Abstract
Background
Proinflammatory cytokines are correlated with the severity of disease in patients with COVID-19. IL6-mediated activation of STAT3 proliferates proinflammatory responses that lead to cytokine storm promotion. Thus, STAT3 inhibitors may play a crucial role in managing the COVID-19 pathogenesis. The present study discusses a method for predicting inhibitors against the STAT3 signaling pathway.
Method
The main dataset comprises 1565 STAT3 inhibitors and 1671 non-inhibitors used for training, testing, and evaluation of models. A number of machine learning classifiers have been implemented to develop the models.
Results
The outcomes of the data analysis show that rings and aromatic groups are significantly abundant in STAT3 inhibitors compared to non-inhibitors. First, we developed models using 2-D and 3-D chemical descriptors and achieved a maximum AUC of 0.84 and 0.73, respectively. Second, fingerprints are used to build predictive models and achieved 0.86 AUC with an accuracy of 78.70% on the validation dataset. Finally, models were developed using hybrid descriptors, which achieved a maximum of 0.87 AUC with 78.55% accuracy on the validation dataset.
Conclusion
We used the best model to identify STAT3 inhibitors in FDA-approved drugs and found few drugs (e.g., Tamoxifen and Perindopril) to manage the cytokine storm in COVID-19 patients. A webserver “STAT3In” (https://webs.iiitd.edu.in/raghava/stat3in/) has been developed to predict and design STAT3 inhibitors.
Keywords: STAT3, COVID-19, FDA-Approved, Cytokine, Inhibitors
Graphical abstract
1. Introduction
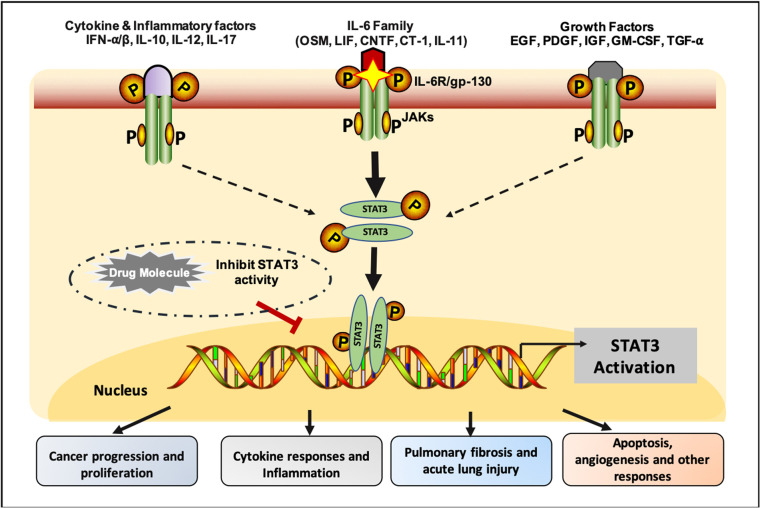
Numerous studies have shown that the elevated level of pro-inflammatory cytokines play a critical role in the COVID-19 pathogenesis [[1], [2], [3], [4]]. The activation of the IL6/STAT3 signaling pathway is associated with the cytokine storm which is responsible for the high mortality rate among COVID-19 patients [5,6]. STAT3 is a cytoplasmic transcription factor, which participates in the normal cellular events, including differentiation, proliferation, and angiogenesis [7]. STAT3 is activated in response to various cytokines, chemokines, and growth factors [8,9]. As shown in Fig. 1 , these factors bind to the Janus kinases and phosphorylate STAT3 monomers to form a homodimer molecule and regulate the gene transcription [9]. However, the upregulation of STAT3 is correlated with pathological events such as cancer proliferation, angiogenesis, and cytokine storm in COVID-19 [10,11]. Various preclinical and clinical evidence has confirmed that STAT3 is a promising potential therapeutic target [12]. Aberration in STAT3 transcription, increases several gene expressions (such as Bcl-xL, Fas, Fas-L, CASP3), responsible for oncogenesis, and apoptosis [[13], [14], [15]]. Evidence indicated that mutations in the STAT3 gene provoked different diseases, such as Type 1 diabetes, pulmonary fibrosis, and acute lung injury [[16], [17], [18]].
Fig. 1.
Schematic representation of STAT3 signaling pathway.
STAT3 hyperactivation promotes COVID-19 pathogenesis via elevation of cytokines storm production [6,[19], [20], [21]]. Thus, it is critical to target IL6 mediated STAT3 activation to manage the pathogenesis of infectious diseases. At present, several STAT3 inhibitors are in clinical trials. For example, pyrrolidinesulphonylaryl molecules (6a), demonstrate promising activity against IL6/STAT3 signaling in breast cancer [22]. In addition, several FDA-approved drugs (e.g., Celecoxib, BBI608, Pyrimethamine) are under clinical trials for cancer immunotherapy [23]. However, the finding of a novel STAT3 inhibitor against the COVID-19 disease remains a major scientific challenge. Thus, it is vital to target the IL6/STAT3 signaling pathway to achieve a better therapeutic candidate against COVID-19.
No computational tool predicts the chemical moieties as potential STAT3 inhibitors accurately. The present study develops a computational model for predicting STAT3 inhibitors. We have used 3236 chemical compounds (STAT3 inhibitors and non-inhibitors) and 16,112 (2-D, 3-D, and FP) descriptors to generate prediction models. To better serve the scientific community, we provide a computational tool “STAT3In” (https://webs.iiitd.edu.in/raghava/stat3in/) to predict and design potential STAT3 inhibitor candidates.
2. Materials and methods
2.1. Dataset collection
The IL-6 mediated STAT3 inhibitors and non-inhibitors were collected from the PubChem repository (https://pubchem.ncbi.nlm.nih.gov). We searched all the assays in PubChem using following keyword “((IL-6 AND STAT3) inhibitors)” and obtained approximately 251 PubChem bioassays. Next, we manually refined the obtained assays based on the number of inhibitors per assay and selected assays that possessed the maximum number of inhibitors. After this rigorous selection criteria without compromising data quality, we selected bioassay AID 862 (see URL: https://pubchem.ncbi.nlm.nih.gov/bioassay/862). The high-throughput bioassay was based on a luciferase experiment performed on STAT-1 deficient human U3A fibrosarcoma cell line, which contained STAT3:luciferase reporter activity. In the assay, 28 nL of the test compound in DMSO (5.5 M final nominal concentration; 0.6% DMSO) was dispensed into sample field wells. However, the control wells received nifuroxazide in DMSO (100 M final concentration; 0.6% DMSO final concentration) or in DMSO only (0.6% final concentration) [24]. IL6 was used as a reaction triggering agent, and the reaction response was monitored via luciferase activity. STAT3 inhibitors were termed as chemicals that induced loss or declined during the luciferase activity in the presence of IL-6.
In this bioassay, a total of 194,698 compounds was tested to identify STAT3 inhibitors. From these compounds, 1724 were reported as STAT3 inhibitors and 192,974 as non-inhibitors. Then, we randomly selected 1724 non-inhibtor compounds to create a balanced dataset. Furthermore, we filtered those compounds whose three-dimensional structures are unavailable. The final dataset contained 1565 inhibitors and 1671 non-inhibitors. We used the standard protocols, for the inhibitors and non-inhibitors classification, frequently implemented by previous studies [[25], [26], [27], [28], [29]]. We divided the entire dataset into 80:20 ratio, where 80% of the data (i.e., 1252 inhibitors, and 1337 non-inhibitors) were used for training. However, the remaining 20% (i.e., 313 inhibitors, and 334 non-inhibitors) of the data were used for validation.
2.2. Descriptors of molecules
Chemical descriptors are the mathematical representations of chemical molecules that transform chemical information into standardized activities. This study used PaDEL software [30] to calculate the chemical descriptors of molecules. The software computed several 1-D/2-D/3-D and binary fingerprints (FP) (e.g., Fingerprinter, Extended, KlekotaRoth count, SubStructure, MACCS keys). From the computation, we obtained 1444 2-D descriptors, 136 3-D descriptors, and 14532 FP descriptors for 1565 positive and 1671 negative compounds. These 2-D, 3-D, and FP descriptors were used to develop various machine learning models.
2.3. Pre-processing of data
The calculated descriptors were lying in a varying range. We normalized each descriptor file using a standard scaler package of Scikit learn to preprocess the dataset. sklearn.preprocessing.StandardScaler is a method that uses a z-score algorithm to normalize the data. Afterward, we removed the null values from each descriptor file. 2-D and FP descriptor files do not have any null values. However, a few null values were found in the 3-D descriptor file. Hence, we were left with 1444 2-D, 116 3-D, and 14532 FP descriptors/features for the entire dataset.
2.4. Selection and ranking of significant descriptors
Previous studies showed that all the calculated descriptors using PaDEL software were irrelevant [25,31]. Thus, selecting the most significant descriptors is a vital step to develop a good prediction model. This study used three major feature selection techniques as follows: VarianceThreshold-based method, correlation-based method, and SVC-L1-based method. We used the VarianceThreshold package of Scikit (sklearn.feature_selection) to remove the low-variance features. Initially, we recorded 1444 2-D, 116 3-D, and 14,532 FP descriptors. After eliminating low variance features, we were left with 622 2-D, 66 3-D, and 2251 FP descriptors. Thus, a correlation-based feature selection method was used to select the features correlated with the coefficient of less than 0.6. Then, we removed the features, in which their correlations were greater than or equal to 0.6. After this process, we were left with 74 2-D, 9 3-D, and 1622 FP descriptors. Finally, we used the SVC-L1 feature selection technique to obtain the most significant feature set. This popular method was effective to minimize the feature vector size. Using the SVC-L1 approach, we obtained the most essential 41 2-D, 5 3-D, and 116 FP descriptors. Furthermore, we used the combination of 2-D, 3-D, FP descriptors to develop a hybrid model. Using the feature-selector program, we ranked 162 features based on their importance to classify the inhibitors/non-inhibitors. The program uses the gradient boosting decision tree, a popular machine learning algorithm, also known as LightGBM. To rank the features through the estimation, the program calculated the number of times a feature split the data across all trees [32]. The selected descriptors were ranked to develop different machine learning models where the performance was computed on top-10, 20, 30, ….162 features.
2.5. Cross-validation techniques
We used the standard five-fold cross-validation technique to train, test, and evaluate our prediction model on the training dataset. In this technique, the training dataset was divided into five sets of the similar size. Of these five sets, four sets were used for training, and the fifth set will be used for testing purposes. The same process is iterated five times so that each of the five sets will be used, at least once, for testing the model. Finally, average performance was computed on five test sets. We tune parameters to optimize the model's performance and achieve the best performance of test sets. To validate the performance of the best model, we selected 20% of data, not used for training or testing of these models. This method was a standard procedure extensively used by the previous studies [[33], [34], [35]].
2.6. Machine learning-based classifiers
This study used several machine learning techniques to develop the prediction models and classify STAT3 inhibitors/non-inhibitors. We implemented Random Forest (RF), Decision Tree (DT), Logistic Regression (LR), Support Vector Classifier (SVC), Gaussian Naive Bayes (GNB), K-nearest neighbour (KNN), and eXtreme Gradient Boosting (XGB) to develop classification models. These machine learning algorithms were implemented in Scikit-learn package [36].
2.7. Performance evaluation parameters
We used the standard evaluation parameters to evaluate the performance of different prediction models. Moreover, we used the standard evaluation parameters. In this study, we have used both threshold-dependent and independent parameters. The model's performance was measured using threshold-dependent parameters such as sensitivity (Sens), specificity (Spec), accuracy (Acc), and Matthews correlation coefficient (MCC). However, the threshold-independent parameter, i.e., the area under the receiver operating characteristic curve (AUC), was used to evaluate the model performance. Some previous studies had used these parameters extensively to evaluate the model's performance [37,38].
| (1) |
| (2) |
| (3) |
| (4) |
Where FP, FN, TP, and TN are false positive, false negative, true positive, and true negative, respectively.
3. Results
3.1. Functional groups analysis
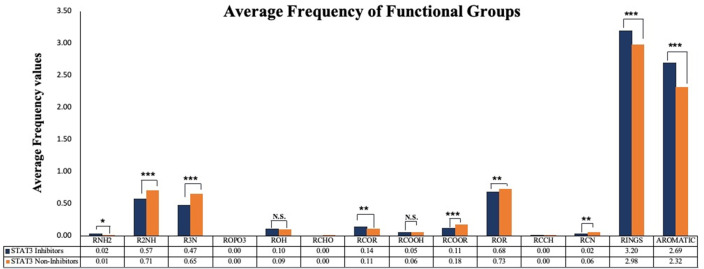
The ChemmineR package was used to calculate the frequency of functional groups of STAT3 inhibitors and non-inhibitors [39]. We analyzed the average frequency values, and found the abundance of rings, and aromatic groups in inhibitors when compared to non-inhibitors. As shown in Fig. 2 , the frequency of secondary amines (R2NH), tertiary amines (R3N), and ester (ROR) groups is significantly elevated in non-inhibitor compounds.
Fig. 2.
Representation of average values of various functional groups in STAT3 inhibitors and non-inhibitors.
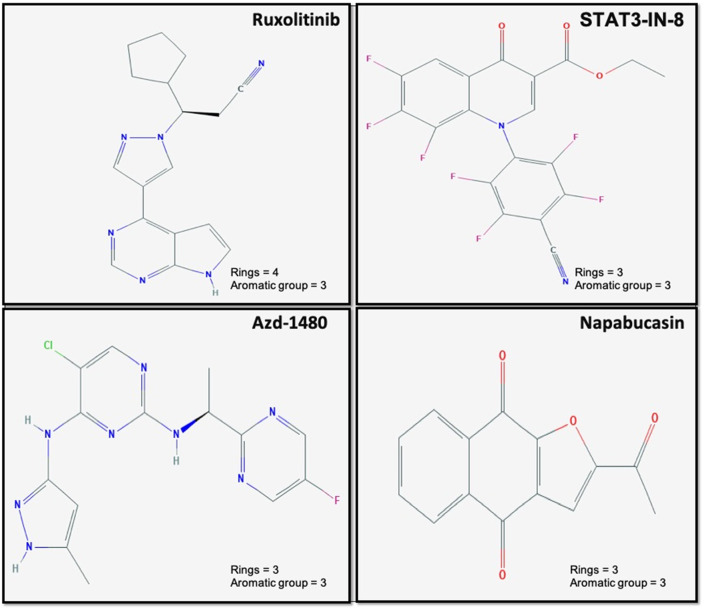
In addition, we observed that the occurrence of the rings and aromatic groups in few existing FDA-approved STAT3 inhibitors such as Napabucasin (BBI608), and STAT3 Inhibitor VII. These drugs are effective to treat advanced malignancies. Some indirect STAT3 inhibitors like AZD-1480 and Ruxolitinib (FDA-approved) also exhibit similar trends. Fig. 3 shows the presence of the functional groups in the chemical 2-D-structures, known as STAT3 inhibitors, i.e., STAT3 Inhibitor VII, Ruxolitinib, AZD-1480, and BBI608. These findings suggest that the analysis can be used to design the novel drug candidates serving as an inhibitor of the STAT3 signaling pathway.
Fig. 3.
Represents the abundance of rings and aromatic functional groups in STAT3 inhibitors (i.e., Ruxolitinib, STAT3 Inhibitor VII, AZD-1480 and B, BI608).
3.2. Prediction models
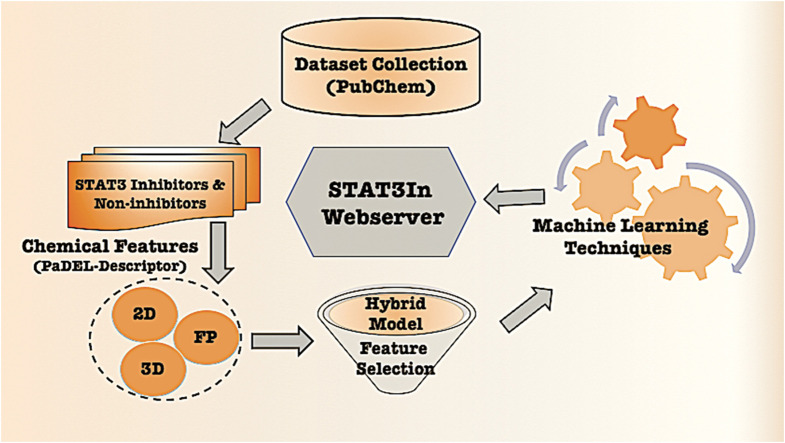
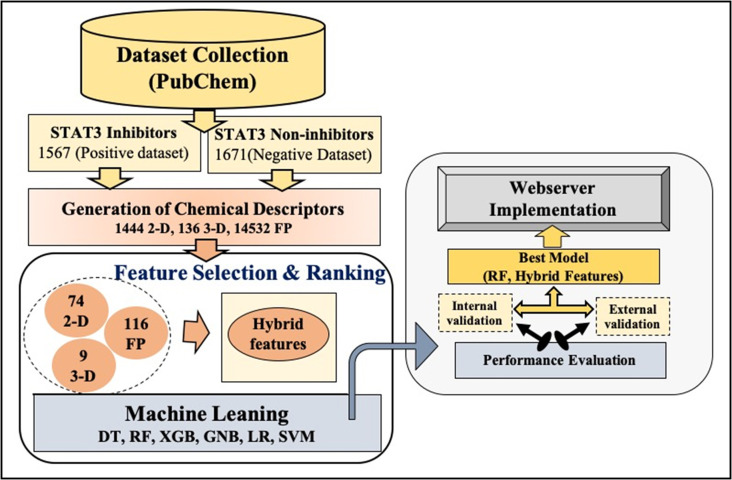
A major challenge is to choose the most appropriate descriptors to classify the chemicals since many of the descriptors are unimportant. Several feature selection techniques are used to select the best features for the classification. After selecting the best features, we developed several prediction models using machine learning-based classifiers such as RF, DT, LR, XGB, SVC, and GBM. Fig. 4 shows the complete architecture.
Fig. 4.
Overall framework of the STAT3In, including creation of the dataset, feature selection and model development.
4. Performance of classification models
4.1. 2-D descriptors
We computed 1444 2-D descriptors initially, after removing low variance and highly correlated features we were left with 74 features. These features were further used to develop classification models for discriminating inhibitors and non-inhibitors. Of all the classifiers, RF attains the maximum performance (0.84 AUC) with balanced sensitivity and specificity; complete information is available in Supplementary Table S1. Furthermore, we obtained 41 2-D descriptors with the help of the SVC-L1 method. A slight alteration is observed after reducing the features with the AUC of 0.83 and 76.35% accuracy on the training dataset and AUC 0.84 with the 75.46% accuracy on the validation datasets (Table 1 ).
Table 1.
The performance of machine-learning models on training and validation dataset with best 41 2-D descriptors.
| Classifier | Training Dataset |
Validation Dataset |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | AUC | MCC | Sensitivity | Specificity | Accuracy | AUC | MCC | |
| DT | 64.15 | 64.29 | 64.22 | 0.69 | 0.28 | 72.24 | 59.74 | 66.20 | 0.73 | 0.32 |
| RF | 76.10 | 76.58 | 76.35 | 0.83 | 0.53 | 74.63 | 76.36 | 75.46 | 0.84 | 0.51 |
| LR | 69.68 | 69.00 | 69.32 | 0.75 | 0.39 | 71.64 | 69.01 | 70.37 | 0.77 | 0.41 |
| XGB | 71.55 | 71.80 | 71.68 | 0.78 | 0.43 | 72.54 | 70.93 | 71.76 | 0.80 | 0.44 |
| KNN | 70.33 | 70.40 | 70.36 | 0.77 | 0.41 | 70.75 | 70.93 | 70.83 | 0.79 | 0.42 |
| GNB | 65.20 | 66.13 | 65.69 | 0.70 | 0.31 | 69.55 | 68.05 | 68.83 | 0.73 | 0.38 |
| SVC | 74.80 | 73.79 | 74.27 | 0.81 | 0.49 | 71.34 | 74.76 | 72.99 | 0.81 | 0.46 |
#DT: Decision Tree; RF: Random Forest; LR: Logistic Regression; XGB: eXtreme Gradient Boosting; KNN: K-nearest neighbour; GNB: Gaussian Naive Bayes; SVC: Support Vector Classifier; AUC: Area under the receiver operating characteristic curve; MCC: Matthews correlation coefficient.
4.2. 3-D descriptors
We performed feature selection on 3-D descriptors and obtained nine 3-D descriptors, which were used for developing classification models. Our RF-based model achieved a maximum AUC of 0.75 on training and an AUC of 0.74 on the validation dataset. Supplementary Table S2 shows the complete information. After removing four features with the help of SVC-L1, the performance is computed on the best five 3-D descriptors. The outcome shows that RF outperforms all other classifiers and obtains the highest AUC of 0.74 on training dataset and an AUC of 0.73 on validation dataset. However, XGB performs excellently by achieving AUC 0.73 on training data and AUC 0.72 on validation data, as shown in Table 2 .
Table 2.
Performance of machine learning models on 5 selected 3-D descriptors on training and validation dataset.
| Classifier | Training Dataset |
Validation Dataset |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | AUC | MCC | Sensitivity | Specificity | Accuracy | AUC | MCC | |
| DT | 64.80 | 62.00 | 63.33 | 0.68 | 0.27 | 67.16 | 51.76 | 59.72 | 0.66 | 0.19 |
| RF | 67.15 | 66.35 | 66.73 | 0.74 | 0.34 | 66.27 | 65.18 | 65.74 | 0.73 | 0.31 |
| LR | 65.77 | 65.54 | 65.65 | 0.71 | 0.31 | 65.67 | 64.54 | 65.12 | 0.70 | 0.30 |
| XGB | 65.29 | 66.94 | 66.15 | 0.73 | 0.32 | 65.67 | 66.13 | 65.90 | 0.72 | 0.32 |
| KNN | 68.21 | 67.01 | 67.58 | 0.74 | 0.35 | 69.85 | 62.62 | 66.36 | 0.73 | 0.33 |
| GNB | 65.85 | 65.69 | 65.77 | 0.71 | 0.32 | 67.46 | 61.98 | 64.82 | 0.70 | 0.30 |
| SVC | 66.91 | 66.50 | 66.69 | 0.73 | 0.33 | 66.87 | 65.18 | 66.05 | 0.71 | 0.32 |
#DT: Decision Tree; RF: Random Forest; LR: Logistic Regression; XGB: eXtreme Gradient Boosting; KNN: K-nearest neighbour; GNB: Gaussian Naive Bayes; SVC: Support Vector Classifier; AUC: Area under the receiver operating characteristic curve; MCC: Matthews correlation coefficient.
4.3. Fingerprints
We developed classification models using fingerprints descriptors, and we obtained 1622 fingerprints after removing low variance and highly correlated descriptors. These selected fingerprints are used for developing prediction models. The RF-based models achieved the maximum performance with AUC 0.86 on both training and validation dataset. The SVC also achieved comparable performance with AUC (training data = 0.84 and testing data = 0.85). Supplementary Table S3 shows the results of other classifiers. In addition, we developed models using 116 features. With the SVC-L1 method, we achieved nearly the same performance (Table 3 ). The results show that fingerprints-based models outperform the classification models based on 2-D and 3-D chemical features.
Table 3.
The performance of machine learning models on 116 FP based features on training and validation dataset.
| Classifier | Training Dataset |
Validation Dataset |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | AUC | MCC | Sensitivity | Specificity | Accuracy | AUC | MCC | |
| DT | 64.96 | 65.24 | 65.11 | 0.71 | 0.30 | 67.46 | 61.66 | 64.66 | 0.70 | 0.29 |
| RF | 78.46 | 77.61 | 78.01 | 0.86 | 0.56 | 79.40 | 77.96 | 78.70 | 0.86 | 0.57 |
| LR | 75.85 | 76.66 | 76.28 | 0.83 | 0.53 | 72.84 | 76.68 | 74.69 | 0.81 | 0.50 |
| XGB | 77.32 | 77.54 | 77.43 | 0.84 | 0.55 | 77.91 | 80.83 | 79.32 | 0.86 | 0.59 |
| KNN | 76.18 | 75.04 | 75.58 | 0.83 | 0.51 | 77.02 | 73.80 | 75.46 | 0.83 | 0.51 |
| GNB | 73.98 | 74.08 | 74.03 | 0.81 | 0.48 | 69.55 | 73.80 | 71.61 | 0.79 | 0.43 |
| SVC | 78.62 | 78.35 | 78.48 | 0.86 | 0.57 | 77.31 | 80.19 | 78.70 | 0.86 | 0.58 |
#DT: Decision Tree; RF: Random Forest; LR: Logistic Regression; XGB: eXtreme Gradient Boosting; KNN: K-nearest neighbour; GNB: Gaussian Naive Bayes; SVC: Support Vector Classifier; AUC: Area under the receiver operating characteristic curve; MCC: Matthews correlation coefficient.
4.4. Hybrid descriptors
To develop the models and improve the performance, we combine the selected 2-D (41 features), 3-D (5 features), and fingerprints (116 features) descriptors. The RF-based model's performance using these combined features was 0.87 and 0.88 AUC on the training and validation dataset, respectively (refer to Supplementary Table S4). Furthermore, we performed feature ranking on the combined 162 features using the feature selector algorithm. Finally, we obtained a minimum set of features that offered nearly similar performance of the above-mentioned models. First, we have ranked the features based on their performances and checked the performance using the top-10, 20, 30, ….162 features. Supplementary Table S5 shows the performance of all the combined 162 features. Finally, we selected the top-49 descriptors (i.e., 14 2-D, 1 3-D and 34 FP) out of 162 feature set as shown in Supplementary Table S4. Models developed on top-49 features performed almost similar as 162 features. The RF-based model obtained the maximum AUC of 0.87, and accuracy >78.5 for training and testing dataset with minimum sensitivity and specificity difference. Table 4 shows the results of all other classifiers i.e., SVC, DT, KNN, LR, XGB, and GBM.
Table 4.
The performance of machine learning based models developed using hybrid descriptors (2-D+3-D + FP) on training and validation dataset.
| Classifier | Training Dataset |
Validation Dataset |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Accuracy | AUC | MCC | Sensitivity | Specificity | Accuracy | AUC | MCC | |
| DT | 68.22 | 68.03 | 68.12 | 0.74 | 0.36 | 66.67 | 72.70 | 69.91 | 0.74 | 0.39 |
| RF | 78.42 | 78.61 | 78.52 | 0.87 | 0.57 | 79.00 | 78.16 | 78.55 | 0.87 | 0.57 |
| LR | 77.00 | 76.34 | 76.66 | 0.84 | 0.53 | 75.67 | 77.87 | 76.85 | 0.83 | 0.54 |
| XGB | 77.31 | 77.10 | 77.20 | 0.85 | 0.54 | 80.00 | 75.29 | 77.47 | 0.85 | 0.55 |
| KNN | 74.94 | 75.89 | 75.43 | 0.83 | 0.51 | 78.00 | 75.58 | 76.70 | 0.83 | 0.53 |
| GNB | 74.23 | 74.00 | 74.11 | 0.81 | 0.48 | 75.33 | 72.99 | 74.07 | 0.80 | 0.48 |
| SVC | 77.71 | 77.55 | 77.63 | 0.86 | 0.55 | 78.33 | 76.72 | 77.47 | 0.85 | 0.55 |
#DT: Decision Tree; RF: Random Forest; LR: Logistic Regression; XGB: eXtreme Gradient Boosting; KNN: K-nearest neighbour; GNB: Gaussian Naive Bayes; SVC: Support Vector Classifier; AUC: Area under the receiver operating characteristic curve; MCC: Matthews correlation coefficient.
5. Repurposing of FDA-approved drugs to target STAT3
We retrieved 1102 FDA-approved drug molecules from the Drug Bank database to identify the potential drug candidates for the inhibition of the STAT3 pathway [40]. From the FDA-approved drugs, we identified the PubChem CID (compound ID). Out of 1102 drugs, a total of 842 drugs comprises the 2-D structures. Furthermore, we use SDF files of 842 molecules, to identify the potential drug candidates. We used the “Predict” module of our web server “STAT3In” (with default parameters, i.e., Random Forest Threshold = 0.48). Our model predicts 19 potential drug candidates for STAT3 inhibition. Numerous previous studies support our findings, showing that these drugs are inhibitors in diseases linked with IL6/STAT3 activation [[41], [42], [43], [44], [45]]. We identify eight potential drugs (warfarin, dexpanthenol, perindopril, tamoxifen, pentagastrin, duloxetine, ledipasvir, and, olopatadine) that are effective to treat severe diseases like tumor progression, angiogenesis, COVID-19 progression, and good to inhibit IL6/STAT3 pathway, as depicted in Table 5 .
Table 5.
Potential FDA-approved drug candidates predicted by our web server (STAT3In) for STAT3 inhibition.
| Drug Bank ID | FDA-Approved Drugs |
STAT3In Prediction |
Functions |
|---|---|---|---|
| DB00682 | Warfarin | Inhibitor | Inhibition of IL6/STAT3-dependent fibrin production in severe listeriosis [42]. |
| DB09357 | Dexpanthenol | Inhibitor | Inhibition of LPS-induced neutrophils influx, protein leakage, and release of TNF-α and IL6 in bronchoalveolar lavage fluid in acute lung injury [43]. |
| DB00790 | Perindopril | Inhibitor | It regulates the inflammatory mediators, NF-κB/TNF-α/IL6, and apoptosis in renal diseases [44] and inhibit the activation of STAT3 [45]. ACE inhibitor perindopril-inhibited tumor growth was associated with the suppression of angiogenesis [46]. |
| DB00675 | Tamoxifen | Inhibitor | Treatment of ER‐positive breast cancer with tamoxifen by inhibiting the IL6/STAT3 signal pathway, inhibition of tumor growth and angiogenesis [47,48]. Anticancer drugs that have shown potential activity in both MERS and SARS-CoV [41]. |
| DB00183 | Pentagastrin | Inhibitor | Anti-malarial, anti-fungal, anti-bacterial, and anti-inflammatory [49]. |
| DB00476 | Duloxetine | Inhibitor | Inhibit overexpression of IL6 mRNA in anxiety- and major depressive disorder, anti-inflammatory action against IL6 [[50], [51], [52]]. |
| DB09027 | Ledipasvir | Inhibitor | Anti-viral activity against COVID-19 [53], (sofosbuvir, and ledipasvir) inhibited STAT3 protein levels to cure HCV infections [54]. |
| DB00768 | Olopatadine | Inhibitor | Inhibit CHMCs activation and release of IL6, tryptase, and histamine and use as anti-allergy drug [55]. |
5.1. Webserver implementation
We developed a webserver STAT3In to classify STAT3 inhibitors and non-inhibitors. The web server is hosted on a Linux (Ubuntu) machine using an Apache HTTP server. The front-end of STAT3In is developed using HTML, PHP, and JavaScript. However, the back-end is developed using Python3.6 and Scikit library. We also used a responsive template to make our website compatible with desktop, tablet, laptop, and smartphones. We implemented the random forest model using hybrid chemical descriptors as the input features, in the back-end of the server. Three major modules are in the webserver, named as “Predict,” “Draw,” and “Analog design”. The comprehensive description of each module is presented below.
5.2. Predict
The predict module helps user to classify the uncharacterized chemical compound as STAT3 inhibitor or non-inhibitor. The module accepts chemical compounds in various formats, such as SDF, SMILES, and MOL, from the users and also allows the user to select the desired threshold. The users can enter either a single molecule, or multiple molecules and can also upload a file consisting of multiple chemical compounds. The output page provides the class(es) of the submitted compound(s) as STAT3 inhibitor or non-inhibitor, along with their machine learning score. The result is presented in comma-separated value (CSV) format to search or sort the output table.
5.3. Draw
In this module, users can draw or alter the chemical molecule structure and transfer it to the prediction model to classify the molecule as a STAT3 inhibitor or non-inhibitor. We implemented Ketcher [56], an open-source web-based chemical structure editor, to carry out the interactive process. The users can select the threshold based on their suitability. The output page shows the predicted class of the molecule in the tabular form, downloadable in CSV format.
5.4. Analog design
In the analog design module, users can generate the analogs using a combination of submitted scaffolds, building blocks, and linkers. We implemented SmiLib [57] software to generate the analogs. Subsequently, the generated analogs are classified into STAT3 inhibitors or non-inhibitors based on the selected threshold. The result page exhibits the class of the generated analogs as inhibitors and non-inhibitors along with their machine learning score in the tabular form, downloadable in CSV format.
6. Discussion and conclusion
STAT3 is one of the most crucial transcription factors and oncogene that plays a significant role in the onset and progression of the tumor. STAT3 is identified as an excellent therapeutic target for various cancer owing to its versatile regulatory pathways and biological roles in cancer [58]. Moreover, many studies confirm that the IL6 concentration is significantly severe in COVID-19 patients globally. Cytokine IL6 mediates its effect via JAK/STAT3 pathway. Therefore, computation methods are crucial to predict a potent chemical molecule to serve as a STAT3 inhibitor. Numerous methods were developed to exploit the structure-activity of the chemical molecules and predict the potential chemical molecule that can serve as an inhibitor, such as EGFRpred [31] which predicts the EGFR inhibitor potential of a molecule. Using machine learning methods, DrugMint [25] predicts if a molecule is a potential drug candidate.
In this study, we attempted to develop a computational method to discriminate the STAT3 inhibitors from non-inhibitors. We observed a high frequency of rings and a low frequency of R2NH, R3N, ROR groups in STAT3 inhibitor compounds. We found the same trend in already existing STAT3 drugs such as AZD-1480, Ruxolitinib, Napabucasin, and STAT3 Inhibitor VII. We consider STAT3 inhibitors and non-inhibitors as the positive and negative datasets to develop the prediction models. Random forest-based models achieve maximum performance (AUC 0.87 with an accuracy of 78.55) on the validation dataset using hybrid descriptors. Furthermore, we used 842 FDA-approved drugs, to identify potential drug candidates against STAT3 activation. As revealed in Table 5, we identify a few drugs that can inhibit IL6/STAT3 activation and can be used as a drug candidate against cytokine storm [59,60] associated with COVID-19. Using machine learning with minimal features derived from chemical molecules, a webserver named STAT3In is developed to predict and design the potential STAT3 inhibitors. We hope that this method will aid researchers working in the field of cancer therapy and infectious diseases, such as COVID-19.
6.1. Limitation of the study
In the current study, we used state-of-the-art techniques to develop a prediction tool and identify STAT3 inhibitor/non-inhibitor chemical compounds. However, the models were built on the chemical compounds tested only on a single cell line “human U3A fibrosarcoma”. Ideally, the study should be performed on animal models or a wider range of cell lines to develop a rigorous method. In the future, we hope to design an upgraded version once we obtained a sufficient experimentally validated data on IL-6/STAT3 inhibition.
Funding source
The current work has not received any specific grant from any funding agencies.
Availability of data and material
All the datasets generated for this study are available at the “STAT3In” webserver, https://webs.iiitd.edu.in/raghava/stat3in/dataset.php.
Authors’ contributions
AD, and SP collected and processed the datasets. AD, SP and GPSR implemented the algorithms and developed the prediction models. AD, SP, NS and GPSR analyzed the results. SP created the back-end of the web server and AD created the front-end user interface. AD, NS, NLD, SP, and GPSR penned the manuscript. GPSR conceived and coordinated the project, and gave overall supervision to the project. All authors have read and approved the final manuscript.
Declaration of competing interest
The authors declare no competing financial and non-financial interests.
Acknowledgements
AD and NS are thankful to the Department of Science and Technology (DST-INSPIRE) and SP is thankful to Department of Biotechnology (DBT) for providing senior research fellowships. NLD is thankful to Department of Biotechnology (DBT) for providing Research Associate fellowship. The authors are thankful to Department of Computational Biology, IIITD New Delhi for infrastructure and facilities.
Research Square DOI: 10.21203/rs.3.rs-495671/v1.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104780.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Merad M., Martin J.C. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mehta P., McAuley D.F., Brown M., Sanchez E., Tattersall R.S., Manson J.J., Hlh U.K. Across Speciality Collaboration, COVID-19: consider cytokine storm syndromes and immunosuppression. Lancet. 2020;395:1033–1034. doi: 10.1016/S0140-6736(20)30628-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore J.B., June C.H. Cytokine release syndrome in severe COVID-19. Science. 2020;368:473–474. doi: 10.1126/science.abb8925. [DOI] [PubMed] [Google Scholar]
- 4.Ye Q., Wang B., Mao J. The pathogenesis and treatment of the 'Cytokine Storm'in COVID-19. J. Infect. 2020;80:607–613. doi: 10.1016/j.jinf.2020.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo W., Li Y.X., Jiang L.J., Chen Q., Wang T., Ye D.W. Targeting JAK-STAT signaling to control cytokine release syndrome in COVID-19. Trends Pharmacol. Sci. 2020;41:531–543. doi: 10.1016/j.tips.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jafarzadeh A., Nemati M., Jafarzadeh S. Contribution of STAT3 to the pathogenesis of COVID-19. Microb. Pathog. 2021;154:104836. doi: 10.1016/j.micpath.2021.104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calo V., Migliavacca M., Bazan V., Macaluso M., Buscemi M., Gebbia N., Russo A. STAT proteins: from normal control of cellular events to tumorigenesis. J. Cell. Physiol. 2003;197:157–168. doi: 10.1002/jcp.10364. [DOI] [PubMed] [Google Scholar]
- 8.Levy D.E., Lee C.K. What does Stat3 do? J. Clin. Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma J.H., Qin L., Li X. Role of STAT3 signaling pathway in breast cancer. Cell Commun. Signal. 2020;18:33. doi: 10.1186/s12964-020-0527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee H., Jeong A.J., Ye S.K. Highlighted STAT3 as a potential drug target for cancer therapy. BMB Rep. 2019;52:415–423. doi: 10.5483/BMBRep.2019.52.7.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corvinus F.M., Orth C., Moriggl R., Tsareva S.A., Wagner S., Pfitzner E.B., Baus D., Kaufmann R., Huber L.A., Zatloukal K., Beug H., Ohlschlager P., Schutz A., Halbhuber K.J., Friedrich K. Persistent STAT3 activation in colon cancer is associated with enhanced cell proliferation and tumor growth. Neoplasia. 2005;7:545–555. doi: 10.1593/neo.04571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong A.L.A., Hirpara J.L., Pervaiz S., Eu J.Q., Sethi G., Goh B.C. Do STAT3 inhibitors have potential in the future for cancer therapy? Expet Opin. Invest. Drugs. 2017;26:883–887. doi: 10.1080/13543784.2017.1351941. [DOI] [PubMed] [Google Scholar]
- 13.Furqan M., Mukhi N., Lee B., Liu D. Dysregulation of JAK-STAT pathway in hematological malignancies and JAK inhibitors for clinical application. Biomark Res. 2013;1:5. doi: 10.1186/2050-7771-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerjee K., Resat H. Constitutive activation of STAT3 in breast cancer cells: a review. Int. J. Canc. 2016;138:2570–2578. doi: 10.1002/ijc.29923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buettner R., Mora L.B., Jove R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin. Canc. Res. 2002;8:945–954. [PubMed] [Google Scholar]
- 16.Gao H., Guo R.F., Speyer C.L., Reuben J., Neff T.A., Hoesel L.M., Riedemann N.C., McClintock S.D., Sarma J.V., Van Rooijen N., Zetoune F.S., Ward P.A. Stat3 activation in acute lung injury. J. Immunol. 2004;172:7703–7712. doi: 10.4049/jimmunol.172.12.7703. [DOI] [PubMed] [Google Scholar]
- 17.Yang X.O., Panopoulos A.D., Nurieva R., Chang S.H., Wang D., Watowich S.S., Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J. Biol. Chem. 2007;282:9358–9363. doi: 10.1074/jbc.C600321200. [DOI] [PubMed] [Google Scholar]
- 18.Shao S., He F., Yang Y., Yuan G., Zhang M., Yu X. Th17 cells in type 1 diabetes. Cell. Immunol. 2012;280:16–21. doi: 10.1016/j.cellimm.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 19.Gubernatorova E.O., Gorshkova E.A., Polinova A.I., Drutskaya M.S. IL-6: relevance for immunopathology of SARS-CoV-2. Cytokine Growth Factor Rev. 2020;53:13–24. doi: 10.1016/j.cytogfr.2020.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jafarzadeh A., Jafarzadeh S., Nozari P., Mokhtari P., Nemati M. Lymphopenia an important immunological abnormality in patients with COVID-19: possible mechanisms. Scand. J. Immunol. 2021;93 doi: 10.1111/sji.12967. [DOI] [PubMed] [Google Scholar]
- 21.Chen X., Tang J., Shuai W., Meng J., Feng J., Han Z. Macrophage polarization and its role in the pathogenesis of acute lung injury/acute respiratory distress syndrome. Inflamm. Res. 2020;69:883–895. doi: 10.1007/s00011-020-01378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zinzalla G., Haque M.R., Basu B.P., Anderson J., Kaye S.L., Haider S., Hasan F., Antonow D., Essex S., Rahman K.M., Palmer J., Morgenstern D., Wilderspin A.F., Neidle S., Thurston D.E. A novel small-molecule inhibitor of IL-6 signalling. Bioorg. Med. Chem. Lett. 2010;20:7029–7032. doi: 10.1016/j.bmcl.2010.09.117. [DOI] [PubMed] [Google Scholar]
- 23.Zou S., Tong Q., Liu B., Huang W., Tian Y., Fu X. Targeting STAT3 in cancer immunotherapy. Mol. Canc. 2020;19:145. doi: 10.1186/s12943-020-01258-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.I N.C.f.B. The Scripps Research Institute Molecular Screening Center; 2021. PubChem Bioassay Record for AID 862, Source. [Google Scholar]
- 25.Dhanda S.K., Singla D., Mondal A.K., Raghava G.P. DrugMint: a webserver for predicting and designing of drug-like molecules. Biol. Direct. 2013;8:28. doi: 10.1186/1745-6150-8-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qureshi A., Thakur N., Kumar M. VIRsiRNApred: a web server for predicting inhibition efficacy of siRNAs targeting human viruses. J. Transl. Med. 2013;11:305. doi: 10.1186/1479-5876-11-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chauhan J.S., Dhanda S.K., Singla D., Open Source Drug Discovery C., Agarwal S.M., Raghava G.P. QSAR-based models for designing quinazoline/imidazothiazoles/pyrazolopyrimidines based inhibitors against wild and mutant EGFR. PloS One. 2014;9 doi: 10.1371/journal.pone.0101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agrawal P., Bhalla S., Chaudhary K., Kumar R., Sharma M., Raghava G.P.S. In silico approach for prediction of antifungal peptides. Front. Microbiol. 2018;9:323. doi: 10.3389/fmicb.2018.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neelam Sharma S.P., Dhall Anjali, Leimarembi Devi Naorem, Gajendra P.S. bioRxiv; 2021. Raghava*, ChAlPred: A Web Server for Prediction of Allergenicity of Chemical Compounds. [DOI] [PubMed] [Google Scholar]
- 30.Yap C.W. PaDEL-descriptor: an open source software to calculate molecular descriptors and fingerprints. J. Comput. Chem. 2011;32:1466–1474. doi: 10.1002/jcc.21707. [DOI] [PubMed] [Google Scholar]
- 31.Singh H., Singh S., Singla D., Agarwal S.M., Raghava G.P. QSAR based model for discriminating EGFR inhibitors and non-inhibitors using Random forest. Biol. Direct. 2015;10:10. doi: 10.1186/s13062-015-0046-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ke G., et al. Lightgbm: a highly efficient gradient boosting decision tree. Adv. Neural Inf. Process. Syst. 2017;30:3146–3154. [Google Scholar]
- 33.Dhall A., Patiyal S., Kaur H., Bhalla S., Arora C., Raghava G.P.S. Computing skin cutaneous melanoma outcome from the HLA-alleles and clinical characteristics. Front. Genet. 2020;11:221. doi: 10.3389/fgene.2020.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma N., Patiyal S., Dhall A., Pande A., Arora C., Raghava G.P.S. Brief Bioinform; 2020. AlgPred 2.0: an Improved Method for Predicting Allergenic Proteins and Mapping of IgE Epitopes. [DOI] [PubMed] [Google Scholar]
- 35.Patiyal S., Agrawal P., Kumar V., Dhall A., Kumar R., Mishra G., Raghava G.P.S. NAGbinder: an approach for identifying N-acetylglucosamine interacting residues of a protein from its primary sequence. Protein Sci. 2020;29:201–210. doi: 10.1002/pro.3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pedregosa F., Varoquaux G., Gramfort A., Michel V., Thirion B., Grisel O., Blondel M., Prettenhofer P., Weiss R., Dubourg V. Scikit-learn: machine learning in Python. J. Mach. Learn. Res. 2011;12:2825–2830. [Google Scholar]
- 37.Kaur H., Dhall A., Kumar R., Raghava G.P.S. Identification of platform-independent diagnostic biomarker panel for hepatocellular carcinoma using large-scale transcriptomics data. Front. Genet. 2019;10:1306. doi: 10.3389/fgene.2019.01306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhalla S., Kaur H., Dhall A., Raghava G.P.S. Prediction and analysis of skin cancer progression using genomics profiles of patients. Sci. Rep. 2019;9:15790. doi: 10.1038/s41598-019-52134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao Y., Charisi A., Cheng L.C., Jiang T., Girke T. ChemmineR: a compound mining framework for R. Bioinformatics. 2008;24:1733–1734. doi: 10.1093/bioinformatics/btn307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant J.R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le D., Pon A., Knox C., Wilson M. DrugBank 5.0: a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J., Rohatgi A., Schneider J., Braunstein M. Considerations for the management of oncology patients during the COVID-19 pandemic. Oncology (Williston Park) 2020;34:432–441. doi: 10.46883/ONC.2020.3410.0432. [DOI] [PubMed] [Google Scholar]
- 42.Nishanth G., Deckert M., Wex K., Massoumi R., Schweitzer K., Naumann M., Schluter D. CYLD enhances severe listeriosis by impairing IL-6/STAT3-dependent fibrin production. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li-Mei W., Jie T., Shan-He W., Dong-Mei M., Peng-Jiu Y. Anti-inflammatory and anti-oxidative effects of dexpanthenol on lipopolysaccharide induced acute lung injury in mice. Inflammation. 2016;39:1757–1763. doi: 10.1007/s10753-016-0410-7. [DOI] [PubMed] [Google Scholar]
- 44.Shalkami A.S., Hassan M.I.A., Abd El-Ghany A.A. Perindopril regulates the inflammatory mediators, NF-kappaB/TNF-alpha/IL-6, and apoptosis in cisplatin-induced renal dysfunction. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2018;391:1247–1255. doi: 10.1007/s00210-018-1550-0. [DOI] [PubMed] [Google Scholar]
- 45.Bhat S.A., Goel R., Shukla R., Hanif K. Angiotensin receptor blockade modulates NFkappaB and STAT3 signaling and inhibits glial activation and neuroinflammation better than angiotensin-converting enzyme inhibition. Mol. Neurobiol. 2016;53:6950–6967. doi: 10.1007/s12035-015-9584-5. [DOI] [PubMed] [Google Scholar]
- 46.Yang Y., Ma L., Xu Y., Liu Y., Li W., Cai J., Zhang Y. Enalapril overcomes chemoresistance and potentiates antitumor efficacy of 5-FU in colorectal cancer by suppressing proliferation, angiogenesis, and NF-kappaB/STAT3-regulated proteins. Cell Death Dis. 2020;11:477. doi: 10.1038/s41419-020-2675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xing J., Li J., Fu L., Gai J., Guan J., Li Q. SIRT4 enhances the sensitivity of ER-positive breast cancer to tamoxifen by inhibiting the IL-6/STAT3 signal pathway. Cancer Med. 2019;8:7086–7097. doi: 10.1002/cam4.2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim J.W., Gautam J., Kim J.E., Kim J.A., Kang K.W. Inhibition of tumor growth and angiogenesis of tamoxifen-resistant breast cancer cells by ruxolitinib, a selective JAK2 inhibitor. Oncol Lett. 2019;17:3981–3989. doi: 10.3892/ol.2019.10059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Balakrishnan V., Lakshminarayanan K. Screening of FDA approved drugs against SARS-CoV-2 main protease: coronavirus disease. Int. J. Pept. Res. Therapeut. 2020:1–8. doi: 10.1007/s10989-020-10115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X., Wang Q., Wang Y., Hu J., Jiang H., Cheng W., Ma Y., Liu M., Sun A., Zhang X., Li X. Duloxetine prevents the effects of prenatal stress on depressive-like and anxiety-like behavior and hippocampal expression of pro-inflammatory cytokines in adult male offspring rats. Int. J. Dev. Neurosci. 2016;55:41–48. doi: 10.1016/j.ijdevneu.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Dionisie V., Filip G.A., Manea M.C., Manea M., Riga S. The anti-inflammatory role of SSRI and SNRI in the treatment of depression: a review of human and rodent research studies. Inflammopharmacology. 2021;29:75–90. doi: 10.1007/s10787-020-00777-5. [DOI] [PubMed] [Google Scholar]
- 52.Jansen van Vuren E., Steyn S.F., Brink C.B., Moller M., Viljoen F.P., Harvey B.H. The neuropsychiatric manifestations of COVID-19: interactions with psychiatric illness and pharmacological treatment. Biomed. Pharmacother. 2021;135:111200. doi: 10.1016/j.biopha.2020.111200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen Y.W., Yiu C.B., Wong K.Y. Prediction of the SARS-CoV-2 (2019-nCoV) 3C-like protease (3CL (pro)) structure: virtual screening reveals velpatasvir, ledipasvir, and other drug repurposing candidates. F1000Res. 2020;9:129. doi: 10.12688/f1000research.22457.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aydin Y., Kurt R., Song K., Lin D., Osman H., Youngquist B., Scott J.W., Shores N.J., Thevenot P., Cohen A., Dash S. Hepatic stress response in HCV infection promotes STAT3-mediated inhibition of HNF4A-miR-122 feedback loop in liver fibrosis and cancer progression. Cancers. 2019:11. doi: 10.3390/cancers11101407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kempuraj D., Huang M., Kandere K., Boucher W., Letourneau R., Jeudy S., Fitzgerald K., Spear K., Athanasiou A., Theoharides T.C. Azelastine is more potent than olopatadine n inhibiting interleukin-6 and tryptase release from human umbilical cord blood-derived cultured mast cells. Ann. Allergy Asthma Immunol. 2002;88:501–506. doi: 10.1016/s1081-1206(10)62389-7. [DOI] [PubMed] [Google Scholar]
- 56.Karulin B., Kozhevnikov M. Ketcher: web-based chemical structure editor. J. Cheminformatics. 2011;3:3. [Google Scholar]
- 57.Schüller A., Hähnke V., Schneider G. SmiLib v2. 0: a Java‐based tool for rapid combinatorial library enumeration. QSAR Comb. Sci. 2007;26:407–410. [Google Scholar]
- 58.Yuan J., Zhang F., Niu R. Multiple regulation pathways and pivotal biological functions of STAT3 in cancer. Sci. Rep. 2015;5:17663. doi: 10.1038/srep17663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Patiyal S., Kaur D., Kaur H., Sharma N., Dhall A., Sahai S., Agrawal P., Maryam L., Arora C., Raghava G.P.S. A web-based platform on coronavirus disease-19 to maintain predicted diagnostic, drug, and vaccine candidates. Monoclon. Antibodies Immunodiagn. Immunother. 2020;39:204–216. doi: 10.1089/mab.2020.0035. [DOI] [PubMed] [Google Scholar]
- 60.Dhall A., Patiyal S., Sharma N., Usmani S.S., Raghava G.P.S. Computer-aided prediction and design of IL-6 inducing peptides: IL-6 plays a crucial role in COVID-19. Briefings Bioinf. 2021;22:936–945. doi: 10.1093/bib/bbaa259. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the datasets generated for this study are available at the “STAT3In” webserver, https://webs.iiitd.edu.in/raghava/stat3in/dataset.php.