Abstract
Background
The nextwave of COVID-19 pandemic is anticipated to be worse than the initial one and will strain the healthcare systems even more during the winter months. Our aim was to develop a novel machine learning-based model to predict mortality using the deep learning Neo-V framework. We hypothesized this novel machine learning approach could be applied to COVID-19 patients to predict mortality successfully with high accuracy.
Methods
We collected clinical and laboratory data prospectively on all adult patients (≥18 years of age) that were admitted in the inpatient setting at Aga Khan University Hospital between February 2020 and September 2020 with a clinical diagnosis of COVID-19 infection. Only patients with a RT-PCR (reverse polymerase chain reaction) proven COVID-19 infection and complete medical records were included in this study. A Novel 3-phase machine learning framework was developed to predict mortality in the inpatients setting. Phase 1 included variable selection that was done using univariate and multivariate Cox-regression analysis; all variables that failed the regression analysis were excluded from the machine learning phase of the study. Phase 2 involved new-variables creation and selection. Phase 3 and final phase applied deep neural networks and other traditional machine learning models like Decision Tree Model, k-nearest neighbor models, etc. The accuracy of these models were evaluated using test-set accuracy, sensitivity, specificity, positive predictive values, negative predictive values and area under the receiver-operating curves.
Results
After application of inclusion and exclusion criteria (n=)1214 patients were selected from a total of 1228 admitted patients. We observed that several clinical and laboratory-based variables were statistically significant for both univariate and multivariate analyses while others were not. With most significant being septic shock (hazard ratio [HR], 4.30; 95% confidence interval [CI], 2.91–6.37), supportive treatment (HR, 3.51; 95% CI, 2.01–6.14), abnormal international normalized ratio (INR) (HR, 3.24; 95% CI, 2.28–4.63), admission to the intensive care unit (ICU) (HR, 3.24; 95% CI, 2.22–4.74), treatment with invasive ventilation (HR, 3.21; 95% CI, 2.15–4.79) and laboratory lymphocytic derangement (HR, 2.79; 95% CI, 1.6–4.86). Machine learning results showed our deep neural network (DNN) (Neo-V) model outperformed all conventional machine learning models with test set accuracy of 99.53%, sensitivity of 89.87%, and specificity of 95.63%; positive predictive value, 50.00%; negative predictive value, 91.05%; and area under the receiver-operator curve of 88.5.
Conclusion
Our novel Deep-Neo-V model outperformed all other machine learning models. The model is easy to implement, user friendly and with high accuracy.
Keywords: COVI-19, Pandemic, Machine Learning, Deep Neural Network, Mortality, SARS-COV-2
1. Key message
During the current COVID-19 pandemic, health systems have been overwhelmed and with the emergence of new strains it has been a challenge for clinicians to triage and classify patients at risk of dying. In this current study, we developed a machine learning-based mortality prediction model with high accuracy that can help millions of patients.
2. Detailed key message
2.1. Viewpoint
There has been limited use of machine learning as a biomarker for outcomes in health care setting. Previously, our group had successfully built powerful biological machine learning models to predict mortality in intensive care units and inpatient settings.
2.2. Commentaries
Many countries like Brazil and India are now experiencing new cases of COVID-19 and are overwhelmed, with limited resources. In such a situations our one-click algorithm can be utilized to predict mortality before it happens. This algorithm can also have the potential to be a screening tool for mortality in future COVID-19-like viral pandemics.
2.3. Innovation
We successfully built an entirely new framework for this unique problem called the Neo-V Framework that is a statistically rigorous training method and has wide applications in the healthcare system, for training models to predict outcomes across the multiple subspecialties.
2.4. Key findings
A total of 1228 participants' records were retrieved; analysis was conducted on 1214 patients after the exclusion criteria were applied.
Clinically the most significant associations with mortality were septic shock (HR, 4.30; 95% 95 %CI: 2.91–6.37), supportive treatment (HR, 3.51; 95 %CI: 2.01–6.14), deranged International Normalized Ratio (INR) (HR, 3.24; 95% CI:2.28–4.63), intensive care unit admission (ICU) (HR, 3.24; 95% CI: 2.22–4.74), treatment with invasive ventilation (HR, 3.21; 95% CI, 2.15–4.79) and blood lymphocytic derangement (HR, 2.79; 95% CI: 1.6–4.86). However, utilization of systemic steroids (n = 430, HR = 0.66, 95 %CI = 0.45–0.97) and presence of fever (n = 768, HR = 0.57, 95 %CI = 0.40–0.81) were associated with better overall survival.
Machine learning results showed that our DNN (Neo-V) model outperformed all conventional machine learning models with a test set accuracy of 99.53%, having Sensitivity 89.87%, Specificity 95.63%; Positive Predictive Value 50.00%; Negative Predictive Value 91.05%; and an Area Under the Receiver-Operator Curve (AUROC) of 88.5.
3. Key implications
Policy makers should direct resources to the development of such triage tools for clinically relevant problems. While in the immediate short term they should facilitate the rolling out of active measures enabling the design and use of such predictive tools.
Program managers should approach clinicians on how they can use these easy-to-use tools in their inpatient setting.
National stakeholders should understand and invest time and effort in creating such tools. Facilitation for the support and use of these predictive tools shall be invested by stakeholders at national level, and efforts and time shall be combined in the formation of such tools with clinical value and public health impact.
4. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2/COVID-19) has caused 60 million infections and 1.4 million deaths worldwide [1] and 800, 000 deaths in the United States [2]. Despite strict measures being deployed in terms of lockdowns, mass gatherings, the second and third wave is anticipated to be far worse than the first [3], [4]. As the COVID-19 virus acquires new mutations in India, United Kingdom and now a more infectious variant in Vietnam [5], [6], [7], the pandemic deepens, and we see significant strain on the healthcare systems in all parts of the world.
With the advent of vaccines, some countries have started to ease those earlier restrictions because of economic implications from the initial lockdown, which may create a further deepening of the current crisis, as cases continue to rise in certain parts of the world (especially developing countries) with emerging new strains, hence increasing the load on current healthcare systems of those with and without stable economies This could overwhelm the already strained healthcare systems across the United States and the world.
Machine learning has been extensively used in the automotive, defense and fin-tech industry over the past couple of years with great success. The use of these systems to predict health outcomes have been limited. The Epidemic Renormalization Group (eRG) has used a machine learning framework to predict the time evolution of the first and second wave based on the data from the first wave in Europe [8].
In the past we were able to develop machine learning algorithms that were capable of predicting mortality in clinical settings and performed better than most clinical scales that are utilized currently to predict mortality [9], [10], [11]. During the current COVID-19 pandemic crisis, our aim was to develop a mortality prediction tool that could predict mortality in COVID-19 patients during hospital admissions. This would help the already strained healthcare systems, physicians and frontline healthcare providers around the world in crucial clinical decision making, resource allocation and family-counselling.
In the current manuscript, we hypothesize that machine learning, specifically deep-learning, could be applied to COVID-19 patients with high accuracy. Using deep learning to predict mortality in these patients may assist in clinical decision making, risk stratification and planning strategies in future for such pandemics at a larger scale. Currently there is not ample work that has been done in mortality prediction in COVID-19 patients in Low Middle Income Countries (LMIC) or developing countries that not only have compromised healthcare systems, but also compromised healthcare status of the population that resides in these countries.
5. Methods
5.1. Clinical setting and dataset
This was a cross sectional study for which the data was extracted retrospectively from electronic medical records (EMR) of the Aga Khan University Hospital (AKUH), Karachi from February 2020 and September 2020, on patients admitted with a primary diagnosis of COVID-19 infection.
The Aga Khan University is a state-of-the-art tertiary care hospital that received the very first COVID-19 patient when the pandemic hit the country for the first time. A COVID-19 Command and Record team was immediately created and all the patients with COVID-19 diagnosis were admitted to specifically designated care units. The electronic data of all patients admitted to the Aga Khan University are only accessible after the study receives Ethical Review Committee (ERC) approval for the conduct. The COVID-19 patient’s data was assigned a specific repository that was accessed after Institutional Review Board (IRB) approval and the main study variables were then extracted based on a structured Performa that was designed to include all relevant baseline, demographic and clinical information of patients that were admitted to the hospital with a confirmed diagnosis of COVID-19.
The dataset was de-identified, and our study complied with the ethical principles recommended by the Helsinki declaration (1964) and its amendments. The study has received IRB/ERC approval from the Aga Khan University Hospital (AKUH), Karachi, Pakistan.
5.2. Data collection and selection criteria
In the current study adult patients (>18 years of age) that were admitted to the hospital with a diagnosis of COVID-19 and were tested positive during their admission on Reverse‐Transcriptase Polymerase Chain Reaction (RT-PCR) based on Center for Disease Control and Prevention (CDC) and College of American Pathologist (CAP) guidelines were included in the study [12], [13]. Data was collected on demographics, comorbidities at admission, first 24-hours laboratory investigations (hematological and blood biochemistry (Table 1 ), imaging, complete clinical characteristics, history, examination, treatment, hospital course and outcomes. All the patients that had RT-PCR negative tests for COVID-19 and those with incomplete records or missing information were excluded from the analysis. Two data collectors separately extracted the relevant information on a google sheet based on a structured questionnaire designed to incorporate all clinical and relevant information related to the study.
Table 1.
Demographics with Univariate and multivariate analysis of clinical variables as part of Phase I of the Neo-V and FLAIM machine learning frameworks.
| Univariate |
Multivariate |
|||||||
|---|---|---|---|---|---|---|---|---|
| n = 1214 (n, affected) and Ranges | p-value | HR | 95 %CI | p-value | HR | 95 %CI | ||
| Demographics | AGE | range (19–96 years) | <0.001* | 1.04 | 1.03–1.06 | – | – | – |
| Age > 50 years | 755 | <0.001* | 3.15 | 1.81–5.50 | – | – | – | |
| Age > 65 years | 336 | <0.001* | 2.43 | 1.72–3.44 | – | – | – | |
| Female | 467 | 0.489 | 0.88 | 0.61–1.27 | – | – | – | |
| Readmission | 46 | 0.147 | 0.05 | 0.01–2.95 | 0.951 | 0.001 | 0.01–3.40E+164 | |
| Mortality | 130 (10.7%) | – | – | – | – | – | – | |
| Blood Grouping | A- | 6 | 0.289 | 2.91 | 0.41–20.90 | 0.553 | 1.822 | 0.26–13.18 |
| A+ | 132 | 0.642 | 0.88 | 0.51–1.51 | 0.561 | 0.852 | 0.5–1.47 | |
| B- | 19 | 0.332 | 1.64 | 0.60–4.45 | 0.858 | 1.098 | 0.4–3.02 | |
| B+ | 233 | <0.05* | 1.5 | 1.03–2.18 | 0.051 | 1.459 | 1–2.13 | |
| O- | 13 | 0.27 | 0.04 | 0.00–11.71 | 0.947 | 0.001 | 0.01–2.07E+130 | |
| O+ | 183 | 0.224 | 1.28 | 0.86–1.90 | 0.25 | 1.262 | 0.85–1.88 | |
| AB- | 6 | <0.05* | 3.88 | 1.23–12.27 | <0.001* | 7.634 | 2.39–24.48 | |
| AB+ | 48 | <0.05* | 2.55 | 1.29–5.04 | <0.001* | 2.483 | 1.26–4.93 | |
| Comorbidities | Chronic kidney disease | 71 | <0.001* | 3.45 | 2.08–5.72 | <0.001* | 2.737 | 1.63–4.61 |
| Chronic liver disease | 13 | 0.15 | 2.32 | 0.74–7.33 | 0.125 | 2.467 | 0.78–7.82 | |
| Chronic obstructive lung disease | 14 | <0.05* | 2.93 | 1.19–7.21 | 0.163 | 1.918 | 0.77–4.79 | |
| Diabetes | 441 | <0.05* | 1.45 | 1.03–2.05 | 0.261 | 1.224 | 0.87–1.74 | |
| Hypertension | 521 | 0.58 | 1.41 | 0.99–2.01 | 0.493 | 1.137 | 0.79–1.64 | |
| Ischemic heart disease | 158 | <0.001* | 2.24 | 1.62–3.62 | <0.05* | 1.801 | 1.19–2.73 | |
| Other Comorbidities | 571 | <0.05* | 1.76 | 1.24–2.48 | <0.05* | 1.721 | 1.21–2.45 | |
| Symptoms | Asymptomatic | 87 | 0.177 | 0.38 | 0.12–1.27 | 0.269 | 0.497 | 0.15–1.72 |
| Chest pain (non-cardiac) | 56 | 0.252 | 1.63 | 0.71–3.71 | 0.434 | 1.395 | 0.61–3.21 | |
| Cough | 522 | 0.110 | 0.76 | 0.54–1.07 | 0.28 | 0.823 | 0.58–1.18 | |
| Fatigue/lethargy | 52 | 0.793 | 1.15 | 0.42–3.12 | 0.63 | 0.78 | 0.29–2.14 | |
| Fever | 768 | <0.001* | 0.56 | 0.39–0.79 | <0.05* | 0.568 | 0.4–0.81 | |
| Gastrointestinal symptoms | 93 | 0.875 | 0.94 | 0.42–2.14 | 0.626 | 0.813 | 0.36–1.88 | |
| Headache | 16 | 0.367 | 0.05 | 0.01–34.53 | 0.953 | 0.001 | 0.01–4.24E+150 | |
| Hemoptysis | 9 | 0.609 | 0.05 | 0.01–5037.35 | 0.961 | 0 | 0–1.48E+166 | |
| Malaise | 83 | 0.976 | 0.99 | 0.41–2.43 | 0.537 | 0.752 | 0.31–1.87 | |
| More than 2 symptoms | 901 | 0.975 | 1.01 | 0.66–1.56 | 0.817 | 0.95 | 0.62–1.47 | |
| Myalgia | 28 | 0.284 | 1.88 | 0.6–5.94 | 0.221 | 2.06 | 0.65–6.54 | |
| Nasal obstruction | 2 | 0.788 | 0.05 | 0.01–157108055.1 | 0.969 | 0.001 | 0.01–4.22E+198 | |
| Other symptoms | 321 | <0.001* | 1.97 | 1.38–2.8 | <0.05* | 1.66 | 1.16–2.38 | |
| Pneumothorax (clinical/radiological) | 22 | <0.05* | 2.00 | 1.17–3.43 | <0.05* | 2.014 | 1.18–3.46 | |
| Rhinorrhea | 7 | 0.502 | 0.05 | 0.01–327.09 | 0.964 | 0.001 | 0.01–1.70E+200 | |
| Sore throat | 58 | 0.240 | 0.51 | 0.16–1.59 | 0.637 | 0.758 | 0.24–2.41 | |
| Sputum | 29 | 0.070 | 2.03 | 0.95–4.35 | 0.247 | 1.576 | 0.73–3.41 | |
| Clinical | Acute Kidney Injury | 179 | <0.001* | 2.95 | 2.08–4.18 | <0.001* | 2.465 | 1.74–3.51 |
| ARDS/Respiratory failure | 147 | <0.001* | 2.81 | 1.96–4.03 | <0.001* | 2.527 | 1.77–3.63 | |
| Septic Shock | 49 | <0.001* | 5.05 | 3.43–7.43 | <0.001* | 4.299 | 2.91–6.37 | |
| Shock Liver | 3 | <0.001* | 11.63 | 2.84–47.73 | <0.001* | 11.476 | 2.77–47.68 | |
| Shortness of breath | 570 | <0.001* | 1.81 | 1.24–2.62 | <0.001* | 1.96 | 1.35–2.86 | |
| Point of Care | Special care unit | 308 | 0.355 | 1.18 | 0.84–1.68 | 0.577 | 1.106 | 0.78–1.58 |
| Days spent in the ICU | Range (0–20 days) | 0.187 | 1.03 | 0.99–1.07 | 0.133 | 1.029 | 1–1.07 | |
| Intensive care unit (ICU) | 106 | <0.001* | 2.71 | 1.88–3.92 | <0.001* | 3.241 | 2.22–4.74 | |
| Number of admissions ICU during current hospitalization | Range (0–2) | <0.001* | 1.78 | 1.3–2.43 | <0.001* | 2.066 | 1.49–2.87 | |
| Radiology | Bilateral chest X-ray abnormalities | 753 | 0.484 | 1.14 | 0.8–1.64 | 0.34 | 1.195 | 0.83–1.72 |
| Unilateral chest X-ray abnormalities | 131 | 0.911 | 0.97 | 0.56–1.7 | 0.997 | 0.999 | 0.58–1.75 | |
| Day One labs | Abnormal blood lymphocyte count | 378 | <0.001* | 3.02 | 1.74–5.26 | <0.001* | 2.786 | 1.6–4.86 |
| Abnormal blood neutrophil count | 1044 | <0.05* | 2.5 | 1.17–5.36 | <0.05* | 2.657 | 1.24–5.7 | |
| Abnormal platelets count | 363 | <0.05* | 1.43 | 1.01–2.03 | <0.05* | 1.413 | 1–2.01 | |
| Abnormal serum albumin | 49 | 0.296 | 1.45 | 0.73–2.87 | 0.207 | 1.552 | 0.79–3.07 | |
| Abnormal serum ALT | 946 | 0.380 | 1.23 | 0.78–1.95 | 0.475 | 1.183 | 0.75–1.88 | |
| Abnormal serum APTT | 347 | 0.217 | 1.26 | 0.88–1.81 | 0.137 | 1.32 | 0.92–1.9 | |
| Abnormal serum bilirubin | 69 | 0.587 | 0.82 | 0.4–1.69 | 0.33 | 0.699 | 0.34–1.42 | |
| Abnormal serum BUN | 382 | <0.001* | 3.15 | 2.19–4.52 | <0.001* | 2.784 | 1.94–4.01 | |
| Abnormal serum calcium | 387 | 0.596 | 0.91 | 0.62–1.33 | 0.633 | 0.91 | 0.62–1.35 | |
| Abnormal serum creatinine | 373 | <0.001* | 3.21 | 2.24–4.6 | <0.001* | 2.73 | 1.9–3.94 | |
| Abnormal serum hematocrit | 505 | <0.001* | 1.87 | 1.33–2.65 | <0.05* | 1.808 | 1.26–2.61 | |
| Abnormal serum hemoglobin | 879 | <0.05* | 1.67 | 1.08–2.58 | <0.05* | 1.63 | 1.05–2.55 | |
| Abnormal serum INR | 198 | <0.001* | 3.38 | 2.38–4.8 | <0.001* | 3.243 | 2.28–4.63 | |
| Abnormal serum LDH | 751 | 0.597 | 1.11 | 0.77–1.58 | 0.806 | 1.047 | 0.74–1.5 | |
| Abnormal serum magnesium | 268 | 0.535 | 0.88 | 0.57–1.35 | 0.841 | 0.957 | 0.63–1.48 | |
| Abnormal serum phosphorus | 32 | 0.893 | 1.08 | 0.4–2.91 | 0.58 | 0.752 | 0.28–2.07 | |
| Abnormal serum potassium | 241 | 0.886 | 0.97 | 0.64–1.49 | 0.678 | 0.914 | 0.6–1.41 | |
| Abnormal serum procalcitonin | 523 | 0.312 | 1.2 | 0.85–1.69 | 0.485 | 1.132 | 0.8–1.61 | |
| Abnormal serum PT | 378 | <0.05* | 1.5 | 1.05–2.15 | <0.05* | 1.787 | 1.24–2.59 | |
| Abnormal serum sodium | 458 | 0.250 | 1.23 | 0.87–1.73 | 0.501 | 1.127 | 0.8–1.6 | |
| Abnormal white cell count | 455 | <0.001* | 2.08 | 1.45–2.96 | <0.001* | 2.089 | 1.47–2.98 | |
| Treatment | Anti-malarial | 2 | 0.385 | 2.4 | 0.34–17.27 | 0.587 | 1.73 | 0.24–12.51 |
| Anti-viral drugs | 99 | 0.715 | 0.9 | 0.51–1.6 | 0.538 | 0.835 | 0.47–1.49 | |
| CRRT | 7 | <0.05* | 2.72 | 1.19–6.23 | 0.054 | 2.287 | 0.99–5.3 | |
| Hydroxychloroquine | 77 | 0.230 | 0.61 | 0.27–1.38 | 0.336 | 0.667 | 0.3–1.53 | |
| Intravenous IgG | 6 | <0.05* | 3.87 | 1.23–12.23 | <0.05* | 3.945 | 1.25–12.49 | |
| Invasive ventilation | 74 | <0.001* | 2.79 | 1.89–4.1 | <0.001* | 3.208 | 2.15–4.79 | |
| Lopinavir/Ritonavir | 1 | 0.616 | 0.05 | 0.01–6488.77 | 0.962 | 0.001 | 0.01–6.44E+175 | |
| Non-invasive ventilation (BiPAP/CPAP) | 243 | <0.001* | 1.96 | 1.38–2.8 | <0.001* | 1.872 | 1.32–2.67 | |
| Plasmapheresis | 96 | 0.317 | 1.25 | 0.82–1.91 | 0.234 | 1.294 | 0.85–1.98 | |
| Supportive treatment | 617 | <0.001* | 3.89 | 2.22–6.81 | <0.001* | 3.507 | 2.01–6.14 | |
| Symptomatic treatment | 1056 | <0.05* | 0.6 | 0.36–0.98 | 0.058 | 0.613 | 0.37–1.02 | |
| Systemic steroids | 430 | 0.238 | 0.8 | 0.55–1.17 | <0.05* | 0.657 | 0.45–0.97 | |
5.3. Neo-V Framework
The new variable framework (Neo-V Framework) was developed to create a pathway for smaller datasets to have additional variables from existing data. The concept is similar to but not the same as a combination of different unrelated variables, the combination of these variables are biologically related to each other (see discussion). More simply Neo V, is a tri-phase bio-statistically rigorous machine learning approach. The framework has better accuracy than the currently used clinical scoring systems in predicting mortality in the intensive care unit (ICU) patients [9], [10]. The three phases of the framework are as follows:
Phase I: Also known as the statistical-phase; in which data was analyzed through univariate and multivariate Cox-regression analysis [14] using IBM SPSS (version 24.0.0.0) [15] for outcome assessment with hazard ratio and confidence intervals. A p-value of <0.05 was considered statistically significant. Frequency analysis was also done on the selected patients. Statistical analysis was carried out on all the variables included in Table 1 .
Phase II: New variables were created from the existing dataset called neo-variables. These variables included a combination of variables with two clinically relevant laboratory investigations that were significant in both the univariate and the multivariate analysis (Table 1). The selection of these variables was done on the basis of clinical and biological relevance to each other and was not a simple combination of the variables. These variables also underwent univariate and multivariate analysis for outcome assessment with hazard ratio and confidence intervals (cox-regression analysis).
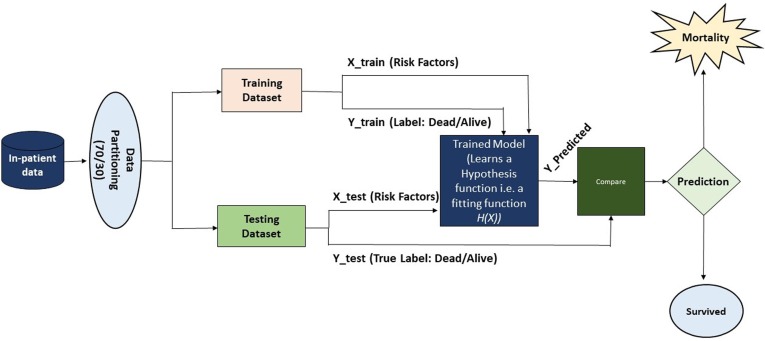
Phase III: Biological datasets are highly imbalanced with respect to the outcomes (i.e., survival is greater than mortality and machine learning models are very sensitive to imbalanced data and can produce variable and non-reproducible results. To address this, optimization was done using Synthetic Minority Over-sampling Technique (SMOTE) algorithm during the training process only [16]. In the machine learning phase, we used all variables that were statistically significant in phase I and II in both the univariate and multivariate analysis (non-significant risk factors were excluded). After applying the data partitioning scheme, the final dataset was randomized and divided into a training and testing set with a 70/30 percent split respectively (30% data left out to test the models) through train-test holdout validation. After partitioning the data, vector features were allocated in the training instances by X_train with the corresponding outcome label as Y_train. Similarly, the test-set was allocated in X_test and Y_test as testing vector instances and corresponding outcomes, respectively. The models were trained on X_train and Y_train. The models tried to learn the behavior/distribution of the data and generate a hypothesis/fitting function. Once the training was concluded the model tested the X_test and produced an output (prediction) called Y_pred. A comparison was done between Y_pred and Y_test (see Fig. 3). Our previous work shows that reduction of the number of irrelevant risk factors can produce better performances and significantly improve classifications. In this study conventional machine learning models were utilized including Random Trees (CART), K-Nearest Neighbor (KNN), Support Vector Classifier - Radial Basis Function (SVC - RBF), Ada-Boost-Classifier (ABC) and Quadratic Discriminant Analysis (QDA) and a deep neural network (DNN).
Fig. 3.
Machine learning workflow.
5.4. Neo-V models
Using the Neo-V Framework, we were able to train a number of different conventional models that included Random Forest (RF), k-nearest neighbors (KNN), Support Vector Machines - Radial Basis Function (SVM-RBF), Decision Trees (DT), Approximate Bayesian Computing (ABC) and Quadratic Discriminant Analysis (QDA). A deep neural network-based model (Deep-Neo-V) was also trained. The Deep-Neo-V model has 3 layers in the following order: input, hidden and output layers.
5.5. Deep-FLAIM model
The Deep-FLAIM model was built using the FLAIM framework (a two-phase statistically rigorous machine learning framework) which our group had previously developed to predict mortality in the inpatient settings, including critical care and non-critical care settings with high accuracy [9,10,17]. The Deep-FLAIM model was developed using this framework and results were compared with the Neo-V model (trained using the Neo-V Framework).
5.6. Performance, primary and secondary outcomes analyses
Performance of all models were evaluated by comparing their respective accuracies (training and test sets) and area under the receiver-operator curves (AUROC). Primary outcomes measures included mortality prediction for the test set using sensitivity and specificity, while secondary outcomes included positive predictive values (PPV) and negative predictive values (NPV).
6. Results
From a total of 1228 adult patients, 14 were excluded based on our selection criteria. The final analyzed dataset had 1214 adult participants with confirmed COVID-19 diagnosis on RT-PCR. Baseline frequency analysis of demographics showed a median age of 55 years (range 19–96 years), of which around 28% (n = 336) of the admitted patients were elderly (>65 years of age). Majority of the admitted patients were male (61.5%). The median length of stay (LOS) for admitted patients was 5 days (range 1–54 days).
Among the study participants the most common comorbid conditions were hypertension (43%, n = 521) and diabetes (36%, n = 441). The presenting symptoms ranged widely from being completely asymptomatic to having shortness of breath. The most significant clinical risk factors associated with in-hospital mortality included chronic kidney disease (CKD) (n = 71, HR = 2.74, 95 %CI = 1.63–4.61), ischemic heart disease (n = 158, HR = 1.80, 95 %CI = 1.19–2.73), other comorbidities (n = 571, HR = 1.72, 95 %CI = 1.21–2.45), shortness of breath (n = 570, HR = 1.96, 95 %CI = 1.35–2.86), other symptoms (non-respiratory and non-gastrointestinal symptoms)(n = 321, HR = 1.66, 95 %CI = 1.16–2.38), acute kidney injury (AKI) (n = 179, HR = 2.47, 95 %CI = 1.74–3.51), acute respiratory distress syndrome(ARDS)/respiratory failure (n = 147, HR = 2.53, 95 %CI = 1.77–3.63), septic shock (n = 49, HR = 4.30, 95 %CI = 2.91–6.37), admission to the intensive care unit (ICU) (n = 106, HR = 3.24, 95 %CI = 2.22–4.74), frequency of ICU admissions during current hospitalization (range = 0–2, HR = 2.01, 95 %CI = 1.49–2.87), mechanical/invasive ventilation (n = 74, HR = 3.21, 95 %CI = 2.15–4.79), non-invasive ventilation (BiPAP/CPAP) (n = 243, HR = 1.87, 95 %CI = 1.32–2.67), supportive treatment (n = 617, HR = 1.87, 95 %CI = 1.32–2.67) and having an AB positive blood group (n = 48, HR = 2.48, 95 %CI = 1.26–4.93). Utilization of systemic steroids (n = 430, HR = 0.66, 95 %CI = 0.45–0.97) and presence of Fever (n = 768, HR = 0.57, 95 %CI = 0.40–0.81) were associated with better overall survival.
The most significant abnormal laboratory findings associated with mortality were white blood cell count (leucocyte count) (n = 455, HR = 2.09, 95 %CI = 1.47–2.59), lymphocyte counts (n = 378, HR = 2.79, 95 %CI = 1.60–4.86), neutrophil count (n = 1044, HR = 2.67, 95 %CI = 1.24–5.70), platelets count (n = 363, HR = 1.41, 95 %CI = 1.00–2.01), hematocrit (n = 505, HR = 1.81, 95 %CI = 1.26–2.61), hemoglobin (n = 879, HR = 1.63, 95 %CI = 1.05–2.55), blood urea nitrogen (BUN, n = 382, HR = 2.78, 95 %CI = 1.94–4.01), creatinine (n = 373, HR = 2.73, 95 %CI = 1.90–3.94), International Normalized Ratio (INR, n = 198, HR = 3.24, 95 %CI = 2.28–4.63) and prothrombin time (PT, n = 378, HR = 1.79, 95 %CI = 1.24–2.59) (Table 1)
There were some risk factors that were significant for mortality but had very few patients including i.e shock liver (n = 3, HR = 11.47, 95 %CI = 2.77–47.68), blood group AB negative (n = 6, HR = 7.63, 95 %CI = 2.39–24.48), rhinorrhea (n = 7, HR = 0.001, 95 %CI = 0.01–1.7e200), treatment with intravenous IgG (immunoglobulin G) (n = 6, HR = 3.95, 95 %CI = 1.25–12.49) and pneumothorax (clinical or radiological, n = 22, HR = 2.01, 95 %CI = 1.18–3.46). Table 1 shows results from univariate and multivariate analysis (hazard ratios, confidence intervals and p-values) of all the clinical and laboratory data.
In phase II (new variable phase) 11 new variables (neo-variables) were computed; all these variables were statistically significant in the univariable and the multivariable analysis except the total number of symptoms (Table 2 ).
Table 2.
Derivative variables and their univariate and multivariate analysis with mortality during hospital stay.
| Univariate | Multivariate | |||||||
| Derivatives for machine learning | Variables | p-value | HR | 95%CI | p-value | HR | 95%CI | |
| Total No. Comorbidities | Range (0–6) | <0.001* | 1.44 | 1.27–1.64 | <0.001* | 1.301 | 1.14–1.49 | |
| More than 2 comorbidities | 499 | <0.001* | 2.28 | 1.59–3.28 | <0.001* | 1.788 | 1.23–2.61 | |
| Total No. symptoms | Range (0–6) | 0.379 | 1.08 | 0.92–1.27 | 0.439 | 1.066 | 0.91–1.26 | |
| Total No. Treatments received | Range (0–6) | <0.001* | 1.31 | 1.14–1.51 | <0.05* | 1.268 | 1.11–1.46 | |
| CR or BUN | 517 | <0.001* | 0.31 | 0.21–0.46 | <0.001* | 0.356 | 0.24–0.54 | |
| HB or HCT | 883 | <0.05* | 0.55 | 0.35–0.87 | <0.05* | 0.562 | 0.36–0.89 | |
| PLT or INR | 460 | <0.001* | 2.21 | 1.55–3.16 | <0.001* | 2.087 | 1.47–2.98 | |
| PT or INR | 502 | <0.001* | 0.46 | 0.33–0.66 | <0.001* | 0.414 | 0.29–0.6 | |
| TLC or LYMP | 882 | <0.001* | 0.18 | 0.08–0.4 | <0.001* | 0.186 | 0.09–0.43 | |
| TLC or NEU | 1085 | <0.05* | 0.2 | 0.07–0.61 | <0.05* | 0.18 | 0.06–0.57 | |
| Total No. laboratory abnormalities | Range (0–17) | <0.001* | 1.22 | 1.16–1.3 | <0.001* | 1.216 | 1.15–1.3 | |
The performance of our previously designed Deep-FLAIM model was compared to the Neo-V framework (including Deep-Neo-V and other conventional machine learning algorithms) see Table 3 . Performance results show Deep-FLAIM (training accuracy = 86.7%, testing accuracy = 84.7%, sensitivity = 68.9, specificity = 86.9, PPV = 42.5, NPV = 95.2, FPR = 13.1 and AUROC = 86.9); while conventional machine learning show: Random Forest (RF, training accuracy = 94.9%, testing accuracy = 85.8%, sensitivity = 28.9, specificity = 93.4, PPV = 32.4, NPV = 90.4, FPR = 6.25 and AUROC = 69.5), k- Nearest Neighbors (k-NN, training accuracy = 92.3%, testing accuracy = 77.8%, sensitivity = 42.2, specificity = 82.8, PPV = 25.7, NPV = 91.1, FPR = 17.2 and AUROC = 66.5), Simple Vector Classifier – Radial Basis Function (SVC – RBF, training accuracy = 69.0%, testing accuracy = 65.8%, sensitivity = 75.6, specificity = 64.4, PPV = 23.0, NPV = 94.9, FPR = 35.6 and AUROC = 80.8), Decision Trees (DT, training accuracy = 98.9%, testing accuracy = 85.2%, sensitivity = 42.2, specificity = 91.3, PPV = 40.4, NPV = 91.8, FPR = 8.8 and AUROC = 66.7), Adaptive Boosted Classifier (ABC, training accuracy = 90.1%, testing accuracy = 85.5%, sensitivity = 48.9, specificity = 90.6, PPV = 42.3, NPV = 82.7, FPR = 9.4 and AUROC = 77.65) and Quadratic discriminant analysis (QDA, training accuracy = 94.6%, testing accuracy = 87.4%, sensitivity = 60.0, specificity = 91.3, PPV = 49.1, NPV = 94.2, FPR = 8.74 and AUROC = 80.84).
Table 3.
Primary and secondary outcomes machine learning results.
| FLAIM Framework | ||||||||
|---|---|---|---|---|---|---|---|---|
| Test set (n = 365) | Deep-FLAIM | Deep-Neo-V | RF (d = 10, e = 2) | KNN (k = 3) | SVC - RBF | DT (d = 10) | ABC | QDA |
| Test set accuracy | 84.66 | 87.67 | 85.75 | 77.81 | 65.75 | 85.21 | 85.48 | 87.4 |
| Training set accuracy | 86.69 | 98.70 | 94.94 | 92.34 | 69.02 | 98.94 | 90.11 | 94.58 |
| Precision | 0.44 | 0.61 | 0.37 | 0.38 | 0.46 | 0.46 | 0.46 | 0.49 |
| Sensitivity | 68.89 | 33.33 | 28.89 | 42.22 | 75.56 | 42.22 | 48.89 | 60 |
| Specificity | 86.88 | 95.31 | 93.75 | 82.81 | 64.38 | 91.25 | 90.63 | 91.25 |
| Positive predictive value | 42.47 | 50.00 | 39.40 | 25.68 | 22.97 | 40.43 | 42.31 | 49.09 |
| Negative predictive value | 95.21 | 91.05 | 90.36 | 91.07 | 94.93 | 91.82 | 92.65 | 94.19 |
| Area Under Receiver-Operator Curve | 86.90 (0.18–0.29) | 88.50 (0.18–0.30) | 69.50 (0.18–0.29) | 66.50 (0.16–0.27) | 80.80 (0.13–0.23) | 66.74 (0.17–0.29) | 77.65 (0.18–0.29) | 80.84 (0.18–0.30) |
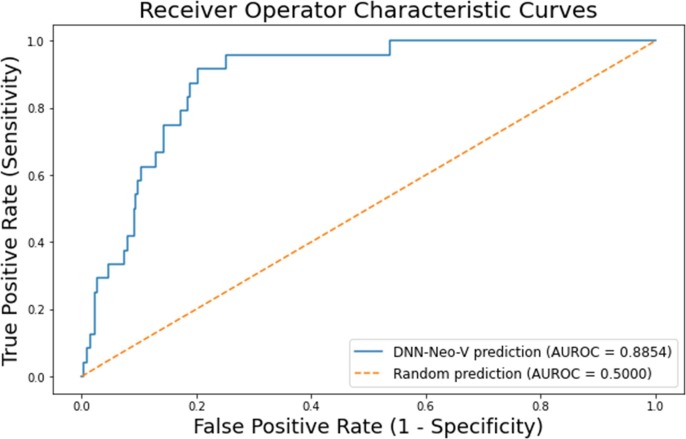
Our best model was a deep neural network (Deep-Neo-V) with training accuracy = 98.7%, testing accuracy = 87.7%, sensitivity = 33.3, specificity = 95.3, PPV = 50.0, NPV = 91.1, FPR = 4.69 and AUROC = 88.5 (see Fig. 2). All receiver operator curves are shown in Supplemental Fig. 1.
Fig. 2.
Receiver Operating Curve for Deep-Neo-V with AUROC.
There were 45 cases that were discordant with the predicted outcomes in our study with 15 cases that were predicted to have mortality but survived their hospital admission. While thirty patients were predicted to survive but died during their hospital stay. More detailed sub-analysis of these patients is presented in Supplemental Table 1.
7. Discussion
As the third wave of COVID-19 has started to unfold, the already stained healthcare systems globally are being pushed to the limit with hospital and intensive care unit (ICU) beds reaching full-capacity. Impact of the virus has been global with developed countries even struggling with infection rates and hospitalization [18]. The second wave is anticipated to be much tougher than the first one [19]. With new vaccines on the horizon infection rates in the United states have skyrocketed to 1.34 million cases being diagnosed in the second week of December 2020 and more than 1100 deaths (weekly) [20].
Multiple vaccines now approved by the FDA in the United States and other developed nations, and high vaccination rates within these countries have helped in controlling the pandemic. However, the new United Kingdom variant (B.1.17 variant), double mutant in India (B.1.617.1 aka Kappa and B.1.617.2 aka Delta variants) and now a new Vietnam variant (a hybrid of the UK and India) threaten to worsen the pandemic even further. Studies are still ongoing if these new variants can be controlled by the original viral vaccine. This still is a major problem for healthcare systems to accommodate these patients such as in India, that is reporting over 3500 deaths a day and an infection rate of over 414,000 per day (May 2021). The losses are even greater owing to the shortage of oxygen, hospital bed, ICU space and ventilators in the country.
However, there is a need for the development of clinical biomarkers and predictive models for mortality among vulnerable patient populations. Machine learning has been used to predict mortality in cancer [21], cardiac disease [22]; similarly our own work on mortality prediction on trauma patients, postoperative ileus cases in the ICU [9], [10] and diverticulitis in the inpatient setting [17] has also predicted the same with good accuracy. A lot of epidemiological studies reporting clinical, laboratory and mortality outcomes have been done worldwide including in developed and developing countries, but very few actually reported or developed a machine learning model for predicting the outcomes with a set.
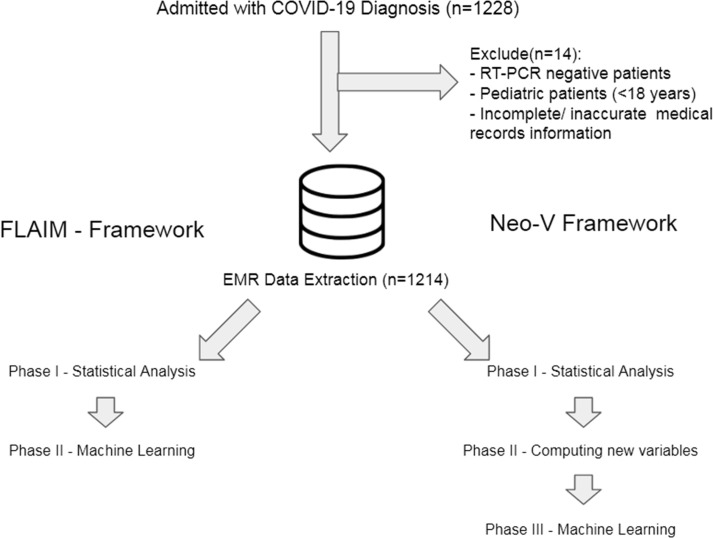
In the current study we were able to develop a new machine learning mortality prediction tool based on a novel framework (Neo-V Framework) that uses a smaller number of cases to train a deep neural network to give better predictions. This model is different from our previously developed FLAIM Framework (two-phase) and has a tri-phase structure (Fig. 1 ). Numerous studies are reported and conducted in various healthcare settings over different parts of the world as the pandemic is amidst us since almost a year and a half.
Fig. 1.
Experimental design of the Framework.
Clinical data analysis showed that with increasing age the patients’ mortality also increases. There are number of clinical risk factors that were associated with worse outcomes and documented in the clinical literature like chronic obstructive lung disease [COPD] [23], chronic kidney disease [CKD] [24], ischemic heart disease [IHD] [25], pneumothorax (radiological or clinical diagnosis) [26], acute respiratory syndrome [ARDS] [27], septic shock [28], shortness of breath [29], ICU admission [30], AB+ Blood group [31] and recurrent admission to the ICU. Hematological labs that were associated with mortality (previously presented in the results) were also seen in other studies [32]. Biochemical laboratory abnormalities like creatinine, blood urea nitrogen [33], INR and PT [34] were also associated with mortality. Patients managed with invasive ventilation [35] and non-invasive ventilation [36] which actually signifies that the patients that were not able to maintain normal respiratory physiology had worse outcomes. Having fever [37] and the use of systemic steroids [38] as an early symptom of COVID-19 had better prognosis in our patient population.
Machine learning has been used to predict mortality in the inpatient and the ICU setting in a number of different clinical conditions including our prior papers. The methods need to continue to evolve to have better outcome predictions, previously our group was able to develop the FLAIM-Framework approach, which was an attempt to build a workflow pipeline to produce more accurate results. The Neo-V Framework builds on our previous work and has the statistical power of FLAIM, but it can be used to apply deep learning to smaller datasets. This technique was named “horizontal expansion” of the dataset in which the dataset is expanded horizontally by combining two or more existing variables to create new-variables (Neo-V), without the need of recollecting the data. The combination was conditional that the variables were clinically relevant e.g., BUN and creatinine. In contrast, vertical expansion of the dataset is adding new patients or cases. There are publications that have used machine learning for rapid diagnosis of COVID-19 at admission [39], [40].
The discordant cases in our study showed there were a number of different risk factors that were distributed unevenly throughout the discordant cases further categorized in Supplemental Table 1.
To present a wholistic picture of the use of machine learning and mortality prediction in COVID-19 we have compared our own model to a number of models that are available at the time of this publication [41], [42], [43], [44], [45]. The comparison is presented in Table 4 . The most comparable of these models is from Vaid et al. that had a much larger dataset from 5 different hospitals across the city of New York, they also used RT-PCR positive adult patients in their study and developed multiple models. Their best model (MLP federated, median) when compared to our Deep-Neo-V model showed an AUROC of 0.809 vs 0.885, test-set accuracy of 87.7% vs 76.2%, sensitivity of 77.8% vs 33.3% and a specificity of 69.9% vs 95.3%. The model presented by Booth et al., had a better NPV and PPV, however our data was normalized before any training was done on the models and has a testset that is much larger.
Table 4.
Comparison of currently available Deep learning models for mortality in the inpatient setting.
| Current | Li et al. (2020)* | Zhu et al. (2020)** | Vaid et al. (2021)Ø | Booth et al. (2021)^ | Chowdhury MEH et al. (2021)*** | |
|---|---|---|---|---|---|---|
| Sample Size (n) | 1214 | 102 | 181 | 4029 | 398 | 375 |
| Endpoints | End of admission | ICU admission and mortality | End of admission | 7-day mortality | End of admission | End of admission |
| Clinical setting | In-patient | Non-ICU and ICU | In-patient | In-patient | In-patient | In-patient |
| Training and Test (set) split | 70/30 | 90/10 | 85/15 | 70/30 | 80/20 | |
| Datapoint/Variables | 47 | 15 | Top 5 lab features | 38 | Top 5 lab features | Top 5 lab features |
| Testing accuracy | 87.7 | 85.3 | nr | 76.2# (median) | nr | nr |
| Training accuracy | 98.7 | 89.2 | nr | nr | nr | nr |
| AUROC | 0.885 | 0.844 | 0.968 | 0.809# (median) | 0.930§ | 0.970 |
| Sensitivity | 33.3 | 75 | nr | 77.8# (median) | 91.0§ | 89.4–94.6 |
| Specificity | 95.3 | 87.2 | nr | 69.9# (median) | 91.0§ | 89.0–95.0 |
| PPV | 50.0 | 52.2 | nr | nr | 62.5§ | 9.1# |
| NPV | 91.1 | 94.9 | nr | nr | 98.4§ | 92.9# |
nr = not reported.
Calculated from data given in paper.
Top 5 serum markers for mortality form 26 only lab parameters, with an unbalanced training set.
Non-normalized AUROC, sensitivity, specificity, PPV, NPV.
MLP federated model (best model, median values were calculated from Supplemental Table 6), some variables have large amount of missing data Supplemental Table 1.
Mortality data used.
Top 5 variables and cut off (very high cutoffs) > 6.7 mg/L for D‐dimer, <94 for O2 index, >10 for NE:LY, >93 mg/L for CRP, and >450 U/L for LDH.
Top 5 feature for mortality prediction: lactate dehydrogenase, neutrophils (%), lymphocytes (%), high-sensitivity C-reactive protein, and age score (TP = 160, FP = 16, FN = 14 and TP = 184).
Our Deep-Neo-V model outperforms all our conventional models and our Deep-FLAIM model. It also outperformed the currently available Deep-learning model by Zhu et al. in terms of training set accuracy, testing set accuracy, AUROC, Precision, specificity and positive predictive value [41]. However, the Deep-Neo-V model underperformed in terms of sensitivity and slightly with negative predictive value. The Deep-Neo-V will continue to improve and develop and will potentially be replaced by a model with better performance parameters (accuracy, PPV and NPV). This model in its current configuration can be used to predict mortality after day-1 (considers labs and clinical characteristics in the first 24 h) of hospital admission and can help in the stratification of patients. It can help clinicians answer a number of questions and aid in decision-making like triaging patients, should an elderly patient receive aggressive treatment, or a younger patient receive life supportive management. These are tough questions, and the model will give clinicians clarity about the course and plan for the patient. This model can also help clinicians with family counselling, appropriate decision making, limit excessive intervention or aggressive treatments and effective resource management. Like most digital tools this current algorithm is user-friendly, can provide results instantaneously and is easy to use. After further validation this model can be incorporated into hospital patient management systems and ready for clinical use. We have also developed a web-based application for testing out the algorithium [46]
Our work does have limitations that include single institution dataset, retrospective nature of the dataset and data from a single hospital, analyzed at admission and day-one data; other observational study confounders may exist and are unaccounted for. In the immediate future our group is actively looking to validate these findings in an external dataset. In the longer term we will continue to develop an algorithm built on the Neo-V Framework approach that has the potential to be implemented, initially in future pandemics because of its ability to accurately predict outcomes using smaller datasets.
8. Conclusion
Deep-Neo-V is a statistically robust machine learning model that is developed for clinical use to predict mortality risk in patients admitted with RT-PCR proven COVID-19 infection. The mortality prediction was modeled based on clinically relevant variables (patient associated risk factors and the first 24-hours labs. Our experimental results show that with a high accuracy and specificity it has the potential to develop as a test of choice for predicting mortality in COVID-19 patients. These findings need further external validation.
9. Author statement
Maleeha Naseem (MN): Data collection lead Study design, data collection protocol design, IRB/ERC approval, data collection supervision, data collection tool and quality control, , medical literature review and article writing.
Hajra Arshad (HA): Data collection, protocol design, data collection tool, statistical analysis review, medical literature review and article writing.
Syeda Amrah Hashmi (SAH): Data collection, statistical analysis review, medical literature review and article writing.
Furqan Irfan (FI). Medical literature review, statistical analysis review, and article writing.
Fahad Shabbir Ahmed (FSA): Analytical, machine learning and web-applicationlead, original concept, study design, methodology, data collection tool and construction of FLAIM, Deep-FLAIM and Neo-V frameworks, statistical analysis and review, python-coding for machine learning, article writing, editing, review, senior and corresponding author.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ijmedinf.2021.104556.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Hsiang S., Allen D., Annan-Phan S., et al. The effect of large-scale anti-contagion policies on the COVID-19 pandemic. Nature. 2020;584(7820):262–267. doi: 10.1038/s41586-020-2404-8. [DOI] [PubMed] [Google Scholar]
- 2.Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter D.J. Trying to “Protect the NHS” in the United Kingdom. N. Engl. J. Med. 2020;383(25):e136. doi: 10.1056/NEJMp2032508. [DOI] [PubMed] [Google Scholar]
- 4.Seong H., Hyun H.J., Yun J.G., et al. Comparison of the second and third waves of the COVID-19 pandemic in South Korea: Importance of early public health intervention. Int. J. Infect. Dis. 2021;104:742–745. doi: 10.1016/j.ijid.2021.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srivastava S., Banu S., Singh P., Sowpati D.T., Mishra R.K. SARS-CoV-2 genomics: An Indian perspective on sequencing viral variants. J. Biosci. 2021;46(1):22. doi: 10.1007/s12038-021-00145-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirby T. New variant of SARS-CoV-2 in UK causes surge of COVID-19. Lancet Respir. Med. 2021;9(2):e20–e21. doi: 10.1016/S2213-2600(21)00005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ang K. Vietnam detects highly contagious new coronavirus variant as infections surge. The Washington Post. 2021 [Google Scholar]
- 8.Cacciapaglia G., Cot C., Sannino F. Second wave COVID-19 pandemics in Europe: a temporal playbook. Sci. Rep. 2020;10(1):15514. doi: 10.1038/s41598-020-72611-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ahmed F.S., Ali L., Joseph B.A., Ikram A., Ul Mustafa R., Bukhari S.A.C. A statistically rigorous deep neural network approach to predict mortality in trauma patients admitted to the intensive care unit. J. Trauma Acute Care Surg. 2020;89(4):736–742. doi: 10.1097/TA.0000000000002888. [DOI] [PubMed] [Google Scholar]
- 10.Ahmad F.S., Ali L., Raza Ul M., et al. A hybrid machine learning framework to predict mortality in paralytic ileus patients using electronic health records (EHRs) J. Ambient Intell. Humaniz. Comput. 2020 [Google Scholar]
- 11.Naseem M., Akhund R., Arshad H., Ibrahim M.T. Exploring the potential of artificial intelligence and machine learning to combat COVID-19 and existing opportunities for LMIC: A scoping review. J. Prim. Care Commun. Health. 2020;11 doi: 10.1177/2150132720963634. 2150132720963634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.CoA P. Guidance for COVID-19 testing for CAP-accredited laboratories. CAP Guidelines, caporg. 2020 [Google Scholar]
- 13.CfDCa P. CDC’s Diagnostic test for COVID-19 only and supplies. CDC Guidelines, cdcgov. 2020 [Google Scholar]
- 14.Cox D.R. Regression models and life-tables. J. Roy. Stat. Soc.: Ser. B (Methodol.) 1972;34(2):187–202. [Google Scholar]
- 15.IBM . IBM Corp.; Armonk NY: 2016. IBM SPSS Statistics for Windows, Version 24.0. [Google Scholar]
- 16.Chawla N.V., Bowyer K.W., Hall L.O., Kegelmeyer W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Int. Res. 2002;16(1):321–357. [Google Scholar]
- 17.Ahmed F.S., Raza-Ul-Mustafa Ali L., et al. Machine learning can predict deaths in patients with diverticulitis during their hospital stay. medRxiv. 2020 2020.2002.2004.20020222. [Google Scholar]
- 18.Keni R., Alexander A., Nayak P.G., Mudgal J., Nandakumar K. COVID-19: Emergence, spread, possible treatments, and global burden. Front Public Health. 2020;8:216. doi: 10.3389/fpubh.2020.00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu S., Li Y. Beware of the second wave of COVID-19. Lancet. 2020;395(10233):1321–1322. doi: 10.1016/S0140-6736(20)30845-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(CDC) CfDCaP, CDC COVID Data Tracker: Maps, charts, and data provided by the CDC. wwwcdcgov (2020).
- 21.Parikh R.B., Manz C., Chivers C., et al. Machine learning approaches to predict 6-month mortality among patients with cancer. JAMA Netw. Open. 2019;2(10):e1915997. doi: 10.1001/jamanetworkopen.2019.15997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motwani M., Dey D., Berman D.S., et al. Machine learning for prediction of all-cause mortality in patients with suspected coronary artery disease: a 5-year multicentre prospective registry analysis. Eur. Heart J. 2017;38(7):500–507. doi: 10.1093/eurheartj/ehw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pranata R., Soeroto A.Y., Huang I., Lim M.A., Santoso P., Permana H., Lukito A.A. Effect of chronic obstructive pulmonary disease and smoking on the outcome of COVID-19. Int. J. Tuberc. Lung Dis. 2020;24(8):838–843. doi: 10.5588/ijtld.20.0278. [DOI] [PubMed] [Google Scholar]
- 24.Ozturk S., Turgutalp K., Arici M., et al. Mortality analysis of COVID-19 infection in chronic kidney disease, haemodialysis and renal transplant patients compared with patients without kidney disease: a nationwide analysis from Turkey. Nephrol. Dial. Transplant. 2020;35(12):2083–2095. doi: 10.1093/ndt/gfaa271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendren N.S., Drazner M.H., Bozkurt B., Cooper L.T. Description and proposed management of the acute COVID-19 cardiovascular syndrome. Circulation. 2020;141(23):1903–1914. doi: 10.1161/CIRCULATIONAHA.120.047349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X.-H., Duan J., Han X., Liu X., Zhou J., Wang X., Zhu L., Mou H., Guo S. High incidence and mortality of pneumothorax in critically Ill patients with COVID-19. Heart Lung. 2021;50(1):37–43. doi: 10.1016/j.hrtlng.2020.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X., Ma X. Acute respiratory failure in COVID-19: is it “typical” ARDS? Crit. Care. 2020;24(1):198. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Coz Yataco A.O., Simpson S.Q. Coronavirus disease 2019 sepsis: A nudge toward antibiotic stewardship. Chest. 2020;158(5):1833–1834. doi: 10.1016/j.chest.2020.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi L.i., Wang Y., Wang Y., Duan G., Yang H. Dyspnea rather than fever is a risk factor for predicting mortality in patients with COVID-19. J. Infect. 2020;81(4):647–679. doi: 10.1016/j.jinf.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Z., Chen A., Hou W., et al. Prediction model and risk scores of ICU admission and mortality in COVID-19. PLoS One. 2020;15(7):e0236618. doi: 10.1371/journal.pone.0236618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hultström M., Persson B., Eriksson O., Lipcsey M., Frithiof R., Nilsson B. Blood type A associates with critical COVID-19 and death in a Swedish cohort. Crit. Care. 2020;24(1):496. doi: 10.1186/s13054-020-03223-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henry B.M., de Oliveira M.H.S., Benoit S., Plebani M., Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): a meta-analysis. Clin. Chem. Lab. Med. 2020;58(7):1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 33.Cheng Y., Luo R., Wang K., Zhang M., Wang Z., Dong L., Li J., Yao Y., Ge S., Xu G. Kidney disease is associated with in-hospital death of patients with COVID-19. Kidney Int. 2020;97(5):829–838. doi: 10.1016/j.kint.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altschul D.J., Unda S.R., Benton J., de la Garza Ramos R., Cezayirli P., Mehler M., Eskandar E.N. A novel severity score to predict inpatient mortality in COVID-19 patients. Sci. Rep. 2020;10(1) doi: 10.1038/s41598-020-73962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becher T., Frerichs I. Mortality in COVID-19 is not merely a question of resource availability. Lancet Respirat. Med. 2020;8(9):832–833. doi: 10.1016/S2213-2600(20)30312-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rahim F., Amin S., Noor M., et al. Mortality of patients with severe COVID-19 in the intensive care unit: An observational study from a major COVID-19 receiving hospital. Cureus. 2020;12(10) doi: 10.7759/cureus.10906. e10906–e10906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leung C. Risk factors for predicting mortality in elderly patients with COVID-19: A review of clinical data in China. Mech. Ageing Dev. 2020;188 doi: 10.1016/j.mad.2020.111255. 111255–111255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sterne J.A.C., Murthy S., Diaz J.V., et al. Association between administration of systemic corticosteroids and mortality among critically Ill patients with COVID-19: A meta-analysis. JAMA. 2020;324(13):1330–1341. doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Soltan A.A.S., Kouchaki S., Zhu T., et al. Rapid triage for COVID-19 using routine clinical data for patients attending hospital: development and prospective validation of an artificial intelligence screening test. Lancet Digit Health. 2021;3(2):e78–e87. doi: 10.1016/S2589-7500(20)30274-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabitza F., Campagner A., Ferrari D., et al. Development, evaluation, and validation of machine learning models for COVID-19 detection based on routine blood tests. Clin. Chem. Lab. Med. 2020;59(2):421–431. doi: 10.1515/cclm-2020-1294. [DOI] [PubMed] [Google Scholar]
- 41.Zhu J.S., Ge P., Jiang C., et al. Deep-learning artificial intelligence analysis of clinical variables predicts mortality in COVID-19 patients. J. Am. College Emerg. Phys. Open. 2020;1(6):1364–1373. doi: 10.1002/emp2.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaid A., Jaladanki S.K., Xu J., et al. Federated learning of electronic health records to improve mortality prediction in hospitalized patients with COVID-19: Machine learning approach. JMIR Med. Inform. 2021;9(1) doi: 10.2196/24207. e24207–e24207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Booth A.L., Abels E., McCaffrey P. Development of a prognostic model for mortality in COVID-19 infection using machine learning. Mod. Pathol. 2021;34(3):522–531. doi: 10.1038/s41379-020-00700-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chowdhury M.E.H., Rahman T., Khandakar A., et al. An early warning tool for predicting mortality risk of COVID-19 patients using machine learning. Cogn. Comput. 2021 doi: 10.1007/s12559-020-09812-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li X., Ge P., Zhu J., et al. Deep learning prediction of likelihood of ICU admission and mortality in COVID-19 patients using clinical variables. PeerJ. 2020;8 doi: 10.7717/peerj.10337. e10337–e10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.2021. SARS-COV-2 (COVID-19) in-patient mortality prediction web-application
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.