Abstract
This work sought to quantify pathologists’ diagnostic bias over time in their evaluation of colorectal polyps to assess how this may impact the utility of statistical process control (SPC). All colorectal polyp specimens(CRPS) for 2011–2017 in a region were categorized using a validated free text string matching algorithm. Pathologist diagnostic rates (PDRs) for high grade dysplasia (HGD), tubular adenoma (TA_ad), villous morphology (TVA + VA), sessile serrated adenoma (SSA) and hyperplastic polyp (HP), were assessed (1) for each pathologist in yearly intervals with control charts (CCs), and (2) with a generalized linear model (GLM). The study included 64,115 CRPS. Fifteen pathologists each interpreted > 150 CRPS/year in all years and together diagnosed 38,813. The number of pathologists (of 15) with zero or one (p < 0.05) outlier in seven years, compared to their overall PDR, was 13, 9, 9, 5 and 9 for HGD, TVA + VA, TA_ad, HP and SSA respectively. The GLM confirmed, for the subset where pathologists/endoscopists saw > 600 CRPS each(total 52,760 CRPS), that pathologist, endoscopist, anatomical location and year were all strongly correlated (all p < 0.0001) with the diagnosis. The moderate PDR stability over time supports the hypothesis that diagnostic rates are amendable to calibration via SPC and outcome data.
Subject terms: Pathology, Statistics, Biomedical engineering, Quality control
Introduction
Diagnostic variance can be measured in several ways. In pathology, it is traditionally measured with kappa values generated by small data sets interpreted by a variable number of pathologists. Inter-rater agreement is inherently dependent on both the tissue type and the clinical diagnosis, with the usual results being poor to moderate. The gold standard diagnosis is commonly determined through the consensus of a panel of experts, rather than hard outcomes-driven data1.
From the perspective of manufacturing industries (where defect rates are commonly measured in parts per thousand or parts per million), significant disagreements/errors in pathology (that change the management) are common2. Such high error rates are often rationalized by the inherently imprecise nature of histomorphology and the innate difficulty in achieving high levels of precision within a complex system such as the human body. The later is a historical and longstanding philosophical debate: anti-reductionism (the body is impossible to subdivide effectively into components that enhance understanding) versus reductionism (the body can be subdivided effectively into components that enhance understanding).
The precision/lack of precision in histomorphology is the topic of this study. In the context of the above-mentioned philosophical debate, we support a reductionist approach of subdivision into component elements can allow for enhanced understanding with sufficient determination, and appropriate models, statistical methods and process management. As such, if diagnostic variation is explained by consistent bias among healthcare providers (as opposed to large swings in the diagnostic rates/diagnostic instability), it should be amendable to an intervention that (deconstructs the diagnostic process), reduces variation and, with calibration (including pathologist rate awareness), may be used to more appropriately stratify patients and improve outcomes.
We, thus, hypothesize that histomorphology is actually much more reproducible than seen in most practise environments, and that a lack of “control” within a practice environment (rather than imprecision in histomorphology) is the basis of the considerable diagnostic variation in pathological reporting encountered. The specific goals of this study are to (1) understand diagnostic variance among pathologists and (2) compare diagnostic variance using a population-based approach.
Population-based comparisons are predicated on the assumptions of (1) population disease stability (i.e. population disease characteristics are stable over time) and (2) the absence of a selection/case assignment bias for the interpreting pathologist. The first assumption can be determined to some degree from the data itself, if the sample is sufficiently large for the time frame. The second assumption depends on the practice environment/case distribution. Prior work in our laboratory found very similar call rates for Helicobacter pylori gastritis3 and endoscopic bronchial ultrasound-fine needle aspirations4. The simplest and most likely explanation is that the case assignment in our practice environment is effectively random.
Statistical process control (SPC) was first applied in medicine by pathologists in the 1960s5, and there is now a substantial literature on SPC in medicine6 and it is used in clinical pathology. Although the SPC methodology has not routinely been applied in anatomical pathology, it should be an effective tool if there is approximate diagnostic stability and approximate disease stability over time7. “Appendix E” briefly explains SPC and why demonstrating disease stability and diagnostic rate stability (in the context of significant diagnostic rate differences) would be sufficient to suggest SPC should be effective.
Colorectal polyp specimens were chosen for study as (1) they are a high-volume specimen familiar to pathology generalists and subspecialists, and (2) interpretative differences are usually of low or moderate consequence; the questions are frequently one benign diagnosis versus another benign diagnosis (rather than benign versus malignant).
Methods
Research ethics board approval was obtained (Hamilton Integrated Regional Ethics Board (HiREB)—HiREB# 2016-2295-C and HiREB# 2018-4445-C) with sign off from the laboratory director. The study was done in accordance with national ethics guidelines and relevant regulations. This study had no research subjects; thus, the requirement for informed consent from subjects is not applicable. All in house surgical pathology reports accessioned from January 2011 to December 2017 were extracted from the Laboratory Information System.
Extracted reports were stripped of all patient identifiers. Custom computer programs written in the Python programming language then reconstructed the report structure, subdivided cases into parts, and allowed complex searches within cases and parts.
Colorectal polyps specimens which met the following criteria were retrieved:
one of the following words: “colon”, “rectum”, “rectal”, “cecum”, “cecal”, “rectosigmoid” in the “source of specimen” section of the report
“polyp” within the “source of specimen” section of the report
The “source of specimen” section is what the endoscopist labels the specimens as. It was chosen as it was deemed to have the most uniformity of the report sections.
It should be noted the “specimens” correspond to bottles/containers submitted. One specimen may in fact contain zero, one or several polyps that are from one or more anatomical sites. Several specimens may be derived from one surgical case; quantification of this is not part of the study. A number of surgical cases may originate from one individual; however, quantification of this is not part of the study.
Retrieved specimens were written to a tab separated file which was then further processed to replace the surgical case number, submitting physicians and pathologists with unique anonymous identifiers.
Specimens were then tabulated.
Cases were classified by fuzzy string matching using an open source library called google-diff-patch-match and several dictionaries of terms into:
one or more 40 diagnostic categories (based on 194 phrases) – see “Appendix A”
one or more 12 anatomical locations (based on 18 phrases or 9 measurement cut points) – see “Appendix B”
Audits were done with randomly selected CRPS to assess the accuracy of the computer’s classification. This involved pathologists comparing the (pathology) report free text with the diagnostic categories (listed in “Appendix A” and “Appendix B”) assigned by the computer.
After categorization and tabulation, the anonymized data set was further processed by a custom GNU/Octave8 program to create funnel plots and control charts. Funnel plots that included data from all pathologists were centred on the group median diagnostic rate (GMDR). The GMDR was chosen, as the reference, as it is (1) not influenced by significant outliers, and (2) not biased by case volume. The funnel edges were defined by two and three standard deviations from the GMDR and calculated via the normal approximation of the binomial distribution as previously described9. Control charts (equivalent to the funnel plots) were created by normalizing to the number of cases read by the highest volume pathologist in the group; details of the normalization are within “Appendix C” 15. Normalization was done to obscure case volume and facilitate ease of interpretation.
Pathologist-specific control charts (showing the year-to-year variation) were created with the individual pathologist’s mean diagnostic rate, if the pathologist interpreted at least 600 specimens. The mean was chosen as the number of cases per year was not equal; using the mean ensured that the cases had equal weight in determining the control chart “centre”. Data points for a given year were plotted only if the pathologist interpreted at least 150 specimens in that year. The thresholds (600 specimens, 150 specimens/year) were chosen to ensure that the PDR estimates are have relatively narrow confidence intervals.
Generalized linear models, with a random intercept for each hospital, were utilized to estimate the association between independent variables (pathologist, submitting MD, anatomical location, and year) and high-grade dysplasia (HGD), villous component (TVA + VA), hyperplastic polyp (HP), tubular adenoma (TA), and sessile serrated adenoma (SSA). These models were implemented using SAS version 9.4 (SAS Institute, Cary, NC).
Prior to this calculation, all pathologists and all submitting physicians interpreting or submitting less than 600 specimens were excluded from the dataset. Uncommon nonspecific/vague anatomical sites (e.g. “anastomosis [not otherwise specified]” or “left colon [not otherwise specified]”) were also excluded from the data set to avoid the possibility of over-fitting.
Ethics approval and consent to participate
Ethics approval was obtained. Consent for publication is not applicable.
Results
In the study period, the program extracted 64,115 colorectal polyp specimens. A small number of polyp specimens (< 1%) may not have been captured, as we previously did an analysis on this (published in abstract form9 in a cohort of 11,457 large bowel polyp specimens, 68 surgical cases could not be parsed (separated into parts/specimens). The 68 cases (not parsed by the computer) were examined in detail and it was determined that 37 had unusual report formatting (e.g. parts were out of order), 24 had a mislabelled part (e.g. “Part D” transcribed as “Part P”), 7 had missing specimen parts (e.g. requisition has Parts A-C, diagnosis sections has Part A-B (Part C is absent)).
In the 64,115 colorectal polyp specimens that were retrieved, the hierarchical free text string matching algorithm (HFTSMA) could classify 63,050 of the specimens with regard to a diagnosis, and 63,508 with regard to the anatomical site. Several individuals independently assessed the accuracy of the computer’s classification via random audits in at least 789 specimens. Prior audits suggested that the overall accuracy is ~ 97%.
The three percent that is not classified correctly is mostly not classifiable; we previously analyzed 55 of 92 unclassified colorectal polyp specimens in a cohort of 11,457 large bowel polyp specimens9. In most cases, the failure was nontechnical/unrelated to the HFTSMA; 19 cases were rare/descriptive diagnoses, 24 vaguely worded diagnoses, 7 failed due to (unusual) report formatting/transcription and 5 failed for an unknown reason.
Since the custom analysis programs have evolved in the past 2 years, we did a further random audit of the computer’s classification. Four hundred polyp specimens were selected at random and the computer-generated diagnostic codes were compared to the text of the diagnosis. In this analysis, 394 cases were correctly classified and 6 not coded; this matched our prior experience. We also recently examined sessile serrated adenomas (SSA) over multiple years in a subset of ~ 7000 colorectal polyp specimens10. In that context, the accuracy of SSA classification was examined; in 400 randomly selected cases there were zero errors in the classification of SSA/not SSA. Report auditing (based on the results) found systematic misclassifications in HGD and TA; these were corrected by adjusting the dictionary of diagnostic terms and re-running the analysis.
Outliers > 7 SDs from the GMDR (seen in SSA, HP and TVA + VA) prompted reviews of 100–200 randomly selected specimen reports for each of the anonymous outlier pathologists, and these confirmed that there is no significant categorization error (due to unusual reporting language) from the HFTSMA that could explain the observed diagnostic rates.
An overview of the colorectal polyp cohort is found within Table S1 (see supplemental materials). ‘Adenoma [not otherwise specified]’ was combined with ‘tubular adenoma’, as these appeared to be used as synonyms by a subset of pathologists.
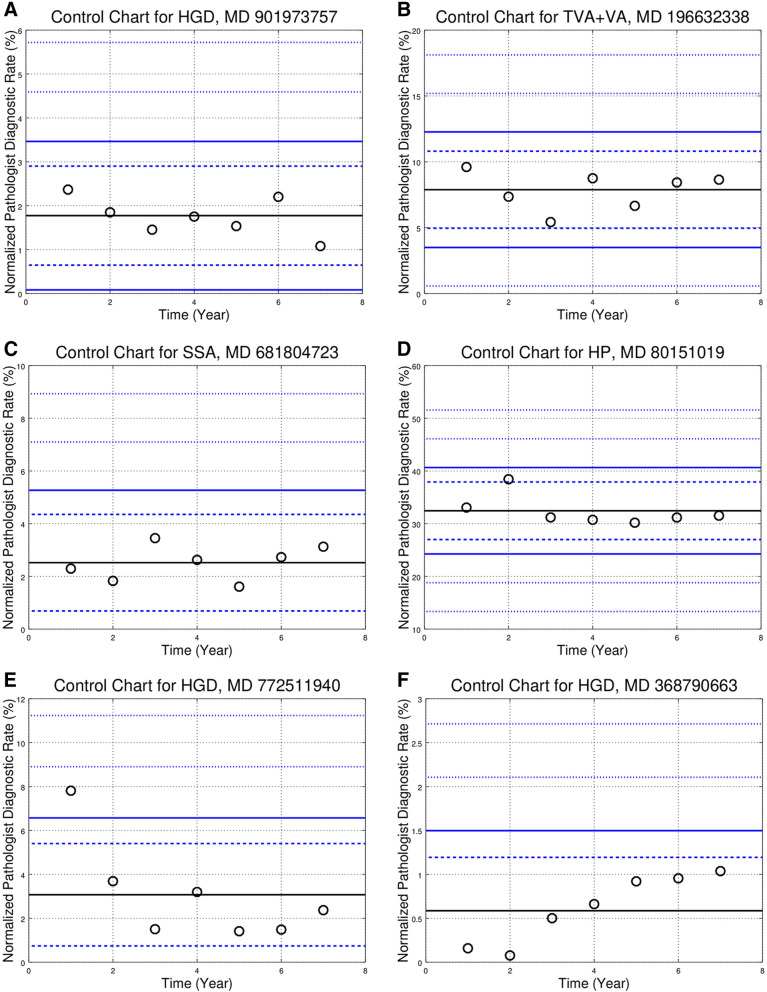
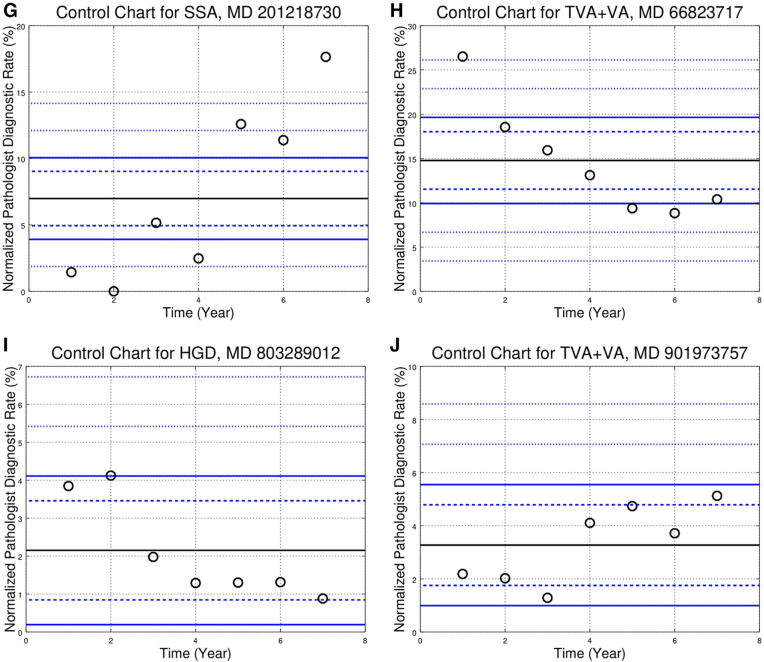
The control chart showed various patterns. The “in control” pattern was common, and is the expected result if (1) the individual pathologist has not changed their practice, (2) the population disease rates are stable. Representative control charts of this type are seen in Fig. 1A–C.
Figure 1.
Control charts for individual pathologists. Each panel (control chart) in this figure is one diagnosis (e.g. SSA) for a pathologist that read > 150 CRPS/year in the seven years of the study. The black circles represent the normalized PDR in different years; the denominator within the normalized PDR is the number of colorectal polyp specimens read by the individual pathologist in the year. The thick solid black line is the pathologist’s average (or mean) call rate (PACR). The dashed blue control lines above and below the PACR are 2 standard deviations (SDs) from the PACR; points outside the range are p < 0.05. The solid blue control lines represent 3 SDs from the PACR; points outside the range are p < 0.003. The outer finely dashed blue control lines above/below the PACR are 5 SDs and 7 SDs from the PACR respectively; points above/below the lines are p < 6e−7 and p < 3e−12 respectively. Controls lines may be absent if the PACR is close to zero (as negative normalized PDRs do not have a physical interpretation). The individual panels (control charts) are (A) high-grade dysplasia (HGD) showing the “in control” condition; (B) tubulovillous adenoma + villous adenoma (TVA + VA) showing the “in control” condition; (C) sessile serrated adenoma (SSA) showing the “in control” condition; (D) hyperplastic polyp (HP) with a “blip” (> 2 SDs/P < 0.05 outlier) in year two of the study; (E) HGD with a “blip” (P < 0.003 outlier) in year one of the study; (F) HGD with a “trend” (non-significant, not crossing control lines); (G) SSA with a “trend” (significant, crossing control lines); (H) TVA + VA with a “trend” (significant, crossing control lines); (I) HGD with a “step” (significant, crossing control lines); (j) TVA + VA with a “step” (significant, crossing control lines).
Some control charts (e.g. Figure 1D,E) showed an outlier in the background of what would otherwise be “in control”; this was the most common pattern (See Table 2). A third type of chart shows a pattern (increasing or decreasing) with or without crossing control lines (e.g. Fig. 1F–H). A fourth type of chart shows a step (upward or downward) with relative stability before and afterward (e.g. Fig. 1I,J).
Table 2.
Comparison of pathologists to self. Number of pathologists by the number of (> 2 standard deviation) outliers in relation to each pathologist’s mean call rate over the seven-year period. This tabulation shows the number of 2 SD outliers they had for each diagnosis, e.g. 8 pathologists had zero outlier years for HGD, 1 pathologist had 5 outlier years for TVA + VA (this is shown in Fig. 1H), 2 pathologists had 4 outlier years for SSA, 2 pathologists had 6 outlier years for SSA (one of the two pathologists is shown in Fig. 1G).
| # Outliers | HGD | TVA + VA | TA_ad | HP | SSA |
|---|---|---|---|---|---|
| 0 | 8 | 6 | 3 | 0 | 2 |
| 1 | 5 | 3 | 6 | 5 | 7 |
| 2 | 2 | 3 | 2 | 5 | 1 |
| 3 | 0 | 1 | 3 | 2 | 0 |
| 4 | 0 | 0 | 1 | 3 | 2 |
| 5 | 0 | 1 | 0 | 0 | 1 |
| 6 | 0 | 0 | 0 | 0 | 2 |
| 7 | 0 | 1 | 0 | 0 | 0 |
| Sum | 15 | 15 | 15 | 15 | 15 |
HGD high-grade dysplasia, HP hyperplastic polyp, SSA sessile serrated adenoma, TVA+VA tubulovillous adenoma + villous adenoma, TA_ad tubular adenoma + adenoma NOS, P number of pathologists.
The control charts constructed around the pathologist’s mean PDR are summarized in Table 1a.
Table 1.
(Subsection a) Comparison of pathologists to self. Number of outlier years for all pathologists with seven years of data. There are 15 pathologists with 7 years of data; thus, there are 105 pathologist x years, (Subsection b) Comparison to pathologists to self (normalized). Fraction that are outliers for control charts centred on the pathologist’s mean diagnostic rate. The numbers in this table are generated by dividing through by the total number of pathologist x years (105), e.g. 9/105 = 0.09 for HGD 2 SD. SD = standard deviation.
| Variation | HGD | TVA + VA | TA_ad | HP | SSA |
|---|---|---|---|---|---|
| Subsection a | |||||
| < 2 SD | 96 | 81 | 82 | 72 | 71 |
| > 2 SD | 9 | 24 | 23 | 33 | 34 |
| > 3 SD | 2 | 10 | 8 | 10 | 18 |
| > 5 SD | 0 | 2 | 1 | 0 | 10 |
| > 7 SD | 0 | 1 | 0 | 0 | 5 |
| PY (Total) | 105 | 105 | 105 | 105 | 105 |
| Variation | HGD | TVA + VA | TA_ad | HP | SSA |
|---|---|---|---|---|---|
| Subsection b | |||||
| < 2 SD | 0.91 | 0.77 | 0.78 | 0.69 | 0.68 |
| > 2 SD | 0.09 | 0.23 | 0.22 | 0.31 | 0.32 |
| > 3 SD | 0.02 | 0.10 | 0.08 | 0.10 | 0.17 |
| > 5 SD | 0.00 | 0.02 | 0.01 | 0.00 | 0.10 |
| > 7 SD | 0.00 | 0.01 | 0.00 | 0.00 | 0.05 |
HGD high-grade dysplasia, HP hyperplastic polyp, SSA sessile serrated adenoma, TVA + VA tubulovillous adenoma + villous adenoma, TA_ad tubular adenoma + adenoma NOS, PY pathologist-years; 15 pathologists × 7 years = 105 pathologist × years.
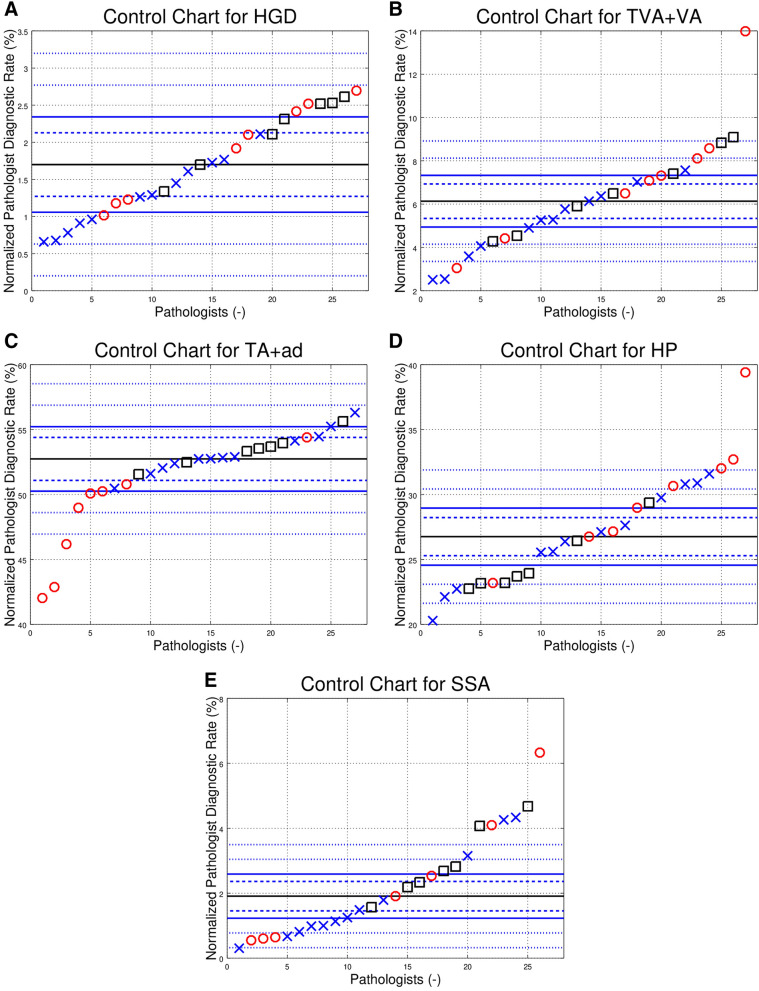
The control charts, centred on the group median diagnostic rate (GMDR), showed many outliers (see Fig. 2A–E), and are summarized in Table 3a. Outliers (p < 0.05) were calculated using the GMDR for each of the hospitals. The results are in Table 3c; specimens from two hospitals are effectively shared by one group of pathologists; thus, this was considered one site for the purpose of the control chart analysis (Table 2).
Figure 2.
Control charts showing the normalized pathologist diagnostic rates (PDRs) for the 27 pathologists reading > 600 CRPS in the seven year study period. Each panel (control chart) in this figure is one diagnosis, e.g. SSA. The different markers (red circles, blue Xs, black boxes) represent individual pathologists from different hospitals. The solid black line is the group median diagnostic rate (GMDR). The dashed blue control lines above and below the GMDR are 2 standard deviations (SDs) from the GDMR; pathologists outside the inner funnel are statistically different than the GMDR (p < 0.05). The solid blue control lines above and below the GMDR represent 3 SDs; pathologist outside the outer funnel are statistically different than the GMDR (p < 0.003). The outer finely dashed blue control lines above/below the GMDR are 5 SDs and 7 SDs from the GMDR respectively; points above/below the lines are p < 6e−7 and p < 3e−12 respectively. Controls lines may be absent if the GMDR is close to zero (as negative normalized PDRs do not have a physical interpretation). The individual panels (control charts) are (A) normalized high-grade dysplasia (HGD) PDRs; (B) normalized tubulovillous adenoma + villous adenoma (TVA + VA) PDRs; (C) normalized tubular adenoma and adenoma not otherwise specified (TA_ad) PDRs; (D) normalized hyperplastic polyp (HP) PDRs; (E) normalized sessile serrated adenoma (SSA) PDRs, only the 26 of the 27 pathologists shown; one pathologist had a normed diagnostic rate > 25% (not shown).
Table 3.
Subsection a Comparison of pathologists to the 27 pathologists that read > 600 CRPS. Number of outlier pathologists based on the group median diagnostic rate. The number of “ < 2 SD” and “ > 2 SD” pathologists total 27. Subsection b Comparison of pathologists to the 27 pathologists that read > 600 CRPS (normalized). Fraction that are outliers for control charts around the group median diagnostic rate. The numbers in this table are generated by dividing through by the total number of pathologists (27). Subsection c Comparison of pathologists to the hospital site. Number of outlier pathologists based on the group median for the individual hospital sites. Subsection d Comparison of pathologists to the hospital site (normalized). Fraction that are outliers for control charts around the group median diagnostic rate for the each of the hospital sites. The numbers in this table are generated by dividing through by the total number of pathologists (27).
| Variation | HGD | TVA + VA | TA_ad | HP | SSA |
|---|---|---|---|---|---|
| Subsection a | |||||
| < 2 SD | 11 | 6 | 14 | 8 | 6 |
| > 2 SD | 16 | 21 | 13 | 19 | 21 |
| > 3 SD | 12 | 16 | 9 | 19 | 19 |
| > 5 SD | 0 | 9 | 3 | 11 | 13 |
| > 7 SD | 0 | 5 | 3 | 4 | 8 |
| P (Total) | 27 | 27 | 27 | 27 | 27 |
| Variation | HGD | TVA + VA | TA_ad | HP | SSA |
|---|---|---|---|---|---|
| Subsection b | |||||
| < 2 SD | 0.41 | 0.22 | 0.52 | 0.30 | 0.22 |
| > 2 SD | 0.59 | 0.78 | 0.48 | 0.70 | 0.78 |
| > 3 SD | 0.44 | 0.59 | 0.33 | 0.70 | 0.70 |
| > 5 SD | 0.00 | 0.33 | 0.11 | 0.41 | 0.48 |
| > 7 SD | 0.00 | 0.19 | 0.11 | 0.15 | 0.30 |
| Variation | HGD | TVA + VA | TA + adN | HP | SSA |
|---|---|---|---|---|---|
| Subsection c | |||||
| < 2 SD | 16 | 9 | 17 | 11 | 11 |
| > 2 SD | 11 | 18 | 10 | 16 | 16 |
| > 3 SD | 6 | 14 | 5 | 14 | 15 |
| > 5 SD | 1 | 10 | 3 | 10 | 12 |
| > 7 SD | 1 | 3 | 2 | 4 | 7 |
| P (Total) | 27 | 27 | 27 | 27 | 27 |
| Variation | HGD | TVA + VA | TA + adN | HP | SSA |
|---|---|---|---|---|---|
| Subsection d | |||||
| < 2 SD | 0.59 | 0.33 | 0.63 | 0.41 | 0.41 |
| > 2 SD | 0.41 | 0.67 | 0.37 | 0.59 | 0.59 |
| > 3 SD | 0.22 | 0.52 | 0.19 | 0.52 | 0.56 |
| > 5 SD | 0.04 | 0.37 | 0.11 | 0.37 | 0.44 |
| > 7 SD | 0.04 | 0.11 | 0.07 | 0.15 | 0.26 |
HGD high-grade dysplasia, HP hyperplastic polyp, SSA sessile serrated adenoma, TVA + VA tubulovillous adenoma + villous adenoma, TA_ad tubular adenoma + adenoma NOS, P number of pathologists.
The control charts based on the individual pathologist’s mean PDR and those based on the GMDR are not directly comparable; however, the summary data (Tables 1a and 3a) does allow some comparison. The fraction of outliers (shown in Tables 1b, 3b,d) were calculated using the total number of elements—105 and 27 respectively. These tables show that there are proportionally less outliers when the data is plotted by the pathologist, suggesting the individual pathologist is a very strong predictor—a result demonstrated with logistic regression. For example, the fraction > 2 SD for HGD is 0.09, 0.41 and 0.59 for comparison to self (Table 1a), comparison to hospital site (Table 3b) and comparison to the group of 27 pathologists (Table 3d) respectively; this is also shown in Table S2 (see supplemental materials).
The outlier frequencies within Table 1a (with the exception of HGD) are highly improbable to be only a consequence of sampling. The cumulative probability of being outside two standard deviations for (1) the number of outliers and (2) all greater number of outliers for HGD is p ~ 0.08 (see “Appendix D” for details). The probability of being outside two standard deviations (for (1) the number of outliers and (2) all greater number of outliers) for all the other diagnoses is p < 0.0001. The outlier frequencies (for two standard deviations) in Table 3a are all significantly in excess of that expected due to sampling.
Table 2 shows the number of pathologists by the number of outlier years for two standard deviations. Stated differently, Table 2 is a tabulation of the 105 control charts; the question answered is: how many > 2 SD outliers do each of the 15 pathologists have for a given diagnosis? Fig. 1A shows one of the 8 pathologists that had zero HGD outliers (all circles between the two dashed blue control lines). Figure 1C is one of the two pathologists that had zero SSA outliers. The outliers-years found in Table 1a, are related to the numbers in Table 2; in Table 2 for HGD: 5 pathologists with 1 outlier year each + 2 pathologists with 2 outlier years each = 9 pathologist-year outliers (> 2 SD) in Table 1a. Table 2 shows that there is good self-consistency for HGD; eight pathologists had zero outlier years. It also shows that SSA had marked changes; two pathologists had six outlier years (one of these two pathologist’s normalized PDRs are shown in Fig. 1G).
The data set cleansed of (1) pathologist interpreting < 600 specimens, (2) submitting physicians submitting < 600 specimens, and (3) rare/ambiguous anatomical sites, contained 52,760 colorectal polyp specimens. Two of the 27 pathologists never called SSA; these were non-zeroed to facilitate numerical convergence.
The random effects models (see Table 4) demonstrated that the pathologist, submitting MD, and anatomical location are all strong predictors (p < 0.0001) of histomorphologic diagnosis of TA, HGD, TVA + VA, HP, and SSA.
Table 4.
Generalized linear model results.
| Factor | DF | F Value | p-value |
|---|---|---|---|
| Outcome: TA_ad | |||
| PATHOLOGIST | 26 | 8.34 | < .0001 |
| CLINICIAN | 45 | 5.63 | < .0001 |
| LOC_FULL_CR | 8 | 646.72 | < .0001 |
| YEAR_VAR | 6 | 6.98 | < .0001 |
| Outcome: TVA | |||
| PATHOLOGIST | 26 | 24.61 | < .0001 |
| CLINICIAN | 45 | 7.1 | < .0001 |
| LOC_FULL_CR | 8 | 47.52 | < .0001 |
| YEAR_VAR | 6 | 12.14 | < .0001 |
| Outcome: HGD | |||
| PATHOLOGIST | 26 | 5.42 | < .0001 |
| CLINICIAN | 45 | 4.26 | < .0001 |
| LOC_FULL_CR | 8 | 11.26 | < .0001 |
| YEAR_VAR | 6 | 7.33 | < .0001 |
| Outcome: HP | |||
| PATHOLOGIST | 26 | 21.96 | < .0001 |
| CLINICIAN | 45 | 5.49 | < .0001 |
| LOC_FULL_CR | 8 | 767.72 | < .0001 |
| YEAR_VAR | 6 | 15.27 | < .0001 |
| Outcome: SSA | |||
| PATHOLOGIST | 26 | 85 | < .0001 |
| CLINICIAN | 45 | 2.69 | < .0001 |
| LOC_FULL_CR | 8 | 161.75 | < .0001 |
| YEAR_VAR | 6 | 60.09 | < .0001 |
“PATHOLOGIST” and “CLINICIAN” are variables that represent individual pathologists and individual submitting physicians/surgeons. “LOC_FULL_CR” is a variable that reperesent the anatomical location; it can be one of nine locations in the colon/rectum (rectum, rectosigmoid colon, sigmoid colon, descending colon, splenic flexure of colon, transverse colon, hepatic flexure of colon, ascending colon, cecum). “YEAR_VAR” is the year in which the specimen was accessioned. “DF” is the degrees of freedom. Tubular adenoma and adenoma NOS (TA_ad) were lumped in this analysis, as a subset of pathologists (early in the study period) signed cases as “adenoma” without further specifying. These are presumed to represent tubular adenomas.
Discussion
The HFTSMA algorithm appears to deliver reliable categorizations that are. sufficient to assess diagnostic variances on the order of 1%. Non-categorized polyps appear to represent a separate group/set of diagnoses that are predominantly descriptive diagnoses or ambiguously-worded reports that cannot be easily classified.
Most of the pathologists in the cohort had relatively stable diagnostic rates over time, but there were apparent outliers. The relative uniformity in some diagnoses (e.g. high-grade dysplasia) provides good evidence against the presence of case assignment bias.
The hospital sites show some clustering of patterns in PDR. This may be mostly explained by the presence of group set-point bias rather than true differences between hospital sites.
The “clinicians” factor (submitting MD) appears to explain less variation in the data than the “pathologist” factor.
Traditional inter-rater studies look at a relatively small set of cases and rarely examine diagnostic bias over a longer period of time. This study examined the reports in an entire region over a period of seven years.
While high-grade dysplasia and villous component are predictive of neoplasia risk in large cohorts, the findings herein suggest risk stratification using high-grade dysplasia and villous component suboptimally risk-stratifies individual patients, due to the consistent (presumptive substantial inter-rater) variation in the pathologist diagnostic rate.
Generally, the findings demonstrate that the histomorphologic interpretation of colorectal polyps could be less varied than seen herein, and imply that (statistical) process control (or an automated analysis), that reproduces the categorization biases of one pathologist (or a panel of pathologists), would deliver more uniformity.
Based on our prior work11 and PDR data (predominantly published as conference abstracts), we are not convinced that more sub-specialization is the only answer. We also note that disagreement among subspecialists may be very high12. The processes changes (independent of training) may significantly improve quality13,14.
Limitations
A few pathologists moved between hospital sites in the 7-year period; however, none of the 15 pathologists interpreted less than 92% of their specimens from their primary site. This is a confounder that was not specifically controlled for in the construction of the control charts; however, the effect is suspected to be small.
It is not possible to determine the ideal rate(s) in this study. Whether large true differences exist between the hospital sites cannot be determined within the context of this study. It is possible that the differences between the hospital sites is totally or partially explained by group bias. A significant number of specimens (~ 300 from each hospital site) would need to be reviewed by an expert panel, as the differences are likely to be small. Normalized plots showing the polyps by lumped anatomical site (left colon, mid colon, right colon) and pathologist suggest there may be differences between the hospitals (see supplemental materials).
Based on how the specimens are submitted and reported in routine practice, it is not possible to do the analysis on the level of the individual polyp.
We did not attempt to make control charts based on the yearly rates for each hospital, as the study set (15 pathologists with data over all years) was deemed to be too small to sub-stratify. This true limitation was explored with the random effects model and logistic regression.
Significant changes over time were identified with the random effect model, thus calling into question the “disease stability” assumption that is a part of the control chart analysis. We are not convinced these changes affect the overall conclusions due to the variation seen in the control charts. It is impossible to determine whether the change over time is (1) diagnostic re-calibration/diagnostic drift by selected pathologists; (2) a change in the population or (3) some combination of drift and population change. The trend data suggests strongly that there is re-calibration. We suspect there was a shift between TA and HP in the population. Supplemental materials show how the diagnoses varied over the seven-year period. There are very clear trends in HP and SSA, which may be rationalized in the context of when SSA was described, and by knowledge dissemination rates in medicine.
Specific healthcare provider characteristics (e.g. training, years in practice, type of practice) were not collected as part of this study. These may be significant predictors.
The study is observational and the data collected is influenced to certain extent by conscious changes to clinical practice. A subgroup of pathologists (due to a quality improvement project/pilot study15 were aware of their diagnostic rates in the last two years of the study period and a subset of those adjusted their practice. This likely decreased consistency with self and thus somewhat decreased the study’s effect size. It is not possible to fully analyze the effect of the subgroup (due the anonymity constraint in the study); however, the control charts show diagnostic rate changes in the early part of the study that are similar in magnitude to changes in the later part of the study period; thus, the overall conclusions are likely unaffected.
Diagnostic rate awareness and improvement
Colorectal polyps are specimens that may be infrequently reviewed at consensus rounds in relation to their volume; thus, call rate harmonization/calibration that takes place within pathology practices may not occur for these specimens. Also, random case reviews are not powered to detect modest call rate differences and would be prohibitively expensive if powered to do so.
SPC is a mechanism that may facilitate greater uniformity in reporting practices through greater dialogue about true population rates (with ideal pathological interpretation), and promote continuous review of outcome data. In the presence of significant differences in interpretation (that are unlikely to result from case assignment/sampling), suboptimal interpretations may be suspected and a process of resolution implemented through consensus guided by outcome data.
Based on a pilot study of ~ 7054 colorectal polyp specimens (interpreted by 9 pathologists (each year) over two years—Sep 2015–Aug 2017) in conjunction with (1) informing each pathologist of their (TA, HP, SSA, TVA + VA) diagnostic rates, and (2) a group review of (SSA cases) with a gastrointestinal pathology expert, it is possible to increase uniformity in sessile serrated adenoma (SSA) diagnostic rates15.We suspect that this process (statistical process control) could be applied more broadly and would lead to further improvements.
Conclusions
Current diagnostic processes for colorectal polyp specimens leave significant room for further improvement. This work suggests that most pathologists have diagnostic rate stability, and that non-stable rates are likely (conscious or unconscious) practice changes.
Statistical process control (SPC) could result in significantly more uniformity, given that many pathologists have moderate diagnostic rate stability. Thus, the further implementation of SPC in pathology should be pursued, as it could substantially optimize and improve care.
Supplementary Information
Acknowledgements
Dr. Sameer Parpia provided high level comments on the modelling approach and the statistical analysis. Dr. Hamid Kazerouni provided input at an early stage of the project. Dr. Jennifer M. Dmetrichuk audited computer categorizations from an early version of the computer program. An earlier version of this work was presented at the European Congress of Pathology 2018 in Bilbao, Spain.
Appendix A (Diagnostic Codes and Search Strings)
| Dx code | String |
|---|---|
| dx1 | Suspicious for adenocarcinoma |
| dx1 | Cannot exclude adenocarcinoma |
| dx1 | Adenocarcinoma can't excluded |
| dx1 | Suspicious for invasive adenocarcinoma |
| dx2 | Invasive adenocarcinoma |
| dx2 | Adenocarcinoma |
| dx2 | Invasive carcinoma |
| dx3 | Intramucosal adenocarcinoma |
| dx3 | Of high-grade dysplasia |
| dx3 | Of high grade dysplasia |
| dx3 | With high-grade dysplasia |
| dx3 | With high grade dysplasia |
| dx3 | Focal high grade dysplasia |
| dx3 | Focal high-grade dysplasia |
| dx3 | Showing high grade dysplasia |
| dx3 | Showing high-grade dysplasia |
| dx4 | Suspicious for lymphoma |
| dx4 | Atypical lymphoid proliferation |
| dx5 | MALT lymphoma |
| dx5 | Mantle cell lymphoma |
| dx6 | Tubular adenoma |
| dx6 | Tubular adenomata |
| dx7 | Hyperplastic polyp |
| dx7 | Hyperplastic polyp |
| dx8 | Hyperplastic changes |
| dx8 | Hyperplastic mucosal changes |
| dx8 | Hyperplastic features |
| dx8 | Hyperplastic-like features |
| dx9 | Tubulovillous adenoma |
| dx9 | Tubularvillous adenoma |
| dx9 | Tubulo-villous adenoma |
| dx9 | Villotubular adenoma |
| dx10 | Villous adenoma |
| dx11 | Sessile serrated adenoma |
| dx11 | Sessile serrated polyp |
| dx11 | Serrated sessile adenoma |
| dx11 | Serrated polyp, favor adenoma |
| dx11 | Serrated polyp, favour adenoma |
| dx11 | Serrated polyps, favor adenomata |
| dx12 | Traditional serrated adenoma |
| dx13 | Serrated adenoma |
| dx13 | Serrated adenomata |
| dx13 | Serrated polyp |
| dx14 | No pathologic finding |
| dx14 | Unremarkable fragment of large bowel mucosa |
| dx14 | Unremarkable large bowel mucosa |
| dx14 | Benign colonic mucosa |
| dx14 | Benign large bowel mucosa |
| dx14 | Normal colonic mucosa |
| dx14 | NO DIAGNOSTIC ABNORMALITY |
| dx14 | Unremarkable colonic mucosa |
| dx14 | Mucosa without significant pathology |
| dx14 | Mucosa within normal limits |
| dx14 | No specific pathology |
| dx14 | No significant histopathologic abnormality |
| dx14 | NEGATIVE for evidence of significant pathology |
| dx14 | No significant pathological changes |
| dx14 | No pathological changes |
| dx14 | No evidence of polyp |
| dx14 | Polypoid mucosa |
| dx14 | Colonic mucosa with prominent lymphoid follicles |
| dx14 | Negative for apparent pathology |
| dx14 | No pathological diagnosis |
| dx14 | CAUTERY ARTIFACT, NOT FURTHER DIAGNOSTIC |
| dx14 | Reactive changes, NEGATIVE |
| dx14 | Colonic mucosa with lymphoid aggregate |
| dx14 | Large bowel mucosa with no definite polyp |
| dx14 | Non-diagnostic polypoid colonic mucosa |
| dx14 | Bowel mucosa with no significant findings |
| dx14 | Bowel mucosa with no evidence of polyp |
| dx14 | No polyp |
| dx14 | No specific polyp |
| dx14 | No definitive polyp |
| dx14 | No definite polyp |
| dx14 | No specific pathologic diagnosis |
| dx14 | No significant pathology identified |
| dx14 | No significant pathological abnormalities |
| dx14 | No findings |
| dx14 | No histopathological abnormality |
| dx14 | Prominent mucosal folds |
| dx14 | Mucosa, likely mucosal fold |
| dx14 | Without diagnostic abnormality |
| dx14 | Unremarkable mucosal tissue |
| dx14 | No significant findings |
| dx14 | Colonic mucosa with no pathology |
| dx14 | No significant inflammation or other findings |
| dx15 | POLYPOID COLONIC MUCOSA |
| dx15 | Prominent lymphoid aggregate |
| dx15 | Lymphoid aggregate |
| dx15 | Large intestinal mucosa slightly polypoid with lymphoid aggregates |
| dx15 | Mucosa with lympho-follicular hyperplasia |
| dx15 | Lymphoid follicle |
| dx15 | Benign lymphoid aggregate |
| dx15 | Mucosal germinal centre |
| dx15 | Lymphoid hyperplasia |
| dx15 | Lymphocytic aggregates |
| dx16 | Inflammatory polyp |
| dx16 | Inflammatory pseudopolyp |
| dx16 | Inflammatory large bowel polyp |
| dx16 | Inflammatory-type polyp |
| dx16 | Inflamed polyp |
| dx17 | Hamartomatous polyp |
| dx18 | Granulation tissue |
| dx19 | Active colitis |
| dx19 | Active proctitis |
| dx19 | Acute cryptitis |
| dx20 | Poorly preserved colonic mucosa |
| dx21 | Solitary rectal ulcer |
| dx21 | Mucosal prolapse syndrome |
| dx21 | Mucosal Prolapse |
| dx21 | Mucosal prolapse-like polyp |
| dx21 | Mucosa with prolapse like changes |
| dx22 | Melanosis coli |
| dx22 | Pseudomelanosis coli |
| dx22 | Slight melanosis |
| dx23 | Juvenile polyp |
| dx23 | Juvenile type polyp |
| dx23 | Retension polyp |
| dx24 | Lipoma |
| dx25 | Granular cell tumour |
| dx25 | Granular cell tumor |
| dx26 | Ischemic colitis |
| dx27 | Leiomyoma |
| dx28 | Xanthoma |
| dx28 | Xanthoma/xanthelasma |
| dx29 | Hemorrhoid |
| dx30 | Prolapse changes |
| dx31 | Fecal material |
| dx31 | Fecal matter only |
| dx31 | Vegetable fibres |
| dx31 | Vegetable matter |
| dx31 | Fecal matter |
| dx31 | Feces |
| dx31 | Food material |
| dx31 | Polypoid vegetable resembling seed |
| dx31 | Degenerated meat fibres |
| dx32 | NEGATIVE for dysplasia |
| dx32 | NEGATIVE for evidence of dysplasia |
| dx32 | No neoplasia present |
| dx32 | Negative for conventional/adenomatous dysplasia |
| dx32 | Negative for adenomatous polyp or dysplasia |
| dx32 | Negative for adenoma |
| dx33 | Tissue not identified |
| dx33 | No tissue is identified |
| dx33 | No tissue present |
| dx33 | No tissue was found |
| dx33 | No microscopic assessment possible |
| dx33 | Tissue did not survive processing |
| dx33 | Did not survive tissue processing |
| dx33 | No material present after processing |
| dx33 | See gross |
| dx33 | No specimen received |
| dx33 | No specimen identified |
| dx33 | Insufficient for evaluation |
| dx33 | Insufficient for assessment |
| dx33 | Insufficient tissue for histologic assessment |
| dx33 | No colon tissue is observed |
| dx33 | Mucosa, not diagnostic |
| dx34 | Negative for high grade dysplasia |
| dx34 | Negative for high-grade dysplasia |
| dx34 | No evidence of high grade dysplasia |
| dx34 | No evidence of high-grade dysplasia |
| dx34 | No evidence of high dysplasia |
| dx34 | No definite evidence of high-grade dysplasia |
| dx34 | No convincing evidence of high grade dysplasia |
| dx34 | Without high grade dysplasia |
| dx34 | Without high-grade dysplasia |
| dx35 | NEGATIVE FOR DYSPLASIA OR MALIGNANCY |
| dx35 | Negative for high-grade dysplasia and malignancy |
| dx35 | Negative for high grade dysplasia and malignancy |
| dx35 | Negative for high-grade dysplasia or invasive malignancy |
| dx35 | Negative for high grade dysplasia or invasive malignancy |
| dx35 | Negative for high-grade dysplasia or invasive carcinoma |
| dx35 | Negative for high grade dysplasia or invasive carcinoma |
| dx35 | Negative for high-grade dysplasia and invasive carcinoma |
| dx35 | Negative for high grade dysplasia and invasive carcinoma |
| dx35 | No convincing evidence of high grade dysplasia or adenocarcinoma |
| dx36 | Cautery/crush artifact |
| dx36 | Cautery artifact |
| dx36 | Cautery artefact |
| dx36 | Cauterized tissue |
| dx36 | Cauterized colonic mucosa |
| dx36 | Polypoid cauterized mucosa |
| dx36 | Crushed fragments of large bowel |
| dx37 | Focal adenomatous changes |
| dx37 | Focal adenomatous change |
| dx37 | Fragments of adenoma |
| dx37 | Possible adenomatous change |
| dx37 | Adenoma |
| dx37 | Suspicious for Adenomatous Changes |
| dx37 | Adenomatous change |
| dx37 | Adenomatous mucosal change |
| dx37 | -ADENOMA(S) |
| dx38 | Chronic inflammation |
| dx38 | Chronic inflammation only |
| dx39 | Dysplasia associated lesion or mass |
| dx39 | Features of DALM |
| dx40 | Carcinoid |
| dx40 | Neuroendocrine tumour |
| dx40 | Neuroendocrine tumor |
Appendix B (Location Codes, Search Strings and Distances)
The location code was based on the location provided in the “source of specimen” section of the report. If the location was given in centimetres from the anal verge, it was converted to named location, based on approximate measures used by NCI.
Appendix B-1: Codes and location dictionary
| Location code | String |
|---|---|
| loc1 | Rectal |
| loc1 | Rectum |
| loc2 | Rectosigmoid |
| loc2 | Rectum—sigmoid |
| loc2 | Rectal—sigmoid |
| loc3 | Sigmoid |
| loc4 | Descending |
| loc5 | Splenic |
| loc6 | Transverse |
| loc7 | Hepatic |
| loc8 | Ascending |
| loc9 | Cecum |
| loc9 | Cecal |
| loc9 | Appendiceal orifice |
| loc10 | Left colon |
| loc11 | Right colon |
| loc12 | Anastomosis |
| loc12 | Anastomotic |
Appendix B-2: Codes and location table
| Location code | Distance (cm) |
|---|---|
| loc1 | < 13 |
| loc2 | < 18, ≥ 13 |
| loc3 | < 58, ≥ 18 |
| loc4 | < 79, ≥ 58 |
| loc5 | < 84, ≥ 79 |
| loc6 | < 130, ≥ 84 |
| loc7 | < 136, ≥ 130 |
| loc8 | < 147, ≥ 136 |
| loc9 | < 151, ≥ 147 |
Appendix C (Control charts/normalized funnel plots)
Based on the normal approximation of the binomial distribution:
| 1 |
where:
SD = standard deviation.
i = ideal (diagnostic) rate †
n = number of specimens interpreted.
† The ideal rate in this study is approximated by the group median rate.
The healthcare provider rate (pathologist diagnostic rate) is normalized as follows:
| 2 |
where:
Nj = healthcare provider rate for healthcare provider “j”.
Mj = measured rate for the healthcare provider “j”.
i = ideal (diagnostic) rate.
SDj = SD for healthcare provider “j”.
Equation (2) can be substituted into Eq. (1):
| 3 |
To normalize we presume that the “SD” is equivalent and that only “n” changes. This amounts to forming two equations from Eq. 3 and solving for the normed Mj. After some rearrangement one can derive a conversion equation:
| 4 |
where
Mj normed = normed (diagnostic) rate for healthcare provider “j”.
Mj measured = measured (diagnostic) rate for healthcare provider “j”.
nj normed = normed number of specimens handled (interpreted) by healthcare provider “j”.
nj measured = number of specimens handled (interpreted) by healthcare provider “j”.
i = ideal (diagnostic) rate.
Appendix D (Probability Calculation)
The probability (Y) of n and < n outliers in k samples is dependent on the probability of the individual outlier (p), and the binomial cumulative distribution function:
| 5 |
The probability (X) of n and > n outliers in this context is the complement of ‘n-1’:
| 6 |
Appendix E: Understanding statistical process control
Overview
Statistical process control is a cyclical process that involves:
Repeated measurement
Assessment of the variation in the measurement (using statistics)
Possible adjustments in the process to ensure that future measurements fall within a prescribed range, as defined by the so-called “control lines”
Statistical process control applied to diagnostic pathology at a conceptual level
In the diagnostic pathology context—if all the following are true:
Cases are assigned randomly to pathologists from a given population
Pathologists see large numbers of a particular type of case (e.g. 200 cases)
Then:
It is likely that the diagnostic rate (of say ‘tubular adenoma’) is similar for different pathologists (e.g. Pathologist ‘A’ diagnoses 102 tubular adenoma in 200 cases, Pathologist ‘B’ diagnosies 105 tubular adenomas in 200 cases) ****
**** Statistically, it can be stated that there is range in which the diagnostic rate (number of diagnoses/total cases interpreted) will fall 95% of the time.
If pathologists have significantly different diagnostic rates:
It is likely that they interpret cases differently and it may be possible to reduce diagnostic variation, via diagnostic calibration.
Diagnostic calibration is not new
Pathologists can change their diagnostic rates over time and may do this when (1) their cases are reviewed and found discrepant, (2) they review cases of (trusted) other (more experienced or subspeciality trained) pathologists, and (3) when new diagnostic entities are discovered or diagnostic criteria revised.
If a pathologist consistently (over time) has a diagnostic rate for tubular adenoma (e.g. 20/200 = 0.2) that is outside the range expected (in relation to the diagnostic rates of all their colleagues (~ 100/200 = 0.5)), one can infer that it would be possible to “re-train” that individual (or all the other pathologists) to arrive at the same diagnostic rate.
SPC in a nutshell is: systematically looking at the data (with statistics) to calibrate a process; in anatomical pathology it would be a process to look at diagnostic rates and feed those diagnostic rates back to the pathologists—such that they can adjust to a target rate/find agreement on what the target rate should be.
Statistical process control is predicated on two conditions
Condition (1) the process that one wants to control is stable over time (such that it is possible to predict the future) or can be made stable.
Condition (2) there is an ability to adjust the control variable * in a meaningful way—in relation to the control lines **.
* a control variable is: a parameter that one wants to control, e.g. the diagnostic rate of ‘sessile serrated adenoma’.
** control lines in SPC are determined by the “expected” statistical variation—when the process is in control/running optimally, e.g. the control parameter falls within a given range 95% of the times. Control limits are confidence intervals and are directly analogous to the funnel lines on funnel plots.
“Ability to adjust the control variable in a meaningful way” implies the following:
the variation (between providers) one expects to see due to chance is smaller than the (actual) variation that is observed; this implies that improvement is possible ***
an intervention can change the control variable in a substantive way, such that the variation is reduced
*** If the variation is less than the variation by chance the process is in control (or one may need a larger sample size).
The conditions for statistical process control and the objective of the manuscript
‘Condition 1’ for SPC is met if there is diagnostic stability.
‘Condition 2’ for SPC is met if there is significant diagnostic variation—that is stable (e.g. one pathologist is a consistent outlier in relation to the median diagnostic rate *****), and it is assumed that pathologists want to improve their practice/can be encouraged to make positive changes (see section “Diagnostic Calibration is Not New”).
‘Condition 1’ and ‘Condition 2’ are sufficient to infer that SPC should be feasible and could be used to improve care.
***** It should be noted that: the ‘median diagnostic rate’ may not be the ideal diagnostic rate for a given population. It is possible that an ‘outlier’ pathologist represents the ideal diagnostic rate.
In SPC, one talks of variation due to an “assignable cause” [a modifiable factor] and “common cause” [un-modifiable factors]. In the language of SPC, the question succinctly is: Is the pathologist an assignable cause?
If diagnostic rates are stable [Condition 1], and the pathologist is an “assignable cause” [Condition 2], SPC should be feasible.
Author contributions
M.B. conceived the study, did the programming, created the control charts and wrote the first draft of the manuscript. A.N., M.R. and M.B. audited the computer categorizations. J.S. and P.A. created the generalized linear model and independently examined the data. All authors interpreted the data and critically reviewed the manuscript. All authors approved the final manuscript.
Funding
The analysis work was supported by funds from St. Joseph’s Healthcare Hamilton Research.
Data availability
The data sets generated and/or analyzed during the current study are not publicly available due confidentially reasons but aggregate data is available from the corresponding author on request.
Competing interests
The first author (MB) retains the copyright on the computer code that was written outside of his employment relationship with McMaster University/St. Joseph’s Healthcare Hamilton/Hamilton Regional Laboratory Medicine Program. There is no financial conflict of interest. There are no conflicts for the other authors.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-95862-2.
References
- 1.Raab SS. Variability of practice in anatomic pathology and its effect on patient outcomes. Semin Diagn Pathol. 2005;22:177–185. doi: 10.1053/j.semdp.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Renshaw AA. Measuring and reporting errors in surgical pathology. Lessons from gynecologic cytology. Am. J. Clin. Pathol. 2001;115:338–341. doi: 10.1309/M2XP-3YJA-V6E2-QD9P. [DOI] [PubMed] [Google Scholar]
- 3.Naqvi A, Mathew J, Bonert M. The automatic extraction and categorisation of 22,760 stomach biopsy specimen parts from 110,970 free text pathology reports to assess Helicobacter pylori diagnostic rates. Virchows Archiv. 2017;471:S33. [Google Scholar]
- 4.Bonert M, Ramadan S, Naqvi A, Cutz J, Finley C. Assessing the healthcare provider factor and anatomical site in 4,381 thoracic lymph node fine needle aspirations using funnel plots/control charts and logistic regression. Virchows Archiv. 2018;473:S39–S40. [Google Scholar]
- 5.Fisher LM, Humphries BL. Statistical quality control of rabbit brain thromboplastin for clinical use in the prothrombin time determination. Am. J. Clin. Pathol. 1966;45:148–152. doi: 10.1093/ajcp/45.2.148. [DOI] [PubMed] [Google Scholar]
- 6.Thor J, et al. Application of statistical process control in healthcare improvement: Systematic review. Qual. Saf. Health Care. 2007;16:387–399. doi: 10.1136/qshc.2006.022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolesar PJ. The relevance of research on statistical process control to the total quality movement. J. Eng. Tech. Manag. 1993;10:317–338. doi: 10.1016/0923-4748(93)90027-G. [DOI] [Google Scholar]
- 8.Octave, G. Scientific Programming Language, http://www.gnu.org/software/octave/ (2018).
- 9.Dmetrichuk, J. M., Bonert, M., Crowther, M., Morgan, D. & Naqvi, A. Next generation quality: an automated assessment of 11,364 large bowel polyp specimens for adenomas with and without a villous component stratified by the endoscopist and pathologist. In CAP-ACP (Charlottetown, PEI, Canada (2017)).
- 10.Naqvi, A., Al-Haddad, S., DMetrichuk, J. M. & Bonert, M. Increasing the consistency of sessile serrated adenoma call a in cohort of 7,054 colorectal polyps using next generation quality with funnel plots and an expert-led group case review. Mod. Pathol.31(2), 784.
- 11.Bonert M, et al. Next generation quality: assessing the physician in clinical history completeness and diagnostic interpretations using funnel plots and normalized deviations plots in 3,854 prostate biopsies. J. Pathol. Inform. 2017;8:43. doi: 10.4103/jpi.jpi_50_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Su HK, et al. Inter-observer variation in the pathologic identification of minimal extrathyroidal extension in papillary thyroid carcinoma. Thyroid. 2016;26:512–517. doi: 10.1089/thy.2015.0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y, Anderson KR, Xu J, Goldsmith JD, Heher YK. Frozen-section checklist implementation improves quality and patient safety. Am. J. Clin. Pathol. 2019;151(6):607–612. doi: 10.1093/ajcp/aqz009. [DOI] [PubMed] [Google Scholar]
- 14.Weiser, T. G. et al. Effect of a 19-item surgical safety checklist during urgent operations in a global patient population. Ann. Surg. 251(5):976–80 (2010) [DOI] [PubMed]
- 15.Bonert M, et al. Application of next generation quality/statistical process control and expert-led case review to increase the consistency of diagnostic rates in precancerous colorectal polyps. Qual. Manag. Health Care. 2021;30:176–183. doi: 10.1097/QMH.0000000000000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets generated and/or analyzed during the current study are not publicly available due confidentially reasons but aggregate data is available from the corresponding author on request.