Abstract
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), the etiologic agent that causes Coronavirus Disease 2019 (COVID-19) pandemic, is a newly emerging respiratory RNA virus with exceptional transmissibility and pathogenicity. Numerous COVID-19 related studies have been fast-tracked, with the ultimate goal to end the pandemic. Here we review the major stages of SARS-CoV-2 infection cycle in cells, with specific emphasis on essential host factors. Insights into the cell biology of SARS-CoV-2 infection have accelerated the development of host-directed therapeutics, as shown by dozens of clinical trials evaluating COVID-19 treatments using host-targeting compounds.
Current Opinion in Virology 2021, 50:159–170
This review comes from a themed issue on Viral immunology
Edited by Matteo Iannacone and Antonio Bertoletti
For complete overview about the section, refer “Viral immunology”
Available online 21st August 2021
https://doi.org/10.1016/j.coviro.2021.08.007
1879-6257/© 2021 Elsevier B.V. All rights reserved.
Introduction
COVID-19 is one of the most grievous pandemics of modern human history. As of 20 months post-outbreak, SARS-CoV-2 has globally infected more than 200 million people causing over 4 million deaths (https://coronavirus.jhu.edu/map.html). A few vaccines have been authorized for emergency usage, but these efforts have yet to contain the pandemic due to limited productions and distributions [1, 2, 3]. COVID-19 research does not seem to be impacted by the stringent lockdowns and restrictions, instead, it has been conducted at an extraordinary pace to demystify the molecular biology of SARS-CoV-2 infection and pathogenesis. In this review, we summarize recently uncovered host factors critical for the various stages of the SARS-CoV-2 infection cycle (Figure 1 ). We further highlight promising druggable targets.
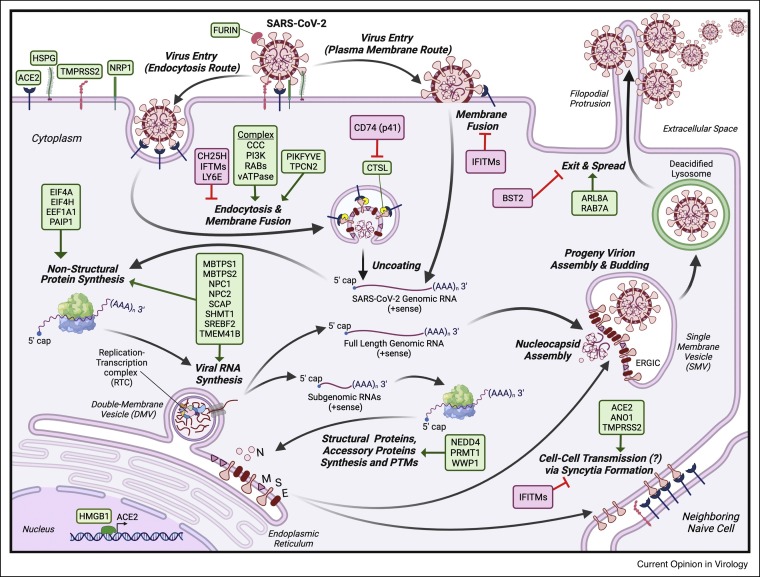
Figure 1.
The SARS-CoV-2 infection cycle.
Upon attachment to human cells, SARS-CoV-2 can fuse at the plasma membrane or internalize and later fuse in the endosome. Uncoated viral RNA genomes are rapidly translated into non-structural proteins to facilitate the full-length and subgenomic viral RNA syntheses. Structural and accessory proteins are translated from the subgenomic RNA transcripts to support the assembly and budding of progeny virions. During egress, SARS-CoV-2 progeny virions hijack a non-canonical lysosomal exocytosis pathway to exit cells. SARS-CoV-2 infected cells can fuse with neighboring naïve cells via syncytia formation.
Abbreviation: Angiotensin Converting Enzyme 2 (ACE2); Heparan sulfate proteoglycans (HSPGs); Transmembrane Serine Protease 2 (TMPRSS2); Neuropilin 1 (NRP1); Furin (FURIN); Cholesterol 25-Hydroxylase (CH25H); Interferon-induced Transmembrane proteins (IFITMs); Lymphocyte Antigen 6 Family Member E (LY6E); COMMD/CCDC22/CCDC93 Complex (CCC); Class III Phosphatidylinositol 3-kinase Complex (PI3K); RAB GTPases; vacuolar ATPase (vATPase) complex; Phosphoinositide Kinase, FYVE-Type Zinc Finger Containing (PIKFYVE); Two Pore Segment Channel 2 (TPCN2); Cathepsin L (CTSL); p41 isoform of the CD74 protein (CD74(p41)); Eukaryotic Translation Initiation Factor 4A1 (EIF4A); Eukaryotic Translation Initiation Factor 4H (EIF4H); Eukaryotic Translation Elongation Factor 1 Alpha 1 (EEF1A1); Poly(A) Binding Protein Interacting Protein 1 (PAIP1); Membrane Bound Transcription Factor Peptidase, Site 1 (MBTPS1); Membrane Bound Transcription Factor Peptidase, Site 2 (MBTPS2); NPC Intracellular Cholesterol Transporter 1 (NPC1); NPC Intracellular Cholesterol Transporter 2 (NPC2); SREBF Chaperone (SCAP); Serine Hydroxymethyltransferase 1 (SHMT1); Sterol Regulatory Element Binding Transcription Factor 2 (SREBF2); Transmembrane Protein 41B (TMEM41B); High Mobility Group Box 1 (HMGB1); NEDD4 E3 Ubiquitin Protein Ligase (NEDD4); WW Domain Containing E3 Ubiquitin Protein Ligase 1 (WWP1); Protein Arginine Methyltransferase 1 (PRMT1); Anoctamin 1 (ANO1); ADP Ribosylation Factor Like GTPase 8A (ARL8A); RAB7A, Member RAS Oncogene Family (RAB7A); Tetherin (BST2). Host dependency factors are boxed in green, and host restriction factors are boxed in red. Lipid bilayer membrane is indicated by double lines in purple. Deacidified lysosome is indicated by double lines in green.
SARS-CoV-2 entry
Similar to SARS-CoV-1 [4], SARS-CoV-2 binds to the cognate proteinaceous receptor, Angiotensin Converting Enzyme 2 (ACE2) at the plasma membrane of permissive cells using the Spike (S) protein [5•,6••,7••,8]. ACE2 is a membrane-bound dipeptidyl carboxypeptidase that may play a physiological role in the regulation of cardiovascular and renal function [9,10]. The S protein comprises two major subunits, S1 and S2, and the former subunit interacts with ACE2 via a receptor-binding domain (RBD) to mediate the attachment process. ACE2 is the key receptor but not the sole factor facilitating SARS-CoV-2 entry (reviewed in Ref. [11]). A few other cell surface factors have been shown to promote the virus entry such as the heparan sulfate proteoglycans (HSPGs) — common attachment factors for many viruses [12••]. The S protein’s RBD can directly interact with heparan sulfate to promote the viral attachment, also likely enhance interaction between the S protein and ACE2 [12••]. Multiple genes involved in the heparan sulfate biosynthesis pathway were implicated in functional genomic screens for SARS-CoV-2, further strengthening the role of heparan sulfate in enhancing SARS-CoV-2 infection [13•,14,15•,16]. Additionally, a few other host proteins such as a secreted isoform of ACE2, AXL Receptor Tyrosine Kinase (AXL, a receptor tyrosine kinase), Basigin (Ok Blood Group) (BSG, a.k.a. CD147, a ubiquitously expressed multifunctional plasma membrane protein), Scavenger Receptor Class B Member 1 (SCARB1, a receptor for high-density lipoprotein cholesterol), and Transmembrane Protein 106B (TMEM106B), have been reported to promote SARS-CoV-2 entry [16, 17, 18, 19, 20]. Intriguingly, enhancement of virus entry could be accomplished without ACE2, suggesting ACE2 may not be the only host factor determining tissue tropism for SARS-CoV-2 [18]. Some cellular factors, such as High Mobility Group Box 1 (HMGB1), a multifunctional DNA-binding nuclear protein, have been shown to indirectly impact SARS-CoV-2 entry by regulating the cell surface expression of ACE2 [21•].
Upon binding to ACE2, the S1–S2 subunits dissociate. The S1–S2 dissociation involves proteolytic cleavage by a ubiquitously expressed serine endoprotease furin (FURIN) at the S1–S2 junction within a multibasic motif, ‘Arg-Arg-Ala-Arg’ (RRAR) [22•,23••]. The multibasic cleavage motif is critical for SARS-CoV-2 to infect human airway cells, and it can be a contributing factor for SARS-CoV-2 tropism and transmissibility [24, 25, 26, 27, 28, 29,30•,31•]. However, it remains unclear whether furin is indispensable during the SARS-CoV-2 entry, as other alternative proteases may be able to cleave the S1–S2 junction in the absence of furin [27, 28, 29]. A few recent reports demonstrated that the untied S1 subunit could bind to Neuropilin 1 (NRP1), a cell surface receptor that recognizes and binds CendR motif, ‘Arg/Lys-X-X-Arg/Lys’ (R/KXXR/K, where X indicates any amino acid), containing ligands [32, 33, 34]. The interaction between NRP1 and the S1 subunit can promote SARS-CoV-2 infection, probably by enhancing the virus internalization process [32,33]. Similar to SARS-CoV-1, the S2 subunit is further processed by either a transmembrane serine protease, TMPRSS2, or lysosomal cysteine proteinases (cathepsins, such as CTSL) to trigger final conformational changes of the S2 subunit, exposing and inserting the viral fusion peptide into the cellular membrane. As a result, the viral membrane fuse with the host membrane to deposit the viral genome into the cytoplasm [29]. Currently, TMPRSS2 inhibitors, such as Camostat Mesylate and Nafamostat Mesylate, are being tested in over a dozen of clinical trials (Table 1 ).
Table 1.
Examples of host-directed therapeutics against COVID-19 undergoing clinical trials(https://clinicaltrials.gov/ct2/home)
| Drug | Pharmacological actions | Targeting host factors | Stage of SARS-CoV-2 infection cycle | Clinical trials | Phase | Reference |
|---|---|---|---|---|---|---|
| Apilimod | Inhibitor | PIKFYVE | Entry | NCT04446377 | 2 | [8,74,117•,118] |
| Camostat mesylate | Inhibitor | TMPRSS2 | Entry | NCT04353284 | 2 | [36,119] |
| NCT04583592 | 2 | |||||
| NCT04608266 | 3 | |||||
| NCT04524663 | 2 | |||||
| NCT04657497 | 3 | |||||
| NCT04681430 | 2 | |||||
| NCT04470544 | 2 | |||||
| NCT04625114 | 2 | |||||
| Nafamostat mesylate | Inhibitor | TMPRSS2 | Entry | NCT04352400 | 2, 3 | [120, 121, 122] |
| NCT04390594 | 3 | |||||
| NCT04418128 | 2, 3 | |||||
| NCT04473053 | 2, 3 | |||||
| NCT04623021 | 2 | |||||
| NCT04628143 | 2 | |||||
| Niclosamide | Inhibitor | ANO1 | SARS-CoV-2 induced syncytia and spread (?) | NCT04399356 | 2 | [114•,123] |
| NCT04603924 | 2, 3 | |||||
| NCT04858425 | 2 | |||||
| NCT04753619 | 2 | |||||
| NCT04558021 | 3 | |||||
| NCT04870333 | 2, 3 | |||||
| NCT04750759 | 2 | |||||
| Plitidepsin | Inhibitor | EEF1A | Translation | NCT04382066 | 1 | [59] |
| NCT04784559 | 3 | |||||
| Statin | Inhibitor | HMGCR | Multiple | NCT04486508 | 3 | [124, 125, 126, 127] |
| NCT04904536 | 3 | |||||
| NCT04472611 | 3 | |||||
| NCT04380402 | 2 | |||||
| Zotatifin | Inhibitor | EIF4A | Translation | NCT04632381 | 1 | [58••,128,129] |
Since proteolytic maturation of the S protein is a prerequisite for the virus entry, the entry site of SARS-CoV-2 is dictated by the subcellular localization of the host proteases. Thus, SARS-CoV-2 can enter cells through two main entry routes: fusing directly at the cell membrane after binding to ACE2, or entering cell through receptor-mediated endocytosis and then fusing to endosomal membrane. In cells with surface expression of ACE2 and TMPRSS2, such as human airway epithelial cells, SARS-CoV-2 readily binds and fuses at the plasma membrane [7••,35,36]. Interestingly, other TMPRSS2-like proteases such as TMPRSS4 and TMPRSS13 may similarly promote the S protein processing [36,37]. Alternatively, the virus can first enter host cells via receptor-mediated endocytosis and subsequently fuses with the endosomal membrane. This entry route allows the virus to access cathepsins in the endosomes, especially when TMPRSS2 is absent from the cell surface [38].
Cathepsin L (CTSL) is the principal endosomal protease responsible for priming the S protein in endosomes. It was observed that a higher level of circulating CTSL in the blood is associated with severe disease in COVID-19 patients [39]. Intriguingly, pharmacological inhibition of CTSL, but not Cathepsin B (CTSB), can reduce SARS-CoV-2 infection in vitro [8,38]. Some other proteases such as Trypsin and Human Airway Trypsin-Like Protease (TMPRSS11D, a.k.a. HAT) have been proposed to play a similar role in the in vitro processing of the S protein [8,27,36]. Notably, the p41 isoform of the CD74 protein (a chaperone that regulates MHC class II antigen processing) can hinder CTSL-mediated activation of the S protein and induce resistance in human cells by arresting viral fusion inside endosomes [40]. In addition to the host proteases, SARS-CoV-2 membrane fusion in endosomes requires a low pH, as neutralization of acidic endosomes inhibits SARS-CoV-2 infection in specific cell types [7••,8,38,39,41]. The impact of the endosomal pH neutralization can be reversed by overexpressing TMPRSS2, supporting the model that SARS-CoV-2 can penetrate the cellular membrane barrier either at the plasma membrane or in the endosome [7••,38]. Some host factors known to play a role in endocytosis and endosomal trafficking pathways, such as subcomponents of COMMD/CCDC22/CCDC93 Complex, Class III Phosphatidylinositol 3-kinase (PI3K) Complex, RAB GTPases, and vacuolar ATPase (vATPase) complex, have been reported to promote SARS-CoV-2 infection [13•,20,42•,43,44]. Chemical perturbations of Two Pore Segment Channel 2 (TPCN2, a nicotinic acid adenine dinucleotide phosphate receptor), and its upstream regulator PIKFYVE, a phosphoinositide kinase that recruits protein complexes to cell- and endosomal-membrane, have been shown to potently block virus entry [8]. PIKFYVE functions can be chemically perturbed using Apilimod — a candidate therapeutic for treating COVID-19 (Table 1).
Regardless of where the S protein-mediated membrane fusion occurs, host cells can intercept the penetration using broad-acting restriction factors such as Cholesterol 25-Hydroxylase (CH25H), Lymphocyte Antigen 6 Family Member E (LY6E), and Interferon-induced Transmembrane proteins (IFITMs, e.g. IFITM1, IFITM2, IFITM3) [45, 46, 47, 48, 49, 50, 51]. CH25H catalyzes the conversion of cholesterol to 25-hydroxycholesterol (25HC). This activates Acetyl-CoA Acetyltransferase 1 at the endoplasmic reticulum (ER) resulting in depletion of plasma membrane cholesterol and accumulation of 25HC in the late endosomes, which blocks the viral membrane fusion [49,50]. Similarly, LY6E, an interferon-stimulated glycosylphosphatidylinositol-anchored protein, can block SARS-CoV-2 entry by impairing the membrane fusion regardless of the S protein maturation stages [45,46]. IFITMs are known to block the membrane fusion of many enveloped viruses, including SARS-CoV-1 [52]. Nonetheless, IFITMs may act as double-edged swords in responding to the SARS-CoV-2 entry. IFITMs can principally inhibit SARS-CoV-2 infection, but the subcellular localization of distinct IFITMs may impose different modulatory effects on the virus entry. For instance, ectopically expressed IFITM3 that traffics between the plasma membrane and endosomes can block the SARS-CoV-2 fusion in the endosome. Conversely, cell surface-immobilized IFITM3 mutant that harbors dysfunctional endocytosis YxxФ motif can enhance SARS-CoV-2 fusion at the plasma membrane [47].
SARS-CoV-2 translation and RNA replication
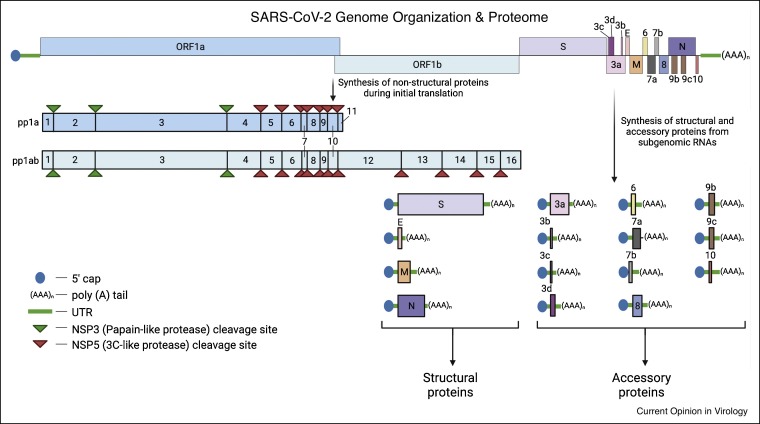
Upon deposition in the cytoplasm, the uncoated viral RNA genome can rapidly recruit cellular translation machinery to synthesize SARS-CoV-2 proteins [53••,54, 55, 56]. SARS-CoV-2 genome (Figure 2 ) is a positive-sense, single-stranded RNA, approximately 30 kb in length, comprising multiple open reading frames (ORFs) that encodes for relatively large numbers of non-structural proteins (NSPs), structural proteins, and accessory proteins [57]. NSPs are directly translated from the full-length viral genome, whereas structural and accessory proteins are encoded by subgenomic RNA transcripts [84]. Each viral transcript mimics host mRNA, that is, decorated with a 5′ cap structure and a 3′ poly (A) tail, enabling the virus to synthesize its proteins by co-opting the cap-dependent mRNA translation machinery. Approximately two-thirds of the viral transcript from the 5′ terminus encodes for two overlapping ORFs, ORF1a, and ORF1b, translated into two polypeptides, pp1a and pp1ab, respectively. Upon cleavages by its viral proteases (NSP3 and NSP5), these polypeptides mature into 16 non-structural proteins (NSPs, NSP1–NSP16). The remainder of the viral transcript towards the 3′ terminus encodes for structural proteins, S protein, envelope (E) protein, membrane (M) protein, and nucleocapsid (N) protein, and multiple accessory proteins.
Figure 2.
SARS-CoV-2 genome organization and proteome.
SARS-CoV-2 genome is a ∼30 kb positive-sense, single-stranded RNA. The 5′ terminal harbors two overlapping ORFs (ORF1a and ORF1b), encoding polyproteins pp1a and pp1ab. These polyproteins are proteolytically processed by viral proteases (NSP3 and NSP5) into 16 non-structural proteins (NSP1–NSP16). The 3′ end of the viral genome encodes for structural and accessory proteins via multiple subgenomic RNAs. Sizes of SARS-CoV-2 genes and proteins are not drawn to scale.
Abbreviation:Spike (S); Envelope (E); Membrane (M); Nucleocapsid (N); accessory proteins (3a, 3b, 3c, 3d, 6, 7a, 7b, 8, 9b, 9c and 10); open reading frame (ORF); untranslated region (UTR).
Overall, in SARS-CoV-2 infected cells, the global host protein translation is reduced via rapid degradation of cytoplasmic mRNA and sequestration of nascent mRNA in the nucleus, allowing viral RNAs to dominate the access to ribosomes [54]. A few translational components, such as translation initiation factors EIF4A and EIF4H, as well as a translation elongation factor EEF1A1, have been demonstrated to be indispensable for the virus infection [58••,59,60]. Excitingly, some of these pro-viral translation factors can be targeted using specific drugs such as Zotatifin and Plitidepsin to inhibit SARS-CoV-2 infection (Table 1). Interestingly, to enhance the translation of viral proteins, the NSP3 protein can interact with human Poly(A) Binding Protein Interacting Protein 1 (PAIP1), which can stimulate translation via interaction with translational initiation components [61]. SARS-CoV-2 NSP1 protein can bind and block the cellular mRNA entry tunnel of the 40S ribosomal subunit, resulting in disruption of the host mRNA translation and rewiring the translation machinery to accommodate biasedly viral protein synthesis [62, 63, 64, 65, 66, 67]. In addition, the NSP1 protein can also occupy the host mRNA nuclear export complex, NXF1-NXT1, to disrupt the mRNA nuclear export [68]. Furthermore, SARS-CoV-2 ORF6 protein can alter the nuclear pore complex via interaction with Ribonucleic Acid Export 1 (RAE1) and Nucleoporin 98 (NUP98) to disrupt the bidirectional nuclear-cytoplasmic trafficking of cellular RNA [69,70]. Additionally, SARS-CoV-2 NSP16 can interact with U1/U2 snRNA, critical components of the major spliceosome, to inhibit global pre-mRNA maturation [67].
Post-translational modifications (PTMs) of SARS-CoV-2 proteins play an essential role in promoting virus infection. The S protein is heavily N- and O-glycosylated, and these glycan decorations are crucial in promoting SARS-CoV-2 entry [71, 72, 73]. Multiple SARS-CoV-2 proteins (e.g. NSP3, NSP9, NSP14, S, M, N, and ORF9b) are phosphorylated, and chemical inhibitions of the PTMs can block virus infection, suggesting these phosphorylations may exhibit a pro-viral role [60,74]. The S protein is ubiquitylated by HECT-E3 ubiquitin ligases, for example, NEDD4 and WWP1, and chemical inhibitions of these E3 ubiquitin ligases can potently decrease SARS-CoV-2 infection and egress [75,76]. The ORF7a protein requires polyubiquitylation to confer the ability to antagonize host innate immunity by blocking STAT2 phosphorylation [77]. SARS-CoV-2 N protein is methylated by Protein Arginine Methyltransferase 1 (PRMT1), and the PTM is required for viral RNA packaging and interaction with cellular stress granules [78].
SARS-CoV-2 NSPs orchestrate the cascade of events during viral RNA synthesis including vast cellular membrane remodeling. The viral Replication and Transcription Complex (RTC), comprising NSP2-16 proteins, is the core component of the viral RNA synthesis machinery [53••]. RTCs are anchored on unique, interconnected compartments called double-membrane vesicles (DMVs) [79, 80, 81]. Like other virus-induced RNA replication compartments, these DMVs serve as hideouts to avoid host innate immune sensing and RNA decay machinery, allowing SARS-CoV-2 to clandestinely synthesize its subgenomic and full-length genomic RNA transcripts [82,83]. Specific host factors co-opted by SARS-CoV-2 to facilitate viral RNA synthesis are yet to be revealed but likely to be similar to those reported for other coronaviruses [85]. In addition, a plethora of SARS-CoV-2 RNA-interacting host proteins have been comprehensively uncovered, and some of these RNA-binding proteins are likely to play a role during virus infection in cells, including viral RNA synthesis [86•,87,88•,89•].
Overall, substantial alterations of cellular organelles and compartments, such as fragmentation of Golgi apparatus, were observed in SARS-CoV-2 infected cells, indicating cell membrane homeostasis and lipid metabolism can play an essential role in accommodating productive propagation of progeny virions [80]. Furthermore, accumulation of lipid droplets and reprogramming of lipid metabolism have been observed in SARS-CoV-2 infected cells and COVID-19 patients’ samples, suggesting a potential causative link between dysregulation of lipid metabolism and viral pathogenesis [90, 91, 92]. Notably, the strategy of treating COVID-19 via pharmacological inhibition of host lipid metabolism using statin and derivatives, a group of commonly used cholesterol-lowering drugs, is currently being evaluated in clinical trials (Table 1). Some vital cellular pathways that regulate cellular lipid metabolism, such as the PI3K and autophagy pathways, have been implicated to play a role during coronavirus infections and pathogenesis [13•,42•,93]. Specific cellular factors that are known to regulate lipid and cholesterol homeostasis, such as Sterol Regulatory Element Binding Transcription Factor 2 (SREBF2, a.k.a. SREBP2), SREBF Chaperone (SCAP), Membrane Bound Transcription Factor Peptidases (MBTPS1, MBTPS2), and NPC Intracellular Cholesterol Transporters (NPC1, NPC2), have been identified as host dependency factors for SARS-CoV-2 via functional genomic screens [15•,42•,43,44]. SARS-CoV-2 ORF3a can directly interact with VPS39 subunits of the Homotypic Fusion and Protein Sorting (HOPS) Complex to inhibit fusion of autophagosomes with lysosomes, preventing the formation of late-stage autophagy organelles, autolysosomes [94,95]. The ORF3a-VPS39 interaction intercepts the subsequent interplay between HOPS-RAB7A, and HOPS-STX17-SNAP29-VAMP8-SNARE, which modulates the autolysosome maturation. Interestingly, depletion of VPS39 can protect human cells from SARS-CoV-2 mediated cytopathic effects, highlighting the role of VPS39 in promoting virus infection [44]. In addition, SARS-CoV-2 infection requires an ER-resident phospholipid scramblase, TMEM41B, that was first known to regulate autophagosome formation and lipid mobilization [15•,16,96, 97, 98]. TMEM41B plays a physiological role in maintaining the ER integrity and cellular lipid homeostasis, especially the distribution of cholesterol and phosphatidylserine [99,100]. In addition to lipid metabolism, SARS-CoV-2 can rewire glycolysis and one-carbon metabolism via hijacking Serine Hydroxymethyltransferase 1 (SHMT1) to induce de novo purine synthesis for supporting mega-scale of viral RNA synthesis [101].
SARS-CoV-2 assembly and exit
Generally, the assembly and exit of enveloped viruses involve a cascade of irreversible events that is well-orchestrated by both viral factors and cellular components [83]. For SARS-CoV-2, the process begins when newly synthesized SARS-CoV-2 RNA genomes are exported from the DMVs via a crown-like, cylindrical membrane pore complex comprising viral proteins (e.g. NSP3) and perhaps host proteins that are yet to be identified [81,102,103]. This channel may also play a role in shuttling host factors and ingredients (e.g. metabolites and nucleotides) into the DMVs to fuel the viral RNA synthesis [80]. Subsequently, via an unknown mode of trafficking, perhaps involving the association of specific viral RNA sequences (e.g. cis-acting elements) with viral proteins and host factors such as cytoskeleton and RNA binding proteins (as suggested in Refs. [86•,87,89•,104]), the viral genomes shuttle to reach the N proteins near the budding site. SARS-CoV-2 RNA genomes can directly interact with the N proteins to form viral ribonucleoprotein complexes (vRNPs) via liquid–liquid phase separation that may involve association with G3BP-mediated stress granules [87,105,106]. vRNPs are frequently seen to accumulate and bud into curved cytosolic surfaces of single-membrane vesicles (SMVs), where viral structural proteins (S, N, E, and M) happen to be co-localized [79,80]. As a result, SARS-CoV-2 progeny virions are assembled in the SMVs that are likely derived from the endoplasmic reticulum-Golgi-intermediate compartment (ERGIC) [79]. The underlying mechanistic details about SARS-CoV-2 assembly and budding remain murky. The budding process is likely promoted by a membrane bending mechanism driven by unknown viral components and host factors [79].
Despite hitch-hiking the canonical exocytosis pathway to exit cells like other enveloped viruses [107], SARS-CoV-2 virions are likely to unconventionally utilize lysosomal exocytosis as an exit strategy [108••]. During exit, SARS-CoV-2 progeny virions may first travel from the SMV to the Golgi intermediate compartments and subsequently re-routed to a deacidified lysosomal compartment before getting released from host cells. A few host factors that are known to regulate lysosomal trafficking, such as RAB7A (a RAS-related GTP-binding protein that regulates endo-lysosomal trafficking), ADP Ribosylation Factor Like GTPase 8A (ARL8A, a regulator of lysosome mobility), and TMEM106B (a regulator of lysosomal pH, morphology trafficking, and exocytosis), may play a role in modulating the lysosomal exocytosis process [13•,16,108••,109]. Intriguingly, colocalization of SARS-CoV-2 virions on cell surface filopodial protrusions was commonly observed on infected cells, suggesting a strategy for the virus to release or spread from cell to cell [80]. The formation of these protrusions involves a Casein kinase II (CK2)-dependent remodeling of the cytoskeleton that is activated by the N protein [58••]. Nevertheless, the exit of viruses is not always smooth sailing; SARS-CoV-2 release from host cells can be blocked by Tetherin (BST2), an IFN-induced restriction factor targeting a broad spectrum of enveloped viruses [110,111•]. Similar to HIV-1’s counteraction of Tetherin using its Vpu accessory protein [112], SARS-CoV-2 has evolved to thwart BST2 inhibition using accessory protein ORF7a [111•].
Although the spread of SARS-CoV-2 virions remains poorly understood, they are likely to transmit via both cell-free and cell-to-cell routes. Syncytia formation is commonly observed in some SARS-CoV-2 infected cells and COVID-19 patients’ tissues, and it has been proposed to play a critical role in cell-to-cell transmission and pathogenesis [113]. The underlying mechanism for the syncytia formation is surprisingly similar to the TMPRSS2-dependent SARS-CoV-2 entry — triggered by the interaction between the S protein (present on the surface of donor cells) and ACE2 (on the surface of neighboring cells) [48,114•,115]. Mechanistically, syncytia formation is regulated by a plasma membrane protein, Anoctamin 1 (ANO1, a.k.a. TMEM16), but can be restricted by IFITMs, especially IFITM1, and can be chemically intervened using Niclosamide (Table 1) [48,114•]. Puzzlingly, the impact of the IFITMs-mediated restriction can be reversed by high levels of TMPRSS2 expression, further clouding the differences between SARS-CoV-2 entry and spread [48].
Host-directed therapeutics against COVID-19
As drug discovery and approval for use entail significant resources and time, drug repurposing represents the primary line of treatment for the COVID-19 pandemic. The strategy is especially advantageous for drugs with a pre-established clinical profile and manufacturing arrangements that could expedite treatment options [116,117•]. A few SARS-CoV-2 host factors have served as promising therapeutic targets to fuel numerous preclinical screens and clinical trials (Table 1). Host-directed therapeutics enjoy the benefits of reduced viral resistance and their broad-spectrum antiviral potential. However, selecting a suitable host target essential for a virus but dispensable for a host remains challenging. In order to avoid undesirable host responses, rigorous validation and extensive monitoring are necessary for the optimal selection of drugs and host factor candidates.
Concluding remarks
Rapid multi-omics studies on SARS-CoV-2 have painted a complex picture of its infection mechanism. This molecular information is crucial for the innovation of novel antiviral strategies including new vaccine designs and host-directed antiviral therapeutics [130,131]. It is perhaps worth noting a few caveats about these discoveries. Since the genuine SARS-CoV-2 must be studied in a contained BSL3 environment with restricted access and limited resources, a substantial number of SARS-CoV-2 studies were performed at BSL2 labs using surrogate systems, such as individual viral protein expression constructs, pseudotyped viruses, viral replicons, and other betacoronaviruses (e.g. HCoV-OC43 and MHV). In addition, non-human cell types (e.g. Vero cells and derivatives) and artificially engineered systems (e.g. human ACE2 overexpressing cell lines and lab animals) have been widely used. These systems have greatly expedited SARS-CoV-2 research, but the outcomes must be carefully interpreted and cross-validated before making conclusions about the genuinevirus. For example, African green monkey kidney epithelial cells, Vero cells, are widely used for COVID-19 research, including virus isolation and propagation. However, Vero cells express only ACE2 but not TMPRSS2 at the cell surface, forcing SARS-CoV-2 to enter these cells solely via the endocytosis route. Endosomal inhibitors such as hydroxychloroquine tend to inhibit SARS-CoV-2 infection in Vero cells potently, but not other cell types (e.g. Calu-3, human lung epithelial cells) that allow the virus to opt for fusion at the plasma membrane and in the endosomes [132••]. Worryingly, repeated passaging of SARS-CoV-2 in Vero cells can result in the loss of the furin-cleavage site on the S protein, altering the authenticity and pathogenicity of the virus [25,31•,133,134]. Therefore, in-depth studies using physiological relevant tools are needed to uncover the bona fide molecular mechanisms underlying the SARS-CoV-2 infection and pathogenesis.
In a nutshell, an unprecedented scale of coronavirus research has been rapidly conducted in response to the COVID-19 pandemic, and significant progress has been achieved to fill up knowledge gaps in some aspects of coronavirus biology. However, these findings may represent the tip of the iceberg. Humankind has witnessed the emergence of three novel coronaviruses in the past two decades, and SARS-CoV-2 is unlikely to be the last. To better prepare for the next pandemic, a comprehensive understanding of the SARS-CoV-2 biology is direly in need to stimulate the development of novel antiviral strategies, such as host-directed therapeutics that can potentially combat a broad spectrum of coronaviruses.
Conflict of interest statement
Nothing declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
We sincerely apologize to all authors whose work could not be cited due to space limitations and rapid updates in COVID-19 research. Research in the YSO lab is supported by Duke-NUS Medical School [Startup Fund, and Pilot Grant Duke/Duke-NUS/RECA(Pilot)/2019/0047] and Ministry of Education [MOE-000095-01] in Singapore. MY is supported by the Duke-NUS Medical School IBM Program PhD Scholarship. Figure 1, Figure 2 were prepared using BioRender.com. We thank reviewers, editors, as well as Jan E. Carette, Alex G. Johnson, Dewei Tan, and Yee Peng Chan for their time and effort to provide constructive comments on the manuscript.
References
- 1.Case J.B., Winkler E.S., Errico J.M., Diamond M.S. On the road to ending the COVID-19 pandemic: are we there yet? Virology. 2021;557:70–85. doi: 10.1016/j.virol.2021.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai L., Gao G.F. Viral targets for vaccines against COVID-19. Nat Rev Immunol. 2021;21:73–82. doi: 10.1038/s41577-020-00480-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 4.Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., et al. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5•.Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]; A key paper (among the firsts) to identify and characterize ACE2 as the functional receptor for SARS-CoV-2.
- 6••.Zhou P., Yang X.-L., Wang X.-G., Hu B., Zhang L., Zhang W., Si H.-R., Zhu Y., Li B., Huang C.-L., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first paper that identify SARS-CoV-2 as the etiologic agent responsible for COVID-19. Also, this is one of the first papers to demonstrate ACE2 is a functional receptor for SARS-CoV-2.
- 7••.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]; In this study the authors confirmed that TMPRSS2 is a key protease required for the maturation of the SARS-CoV-2 Spike protein.
- 8.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., et al. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11 doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donoghue M., Hsieh F., Baronas E., Godbout K., Gosselin M., Stagliano N., Donovan M., Woolf B., Robison K., Jeyaseelan R., et al. A novel angiotensin-converting enzyme-related carboxypeptidase (ACE2) converts angiotensin I to angiotensin 1-9. Circ Res. 2000;87:E1–E9. doi: 10.1161/01.res.87.5.e1. [DOI] [PubMed] [Google Scholar]
- 10.Paz Ocaranza M., Riquelme J.A., García L., Jalil J.E., Chiong M., Santos R.A.S., Lavandero S. Counter-regulatory renin-angiotensin system in cardiovascular disease. Nat Rev Cardiol. 2020;17:116–129. doi: 10.1038/s41569-019-0244-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Evans J.P., Liu S.-L. Role of host factors in SARS-CoV-2 entry. J Biol Chem. 2021;297 doi: 10.1016/j.jbc.2021.100847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., et al. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057.e15. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; This is the first paper to provide structural evidence showing heparan sulfate as a cellular attachment factors of SARS-CoV-2.
- 13•.Wang R., Simoneau C.R., Kulsuptrakul J., Bouhaddou M., Travisano K.A., Hayashi J.M., Carlson-Stevermer J., Zengel J.R., Richards C.M., Fozouni P., et al. Genetic screens identify host factors for SARS-CoV-2 and common cold coronaviruses. Cell. 2021;184:106–119.e14. doi: 10.1016/j.cell.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of the key papers to study the cell biology of SARS-CoV-2 using genome-scale CRISPR screens.
- 14.Chu H., Hu B., Huang X., Chai Y., Zhou D., Wang Y., Shuai H., Yang D., Hou Y., Zhang X., et al. Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat Commun. 2021;12 doi: 10.1038/s41467-020-20457-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Schneider W.M., Luna J.M., Hoffmann H.-H., Sánchez-Rivera F.J., Leal A.A., Ashbrook A.W., Pen J.L., Ricardo-Lax I., Michailidis E., Peace A., et al. Genome-scale identification of SARS-CoV-2 and pan-coronavirus host factor networks. Cell. 2021;184:120–132.e14. doi: 10.1016/j.cell.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of the key papers to study the cell biology of SARS-CoV-2 using genome-scale CRISPR screens.
- 16.Baggen J., Persoons L., Vanstreels E., Jansen S., Van Looveren D., Boeckx B., Geudens V., De Man J., Jochmans D., Wauters J., et al. Genome-wide CRISPR screening identifies TMEM106B as a proviral host factor for SARS-CoV-2. Nat Genet. 2021;53:435–444. doi: 10.1038/s41588-021-00805-2. [DOI] [PubMed] [Google Scholar]
- 17.Wei C., Wan L., Yan Q., Wang X., Zhang J., Yang X., Zhang Y., Fan C., Li D., Deng Y., et al. HDL-scavenger receptor B type 1 facilitates SARS-CoV-2 entry. Nat Metab. 2020;2:1391–1400. doi: 10.1038/s42255-020-00324-0. [DOI] [PubMed] [Google Scholar]
- 18.Wang S., Qiu Z., Hou Y., Deng X., Xu W., Zheng T., Wu P., Xie S., Bian W., Zhang C., et al. AXL is a candidate receptor for SARS-CoV-2 that promotes infection of pulmonary and bronchial epithelial cells. Cell Res. 2021;31:126–140. doi: 10.1038/s41422-020-00460-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang K., Chen W., Zhang Z., Deng Y., Lian J.-Q., Du P., Wei D., Zhang Y., Sun X.-X., Gong L., et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Sig Transduct Target Ther. 2020;5:1–10. doi: 10.1038/s41392-020-00426-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeung M.L., Teng J.L.L., Jia L., Zhang C., Huang C., Cai J.-P., Zhou R., Chan K.-H., Zhao H., Zhu L., et al. Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell. 2021;184:2212–2228.e12. doi: 10.1016/j.cell.2021.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21•.Wei J., Alfajaro M.M., DeWeirdt P.C., Hanna R.E., Lu-Culligan W.J., Cai W.L., Strine M.S., Zhang S.-M., Graziano V.R., Schmitz C.O., et al. Genome-wide CRISPR screens reveal host factors critical for SARS-CoV-2 infection. Cell. 2021;184:76–91.e13. doi: 10.1016/j.cell.2020.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of the key papers to study the cell biology of SARS-CoV-2 using genome-scale CRISPR screens.
- 22•.Coutard B., Valle C., de Lamballerie X., Canard B., Seidah N.G., Decroly E. The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade. Antiviral Res. 2020;176 doi: 10.1016/j.antiviral.2020.104742. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper [22•] describes the multibasic cleavage site in the S1–S2 junction of SARS-CoV-2 S glycoprotein, which was absent in SARS-CoV-1 sequence.
- 23••.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell. 2020;78:779–784.e5. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper [22•] describes the multibasic cleavage site in the S1–S2 junction of SARS-CoV-2 S glycoprotein, which was absent in SARS-CoV-1 sequence.
- 24.Mykytyn A.Z., Breugem T.I., Riesebosch S., Schipper D., van den Doel P.B., Rottier R.J., Lamers M.M., Haagmans B.L. SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. eLife. 2021;10 doi: 10.7554/eLife.64508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamers M.M., Mykytyn A.Z., Breugem T.I., Wang Y., Wu D.C., Riesebosch S., van den Doel P.B., Schipper D., Bestebroer T., Wu N.C., et al. Human airway cells prevent SARS-CoV-2 multibasic cleavage site cell culture adaptation. eLife. 2021;10 doi: 10.7554/eLife.66815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau S.-Y., Wang P., Mok B.W.-Y., Zhang A.J., Chu H., Lee A.C.-Y., Deng S., Chen P., Chan K.-H., Song W., et al. Attenuated SARS-CoV-2 variants with deletions at the S1/S2 junction. Emerging Microbes Infect. 2020;9:837–842. doi: 10.1080/22221751.2020.1756700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xia S., Lan Q., Su S., Wang X., Xu W., Liu Z., Zhu Y., Wang Q., Lu L., Jiang S. The role of furin cleavage site in SARS-CoV-2 spike protein-mediated membrane fusion in the presence or absence of trypsin. Signal Transduct Targeted Ther. 2020;5:1–3. doi: 10.1038/s41392-020-0184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Papa G., Mallery D.L., Albecka A., Welch L.G., Cattin-Ortolá J., Luptak J., Paul D., McMahon H.T., Goodfellow I.G., Carter A., et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30•.Peacock T.P., Goldhill D.H., Zhou J., Baillon L., Frise R., Swann O.C., Kugathasan R., Penn R., Brown J.C., Sanchez-David R.Y., et al. The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol. 2021 doi: 10.1038/s41564-021-00908-w. [DOI] [PubMed] [Google Scholar]; This paper [30•] demonstrates the multibasic furin cleavage site is critical for SARS-CoV-2 transmission.
- 31•.Johnson B.A., Xie X., Bailey A.L., Kalveram B., Lokugamage K.G., Muruato A., Zou J., Zhang X., Juelich T., Smith J.K., et al. Loss of furin cleavage site attenuates SARS-CoV-2 pathogenesis. Nature. 2021;591:293–299. doi: 10.1038/s41586-021-03237-4. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper [30•] demonstrates the multibasic furin cleavage site is critical for SARS-CoV-2 transmission and pathogenesis.
- 32.Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daly J.L., Simonetti B., Klein K., Chen K.-E., Williamson M.K., Antón-Plágaro C., Shoemark D.K., Simón-Gracia L., Bauer M., Hollandi R., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370:861–865. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kielian M. Enhancing host cell infection by SARS-CoV-2. Science. 2020;370:765–766. doi: 10.1126/science.abf0732. [DOI] [PubMed] [Google Scholar]
- 35.Wettstein L., Weil T., Conzelmann C., Müller J.A., Groß R., Hirschenberger M., Seidel A., Klute S., Zech F., Prelli Bozzo C., et al. Alpha-1 antitrypsin inhibits TMPRSS2 protease activity and SARS-CoV-2 infection. Nat Commun. 2021;12 doi: 10.1038/s41467-021-21972-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann M., Hofmann-Winkler H., Smith J.C., Krüger N., Arora P., Sørensen L.K., Søgaard O.S., Hasselstrøm J.B., Winkler M., Hempel T., et al. Camostat mesylate inhibits SARS-CoV-2 activation by TMPRSS2-related proteases and its metabolite GBPA exerts antiviral activity. EBioMedicine. 2021;65 doi: 10.1016/j.ebiom.2021.103255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zang R., Castro M.F.G., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B., et al. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci Immunol. 2020;5 doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ou T., Mou H., Zhang L., Ojha A., Choe H., Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao M.-M., Yang W.-L., Yang F.-Y., Zhang L., Huang W.-J., Hou W., Fan C.-F., Jin R.-H., Feng Y.-M., Wang Y.-C., et al. Cathepsin L plays a key role in SARS-CoV-2 infection in humans and humanized mice and is a promising target for new drug development. Signal Transduct Targeted Ther. 2021;6:1–12. doi: 10.1038/s41392-021-00558-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bruchez A., Sha K., Johnson J., Chen L., Stefani C., McConnell H., Gaucherand L., Prins R., Matreyek K.A., Hume A.J., et al. MHC class II transactivator CIITA induces cell resistance to Ebola virus and SARS-like coronaviruses. Science. 2020;370:241–247. doi: 10.1126/science.abb3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci U S A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Daniloski Z., Jordan T.X., Wessels H.-H., Hoagland D.A., Kasela S., Legut M., Maniatis S., Mimitou E.P., Lu L., Geller E., et al. Identification of required host factors for SARS-CoV-2 infection in human cells. Cell. 2021;184:92–105.e16. doi: 10.1016/j.cell.2020.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]; One of the key papers to study the cell biology of SARS-CoV-2 using genome-scale CRISPR screens.
- 43.Zhu Y., Feng F., Hu G., Wang Y., Yu Y., Zhu Y., Xu W., Cai X., Sun Z., Han W., et al. A genome-wide CRISPR screen identifies host factors that regulate SARS-CoV-2 entry. Nat Commun. 2021;12 doi: 10.1038/s41467-021-21213-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffmann H.-H., Sánchez-Rivera F.J., Schneider W.M., Luna J.M., Soto-Feliciano Y.M., Ashbrook A.W., Le Pen J., Leal A.A., Ricardo-Lax I., Michailidis E., et al. Functional interrogation of a SARS-CoV-2 host protein interactome identifies unique and shared coronavirus host factors. Cell Host Microbe. 2021;29:267–280.e5. doi: 10.1016/j.chom.2020.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfaender S., Mar K.B., Michailidis E., Kratzel A., Boys I.N., V’kovski P., Fan W., Kelly J.N., Hirt D., Ebert N., et al. LY6E impairs coronavirus fusion and confers immune control of viral disease. Nat Microbiol. 2020;5:1330–1339. doi: 10.1038/s41564-020-0769-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao X., Zheng S., Chen D., Zheng M., Li X., Li G., Lin H., Chang J., Zeng H., Guo J.-T. LY6E restricts entry of human coronaviruses, including currently pandemic SARS-CoV-2. J Virol. 2020;94 doi: 10.1128/JVI.00562-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi G., Kenney A.D., Kudryashova E., Zani A., Zhang L., Lai K.K., Hall-Stoodley L., Robinson R.T., Kudryashov D.S., Compton A.A., et al. Opposing activities of IFITM proteins in SARS-CoV-2 infection. EMBO J. 2021;40 doi: 10.15252/embj.2020106501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buchrieser J., Dufloo J., Hubert M., Monel B., Planas D., Rajah M.M., Planchais C., Porrot F., Guivel-Benhassine F., Van der Werf S., et al. Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 2020;39 doi: 10.15252/embj.2020106267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zang R., Case J.B., Yutuc E., Ma X., Shen S., Castro M.F.G., Liu Z., Zeng Q., Zhao H., Son J., et al. Cholesterol 25-hydroxylase suppresses SARS-CoV-2 replication by blocking membrane fusion. PNAS. 2020;117:32105–32113. doi: 10.1073/pnas.2012197117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang S., Li W., Hui H., Tiwari S.K., Zhang Q., Croker B.A., Rawlings S., Smith D., Carlin A.F., Rana T.M. Cholesterol 25-hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020;39 doi: 10.15252/embj.2020106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Winstone H., Lista M.J., Reid A.C., Bouton C., Pickering S., Galao R.P., Kerridge C., Doores K.J., Swanson C.M., Neil S.J.D. The polybasic cleavage site in SARS-CoV-2 spike modulates viral sensitivity to type I interferon and IFITM2. J Virol. 2021;95 doi: 10.1128/JVI.02422-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diamond M.S., Farzan M. The broad-spectrum antiviral functions of IFIT and IFITM proteins. Nat Rev Immunol. 2013;13:46–57. doi: 10.1038/nri3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53••.V’kovski P., Kratzel A., Steiner S., Stalder H., Thiel V. Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol. 2021;19:155–170. doi: 10.1038/s41579-020-00468-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review about the biology of SARS-CoV-2.
- 54.Finkel Y., Gluck A., Nachshon A., Winkler R., Fisher T., Rozman B., Mizrahi O., Lubelsky Y., Zuckerman B., Slobodin B., et al. SARS-CoV-2 uses a multipronged strategy to impede host protein synthesis. Nature. 2021;594:240–245. doi: 10.1038/s41586-021-03610-3. [DOI] [PubMed] [Google Scholar]
- 55.Irigoyen N., Firth A.E., Jones J.D., Chung B.Y.-W., Siddell S.G., Brierley I. High-resolution analysis of coronavirus gene expression by RNA sequencing and ribosome profiling. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Münch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim D., Lee J.-Y., Yang J.-S., Kim J.W., Kim V.N., Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. 2020;181:914–921.e10. doi: 10.1016/j.cell.2020.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58••.Gordon D.E., Jang G.M., Bouhaddou M., Xu J., Obernier K., White K.M., O’Meara M.J., Rezelj V.V., Guo J.Z., Swaney D.L., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first proteome-wide study of protein–protein interactions between SARS-CoV-2 and human cells.
- 59.White K.M., Rosales R., Yildiz S., Kehrer T., Miorin L., Moreno E., Jangra S., Uccellini M.B., Rathnasinghe R., Coughlan L., et al. Plitidepsin has potent preclinical efficacy against SARS-CoV-2 by targeting the host protein eEF1A. Science. 2021;371:926–931. doi: 10.1126/science.abf4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Klann K., Bojkova D., Tascher G., Ciesek S., Münch C., Cinatl J. Growth factor receptor signaling inhibition prevents SARS-CoV-2 replication. Mol Cell. 2020;80:164–174.e4. doi: 10.1016/j.molcel.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lei J., Ma-Lauer Y., Han Y., Thoms M., Buschauer R., Jores J., Thiel V., Beckmann R., Deng W., Leonhardt H., et al. The SARS-unique domain (SUD) of SARS-CoV and SARS-CoV-2 interacts with human Paip1 to enhance viral RNA translation. EMBO J. 2021;40:e102277. doi: 10.15252/embj.2019102277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.-A., Leibundgut M., Thiel V., Mühlemann O., Ban N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 63.Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yuan S., Peng L., Park J.J., Hu Y., Devarkar S.C., Dong M.B., Shen Q., Wu S., Chen S., Lomakin I.B., et al. Nonstructural protein 1 of SARS-CoV-2 is a potent pathogenicity factor redirecting host protein synthesis machinery toward viral RNA. Mol Cell. 2020;80:1055–1066.e6. doi: 10.1016/j.molcel.2020.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lapointe C.P., Grosely R., Johnson A.G., Wang J., Fernández I.S., Puglisi J.D. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. PNAS. 2021;118 doi: 10.1073/pnas.2017715118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tidu A., Janvier A., Schaeffer L., Sosnowski P., Kuhn L., Hammann P., Westhof E., Eriani G., Martin F. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA. 2021;27:253–264. doi: 10.1261/rna.078121.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., et al. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339.e21. doi: 10.1016/j.cell.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang K., Miorin L., Makio T., Dehghan I., Gao S., Xie Y., Zhong H., Esparza M., Kehrer T., Kumar A., et al. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci Adv. 2021;7 doi: 10.1126/sciadv.abe7386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Addetia A., Lieberman N.A.P., Phung Q., Hsiang T.-Y., Xie H., Roychoudhury P., Shrestha L., Loprieno M.A., Huang M.-L., Gale M., et al. SARS-CoV-2 ORF6 disrupts bidirectional nucleocytoplasmic transport through interactions with Rae1 and Nup98. mBio. 2021;12 doi: 10.1128/mBio.00065-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Miorin L., Kehrer T., Sanchez-Aparicio M.T., Zhang K., Cohen P., Patel R.S., Cupic A., Makio T., Mei M., Moreno E., et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. PNAS. 2020;117:28344–28354. doi: 10.1073/pnas.2016650117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang Q., Hughes T.A., Kelkar A., Yu X., Cheng K., Park S., Huang W.-C., Lovell J.F., Neelamegham S. Inhibition of SARS-CoV-2 viral entry upon blocking N- and O-glycan elaboration. eLife. 2020;9 doi: 10.7554/eLife.61552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhao P., Praissman J.L., Grant O.C., Cai Y., Xiao T., Rosenbalm K.E., Aoki K., Kellman B.P., Bridger R., Barouch D.H., et al. Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe. 2020;28:586–601.e6. doi: 10.1016/j.chom.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Correa Marrero M., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., et al. The global phosphorylation landscape of SARS-CoV-2 infection. Cell. 2020;182:685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang H., Zheng H., Zhu J., Dong Q., Wang J., Fan H., Chen Y., Zhang X., Han X., Li Q., et al. Ubiquitin-modified proteome of SARS-CoV-2-infected host cells reveals insights into virus–host interaction and pathogenesis. J Proteome Res. 2021;20:2224–2239. doi: 10.1021/acs.jproteome.0c00758. [DOI] [PubMed] [Google Scholar]
- 76.Novelli G., Liu J., Biancolella M., Alonzi T., Novelli A., Patten J.J., Cocciadiferro D., Agolini E., Colona V.L., Rizzacasa B., et al. Inhibition of HECT E3 ligases as potential therapy for COVID-19. Cell Death Dis. 2021;12:1–18. doi: 10.1038/s41419-021-03513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Xia H., Cao Z., Xie X., Zhang X., Chen J.Y.-C., Wang H., Menachery V.D., Rajsbaum R., Shi P.-Y. Evasion of type I interferon by SARS-CoV-2. Cell Rep. 2020;33 doi: 10.1016/j.celrep.2020.108234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cai T., Yu Z., Wang Z., Liang C., Richard S. Arginine methylation of SARS-Cov-2 nucleocapsid protein regulates RNA binding, its ability to suppress stress granule formation, and viral replication. J Biol Chem. 2021;297:100821. doi: 10.1016/j.jbc.2021.100821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Klein S., Cortese M., Winter S.L., Wachsmuth-Melm M., Neufeldt C.J., Cerikan B., Stanifer M.L., Boulant S., Bartenschlager R., Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun. 2020;11 doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cortese M., Lee J.-Y., Cerikan B., Neufeldt C.J., Oorschot V.M.J., Köhrer S., Hennies J., Schieber N.L., Ronchi P., Mizzon G., et al. Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe. 2020;28:853–866.e5. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wolff G., Limpens R.W.A.L., Zevenhoven-Dobbe J.C., Laugks U., Zheng S., de Jong A.W.M., Koning R.I., Agard D.A., Grünewald K., Koster A.J., et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395–1398. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dickson A.M., Wilusz J. Strategies for viral RNA stability: live long and prosper. Trends Genet. 2011;27:286–293. doi: 10.1016/j.tig.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hernandez-Gonzalez M., Larocque G., Way M. Viral use and subversion of membrane organization and trafficking. J Cell Sci. 2021;134 doi: 10.1242/jcs.252676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yan L., Zhang Y., Ge J., Zheng L., Gao Y., Wang T., Jia Z., Wang H., Huang Y., Li M., et al. Architecture of a SARS-CoV-2 mini replication and transcription complex. Nat Commun. 2020;11 doi: 10.1038/s41467-020-19770-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.V’kovski P., Gerber M., Kelly J., Pfaender S., Ebert N., Braga Lagache S., Simillion C., Portmann J., Stalder H., Gaschen V., et al. Determination of host proteins composing the microenvironment of coronavirus replicase complexes by proximity-labeling. eLife. 2019;8 doi: 10.7554/eLife.42037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86•.Flynn R.A., Belk J.A., Qi Y., Yasumoto Y., Wei J., Alfajaro M.M., Shi Q., Mumbach M.R., Limaye A., DeWeirdt P.C., et al. Discovery and functional interrogation of SARS-CoV-2 RNA-host protein interactions. Cell. 2021;184:2394–2411.e16. doi: 10.1016/j.cell.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper [86•] comprehensively identified host proteins that interact with SARS-CoV-2 RNAs.
- 87.Sun L., Li P., Ju X., Rao J., Huang W., Ren L., Zhang S., Xiong T., Xu K., Zhou X., et al. In vivo structural characterization of the SARS-CoV-2 RNA genome identifies host proteins vulnerable to repurposed drugs. Cell. 2021;184:1865–1883.e20. doi: 10.1016/j.cell.2021.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88•.Lee S., Lee Y.S., Choi Y., Son A., Park Y., Lee K.M., Kim J., Kim J.S., Kim V.N. The SARS-CoV-2 RNA interactome. Mol Cell. 2021;8:2838–2850.e6. doi: 10.1016/j.molcel.2021.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper [88•] comprehensively identified host proteins that interact with SARS-CoV-2 RNAs.
- 89•.Schmidt N., Lareau C.A., Keshishian H., Ganskih S., Schneider C., Hennig T., Melanson R., Werner S., Wei Y., Zimmer M., et al. The SARS-CoV-2 RNA–protein interactome in infected human cells. Nat Microbiol. 2021;6:339–353. doi: 10.1038/s41564-020-00846-z. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper [89•] comprehensively identified host proteins that interact with SARS-CoV-2 RNAs.
- 90.Nardacci R., Colavita F., Castilletti C., Lapa D., Matusali G., Meschi S., Del Nonno F., Colombo D., Capobianchi M.R., Zumla A., et al. Evidences for lipid involvement in SARS-CoV-2 cytopathogenesis. Cell Death Dis. 2021;12:263. doi: 10.1038/s41419-021-03527-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dias S.S.G., Soares V.C., Ferreira A.C., Sacramento C.Q., Fintelman-Rodrigues N., Temerozo J.R., Teixeira L., Nunes da Silva M.A., Barreto E., Mattos M., et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Thomas T., Stefanoni D., Dzieciatkowska M., Issaian A., Nemkov T., Hill R.C., Francis R.O., Hudson K.E., Buehler P.W., Zimring J.C., et al. Evidence of structural protein damage and membrane lipid remodeling in red blood cells from COVID-19 patients. J Proteome Res. 2020;19:4455–4469. doi: 10.1021/acs.jproteome.0c00606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Miller K., McGrath M.E., Hu Z., Ariannejad S., Weston S., Frieman M., Jackson W.T. Coronavirus interactions with the cellular autophagy machinery. Autophagy. 2020;16:2131–2139. doi: 10.1080/15548627.2020.1817280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Miao G., Zhao H., Li Y., Ji M., Chen Y., Shi Y., Bi Y., Wang P., Zhang H. ORF3a of the COVID-19 virus SARS-CoV-2 blocks HOPS complex-mediated assembly of the SNARE complex required for autolysosome formation. Dev Cell. 2021;56:427–442.e5. doi: 10.1016/j.devcel.2020.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang Y., Sun H., Pei R., Mao B., Zhao Z., Li H., Lin Y., Lu K. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 2021;7:1–12. doi: 10.1038/s41421-021-00268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moretti F., Bergman P., Dodgson S., Marcellin D., Claerr I., Goodwin J.M., DeJesus R., Kang Z., Antczak C., Begue D., et al. TMEM41B is a novel regulator of autophagy and lipid mobilization. EMBO Rep. 2018;19 doi: 10.15252/embr.201845889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Morita K., Hama Y., Izume T., Tamura N., Ueno T., Yamashita Y., Sakamaki Y., Mimura K., Morishita H., Shihoya W., et al. Genome-wide CRISPR screen identifies TMEM41B as a gene required for autophagosome formation. J Cell Biol. 2018;217:3817–3828. doi: 10.1083/jcb.201804132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Trimarco J.D., Heaton B.E., Chaparian R.R., Burke K.N., Binder R.A., Gray G.C., Smith C.M., Menachery V.D., Heaton N.S. TMEM41B is a host factor required for the replication of diverse coronaviruses including SARS-CoV-2. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang D., Xu B., Liu L., Wu L., Zhu Y., Ghanbarpour A., Wang Y., Chen F.J., Lyu J., Hu Y., et al. TMEM41B acts as an ER scramblase required for lipoprotein biogenesis and lipid homeostasis. Cell Metab. 2021;33:1655–1670.e8. doi: 10.1016/j.cmet.2021.05.006. [DOI] [PubMed] [Google Scholar]
- 100.Li Y.E., Wang Y., Du X., Zhang T., Mak H.Y., Hancock S.E., McEwen H., Pandzic E., Whan R.M., Aw Y.C., et al. TMEM41B and VMP1 are scramblases and regulate the distribution of cholesterol and phosphatidylserine. J Cell Biol. 2021;220 doi: 10.1083/jcb.202103105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang Y., Guo R., Kim S.H., Shah H., Zhang S., Liang J.H., Fang Y., Gentili M., Leary C.N.O., Elledge S.J., et al. SARS-CoV-2 hijacks folate and one-carbon metabolism for viral replication. Nat Commun. 2021;12 doi: 10.1038/s41467-021-21903-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Unchwaniwala N., Ahlquist P. Coronavirus dons a new crown. Science. 2020;369:1306–1307. doi: 10.1126/science.abe0322. [DOI] [PubMed] [Google Scholar]
- 103.Iserman C., Roden C.A., Boerneke M.A., Sealfon R.S.G., McLaughlin G.A., Jungreis I., Fritch E.J., Hou Y.J., Ekena J., Weidmann C.A., et al. Genomic RNA elements drive phase separation of the SARS-CoV-2 nucleocapsid. Mol Cell. 2020;80:1078–1091.e6. doi: 10.1016/j.molcel.2020.11.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ziv O., Price J., Shalamova L., Kamenova T., Goodfellow I., Weber F., Miska E.A. The short- and long-range RNA-RNA interactome of SARS-CoV-2. Mol Cell. 2020;80:1067–1077.e5. doi: 10.1016/j.molcel.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Carlson C.R., Asfaha J.B., Ghent C.M., Howard C.J., Hartooni N., Safari M., Frankel A.D., Morgan D.O. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. Mol Cell. 2020;80:1092–1103.e4. doi: 10.1016/j.molcel.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lu S., Ye Q., Singh D., Cao Y., Diedrich J.K., Yates J.R., Villa E., Cleveland D.W., Corbett K.D. The SARS-CoV-2 nucleocapsid phosphoprotein forms mutually exclusive condensates with RNA and the membrane-associated M protein. Nat Commun. 2021;12 doi: 10.1038/s41467-020-20768-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen Y.-J., Bagchi P., Tsai B. ER functions are exploited by viruses to support distinct stages of their life cycle. Biochem Soc Trans. 2020;48:2173–2184. doi: 10.1042/BST20200395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108••.Ghosh S., Dellibovi-Ragheb T.A., Kerviel A., Pak E., Qiu Q., Fisher M., Takvorian P.M., Bleck C., Hsu V.W., Fehr A.R., et al. β-coronaviruses use lysosomes for egress instead of the biosynthetic secretory pathway. Cell. 2020;183:1520–1535.e14. doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first paper showing betacoronaviruses exit host cells via lysosomal exocytosis.
- 109.Feng T., Lacrampe A., Hu F. Physiological and pathological functions of TMEM106B: a gene associated with brain aging and multiple brain disorders. Acta Neuropathol. 2021;141:327–339. doi: 10.1007/s00401-020-02246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sauter D. Counteraction of the multifunctional restriction factor tetherin. Front Microbiol. 2014;5:163. doi: 10.3389/fmicb.2014.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111•.Martin-Sancho L., Lewinski M.K., Pache L., Stoneham C.A., Yin X., Becker M.E., Pratt D., Churas C., Rosenthal S.B., Liu S., et al. Functional landscape of SARS-CoV-2 cellular restriction. Mol Cell. 2021;81:2656–2668.e8. doi: 10.1016/j.molcel.2021.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]; A key paper that systematically identified host restriction factors for SARS-CoV-2.
- 112.Neil S.J.D., Zang T., Bieniasz P.D. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature. 2008;451:425–430. doi: 10.1038/nature06553. [DOI] [PubMed] [Google Scholar]
- 113.Bussani R., Schneider E., Zentilin L., Collesi C., Ali H., Braga L., Volpe M.C., Colliva A., Zanconati F., Berlot G., et al. Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine. 2020;61 doi: 10.1016/j.ebiom.2020.103104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114•.Braga L., Ali H., Secco I., Chiavacci E., Neves G., Goldhill D., Penn R., Jimenez-Guardeño J.M., Ortega-Prieto A.M., Bussani R., et al. Drugs that inhibit TMEM16 proteins block SARS-CoV-2 spike-induced syncytia. Nature. 2021;594:88–93. doi: 10.1038/s41586-021-03491-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; A key paper investigating the mechanism of syncytia formation in SARS-CoV-2 infected cells.
- 115.Papa G., Mallery D.L., Albecka A., Welch L.G., Cattin-Ortolá J., Luptak J., Paul D., McMahon H.T., Goodfellow I.G., Carter A., et al. Furin cleavage of SARS-CoV-2 Spike promotes but is not essential for infection and cell-cell fusion. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dittmar M., Lee J.S., Whig K., Segrist E., Li M., Kamalia B., Castellana L., Ayyanathan K., Cardenas-Diaz F.L., Morrisey E.E., et al. Drug repurposing screens reveal cell-type-specific entry pathways and FDA-approved drugs active against SARS-Cov-2. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.108959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117•.Riva L., Yuan S., Yin X., Martin-Sancho L., Matsunaga N., Pache L., Burgstaller-Muehlbacher S., De Jesus P.D., Teriete P., Hull M.V., et al. Discovery of SARS-CoV-2 antiviral drugs through large-scale compound repurposing. Nature. 2020;586:113–119. doi: 10.1038/s41586-020-2577-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive drug repurposing studies with validations in multiple cell lines and tissue model.
- 118.Kang Y.-L., Chou Y., Rothlauf P.W., Liu Z., Soh T.K., Cureton D., Case J.B., Chen R.E., Diamond M.S., Whelan S.P.J., et al. Inhibition of PIKfyve kinase prevents infection by Zaire ebolavirus and SARS-CoV-2. PNAS. 2020;117:20803–20813. doi: 10.1073/pnas.2007837117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gunst J.D., Staerke N.B., Pahus M.H., Kristensen L.H., Bodilsen J., Lohse N., Dalgaard L.S., Brønnum D., Fröbert O., Hønge B., et al. Efficacy of the TMPRSS2 inhibitor camostat mesilate in patients hospitalized with Covid-19-a double-blind randomized controlled trial. EClinicalMedicine. 2021;35 doi: 10.1016/j.eclinm.2021.100849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Doi K., Ikeda M., Hayase N., Moriya K., Morimura N. Nafamostat mesylate treatment in combination with favipiravir for patients critically ill with Covid-19: a case series. Crit Care. 2020;24:1–4. doi: 10.1186/s13054-020-03078-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jang S., Rhee J.Y. Three cases of treatment with nafamostat in elderly patients with COVID-19 pneumonia who need oxygen therapy. Int J Infect Dis. 2020;96:500–502. doi: 10.1016/j.ijid.2020.05.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hoffmann M., Schroeder S., Kleine-Weber H., Müller M.A., Drosten C., Pöhlmann S. Nafamostat mesylate blocks activation of SARS-CoV-2: new treatment option for COVID-19. Antimicrob Agents Chemother. 2020;64 doi: 10.1128/AAC.00754-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Jeon S., Ko M., Lee J., Choi I., Byun S.Y., Park S., Shum D., Kim S. Identification of antiviral drug candidates against SARS-CoV-2 from FDA-approved drugs. Antimicrob Agents Chemother. 2020;64 doi: 10.1128/AAC.00819-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Masana L., Correig E., Rodríguez-Borjabad C., Anoro E., Arroyo J.A., Jericó C., Pedragosa A., la Miret M., Näf S., Pardo A., et al. Effect of statin therapy on SARS-CoV-2 infection-related mortality in hospitalized patients. Eur Heart J Cardiovasc Pharmacother. 2020 doi: 10.1093/ehjcvp/pvaa128. pvaa128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Tan W.Y.T., Young B.E., Lye D.C., Chew D.E.K., Dalan R. Statin use is associated with lower disease severity in COVID-19 infection. Sci Rep. 2020;10:17458. doi: 10.1038/s41598-020-74492-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rossi R., Talarico M., Coppi F., Boriani G. Protective role of statins in COVID 19 patients: importance of pharmacokinetic characteristics rather than intensity of action. Intern Emerg Med. 2020;15:1573–1576. doi: 10.1007/s11739-020-02504-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Zhang X.-J., Qin J.-J., Cheng X., Shen L., Zhao Y.-C., Yuan Y., Lei F., Chen M.-M., Yang H., Bai L., et al. In-hospital use of statins is associated with a reduced risk of mortality among individuals with COVID-19. Cell Metab. 2020;32:176–187.e4. doi: 10.1016/j.cmet.2020.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Thompson P.A., Eam B., Young N.P., Fish S., Chen J., Barrera M., Howard H., Sung E., Parra A., Staunton J., et al. Targeting oncogene mRNA translation in B-cell malignancies with eFT226, a potent and selective inhibitor of eIF4A. Mol Cancer Ther. 2021;20:26–36. doi: 10.1158/1535-7163.MCT-19-0973. [DOI] [PubMed] [Google Scholar]
- 129.Sauvat A., Ciccosanti F., Colavita F., Di Rienzo M., Castilletti C., Capobianchi M.R., Kepp O., Zitvogel L., Fimia G.M., Piacentini M., et al. On-target versus off-target effects of drugs inhibiting the replication of SARS-CoV-2. Cell Death Dis. 2020;11:656. doi: 10.1038/s41419-020-02842-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kaufmann S.H.E., Dorhoi A., Hotchkiss R.S., Bartenschlager R. Host-directed therapies for bacterial and viral infections. Nat Rev Drug Discov. 2018;17:35–56. doi: 10.1038/nrd.2017.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nyakatura E.K., Soare A.Y., Lai J.R. Bispecific antibodies for viral immunotherapy. Hum Vaccin Immunother. 2017;13:836–842. doi: 10.1080/21645515.2016.1251536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132••.Hoffmann M., Mösbauer K., Hofmann-Winkler H., Kaul A., Kleine-Weber H., Krüger N., Gassen N.C., Müller M.A., Drosten C., Pöhlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585:588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]; This paper was the first to address why Chloroquine and its derivatives are ineffective to block SARS-CoV-2 entry in human cells and limitations in SARS-CoV-2 studies using Vero cells due to their lack of TMPRSS2.
- 133.Liu Z., Zheng H., Lin H., Li M., Yuan R., Peng J., Xiong Q., Sun J., Li B., Wu J., et al. Identification of common deletions in the spike protein of severe acute respiratory syndrome coronavirus 2. J Virol. 2020;94 doi: 10.1128/JVI.00790-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Klimstra W.B., Tilston-Lunel N.L., Nambulli S., Boslett J., McMillen C.M., Gilliland T., Dunn M.D., Sun C., Wheeler S.E., Wells A., et al. SARS-CoV-2 growth, furin-cleavage-site adaptation and neutralization using serum from acutely infected hospitalized COVID-19 patients. J Gen Virol. 2020;101:1156–1169. doi: 10.1099/jgv.0.001481. [DOI] [PMC free article] [PubMed] [Google Scholar]