug-resistant malaria. Chloroquine (CQ) and hydroxychloroquine (HCQ) are antimalarial drugs that have also been researched and used for autoimmune conditions such as RA and SLE [2]. Research on the antiviral activity of CQ started in the late 1960s [4]. After the 2002 severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1) and the 2012 Middle East respiratory syndrome coronavirus (MERS-CoV) outbreaks, the research on antimalarials for coronaviruses intensified. CQ and HCQ were studied for their use against SARS-CoV-1, MERS-CoV, and other viruses through in vitro and clinical trials [5]. The investigations pointed to the beneficial antiviral properties of CQ and HCQ against a variety of viruses, such as human coronavirus, enterovirus EV-A71, Zica virus, influenza virus A H5N1, chikungunya virus, dengue virus, and modestly for hepatitis C virus (HCV) [4], [5]. These studies paved the way for the development of similar research models for SARS-CoV-2, which causes coronavirus disease 2019 (COVID-19). Some in vitro, in silico, and pharmacokinetic studies showed favorable results for the use of CQ or HCQ for COVID-19 patients [6], [7], [8]. However, other researchers pointed to the high toxicity and side effects of CQ and HCQ, including possible cardiomyopathy, retinopathy, hypoglycemia, neuropsychiatric effects, hematologic toxicities, idiosyncratic hypersensitivity reaction, immune-mediated adverse reactions, teratogenicity, and drug-drug reactions (e.g., when combined with azithromycin) [9]. A review showed that there is a shortage of good-quality clinical evidence to support the use of CQ and HCQ to prevent COVID-19 [10]. Another recent systematic review examined the role of these two drugs and concluded that at this point it is uncertain if they have benefits for the treatment of COVID-19 [11]. Thus, the use of CQ and HCQ for COVID-19 remains controversial. Nonetheless, the study of antimalarial agents for viruses, such as coronaviruses, has a long history with many publications, as reviewed by Touret and de Lamballerie [4]. Likewise, A. annua is an antimalarial agent with a successful track record against different viruses and often demonstrating low cytotoxicity [12]. Thus, the hypothesis that A. annua and its phytoconstituents can provide anti-SARS-CoV-2 properties was raised.
The main bioactive compounds of A. annua are the sesquiterpene lactones, monoterpenes, triterpenoids, steroids, flavonoids, and essential oils [13]. The principal sesquiterpene of A. annua is artemisinin. Artemisinin derivatives, collectively called artemisinins, include artesunate, dihydroartemisinin (DHA; aka. artenimol), artemether, arteether, arteannuin B, and artemisone, among others. DHA, artesunate, artemether, and arteether are ten times more potent than artemisinin [2]. Also, the essential oils derived from A. annua vary in percentage and by the geographical region of their cultivation, and include artemisia alcohol, artemisia ketone, borneol, camphene, camphene hydrate, camphor, trans-caryophyllene, β-caryophyllene, caryophyllene oxide, chrysanthenone, 1,8-cineole (eucalyptol), β-farnesene, germacrene D, α-guaiene, linalool, linalool acetate, myrcene, α-pinene, (trans)-pinocarveol, sabinene, and spathulenol [14]. In different studies, A. annua and its compounds exhibited antiviral, anticancer, pro-apoptotic, anti-inflammatory, antipyretic, antioxidant, and immunomodulatory activities [2], [15], [16]. The essential oils of A. annua have shown activity against both gram-positive and gram-negative bacteria, as well as antifungal actions [14]. Different essential oils and their components have also demonstrated antiviral activity against RNA and DNA viruses, including herpes simplex virus types 1 and 2 (HSV-1 and HSV-2), dengue virus, influenza viruses (H1N1 and H5N1), human immunodeficiency virus (HIV), coxsackieviruses, respiratory syncytial virus, Zica virus and adenovirus [17]. Importantly, several reviews have identified the role of artemisinin and artesunate in the inhibition of hepatitis B virus (HBV), HCV, bovine viral diarrhea, human cytomegalovirus (HCMV), and members of the Herpesviridae family [3], [12]. Of note, artesunate is especially potent against herpesviruses such as HSV-1 and HSV-2, human herpesvirus 6A, and Epstein-Barr virus [3], [12], [18]. Moreover, the tea infusion of A. annua showed significant and potent anti-HIV activity, with a half maximal inhibitory concentration (IC50) of 2.0–14.8 μg/mL in vitro. Yet, its antiviral effect was not associated with artemisinin, suggesting that other compounds in A. annua may inhibit HIV [19]. Coronaviruses are very contagious pathogens that often require a biosafety level 3 (BSL-3) laboratory to handle them [20]. Due to the high risk of viral manipulation, the inability to access a BSL-3 laboratory or adapt a BSL-2 laboratory, and other factors, researchers must use other viral models for coronavirus research. Some examples are pseudotyped viruses that do not replicate, viruses that can be researched in a BSL-2 laboratory, and RNA viruses that may provide insights and results relevant to the RNA-based coronaviruses. For instance, HCV, HIV, human coronavirus-OC43, mouse hepatitis virus, and HIV-luc/SARS pseudotyped virus are some of the commonly used viruses [21]. RNA viruses include SARS, MERS, HIV, HCV, Ebola virus, influenza, polio measles, dengue virus, and adult human T-cell lymphotropic virus type 1 [22]. Thus, the previous research on viral infections provided a strong background to further the SARS-CoV-2 research. Knowledge translation from academic research to the uptake of evidence into policy making and clinical practice can take many years. Given the urgency of the current pandemic, the existence of previous pharmacokinetic, pharmacodynamic, and cytotoxicity studies of A. annua and its compounds are additional factors that make them interesting candidates for drug repurposing [23], [24], [25]. Thus, due to the multifunctional properties of A. annua and artemisinins against different viruses and their good safety profiles, their applicability as an anti-SARS-CoV-2 seems promising and should be further investigated.
At this time (as of Aug 24, 2020), the WHO has reported a total of 23 million people confirmed with COVID-19 and 800,000 deaths worldwide, with a rapid increase in cases (1.7 million new cases and 39,000 deaths during the past week) [26]. To date, no drugs or phytochemicals have been shown to eradicate the SARS-CoV-2. The COVID-19 pathogenesis involves the upper respiratory tract and other organ systems, and the common and often mild symptomatology consists of fever, myalgia, fatigue, headache, dyspnea, and cough [27]. In a retrospective cohort study involving 201 COVID-19 patients, 41.8% developed acute respiratory distress syndrome (ARDS) triggered by a cytokine storm [27]. ARDS is characterized by pulmonary infiltration, thick mucus secretions, elevated pro-inflammatory cytokines, extensive lung damage and fibrosis, and micro-thrombosis [28]. This severe clinical presentation, if untreated, often evolves to multiple-organ failure and death. Recently, we have seen investigations on the possibility of SARS-COV-2 reinfection, relapse, inflammatory rebound [29], [30], and the prospect of COVID-19 neuropsychiatric sequelae [31], [32]. Also, autoimmunity is being researched as an immune response to this viral infection, and published studies on the cross-reactivity between SARS-CoV-2 proteins and self-tissues are raising concerns that vaccines could trigger autoimmunity [33], [34], [35], [36]. Molecular mimicry between the viral proteins and self-antigens could be one explanation for the wide range of symptoms in different organ systems seen in many cases [33]. With the vaccine research still underway and the complexity of the COVID-19 pathogenesis, it is imperative to further the study of antiviral, anti-inflammatory, immunomodulatory, and anti-pulmonary fibrosis agents that could support the healing from this viral infection and its consequences. In unvaccinated populations (e.g., due to medical exemptions), managing the COVID-19 symptomatology will continue to be an issue. Thus, the need for effective, safe, and inexpensive antiviral remedies to assist in the treatment of SARS-CoV-2/COVID-19 is urgent. With all the above-mentioned justifications in mind, the goal of this article was to examine the evidence for the antiviral properties of A. annua and its phytocompounds against SARS-CoVs. Special emphasis was placed on identifying potential mechanisms that would interfere in the replication in different phases of the viral life cycle, as well as in the pathogenesis of the COVID-19.
2. Methodological approach
The main databases of PubMed/MEDLINE, and Scopus were searched using the following terms: (“Artemisia annua” OR artemisinin OR artesunate OR “artemisinin derivatives”) AND (coronavirus OR COVID-19 OR SARS-CoV-2). Given the very few results from the main databases, Google Scholar was added to the search to locate additional relevant papers. The first 30 pages returned from the Google Scholar query were evaluated for relevance to this review. The included studies were peer-reviewed experimental or clinical studies that investigated the antiviral or action mechanisms of A. annua against SARS-CoVs in the English language. Particularly, the studies that supported the role of A. annua in suppressing the viral infection through interaction with host or coronavirus proteins were favored. Also, articles that could provide proof of the direct influence of artemisinins on the signaling pathways involved in the COVID-19 were highlighted. Non-peer-reviewed preprint articles were not considered.
3. Results and discussion
PubMed/MEDLINE and Scopus identified 28 results, and 300 articles returned from the Google Scholar search were examined. After screening all articles for relevance, the combined searches resulted in a total of 19 studies. Four studies were in vitro, fourteen were in silico, and one article was a controlled clinical trial. The results of the antiviral properties and target proteins tested against SARS-CoVs are summarized in Table 1 . This present article provides a first comprehensive and up-to-date overview of all protein-associated mechanisms of A. annua and its phytocompounds that can potentially act as anti-SARS-CoV-2. No articles on the effects of A. annua or artemisinins on the COVID-19 pathogenesis in its mild, moderate, severe, or critical forms were found. However, additional data and insights are offered in section four, regarding the possible intervention of A. annua and its compounds on the signaling pathways affected by the COVID-19.
Table 1.
Published research on the antiviral activity of Artemisia annua and artemisinin derivatives against SARS-CoV-2 and the coronavirus/host protein-associated mechanisms.
| Research | Herbs/compounds/herbal preparation | Type of research | Antiviral properties and protein-associated mechanisms against SARS-CoVs |
|---|---|---|---|
| Li et al., 2005 [37] | Ethanolic extract of A. annua. Positive control: IFN-α. | In vitro (Vero E6 cells and HepG2 cells), and CPE/MTS assays | Antiviral activity against SARS-CoV-1 BJ001 strain: EC50 (34.5 ± 2.6) μg/mL, CC50 (1053.0 ± 92.8) μg/mL, SI50 31. IFN-α: EC50 (660.3 ± 119.1) μg/mL, CC50 (100,000.0 ± 710.1) μg/mL, SI50 151. |
| Wang et al., 2007 [38] | MOL736 extracted from A. annua. Template: MDL28170. | In silico molecular docking study | MOL736 inhibited CTSL and presented with a better docking score than MDL28170. Docking binding energy: MOL736, −50.767 kcal/mol (Etotal); MDL21870, −47.887 kcal/mol (Etotal). |
| Sehailia & Chemat, 2020 [7] | Artemisinin and 10 derivatives were screened against the SARS-CoV-2 S protein RBD: artelinic acid, artesunate, artemisone, artemisinin, artemiside, artenimol (DHA), artemotil, artemether, and PubChem (ID 90667934, 122185220, 10380074). | In silico molecular docking study | All artemisinin and derivatives showed better docking scores than HCQ. Artelinic acid: −7.1 kcal/mol; artesunate: −6.8 kcal/mol; artemisone: −6.6 kcal/mol; artemisinin: −6.5 kcal/mol; artemiside: −6.4 kcal/mol; artenimol (DHA): −6.4 kcal/mol; artemotil: −6.3 kcal/mol; PubChem CID 90667934: −6.3 kcal/mol; PubChem CID 122185220: −6.3 kcal/mol; artemether: −6.0 kcal/mol; PubChem CID 10380074 kcal/mol; HCQ: −5.5 kcal/mol. Artemisinin, artesunate, and artenimol inhibited the SARS-CoV-2 S protein RBD at the Lys353 and Lys31 hotspot-binding sites of ACE-2. |
| Das et al., 2020 [39] | 33 compounds were screened as SARS-CoV-2 3CLPRO (or MPRO) inhibitors, including artemisinin. | In silico molecular docking study | Artemisinin interacted with 3CLPRO-binding sites MET49, CYS145, and HIS163. ΔG −7.15 kcal/mol; FFS −1233.81 kcal/mol. Other potential inhibitors (drugs/natural compounds): HCQ (ΔG −7.75; FFS −1236.76 kcal/mol), ritonavir (ΔG −9.52; FFS −1313.38 kcal/mol), rutin (ΔG −9.55; FFS −1100.25 kcal/mol), curcumin (ΔG −8.15; FFS −1236.50 kcal/mol), emetine (ΔG −9.07; FFS −1230.03 kcal/mol). |
| Alsaffar et al., 2020 [40] | 36 phytochemicals selected from plants growing in the Arabic area and exerting antiviral properties were tested as SARS-CoV-2 3CLPRO inhibitors. | In silico molecular docking study. Positive controls: CQ and HCQ. | Artesunate: −6.46 kcal/mol; KI 18.40 µmol/L. Artemisinin: −7.78 kcal/mol; KI 1.97 µmol/L. CQ: −7.12 kcal/mol; KI 6.04 µmol/L. HCQ: −7.35 kcal/mol; KI 4.13 µmol/L. Other interactions: betulinic acid, −10.0 kcal/mol; silibinin, −9.13 kcal/mol; oleanolic acid, −9.08 kcal/mol; epigallocatechin-3-gallate, −8.51 kcal/mol. |
| Peele et al., 2020 [41] | 24 natural compounds, 22 US Food and Drug Administration-approved antiviral drugs, and 16 antimalarial drugs were investigated for their anti-SARS-CoV-2 3CLPRO. | In silico molecular docking study | The artemisinin derivatives reached the docking scores of: artenimol, −5.178 kcal/mol; artesunate, −4.862 kcal/mol; artemether, −4.764 kcal/mol. Examples of lower scores (more potent interactions): amodiaquine, −7.429 kcal/mol; CQ, −6.075 kcal/mol; lopinavir, −9.918 kcal/mol; theaflavin digallate, −10.574 kcal/mol. |
| Cao et al., 2020 [42] | Nine artemisinin-related compounds: artemisinin, arteether, artemether, artemisic acid, artemisone, artesunate, DHA, arteannuin B, and lumefantrine were tested for their antiviral properties. | In vitro (African green monkey kidney Vero E6 cells), cytotoxicity assay, qRT-PCR, IFA, time-of-drug-addition assay, and pharmacokinetic prediction model | Arteannuin B showed the highest inhibition of SARS-CoV-2 (EC50 [10.28 ± 1.12] μmol/L; CC50 [71.13 ± 2.50] μmol/L; SI [7.00 ± 0.76]). Artesunate: EC50 (12.98 ± 5.30) μmol/L; SI (5.10 ± 2.08). DHA: EC50 (13.31 ± 1.24) μmol/L; SI (2.38 ± 0.22). Arteannuin B, artesunate, DHA, and lumefantrine: reduced the production of SARS-CoV-2 N protein. |
| Gendrot et al., 2020 (1) [43] | ACTs were tested (same drug combination and doses used for uncomplicated malaria). Combinations: artemether-lumefantrine, artesunate-amodiaquine, DHA-piperaquine, artesunate-mefloquine, and artesunate-pyronaridine. | In vitro study: Vero E6 cells infected with isolated SARS-CoV-2 strain (IHUMI-3) | Highest viral inhibition: mefloquine-artesunate (72.1% ± 18.3%). All other combinations also showed viral inhibition (27.1% to 34.1%). |
| Gendrot et al., 2020 (2) [44] | Antimalarial drugs were tested for their anti-SARS-CoV-2 activity: CQ, HCQ, ferroquine, quinine, mefloquine, desethylamodiaquine, pyronaridine, piperaquine, DHA, and lumefantrine. | In vitro study: Vero E6 cells infected with clinically isolated SARS-CoV-2 strain (IHUMI-3) | DHA showed low antiviral activity: EC50 20.1 μmol/L; EC90 41.9 μmol/L; CC50 58.9 μmol/L; SI 3. |
| da Silva et al., 2020 [45] | 171 essential oil components against SARS-CoV-2 3CLPRO, RdRp, S protein, hACE-2, nsp15/NendoU, and ADRP were tested. | In silico molecular docking study | Most of the A. annua essential oil components were screened as target protein inhibitors. The binding energy of the strongest binders was not enough to elect them as SARS-CoV-2 and host protein inhibitors. However, the combination of several essential oils may provide synergistic action to oppose the virus. |
| Sudeep et al., 2020 [46] | 25 natural compounds were screened for their anti-SARS-CoV-2 3CLPRO and anti-host receptor GRP78. | In silico molecular docking study | Molecular docking against GRP78-SBD. Binding energy: withaferin, −8.7 kcal/mol; artemisinin, −7.89 kcal/mol; curcumin, −6.21 kcal/mol; andrographolide, −6.17 kcal/mol. Molecular docking against 3CLPRO. Binding energy: withaferin, −9.83 kcal/mol; artemisinin, −8.06 kcal/mol; curcumin, −6.58 kcal/mol; andrographolide, −6.49 kcal/mol. |
| Li et al., 2021 [47] | Two groups of patients with COVID-19: the artemisinin-piperaquine (AP) group with AP, and the control patients with HCQ-arbidol. | Controlled clinical trial: AP group (n = 23); control group (n = 18) | The mean time to reach undetectable SARS-CoV-2 RNA: AP group, (10.6 ± 1.1) d; control group, (19.3 ± 2.1) d. The percentage of patients with undetectable viral RNA on days 7, 10, 14, 21, and 28: AP group, 26.1%, 43.5%, 78.3%, 100%, and 100%; control group, 5.6%, 16.7%, 44.4%, 55.6%, and 72.2%. Length of hospital stay: AP group, (13.3 ± 4.8) d; control group, (21.3 ± 9.1) d. CT image within 10 days post-treatment: lung improvements but no differences between the two groups (P > 0.05). |
| Sharma & Deep, 2020 [48] | 125 FDA-approved antiviral and antimalarial drugs (Drug Bank), Ayurveda compounds (PubChem), natural polyphenols (Marvin Sketch 17.23.0 ChemAxon), and EGCG (Zinc Database) were screened against SARS-CoV-2 3CLPRO. | In silico study by molecular docking, MD simulation (stability of docking complexes), and MM-GBSA binding method (binding energy). Compounds with binding energy −8 kcal/mol or more potent were selected for MD simulation. | Potential anti-SARS-CoV-2 3CLPRO were artesunate, EGCG, withaferin A, and dolutegravir. Artesunate: binging energy, −8.0 kcal/mol; interacting residues, 24–27, 41, 46, 49, 140–145, 163, 165, 166; MD simulation, RMSD = 0.59 nm (control N3 inhibitor, RMSD = 0.34 nm); MM-GBSA, (−24.72 ± 5.01) kcal/mol (control N3 inhibitor, [−28.79 ± 3.73] kcal/mol). |
| Prashantha et al., 2021 [49] | Drugs and compounds were tested as potential inhibitors of SARS-CoV-2 S protein. Tested: CQ, HCQ, pyrimethamine, artemisinin, mefloquine, lopinavir, ritonavir, darunavir, cobicistat, baricitinib, ruxolitinib, thalidomide, azithromycin, clarithromycin, erythromycin, spiramycin, camostat, fingolimod, and umifenovir (Drug Bank Database) | Molecular docking, virtual screening, drug-like, and ADMET predictions. Results were based on docking scores, H-bonds, and amino acid interactions. | Antimalarial drugs had a poor affinity to S protein compared with other drug types. Artemisinin: binding energy, −6.8 kcal/mol; H-bonds, 2; KI, 15.37 μmol/L; amino acids, Asn460, Lys462. Other antimalarials: CQ, −6.2 kcal/mol; HCQ, −5.2 kcal/mol; mefloquine, −6.7 kcal/mol; pyrimethamine, −5.8 kcal/mol. |
| Roy Chattopadhyay et al., 2020 [50] | FDA-approved drugs and chemicals were screened and tested against several SARS-CoV-2-host target proteins such as RdRp, helicase protein, NC, S protein RBD, E protein, nsp10, nsp14, and nsp15. | Virtual screening and molecular docking. Selection: drugs with at least one viral protein interaction; minimum binding energy: –0.7 kcal/mol. | Artesunate binding to proteins: E protein, −7.2 kcal/mol; helicase protein, −7.1 kcal/mol; nsp10, −7.6 kcal/mol; nsp14, −8.4 kcal/mol; nsp15, −8.2 kcal/mol; NC protein, −8.8 kcal/mol. Artemether binding to proteins: helicase protein, −7.5 kcal/mol; Nsp10, −7.0 kcal/mol; Nsp15, −7.4 kcal/mol; NC, −8.0 kcal/mol. Other examples: proguanil-E protein and proguanil-NC protein, −9.5 kcal/mol; CQ-nsp14, −7.3 kcal/mol; remdesivir-nsp15, −9.5 kcal/mol; mefloquine-NC protein, −8.9 kcal/mol. |
| Alazmi & Motwalli, 2020 [51] | 203,458 natural compounds (Zinc library) were screened and tested as ACE-2/S protein complex inhibitors. | Virtual screening, molecular docking study, toxicity analysis, and MD simulations (docking stability). Positive control: HCQ. | The 4 final compounds found by blind docking against ACE-2/S protein complex: andrographolide, artemisinin, pterostilbene, and resveratrol. Toxicity: artemisinin and andrographolide were the least toxic. Binding scores: andrographolide, −9.1 kcal/mmol; artemisinin, −6.2 kcal/mol; pterostilbene, −8.9 kcal/mol; resveratrol, −8.7 kcal/mol; HCQ, −7.1 kcal/mol. Structural analysis best results: andrographolide and artemisinin. MD simulations overall best results: andrographolide and pterostilbene. |
| Khan et al., 2020 [52] | 123 antiviral drugs (Drug Bank Database) were screened as inhibitors of the nsp15. | In silico study: binding affinity, estimated KI, orientation of molecules in the active sites, and key interacting with residues of nsp15. | Three promising candidates were identified: artesunate, simeprevir, and paritaprevir. Simeprevir: −8.4 kcal/mol; KI 0.696 μmol/L. Paritaprevir: −7.5 kcal/mol; KI 3.179 μmol/L. Artesunate: −7.2 kcal/mol; KI 5.275 μmol/L. Artesunate-nsp15 residue interactions: Lys290, Ser294, Thr341, and Tyr343 (hydrogen bonds); His250, His235 and Trp33 (π interactions): Gly247, Gly248, Val292, Cys293, Glu340, and Lys345 (Van der Waals interactions). MS analysis: overall, the nsp-15 drug complexes showed structural integrity and stability. However, simeprevir and paritaprevir were more stable than artesunate. Binding free energy (MM/PBSA): Nsp-15/Simeprevir, −259.522 kJ/mol; Nsp-15/Paritaprevir, −154.051 kJ/mol; Nsp-15/Artesunate, 75.449 kJ/mol. |
| Marak et al., 2020 [53] | 108 FDA-approved anti-inflammatory and antiparasitic drugs were screened and tested to access the binding affinity against SARS-CoV-2/host target proteins: 3CLPRO, PLPRO, RdRp, S protein, helicase protein, nsp1, nsp3, nsp4, nsp9, and nsp16–nsp10. Drugs included: artesunate, ivermectin B1a, ivermectin B1b, spiramycin, moxidectin, amphotericin B, posaconazole, chlorotetracycline, doxycycline, parecoxib, etoricoxib, and sulfasalazine. | In silico study: molecular docking. | Example of results of drug-protein complexes: artesunate-nsp3, −8.1 kcal/mol. Antiparasitic drugs showed higher inhibitory action. Examples of strong interactions: ivermectin B1b-3CLPRO, −9.3 kcal/mol; ivermectin B1a-RdRp, −9.3 kcal/mol; ivermectin B1b-nsp16-nsp10, −9.8 kcal/mol; amphotericin B-RdRp, −10.5 kcal/mol; atovaquone-S protein, −9.6 kcal/mol; parecoxib-nsp3, −10.17 kcal/mol; chlortetracycline-nsp4, −9.3 kcal/mol. |
| Gupta et al., 2020 [54] | Secondary metabolites from Ayurvedic medicine of 69 herbs and spices, and 10 pharmacological drugs were screened and tested for their potential inhibition of SARS-CoV-2/host proteins: 3CLPRO (nsp5), PLPRO (nsp3), RdRp, helicase (Hel) protein, S protein, M protein, NC protein, E protein, hACE-2 receptor, nsp1, nsp2, nsp4, nsp6, nsp7-nsp8, nsp9, nsp10-nsp16, and ExoN, and NendoU. Drugs: arbidol, CQ, colchicine, HCQ, dexamethasone, losartan, remdesivir, ribavirin, oseltamivir, and quinazoline. | In silico study: molecular docking and MD simulations; ADMET analysis. | Artemisinin-nsp2: −5.174 kcal/mol; artemisinin- PLPRO: −6.134 kcal/mol (formed only one H-bold with Asp108). Hesperidin and epicatechin were the best inhibitors of a series of proteins. |
SARS-CoV: severe acute respiratory syndrome coronavirus; IFN-α: interferon-α; CPE: cytopathic effect; IC50: half-maximal inhibitory concentration; EC50: half-maximal effective concentration; CC50: 50% cytotoxic concentration; SI: selectivity index; KI: inhibitor constant; MOL736: aurantiamide acetate; CTSL: cathepsin-L; RBD: receptor-binding domain; CQ: chloroquine; HCQ: hydroxychloroquine; 3CLPRO or MPRO: 3C-like protease or main protease; ΔG: free binding energy; FFS: fullfitness score; qRT-PCR: real-time quantitative reverse transcription polymerase chain reaction; IFA: immunofluorescence assay; ACTs: artemisinin-based combination therapies; DHA: dihydroartemisinin; RdRp: RNA-dependent RNA polymerase; hACE-2: human angiotensin-converting enzyme 2; nsp: nonstructural protein; nsp15/NendoU: nonstructural protein 15; ADRP: ADP-ribose-1″-phosphatase; GRP78: glucose-regulated protein 78; SBD: substrate-binding domain; EGCG: epigallocatechin gallate; MD: molecular dynamics; MM-GBSA: molecular mechanics/generalized born surface area; RMSD: root-mean square deviation; ADMET: absorption, distribution, metabolism, excretion, and toxicity; NC: nucleocapsid protein.
3.1. Antiviral properties of A. annua in vitro and clinical trial
The first study that included A. annua as an anti-SARS-CoV-1 was published in 2005 [37]. It reported that A. annua is one of the four herbs that significantly inhibited the in vitro activity of the SARS-CoV-1 (strain BJ001) in a dose-dependent manner, with interferon-α (IFN-α) used as a positive control [37]. However, this study did not investigate the mechanism of antiviral action. In another study, the antiviral properties of nine artemisinin-related compounds were tested against the SARS-CoV-2 in vitro [42]. The authors concluded that artemisinins, especially artesunate, arteannuin B, and DHA inhibited the virus and exhibited potential as anti-SARS-CoV-2 agents [42]. Arteannuin B, for instance, showed the highest inhibition of SARS-CoV-2. Several artemisinins reduced the production of the SARS-CoV-2 nucleocapsid (N) proteins in a dose-dependent manner [42]. N proteins play an important role in incorporating genomic RNA in the virions and replication and transcription of CoVs [55]. Therefore, targeting N proteins may be one option for inhibiting the viral infection. Also, both arteannuin B and lumefantrine blocked the infection after the viral entry into the host cells and may work similarly by hindering intracellular signaling pathways not yet identified [42]. Another group of researchers investigated whether the ACTs that are extensively used for malaria in African countries could be responsible for the later emergence and lower than expected number of confirmed COVID-19 cases in these countries [43]. This in vitro study revealed that all artemisinin-drug combinations inhibited the SARS-CoV-2 at expected maximum blood concentration at doses equivalent to those used for uncomplicated malaria [43]. Mefloquine-artesunate exerted the highest anti-coronavirus activity at 72.1% ± 18.3% [43]. The same group of scientists examined the ability of antimalarial drugs to inhibit the SARS-CoV-2 in vitro, including DHA, CQ, HCQ, ferroquine, desenthyamodiaquine, quinine, mefloquine, pyronaridine, lumenfantrine, and piperaquine [44]. DHA showed low antiviral activity ([20.1 ± 4.5] μmol/L) while the highest inhibition was exerted by desenthyamodiaquine ([0.5 ± 0.2]) μmol/L) [44]. Data were also reported for mefloquine ([1.8 ± 1.0] μmol/L), CQ ([2.1 ± 0.7] μmol/L), and lumefantrine ([24.7 ± 3.6] μmol/L). Thus, the authors hypothesized that countries using antimalarial ACTs with the combination of artesunate-mefloquine or artesunate-amodiaquine would have fewer COVID-19 cases and deaths than those using artemether-lumefantrine or DHA-piperaquine [44]. However, a recent controlled clinical trial investigated the anti-SARS-CoV-2 effects of artemisinin-piperaquine (AP) [47]. Patients diagnosed with mild to moderate COVID-19 were divided into two groups and the majority of the patients had a moderate form of the disease (82.6% in the AP group and 88.9% in the control group). One group of 23 patients received AP while the control group with 18 patients received a combination of HCQ-arbidol [47]. Both drug combinations were used as antiviral and symptomatic treatments. The AP group took significantly less time to reach undetectable levels of SARS-CoV-2 than the controls, requiring (10.6 ± 1.1) d and (19.3 ± 2.1) d, respectively (P = 0.001, 0.005) [47]. Considerable reduction was also found in the percentage of undetectable RNA on days 7, 10, 14, 21, and 28 and in the length of hospital stay in the AP group. No patients progressed to a severe or critical disease stage, and adverse reactions in both groups were mild. The authors pointed to research limitations, such as sample size and trial design and advised the monitoring of electrocardiogram (ECG) and liver enzymes. Nevertheless, they recommended the use of AP for COVID-19 prevention and treatment of mild to moderate cases (8 tablets—artemisinin 500 mg/piperaquine 3000 mg during 7 d) [47]. Hence, ACTs could be a viable antiviral resource to assist in the treatment of SARS-CoV-2 infection, but more research is needed.
3.2. In silico docking studies and the coronavirus-host target protein mechanisms
According to our recently published systematic review, 31 studies identified herbs and natural compounds that are able to inhibit coronavirus/host protein pathways [21]. These proteins included the spike (S) protein, angiotensin-converting enzyme-2 (ACE-2), nucleocapsid (N) protein, 3-chymotrypsin-like protease (3CLPRO), papain-like protease (PLPRO), RNA-dependent RNA polymerase (RdRp), helicase, cathepsin-L (CTSL), nonstructural protein 1 (nsp1), nsp3C, ORF7a, transmembrane protease serine-type 2, and 3a protein [21]. Drugs, herbs, or natural compounds able to inhibit or block one or more of these target proteins could interfere in the natural life cycle of the SARS-CoV-2, providing antiviral properties. Molecular docking simulations were used to predict the interrelation between a small molecule ligand (e.g., drugs, herbs, or phytochemicals) and viral/host proteins [56]. The docking process examines the ligand conformation, position, and orientation in relation to the docking site, and assesses the binding affinity [56]. It is important to note that, conventional drugs, herbs and natural compounds go through the same or similar testing methods to discover possible antiviral agents against the SARS-CoV-2 [21]. In silico studies often screen and test hundreds or even thousands of pharmaceutical drugs, herbs, and phytocompounds to identify drug candidates. One study, for example, detected possible drugs and natural substances as anti-SARS-CoV-2 options from a series of drug classes including antimalarial, antibacterial, anti-hypertensive, anti-psychotic, antitumor, antitussive, muscle relaxant, anti-erectile dysfunction, anti-inflammatory, anti-HBV, anti-HSV-1, antioxidant, anti-ulcerative, anti-epileptic, anti-HIV-1 protease inhibitor, antidiabetic, and antifungal drugs [57]. Thus, for this type of drug discovery and repurposing research, scientists are not looking for similarities between pathogens (e.g., malaria parasite and SARS-CoV-2). Below, the computational simulations that investigated the A. annua phytocompounds and their effects on the coronavirus and host proteins are reviewed. Also, due to different methodologies and data interpretation of the studies, for the present review, the binding score of ≤ −7.0 kcal/mol will be considered appreciable to provide a baseline for reporting and comparing. However, other examined compounds-drugs and their scores can be visualized in Table 1. A summary of the A. annua compounds that reached the score of ≤ −7.0 kcal/mol is shown in Fig. 1 .
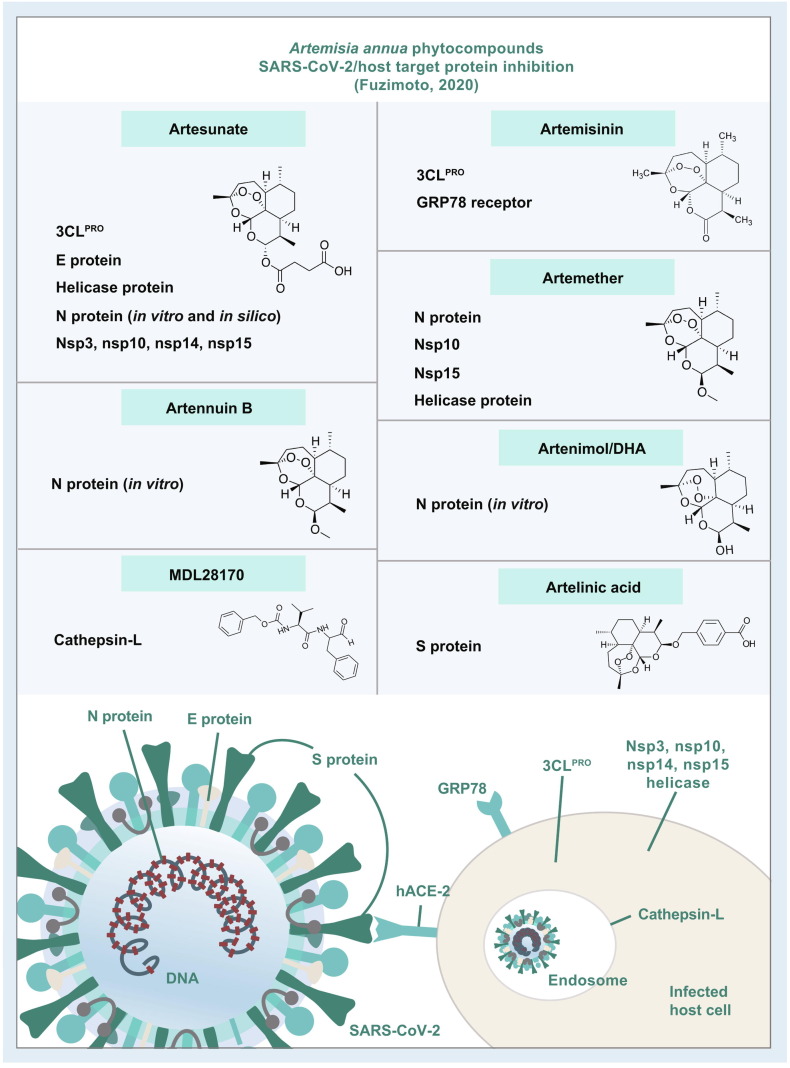
Fig. 1.
Summary of the Artemisia annua phytocompounds as potential SARS-CoV-2/host protein inhibitors. These compounds attained appreciable antiviral activity by inhibiting SARS-CoV-2/host proteins with binding scores of ≤ −7.0 kcal/mol. The stronger binding interaction among the artemisinins was reached by artesunate-N protein (−8.8 kcal/mol). The results were attained by in silico studies except those that are highlighted as in vitro studies (e.g., arteannuin B and dihydroartemisinin inhibited the N protein in vitro; artesunate inhibited N protein in vitro and in silico; and the rest of the compounds were in silico). For the A. annua phytocompounds and a full list of scores and other results, see Table 1. SARS-CoV-2: severe acute respiratory syndrome coronavirus-2.
3.2.1. Studies that only investigated the SARS-CoV-2 3CLPRO
In an in silico study, 33 molecules, including natural products, antivirals, antifungals, anti-nematodes, and antiprotozoals, were screened as possible inhibitors of SARS-CoV-2 3CLPRO [39]. Artemisinin was one of the compounds that interacted with the 3CLPRO active binding sites [39]. SARS-CoV 3CLPRO, or the main protease (MPRO), is a viral enzyme responsible for the cleavage of polyproteins (pp1a and pp1ab) into functional proteins important for the coronavirus replication. 3CLPRO is considered a promising drug target and its inhibition would hinder viral replication. Although artemisinin was not the best possible 3CLPRO inhibitor among the compounds, it still presented a good estimated free binding energy (ΔG) of −7.15 kcal/mol [39]. The free binding energy represents how strongly the binding happens, as opposed to how accurately the fit occurs. Other investigated drugs and compounds were better binders, such as rutin, curcumin, emetine, HCQ, ritonavir, and lopinavir. Rutin was the most potent inhibitor, with ΔG of −9.55 kcal/mol [39]. Nevertheless, HCQ had similar docking energy to artemisinin, with a ΔG of −7.75 kcal/mol. In the simulation, artemisinin bound to the SARS-CoV-2 3CLPRO through an alkyl hydrophobic interaction with MET49 and CYS145 residues and a π-alkyl interface with HIS163 [39]. This result was somewhat corroborated by another in silico study that examined 36 compounds for their SARS-CoV-2 3CLPRO inhibition potential [40]. Artesunate had a docking score of −6.46 kcal/mol, while artemisinin had a stronger docking score of −7.78 kcal/mol [40]. Both CQ and HCQ were used as positive controls, with binding scores of −7.12 kcal/mol and −7.35 kcal/mol, respectively. Although other phytocompounds were better binders such as betulinic acid (−10.0 kcal/mol), artemisinin had a better binding score than the controls.
Another molecular docking and dynamics simulation investigated 24 natural compounds, 22 US FDA-approved drugs, and 16 antimalarial drugs for their potential as SARS-CoV-2 3CLPRO inhibitors, including artemisinin derivatives [41]. In this study, lopinavir, amodiaquine, theaflavin digallate, CQ, and quinine, among other substances, had better docking scores than artenimol, artesunate, and artemether [41]. Yet, another docking and molecular dynamics simulation utilizing the Michael acceptor N3 inhibitor as a control investigated artesunate as a potential anti-SARS-CoV-2 3CLPRO [48]. The binding energy for the protein-artesunate complex was −8.0 kcal/mol and the binding energy using the MM-GBSA method was (−24.72 ± 5.01) kcal/mol, compared with N3, at (−28.79 ± 3.73) kcal/mol [48]. Overall, the simulations revealed that artesunate had significant binding energy, a stable and tight docking to residues, and exhibited five active site interactions. The authors concluded that artesunate is one of the four agents, together with epigallocatechin gallate, withaferin, and dolutegravir, that may act as anti-SARS-CoV-2 3CLPRO [48].
3.2.2. Studies that only investigated SARS-CoV-2 S protein and S/ACE-2 complex
In a more recent in silico study, all eleven artemisinin derivatives were able to block the SARS-CoV S protein receptor-binding domain (RBD) [7]. Particularly, artemisinin, artesunate, and artenimol prevented the SARS-CoV-2 S protein RBD from binding to the Lys353 and Lys31 hotspots of the human angiotensin-converting enzyme 2 (hACE-2) and could be good S protein inhibitor candidates [7]. The coronavirus S protein binds to the hACE-2, and substances that prevent the coronavirus S protein from docking to the hACE-2 receptor may inhibit the infection [7]. After further analysis, the authors recommended the prioritization of artenimol as a candidate for future clinical trials, as most artemisinin derivatives end up being converted to artenimol in the body. However, the binding scores of most artemisinins were below what many researchers would consider appreciable (from −6.0 to −6.8 kcal/mol) and artelinic acid showed the best score among the compounds (−7.1 kcal/mol). Still, artemisinin and its derivatives were less toxic and more efficient at docking at the SARS-CoV-2 S protein than HCQ (−5.5 kcal/mol) [7]. Moreover, another computational study examined if antimalarial, HIV-protease inhibitor, anti-inflammatory, and antibiotic drugs had a good affinity to the S protein [49]. They verified that the antimalarial agents, artemisinin, CQ, HCQ, mefloquine, and pyrimethamine, had poor anti-SARS-CoV-2 S glycoprotein activity when compared with other drugs [49]. Artemisinin had a binding score of −6.8 kcal/mol and an estimated inhibitor constant (KI) of 15.37 μmol/L, and formed two bonds with the amino-acid residues Asn460 and Lys462 [49]. For this study, other drugs had more potential to hinder the S protein from binding to the ACE-2 receptor, such as lopinavir (−9.1 kcal/mol), ritonavir (−8.0 kcal/mol), cobicistat (−8.3 kcal/mol), erythromycin (−9.0 kcal/mol), and spiramycin (−8.5 kcal/mol) [49]. Also, the HIV protease inhibitors, anti-inflammatory, and antimicrobial drugs formed more bonds with a series of other S protein amino acid residues (e.g., from 3 to 6 hydrogen bonds) [49].
Although the S glycoprotein is one of the most researched target proteins to act as anti-SARS-CoV-2, the interaction between the drug candidates and the ACE-2/S protein complex is also important to consider. An in silico investigation screened and tested 203,458 natural compounds against this protein complex using HCQ as a positive control [51]. After the first round of blind docking, four possible inhibitor candidates emerged: artemisinin (−6.2 kcal/mol), andrographolide (−9.1 kcal/mol), pterostilbene (−8.9 kcal/mol), and resveratrol (−7.1 kcal/mol) [51]. Further structural analysis showed that, although artemisinin had a low binding capacity, it had the second-best interaction and a good fit between the interface of ACE-2/S protein. Andrographolide had the best binding score, with six residue interactions (Asp-30, Asn-33, His-34, Pro-389, Arg-393, and Tyr-505), while artemisinin was bound to four residues (His-34, Ala-387, Pro-389, and Tyr-505) [51]. Also, upon the toxicity testing, andrographolide and artemisinin exerted the lowest toxicity levels of the compounds. However, additional molecular dynamics simulation to predict the stability of the interactions showed that artemisinin had the least preferable docking score. Overall, the authors concluded that the more stable compounds andrographolide and pterostilbene would be the best options for further studies [51].
3.2.3. Study that investigated CTSL
In another molecular docking study from 2007, 26 compounds (10,458 natural product molecules) from the Traditional Chinese Medicine Database (TCMD) were screened using MDL28170 as a template [38]. MDL28170 was recognized as an effective CTSL inhibitor. CTSL is an endosomal protease that plays an essential role in membrane fusion and the internalization of the coronavirus. Thus, CTSL is an important target protein for therapeutic interventions in SARS-CoV infections. The compound MOL736 (aurantiamide acetate) extracted from A. annua was detected as an efficient inhibitor of the host CTSL protein and it was more “matchable” than MDL28170 [38]. Thus, this study showed this A. annua’s isolate as a potential therapeutic resource in the fight against SARS-CoVs.
3.2.4. Studies that investigated multiple target proteins
Another research group examined the in silico ability of 25 natural compounds and their potential as inhibitors of SARS-CoV-2 3CLPRO and host glucose-regulated protein 78 (GRP78) [46]. The GRP78 is a master chaperone protein that responds to accumulated unfolded or misfolded proteins in the cells [58]. When translocated to the cell membrane, it can bind to region IV of the SARS-CoV S protein (the strongest binding affinity of −9.8 kcal/mol), thus mediating viral entry into the host cells [58]. Agents that inhibit the GRP78 cell receptor could offer another antiviral therapeutic option. Four compounds displayed promising activity as antiviral agents: withaferin, artemisinin, curcumin, and andrographolide. All four phytochemicals showed the ability to bind to both SARS-CoV-2 3CLPRO and GRP78 substrate-binding domain, with withaferin displaying the strongest binding energy (−9.83 kcal/mol). Also, all four compounds satisfied the “rule of five” in drug-likeness, according to SwissADME, and the selected proteins demonstrated a high probability of druggability [46]. Yet, for our analysis here, artemisinin exhibited a favorable binding score of −8.06 and −7.89 kcal/mol for 3CLPRO and GRP78, respectively [46]. In another investigation, the researchers performed a molecular docking analysis of 171 essential oil components against SARS-CoV-2 3CLPRO, RdRp, the S protein binding domain, and hACE-2. Additionally, they tested the SARS-CoV-2 endoribonuclease (SARS-CoV-2 nsp15/NendoU) and the SARS-CoV-2 ADP-ribose-1’-phosphatase (SARS-CoV-2 ADRP) [45]. The non-structural protein, nsp15, is an endoribonuclease that cleaves RNA at uridylate and is required for viral infection. The inhibition of the coronavirus ADRP may reduce the multiplication of the virus, providing another target for therapeutic intervention. The study included most of the A. annua essential oil constituents and showed the binding scores for each target protein. The best docking scores were reached by (E,E)-α-farnesene, (E)-β-farnesene, and (E,E)-farnesol [45]. However, the docking energies of these components were somewhat weak, compared with other interactions. The authors concluded that the essential oil components would not make good coronavirus/host protein inhibitors, but they could work synergistically with other antiviral agents to provide relief for the COVID-19 symptoms [45]. As mentioned by the authors, linalool, β-caryophyllene, and 1,8-cineole provide anti-inflammatory and antinociceptive properties, and camphor has an antitussive action that could add up to the benefits of A. annua for the COVID-19. Additionally, the authors listed the in vitro inhibitory effects of several Artemisia species and their essential oil constituents against different viruses. A. vulgaris L. essential oils produced 100% reduction of yellow fever virus at 100 μg/mL; A. princeps var. orientalis reduced 64% of the murine norovirus-1 (MNV-1) at 0.01% of the essential oil; and A. arborescens L. inhibited HSV-1 and HSV-2 infected Vero-6 cells with IC50 of 2.4 and 4.1 μg/mL, respectively [45].
In another study, the binding capacity of FDA-approved drugs and chemicals against a series of SARS-CoV-2/host proteins such as RdRp, helicase protein, nucleocapsid (NC or N) protein, S protein RBD, E protein, nsp10, nsp14, and nsp15 were examined [50]. The screened drugs included artesunate, artemether, antimalarial pharmaceutical drugs (CQ, quinine, primaquine, amodiaquine, mefloquine, and proguanil), the antiviral drugs galidesivir, remdesivir, pirodavir, and others. Artesunate exhibited binding activity with E protein, helicase protein, nucleocapsid (NC) protein, and with the non-structural proteins nsp10, nsp14, and nsp15 [50]. Artesunate did not interact with S protein RBD or RdRp. Also, artemether interacted with NC protein, helicase protein, nsp10, and nsp15 [50]. The corresponding author of this study was contacted to clarify some aspects of the methodology and parameters of data interpretation. In this case, artesunate was classified as “may be chosen” and artemether was “not chosen” as SARS-CoV-2/host inhibitors even though all binding scores were ≤ −7.0 kcal/mol [50]. From these two, the best binding score was attained by the artesunate-N protein interaction (−8.8 kcal/mol). Regardless, the reported results on artesunate and artemether showed potential antiviral activity associated with proteins never before reported. Also, the authors proposed four drug combinations for “proper and effective management of COVID-19” and two of them included artesunate. The rationale for their proposed combination of artesunate-drug or drug-drug regimens that would include “immune boosters” also requires more investigation and careful analysis. On the other hand, another research study screened and tested 123 antiviral drugs as nsp15 inhibitors [52]. The top three candidates that reached significant binding scores of ΔG ≤ −7.0 kcal/mol were simeprevir (−8.4 kcal/mol), paritaprevir (−7.5 kcal/mol), and artesunate (−7.2 kcal/mol) [56]. The molecular dynamics simulations revealed that the nsp15 drug complexes were stable and had structural integrity. Nonetheless, in some evaluations, simeprevir and paritaprevir were more stable than artesunate, and the binding free energy utilizing the MM/PBSA approach showed that the interaction of artesunate with nsp15 was non-spontaneous and infeasible [52]. Thus, in this instance artesunate was not considered a good candidate as a nsp15 inhibitor.
One more study screened 108 FDA-approved anti-inflammatory and antiparasitic drugs against SARS-CoV-2/host target proteins 3CLPRO, PLPRO, RdRp, S protein, helicase protein, nsp1, nsp3, nsp4, nsp9, and nsp16-nsp10 [53]. Although some researchers do not make a clear distinction between PLPRO and nsp3, the latter is a multi-domain protein and PLPRO is one of the catalytic domains of nsp3 [53]. Artesunate inhibited the nsp3 and formed H-bonds with Ala154, Phe156, Asp157, and Leu126, and interacted with residues Val49, IIe23, Ala52, and Phe156 in other interface types [53]. The antiparasitic drugs showed higher inhibitory actions. Although artesunate was not the most potent (−8.1 kcal/mol), it did exert significant inhibition [53]. Also, another in silico study tested Ayurvedic herbs and spices, and pharmaceutical drugs for their potential uses against SARS-CoV-2/host proteins, including 3CLPRO (nsp5), PLPRO (nsp3), RdRp, helicase protein (Hel), S protein, M protein, NC protein, E protein, hACE-2 receptor, nsp1, nsp2, nsp4, nsp6-nsp16, ExoN, and NendoU [54]. Artemisinin which was included in this study only inhibited nsp2 and PLPRO with binding energies of −5.174 and −6.134 kcal/mol, respectively [54]. However, other compounds, such as epicatechin and hesperidin reached a multi-protein higher inhibition [54].
4. Considerations and perspectives
As mentioned, no primary data on the possible role of A. annua and artemisinins on the pathogenesis of COVID-19 were found in the literature search. However, in this section, the biological properties of artemisinins, their influence on signaling pathways of COVID-19, advantages and disadvantages of their administration and repurposing, and the use of the whole plant versus the isolates are explored. Fig. 2 shows an overview of the potential mechanisms of A. annua and phytocompounds against SARS-CoV-2/COVID-19.
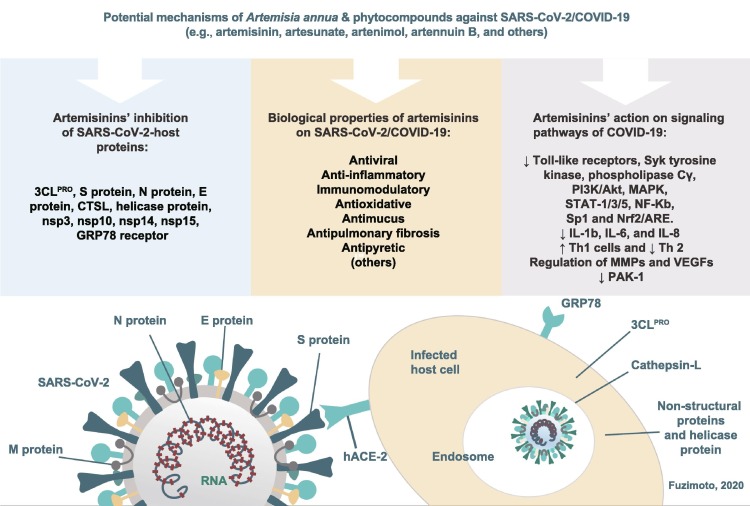
Fig. 2.
Potential mechanisms of Artemisia annua and its phytocompounds against SARS-CoV-2/COVID-19. The combination of A. annua’s biological properties, action on different signaling pathways, and target proteins may synergistically inhibit the SARS-CoV-2, decrease inflammation, modulate the host immune response, and alleviate the COVID-19 symptomatology. SARS-CoV-2: severe acute respiratory syndrome coronavirus-2; COVID-19: coronavirus disease 2019.
4.1. A. annua’s biological properties and signaling pathways involved in the COVID-19
Scientists are actively investigating the pathogenesis of COVID-19 and trying to understand how the disease progresses. Part of the assumptions come from previous research on the 2002 coronavirus epidemic in China. Characteristically, SARS-CoV-1 infects the airway and alveolar epithelial cells, vascular endothelial cells, and macrophages [59]. The recognition of the SARS-CoV-2 nucleic acids, as with other coronaviruses, would be mediated by pattern recognition receptors that signal the release of type 1 IFN [59]. Patients with COVID-19 had a high infiltration of macrophages into the lung tissues, which caused a significant amount of interleukin-6 (IL-6) expression. Macrophage activation syndrome and high levels of IL-6 may contribute to the excessive inflammation during the COVID-19 [59]. However, coronaviruses encode proteins that interfere in signaling pathways to weaken the host immune response. Examples of the coronavirus strategies to evade the innate immune system are the blockage of the pathogen-associated molecular pattern recognition, the downregulation of major histocompatibility complex I and II class molecules in the antigen-presenting cells, and the viral M protein inhibiting IFN production [59]. Preliminary evaluations of COVID-19 pathogenesis indicate that patients present with activation of the nuclear factor-κB (NF-κB) and upregulation of pro-inflammatory cytokines and chemokines including IL-6, IL-1β, IFN-γ, IL-4, IL-10, IFN-γ, C–C motif chemokine ligand 2 (CCL2), CCL5, IFN-γ inducible protein 10 (IP-10), CCL3, and monocyte chemoattractant protein-1 [3], [60]. The pro-inflammatory cytokines may activate the Th1 cell response, but COVID-19 patients may also have an upregulation of cytokines secreted by Th2 cells, such as IL-4 and IL-10, which inhibit inflammation [60]. COVID-19 is believed to be a Th1-driven disease, but CD8+ and CD4+ T lymphocytes and regulatory T cells are often downregulated, and cytotoxic T cells and natural killer (NK) cells suffer functional exhaustion in severe clinical presentations [59]. Thus, the SARS-CoV-2 exerts strong immunosuppressive action on the adaptive immune system of the host [59]. Regardless, the serum levels of IL-2R and IL-6 are correlated with disease severity. More critical patients may have higher levels of IL-2, IL-6, IL-7, IL-10, granulocyte colony-stimulating factor, IP-10, MCP1, macrophage inflammatory protein 1-α (MIP1α), and TNF-α [3]. One study suggested that the matrix metalloproteinase-9 (MMP-9) may be an early indicator of respiratory failure in COVID-19, and treatments that address MMP-9 could assist healing the lung tissue [61]. Also, vascular endothelial growth factors (VEGFs) play an important role in brain inflammation during the COVID-19 through the recruitment of inflammatory cells and the regulation of the angiopoietin II levels [62]. Although it would be tempting to select therapies to upregulate the host immune function due to the immune suppression and possible exhaustion caused by the SARS-CoV-2, an overdrive of Th1 cell activity could increase the systemic inflammation and trigger a worsening of the COVID-19 symptoms. The promotion of immune regulation would likely be a safer and gentler treatment approach. Thus, therapies that can decrease leukocyte activation (e.g., macrophage hyperactivity), restore lymphocyte balance, dampen pro-inflammatory cytokines, and assist in additional pathways involved in the COVID-19 are likely to benefit the treatment. Future research will clarify the intricacies of this disease pathogenesis and the appropriate therapeutic strategies according to disease stages.
The effects of artemisinins have been investigated in models of asthma, chronic obstructive pulmonary diseases, lung cancer, nasopharyngeal carcinoma, and acute lung injury [2]. A. annua derivatives have shown antioxidant, anti-inflammatory, anti-pulmonary fibrosis, antimetastatic, anti-angiogenic, antimucus, and anti-tumor proliferation activities that happen through several signaling pathways benefitting these respiratory diseases [2]. Notably, artemisinins can regulate the expression of pro-inflammatory cytokines, NF-κB, MMPs, and VEGFs [2]. In general, the anti-inflammatory properties of the artemisinins are due to inhibition of toll-like receptors, Syk tyrosine kinase, phospholipase C-γ, phosphoinositide 3-kinase/protein kinase B (PI3K/Akt), mitogen-activated protein kinase (MAPK), signal transducer and activator of transcription (STAT)-1/3/5, NF-κB, specificity protein 1 (Sp1), and nuclear erythroid 2-related factor 2/antioxidant response element (Nrf2/ARE) signaling pathways [63]. In an RA study, artesunate downregulated the secretion of IL-1β, IL-6, and IL-8 through the inhibition of NF-κB and the regulation of the PI3K signaling pathway [64]. Also, the flavonoids casticin and chrysosphenol-D, the monoterpene 1,8-cineol, and rosmarinic and chlorogenic acids of A. annua have shown anti-inflammatory properties [65]. On another note, in a murine model of ulcerative colitis, the artemisinin analogue SM934 suppressed neutrophils and macrophages in the colon tissues, inhibited NF-κB, and decreased pro-inflammatory cytokines (IL-1β, IL-6, and TNF-α) [66]. Artesunate affected innate immunity in vivo and in vitro by suppressing macrophages, dendritic cells, IL-12, and TNF-α in another study of colitis [67]. Also, dihydroarteannuin improved the symptoms of SLE by inhibiting TNF-α in the macrophages of mice in vitro and in vivo and by blocking the NF-κB nuclear translocation in vivo [68]. Thus, these studies exemplify the potentially beneficial influence of A. annua and its phytochemicals on the innate immune response of the host. In another animal model of leishmaniasis, A. annua leaves and seeds were administered for ten days. The results showed immunomodulation biased toward an increase of IFN-γ, CD4+, and CD8+ T cell activity, and a decrease of Th2 cytokines (IL-4 and IL-10), compared with the untreated infected controls [69]. In an animal model of protozoan infection (Toxoplasma gondii and Plasmodium berghei), DHA increased the circulating T helper and splenic CD8+ T cells while decreasing the splenic and circulatory B cells, thus providing an immunomodulatory response [70]. Also, the expression of TNF-α, IFN-γ, IL-2, IL-6, IL-4, and IL-13 was significantly reduced by DHA, while Th2 cytokines (e.g., IL-5) were increased in this study [70]. Although these investigated pathogens are not viruses, these studies point to the potential of A. annua and artemisinins as modulators of the host immune response to give patients a better fighting chance against the COVID-19. Factors such as dosage, concentration, and frequency of drug administration may influence the final therapeutic effect and need to be examined [59]. Given the favorable effects of artemisinins in inflammation, immunomodulation, and models of respiratory diseases, their role in the management of COVID-19 should be explored. A. annua and artemisinins may provide support in alleviating the cytokine storm and ARDS during the COVID-19.
4.2. Artemisinin as a p21-activated kinase-1 inhibitor
One review article examined the role of the p21-activated kinase-1 (PAK-1) in the coronavirus pathogenesis and proposed artemisinin as a PAK-1 blocker to act against coronaviruses, suppress lung fibrosis and inflammation, and boost immune function [71]. The authors also looked at the effects of CQ as a PAK-1 inhibitor and suggested that PAK-1-blockers could assist in the current pandemic [71]. PAK-1 is an enzyme encoded by the PAK-1 gene, which belongs to the serine/threonine PAK family and is activated by RAC/CDC42, the RAS-related GTPases (p21) [72], [73]. PAKs are distributed throughout body tissues and are essential for different cellular functions, such as cytoskeletal modeling, focal adhesion assembly, cell migration, survival, mitosis, and transcriptional modulation and protein synthesis involving ERK and NF-κB [72]. The PAK-1 activity has been linked to a variety of diseases, such as cancer and tumors, viral and bacterial infections, inflammatory diseases, such as asthma and arthritis, acquired immune deficiency, type 2 diabetes, hypertension, neuronal diseases, such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, depression, and epilepsy, and others [73]. PAK-1 gets activated during the infection in patients with malaria, influenza A, HIV, and HPV and contributes to the replication of these pathogens [73]. Thus, PAK-1 could also offer another route for therapeutic intervention in coronavirus infection. According to the authors, one of the advantages of using PAK1-blockers is that they are not needed for normal cell growth, thus they would cause no side effects to people or animals when treating PAK-1-dependent diseases [73]. Interestingly, another PAK-1 blocker, curcumin, has also shown anti-COVID-19 activity [74]. Therefore, the hypothesis that artemisinin may act as a PAK-1 blocker offers another mode of action against COVID-19.
4.3. Advantages of repurposing of A. annua and its phytocompounds for the SARS-CoV-2
A series of docking simulations reviewed herein demonstrated the ability of A. annua phytocompounds to bind SARS-CoV-2/host proteins to hinder the viral replication process. The docking binding strength between the drug or natural compound and proteins, their interaction, fit, and stability are some of the factors that suggest a molecule’s potential as an efficient inhibitor. However, other factors influence which molecules may be good antiviral candidates. The advantages of repurposing artemisinins include low toxicity, safer higher dosages, few side effects, low cost, and easy production [2], [12]. Many artemisinins have gone through pharmacokinetic and pharmacodynamic studies demonstrating good profiles [2]. These phytocompounds have been tested for malaria and are being used through different modes of administration including oral, intravenous, intramuscular, and rectal [2]. Artemisinins can also sensitize other drugs, providing an attractive motive to combine them with other drugs such as the current ACT protocols [2]. Drug-combination therapy targeting different mechanisms may delay drug resistance and increase treatment effectiveness [12]. For instance, the concentration of artesunate that showed effectiveness against HBV, HCMV, and HepG2 2.2.15 cells is similar to the concentration used for malaria [12].
4.4. The use of the A. annua whole plant vs. its isolates
Since A. annua has a variety of phytochemicals that may target different proteins and signaling pathways, the questions of whether using the whole plant instead of its separate isolates would attain better results and if it would reach the desired bioavailability to address the infection are important ones. In a very informative study, researchers examined the absorption, distribution, metabolism, and excretion of the dried leaves of A. annua (DLA) vs. pure artemisinin in vitro and in vivo [65]. They also used a rat model to investigate the effects of these herbal agents on systemic inflammation. The results showed that the whole plant taken orally in the form of extracts and teas made from DLA was more bioavailable than pure artemisinin [65]. These preparations inhibited artemisinin-metabolizing enzymes (CYP2B6 and CYP3A4) which made the DLA-delivered artemisinin pass through the liver unmetabolized, causing it to be more available in the tissues [65]. Also, the artemisinin contained in the DLA had better absorption in the tissues of the female rats. Furthermore, after administration to the lipopolysaccharide-challenged rats, which exhibited systemic inflammation, both DLA and pure artemisinin significantly reduced the serum TNF-α in females, while DLA alone reduced TNF-α and IL-6 in the males. No effects were noticed on the anti-inflammatory cytokine IL-10, suggesting that other signaling pathways may be involved [65]. The authors hypothesized that DLA provided better absorption and bioavailability of its artemisinin due to reduced liver enzyme activity, or it worked through the additive effect of several compounds providing a better outcome. Regardless, it was evident that the use of the whole plant was more effective than the isolated compound [65]. In a Caco-2 permeability assay, the same group of researchers indicated that the intestinal permeability of the DLA-delivered artemisinin was significantly greater by 37% compared with pure artemisinin [75]. This suggested that one or more phytocompounds in A. annua may increase intestinal absorption of artemisinin [75]. In a previous study on the use of DLA for malaria, the authors also showed that artemisinin delivered through DLA was about four times more soluble than the pure artemisinin, and that the essential oil fraction of A. annua contributed to the increase in artemisinin solubility in the DLA [76]. Remarkably, 18 severe malaria patients who were unresponsive to the WHO-approved ACT protocol (Coartem®) and IV artesunate after six months were given DLA tablets (0.5 g DLA per os, twice daily for 5 d), as last resort compassionate care [77]. For those in a coma or too young to swallow, the tablets were crushed, mixed with water, and delivered by nasogastric tube. Of the 18 ACT-resistant patients, all recovered completely with the use of DLA and experienced no side effects. Thus, due to the known low bioavailability and possibility of resistance of artemisinin, which poses some disadvantages to its use, the A. annua whole-plant could be a valuable and inexpensive resource to assist with the coronavirus pandemic. Not surprisingly, traditional medicines have been using whole plant extracts for thousands of years, and science has been re-discovering its benefits. The compounds in the DLA tablet analyzed in this study consisted of artemisinin (10.97 mg), deoxyartemisinin (2.54 mg), arteannuin B (0.89 mg), artemisinin acid (1.30 mg), and dihydroartemisinic acid (3.91 mg). It also contained chlorogenic acid 10.94 mg, coumarins (scopoletin 2.47 mg), flavonoids (18.32 mg), and non-artemisinic terpenes (camphor 0.27 mg, phytol 0.56 mg) [77]. Some of these compounds may work synergistically with artemisinin, the target proteins of the SARS-CoV-2/host, and signaling pathways of COVID-19.
The combination of A. annua’s biological properties, action on different signaling pathways and target proteins, and a combined therapy approach may synergistically inhibit the SARS-CoV-2 and assist in the elaboration of therapies to attenuate the COVID-19 symptoms. The addition of other herbs and phytochemicals that have also shown antiviral activity, anti-coronavirus and host protein inhibition, and other properties may contribute to designing therapies that modulate the host immune system to better fight the infection [21]. A big limitation of the present review is the small number of retrieved peer-reviewed studies. The different methodologies, algorithms, and software used by the docking studies also limited the comparison of results.
5. Conclusion
A. annua and its phytocompounds may be able to inhibit the SARS-CoV-2/host proteins 3CLPRO, S protein, CTSL, N protein, E protein, helicase protein, nsp3, nsp10, nsp14, nsp15, and GRP78 receptor. Thus, collectively, the A. annua constituents may impede the SARS-CoV-2 attachment, membrane fusion, internalization into the host cell, and hinder the viral replication and transcription process. The best inhibitions were attained by the interactions of artesunate-3CLPRO (−8.0 kcal/mol), artemisinin-3CLPRO (−8.06 kcal/mol), artemether-N protein (−8.0 kcal/mol), artesunate-N protein (−8.8 kcal/mol), artesunate-nsp3 (−8.1 kcal/mol), artesunate-nsp14 (−8.4 kcal/mol), and artesunate-nsp15 (−8.2 kcal/mol). Other appreciable scores of ≤ −7.0 kcal/mol were also reported (Table 1/Fig. 1). To date, no peer-reviewed articles confirmed a significant binding of A. annua and its compounds directly with RdRp, PLPRO, hACE-2 receptor, S/ACE-2 complex, 3a protein, or additional non-structural proteins that are important for the coronavirus replication. Nonetheless, the research on the SARS-CoV-2/COVID-19 is emerging rapidly with new peer-reviewed papers being published weekly, and other yet unknown target proteins may be involved. Not only the binding strength of an herb-compound to the viral proteins is important, but also the type of bond, fit, and stability should be considered when electing anti-SARS-CoV-2 options. However, the in vitro and in silico studies alone are not enough to determine the best antiviral candidates, and other factors need to be considered. Although in some studies the artemisinins did not show the highest binding capacity, the combination of their antiviral action, target-protein inhibition, and biological properties may synergistically contribute to the treatment of SARS-CoV-2 and intervene in additional signaling pathways to favorably influence the COVID-19 pathogenesis. Importantly, A. annua may re-modulate the host’s innate and adaptive immune system and assist in reducing the cytokine storm, ARDS, and COVID-19 symptoms. Advantages for repurposing artemisinins include low toxicity, safer higher dosages, few side-effects, cost-effectiveness, easy production, pre-existing pharmacokinetic and pharmacodynamic studies showing good profiles, well-understood modes of administration, drug sensitization to design drug and/or herbal-combination therapies, reduction of drug resistance (when drugs are associated), and dosage predictability based on other disease protocols. The evidence reviewed here supports future research and clinical trials to further understand the use of A. annua for the SARS-CoV-2 infection, COVID19 prevention, reduction of severity, treatment of different phases of the disease, and management of symptoms.
Funding
No funding was received for this study.
Declaration of competing interest
The author declares that there is no conflict of interest.
Acknowledgements
I would like to thank Dr. Atanu Singha Roy, B.Sc., M.Sc., Ph.D., Assistant Professor and Head of the Department of Chemistry, National Institute of Technology Meghalaya, Shillong, India, for carefully reading the manuscript especially regarding the results of the in silico molecular docking studies and suggesting additional improvements to the manuscript.
References
- 1.Aftab T., Ferreira J.S., Khan M., Naeem M. Springer; New York: 2014. Artemisia annua pharmacology and biotechnology. [Google Scholar]
- 2.Cheong D.H.J., Tan D.W.S., Wong F.W.S., Tran T. Antimalarial drug, artemisinin and its derivatives for the treatment of respiratory diseases. Pharmacol Res. 2020;158:104901. doi: 10.1016/j.phrs.2020.104901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uzun T., Toptas O. Artesunate: could be an alternative drug to chloroquine in COVID-19 treatment? Chin Med. 2020;15:54. doi: 10.1186/s13020-020-00336-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Touret F., de Lamballerie X. Of chloroquine and COVID-19. Antivir Res. 2020;177:104762. doi: 10.1016/j.antiviral.2020.104762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dyall J., Gross R., Kindrachuk J., Johnson R.F., Olinger G.G., Hensley L.E., et al. Middle East respiratory syndrome and severe acute respiratory syndrome: current therapeutic options and potential targets for novel therapies. Drugs. 2017;77(18):1935–1966. doi: 10.1007/s40265-017-0830-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao X, Ye F, Zhang M, Cui C, Huang B, Niu P, et al. In vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis 2020; 71(15):732–9. [DOI] [PMC free article] [PubMed]
- 7.Sehailia M., Chemat S. Antimalarial-agent artemisinin and derivatives portray more potent binding to Lys353 and Lys31-binding hotspots of SARS-CoV-2 spike protein than hydroxychloroquine: potential repurposing of artenimol for COVID-19. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1796809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fantini J., Di Scala C., Chahinian H., Yahi N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int J Antimicrob Agents. 2020;55(5):105960. doi: 10.1016/j.ijantimicag.2020.105960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juurlink D.N. Safety considerations with chloroquine, hydroxychloroquine and azithromycin in the management of SARS-CoV-2 infection. CMAJ. 2020;192(17):E450–E453. doi: 10.1503/cmaj.200528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah S., Das S., Jain A., Misra D.P., Negi V.S. A systematic review of the prophylactic role of chloroquine and hydroxychloroquine in coronavirus disease-19 (COVID-19) Int J Rheuma Dis. 2020;23(5):613–619. doi: 10.1111/1756-185X.13842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rodrigo C., Fernando S.D., Rajapakse S. Clinical evidence for repurposing chloroquine and hydroxychloroquine as antiviral agents: a systematic review. Clin Microbiol Infect. 2020;26(8):979–987. doi: 10.1016/j.cmi.2020.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Efferth T., Romero M., Wolf D., Stamminger T., Marin J.G., Marschall M. The antiviral activities of artemisinin and artesunate. Clin Infect Dis. 2008;47(6):804–811. doi: 10.1086/591195. [DOI] [PubMed] [Google Scholar]
- 13.Kapepula PM, Kabengele JK, Kingombe M, Bambeke FV, Tulkens PM, Kishabongo AS, et al. Artemisia Spp. derivatives for COVID-19 treatment: anecdotal use, political hype, treatment potential, challenges, and road map to randomized clinical trials. Am J Trop Med Hyg 2020; 103(3): 960–4. [DOI] [PMC free article] [PubMed]
- 14.Bilia A.R., Santomauro F., Sacco C., Bergonzi M.C., Donato R. Essential oil of Artemisia annua L.: an extraordinary component with numerous antimicrobial properties. Evid Based Complement Altern Med. 2014;2014:1–7. doi: 10.1155/2014/159819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Septembre-Malaterre A., Lalarizo Rakoto M., Marodon C., Bedoui Y., Nakab J., Simon E., et al. Artemisia annua, a traditional plant brought to light. Int J Mol Sci. 2020;21(14):4986. doi: 10.3390/ijms21144986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira J.F.S., Luthria D.L., Sasaki T., Heyerick A. Flavonoids from Artemisia annua L. as antioxidants and their potential synergism with artemisinin against malaria and cancer. Molecules. 2010;15(5):3135–3170. doi: 10.3390/molecules15053135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma L., Yao L. Antiviral effects of plant-derived essential oils and their components: an updated review. Molecules. 2020;25(11):2627. doi: 10.3390/molecules25112627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Auerochs S., Korn K., Marschall M. A reporter system for Epstein-Barr virus (EBV) lytic replication: anti-EBV activity of the broad anti-herpesviral drug artesunate. J Virol Methods. 2011;173(2):334–339. doi: 10.1016/j.jviromet.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 19.Lubbe A., Seibert I., Klimkait T., van der Kooy F. Ethnopharmacology in overdrive: the remarkable anti-HIV activity of Artemisia annua. J Ethnopharmacol. 2012;141(3):854–859. doi: 10.1016/j.jep.2012.03.024. [DOI] [PubMed] [Google Scholar]
- 20.Li L., Gu J., Shi X., Gong E., Li X., Shao H., et al. Biosafety level 3 laboratory for autopsies of patients with severe acute respiratory syndrome: principles, practices, and prospects. Clin Infect Dis. 2005;41(6):815–821. doi: 10.1086/432720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuzimoto A.D., Isidoro C. The antiviral and coronavirus-host protein pathways inhibiting properties of herbs and natural compounds—additional weapons in the fight against the COVID-19 pandemic? J Tradit Complement Med. 2020;10(4):405–419. doi: 10.1016/j.jtcme.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poltronieri P., Sun B., Mallardo M. RNA viruses: RNA roles in pathogenesis, coreplication and viral load. Curr Genomics. 2015;16(5):327–335. doi: 10.2174/1389202916666150707160613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J., Zhang C., Gong M., Wang M. Combination of artemisinin-based natural compounds from Artemisia annua L. for the treatment of malaria: pharmacodynamic and pharmacokinetic studies. Phytother Res. 2018;32(7):1415–1420. doi: 10.1002/ptr.6077. [DOI] [PubMed] [Google Scholar]
- 24.Rath K., Taxis K., Walz G., Gleiter C.H., Li S.M., Heide L. Pharmacokinetic study of artemisinin after oral intake of a traditional preparation of Artemisia annua L (annual wormwood) Am Trop Med Hyg. 2004;70(2):128–132. [PubMed] [Google Scholar]
- 25.Radulovic N.S., Randjelovic P.J., Stojanovic N.M., Blagojevic P.D., Stojanovic-Radic Z.Z., Ilic I.R., et al. Toxic essential oils. Part II: chemical, toxicological, pharmacological and microbiological profiles of Artemisia annua L. volatiles. Food Chemm Toxicol. 2013;58:37–49. doi: 10.1016/j.fct.2013.04.016. [DOI] [PubMed] [Google Scholar]
- 26.World Health Organization. Coronavirus disease (COVID-19), weekly epidemiological update 1. (2020-08-24) [2021-01-01]. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200824-weekly-epi-update.pdf?sfvrsn=806986d1_4.
- 27.Wu C., Chen X., Cai Y., Xia J., Zhou X., Xu S., et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan. China. JAMA Intern Med. 2020;180(7):934. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barnes BJ, Adrover JM, Baxter-Stolzfus A, Borczuk A, Cools-Lartigue J, Crawford JM, et al. Targeting potential drivers of COVID-19: neutrophil extracellular traps. J Exp Med 2020; 217(6): e20200652. [DOI] [PMC free article] [PubMed]
- 29.Gousseff M., Penot P., Gallay L., Batisse D., Benech N., Bouiller K., et al. Clinical recurrences of COVID-19 symptoms after recovery: viral relapse, reinfection or inflammatory rebound? J Infect. 2020;81(5):816–846. doi: 10.1016/j.jinf.2020.06.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kang H., Wang Y., Tong Z., Liu X. Retest positive for SARS-CoV-2 RNA of “recovered” patients with COVID-19: persistence, sampling issues, or re-infection? J Med Virol. 2020;92(11):2263–2265. doi: 10.1002/jmv.26114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cothran T.P., Kellman S., Singh S., Beck J.S., Powell K.J., Bolton C.J., et al. A brewing storm: the neuropsychological sequelae of hyperinflammation due to COVID-19. Brain Behav Immun. 2020;88:957–958. doi: 10.1016/j.bbi.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Troyer E.A., Kohn J.N., Hong S. Are we facing a crashing wave of neuropsychiatric sequelae of COVID-19? Neuropsychiatric symptoms and potential immunologic mechanisms. Brain Behav Immun. 2020;87:34–39. doi: 10.1016/j.bbi.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vojdani A., Kharrazian D. Potential antigenic cross-reactivity between SARS-CoV-2 and human tissue with a possible link to an increase in autoimmune diseases. Clin Immunol. 2020;217:108480. doi: 10.1016/j.clim.2020.108480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marino Gammazza A., Légaré S., Lo Bosco G., Fucarino A., Angileri F., Conway de Macario E., et al. Human molecular chaperones share with SARS-CoV-2 antigenic epitopes potentially capable of eliciting autoimmunity against endothelial cells: possible role of molecular mimicry in COVID-19. Cell Stress Chaperones. 2020;25(5):737–741. doi: 10.1007/s12192-020-01148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cappello F., Gammazza A.M., Dieli F., de Macario E., Macario A.J. Does SARS-CoV-2 trigger stress-induced autoimmunity by molecular mimicry? A hypothesis. J Clin Med. 2020;9(7):2038. doi: 10.3390/jcm9072038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lucchese G., Flöel A. SARS-CoV-2 and Guillain-Barré syndrome: molecular mimicry with human heat shock proteins as potential pathogenic mechanism. Cell Stress Chaperones. 2020;25(5):731–735. doi: 10.1007/s12192-020-01145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li S., Chen C., Zhang H., Guo H., Wang H., Wang L., et al. Identification of natural compounds with antiviral activities against SARS-associated coronavirus. Antivir Res. 2005;67(1):18–23. doi: 10.1016/j.antiviral.2005.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang S.Q., Du Q.S., Zhao K., Li A.X., Wei D.Q., Chou K.C. Virtual screening for finding natural inhibitor against cathepsin-L for SARS therapy. Amino Acids. 2007;33(1):129–135. doi: 10.1007/s00726-006-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Das S., Sarmah S., Lyndem S., Roy A.S. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. J Biomol Struct Dyn. 2020;13:1–11. doi: 10.1080/07391102.2020.1763201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alsaffar D., Yaseen A., Jabal G. In silico molecular docking studies of medicinal Arabic plant-based bioactive compounds as a promising drug candidate against COVID-19. Int J Innov Sci Res Technol. 2020;5(5):876–896. [Google Scholar]
- 41.Peele K.A., Potla Durthi C., Srihansa T., Krupanidhi S., Ayyagari V.S., Babu D.J., et al. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Inform Med Unlocked. 2020;19:100345. doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cao R., Hu H., Li Y., Wang X.i., Xu M., Liu J., et al. Anti-SARS-CoV-2 potential of artemisinins in vitro. ACS Infect Dis. 2020;6(9):2524–2531. doi: 10.1021/acsinfecdis.0c00522. [DOI] [PubMed] [Google Scholar]
- 43.Gendrot M., Duflot I., Boxberger M., Delandre O., Jardot P., Le Bideau M., et al. Antimalarial artemisinin-based combination therapies (ACT) and COVID-19 in Africa: in vitro inhibition of SARS-CoV-2 replication by mefloquine-artesunate. Int J Infect Dis. 2020;99:437–440. doi: 10.1016/j.ijid.2020.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gendrot M., Andreani J., Boxberger M., Jardot P., Fonta I., Le Bideau M., et al. Antimalarial drugs inhibit the replication of SARS-CoV-2: an in vitro evaluation. Travel Med Infect Dis. 2020;37:101873. doi: 10.1016/j.tmaid.2020.101873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.da Silva J.K.R., Figueiredo P.L.B., Byler K.G., Setzer W.N. Essential oils as antiviral agents, potential of essential oils to treat SARS-CoV-2 infection: an in silico investigation. Int J Mol Sci. 2020;21(10):3426. doi: 10.3390/ijms21103426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sudeep H.V., Gouthamchandra K., Shyamprasad K. Molecular docking analysis of withaferin A from Withania somnifera with the glucose regulated protein 78 (GRP78) receptor and the SARS-CoV-2 main protease. Bioinformation. 2020;16(5):411–417. doi: 10.6026/97320630016411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li G., Yuan M., Li H., Deng C., Wang Q.i., Tang Y., et al. Safety and efficacy of artemisinin-piperaquine for treatment of COVID-19: an open-label, non-randomised and controlled trial. Int J Antimicrob Agents. 2021;57(1):106216. doi: 10.1016/j.ijantimicag.2020.106216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharma S., Deep S. In silico drug repurposing for targeting SARS-CoV-2 main protease (Mpro) J Biolol Struct Dyn. 2020:1–8. doi: 10.1080/07391102.2020.1844058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prashantha C.N., Gouthami K., Lavanya L., Bhavanam S., Jakhar A., Shakthiraju R.G., et al. Molecular screening of antimalarial, antiviral, anti-inflammatory and HIV protease inhibitors against spike glycoprotein of coronavirus. J Mol Graph Model. 2021;102:107769. doi: 10.1016/j.jmgm.2020.107769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roy Chattopadhyay N., Chatterjee K., Banerjee A., Choudhuri T. Combinatorial therapeutic trial plans for COVID-19 treatment armed up with antiviral, antiparasitic, cell-entry inhibitor, and immune-boosters. Virusdisease. 2020;31(4):479–489. doi: 10.1007/s13337-020-00631-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alazmi M., Motwalli O. Molecular basis for drug repurposing to study the interface of the S protein in SARS-CoV-2 and human ACE2 through docking, characterization, and molecular dynamics for natural drug candidates. J Mol Model. 2020;26(12):338. doi: 10.1007/s00894-020-04599-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Khan R.J., Jha R.K., Singh E., Jain M., Amera G.M., Singh R.P., et al. Identification of promising antiviral drug candidates against non-structural protein 15 (NSP15) from SARS-CoV-2: an in silico assisted drug-repurposing study. J Biomol Struct Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1814870. [DOI] [PubMed] [Google Scholar]
- 53.Marak B.N., Dowarah J., Khiangte L., Singh V.P. Step toward repurposing drug discovery for COVID-19 therapeutics through in silico approach. Drug Dev Res. 2021;82(3):374–392. doi: 10.1002/ddr.21757. [DOI] [PubMed] [Google Scholar]
- 54.Gupta S, Singh V, Varadwaj PK, Chakravartty N, Katta AVSKM, Lekkala SP, et al. Secondary metabolites from spice and herbs as potential multitarget inhibitors of SARS-CoV-2 proteins. J Biomol Struct Dyn 2020; 1–20. [DOI] [PMC free article] [PubMed]
- 55.Cong Y., Ulasli M., Schepers H., Mauthe M., V’kovski P., Kriegenburg F., et al. Nucleocapsid protein recruitment to replication-transcription complexes plays a crucial role in coronaviral life cycle. J Virol. 2020;94(4) doi: 10.1128/JVI.01925-19. e01925–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meng X.Y., Zhang H.X., Mezei M., Cui M. Molecular docking: a powerful approach for structure-based drug discovery. Curr Comput Aided Drug Des. 2011;7(2):146–157. doi: 10.2174/157340911795677602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu C., Liu Y., Yang Y., Zhang P., Zhong W.u., Wang Y., et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pham Sin B. 2020;10(5):766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ibrahim I.M., Abdelmalek D.H., Elshahat M.E., Elfiky A.A. COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect. 2020;80(5):554–562. doi: 10.1016/j.jinf.2020.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paces J., Strizova Z., Smrz D., Cerny J. COVID-19 and the immune system. Physiol Res. 2020;69(3):379–388. doi: 10.33549/physiolres.934492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye Q., Wang B., Mao J. The pathogenesis and treatment of the “Cytokine Storm” in COVID-19. J Infect. 2020;80(6):607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ueland T., Holter J.C., Holten A.R., Müller K.E., Lind A., Bekken G.K., et al. Distinct and early increase in circulating MMP-9 in COVID-19 patients with respiratory failure. J Infect. 2020;81(3):e41–e43. doi: 10.1016/j.jinf.2020.06.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yin X.X., Zheng X.R., Peng W., Wu M.L., Mao X.Y. Vascular endothelial growth factor (VEGF) as a vital target for brain inflammation during the COVID-19 outbreak. ACS Chem Neurosci. 2020;11(12):1704–1705. doi: 10.1021/acschemneuro.0c00294. [DOI] [PubMed] [Google Scholar]
- 63.Ho W.E., Peh H.Y., Chan T.K., Wong W.S.F. Artemisinins: pharmacological actions beyond anti-malarial. Pharmacol Ther. 2014;142(1):126–139. doi: 10.1016/j.pharmthera.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 64.Xu H., He Y., Yang X., Liang L., Zhan Z., Ye Y., et al. Antimalarial agent artesunate inhibits TNF-α-induced production of proinflammatory cytokines via inhibition of NF-κB and PI3 kinase/Akt signal pathway in human rheumatoid arthritis fibroblast-like synoviocytes. Rheumatology (Oxford) 2007;46(6):920–926. doi: 10.1093/rheumatology/kem014. [DOI] [PubMed] [Google Scholar]
- 65.Desrosiers M.R., Mittelman A., Weathers P.J. Dried leaf Artemisia annua improves bioavailability of artemisinin via cytochrome P450 inhibition and enhances artemisinin efficacy downstream. Biomolecules. 2020;10(2):254. doi: 10.3390/biom10020254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yan Y.X., Shao M.J., Qi Q., Xu Y.S., Yang X.Q., Zhu F.H., et al. Artemisinin analogue SM934 ameliorates DSS-induced mouse ulcerative colitis via suppressing neutrophils and macrophages. Acta Pharmacol Sin. 2018;39(10):1633–1644. doi: 10.1038/aps.2017.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sun W., Han X., Wu S., Wu J., Yang C., Li X. Unexpected mechanism of colitis amelioration by artesunate, a natural product from Artemisia annua L. Inflammopharmacology. 2020;28(4):851–868. doi: 10.1007/s10787-019-00678-2. [DOI] [PubMed] [Google Scholar]
- 68.Li W.D., Dong Y.J., Tu Y.Y., Lin Z.B. Dihydroarteannuin ameliorates lupus symptom of BXSB mice by inhibiting production of TNF-α and blocking the signaling pathway NF-κB translocation. Int Immunopharmacol. 2006;6(8):1243–1250. doi: 10.1016/j.intimp.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 69.Islamuddin M., Chouhan G., Farooque A., Dwarakanath B.S., Sahal D., Afrin F., et al. Th1-biased immunomodulation and therapeutic potential of Artemisia annua in murine visceral leishmaniasis. PLoS Negl Trop Dis. 2015;9(1):e3321. doi: 10.1371/journal.pntd.0003321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang T., Zhang Y., Jiang N., Zhao X.u., Sang X., Yang N.a., et al. Dihydroartemisinin regulates the immune system by promotion of CD8+ T lymphocytes and suppression of B cell responses. Sci China Life Sci. 2020;63(5):737–749. doi: 10.1007/s11427-019-9550-4. [DOI] [PubMed] [Google Scholar]
- 71.Maruta H., He H. PAK1-blockers: potential therapeutics against COVID-19. Med Drug Discov. 2020;6:100039. doi: 10.1016/j.medidd.2020.100039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kichina J.V., Goc A., Al-Husein B., Somanath P.R., Kandel E.S. PAK1 as a therapeutic target. Expert Opin Ther Targets. 2010;14(7):703–725. doi: 10.1517/14728222.2010.492779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Maruta H. Herbal therapeutics that block the oncogenic kinase PAK1: a practical approach towards PAK1-dependent diseases and longevity. Phytother Res. 2014;28(5):656–672. doi: 10.1002/ptr.5054. [DOI] [PubMed] [Google Scholar]
- 74.Zahedipour F., Hosseini S.A., Sathyapalan T., Majeed M., Jamialahmadi T., Al-Rasadi K., et al. Potential effects of curcumin in the treatment of COVID-19 infection. Phytother Res. 2020;34(11):2911–2920. doi: 10.1002/ptr.6738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Desrosiers M.R., Weathers P.J. Artemisinin permeability via Caco-2 cells increases after simulated digestion of Artemisia annua leaves. J Ethnopharmacol. 2018;210:254–259. doi: 10.1016/j.jep.2017.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Desrosiers M.R., Weathers P.J. Effect of leaf digestion and artemisinin solubility for use in oral consumption of dried Artemisia annua leaves to treat malaria. J Ethnopharmacol. 2016;190:313–318. doi: 10.1016/j.jep.2016.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Daddy N.B., Kalisya L.M., Bagire P.G., Watt R.L., Towler M.J., Weathers P.J. Artemisia annua dried leaf tablets treated malaria resistant to ACT and i.v. artesunate: case reports. Phytomedicine. 2017;32:37–40. doi: 10.1016/j.phymed.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]