Supplemental Digital Content is available in the text.
Keywords: artificial intelligence, coronavirus disease 2019, intensive care unit, machine learning
OBJECTIVES:
Critically ill patients with coronavirus disease 2019 have variable mortality. Risk scores could improve care and be used for prognostic enrichment in trials. We aimed to compare machine learning algorithms and develop a simple tool for predicting 28-day mortality in ICU patients with coronavirus disease 2019.
DESIGN:
This was an observational study of adult patients with coronavirus disease 2019. The primary outcome was 28-day inhospital mortality. Machine learning models and a simple tool were derived using variables from the first 48 hours of ICU admission and validated externally in independent sites and temporally with more recent admissions. Models were compared with a modified Sequential Organ Failure Assessment score, National Early Warning Score, and CURB-65 using the area under the receiver operating characteristic curve and calibration.
SETTING:
Sixty-eight U.S. ICUs.
PATIENTS:
Adults with coronavirus disease 2019 admitted to 68 ICUs in the United States between March 4, 2020, and June 29, 2020.
INTERVENTIONS:
None.
MEASUREMENTS AND MAIN RESULTS:
The study included 5,075 patients, 1,846 (36.4%) of whom died by day 28. eXtreme Gradient Boosting had the highest area under the receiver operating characteristic curve in external validation (0.81) and was well-calibrated, while k-nearest neighbors were the lowest performing machine learning algorithm (area under the receiver operating characteristic curve 0.69). Findings were similar with temporal validation. The simple tool, which was created using the most important features from the eXtreme Gradient Boosting model, had a significantly higher area under the receiver operating characteristic curve in external validation (0.78) than the Sequential Organ Failure Assessment score (0.69), National Early Warning Score (0.60), and CURB-65 (0.65; p < 0.05 for all comparisons). Age, number of ICU beds, creatinine, lactate, arterial pH, and Pao2/Fio2 ratio were the most important predictors in the eXtreme Gradient Boosting model.
CONCLUSIONS:
eXtreme Gradient Boosting had the highest discrimination overall, and our simple tool had higher discrimination than a modified Sequential Organ Failure Assessment score, National Early Warning Score, and CURB-65 on external validation. These models could be used to improve triage decisions and clinical trial enrichment.
Coronavirus disease 2019 (COVID-19) has infected over 30 million people and killed more than 500,000 in the United States as of June 2021 (1). Most deaths have occurred in the approximate 11% of patients requiring ICU admission (2). In some locations, the number of critically ill patients has exceeded ICU capacity, and frameworks have been proposed that allocate scarce resources based on maximizing benefit (3–5). This includes some states incorporating severity of illness scores, such as the Sequential Organ Failure Assessment (SOFA) score, into triage criteria for ventilator allocation (6). Resource utilization, ICU triage, and goals of care discussions could be facilitated by an objective model that accurately predicts mortality. Although prior studies have identified risk factors for severe disease and a growing number of studies have published prognostic models, most have been limited by modest sample sizes from single health systems and lack of both external and temporal generalizability assessment (7–12). As a result, no validated prognostic models are in widespread use today.
Prediction models in medicine have historically relied on logistic regression (13). However, more flexible machine learning methods have led to the development of highly accurate models. For example, recent studies have demonstrated that machine learning algorithms, such as random forests and deep neural networks, can predict acute conditions in hospitalized patients more accurately than traditional logistic regression (14, 15). Nevertheless, it is difficult to determine in advance which method will work best for a specific problem (16).
Given the paucity of rigorously validated prediction models for mortality in critically ill patients with COVID-19, we aimed to develop and both externally and temporally validate a simple tool for predicting 28-day mortality that could be calculated at the bedside using a dataset of patients admitted to the ICU in 68 hospitals across the United States as part of the Study of the Treatment and Outcomes in critically ill Patients with COVID-19 (STOP-COVID) (9). We also aimed to compare different machine learning approaches for predicting 28-day mortality. Finally, we aimed to identify risk factors for death using interpretable machine learning approaches.
MATERIALS AND METHODS
Study Design and Patient Population
STOP-COVID is a multicenter cohort study that enrolled consecutive adult ICU patients with COVID-19 from 68 U.S. hospitals, including a variety of hospital sizes and types across a wide geographic range (9). The list of participating sites is shown in eTable 1 (http://links.lww.com/CCX/A759). Adult patients (≥ 18 yr) with laboratory-confirmed COVID-19 admitted to an ICU at a participating site between March 4, 2020, and June 29, 2020, were eligible for inclusion. Patients were followed until death, hospital discharge, or at least 28 days after ICU admission. The Institutional Review Boards at Partners Human Research Committee (2007P000003) and the University of Wisconsin (2019-1124) approved the study with waiver of informed consent. Additional details are found in the eMethods (http://links.lww.com/CCX/A759).
Data Collection
Study personnel at each site performed manual chart review using a standardized case report form. These data included demographic information, comorbidities, symptoms, vital signs on ICU admission, longitudinal laboratory values and physiologic parameters, and outcomes. Hospital-level data included the number of pre-COVID ICU beds. Definitions of key variables and outcomes are shown in eTable 2 (http://links.lww.com/CCX/A759), and the complete list of variables is shown in the Case Report Form in the Supplemental Material (http://links.lww.com/CCX/A758).
Outcomes
The primary outcome was inhospital death within 28 days of ICU admission. Patients discharged alive before day 28 were assumed to be alive at day 28. We confirmed the validity of this assumption in a subset of patients (eMethods, http://links.lww.com/CCX/A759).
Predictor Variables
Variables from the first 2 days of ICU admission were included based on missing data considerations in the training data. Because data collection days were based on calendar dates, utilizing 2 days of data ensured that all patients would have at least 24 hours of data for use in the models. Candidate variables excluded from consideration due to greater than 50% missing values included interleukin-6 and fibrinogen. Variables with low frequency categories (< 5% positive) were also excluded (e.g., mechanical cardiac support devices). The final variable list is shown in eTable 3 (http://links.lww.com/CCX/A759) and included age, vital signs and respiratory support on ICU admission, Fio2 among patients requiring invasive mechanical ventilation, laboratory values, and organ support. If values were collected on both days 1 and 2, the worst value was used. Bagged trees were used for missing value imputation for the machine learning models using the caret package in R (The R Foundation for Statistical Computing, Vienna, Austria), which takes into account interactions and nonlinearity automatically (17).
Splitting Into Training and Testing Data
To estimate external generalizability of the models (primary analysis), the 68 ICUs were randomly separated into two groups, with the models developed using training data from admissions during March 2020 and April 2020 in 75% of the hospitals and tested in 25% of the hospitals. A temporal validation was also performed as a secondary analysis by validating these same trained models in data from all admissions from May 2020 to June 2020. Performance metrics were based on the out-of-sample test set predictions for both the external and temporal validations using the same models developed in the March and April training data.
Machine Learning Methods
Several machine learning algorithms were compared, which are described briefly below and in more detail in the eMethods (http://links.lww.com/CCX/A759) (18). Hyperparameters were selected using 10-fold cross-validation in the training data to maximize the area under the receiver operating characteristic curve (AUC).
Elastic Net Logistic Regression.
This approach combines logistic regression with lasso and ridge regression penalty terms. These penalty terms shrink model coefficients to decrease overfitting and improve generalizability. To account for potential nonlinearity of predictor variables, restricted cubic splines were used (18). This allows the risk of mortality to vary nonlinearly across values of a variable.
eXtreme Gradient Boosting.
Gradient boosted machines (GBMs) are based on decision trees that separate patients with and without the outcome of interest using simple yes-no splits, which can be visualized in the form of a tree (18). GBM builds trees sequentially such that each tree improves model fit by more highly weighting the difficult-to-predict patients. A popular implementation of GBM, called eXtreme Gradient Boosting (XGBoost), was used in this work.
Random Forests.
The random forests algorithm is similar to XGBoost in that it builds an ensemble of decision trees, but instead of building them sequentially, it builds each tree separately based on a random sample of the training data (18). Within each tree, only a random number of predictor variables are available for each yes-no split, which results in trees that are different from each other.
Neural Networks.
Neural networks are flexible, nonlinear models that were initially inspired by how the brain works (19). These models are composed of a combination of individual neuron-like units that take the predictor variables as inputs, combine them in hidden layers, transform them through activation functions, and then output predictions (18).
Support Vector Machines.
Support vector machines (SVMs) project the data into multidimensional space based on the variable values for each patient and then create a boundary that attempts to maximize the margin between the patients with and without the outcome (18).
K-Nearest Neighbors.
K-nearest neighbor (KNN) also projects the patients into multidimensional space based on their variable values, but instead of identifying a separating boundary, KNN assigns the outcome of a new patient based on the majority outcome of the K closest training patients (18).
Published Scoring Systems
These machine learning models were compared with the SOFA score, the National Early Warning Score (NEWS), and CURB-65. The SOFA score is commonly used for predicting death in critically ill patients (20, 21). In this study, the SOFA score was modified due to variable availability (eTable 4, http://links.lww.com/CCX/A759), which has been used in prior work (22). NEWS is a vital sign-based early warning score and was modified for this study (eTable 5, http://links.lww.com/CCX/A759) (23). CURB-65 was originally developed for patients with pneumonia and incorporates confusion, blood urea nitrogen greater than 19 mg/dL, respiratory rate greater than or equal to 30 breaths per minute, systolic blood pressure less than 90 mm Hg or diastolic blood pressure less than or equal to 60 mm Hg, and age greater than 65 years (24). The score was modified due to the absence of blood urea nitrogen in the database, a version that has been previously validated to predict outcomes in patients with community-acquired pneumonia (25). For these published scores, missing values were assumed to be normal, as is typically done when they are calculated clinically.
Development of a Simple Clinical Tool
A simple clinical tool was developed by choosing the top 20 variables from the XGBoost model because it had the best discrimination and then predefining variable cutpoints a priori based on clinical knowledge and prior literature (9). Alanine transaminase was excluded due to its high correlation with aspartate transaminase (> 0.85). Missing values were imputed using the mode of each variable category in the training data to make it easier to operationalize at the bedside. Lasso regression was then used for variable selection to develop a parsimonious model with less than or equal to 10 variables, with 10-fold cross-validation in the training data used to optimize the lasso penalty. Model performance was calculated in the out-of-sample external and temporal test data in the same manner as the machine learning models. The final model coefficients were multiplied by a factor of fifteen and rounded for ease of use.
Statistical Analysis
Patient characteristics were compared between those alive versus dead at 28 days using Wilcoxon rank-sum and chi-square tests. Model discrimination was calculated using the AUC and compared using the method of DeLong et al (26). Permutation importance was calculated for each model to determine the most important variables, with the exception of Elastic Net, which used the absolute value of the model coefficients (18). Partial dependence plots were used to illustrate the relationship between variable values and mortality in the model with the highest discrimination (18). Model calibration, which compares the true probability of the outcome versus a model’s predictions, was compared using calibration intercept, slope, and unreliability index p value, with p value of less than 0.05 denoting poor calibration (27). Model Brier scores were also calculated in the test sets, which are defined as the mean squared difference between the model predicted probabilities and the outcome. Two-sided p values less than 0.05 were considered statistically significant. All models were developed using the caret package in R (The R Foundation for Statistical Computing).
RESULTS
Patient Characteristics and Comparisons
A total of 5,075 patients were included in the study, 1,846 (36.4%) of whom died by day 28. Baseline characteristics for patients who died versus survived by day 28 are shown in eTable 6 (http://links.lww.com/CCX/A759) and eTable 7 (http://links.lww.com/CCX/A759). Patients who died were older (median [interquartile range (IQR)] age 67 yr [58–76 yr] vs 59 yr [49–68 yr]), more likely to be male (66% vs 61%), had a lower Pao2/Fio2 (P/F) ratio on ICU admission (median [IQR] 106 mm Hg [76–159 mm Hg] vs 122 mm Hg [84–178 mm Hg]), were more likely to have chronic kidney disease (17% vs 11%), and were more likely to be receiving invasive mechanical ventilation within the first 2 days of ICU admission (71% vs 53%). The distribution of the number of patients per site is shown in eFigure 1 (http://links.lww.com/CCX/A759).
Machine Learning Model and Published Scoring Systems Performance
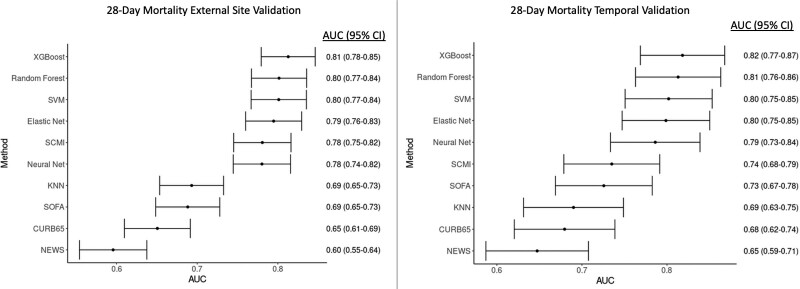
For the primary external validation analysis, 51 sites (n = 3,825 admission) were included as training data and 17 sites were included for independent validation (n = 810 admissions) using data from March 2020 to April 2020. Missing data percentages for the variables included in the study are shown in eTable 3 (http://links.lww.com/CCX/A759). During external validation, the XGBoost model had the highest AUC (0.81; 95% CI, 0.78–0.85), followed by random forests (AUC, 0.80; 95% CI, 0.77–0.84), SVM (AUC, 0.80; 95% CI, 0.77–0.84), Elastic Net (AUC, 0.79; 95% CI, 0.76–0.83), neural network (AUC, 0.78; 95% CI, 0.74–0.82), and KNN (AUC, 0.69; 95% CI, 0.65–0.73; Fig. 1). In addition, the hyperparameter values searched and chosen for each model, software packages used, and methods to calculate probabilities from the models are shown in eTable 8 (http://links.lww.com/CCX/A759). The modified SOFA score had an AUC of 0.69 (95% CI, 0.65–0.73), NEWS had an AUC of 0.60 (95% CI, 0.55–0.64), and CURB-65 had an AUC of 0.65 (95% CI, 0.61–0.69). Pairwise comparisons between XGBoost and the other models using the method of DeLong et al (26) demonstrated statistical improvement in discrimination over the neural network and KNN models (p < 0.05 for AUC comparisons to XGBoost). Results were similar when using inhospital mortality as the model outcome (eFig. 2, http://links.lww.com/CCX/A759). XGBoost, SVM, and Elastic Net were all well-calibrated (unreliability index p > 0.05), in contrast with all of the other models (calibration p < 0.01; eFig. 3, http://links.lww.com/CCX/A759). XGBoost also had the lowest Brier score (0.16; eFig. 3, http://links.lww.com/CCX/A759). Risk of 28-day mortality for SOFA score, NEWS, and CURB-65 values in the external validation dataset are shown in eFigures 4–6 (http://links.lww.com/CCX/A759).
Figure 1.
Comparison of model discrimination between the different models in both the external and temporal validation cohorts. As shown, with point estimates and 95% CIs for the area under the receiver operating characteristic curve (AUC), the eXtreme Gradient Boosting (XGBoost) model had the highest discrimination in both validation datasets. KNN = K-nearest neighbors, NEWS = National Early Warning Score, SCMI = Study of the Treatment and Outcomes in Critically Ill Patients With Coronavirus Disease 2019 Mortality Index, SOFA = Sequential Organ Failure Assessment, SVM = support vector machine.
Results for the secondary temporal validation, which used the same models from the training dataset above (n = 3,825 admission) and validated the models in 440 admissions from May 2020 to June 2020, were similar (Fig. 1), with XGBoost having the highest discrimination (AUC 0.82) and KNN having the lowest (AUC 0.69). XGBoost, SVM, and Elastic Net were all well-calibrated and XGBoost had the lowest Brier score (0.14; eFig. 2, http://links.lww.com/CCX/A759).
Variable Importance
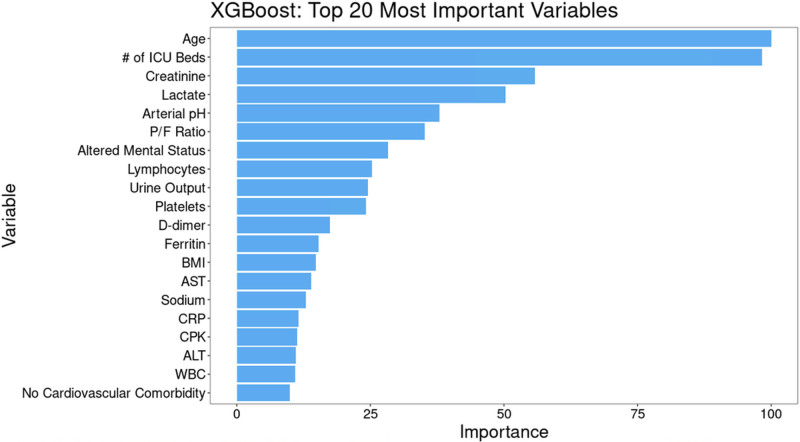
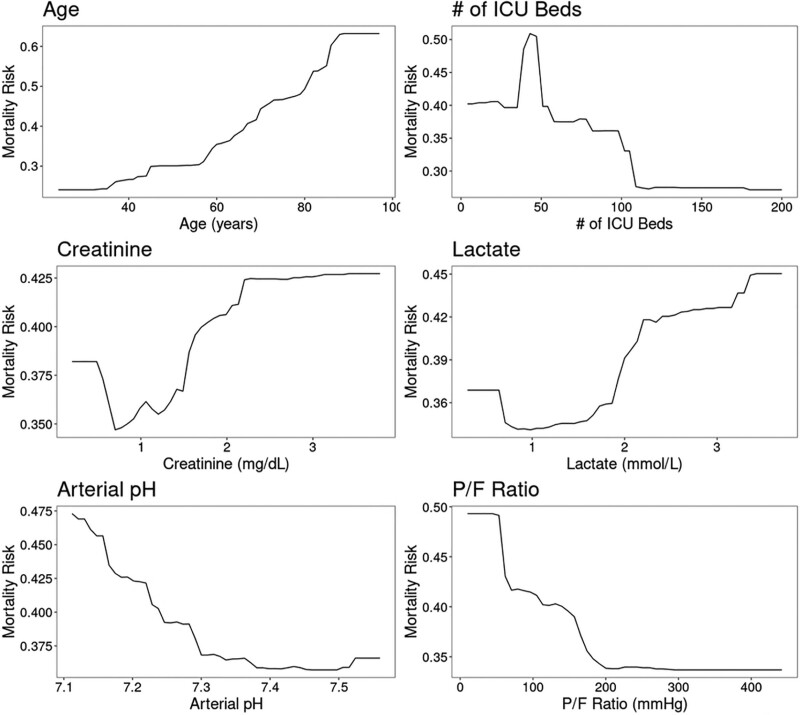
The most important variables in the XGBoost model were age, number of ICU beds, serum creatinine, lactate, arterial pH, and P/F ratio (Fig. 2). This was similar to the other models, with the exception of Elastic Net (eFig. 7, http://links.lww.com/CCX/A759). Partial dependence plots for the most important continuous variables in the XGBoost model are shown in Figure 3. Age showed a relatively flat risk profile up to age 40 after which risk of death increased linearly with increasing age. Risk of death also increased with smaller ICU size, with a rapid increase in risk for hospitals with less than 100 ICU beds.
Figure 2.
Variable importance for the eXtreme Gradient Boosting (XGBoost) model. Permutation variable importance for the most accurate model (XGBoost) scaled to a maximum of 100, which shows that age, number of ICU beds, creatinine, and lactate were the most important variables for predicting 28-d mortality when using data from the first 2 d of ICU admission. ALT = alanine transaminase, AST = aspartate aminotransferase, BMI = body mass index, CPK = creatine phosphokinase, CRP= c-reactive protein, P/F ratio = Pao2/Fio2 ratio.
Figure 3.
Partial dependence plots for eXtreme Gradient Boosting (XGBoost) illustrating the relationship between 28-d mortality and values of the six most important predictor variables. As shown, the risk of mortality increases with age greater than 40 yr, fewer than 100 (and especially < 50) ICU beds, serum creatinine greater than 1 mg/dL, arterial pH less than 7.30, lactate greater than or equal to 2 mmol/L, and Pao2/Fio2 ratio (P/F ratio) less than 150 mm Hg.
Simple Tool
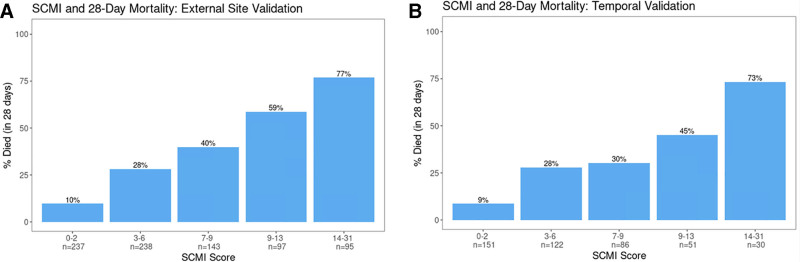
The developed simple tool, hereafter referred to as the STOP-COVID Mortality Index (SCMI), was created using the most important variables from the XGBoost model and is shown in Table 1. The SCMI includes 10 variables, with risk denoted using a point system, and the final score can be calculated as the sum of the individual values for each variable. Figure 4 shows the risk of 28-day mortality across different values of the total sum score, and Table 2 shows accuracy at different score threshold cutoffs. During external validation, the tool had an AUC of 0.78, which was significantly higher than the modified SOFA score (AUC 0.69), NEWS (AUC 0.60), and CRB-65 (AUC 0.65; p < 0.05 for all comparisons). For the secondary temporal validation, the discrimination of SCMI was lower (AUC 0.74) and similar to the SOFA score (AUC 0.73; p = 0.78) but significantly higher than NEWS (AUC 0.65; p < 0.01) and CURB-65 (AUC 0.68; p = 0.04). In both validations, scores between 0 and 2 had a risk of mortality of 9–10%, whereas scores greater than or equal to 14 had very high mortality rates at 28 days (> 70%; Fig. 4). The positive predictive value (PPV) and specificity of a score greater than 10 was 70% and 90%, which increased to 77% and 96% for scores greater than 13 (Table 2).
TABLE 1.
Study of the Treatment and Outcomes in Critically Ill Patients With Coronavirus Disease 2019 Mortality Index—A Simple Scoring System for Predicting 28-Day Mortality
| Risk Factor | Points |
|---|---|
| Age (yr) | |
| 18–69 | 0 |
| 70–79 | 2 |
| ≥ 80 | 5 |
| Altered mental status | |
| Yes | 5 |
| No | 0 |
| Number of ICU beds | |
| ≤ 50 | 6 |
| > 50 | 0 |
| Arterial pH | |
| ≤ 7.2 | 6 |
| > 7.2 | 0 |
| Creatinine (mg/dL) | |
| < 2 | 0 |
| ≥ 2 or renal replacement therapy | 2 |
| Lactate (mmol/L) | |
| ≤ 2.0 | 0 |
| > 2.0 | 2 |
| Pao2/Fio2 ratio (mm Hg) | |
| ≤ 100 | 1 |
| > 100 | 0 |
| Sodium (mEq/L) | |
| ≤ 145 | 0 |
| > 145 | 1 |
| Urine output (mL) | |
| ≤ 200 | 1 |
| > 200 | 0 |
| Cardiovascular comorbidity | |
| None | 0 |
| Any | 2 |
Figure 4.
Calibration results for Study of the Treatment and Outcomes in Critically Ill Patients With Coronavirus Disease 2019 Mortality Index (SCMI) in the external and temporal validation cohorts. Bar chart showing the percentage of patients who died across values of the SCMI scoring system in both the external (A) and temporal (B) validation cohorts.
TABLE 2.
Accuracy of Score Cutoffs for Detecting 28-Day Mortality Using the Study of the Treatment and Outcomes in Critically Ill Patients With Coronavirus Disease 2019 Mortality Index in the External Site Validation
| Score Cutoff | Sensitivity (%) | Specificity (%) | Positive Predictive Value | Negative Predictive Value |
|---|---|---|---|---|
| > 0 | 99 | 11 | 36 | 93 |
| > 1 | 98 | 13 | 37 | 93 |
| > 2 | 92 | 40 | 44 | 90 |
| > 3 | 87 | 49 | 47 | 88 |
| > 4 | 80 | 62 | 52 | 86 |
| > 5 | 75 | 67 | 55 | 84 |
| > 6 | 68 | 72 | 56 | 81 |
| > 7 | 61 | 78 | 60 | 79 |
| > 8 | 58 | 82 | 62 | 79 |
| > 9 | 47 | 88 | 68 | 76 |
| > 10 | 42 | 90 | 70 | 75 |
| > 11 | 38 | 93 | 74 | 74 |
| > 12 | 32 | 95 | 76 | 73 |
| > 13 | 26 | 96 | 77 | 71 |
| > 14 | 21 | 97 | 78 | 70 |
| > 15 | 18 | 98 | 80 | 70 |
| > 16 | 17 | 98 | 85 | 69 |
| > 17 | 13 | 99 | 85 | 69 |
| > 18 | 10 | 99 | 90 | 68 |
| > 19 | 8 | 100 | 92 | 68 |
| > 20 | 6 | 100 | 94 | 67 |
| > 21 | 4 | 100 | 100 | 67 |
| > 22 | 3 | 100 | 100 | 66 |
| > 23 | 2 | 100 | 100 | 66 |
| > 24 | 2 | 100 | 100 | 66 |
| > 25 | 1 | 100 | 100 | 66 |
| > 26 | 1 | 100 | 100 | 66 |
| > 27 | 1 | 100 | 100 | 66 |
| > 28 | 0 | 100 | 100 | 66 |
DISCUSSION
In this multicenter study of 5,075 critically ill patients with COVID-19 admitted to ICUs at 68 geographically diverse hospitals across the United States, we developed and validated a simple tool (SCMI), which could be calculated at the bedside using data from the first 2 days of ICU admission to estimate a patient’s risk of mortality. We also found that XGBoost had the highest discrimination and best calibration of all the machine learning models we tested. The machine learning models, except for KNN, had higher discrimination than a modified SOFA score, NEWS, and CURB-65. Our results provide a simple bedside tool and highlight risk factors that can provide important information for goals of care discussions, triage decisions, and prognostic enrichment in clinical trials.
Our developed tool, the SCMI, includes 10 commonly collected variables found to be important in the XGBoost model and had higher discrimination than a modified SOFA score, NEWS, and CURB-65 on external validation and similar discrimination to some of the machine learning methods that would be more difficult to deploy. Its AUC was higher than the SOFA score on temporal validation, but this difference was not statistically significant. This tool could be used for prognostication to help clinicians better prepare for the likely future trajectory of their patients. It could also be used to help triage patients in times of limited resources, as it could identify patients at very high and very low risk of mortality (4, 5). For example, scores greater than 13 had a PPV of 77% and a specificity of 96% for 28-day mortality. However, further validation in patients outside the ICU is needed to determine if it can predict risk of mortality in patients prior to their transfer to the ICU. Similar severity of illness scores, such as the SOFA score, are included in the triage guidelines in some states (6). Furthermore, this tool could identify high-risk patients for clinical trials for prognostic enrichment. Although the SCMI tool is simpler to calculate, this does come at the cost of decreased performance compared with the more complex machine learning algorithms. As with any tool designed to predict the complex outcome of mortality in critically ill patients, choosing an optimal threshold for use involves a trade-off between PPV and sensitivity. Thus, the choice of threshold should be individualized to the planned use, with the understanding that these scores can identify patients at high (or low) risk of mortality, which could allow for more personalized prognosis and treatment than without the score, but these predictions are imperfect. Importantly, using the tool outside of data collection processes similar to the STOP-COVID cohort requires validation to ensure that the tool retains similar accuracy before use.
Our finding that XGBoost was the most accurate algorithm is consistent with prior studies. For example, we previously found that random forests and GBM had the highest discrimination for predicting clinical deterioration (14). Our group and others have used these algorithms to develop models for predicting acute kidney injury, respiratory failure, ICU readmission, and mortality (28–31). However, it is important to note that more advanced algorithms will not always outperform logistic regression (16). Furthermore, deep neural networks are even more flexible, and it is possible that with a larger sample size the neural network model could have outperformed other algorithms, as it has in other studies (15, 32). Importantly, the developed models performed similarly in the external and temporal validations. This included similar model discrimination as well as calibration (i.e., agreement between predicted and actual mortality). Although reports of improving outcomes of COVID-19 patients over time have suggested that this might be due to improvements in the quality of care, our findings suggest that at least in patients admitted to the ICU that models developed using acute physiologic and demographic variables are robust to these time trends early in the pandemic.
Flexible machine learning models are more difficult to interpret. However, interpretable machine learning approaches have been developed to better understand these models (33, 34). In this work, we investigated the most important variables in the XGBoost model and found that older age, higher serum creatinine, and admission to a hospital with fewer ICU beds were most important. Risk factors for severe disease in patients with COVID-19 have been also described by others. For example, Wang et al (11) investigated risk factors for death in 296 hospitalized patients and found that older age and impaired kidney function were among the most important risk factors. Others have identified risk factors for mortality, such as comorbidities, liver function test abnormalities, platelet count, ferritin, and other inflammatory markers (7, 10, 12, 35, 36). Our work highlights the fact that, even in a granular dataset with many variables, age remains one of the strongest predictors of death. Mortality also increases sharply with worsening renal function. Finally, we discovered an increase in mortality in hospitals with fewer than 100 ICU beds. This could relate to lack of capacity and hospital strain or to more favorable outcomes of critically ill patients admitted to larger volume hospitals (37–39). However, this finding should be interpreted with caution because resource availability and other hospital-level variables were not collected.
This study has several strengths, including its large sample size and the fact that we presented both external and temporal validation results across patients admitted to 68 ICUs at geographically diverse hospitals. Although performance often degrades with external and temporal validation as opposed to internal validation, we used this approach to estimate the expected model performance when used on new patients from other centers or future patients from the same centers.
There are also several limitations to this work. First, the data used for model development were collected using manual chart review. This may result in data entry errors, although we performed data quality checks to ensure accuracy. In addition, we assumed that patients discharged alive from the hospital before 28 days were still alive at 28 days, which may not be true. However, we did validate this at six participating hospitals, and results were similar when using inhospital mortality as the outcome. Furthermore, the scope of variables is limited compared with electronic health records, and we were unable to investigate the predictive value of some variables due to high rates of missingness. This also required us to modify previously published tools, which may have decreased their performance. Finally, we were unable to account for ICU structural variables, such as the presence of an intensivist and nurse staffing ratios, which may influence patient outcomes.
CONCLUSIONS
In conclusion, we found that age, serum creatinine, lactate, and number of ICU beds were the most important predictors of 28-day mortality in ICU patients with COVID-19, and XGBoost was the most accurate machine learning model for predicting this outcome. A simple tool we developed, the SCMI, could be calculated at the bedside of critically ill COVID-19 patients to provide prognostic information for patients and providers, resource utilization, and clinical trial enrichment.
ACKNOWLEDGMENTS
We would like to thank the research and clinical staff from the participating sites.
Supplementary Material
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccejournal).
Drs. Churpek and Gupta contributed equally.
Drs. Churpek, Gupta, Spicer, and Leaf involved in study concept and design; administrative, technical, and material support; and had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Dr. Churpek involved in first drafting of the article. Drs. Churpek and Spicer involved in statistical analysis. Drs. Churpek and Leaf involved in obtained funding and study supervision. Drs. Gupta, Hayek, Srivastava, Chan, Melamed, Brenner, Radbel, Bhatraju, Bansal, Green, Goyal, Shaefi, Parikh, Semler, and Leaf involved in acquisition of data. Dr. Churpek takes full responsibility for the content of this article, including the data analysis, draft, and final version. All authors involved in analysis and interpretation of data, and critical revision of the article for important intellectual content.
Dr. Churpek is supported by an R01 from National Institute of General Medical Sciences (NIGMS) (R01 GM123193), has a patent pending (ARCD. P0535US.P2) for risk stratification algorithms for hospitalized patients, and has received research support from EarlySense (Tel Aviv, Israel). Dr. Gupta is a scientific coordinator for the A Study of Cardiovascular Events in Diabetes trial (GlaxoSmithKline). Dr. Shaefi is supported by a K08 from NIGMS (K08GM134220) and an R03 from National Institute of Aging (R03AG060179). Dr. Leaf is supported by an R01 from National Heart, Lung, and Blood Institute (R01HL144566). The remaining authors have disclosed that they do not have any potential conflicts of interest.
This study was performed at all sites (data collection) and University of Wisconsin-Madison (statistical analysis).
A full list of the STOP-COVID investigators is provided in the Supplemental Appendix (http://links.lww.com/CCX/A759).
Contributor Information
Collaborators: Carl P. Walther, Samaya J. Anumudu, Justin Arunthamakun, Kathleen F. Kopecky, Gregory P. Milligan, Peter A. McCullough, ThuyDuyen Nguyen, Shahzad Shaefi, Megan L. Krajewski, Sidharth Shankar, Ameeka Pannu, Juan D. Valencia, Sushrut S. Waikar, Zoe A. Kibbelaar, Ambarish M. Athavale, Peter Hart, Oyintayo Ajiboye, Matthew Itteera, Adam Green, Jean-Sebastien Rachoin, Christa A. Schorr, Lisa Shea, Daniel L. Edmonston, Christopher L. Mosher, Alexandre M. Shehata, Zaza Cohen, Valerie Allusson, Gabriela Bambrick-Santoyo, Noor ul aain Bhatti, Bijal Metha, Aquino Williams, Samantha K. Brenner, Patricia Walters, Ronaldo C. Go, Keith M. Rose, Miguel A. Hernán, Amy M. Zhou, Ethan C. Kim, Rebecca Lisk, Lili Chan, Kusum S. Mathews, Steven G. Coca, Deena R. Altman, Aparna Saha, Howard Soh, Huei Hsun Wen, Sonali Bose, Emily Leven, Jing G. Wang, Gohar Mosoyan, Girish N. Nadkarni, Allon N. Friedman, John Guirguis, Rajat Kapoor, Christopher Meshberger, Chirag R. Parikh, Brian T. Garibaldi, Celia P. Corona-Villalobos, Yumeng Wen, Steven Menez, Rubab F. Malik, Carmen Elena Cervantes, Samir C. Gautam, Crystal Chang, H. Bryant Nguyen, Afshin Ahoubim, Leslie F. Thomas, Pramod K. Guru, Paul A. Bergl, Yan Zhou, Jesus Rodriguez, Jatan A. Shah, Mrigank S. Gupta, Princy N. Kumar, Deepa G. Lazarous, Seble G. Kassaye, Michal L. Melamed, Tanya S. Johns, Ryan Mocerino, Kalyan Prudhvi, Denzel Zhu, Rebecca V. Levy, Yorg Azzi, Molly Fisher, Milagros Yunes, Kaltrina Sedaliu, Ladan Golestaneh, Maureen Brogan, Jyotsana Thakkar, Neelja Kumar, Michael J. Ross, Michael Chang, Ritesh Raichoudhury, Edward J. Schenck, Soo Jung Cho, Maria Plataki, Sergio L. Alvarez-Mulett, Luis G. Gomez-Escobar, Di Pan, Stefi Lee, Jamuna Krishnan, William Whalen, David Charytan, Ashley Macina, Daniel W. Ross, Anand Srivastava, Alexander S. Leidner, Carlos Martinez, Jacqueline M. Kruser, Richard G. Wunderink, Alexander J. Hodakowski, Juan Carlos Q. Velez, Eboni G. Price-Haywood, Luis A. Matute-Trochez, Anna E. Hasty, Muner MB. Mohamed, Rupali S. Avasare, David Zonies, David E. Leaf, Shruti Gupta, Rebecca M. Baron, Meghan E. Sise, Erik T. Newman, Samah Abu Omar, Kapil K. Pokharel, Shreyak Sharma, Harkarandeep Singh, Simon Correa Gaviria, Tanveer Shaukat, Omer Kamal, Wei Wang, Heather Yang, Jeffery O. Boateng, Meghan Lee, Ian A. Strohbehn, Jiahua Li, Saif A. Muhsin, Ernest I. Mandel, Ariel L. Mueller, Nicholas S. Cairl, Farah Madhani-Lovely, Chris Rowan, Farah Madhai-Lovely, Vasil Peev, Jochen Reiser, John J. Byun, Andrew Vissing, Esha M. Kapania, Zoe Post, Nilam P. Patel, Joy-Marie Hermes, Anne K. Sutherland, Amee Patrawalla, Diana G. Finkel, Barbara A. Danek, Sowminya Arikapudi, Jeffrey M. Paer, Jared Radbel, Sonika Puri, Jag Sunderram, Matthew T. Scharf, Ayesha Ahmed, Ilya Berim, Jayanth Vatson, Shuchi Anand, Joseph E. Levitt, Pablo Garcia, Suzanne M. Boyle, Rui Song, Jingjing Zhang, Moh’d A. Sharshir, Vadym V. Rusnak, Anip Bansal, Amber S. Podoll, Michel Chonchol, Sunita Sharma, Ellen L. Burnham, Arash Rashidi, Rana Hejal, Eric Judd, Laura Latta, Ashita Tolwani, Timothy E. Albertson, Jason Y. Adams, Steven Y. Chang, Rebecca M. Beutler, Carl E. Schulze, Etienne Macedo, Harin Rhee, Kathleen D. Liu, Vasantha K. Jotwani, Jay L. Koyner, Chintan V. Shah, Vishal Jaikaransingh, Stephanie M. Toth-Manikowski, Min J. Joo, James P. Lash, Javier A. Neyra, Nourhan Chaaban, Alfredo Iardino, Elizabeth H. Au, Jill H. Sharma, Marie Anne Sosa, Sabrina Taldone, Gabriel Contreras, David De La Zerda, Hayley B. Gershengorn, Salim S. Hayek, Pennelope Blakely, Hanna Berlin, Tariq U. Azam, Husam Shadid, Michael Pan, Patrick O’ Hayer, Chelsea Meloche, Rafey Feroze, Kishan J. Padalia, Jeff Leya, John P. Donnelly, Andrew J. Admon, Jennifer E. Flythe, Matthew J. Tugman, Brent R. Brown, Amanda K. Leonberg-Yoo, Ryan C. Spiardi, Todd A. Miano, Meaghan S. Roche, Charles R. Vasquez, Amar D. Bansal, Natalie C. Ernecoff, Csaba P. Kovesdy, Miklos Z. Molnar, S. Susan Hedayati, Mridula V. Nadamuni, Sadaf S. Khan, Duwayne L. Willett, Samuel A.P. Short, Amanda D. Renaghan, Pavan Bhatraju, A. Bilal Malik, Matthew W. Semler, Anitha Vijayan, Christina Mariyam Joy, Tingting Li, Seth Goldberg, Patricia F. Kao, Greg L. Schumaker, Nitender Goyal, Anthony J. Faugno, Greg L. Schumaker, Caroline M. Hsu, Asma Tariq, Leah Meyer, Marta Christov, Francis P. Wilson, Tanima Arora, and Ugochukwu Ugwuowo
REFERENCES
- 1.Centers for Disease Control and Prevention: COVID Data Tracker. 2020. Available at: https://www.cdc.gov/covid-data-tracker/#cases. Accessed June 10, 2021
- 2.Centers for Disease Control and Prevention: Severe Outcomes Among Patients With Coronavirus Disease 2019. 2020. Available at: https://www.cdc.gov/mmwr/volumes/69/wr/mm6912e2.htm. Accessed June 20, 2020
- 3.Emanuel EJ, Persad G, Upshur R, et al. Fair allocation of scarce medical resources in the time of Covid-19. N Engl J Med. 2020; 382:2049–2055 [DOI] [PubMed] [Google Scholar]
- 4.Savulescu J, Vergano M, Craxì L, et al. An ethical algorithm for rationing life-sustaining treatment during the COVID-19 pandemic. Br J Anaesth. 2020; 125:253–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Truog RD, Mitchell C, Daley GQ. The toughest triage - allocating ventilators in a pandemic. N Engl J Med. 2020; 382:1973–1975 [DOI] [PubMed] [Google Scholar]
- 6.Wunsch H, Hill AD, Bosch N, et al. Comparison of 2 triage scoring guidelines for allocation of mechanical ventilators. JAMA Netw Open. 2020; 3:e2029250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Henry BM, de Oliveira MHS, Benoit S, et al. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): A meta-analysis. Clin Chem Lab Med. 2020; 58:1021–1028 [DOI] [PubMed] [Google Scholar]
- 8.Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of Covid-19: Systematic review and critical appraisal. BMJ. 2020; 369:m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta S, Hayek SS, Wang W, et al. Risk factors for death in critically ill patients with COVID-19 in the United States. JAMA Intern Med. 2020; 180:1436–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020; 180:934–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang K, Zuo P, Liu Y, et al. Clinical and laboratory predictors of in-hospital mortality in patients with COVID-19: A cohort study in Wuhan, China. Clin Infect Dis. 2020; 71:2079–2088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruan Q, Yang K, Wang W, et al. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med. 2020; 46:846–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldstein BA, Navar AM, Pencina MJ, et al. Opportunities and challenges in developing risk prediction models with electronic health records data: A systematic review. J Am Med Inform Assoc. 2017; 24:198–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Churpek MM, Yuen TC, Winslow C, et al. Multicenter comparison of machine learning methods and conventional regression for predicting clinical deterioration on the wards. Crit Care Med. 2016; 44:368–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomašev N, Glorot X, Rae JW, et al. A clinically applicable approach to continuous prediction of future acute kidney injury. Nature. 2019; 572:116–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christodoulou E, Ma J, Collins GS, et al. A systematic review shows no performance benefit of machine learning over logistic regression for clinical prediction models. J Clin Epidemiol. 2019; 110:12–22 [DOI] [PubMed] [Google Scholar]
- 17.Kuhn M, Johnson K: Feature Engineering and Selection: A Practical Approach for Predictive Models. Boca Raton, FL, CRC Press, 2019 [Google Scholar]
- 18.Hastie T, Tibshirani R, Friedman JH: The Elements of Statistical Learning: Data Mining, Inference, and Prediction. New York, NY, Springer, 2009, pp xxii, 745 [Google Scholar]
- 19.Goodfellow I, Bengio Y, Courville A. Deep Learning. Cambridge, MA, The MIT Press, 2016, pp xxii, 775 [Google Scholar]
- 20.Vincent JL, Moreno R, Takala J, et al. The SOFA (Sepsis-related Organ Failure Assessment) score to describe organ dysfunction/failure. On behalf of the Working Group on Sepsis-Related Problems of the European Society of Intensive Care Medicine. Intensive Care Med. 1996; 22:707–710 [DOI] [PubMed] [Google Scholar]
- 21.Seymour CW, Liu VX, Iwashyna TJ, et al. Assessment of clinical criteria for sepsis: For the third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA. 2016; 315:762–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayek SS, Brenner SK, Azam TU, et al. ; STOP-COVID Investigators: In-hospital cardiac arrest in critically ill patients with Covid-19: Multicenter cohort study. BMJ. 2020; 371:m3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith GB, Prytherch DR, Meredith P, et al. The ability of the National Early Warning Score (NEWS) to discriminate patients at risk of early cardiac arrest, unanticipated intensive care unit admission, and death. Resuscitation. 2013; 84:465–470 [DOI] [PubMed] [Google Scholar]
- 24.Lim WS, van der Eerden MM, Laing R, et al. Defining community acquired pneumonia severity on presentation to hospital: An international derivation and validation study. Thorax. 2003; 58:377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bauer TT, Ewig S, Marre R, et al. ; CAPNETZ Study Group: CRB-65 predicts death from community-acquired pneumonia. J Intern Med. 2006; 260:93–101 [DOI] [PubMed] [Google Scholar]
- 26.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics. 1988; 44:837–845 [PubMed] [Google Scholar]
- 27.Fenlon C, O’Grady L, Doherty ML, et al. A discussion of calibration techniques for evaluating binary and categorical predictive models. Prev Vet Med. 2018; 149:107–114 [DOI] [PubMed] [Google Scholar]
- 28.Koyner JL, Carey KA, Edelson DP, et al. The development of a machine learning inpatient acute kidney injury prediction model. Crit Care Med. 2018; 46:1070–1077 [DOI] [PubMed] [Google Scholar]
- 29.Dziadzko MA, Novotny PJ, Sloan J, et al. Multicenter derivation and validation of an early warning score for acute respiratory failure or death in the hospital. Crit Care. 2018; 22:286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allyn J, Allou N, Augustin P, et al. A comparison of a machine learning model with EuroSCORE II in predicting mortality after elective cardiac surgery: A decision curve analysis. PLoS One. 2017; 12:e0169772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rojas JC, Carey KA, Edelson DP, et al. Predicting intensive care unit readmission with machine learning using electronic health record data. Ann Am Thorac Soc. 2018; 15:846–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon JM, Lee Y, Lee Y, et al. An algorithm based on deep learning for predicting in-hospital cardiac arrest. J Am Heart Assoc. 2018; 7:e008678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayampurath A, Sanchez-Pinto LN, Carey KA, et al. Combining patient visual timelines with deep learning to predict mortality. PLoS One. 2019; 14:e0220640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molnar C: Interpretable Machine Learning. Victoria, BC, Canada, Leanpub, 2020 [Google Scholar]
- 35.Liang W, Liang H, Ou L, et al. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020; 180:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Xu S, Yu M, et al. Risk factors for severity and mortality in adult COVID-19 inpatients in Wuhan. J Allergy Clin Immunol. 2020; 146:110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kahn JM, Goss CH, Heagerty PJ, et al. Hospital volume and the outcomes of mechanical ventilation. N Engl J Med. 2006; 355:41–50 [DOI] [PubMed] [Google Scholar]
- 38.Zuber B, Tran TC, Aegerter P, et al. ; CUB-Réa Network: Impact of case volume on survival of septic shock in patients with malignancies. Crit Care Med. 2012; 40:55–62 [DOI] [PubMed] [Google Scholar]
- 39.Peelen L, de Keizer NF, Peek N, et al. The influence of volume and intensive care unit organization on hospital mortality in patients admitted with severe sepsis: A retrospective multicentre cohort study. Crit Care. 2007; 11:R40. [DOI] [PMC free article] [PubMed] [Google Scholar]