Abstract
A wide range of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) neutralizing monoclonal antibodies (mAbs) have been reported, most of which target the spike glycoprotein. Therapeutic implementation of these antibodies has been challenged by emerging SARS-CoV-2 variants harboring mutated spike versions. Consequently, re-assessment of previously identified mAbs is of high priority. Four previously selected mAbs targeting non-overlapping epitopes are now evaluated for binding potency to mutated RBD versions, reported to mediate escape from antibody neutralization. In vitro neutralization potencies of these mAbs, and two NTD-specific mAbs, are evaluated against two frequent SARS-CoV-2 variants of concern, the B.1.1.7 Alpha and the B.1.351 Beta. Furthermore, we demonstrate therapeutic potential of three selected mAbs by treatment of K18-human angiotensin-converting enzyme 2 (hACE2) transgenic mice 2 days post-infection with each virus variant. Thus, despite the accumulation of spike mutations, the highly potent MD65 and BL6 mAbs retain their ability to bind the prevalent viral mutants, effectively protecting against B.1.1.7 and B.1.351 variants.
Keywords: SARS-CoV-2, variants, mAbs, neutralizing antibodies, VOCs, escape mutants, K18-hACE2 mice
Graphical abstract
Novel SARS-CoV-2 antigenic variants jeopardize the efficacy of immunotherapies. Makdasi et al. re-evaluate anti-SARS-CoV-2 Abs previously shown to be highly effective against the original version of the virus. Some of the inspected antibodies retain their neutralization ability and in vivo protective efficacy against various viral variants.
Introduction
Unprecedented worldwide research and development efforts resulted in the rapid development of prophylactic and therapeutic immune tools to combat the coronavirus disease 2019 (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). These tools predominantly target the virus spike glycoprotein, which is essential for attachment of the virus to target cells, and hence plays an essential role in virus infectivity (Walls et al., 2020). Emergency-authorized vaccines against the SARS-CoV-2 spike are already used in mass vaccination campaigns (https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines) (Krammer, 2020). Additionally, passive immunity was investigated by the administration of convalescent plasma or recombinant neutralizing monoclonal antibodies (mAbs) (Alam et al., 2021; Weinreich et al., 2021; Wu et al., 2020b). Such countermeasure strategies rely on the concept that neutralization may involve interference with the entry of the virus to the host cells and its subsequent propagation. This therapeutic avenue accelerated the development of many potent neutralizing mAbs, primarily targeting the receptor binding domain (RBD) and the N-terminal domain (NTD) of the spike-S1 subunit (reviewed by Xiaojie et al., 2020). A single therapeutic mAb, generated by Eli Lilly and Company, and a dual-antibody combination, generated by Regeneron Pharmaceuticals, recently received emergency use authorization (Chen et al., 2021a; Weinreich et al., 2021).
Prior to its global expansion, SARS-CoV-2 was expected to exhibit relatively low mutation rates as compared with other RNA viruses, because its genome encodes a proofreading exoribonuclease (Robson et al., 2020). Nevertheless, the rapid global spread of the SARS-CoV-2, possibly combined with selective pressure for immune escape (Kemp et al., 2021), enabled emergence of new SARS-CoV-2 variants. Specifically, multiple mutations in the spike glycoprotein are evolving, including mutations residing in the antigenic supersite of the NTD (Cerutti et al., 2021; McCallum et al., 2021; Noy-Porat et al., 2021) or in the RBD (human angiotensin-converting enzyme 2 [hACE2] binding site; Baum et al., 2020; Chen et al., 2020; Noy-Porat et al., 2020), both representing major targets of potent virus-neutralizing antibodies.
The impact of accumulated mutations is closely monitored, yet only a minor fraction, which are selectively favorable, might spread and reach high frequency, and more importantly, become fixed in the population. Emergence of such genetic variants has important epidemiological consequences because they may exhibit increased transmissibility, reinfection of vaccinated or convalescent individuals, or increased disease severity. One of the major variants of concern (VOCs; https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/) identified and monitored is denoted 20I/501Y.V1 belonging to the B.1.1.7 lineage, labeled Alpha variant, which has a total of 18 nonsynonymous mutations relative to the original Wuhan strain. In this variant, 7 replacements and 2 deletions reside in the spike protein (see Figure S1 for schematic presentation; Rambaut et al., 2020b). Since its first emergence in the United Kingdom in September 2020 (Rambaut et al., 2020a), the B.1.1.7 variant is rapidly spreading globally. As of June 2021, it was detected in over 140 countries, with an apparent cumulative prevalence of 44% worldwide (for instance, 58%, 33%, and 68% in the United Kingdom, United States, and Israel, respectively) and a worldwide current prevalence of ∼75% (https://outbreak.info). Two additional VOCs were reported: B.1.351, labeled Beta variant (also known as 20H/501Y.V2; schematically depicted in Figure S1), identified for the first time in October 2020 in South Africa (Tegally et al., 2021); and P.1, labeled Gamma variant (also known as 501Y.V3), identified in December 2020 in Brazil (Faria et al., 2021). Both variants are less abundant worldwide (up to 2%) and mostly contained in the geographic surrounding of their originating site. The most recent variant determined by the World Health Organization (WHO) as VOC is B.1.617.2, labeled Delta variant, identified in India, with an apparent cumulative prevalence of 3% worldwide (as of June 2021), and recently also detected in Israel. This variant is associated with higher transmissibility (Saito et al., 2021) and potential immune escape (Planas et al., 2021; Yadav et al., 2021). The full biological and clinical implications of the new SARS-CoV-2 variants are yet to be determined. Nevertheless, the careful immunological assessment of known mutations, in particular in the RBD, is essential due to possible impact on vaccines and therapeutic countermeasures, such as mAbs. Of the multitude of possible genomic loci, mutations at several positions were already reported at relatively high frequency in the ∼2 × 106 sequences available to date (GISAID initiative, https://www.gisaid.org; Elbe and Buckland-Merrett, 2017). The most frequent mutation, N501Y, representing the hallmark of three circulating VOCs (B.1.1.7, B.1.351, and P.1), was first detected in February 2020, and as of June 2021 is present in over 70% of the global cases in more than 160 countries. The mutation S477N was reported in 43% of the cases worldwide since its emergence in February 2020. Its cumulative prevalence (percentage of sequences containing the variant to all sequences collected since the identification of this variant) is most prominent in Australia (56% as of June 2021). The mutation E484K was detected in more than 120 countries, exhibiting a worldwide cumulative prevalence of 6%. This mutation was detected in the B.1.351, P.1, and UK “B.1.1.7+E484K” variants. The N439K, a sentinel receptor binding motif mutation (Thomson et al., 2021), has an apparent worldwide cumulative prevalence of 2%, reported in at least 79 countries. This mutation has emerged in multiple SARS-CoV-2 clades and is mostly associated with the B.1.258 derivatives circulating in central Europe. The K417N mutation was reported in over 1% of the cases worldwide in at least 105 countries. This mutation represents one of the hallmarks of the B.1.351 and is exhibited in approximately 50% of South African cases. The replacement Y453F was detected in at least 15 countries, predominantly in Denmark. In late 2020, this mutation raised substantial concern when it was detected in a variant found in the mink population (Thomson et al., 2021).
Both predictive-theoretical and experimental approaches revealed that escape mutants can rapidly occur when SARS-CoV-2 is exposed to selective pressures mediated by neutralizing polyclonal sera or individual mAbs (Andreano et al., 2020; Liu et al., 2021; Starr et al., 2021a; Weisblum et al., 2020). Escape mutations within the RBD were predicted and experimentally confirmed to affect its function (mainly with respect to hACE2 binding) and recognition by mAbs. Substitutions N501Y, E484K, K417N, Y453F, N439K, and S477N were among the most frequent mutations mediating immune escape and were shown to reduce and even completely abrogate the neutralizing activity of several highly potent mAbs, including those that are already in clinical use (Andreano et al., 2020; Chen et al., 2021b; Liu et al., 2021; Starr et al., 2021a; Weisblum et al., 2020). Most of these substitutions naturally occurred in infected individuals and are now represented by emerging SARS-CoV-2 genetic variants, which spread worldwide.
We previously reported the isolation of RBD- and NTD-specific highly neutralizing mAbs (Barlev-Gross et al., 2021; Noy-Porat et al., 2020, 2021; Rosenfeld et al., 2021). In the current study, we present the re-evaluation of four SARS-CoV-2 neutralizing mAbs (MD65, MD62, MD29, and BL6), directed against four distinct epitopes in the spike RBD, for their ability to bind recombinant RBD (rRBD) variants that represent individual substitutions encountered in the VOCs. Additionally, we assessed the in vitro neutralization capacity of these four anti-RBD and two anti-NTD mAbs to counteract the SARS-CoV-2 B.1.1.7 and B.1.351 variants. Comparative structural modeling was conducted to determine the possible impact of mutations on the binding efficiency of the MD65 mAb. Finally, we evaluated the in vivo therapeutic potential of three selected mAbs by treatment of K18-hACE2 transgenic mice 2 days post-infection (dpi) with each of the virus strains.
Results and Discussion
Binding SARS-CoV-2 single-mutated RBD versions by specific mAbs
We re-evaluated the antigen binding of the recently reported mAbs, MD65, MD62, MD29, and BL6, targeting four distinct RBD epitopes (Noy-Porat et al., 2020) (Figure S2). Binding capability of these mAbs was tested with respect to six individual mutations in the SARS-CoV-2 spike rRBD, identified in circulating variants, including VOCs. These inspected mutations are N501Y, S477N, E484K, N439K, K417N, and Y453F (schematically described in Figure S3A; for details, see https://outbreak.info).
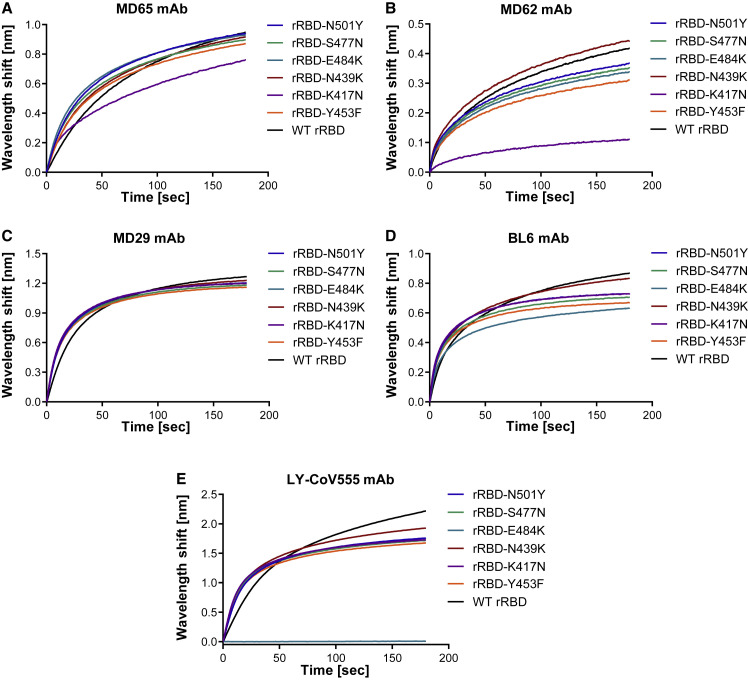
Biolayer interferometry (BLI) analysis was applied to evaluate the ability of the RBD-specific mAbs to bind the SARS-CoV-2 single-mutated RBD variants. The binding of these mAbs (Figures 1 A–1D) was only slightly affected (5%–22% loss of binding) by five of the six substitutions in the RBD. The only significant reduction in binding capacity, compared with the wild-type (WT) rRBD, was observed for the K417N mutant by the MD62 mAb (∼74% reduction) and to a lesser extent by the MD65 mAb (17% reduced binding). In light of these results, it is anticipated that these mAbs maintain their potency against variant strains carrying these mutations.
Figure 1.
mAb binding of singly mutated rRBDs
Binding of the WT and the six indicated singly mutated rRBDs by MD65 (A), MD62 (B), MD29 (C), BL6 (D), and LY-CoV555 (E) mAbs was evaluated by biolayer interferometry (BLI). Each mAb was immobilized on a protein A sensor and incubated with each of the rRBD mutants (N501Y, S477N, E484K, N439K, K417N, and Y453F; depicted in Figure S3A) or WT rRBD as a control for 180 s. The figure includes representative graphs of at least two independent repeats of each experiment, yielding similar results.
Comparison of binding to the SARS-CoV-2 spike protein by MD65 and the commercial licensed LY-CoV555 mAbs
Among the four RBD-specific mAbs studied here, MD65 is the most potent antibody in terms of in vitro neutralization. Additionally, in vivo studies demonstrated that MD65 effectively elicited post-exposure protection in mice at relatively low doses (Rosenfeld et al., 2021). MD65 (whose variable regions are encoded by the IGHV3-66 and IGKV3-20 germline heavy and light chain alleles, respectively), which belongs to a public clonotype (frequently encoded by IGHV3-53/3-66) that was extensively characterized in the context of SARS-CoV-2 neutralizing human antibodies (Barnes et al., 2020; Fagiani et al., 2020; Tan et al., 2021; Yuan et al., 2020), specifically targets the receptor binding motif, competing with hACE2 binding. Noteworthy, recent studies showed that binding and neutralization by antibodies belonging to this public clonotype are weakened by either the K417N or E484K replacements (Andreano et al., 2020; Yuan et al., 2021). Specifically, the E484K mutation, which is present in several SARS-CoV-2 natural isolates (including B.1.351, P.1 and “B.1.1.7+E484K” VOCs), was reported to be associated with lower susceptibility to neutralization by some mAbs, convalescent plasma, and sera collected from vaccinated individuals (Chen et al., 2021b; Wang et al., 2021a, 2021b).
The RBD-specific therapeutic mAb LY-CoV555 (bamlanivimab; Chen et al., 2021a), encoded by IGHV1-69 and IGKV1-39 germline alleles, was also shown to block hACE2 binding by SARS-CoV-2 (Jones et al., 2021) and, accordingly, possibly compete with MD65. However, although they may target close epitopes, their recognition pattern may differ. In order to test whether LY-CoV555 functionality is affected in a similar manner as MD65, the commercially available LY-CoV555 mAb was used in binding experiments. ELISA against SARS-CoV-2 spike protein revealed similar binding profiles for LY-CoV555 and MD65 (Figure S4A), and BLI epitope binning clearly indicated that LY-CoV555 and MD65 target overlapping epitopes (Figure S4B). The LY-CoV555 binding capacity toward the panel of RBD mutants was evaluated (Figure 1E), demonstrating equivalent binding of rRBD-N439K, Y453Y, S477N, and N501Y, compared with the WT rRBD. However, LY-CoV555 binding was completely obstructed by the E484K substitution. This observation is in line with recently reported studies suggesting that the E484K substitution is accountable for the abolishment of neutralization of SARS-CoV-2 natural variants that carry this mutation by LY-CoV555 mAb (Starr et al., 2021b; Wang et al., 2021a).
Structural modeling of anti-RBD antibodies
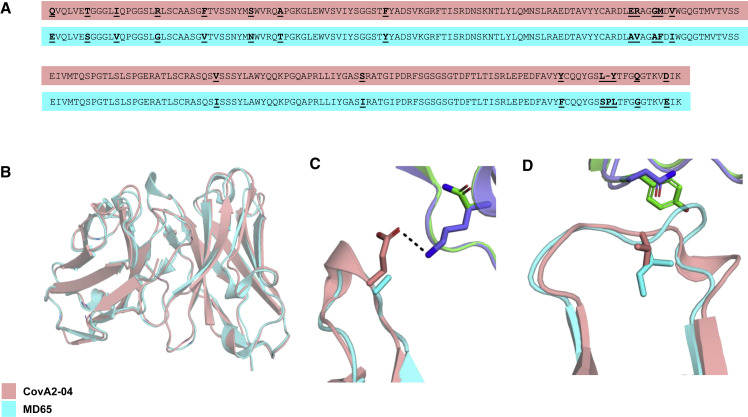
To define the molecular basis for the observed high cross-reactivity of MD65 mAb against all inspected mutant RBDs, we modeled its variable domain structure using AbPredict2 (Lapidoth et al., 2019; Figure 2 ). AbPredict2 uses Rosetta energy calculations as the sole criterion to predict a model structure on the basis of the variable domain sequence, ignoring sequence homology to existing antibody structures (Norn et al., 2017), producing energy-relaxed models. We noted that the recently solved structure of antibody CovA2-04 in complex with the RBD (PDB [Protein Data Bank]: 7jmo; Wu et al., 2020a) is very similar to the top-ranked AbPredict2 model of MD65 (Cα and carbonyl oxygen root mean square deviation < 1.0 Å). The two antibodies are also highly similar in their primary amino acid sequence (93% V gene sequence identity; Figure 2A), and both derive from the same heavy and light chain germline genes (IGHV3-53/66 and IGKV3-20, respectively), and hence are assigned to the same public clonotype. The differences between the two antibodies are mostly restricted to a diverged amino acid residue in L1, a deletion in L3, and different H3 sequences. The high sequence and structure similarity suggests that the bound state observed for CovA2-04 provides a reliable structural framework for analyzing MD65 binding to RBD variants. Therefore, the MD65 model structure was aligned to the CovA2-04 structure (Figure 2B) obtaining a model of interaction of MD65 with RBD.
Figure 2.
Structural basis of MD65 binding to COVID-19 spike variants determined by comparative modeling to the CovA2-04 mAb
(A) Alignment of the primary amino acid sequences of the heavy (two upper sequences) and light (two lower sequences) chain variable domains of the two antibodies compared (CovA2-04 in pink and MD65 in cyan). Diverged residues are indicated by bold and underlined letters.
(B–D) MD65 model structure (cyan) aligned with CovA2-04 crystal structure (pink; WT [PDB: 7jmo] and B.1.351 spike [PDB: 7nxa], violet and green, respectively). (B) A view of the superimposed variable domain models of the two mAbs, indicating the close correspondence of the MD65 model structure and the experimentally determined structure of CovA2-04. (C) The Lys residue at position 417 of the WT spike protein forms a stabilizing hydrogen bond (dashed line) with the Glu residue at position 100 of CovA2-04. The K417N present in B.1.351 abrogates this stabilizing interaction leading to potential strain in binding to the negative surface on the complementarity-determining region (CDR) H3 loop of CovA2-04. The Ala at the analogous position in MD65 may relieve this strain. (D) The small-to-large N501Y substitution on the B.1.351 spike may physically overlap with the CDR L1 of CovA2-04. The V29I difference in MD65 (compared with CovA2-04) modifies the CDR L1 backbone conformation, expanding the space in this region for the bulkier Tyr residue of the spike.
We initially focused on the K417N mutation located on the B.1.351 spike (Figure 2C). Despite the high sequence similarity of CovA2-04 and MD65, this mutation abrogates binding to the former while minimally perturbing binding to the latter (Yuan et al., 2021). Whereas complementarity-determining region (CDR) H3 in CovA2-04 presents a negatively charged side chain (Glu100; annotated as Glu97 according to Kabat numbering scheme) to counter the positive charge on the Lys at position 417 in RBD, the model shows that the H3 loop of MD65 is neutral. Thus, the K417N mutation may lead to electrostatic strain in binding the CovA2-04 antibody, but not in that of MD65. Negative charges are also observed in this position in other RBD-binding antibodies derived from the IGHV3-53/66 germline gene (Yuan et al., 2021), and interactions with these residues may be similarly affected by the K417N mutation.
Structural modeling may also provide an explanation for the ability of MD65 to neutralize RBD variants that exhibit the N501Y mutation (Figure 2D). RBD position 501 is proximate to the tip of CDR L1 of CovA2-04. The AbPredict2 model predicts that the CDR L1 adopts a different backbone conformation than CovA2-04 because of the CDR L1 mutation V29I (refers to position 28 according to the Kabat numbering scheme). In this altered backbone conformation, the CDR L1 of MD65 provides more space for the bulky Tyr at RBD position 501. Finally, the structure model suggests that RBD Glu at position 484 is distant from the interaction with MD65. Therefore, even radical mutations at this position would not impact antibody binding, as is indeed observed in the binding and neutralization experiments. Thus, it may be concluded that electrostatic strain at CDR H3 and steric hindrance in CDR L1 provide a likely mechanistic basis for understanding the differential effects of RBD mutations on binding affinity and neutralization in this class of antibodies. It remains to be seen whether this explanation extends to additional antibodies belonging to this class and other spike variants that may emerge in the future.
Binding SARS-CoV-2 multiply mutated S1 versions by specific mAbs
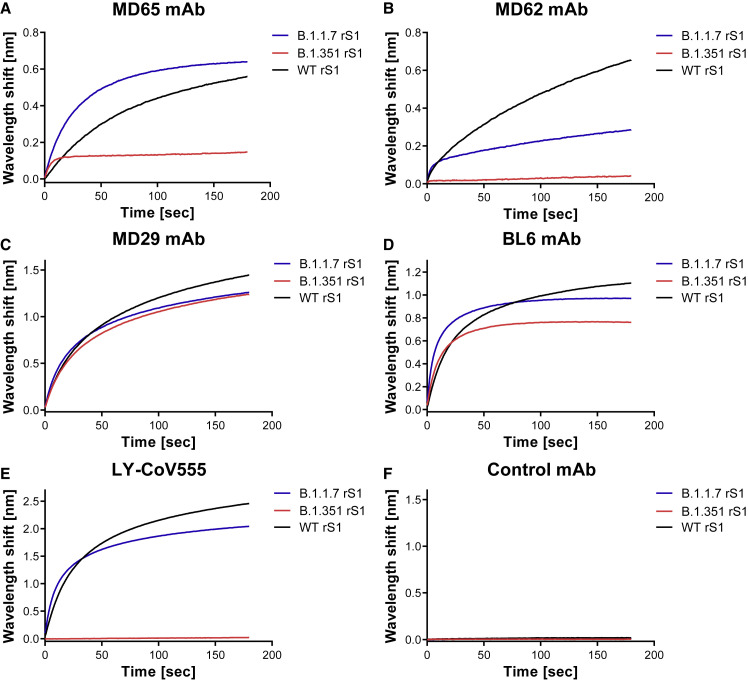
The retained binding capabilities, observed for the tested mAbs toward the individual RBD mutations, may not necessarily predict their interaction in the context of multiple mutations present in emerged VOCs. Therefore, we studied the ability of the four RBD-specific mAbs to bind recombinant mutated spike S1 subunit proteins, representing the RBD accumulated mutations associated with the B.1.1.7 and B.1.351 genetic variants (schematically depicted in Figures S3B and S3C, respectively). BLI analyses (Figure 3 ) were applied for binding evaluation of recombinant S1:Δ 69–70; Δ 144; N501Y; A570D; D614G; P681H, representing all the modifications encountered in the S1 of the B.1.1.7 genetic variant (Figure 3, B.1.1.7 rS1, blue curves) and of recombinant S1:K417N; E484K; N501Y; D614G, representing the RBD-related substitutions of B.1.351 (B.1.351 rS1; Figure 3, red curves). Recombinant WT S1 subunit (WT rS1; Figure 3, black curves) was used for comparison. Although binding of B.1.1.7 rS1 by MD65, MD29, BL6, and LY-CoV555 was not impaired by the mutations (Figures 3A and 3C–3E, respectively), MD62 binding was reduced by ∼50% (Figure 3B). This observation may not be attributed to the minor reduction (8% loss of binding) observed in binding of N501Y-RBD mutant by this antibody (Figure 1B) because it represents the only substitution in the RBD of B.1.1.7. Therefore, it can be speculated that structural changes in the B.1.1.7 rS1 involving the mutated NTD allosterically affected MD62 binding.
Figure 3.
Binding of rS1 variants by RBD-specific mAbs
(A–E) BLI was applied to evaluate the ability of each tested mAb to bind the indicated recombinant multiply mutated spike S1 subunit proteins: B.1.1.7 rS1 (Δ 69–70; Δ 144; N501Y; A570D; D614G; P681H; Figure S3B) and B.1.351 rS1 (K417N; E484K; N501Y; D614G; Figure S3C). Each of the indicated mAbs (A–E) was immobilized on protein A sensor and incubated for 180 s with B.1.1.7 (blue) or B.1.351 (red) rS1 variants or with the WT rS1 (black).
(F) Non-specific control antibody (anti-ricin MH75) was included.
The figure includes representative graphs of at least two independent repeats of each experiment, yielding similar results.
The B.1.351 rS1 includes the N501Y substitution, as well as the K417N and E484K replacements that were previously shown to substantially impair binding by various mAbs (Chen et al., 2021b; Cheng et al., 2021; Starr et al., 2021b; Wang et al., 2021b). Thus, in line with the individual mutation binding results (see Figure 1E for LY-CoV555 and Figure 1D for BL6), the observed binding abrogation of the B.1.351 rS1 by LY-CoV555 mAb (Figure 3E) and the mild (18%) reduction observed for BL6 (Figure 3D) can be attributed mainly to the E484K substitution. Similarly, the complete loss of binding by MD62 (Figure 3B) and significant loss of binding by MD65 (∼65%; Figure 3A) may have been mediated by the K417N substitution (see Figures 1B and 1A for MD62 and MD65, respectively).
Antibody-mediated neutralization evaluated by a cell culture plaque reduction test
In order to conclusively determine the potential of the B.1.1.7 and B.1.351 SARS-CoV-2 genetic variants (see Figure S1 for their spike mutations) to escape immune neutralization, we evaluated the ability to countermeasure the B.1.1.7 and B.1.351 live variants of the four RBD-specific mAbs, LY-CoV555, and two additional mAbs, targeting separate epitopes of the NTD (BLN14 and BLN12; see also Figure S5 for data pertaining to these two anti-NTD antibodies to B.1.1.7 rS1; Noy-Porat et al., 2021). A plaque reduction neutralization test (PRNT) was applied in which each mAb was tested against either the B.1.1.7 or B.1.351 variants or the parental WT SARS-CoV-2 strain.
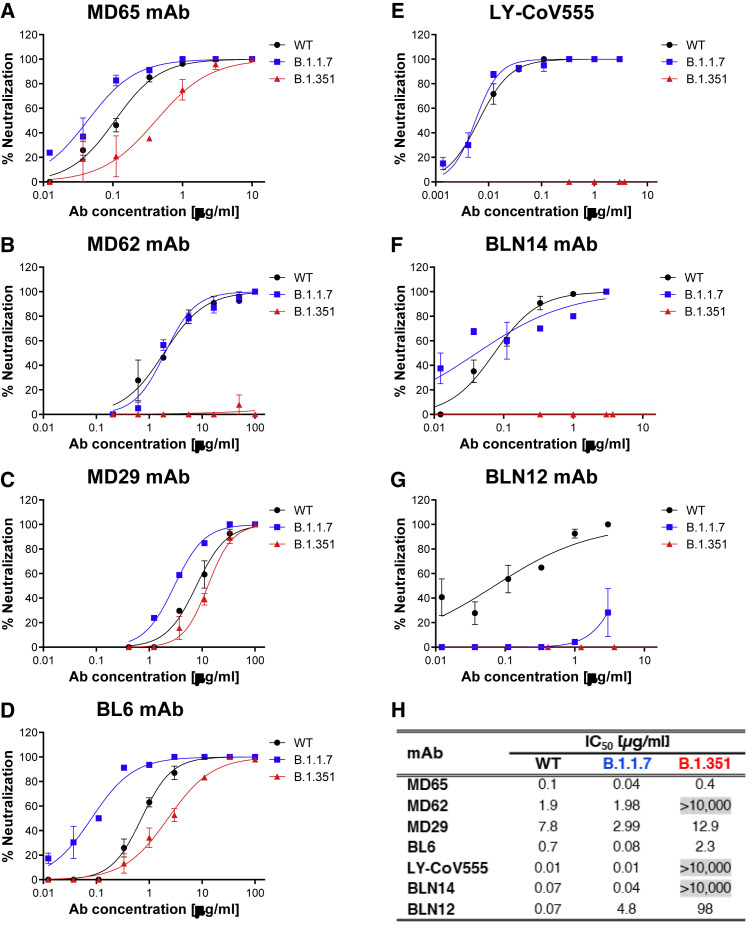
The results presented in Figure 4 indicated potent neutralization of the B.1.1.7 variant (blue curves) by anti-RBD MD65 (Figure 4A), MD62 (Figure 4B), MD29 (Figure 4C), BL6 (Figure 4D), as well as LY-CoV555 (Figure 4E), a neutralization that was not impaired compared with the WT SARS-CoV-2 strain (black curves). Interestingly, although MD62 revealed reduced binding to the B.1.1.7 rS1 compared with the WT rS1 (Figure 3B), its neutralization capacity of both viral strains was similar (Figure 4B). It can be concluded that all anti-RBD mAbs studied here fully retained their potency toward the B.1.1.7 variant.
Figure 4.
Neutralization of SARS-CoV-2 B.1.1.7 and B.1.351 by RBD and NTD-specific mAbs
(A–G) Neutralization capacity of the RBD-specific mAbs, MD65 (A), MD62 (B), MD29 (C), BL6 (D), and LY-CoV555 (E) and of the NTD-specific BLN14 (F) and BLN12 (G) was evaluated by plaque reduction neutralization test (PRNT). The in vitro neutralization of each of the listed mAbs was assessed against both SARS-CoV-2 B.1.1.7 (blue) and B.1.351 (red) variant, compared with WT SARS-CoV-2 strain (black). Neutralization potency was determined by the ability of each antibody (at indicated concentrations) to reduce plaque formation. Results are expressed as percent inhibition of control without Ab. Values along the curve depict averages of triplicates ± SEM. The figure includes a representative graph of at least two independent repeats of each experiment, yielding similar results.
(H) Summary of the calculated IC50 values (μg/ml). IC50 > 10,000 indicates complete loss of neutralization capacity, emphasized by gray shading.
The neutralization results, together with previously published biochemical data of the six inspected mAbs, are summarized in Table S1.
As could be anticipated, the B.1.351 variant (Figure 4, red curves) manifested a higher degree of immune escape than B.1.1.7. In line with the complete loss of binding of the respective viral rS1 by MD62 (Figure 3B) and LY-CoV555 (Figure 3E), the neutralization capacity of the two mAbs (MD62 and LY-CoV555) against the B.1.351 variant was completely abolished (Figures 4B and 4E, respectively). By contrast, MD65, MD29, and BL6 effectively neutralized the B.1.351 variant (Figures 4A, 4C, and 4D), albeit with a partial decrease in potency.
The two NTD-specific mAbs, BLN14 and BLN12, differed in their potency against the B.1.1.7 variant (Figures 4F and 4G, respectively), with BLN14 showing comparable neutralization to that of the WT, while BLN12 neutralization activity was markedly hampered. These results are consistent with binding experiments showing strong binding of B.1.1.7 rS1 by BLN14 and no binding by BLN12 (Figure S5). Epitope mapping previously revealed that BLN12 binds a linear epitope that resides between amino acids 141–155 and also recognizes an N-glycan at position 149 (Noy-Porat et al., 2021). It is therefore speculated that the deletion of a Tyr residue at position 144 in the B.1.1.7 variant is responsible for the loss of neutralization of this mAb. BLN14 recognizes a conformational epitope that apparently was not significantly altered by the Y144 deletion. However, the neutralization capability of both mAbs was dramatically reduced in the case of the B.1.351 variant, suggesting a considerable structural change at its NTD. This observation is in agreement with previous studies that indicated a frequent loss of functionality among NTD-specific mAbs (Andreano et al., 2020; Wang et al., 2021b), especially toward variants containing modifications in NTD supersite (Cerutti et al., 2021; McCallum et al., 2021) associated with significant structural alterations. The calculated half-maximum inhibitory concentration (IC50) values, characterizing the neutralization potency of each inspected antibody with respect to the three tested viral strains, are tabulated in Figure 4H.
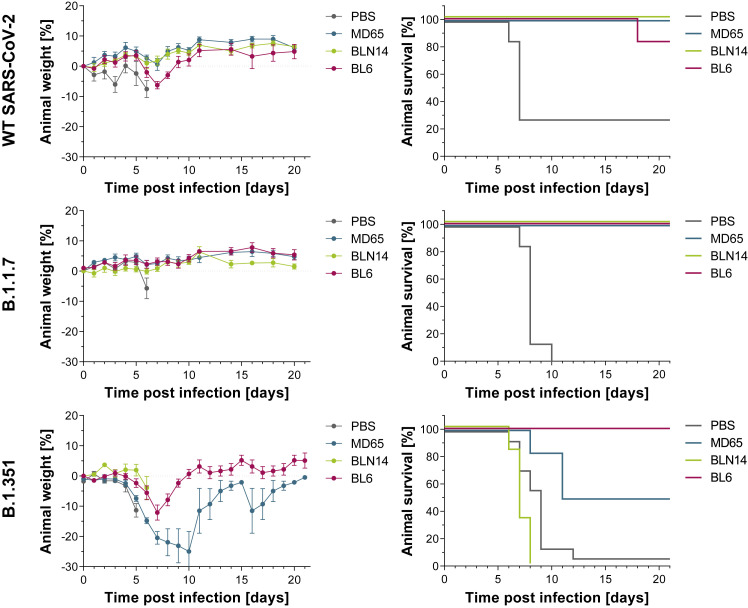
Antibody-mediated protection evaluated by post-exposure administration in a transgenic murine model of COVID-19
In line with the in vitro neutralization performance of the studied mAbs, MD65, BL6, and BLN14 mAbs were selected for further assessment of their therapeutic potential in vivo against various SARS-CoV-2 variants. Accordingly, K18-hACE2 transgenic mice were intranasally infected with a lethal dose of either one of the SARS-CoV-2 tested strains. Infection was characterized by body weight loss accompanied by high mortality (70%–100%, i.e., animals administered with PBS; Figure 5 , gray lines) within 6–12 days following infection. One milligram of each of the mAbs was administered intraperitoneally (i.p.) 2 dpi. Body weight and survival of experimental animals were monitored daily for 21 days. The data depicted in Figure 5 clearly demonstrate that antibody administration to infected animals resulted in in vivo protection against the three variants (for the effect of the MD65 and BLN14 antibodies against the WT version of SARS-CoV-2, see also Noy-Porat et al., 2021; Rosenfeld et al., 2021). Notably, the RBD-specific antibodies MD65 and BL6 exhibited therapeutic ability against all three viral variants inspected. Treatment with BL6 rescued 83%–100% of infected animals regardless of the infective strain. MD65 afforded complete protection against the WT and B.1.1.7 strains and 50% protection against the B.1.351 strain. It is important to note that a higher dose of this variant was administrated to the experimental animals (see Method details section). The anti-NTD antibody BLN14 demonstrated high potency against the WT and B.1.1.7 variant, yet post-exposure treatment with BLN14 mAb failed to rescue mice from infection with the B.1.351 strain. Taken together, these results are in good agreement with the in vitro BLI binding and PRNT data establishing that these in vitro tests may provide a reliable predictive means for evaluation of therapeutic antibodies.
Figure 5.
Post-exposure therapeutic potency of MD65, BL6, and BLN14 in K18-hACE2 mice infected with various SARS-CoV-2 strains
The SARS-CoV-2 strains are indicated in the left vertical lane and the administered antibodies in the legends within each individual panel (colored differently). A single dose of 1 mg Ab/animal of each indicated mAb (n = 6 per experimental group) or PBS for the control groups (n ≥ 7) was administered at day 2 after viral infection. Left panels: body weight profiles. Body weight is displayed as percentage change of initial weight (average ± SEM). Only data of the first 6 days are presented in the control groups exhibiting significant mortality. Right panels: Kaplan-Meier survival curves.
Conclusions
The unprecedented scale of the COVID-19 pandemic combined with selective pressure for escaping immune responses boosted the rapid evolution of SARS-CoV-2 virus, resulting in antigenic variability that might jeopardize the potency of pre- and post-exposure immunotherapies. Consequently, attention must be given to the development of mAb treatments that may combat emerging variants. In this perspective, it is of high importance to re-evaluate anti-SARS-CoV-2 mAbs previously shown to exhibit therapeutic potential against the original version of the virus. Furthermore, the impact of individual mutations on the neutralization potency of mAbs may provide important information impacting the preparedness for future anticipated antigenic drifts of the virus.
In the current report, we document the neutralization of the most abundant B.1.1.7 variant (as of today) by four anti-RBD mAbs and one anti-NTD mAb that we recently generated and determined their therapeutic potential against the original version of the virus. Furthermore, three RBD-specific mAbs (MD65, MD29, and BL6) retained neutralization against the B.1.351 VOC. These findings are supported by binding experiments conducted with individual and combined mutations derived from various variants. The binding and neutralizing data of the six investigated mAbs targeting distinct epitopes within S1 are summarized in Table S1. The E484K and K417N substitutions in the RBD were reported to mediate the lower susceptibility to neutralization by a significant proportion of reported mAbs, including clinically used LY-CoV555 and REGN10933, as well as by immune post-vaccination sera (Chen et al., 2021b; Cheng et al., 2021; Dejnirattisai et al., 2021; Hu et al., 2021; Rees-Spear et al., 2021; Starr et al., 2021b; Wang et al., 2021a, 2021b; Yuan et al., 2021). Of note, the anti-RBD antibody MD65, shown here to retain its neutralizing potential against emerging variants, was recently suggested by extensive pre-clinical studies to be an important therapeutic for efficient clinical intervention in COVID-19 cases (Rosenfeld et al., 2021). Structural modeling identified specific residues in the sequence of the MD65 mAb, which may explain the potency of this antibody against all the viral variants inspected. Finally, the binding and neutralizing data were confirmed by in vivo protection of infected transgenic mice administered with the various antibodies 2 dpi.
In conclusion, the present study substantiates the ability of recently reported mAbs to serve, individually or possibly in combination as a cocktail formulation, for designing efficient therapeutic approaches against emerging SARS-CoV-2 variants.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AP-conjugated Donkey anti-human IgG | Jackson ImmunoResearch | Cat#709-055-149, Lot: 130049; RRID:AB_2340501 |
| LY-CoV555 (Bamlanivimab) | Chen et al., 2021a | N/A |
| Bacterial and virus strains | ||
| SARS-CoV-2 | Bundeswehr Institute of Microbiology | GISAID accession EPI_ISL_406862 |
| SARS-CoV-2, isolate Human 2019-nCoV ex China strain BavPat1/2020 | Prof. Dr. Christian Drosten (through the European Virus Archive – Global) | EVAg Ref-SKU: 026V-03883 |
| SARS-CoV-2 B.1.1.7 (501Y.V1) variant | This paper | N/A |
| SARS-CoV-2 B.1.351 (501Y.V2) variant | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| SARS-CoV-2 spike (S) stabilized soluble ectodomain | Noy-Porat et al., 2020 | GenPept: QHD43416 ORF |
| Human Angiotensin-Converting Enzyme 2 (hACE2) | Rosenfeld et al., 2021 | GenPept: NP_068576.1 ORF |
| B.1.1.7 rS1-SARS-CoV-2 spike S1 [Δ 69-70; Δ 144; N501Y; A570D; D614G; P681H] | Sino Biologicals | Cat#40591-V08H12 |
| B.1.351 rS1-SARS-CoV-2 spike S1 [K417N; E484K; N501Y; D614G] | Sino Biologicals | Cat#40591-V08H10 |
| Spike RBD[N501Y] | Sino Biologicals | Cat#40592-V08H82 |
| Spike RBD[S477N] | Sino Biologicals | Cat#40592-V08H46 |
| Spike RBD[E484K] | Sino Biologicals | Cat#40592-V08H84 |
| Spike RBD[N439K] | Sino Biologicals | Cat#40592-V08H14 |
| Spike RBD[K417N] | Sino Biologicals | Cat#40592-V08H59 |
| Spike RBD[Y453F] | Sino Biologicals | Cat#40592-V08H80 |
| Experimental models: Cell lines | ||
| Vero E6 | ATCC | CRL-1586™ |
| ExpiCHO-S | Thermoscientific | Cat#A29127 |
| Experimental models: Organisms/strains | ||
| Mouse: B6.CgTg (K18hACE2)2Prlmn/J HEMI | Jackson Laboratories | JAX: 034860 |
| Recombinant DNA | ||
| pcDNA3.1+_YTE_Heavy/Light vectors | Rosenfeld et al., 2021 | N/A |
| Software and algorithms | ||
| Octet Data analysis software Version 8.1 | ForteBio | https://www.sartorius.com/en/products/protein-analysis/octet-systems-software |
| Prism Version 5 | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| AbPredict2 | Lapidoth et al., 2019 | http://AbPredict.weizmann.ac.il/bin/steps |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Ohad Mazor from the Israel Institute for Biological Research; ohadm@iibr.gov.il.
Materials availability
Antibodies are available for research purposes only under an MTA, which allows the use of the antibodies for non-commercial purposes but not their disclosure to third parties.
All other data are available from the Lead contact upon reasonable requests.
Experimental model and subject details
Animals
Female K18-hACE2 transgenic (B6.Cg-Tg (K18-hACE2)2Prlmn/J HEMI; Jackson Laboratories, USA) mice, age 8-16 weeks, were maintained at 20-22°C and a relative humidity of 50 ± 10% on a 12 hours light/dark cycle, fed with commercial rodent chow (Koffolk Inc.) and provided with tap water ad libitum. Treatment of animals was in accordance with regulations outlined in the U.S. Department of Agriculture (USDA) Animal Welfare Act and the conditions specified in the Guide for Care and Use of Laboratory Animals, National Institute of Health, 2011. Animal studies were approved by the local ethical committee on animal experiments (protocol number M-57-20).
Cells and virus strains
ExpiCHO-S (Thermoscientific, USA, Cat# A29127) were used for expression of recombinant proteins as described above.
Vero E6 (ATCC® CRL-1586™) were obtained from the American Type Culture Collection. Cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), MEM non-essential amino acids (NEAA), 2 mM L-glutamine, 100 Units/ml penicillin, 0.1 mg/ml streptomycin and 12.5 Units/ml Nystatin (P/S/N) (Biological Industries, Israel). Cells were cultured at 37°C, 5% CO2 at 95% air atmosphere.
Wild-type (WT) SARS-CoV-2 strain (GISAID accession EPI_ISL_406862) was kindly provided by Bundeswehr Institute of Microbiology, Munich, Germany.
WT SARS-CoV-2, isolate Human 2019-nCoV ex China strain BavPat1/2020, was kindly provided by Prof. Dr. Christian Drosten (Charité, Berlin, Germany) through the European Virus Archive – Global (EVAg Ref-SKU:026V-03883).
SARS-CoV-2 B.1.1.7 (501Y.V1) variant was isolated on Dec 2020 from a person who came back from the UK. The identity of the B.1.1.7 strain was confirmed using NGS.
SARS-CoV-2 B.1.351 (501Y.V2) variant was isolated on Jan 2021 from a person who was in contact with a patient who came back from South Africa. The identity of the B.1.351 strain was confirmed using NGS.
Stocks were prepared by infection of Vero E6 cells for two days. When viral cytopathic effect (CPE) was observed, media were collected, clarified by centrifugation, aliquoted and stored at −80°C. Titer of stock was determined by plaque assay using Vero E6 cells.
Handling and working with SARS-CoV-2 was conducted in BL3 facility in accordance with the biosafety guidelines of the IIBR.
Method details
Recombinant Proteins
The SARS-CoV-2 spike (S) stabilized soluble ectodomain, S1 subunit (WT rS1) and receptor binding domain (WT rRBD) were produced as previously described (Noy-Porat et al., 2020).
The following His-tagged recombinant proteins were purchased from Sino Biologicals: B.1.1.7 rS1-SARS-CoV-2 spike S1 [Δ 69-70; Δ 144; N501Y; A570D; D614G; P681H], cat#40591-V08H12; B.1.351 rS1-SARS-CoV-2 spike S1 [K417N; E484K; N501Y; D614G], cat#40591-V08H10; spike RBD[N501Y] cat#40592-V08H82; spike RBD[S477N] cat#40592-V08H46; spike RBD[E484K] cat#40592-V08H84; spike RBD[N439K] cat#40592-V08H14; spike RBD[K417N] cat#40592-V08H59; spike RBD[Y453F] cat#40592-V08H80.
All antibodies (except LY-CoV555) were produced as full IgG1 antibodies as described (Barlev-Gross et al., 2021; Rosenfeld et al., 2021), expressed using ExpiCHO™ Expression system (Thermoscientific, USA) and purified on HiTrap Protein-A column (GE healthcare, UK). The integrity and purity of the antibodies were analyzed using SDS-PAGE. Isolation and characterization of the MD29, MD65 and MD62 mAbs, targeting epitopes I-III on the RBD including their full amino acid sequences as previously reported (Noy-Porat et al., 2021). The BL6 mAb was isolated as described (Barlev-Gross et al., 2021) and is representing epitope IV on the RBD (competing with the MD47 mAb (Noy-Porat et al., 2020). For BL6 full amino acid sequence see Figure S2C. BLN12 and BLN14 mAbs, targeting two distinct epitopes on the NTD, isolation, characterization and amino acid sequence as previously reported (Noy-Porat et al., 2021).
LY-CoV555 (Bamlanivimab) (∼2.5 mg Ab/ml in 0.9% Sodium Cholride), was obtained as a remnant from an infusion bag and its set following administration to a COVID-19 patient at Kaplan Medical Center.
All antibodies were extensively dialyzed against PBS and filter-sterilized prior to any in vitro or in vivo experimentation.
ELISA
Direct ELISA (Noy-Porat et al., 2016) consisted of coating microtiter plates with 1 μg/ml of recombinant SARS-CoV-2 spike. ELISA was applied with AP-conjugated Donkey anti-human IgG (Jackson ImmunoResearch, USA, Cat# 709-055-149 lot 130049; used at 1:2000 working dilution) following detection using p-nitrophenyl phosphate (pNPP) substrate (Sigma, Israel).
Biolayer interferometry (BLI)
Binding studies were carried out using the Octet system (ForteBio, USA, Version 8.1, 2015) that measures biolayer interferometry (BLI). All steps were performed at 30°C with shaking at 1500 rpm in a black 96-well plate containing 200 μL solution in each well. For assessment of binding to S1 variants or mutated RBD, antibodies were captured on Protein-A or anti-Fab CH1 sensors (FAB2G) and incubated with recombinant S1 (WT, B.1.1.7 or B.1.351) or recombinant RBD (WT or mutated) at a constant concentration of 10 μg/ml for 180 s and then transferred to buffer containing wells for additional 60 s. Binding was measured as changes over time in light interference. Parallel measurements from unloaded biosensors were used as control. The anti-ricin MH75 mAb, used as isotype control (Figure 3F). For the comparison of the binding capacity of each tested mAb to a constant concentration of recombinant RBD or S1, the area under curve (AUC) was calculated for each binding curve, using GraphPad Prism 5, and percent binding was calculated compared to the WT protein, representing 100% binding.
For epitope binning, MD65 antibody was biotinylated, immobilized on streptavidin sensor, incubated with a fixed concentration of WT rS1 (20 μg/ml) to reach saturation, washed and incubated with non-labeled LY-CoV555 for 180 s. MD29 and MD65 were used as positive and negative controls, respectively.
Plaque reduction neutralization test (PRNT)
Plaque reduction neutralization test (PRNT), performed essentially as described (Makdasi et al., 2021). Vero E6 cells were seeded overnight at a density of 0.5e6 cells/well in 12-well plates. Antibody samples were 3-fold serially diluted (ranging from 200 to 0.002 μg/ml) in 400 μL of MEM supplemented with 2% FBS, MEM non-essential amino acids, 2 mM L-glutamine, 100 Units/ml penicilin, 0.1 mg/ml streptomycin and 12.5 Units/ml Nystatin (Biological Industries, Israel). 400 μL containing 300 PFU/ml of each SARS-CoV-2 strain, were then added to the mAb solution supplemented with 0.25% guinea pig complement sera (Sigma, Israel) and the mixture incubated at 37°C, 5% CO2 for 1 h. Two hundred μl of each mAb-virus mixture was added in duplicates to the cells for 1 h. Virus mixture w/o mAb was used as control. 2 mL overlay [supplemented MEM containing 0.4% tragacanth (Sigma, Israel)] were added to each well and plates were further incubated at 37°C, 5% CO2 for 48 h for WT and B.1.351 strains or 5 days for the B.1.1.7 strain. The number of plaques in each well was determined following media aspiration, cells fixation and staining with 1 mL of crystal violet (Biological Industries, Israel). Half-maximum inhibitory concentration (IC50) was defined as mAb concentration at which the plaque number was reduced by 50%, compared to plaque number of the control (in the absence of Ab).
Animal experiments
All animal experiments involving SARS-CoV-2 were conducted in a BSL3 facility.
Infection experiments were carried out using SARS-CoV-2 BavPat1/2020 (WT), B.1.1.7 and B.1.351 strains. SARS-CoV-2 virus diluted in PBS supplemented with 2% FBS (Biological Industries, Israel) was used to infect anesthetized mice by intranasal instillation. For mAbs protection evaluation, mice (n = 6) were treated intraperitoneally by single administration of 1 mL volume, containing 1 mg Ab/mouse, two days following infection with 500, 10 or 10,000 PFU of the WT, B.1.1.7 and B.1.351 SARS-CoV-2 strains, respectively. Control groups were administered with PBS (n = 7 for WT and B.1.1.7 and n = 14 for the B.1.351). Body weight was monitored daily throughout the follow-up period post infection.
Antibody structure prediction
Antibody structure prediction was done using AbPredict2 with default settings. RMSD calculation and alignments were done using Pymol. AbPredict2 is available for academic use at http://AbPredict.weizmann.ac.il/bin/steps.
Quantification and statistical analysis
All Biolayer Interferometry assays were analyzed using the Octet Data analysis software (ForteBio, Version 8.1) and visualized using GraphPad Prism 5.
ELISA results were analyzed using GraphPad Prism 5. Mean and SEM were calculated where appropriate and are presented in the relevant figures.
All the following statistical analyses were conducted using GraphPad Prism 5. For In vitro neutralization experiments, mean and SEM were calculated for each concentration, curves were fitted using nonlinear regression and IC50 values were extrapolated from the resulting curves. Results are presented in the relevant figures, in the results section and in Table S1.
Area under curve (AUC) was calculated for each binding curve presented in Figure 3. Results are presented in Table S1.
For all experiments, exact value and meaning of n are presented in the figure legends.
For antibody structure modeling, the amino acid sequence of the variable domain of MD65 was submitted to the AbPredict2 webserver which is available freely for non-commercial use (http://AbPredict.weizmann.ac.il/bin/steps).
Acknowledgments
We thank Prof. Dr. Christian Drosten at the Charité Universitätsmedizin, Institute of Virology, Berlin, Germany, for providing the SARS-CoV-2 BavPat1/2020 strain. We wish to express our gratitude to our colleagues Dr. Sharon Melamed, Boaz Politi, Dr. Liat Bar-On, Yfat Yahalom-Ronen, and Dr. Emanuelle Mamroud for fruitful discussions and support. The graphical abstract was created with BioRender.com. Research in the Fleishman lab was supported by the Weizmann Institute CoronaVirus Fund and by a charitable donation in memory of Sam Switzer.
Author contributions
E.M., A.Z., R.A., T.N.-P., E.P., A.M., Y.L., E.E., A.T., M.A., D.G., S.J.F., O.M., and R.R. designed, carried out, and analyzed the data. M.M., E.M., N.Z., I.N., and L.K. isolated, sequenced, and provided the SARS-CoV-2 variant strains. N.P., H.T., and T.I. cultured and prepared SARS-CoV-2 viruses for the neutralization experiments. O.Z. and S.W. provided crucial reagents. S.Y. and S.C.S. added fruitful discussions. R.R., O.M., A.Z., and T.C. wrote the manuscript. O.M. and R.R. supervised the project. All authors have reviewed and approved the final manuscript.
Declaration of interests
Patent application for the described antibodies was filed by the Israel Institute for Biological Research. None of the authors declared any additional competing interests.
Published: August 21, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109679.
Supplemental information
Data and code availability
-
•
The published paper includes all dataset generated or analyzed during this study.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
References
- Alam M.M., Mahmud S., Aggarwal S., Fathma S., Al Mahi N., Shibli M.S., Haque S.M., Mahmud S., Ahmed Z. Clinical Impact of the Early Use of Monoclonal Antibody LY-CoV555 (Bamlanivimab) on Mortality and Hospitalization Among Elderly Nursing Home Patients: A Multicenter Retrospective Study. Cureus. 2021;13:e14933. doi: 10.7759/cureus.14933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A., et al. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlev-Gross M., Weiss S., Ben-Shmuel A., Sittner A., Eden K., Mazuz N., Glinert I., Bar-David E., Puni R., Amit S., et al. Spike vs nucleocapsid SARS-CoV-2 antigen detection: application in nasopharyngeal swab specimens. Anal. Bioanal. Chem. 2021;413:3501–3510. doi: 10.1007/s00216-021-03298-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., West A.P., Jr., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842.e16. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Ajithdoss D., Copin R., Zhou A., Lanza K., Negron N., Ni M., Wei Y., Mohammadi K., Musser B., et al. REGN-COV2 antibodies prevent and treat SARS-CoV-2 infection in rhesus macaques and hamsters. Science. 2020;370:1110–1115. doi: 10.1126/science.abe2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti G., Guo Y., Zhou T., Gorman J., Lee M., Rapp M., Reddem E.R., Yu J., Bahna F., Bimela J., et al. Potent SARS-CoV-2 neutralizing antibodies directed against spike N-terminal domain target a single supersite. Cell Host Microbe. 2021;29:819–833.e7. doi: 10.1016/j.chom.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Li R., Pan Z., Qian C., Yang Y., You R., Zhao J., Liu P., Gao L., Li Z., et al. Human monoclonal antibodies block the binding of SARS-CoV-2 spike protein to angiotensin converting enzyme 2 receptor. Cell. Mol. Immunol. 2020;17:647–649. doi: 10.1038/s41423-020-0426-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., Huhn G., Cardona J., Mocherla B., Stosor V., et al. BLAZE-1 Investigators SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:229–237. doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., et al. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng M.H., Krieger J.M., Kaynak B., Arditi M., Bahar I. Impact of South African 501.V2 Variant on SARS-CoV-2 Spike Infectivity and Neutralization: A Structure-based Computational Assessment. bioRxiv. 2021 doi: 10.1101/2021.01.10.426143. [DOI] [Google Scholar]
- Dejnirattisai W., Zhou D., Supasa P., Liu C., Mentzer A.J., Ginn H.M., Zhao Y., Duyvesteyn H.M.E., Tuekprakhon A., Nutalai R., et al. Antibody evasion by the P.1 strain of SARS-CoV-2. Cell. 2021;184:2939–2954.e9. doi: 10.1016/j.cell.2021.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbe S., Buckland-Merrett G. Data, disease and diplomacy: GISAID’s innovative contribution to global health. Glob. Chall. 2017;1:33–46. doi: 10.1002/gch2.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagiani F., Catanzaro M., Lanni C. Molecular features of IGHV3-53-encoded antibodies elicited by SARS-CoV-2. Signal Transduct. Target. Ther. 2020;5:170. doi: 10.1038/s41392-020-00287-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faria N.R., Mellan T.A., Whittaker C., Claro I.M., Candido D.D.S., Mishra S., Crispim M.A.E., Sales F.C.S., Hawryluk I., McCrone J.T., et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science. 2021;372:815–821. doi: 10.1126/science.abh2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Peng P., Wang K., Fang L., Luo F.Y., Jin A.S., Liu B.Z., Tang N., Huang A.L. Emerging SARS-CoV-2 variants reduce neutralization sensitivity to convalescent sera and monoclonal antibodies. Cell. Mol. Immunol. 2021;18:1061–1063. doi: 10.1038/s41423-021-00648-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne J.L., Heinz B.A., Foster D., et al. The neutralizing antibody, LY-CoV555, protects against SARS-CoV-2 infection in nonhuman primates. Sci. Transl. Med. 2021;13:eabf1906. doi: 10.1126/scitranslmed.abf1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp S.A., Collier D.A., Datir R.P., Ferreira I.A.T.M., Gayed S., Jahun A., Hosmillo M., Rees-Spear C., Mlcochova P., Lumb I.U., et al. CITIID-NIHR BioResource COVID-19 Collaboration. COVID-19 Genomics UK (COG-UK) Consortium SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021;592:277–282. doi: 10.1038/s41586-021-03291-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Lapidoth G., Parker J., Prilusky J., Fleishman S.J. AbPredict 2: a server for accurate and unstrained structure prediction of antibody variable domains. Bioinformatics. 2019;35:1591–1593. doi: 10.1093/bioinformatics/bty822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J., et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makdasi E., Levy Y., Alcalay R., Noy-Porat T., Zahavy E., Mechaly A., Epstein E., Peretz E., Cohen H., Bar-On L., et al. Neutralizing Monoclonal Anti-SARS-CoV-2 Antibodies Isolated from Immunized Rabbits Define Novel Vulnerable Spike-Protein Epitope. Viruses. 2021;13:566. doi: 10.3390/v13040566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., et al. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347.e16. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norn C.H., Lapidoth G., Fleishman S.J. High-accuracy modeling of antibody structures by a search for minimum-energy recombination of backbone fragments. Proteins. 2017;85:30–38. doi: 10.1002/prot.25185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy-Porat T., Rosenfeld R., Ariel N., Epstein E., Alcalay R., Zvi A., Kronman C., Ordentlich A., Mazor O. Isolation of Anti-Ricin Protective Antibodies Exhibiting High Affinity from Immunized Non-Human Primates. Toxins (Basel) 2016;8:64. doi: 10.3390/toxins8030064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy-Porat T., Makdasi E., Alcalay R., Mechaly A., Levy Y., Bercovich-Kinori A., Zauberman A., Tamir H., Yahalom-Ronen Y., Israeli M., et al. A panel of human neutralizing mAbs targeting SARS-CoV-2 spike at multiple epitopes. Nat. Commun. 2020;11:4303. doi: 10.1038/s41467-020-18159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noy-Porat T., Mechaly A., Levy Y., Makdasi E., Alcalay R., Gur D., Aftalion M., Falach R., Leviatan Ben-Arye S., Lazar S., et al. Therapeutic antibodies, targeting the SARS-CoV-2 spike N-terminal domain, protect lethally infected K18-hACE2 mice. iScience. 2021;24:102479. doi: 10.1016/j.isci.2021.102479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas D., Veyer D., Baidaliuk A., Staropoli I., Guivel-Benhassine F., Rajah M.M., Planchais C., Porrot F., Robillard N., Puech J., et al. Reduced sensitivity of infectious SARS-CoV-2 variant B.1.617.2 to monoclonal antibodies and sera from convalescent and vaccinated individuals. bioRxiv. 2021 doi: 10.1101/2021.05.26.445838. [DOI] [Google Scholar]
- Rambaut A., Holmes E.C., O’Toole Á., Hill V., McCrone J.T., Ruis C., du Plessis L., Pybus O.G. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A., Loman N., Pybus O., Barclay W., Barrett J., Carabelli A., Connor T., Peacock T., Robertson D.L., Volz E. Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. Virological.org. 2020 https://virological.org/teliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/ [Google Scholar]
- Rees-Spear C., Muir L., Griffith S.A., Heaney J., Aldon Y., Snitselaar J.L., Thomas P., Graham C., Seow J., Lee N., et al. SAFER Investigators The effect of spike mutations on SARS-CoV-2 neutralization. Cell Rep. 2021;34:108890. doi: 10.1016/j.celrep.2021.108890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robson F., Khan K.S., Le T.K., Paris C., Demirbag S., Barfuss P., Rocchi P., Ng W.L. Coronavirus RNA Proofreading: Molecular Basis and Therapeutic Targeting. Mol. Cell. 2020;80:1136–1138. doi: 10.1016/j.molcel.2020.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld R., Noy-Porat T., Mechaly A., Makdasi E., Levy Y., Alcalay R., Falach R., Aftalion M., Epstein E., Gur D., et al. Post-exposure protection of SARS-CoV-2 lethal infected K18-hACE2 transgenic mice by neutralizing human monoclonal antibody. Nat. Commun. 2021;12:944. doi: 10.1038/s41467-021-21239-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito A., Nasser H., Uriu K., Kosugi Y., Irie T., Shirakawa K., Sadamasu K., Kimura I., Ito J., Wu J., et al. SARS-CoV-2 spike P681R mutation enhances and accelerates viral fusion. bioRxiv. 2021 doi: 10.1101/2021.06.17.448820. [DOI] [Google Scholar]
- Starr T.N., Greaney A.J., Addetia A., Hannon W.W., Choudhary M.C., Dingens A.S., Li J.Z., Bloom J.D. Prospective mapping of viral mutations that escape antibodies used to treat COVID-19. Science. 2021;371:850–854. doi: 10.1126/science.abf9302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Dingens A.S., Bloom J.D. Complete map of SARS-CoV-2 RBD mutations that escape the monoclonal antibody LY-CoV555 and its cocktail with LY-CoV016. Cell Rep. Med. 2021;2:100255. doi: 10.1016/j.xcrm.2021.100255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T.J.C., Yuan M., Kuzelka K., Padron G.C., Beal J.R., Chen X., Wang Y., Rivera-Cardona J., Zhu X., Stadtmueller B.M., et al. Sequence signatures of two public antibody clonotypes that bind SARS-CoV-2 receptor binding domain. Nat. Commun. 2021;12:3815. doi: 10.1038/s41467-021-24123-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N., et al. Emergence of a SARS-CoV-2 variant of concern with mutations in spike glycoprotein. Nature. 2021;592:438–443. doi: 10.1038/s41586-021-03402-9. [DOI] [PubMed] [Google Scholar]
- Thomson E.C., Rosen L.E., Shepherd J.G., Spreafico R., da Silva Filipe A., Wojcechowskyj J.A., Davis C., Piccoli L., Pascall D.J., Dillen J., et al. ISARIC4C Investigators. COVID-19 Genomics UK (COG-UK) Consortium Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e20. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;183:1735. doi: 10.1016/j.cell.2020.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., Ho D.D. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 2021;29:747–751.e4. doi: 10.1016/j.chom.2021.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Weinreich D.M., Sivapalasingam S., Norton T., Ali S., Gao H., Bhore R., Musser B.J., Soo Y., Rofail D., Im J., et al. Trial Investigators REGN-COV2, a Neutralizing Antibody Cocktail, in Outpatients with Covid-19. N. Engl. J. Med. 2021;384:238–251. doi: 10.1056/NEJMoa2035002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., et al. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.C., Yuan M., Liu H., Lee C.D., Zhu X., Bangaru S., Torres J.L., Caniels T.G., Brouwer P.J.M., van Gils M.J., et al. An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain. Cell Rep. 2020;33:108274. doi: 10.1016/j.celrep.2020.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Hong K., Ruan L., Yang X., Zhang J., Xu J., Pan S., Ren L., Chen L., Huang C., Shang Y. Patients with Prolonged Positivity of SARS-CoV-2 RNA Benefit from Convalescent Plasma Therapy: A Retrospective Study. Virol. Sin. 2020;35:768–775. doi: 10.1007/s12250-020-00281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiaojie S., Yu L., Lei Y., Guang Y., Min Q. Neutralizing antibodies targeting SARS-CoV-2 spike protein. Stem Cell Res. (Amst.) 2020;50:102125. doi: 10.1016/j.scr.2020.102125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav P.D., Sapkal G.N., Ella R., Sahay R.R., Nyayanit D.A., Patil D.Y., Deshpande G., Shete A.M., Gupta N., Mohan V.K., et al. Neutralization of Beta and Delta variant with sera of COVID-19 recovered cases and vaccinees of inactivated COVID-19 vaccine BBV152/Covaxin. J. Travel Med. 2021 doi: 10.1093/jtm/taab104. Published online July 6, 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H., et al. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Huang D., Lee C.D., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W., et al. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. Science. 2021;373:818–823. doi: 10.1126/science.abh1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The published paper includes all dataset generated or analyzed during this study.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request