Abstract
The novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), is causing a severe global health emergency owing to its highly infectious nature. Although the symptoms of SARS-CoV-2 are well known but its impact on nasopharyngeal microbiome is poorly studied. The present cross-sectional study was intended to understand the perturbation in the nasopharyngeal microbiome composition within the infected (n = 63) and non-infected (n = 26) individuals using 16S rRNA gene based targeted amplicon sequencing and their association with host types and the prevalence of opportunistic pathogens at the stage of infection. The results confirmed that number of OTUs were significantly (p < 0.05) decreased in the SARS-CoV-2 infected individuals in comparison to non-infected individuals. Pairwise Wilcoxon test showed a significant (p < 0.05) increase in the abundance of Proteobacteria in infected individuals compared to non-infected ones and vice-versa for Fusobacteria and Bacteroidetes. Similarity percentage (SIMPER) analysis showed the increment in the abundance of opportunistic pathogens (Haemophilus, Stenotrophomonas, Acinetobacter, Moraxella, Corynebacterium 1, Gemella, Ralstonia, and Pseudomonas) involved in secondary infection. Furthermore, this study highlighted the microbial community structure of individuals within and across the families. In this study, we also performed the assesment of microbiome associated with host types (age and genders) and COVID-19 conditions (symptomatic and asymptomatic). The data suggested that the host types/conditions during the COVID-19 infection are potential factors in enrichment of specific bacterial communities in upper respiratory tract.
Keywords: SARS-CoV-2, Nasopharyngeal microbiome, Symptomatic, Asymptomatic, Host types
Severe acute respiratory syndrome coronavirus (SARS-CoV-2) is a novel highly transmissible and pathogenic coronavirus, causing the respiratory illness named as coronavirus diseases 2019 (COVID-19) [[1], [2], [3]]. It has infected more than 180 million people with several million deaths across the globe until 1st July 2021 (https://www.worldometers.info/coronavirus). SARS-CoV-2 spreads globally within short span due to its high transmissible rate causing severe threat to global public health and economy [2,4]. Before the outbreak of SARS-CoV-2, two other pathogenic coronavirus known as SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) with zoonotic origin have emerged in 2002 and 2012, respectively [[5], [6], [7], [8]]. There is no concrete evidence how and when this SARS-CoV-2 (animal origin) contracted the humans and causes respiratory illness. SARS-CoV-2 infection causes pneumonia, fever, cough, chest discomfort, and in severe cases dyspnea and bilateral lung infiltration [9]. SARS-CoV-2 causes mild symptoms in the children and young adults and most of them are asymptomatic. Older people and people with comorbidities (including cancer, diabetes, high blood pressure, obesity, etc.) are at a higher risk of respiratory failure and death [9]. Recent studies are mainly focussing on the etiology, epidemiology, pathophysiology, evolution, and genomics of SARS-CoV-2 but less is known about the impact of SARS-CoV-2 on nasal microbiome [[10], [11], [12], [13], [14], [15], [16]]. It is well established now that human microbiome plays an important role in host genetics, physiology, health, and disease [[17], [18], [19], [20], [21]]. Hence, there is an urgent need to understand the nasal microbiome of the SARS-CoV-2 infected patients, as it is well established that nasal microbiota plays a significant role in host immune response and helps in maintaining the health status of the individuals.
Nasal microbiome of upper and lower respiratory tracts is constituted of both commensal and opportunistic pathogens (pathobionts) and plays an important role in host immune response [[18], [19], [20], [21]]. Growing evidence supports that respiratory viruses cause alteration in the nasal microbiome during infection [20,22]. Recent development in the omics approaches has enabled us to get deeper insights in to the microbiome profile of human health and their association with various environmental and biotic factors. Several nasal microbiome studies have been conducted mostly on the influenza-infected patients with acute respiratory illness or with other respiratory diseases [18,[23], [24], [25], [26], [27], [28], [29], [30]]. These studies offer a better understanding about the alteration in the nasal microbiome of infected patient's during the disease course and their impact on host types (age, gender, behaviour, etc.). It has been reported that viral infection causes enrichment of various pathogenic microbes for secondary infection, hence, enhancing the severity of the disease [31,32]. Most of the nasal microbiome studies have been conducted on the influenza-infected patients or other respiratory diseases while less is known about the alteration of nasal microbiome during coronavirus infections. Recently, few studies have been conducted to understand the impact of SARS-CoV-2 infection and/or disease severity on the nasal and gut microbiome [[33], [34], [35], [36], [37], [38], [39]]. Since SARS-CoV-2 infection is associated with the respiratory illness, hence it is imperative to have a better understanding about the nasal microbiome profile of the infected patients, as it is the first defensive line to avoid infection. Along with this, interplay between virus and bacteria during SAR-CoV-2 infection is less understood. It is intriguing to understand which microbial taxa are enriched during the SARS-CoV-2 infection as different viruses are known to enrich distinct microbial groups, which may help in developing strategies to combat SARS-CoV-2 infection via altering the nasal microbial community.
The present cross-sectional study is aimed to understand the nasopharyngeal microbiota in the SARS-CoV-2 infected (n = 63) and non-infected (n = 26) individuals using 16S rRNA gene based targeted metagenome. Furthermore, this study captures the nasopharyngeal microbiome profile of the infected individuals and non-infected individuals to understand the alteration in the microbial community composition within and across the distinct families. The association of nasal microbiome with host types (age and gender) and symptomatic and asymptomatic conditions is also investigated. The current study represents the first report of nasal microbiome of SARS-CoV-2 infected and non-infected individuals within the families from India.
1. Materials and methods
1.1. Sample collection
A total of 89 nasopharyngeal swabs were collected from clinically suspected patients of SARS-CoV-2 infection and their family contacts. The sample collection was performed as per the standard guidelines. The collected sample was immediately put in Viral Transport Medium (VTM) and was transported to the laboratory of B J Government Medical College, Pune for COVID-19 real time Polymerase Chain Reaction (PCR) testing ensuring proper cold chain conditions and triple layer packaging. The detailed list of recruiting samples and their clinical data are presented in Table 1 and Supplementary Table 1.
Table 1.
Details of recruiting samples.
| Total no of subjects (n = 89) | |
|---|---|
| SARS-CoV-2 infected individuals | n = 63 |
| Non-infected individuals | n = 26 |
| Male | n = 54 |
| Female |
n = 35 |
| Total no of Families (n = 11) | |
| No of subjects | n = 46 |
| SARS-CoV-2 infected individuals | n = 20 |
| Non-infected individuals | n = 26 |
| Male | n = 27 |
| Female | n = 19 |
| Age group 1 (0–15 years) | n = 14 |
| Age group 2 (16–30 years) | n = 13 |
| Age group 3 (31–46 years) | n = 8 |
| Age group 4 (46 & above) | n = 11 |
| SARS-CoV-2 infected- Asymptomatic | n = 9 |
| SARS-CoV-2 infected- Symptomatic | n = 11 |
1.2. RNA extraction and real time PCR testing
For RNA extraction, 200 uL of sample was used and was extracted using magnetic bead-based MagRNA-II Viral RNA Extraction Kit (Genes2me, Gurgaon, Haryana, India) following manufacturer instructions. RNA was eluted in 50 uL of elution buffer. Extracted RNA was subjected to real-time PCR testing for COVID-19 using Multiplex Single Tube Real time PCR kit, Version 3 developed by ICMR-NIV, Pune, India. PCR was performed using Applied Biosystems™ 7500 Fast Dx Real-Time PCR instrument (Thermo Fisher Scientific, Waltham, U.S). The target genes used for detection of SARS-CoV-2 were Envelope gene (E gene) and Open Reading Frame-1ab (ORF-1ab gene). The amplification data was interpreted based on cut–off cycle threshold (Ct) values recommended by ICMR-NIV, Pune, India.
1.3. DNA extraction, targeted 16S rRNA gene amplification and sequencing
Genomic DNA was extracted using MagMAX Viral and Pathogen Nucleic Acid Isolation Kit (Thermo Fisher Scientific, Waltham, U.S.A) following the manufacturer instructions. Extracted DNA samples were transported to National Centre for Cell Sciences (NCCS), Pune for the microbiome study. The quality of DNA was assessed as described previously [40]. DNA of respective samples was subjected to amplification of V4 region of 16S rRNA gene using the V4 specific primer set 515F/806R [40]. The amplification of V4 region was performed as described previously [40] and the resultant amplicons were processed for library preparation by following Illumina 16S metagenomics protocol (Illumina, USA). The barcoded libraries were pooled in equimolar concentration and sequenced on the Illumina MiSeq platform using 2 × 250 bp v2 chemistry.
1.4. Bioinformatics analysis and statistics
The obtained raw reads were subjected to quality trimming. BBduk [41] was used to trim the last 20 bases from both sides of the reads. The pair end Illumina reads were merged with FLASH-1.2.11 [42]. at minimum overlap of 10 bp and maximum overlap of 65 bp. The obtained merged reads were analysed with QIIME 1.9.1 pipeline [43]. Quality filtering was performed by removing barcode, primers, sequences with homopolymers run of >6bp and read length of sequences <230 bp using split_libraries.py command. Denovo operational taxonomic unit (OTU) was assigned at the sequence identity level of 97% using UCLUST algorithm [44]. The taxonomic assignment was performed with SILVA 132 database (www.arb-silva.de/documentation/release-132) with a minimum confidence of 80%. Chimeric sequences were checked and removed using ChimeraSlayer [45]. Alpha diversity indices taxonomic distribution were calculated with rarefied reads sampled to even depth (n = 25,000). Pairwise Wilcoxon test was performed in R software using ggpubr package to assess the difference between the microbial community composition of the infected and non-infected individuals. Various R packages such as ggplot2, reshape2, and RColorBrewer were used for data visualization. Non-Metric multidimensional scaling (NMDS) was performed using Bray–Curtis similarity matrix at family and genera level datasets in PAST3.0 software [46] Permutational analysis of variance (PERMANOVA) and analysis of similarity (ANOSIM) and similarity percentage (SIMPER) analysis were performed in PAST 3.0.
1.5. Data submission
The raw 16S rRNA gene amplicon sequencing data generated in this study was submitted to NCBI SRA database and it is available under the BioProject ID: PRJNA707350.
2. Results
2.1. Microbiome profile of SARS-CoV-2 infected and non-infected individuals
Nasopharyngeal microbiome of SARS-CoV-2 infected (n = 63) and non-infected individuals (n = 26) was ascertained through 16S rRNA gene based targeted amplicon sequencing. In total, ∼8.1 million useable reads were obtained after quality filtering. Detailed taxonomic and alpha diversity analyses was performed on the rarefied reads (n = 25,000) and the number of OTUs observed in the samples ranged from 919 to −2212 OTUs. Alpha diversity parameters such as Simpson and Shannon indices did not show significant differences between the infected and non-infected individuals (Fig. 1 a). In contrast, observed OTUs and Chao1 values were found to be significantly (p < 0.05) decreased in the infected individuals (Fig. 1a). Goods coverage value (>0.92) indicated the substantial coverage of the microbial diversity in both the conditions (Fig. 1a).
Fig. 1.
Comparison of alpha diversity parameters and taxonomic distribution (at phylum-level) across the SARS-CoV-2 infected and non-infected individuals. (a) Box-whisker plots of alpha diversity indices and its comparison using Wilcoxon signed-rank test between SARS-CoV-2 infected and non-infected individuals. (b) Relative abundance (%) of major phyla between the infected and non-infected individuals.
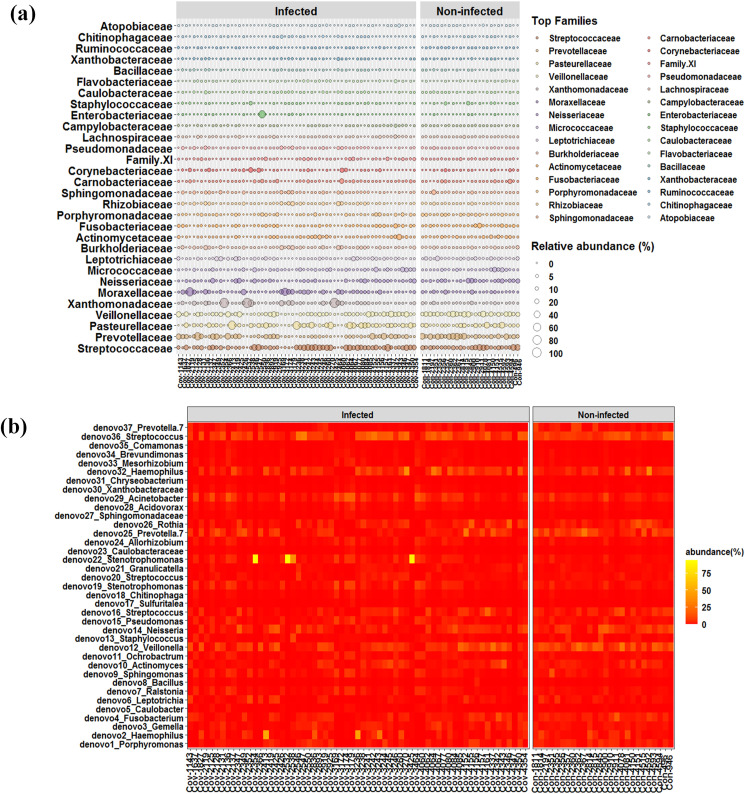
In total, 28 distinct bacterial phyla were observed in the nasal microbiome with a minimum of ∼10 phyla in each individual. Most of the reads (>90%) were assigned to the Proteobacteria, Firmicutes, Bacteroidetes, Actinobacteria, and Fusobacteria in each of the infected and non-infected individuals (Fig. 1b). In case of non-infected individuals, Firmicutes (average: ∼36%) was found to be the dominant phyla followed by Proteobacteria (average: ∼28%), and Bacteroidetes (average ∼20%). In contrast, Proteobacteria accounted for an average of ∼43% of the reads in infected individuals followed by Firmicutes (average: ∼29%) and Bacteroidetes (average: ∼12%). Pairwise Wilcoxon test showed the significant (p < 0.05) increase in the abundance of Proteobacteria in SARS-CoV-2 infected individuals compared to non-infected ones and vice versa for Fusobacteria and Bacteroidetes (Supplementary Fig. 1). Streptococcaeae, Prevotellaceae, Pasteurellaceae, Veilloellaceae, Xanthomonadaceae, Neisseriaceae, Micrococaceae, Leptotrichiaceae, Burkholderiaceae, Fusobacteriaceae, Actinomycetaceae, and Porphyromonadaceae constituted the major part of the nasal microbiome in both infected and non-infected individuals (Fig. 2 a). Interestingly, abundance of Prevetolleceae, Veillonellaceae, Neisseriaceae, Leptotrichiaceae, and Fusobacteriaceae was found to be significantly (p < 0.05) decreased in the infected individuals (Supplementary Fig. 2a). Additionally, core microbiome (OTUs present in minimum 90% of the total samples) between the infected and non-infected individuals was investigated (Fig. 2b). Only 37 OTUs were found to be the part of core microbiome (Fig. 2b). In most of the samples, these core OTUs accounted for more than 50% of the total microbial community. In order to understand the difference between the shift in these core OTUs, pairwise Wilcoxcon test was performed. It was found that out of the 37 core OTUs, only 7 OTUs [denovo 2 (Haemophilus), denovo 4 (Fusobacterium), denovo 12 (Villonella), denovo 14 (Niesseria), denovo 16 (Streptococcus), denovo 25 (Prevotella 7), and denovo 37 (Prevotella 7)] decreased significantly (p < 0.05) decreased in the infected individuals (Supplementary Fig. 2b). Furthermore, SIMPER analysis was performed on the amplicon dataset to investigate the contribution of the taxa in nasopharyngeal microbiome of infected and non-infected individuals (Table 2 ). It was observed that Haemophilus, Stenotrophomonas, Acine t obacter, Moraxella, Corynebacterium I, Gemella, Ralstonia, and Pseudomonas contributed substantially in nasal microbiome of infected individuals (Table 2). In contrast, Prevotella7, Veillonella, Neisseria, Rothia, Leptotrichia, Fusobacterium, Alloprevotella, Meagsphaera, and Dolosigranulum were dominant in non-infected individuals (Table 2). Multidimensional scaling using Bray–Curtis dissimilarity matrix at family and genus level data showed no distinct clustering between the infected and non-infected individuals, indicating there was no significant difference in the microbial community composition between these two states which was well supported by ANOSIM (p > 0.05) and PERMANOVA (P > 0.05) analyses (Supplementary Fig. 3a and 3b).
Fig. 2.
Distribution of major microbial families and core microbiome between SARS-CoV-2 infected and non-infected individuals. (a) Bubble plot showing abundance pattern of top 30 families between SARS-CoV-2 infected and non-infected individuals. (c) Heat map based distribution of abundance of core microbiome (core OTUs).
Table 2.
Summary of similarity percentage (SIMPER) of various taxa in infected and non-infected individuals.
| Taxon | Average dissimilarity | Contribution % | Cumulative % | Non-infected (mean value) | Infected (mean value) |
|---|---|---|---|---|---|
| Streptococcus | 5.33 | 8.81 | 8.81 | 13.60 | 13.40 |
| Haemophilus | 5.14 | 8.49 | 17.31 | 7.58 | 9.69 |
| Prevotella 7 | 4.47 | 7.40 | 24.70 | 10.00 | 4.81 |
| Veillonella | 4.43 | 7.33 | 32.03 | 11.80 | 6.89 |
| Stenotrophomonas | 4.34 | 7.17 | 39.20 | 2.15 | 9.05 |
| Neisseria | 2.81 | 4.64 | 43.83 | 6.11 | 3.71 |
| Acinetobacter | 2.38 | 3.93 | 47.77 | 2.69 | 4.90 |
| Rothia | 2.05 | 3.39 | 51.15 | 3.41 | 3.08 |
| Leptotrichia | 1.70 | 2.81 | 53.97 | 3.53 | 2.50 |
| Fusobacterium | 1.49 | 2.46 | 56.42 | 3.29 | 2.08 |
| Actinomyces | 1.46 | 2.42 | 58.84 | 2.60 | 2.48 |
| Porphyromonas | 1.26 | 2.09 | 60.93 | 2.69 | 1.90 |
| Moraxella | 1.12 | 1.85 | 62.78 | 0.43 | 1.95 |
| Prevotella | 1.10 | 1.82 | 64.60 | 2.51 | 1.18 |
| Corynebacterium 1 | 1.06 | 1.75 | 66.35 | 0.89 | 1.70 |
| Sphingomonas | 1.05 | 1.73 | 68.08 | 1.66 | 1.44 |
| Megasphaera | 0.99 | 1.64 | 69.71 | 1.94 | 0.72 |
| Alloprevotella | 0.96 | 1.58 | 71.30 | 1.88 | 1.02 |
| Prevotella 6 | 0.90 | 1.49 | 72.79 | 1.45 | 0.91 |
| Dolosigranulum | 0.87 | 1.44 | 74.23 | 1.16 | 0.83 |
| Gemella | 0.77 | 1.27 | 75.50 | 1.08 | 1.59 |
| Pseudomonas | 0.75 | 1.24 | 76.74 | 0.94 | 1.51 |
| Ralstonia | 0.61 | 1.00 | 77.74 | 0.71 | 1.28 |
| Actinobacillus | 0.57 | 0.95 | 78.69 | 0.58 | 0.98 |
| Staphylococcus | 0.52 | 0.87 | 79.56 | 0.81 | 0.53 |
| Rhizobium | 0.45 | 0.75 | 80.31 | 0.61 | 0.89 |
| Ochrobactrum | 0.44 | 0.73 | 81.03 | 0.54 | 0.88 |
| Enterobacter | 0.44 | 0.73 | 81.76 | 0.09 | 0.87 |
| Campylobacter | 0.39 | 0.65 | 82.41 | 0.80 | 0.74 |
| Granulicatella | 0.39 | 0.64 | 83.05 | 0.61 | 0.80 |
| Capnocytophaga | 0.36 | 0.60 | 83.65 | 0.49 | 0.56 |
| Lautropia | 0.31 | 0.51 | 84.16 | 0.47 | 0.25 |
| Selenomonas 3 | 0.31 | 0.51 | 84.67 | 0.44 | 0.43 |
| Acidovorax | 0.28 | 0.46 | 85.14 | 0.37 | 0.59 |
| Ruminococcaceae UCG-014 | 0.27 | 0.45 | 85.59 | 0.48 | 0.31 |
| Atopobium | 0.26 | 0.43 | 86.02 | 0.42 | 0.38 |
| Bacillus | 0.26 | 0.43 | 86.45 | 0.33 | 0.51 |
| Lachnoanaerobaculum | 0.25 | 0.42 | 86.87 | 0.40 | 0.43 |
| Streptobacillus | 0.24 | 0.39 | 87.66 | 0.25 | 0.28 |
| Erysipelotrichaceae UCG-007 | 0.23 | 0.38 | 88.04 | 0.39 | 0.36 |
| Mesorhizobium | 0.23 | 0.38 | 88.42 | 0.27 | 0.47 |
| Oribacterium | 0.21 | 0.36 | 88.78 | 0.37 | 0.36 |
| Prevotella 2 | 0.21 | 0.35 | 89.13 | 0.18 | 0.39 |
| Absconditabacteriales (SR1)_uncultured | 0.19 | 0.32 | 89.45 | 0.23 | 0.19 |
| Chitinophaga | 0.18 | 0.30 | 89.75 | 0.21 | 0.37 |
| Peptostreptococcus | 0.17 | 0.29 | 90.04 | 0.33 | 0.19 |
| Xanthobacteraceae | 0.16 | 0.27 | 90.31 | 0.16 | 0.32 |
| Brevundimonas | 0.16 | 0.26 | 90.57 | 0.19 | 0.31 |
| Eubacterium nodatum group | 0.16 | 0.26 | 90.83 | 0.30 | 0.13 |
| Absconditabacteriales (SR1)_uncultured | 0.15 | 0.25 | 91.08 | 0.21 | 0.16 |
| Tepidiphilus | 0.14 | 0.24 | 91.31 | 0.14 | 0.20 |
2.2. Bacterial community pattern within and across the family members
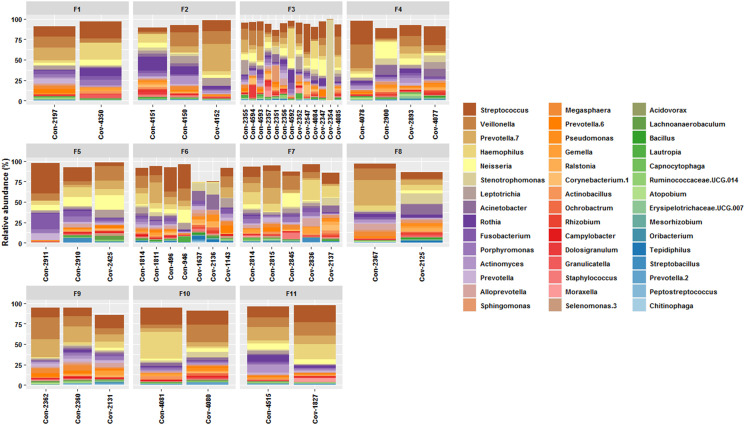
Out of the total 89 individuals, 46 individuals were found to be the part of 11 distinct families with minimum of one infected and one non-infected members (Fig. 3 ). In order to understand the difference between the nasal microbiome of the infected and non-infected individuals within and across the families, a thorough analysis was performed on the microbiome datasets. Nasal microbiome of the members of these families comprised of similar microbial taxa with varied abundance (Fig. 3). Difference between the microbial abundance of the few taxa in the infected and non-infected individuals was noted within and across the families. In most of the cases, abundance of Streptococcus increased while abundance of Veillonella, Rothia, and Prevotella 7 decreased in infected individuals. In addition, few microbial taxa (Haemophilus, Ne i sseria, Stenotrophomonas, Leptotrichia, Fusobacterium, Acinetobacter, Prevotella 6, Alloprevotella, Megasphaera, etc.) which constituted the major part of the nasal microbiome showed abrupt change in their abundance pattern within the infected and non-infected individuals across the families.
Fig. 3.
Taxonomic distribution of major genera across members of 11 distinct families of SARS-CoV-2 infected and non-infected individuals. Note: Infected individuals: sample names with prefix Cov. Non-infected individuals: sample names with prefix Con. F1–F11 denote distinct families.
2.2.1. Bacterial communities associated with host types (age and gender) across the families
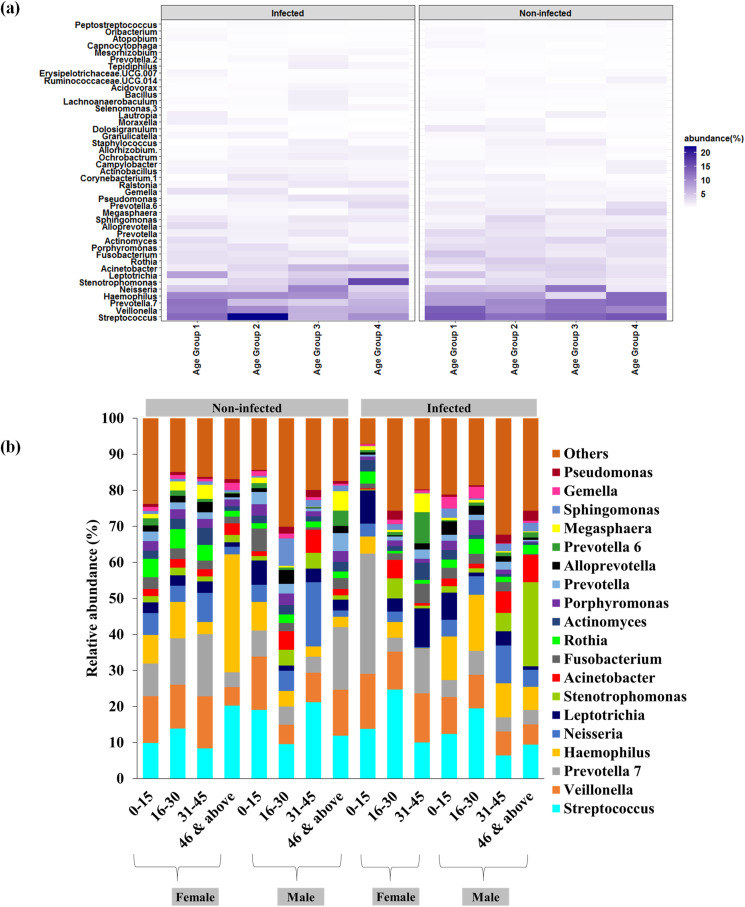
In order to get deeper insights into the microbial differences across the families, members of the families were classified based on their age and gender. Four distinct age groups (Age Group 1: 0–15 years, Age Group 2: 16–30; Age Group 3: 31–45, Age Group 4: 46& above) and two gender (male and female) were considered for the analysis. Heatmap based distribution of the major microbial taxa showed the changes in their abundance pattern (Fig. 4 a). In the Age Group 1 (0–15 years), abundance of Prevotella 7, Haemophilus, Leptotrichia, Alloprevotella, Gemella, Granulicatellata, Moraxella, and Lautropia increased in infected individual's comparison to non-infected ones. In Age Group 2 (16–30 years), enhanced abundance of Streptococcus was observed in infected individuals along with Haemophilus, Corynebacterium 1, Gemella, Granulicatellata, and Prevotella 2. In Age Group 3 (31–45years), higher abundance of Haemophilus, Stenotrophomonas, Leptotrichia, Acinetobacter, Fusobacterium, Prevotella, Pseudomonas, Staphylococcus, Lachnoaerobaculum, etc was noted in the infected individuals. Stenotrophomas, Leptotrichia, Acinetobacter, N e isseria, Alloprevotella, Pseudomonas, Granulicatellata, Ochrobactrum, etc. were enriched in the infected individuals of Age Group 4 (46&above) (Fig. 4a). Irrespective of these changes, multidimensional ordination plot did not show distinct clustering of the age groups based on the microbial community composition (Supplementary Fig. 4). PERMANOVA analysis further confirmed that no significant differences (p > 0.05) were observed between infected and non-infected individuals of each age group. In addition to this, microbiome profile of the male and female infected and non-infected individuals was analysed (Fig. 4b). A shift in the microbiome of the male and female individuals was observed across all the age group categories (Fig. 4b).
Fig. 4.
Taxonomic distribution of major genera across the family members based on host types (age and gender). (a) Heat map based distribution of major taxa at various age groups. (b) Microbiome profile of the male and female infected and non-infected individuals.
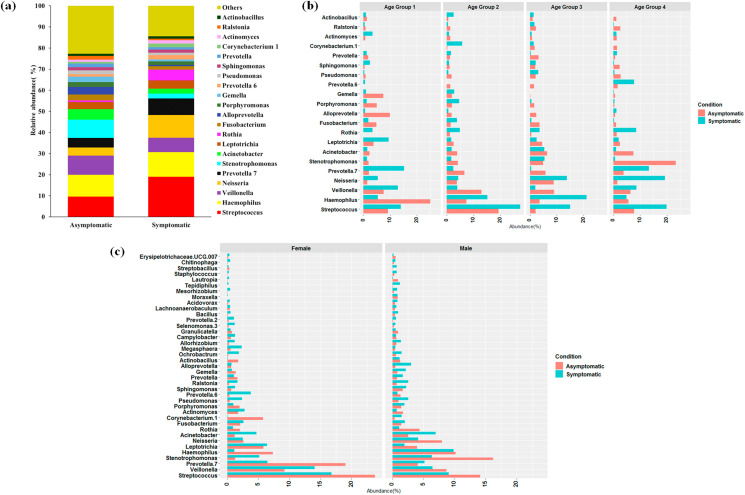
2.2.2. Microbiome of SARS-CoV-2 infected asymptomatic and symptomatic individuals within the families
Based on the above observations, it was intriguing to look into the microbiome shift in the asymptomatic and symptomatic SARS-CoV-2 infected individuals of the families. Of the total 46 individuals from 11 families, 20 were found to be COVID-19 positive and among them, nine were asymptomatic while eleven were symptomatic. Taxonomic differences were observed between symptomatic and asymptomatic individuals (Fig. 5 a). Abundance of Streptoco c cus, Haemophilus, N ei sseria, Prevotella, Rothia, Leptotrichia, Corynebacterium 1 enhanced in the symptomatic SARS-CoV-2 infected individuals compared to non-infected ones (Fig. 5a). These symptomatic and asymptomatic individuals were further classified based on the above mentioned age group categories and gender. It was interesting to note that contrasting change in the abundance of the microbial taxa within the asymptomatic and symptomatic individuals was observed in all the age groups (Fig. 5b). Among the most abundant taxa (top 40), Streptococcus, N e isseria, Acinetobacter, and Rothia were found to be increased in symptomatic individuals across all the age groups (Fig. 5b). Similarly, distinction in the microbial community composition between female–female and male–male symptomatic versus asymptomatic individuals were noted. Decrease in abundance of Streptococcus, Prevotella 7, Haemophilus, Rothia, Corynebacterium 1, Porphyromonas, Gemmella, and Actinobacillus was noted in symptomatic females (Fig. 5c). However, abundance of Streptococcus, Veillonella, Stenotrophomonas, Leptotrichia, N e isseria, Rothia, Actinomyces, Prevotella 6, Actinobacillus, and Granulicatellata was found to be decreased in symptomatic males (Fig. 5c).
Fig. 5.
Microbiome profile of asymptomatic versus symptomatic SARS-CoV-2 infected individuals. (a) Relative abundance (%) of major genera between asymptomatic and symptomatic SARS-CoV-2 infected individuals. (b) Changes of microbial taxa between different age groups of symptomatic and asymptomatic SARS-CoV-2 infected individuals. (c) Changes of microbial taxa between male and female asymptomatic and symptomatic SARS-CoV-2 infected individuals.
3. Discussion
Upper respiratory tract is the entry point of various pathogenic microbes and respiratory viruses and it provides numerous niches for microbial communities to grow and adapt in the nasal environment [19,21]. It harbours various commensals and opportunistic pathogens such as Streptococcus, Acinetobacter, Moraxella, Dolosigranulum, Corynebacterium, Staphylococcus, etc. [47,48] Various intrinsic and extrinsic factors alter the human associated microbiome including nasal, gut, and other body sites [21,[49], [50], [51]]. In recent years, several cross sectional and longitudinal studies have been conducted to understand the effect of influenza virus infections and other respiratory diseases on the nasal microbiota [25,30]. Respiratory viruses are known to alter the microbiota of the upper and lower respiratory tracts and enrich the abundance of the opportunistic pathogens, which ultimately promote the severity of the disease in the infected individuals [[20], [21], [22]]. The present study has elucidated the nasopharyngeal microbial community composition in SARS-CoV-2 infected and non-infected individuals. Furthermore, we have delineated the microbiome shift within and across the family members and host types (age and gender) as our cohort is comprised of 11 distinct families (with at least one infected and non-infected individuals in each family). Our amplicon data suggest that SARS-CoV-2 infected individuals have shown no significant difference in the bacterial richness (Shannon and Simpson diversity indices) in comparison to non-infected ones which is in agreement with the previous study on nasal microbiome of COVID-19 positive patients [33,38]. Interestingly, significant decrease in the number of OTUs in the SARS-CoV-2 infected individuals has been observed in the present study indicating the alteration of the microbiome during initial days of diagnosis. It has been observed that the altered microbiome within adults having mild SARS-CoV-2 infection is restored from early dysbiosis within a short span of time or before their clinical recovery, while in the children it takes longer period due to their vulnerable and less resilient microbiome [35]. Our study further evaluated the changes in the nasopharyngeal microbial community composition in the infected individuals compared to the non-infected individuals. Abundance of Proteobacteria, Fusobacteria, and Bacteroidetes is found to be significantly (P < 0.05) altered in the infected individuals. Nardelli et al. [38] has reported that COVID-19 patients show a significant reduction in abundance of Fusobacteria as compared to controls. It has been reported that Bacteroidetes (Bacteroides spp.) plays an important role as gut microbiota member against COVID-19 infection by downregulating ACE2 and reduction of SARS-CoV-2 entry [37]. Previous studies have reported that these members constitute the core microbiome of nasal community and harbour various opportunistic pathogens, which alter the host immune response during influenza infections [20,28]. Moreover, multivariate analysis suggests no significant difference in the overall microbial community composition between the infected and non-infected individuals, which is in line with the De Maio et al. [20] study, indicating that SARS-CoV-2 is unable to induce the alteration in the microbiome during early stage of COVID-19. However we observed higher abundance of specific opportunistic pathogens such as Haemophilus, Stenotrophomonas, Acine t obacter, Moraxella, Corynebacterium 1, Gemella, Ralstonia, Granulicatella, Acidovorax, and Pseudomonas in the infected individuals suggests that the inflammatory environment caused by SARS-CoV-2 infection leads to the prevalence of bacterial pathogens that may result in secondary infection. Increment in pathogenic bacteria such as Acineobacter, Moraxella, and Pseudomonas in the nasal microbiome of COVID-19 patients is due to the accumulation of mucus, known to favour the growth of these organisms, and hyperinflammatory environment that supports their growth [52]. It has been previously reported that increment of Haemophilus and Streptococcus species during viral infection lead to the upregulation of the adhesion receptors for viral entry [19]. It is well known that the commensal bacteria play an important role in host immunity however part of these commensal bacteria become opportunistic pathogens during dysbiosis or impaired host immunity and lead to the secondary infection [53]. In the present study, enrichment of various opportunistic pathogen might promote the entry of virus via this mechanism, but need to be further validated via experimental evidence. It is evident from the data that the short-chain fatty acid producing bacteria with known immunomodulatory potential have been reduced in the COVID-19 patients due to hyperinflammatory environment which leads to the increment in opportunistic pathogen [53]. In future longitudinal studies are required to better comprehend the disturbance/dysbiosis in the microbial composition in the infected individuals over the course of the disease (from onset until recovery). This will help in understanding how the SARS-Cov-2 promotes the enrichment of opportunistic pathogens and enhances the severity of the diseases along with the interplay among host-microbes-virus.
Moreover, the present study is cross-sectional and our cohort is comprised of distinct families, hence it is interesting to investigate the difference between the nasopharyngeal microbiota between the infected and non-infected individuals within and across the families. The results reveal that each of the non-infected individuals of the families comprise of more or less similar microbial community with varied abundance of few of the microbial taxa. It is well reported that individuals cohabiting together or share common ecological niches contain similar meta-community which can be further selected upon by host genetics, diet, lifestyle, etc. [54]. The nasal microbiome of the non-infected individuals of these families is comprised of the Streptococcus, Veillonella, Rothia, Prevotella 7, Haemophilus, Ne i sseria, Stenotrophomonas, Leptotrichia, Fusobacterium, Acinetobacter, Prevotella 6, Alloprevotella, Megasphaera, etc. which are known to be a part of the upper respiratory microbial community and modulate the host response against viral infection [20]. These bacterial taxa are known to be associated with the various acute viral infection [25,55,56]. The shift in the abundance pattern of these microorganisms in the SARS-CoV-2 infected individuals suggests that viral infection increases the colonization of the pathobionts such as Streptococcus, Dolosigranulum, Pseudomonas, etc. In contrast, abundance of the Veillonella, Rothia, and Prevotella 7 is decreased in most of the infected individuals, while abundance pattern of the other opportunistic pathogens such as Acientobacter, N e iss eria, Haemophilus, etc. differs substantially, which reveals that each individual may harbour a distinct nasal microbiome or they have different response towards the SARS-CoV-2 infection. However, it is reported that specific viruses are linked with the enrichment of the certain bacterial taxa [31,32]. There are several studies where colonization of the opportunistic pathogens such as Streptococcus, Haemophilus, N e isseria, Pseudomonas, Acinetobacter, etc. has been observed during pneumonia or influenza infections in the upper respiratory tracts [20,29,57].
Furthermore, we investigated the relationship between host types (age and gender) and nasal microbial community composition. Enrichment of various opportunistic pathogens (Acinetobacter, Pseudomonas, Corynebacterium 1, Staphylococcus, etc.) in the Age group 3 (31–45 years) and Age group 4 (46 & above) were observed in the SARS-CoV-2 infected individuals. In contrast to this, Age Group 1 (0–15 years) and 2 (16–30 years) categories infected individuals show distinct response towards the SARS-CoV-2 infection. Hence, it can be suggested that distinct microbial groups are associated with distinct age groups and respond differently to SARS-CoV-2 infection. Increased abundance of these pathogens has been previously reported from the oropharyngeal microbiome of H1N1 influenza infected patients [24]. Besides age, a thorough investigation on the nasal microbiota linked with gender reveals that a marked difference in the microbial community structure within and between the male and female individuals have not been observed. Nevertheless, few microbial groups have been found to enrich in male or female infected individuals compared to non-infected ones, supporting the hypothesis that male and female individuals may respond differently during SARS-CoV-2 infection. Our results are in agreement with the previous study conducted on influenza A virus infected patients where no significant association of age and gender on temporal nature of microbiome elasticity has been found [30]. Overall, it could be suggested that change in nasal microbiome during SARS-CoV-2 infection promotes the colonization of distinct bacterial taxa across the diverse age groups and gender. Two categories (symptomatic and asymptomatic) of SARS-CoV-2 infected individuals are detected across the families. Nasal microbiome of the symptomatic and asymptomatic individuals is found to be different for different age groups and gender. Irrespective of age, abundance of Streptococcus, Rothia, and N e isseria is enhanced in the symptomatic patients while other bacterial taxa show difference in the abundance pattern across the diverse age groups. This suggests that symptomatic patients of different age groups are colonized by distinct bacterial taxa and modulates the host immune response. Difference in the microbial community composition of symptomatic and asymptomatic patients has already been observed in the patients with influenza virus, parainfluenza virus, coronavirus, adenovirus or metapneumovirus [18]. Severity of the infection of the upper respiratory tracts depends upon the complex interplay involving bacteria-virus-host interactions [58]. During virus infection, the nasal microbiome has influence on the virus load and host innate immune response along with clinical symptoms [59]. Hence, it can be suggested that difference in the microbiome amongst individuals could explain the variation in symptoms of SARS-CoV-2 infections [3]. Furthermore, it suggests that the asymptomatic individuals may harbor such bacterial communities, which could directly or indirectly elicit the immune response from the host, preventing full-fledged infection of SARS-CoV-2. Several studies have highlighted that nasal microbiota regulates the immunity in respiratory mucosa via proper activation of inflammasomes to reduce the viral loads/infections [59,60] whereas certain microorganisms induce the expression of various binding receptors or increase the viral replication [61,62].
4. Conclusion
Overall, this study demonstrates the nasopharyngeal microbiome of SARS-CoV-2 infected and non-infected individuals at the time of early diagnosis. The study further highlights the nasal microbial community structure of the individuals belonging to different families and how the nasal microbiome responds to the host types (age and gender) and COVID-19 conditions (symptomatic and asymptomatic). Our results suggest enrichment of specific opportunistic pathogenic microorganisms in the infected individuals due to the hyperinflammatory environment associated with viral infection. The data generated in the present study would be helpful in devising the strategies to perform future microbiome studies to understand the role of microbiome in infected individuals and severity of the disease to further manage the effect of virus on the host immunity and reduce the infection rate in the patient.
Ethical clearance
The study was approved by the Institutional Ethical Committees of both National Centre for Cell Science, Pune, India and BJ Medical College, Pune, India.
Funding
This work was supported by the ‘Department of Biotechnology (DBT), Government of India’ (by Grant No. BT/Coord.II/01/03/2016), under the project Establishment of Centre for excellence National Centre for Microbial Resource (NCMR). Avinash Sharma acknowledges the support from the DBT/Wellcome Trust India Alliance fellowship under the project grant (IA/E/17/1/503700).
Declaration of competing interest
Authors declare no competing interest.
Acknowledgement
Authors are thankful to BJ medical staff for helping in sample collection and Director NCCS for their support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.micinf.2021.104880.
Appendix A. Supplementary data
The following are the supplementary data to this article:
References
- 1.Zhu Z., Lian X., Su X., Wu W., Marraro G.A., Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir Res. 2020;21(1):1–4. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abdelrahman Z., Li M., Wang X. Comparative review of SARS-CoV-2, SARS-CoV, MERS-CoV, and influenza a respiratory viruses. Front Immunol. 2020;11:2309. doi: 10.3389/fimmu.2020.552909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shah V. Letter to the editor: microbiota in the respiratory system—a possible explanation to age and sex variability in susceptibility to SARS-CoV-2. Microbiol Insights. 2021;14 doi: 10.1177/1178636120988604. 1178636120988604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shrestha N., Shad M.Y., Ulvi O., Khan M.H., Karamehic-Muratovic A., Nguyen U.S., et al. The impact of COVID-19 on globalization. One Health. 2020:100180. doi: 10.1016/j.onehlt.2020.100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peiris J.S., Yuen K.Y., Osterhaus A.D., Stöhr K. The severe acute respiratory syndrome. N Engl J Med. 2003 Dec 18;349(25):2431–2441. doi: 10.1056/NEJMra032498. Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet 2015;386:995–1007. [DOI] [PubMed] [Google Scholar]
- 6.Hilgenfeld R., Peiris M. From SARS to MERS: 10 years of research on highly pathogenic human coronaviruses. Antivir Res. 2013;100(1):286–295. doi: 10.1016/j.antiviral.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan J.F., Lau S.K., To K.K., Cheng V.C., Woo P.C., Yuen K.Y. Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev. 2015;28(2):465–522. doi: 10.1128/CMR.00102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Wit E., Van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat Rev Microbiol. 2016;14(8):523. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu B., Guo H., Zhou P., Shi Z.L. Characteristics of SARS-CoV-2 and COVID-19. Nat Rev Microbiol. 2020:1–4. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moustafa A.M., Planet P. Emerging SARS-CoV-2 diversity revealed by rapid whole genome sequence typing. bioRxiv. 2020 doi: 10.1093/gbe/evab197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pipes L., Wang H., Huelsenbeck J.P., Nielsen R. Assessing uncertainty in the rooting of the SARS-CoV-2 phylogeny. Mol Biol Evol. 2021;38(4):1537–1543. doi: 10.1093/molbev/msaa316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fadaka A.O., Sibuyi N.R., Adewale O.B., Bakare O.O., Akanbi M.O., Klein A., et al. Understanding the epidemiology, pathophysiology, diagnosis and management of SARS-CoV-2. J Int Med Res. 2020;48(8) doi: 10.1177/0300060520949077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rothan H.A., Byrareddy S.N. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salzberger B., Buder F., Lampl B., Ehrenstein B., Hitzenbichler F., Holzmann T., et al. Epidemiology of SARS-CoV-2. Infection. 2020:1–7. doi: 10.1007/s15010-020-01531-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pachetti M., Marini B., Benedetti F., Giudici F., Mauro E., Storici P., et al. Emerging SARS-CoV-2 mutation hot spots include a novel RNA-dependent-RNA polymerase variant. J Transl Med. 2020;18:1–9. doi: 10.1186/s12967-020-02344-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Dorp L., Acman M., Richard D., Shaw L.P., Ford C.E., Ormond L., et al. Emergence of genomic diversity and recurrent mutations in SARS-CoV-2. Infect Genet Evol. 2020;83:104351. doi: 10.1016/j.meegid.2020.104351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosch A.A., Biesbroek G., Trzcinski K., Sanders E.A., Bogaert D. Viral and bacterial interactions in the upper respiratory tract. PLoS Pathog. 2013;9(1) doi: 10.1371/journal.ppat.1003057. 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yi H., Yong D., Lee K., Cho Y.-J., Chun J. Profiling bacterial community in upper respiratory tracts. BMC Infect Dis. 2014;14:583. doi: 10.1186/s12879-014-0583-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Man W.H., de Steenhuijsen Piters W.A.A., Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanada S., Pirzadeh M., Carver K.Y., Deng J.C. Respiratory viral infection-induced microbiome alterations and secondary bacterial pneumonia. Front Immunol. 2018;9:2640. doi: 10.3389/fimmu.2018.02640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumpitsch C., Koskinen K., Schöpf V., Moissl-Eichinger C. The microbiome of the upper respiratory tract in health and disease. BMC Biol. 2019;17(1):1–20. doi: 10.1186/s12915-019-0703-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Santacroce L., Charitos I.A., Ballini A., Inchingolo F., Luperto P., De Nitto E., et al. The human respiratory system and its microbiome at a glimpse. Biology. 2020;9(10):318. doi: 10.3390/biology9100318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abreu N.A., Nagalingam N.A., Song Y., Roediger F.C., Pletcher S.D., Goldberg A.N., et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4(151):151ra124. doi: 10.1126/scitranslmed.3003783. 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leung R.K.-K., Zhou J.-W., Guan W., Li S.-K., Yang Z.-F., Tsui S.K.-W. Modulation of potential respiratory pathogens by pH1N1 viral infection. Clin Microbiol Infect. 2013;19:930–935. doi: 10.1111/1469-0691.12054. [DOI] [PubMed] [Google Scholar]
- 25.Safaeyan F., Nahaei M.R., Seifi S.J., Kafil H.S., Sadeghi J. Quantitative detection of Staphylococcus aureus, Streptococcus pneumoniae and Haemophilus influenzae in patients with new influenza A (H1N1)/2009 and influenza A/2010 virus infection. GMS Hyg Infect Control. 2015;10 doi: 10.3205/dgkh000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Coburn B., Wang P.W., Diaz Caballero J., Clark S.T., Brahma V., Donaldson S., et al. Lung microbiota across age and disease stage in cystic fibrosis. Sci Rep. 2015;5:10241. doi: 10.1038/srep10241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wen Z., Xie G., Zhou Q., Qiu C., Li J., Hu Q., et al. Distinct nasopharyngeal and oropharyngeal microbiota of children with influenza A virus compared with healthy children. BioMed Res Int. 2018;2018:9. doi: 10.1155/2018/6362716. 6362716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ding T., Song T., Zhou B., Geber A., Ma Y., Zhang L., et al. Microbial composition of the human nasopharynx varies according to influenza virus type and vaccination status. mBio. 2019;10(4) doi: 10.1128/mBio.01296-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee K.H., Gordon A., Shedden K., Kuan G., Ng S., Balmaseda A., et al. The respiratory microbiome and susceptibility to influenza virus infection. PloS One. 2019;14(1) doi: 10.1371/journal.pone.0207898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaul D., Rathnasinghe R., Ferres M., Tan G.S., Barrera A., Pickett B.E., et al. Microbiome disturbance and resilience dynamics of the upper respiratory tract during influenza A virus infection. Nat Commun. 2020;11(1):1–2. doi: 10.1038/s41467-020-16429-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacoby P., Watson K., Bowman J., Taylor A., Riley T.V., Smith D.W., et al. Kalgoorlie Otitis Media Research Project Team. Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine. 2007;25:2458–2464. doi: 10.1016/j.vaccine.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van den Bergh M.R., Biesbroek G., Rossen J.W., de Steenhuijsen Piters W.A., Bosch A.A., van Gils E.J., et al. Associations between pathogens in the upper respiratory tract of young children: interplay between viruses and bacteria. PloS One. 2012;7(10) doi: 10.1371/journal.pone.0047711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.De Maio F., Posteraro B., Ponziani F.R., Cattani P., Gasbarrini A., Sanguinetti M. Nasopharyngeal microbiota profiling of SARS-CoV-2 infected patients. Biol Proced Online. 2020 Dec;22(1):1–4. doi: 10.1186/s12575-020-00131-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mostafa H.H., Fissel J.A., Fanelli B., Bergman Y., Gniazdowski V., Dadlani M., et al. Metagenomic next-generation sequencing of nasopharyngeal specimens collected from confirmed and suspect COVID-19 patients. mBio. 2020;11(6) doi: 10.1128/mBio.01969-20. e01969-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yeoh Y.K., Zuo T., Lui G.C., Zhang F., Liu Q., Li A.Y., et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut. 2021;70:698–706. doi: 10.1136/gutjnl-2020-323020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu R., Liu P., Zhang T., Wu Q., Zeng M., Ma Y., et al. Progressive worsening of the respiratory and gut microbiome in children during the first two months of COVID-19. medRxiv. 2020 [Google Scholar]
- 37.Zuo T., Zhang F., Lui G.C., Yeoh Y.K., Li A.Y., Zhan H., et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020;159(3):944–955. doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nardelli C., Gentile I., Setaro M., Di Domenico C., Pincher B., Buonomo A.R., et al. Nasopharyngeal microbiome signature in COVID-19 positive patients: can we definitively get a role to fusobacterium periodonticum? Front Cell Infect Microbiol. 2021;11:18. doi: 10.3389/fcimb.2021.625581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rueca M., Fontana A., Bartolini B., Piselli P., Mazzarelli A., Copetti M., et al. Investigation of nasal/oropharyngeal microbial community of COVID-19 patients by 16S rDNA sequencing. Int J Environ Res Publ Health. 2021;18(4):2174. doi: 10.3390/ijerph18042174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jani K., Dhotre D., Bandal J., Shouche Y., Suryavanshi M., Rale V., et al. World's largest mass bathing event influences the bacterial communities of Godavari, a holy river of India. Microb Ecol. 2018;76(3):706–718. doi: 10.1007/s00248-018-1169-1. [DOI] [PubMed] [Google Scholar]
- 41.Bushnell B. BBDuk: adapter/quality trimming and filtering. https://sourceforgenetprojectsbbmap
- 42.Magoč T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27(21):2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caporaso J.G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F.D., Costello E.K. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Edgar R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26(19):2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 45.Haas B.J., Gevers D., Earl A.M., Feldgarden M., Ward D.V., Giannoukos G., et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21(3):494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammer Ø., Harper D.A., Ryan P.D. PAST: paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4(1):9. [Google Scholar]
- 47.Stearns J.C., Davidson C.J., McKeon S., Whelan F.J., Fontes M.E., Schryvers A.B., et al. Culture and molecular-based profiles show shifts in bacterial communities of the upper respiratory tract that occur with age. ISME J. 2015;9(5):1246–1259. doi: 10.1038/ismej.2014.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Unger S.A., Bogaert D. The respiratory microbiome and respiratory infections. J Infect. 2017;74:S84–S88. doi: 10.1016/S0163-4453(17)30196-2. [DOI] [PubMed] [Google Scholar]
- 49.Costello E.K., Lauber C.L., Hamady M., Fierer N., Gordon J.I., Knight R. Bacterial community variation in human body habitats across space and time. Science. 2009;326:1694–1697. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Camarinha-Silva A., Jáuregui R., Pieper D.H., Wos-Oxley M.L. The temporal dynamics of bacterial communities across human anterior nares. Environ Microbiol Rep. 2012;4:126–132. doi: 10.1111/j.1758-2229.2011.00313.x. [DOI] [PubMed] [Google Scholar]
- 51.Dimitriu P.A., Iker B., Malik K., Leung H., Mohn W.W., Hillebrand G.G. New insights into the intrinsic and extrinsic factors that shape the human skin microbiome. mBio. 2019;10(4) doi: 10.1128/mBio.00839-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rhoades N.S., Pinski A., Monsibais A.N., Jankeel A., Doratt B.M., Cinco I.R., et al. Acute SARS-CoV-2 infection is associated with an expansion of bacteria pathogens in the nose including Pseudomonas Aeruginosa. bioRxiv. 2021 doi: 10.1016/j.celrep.2021.109637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto S., Saito M., Tamura A., Prawisuda D., Mizutani T., Yotsuyanagi H. The human microbiome and COVID-19: a systematic review. PloS One. 2021;16(6) doi: 10.1371/journal.pone.0253293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song S.J., Lauber C., Costello E.K., Lozupone C.A., Humphrey G., Berg-Lyons D., et al. Cohabiting family members share microbiota with one another and with their dogs. eLife. 2013;2 doi: 10.7554/eLife.00458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Langevin S., Pichon M., Smith E., Morrison J., Bent Z., Green R., et al. Early nasopharyngeal microbial signature associated with severe influenza in children: a retrospective pilot study. J Gen Virol. 2017;98(10):2425. doi: 10.1099/jgv.0.000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Edouard S., Million M., Bachar D., Dubourg G., Michelle C., Ninove L., et al. The nasopharyngeal microbiota in patients with viral respiratory tract infections is enriched in bacterial pathogens. Eur J Clin Microbiol Infect Dis. 2018;37(9):1725–1733. doi: 10.1007/s10096-018-3305-8. [DOI] [PubMed] [Google Scholar]
- 57.Morris D.E., Cleary D.W., Clarke S.C. Secondary bacterial infections associated with influenza pandemics. Front Microbiol. 2017;8:1041. doi: 10.3389/fmicb.2017.01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Borges L.G., Giongo A., Pereira L.D., Trindade F.J., Gregianini T.S., Campos F.S., et al. Comparison of the nasopharynx microbiome between influenza and non-influenza cases of severe acute respiratory infections: a pilot study. Health Sci Rep. 2018;1(6) doi: 10.1002/hsr2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lehtinen M.J., Hibberd A.A., Männikkö S., Yeung N., Kauko T., Forssten S., et al. Nasal microbiota clusters associate with inflammatory response, viral load, and symptom severity in experimental rhinovirus challenge. Sci Rep. 2018;8(1):1–2. doi: 10.1038/s41598-018-29793-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen H.W., Liu P.F., Liu Y.T., Kuo S., Zhang X.Q., Schooley R.T., et al. Nasal commensal Staphylococcus epidermidis counteracts influenza virus. Sci Rep. 2016;6(1):1–2. doi: 10.1038/srep27870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ichinohe T., Pang I.K., Kumamoto Y., Peaper D.R., Ho J.H., Murray T.S., et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci USA. 2011;8(13):5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.de Steenhuijsen Piters W.A., Sanders E.A., Bogaert D. The role of the local microbial ecosystem in respiratory health and disease. Philos Trans R Soc B. 2015;370(1675):20140294. doi: 10.1098/rstb.2014.0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.