Abstract
Stenotrophomonas sepilia strain SM16975 (= JCM 32102; = KCTC 62052) is a new species isolated from the blood culture of a hospitalized patient. The biochemical characterization, phenotypic criteria, phylogenomic reconstruction, and genomic analysis were carried out to differentiate it from its phylogenetic neighbours, establishing novel species status in the genus Stenotrophomonas and within Stenotrophomonas maltophilia complex (Smc).
Keywords: Blood culture isolate, Stenotrophomonas sepilia sp. nov., Taxonogenomics
Abbreviations: ANI, average nucleotide identity; dDDH, Digital DNA-DNA hybridization; Smc, Stenotrophomonas maltophilia complex
Introduction
The genus Stenotrophomonas was described with S. maltophilia as type species, which was earlier isolated from the pleural fluid and named as Bacterium bookeri, which was further classified as Pseudomonas maltophilia [1,2]. Further, it was transferred to the genus Xanthomonas [3] and finally to the new genus Stenotrophomonas [1]. At the time of writing, the genus Stenotrophomonas comprises 18 species with validly published names isolated from a large range of natural and artificial environments and geographical regions (https://lpsn.dsmz.de/genus/stenotrophomonas). These 18 species include S. maltophilia, S. africana, S. nitritireducens [4], S. acidaminiphila [5], S. rhizophila [6], S. koreensis [7], S. humi, S. terrae [8], S. chelatiphaga [9], S. ginsengisoli [10], S. panacihumi [11], S. daejeonensis [12], S. pavanii [13], S. tumulicola [14], S. bentonitica [15], S. pictorum [16], S. lactitubi and S. indicatrix [17].
The genus Stenotrophomonas is one of the rapidly expanding genera having biotechnological importance with only the exception of S. maltophilia, which has globally emerged as a multi-drug resistant opportunistic pathogen [18,19]. Due to 16S rRNA gene sequence conservation, taxonomy of S. maltophilia is complicated. Apart from the validly described species, there are misclassified species also that are associated with the genus Stenotrophomonas and include Pseudomonas hibiscicola, P. beteli, and P. geniculata, which are considered as synonyms of S. maltophilia [20]. Along with these misclassified species, S. maltophilia, S. africana, and S. pavanii belong to the S. maltophilia complex (Smc), which is a group of closely related species that cannot be resolved by 16S rRNA gene-based phylogeny. Although rep-PCR, gyrB and multi-locus sequence typing have revealed high diversity in strains of S. maltophilia [[20], [21], [22], [23]], they lack in resolving true phylogeny of Stenotrophomonas.
The advent of the genomic era has radically transformed our understanding of bacterial taxonomy and evolution. Taxonogenomics and phylogenomics provide us intra-species and strain level resolution. Genome-derived criteria such as average nucleotide identity (ANI), average amino acid identity (AAI), and digital DNA-DNA hybridization (dDDH) with species-level cut-offs of 95%, 95%, and 70%, respectively [[24], [25], [26], [27]]. Genome resources of the family Lysobacteraceae can help better to resolve and demarcate species of the genus Stenotrophomonas [28,29]. As a part of our previous study, we aimed to study the strain level diversity of clinical isolates of S. maltophilia from hospitalized patients by whole-genome sequencing and taxonogenomic studies using reference strains of the genus Stenotrophomonas [30]. On the basis of species delineation whole-genome similarity parameters that included ANI and dDDH cut-offs, we found five cryptic novel genomospecies (genomospecies 2–6) clustering along with S. maltophilia (genomospecies 1) that further constitutes the S. maltophilia complex (Smc). The strain SM16975T was part of that study, and based on the phylogenomics, ANI, and dDDH, it is considered as the putative novel genomospecies of the Smc along with the eight other isolates, which were designated under genomospecies 3. Genomospecies 3 is the second most dominant after genomospecies 1, which includes S. maltophilia among the total isolates under study. We have also generated the whole-genome resource of type strains of the genus Stenotrophomonas [31], which can be used for genome-based classification. Herein, we are describing genomospecies 3 as novel species of the genus Stenotrophomonas.
Isolation and growth conditions
The strain SM16975T was isolated from the blood specimen of a 28-year-old male patient admitted in cardiothoracic vascular surgery intensive care unit of a tertiary care hospital, Post Graduate Institute of Medical Education and Research (PGIMER), Chandigarh, Northern part of India (30.7650° N, 76.7750° E), in September 2012. The strain SM16975T was recovered from a blood specimen by using a continuous monitoring blood culture system BACTEC 9240 (BD Diagnostics, USA). Out of 46,243 blood cultures performed from April 2012 to March 2013, a total of 33 isolates of S. maltophilia were obtained [32].
Phenotypic and biochemical characteristics
Colonies were large, smooth, convex, glistening, circular with lavender-green pigment on blood agar. Colourless colonies were obtained on MacConkey agar as it is a non-lactose fermenting organism [33].
Bacterial cells were Gram-negative, rod-shaped, motile, catalase-positive, and oxidase-negative. Antibiotic susceptibility profile was determined on Mueller–Hinton agar (Oxoid) by Kirby Bauer disk diffusion method according to the breakpoints of the Clinical and Laboratory Standards Institute (CLSI) (http://www.clsi.org) approved standards. [CLSI. 2020. Performance Standards for Antimicrobial Susceptibility Testing. 30th ed. CLSI M100. Clinical and Laboratory Standards Institute, Wayne, PA.] The strain SM16975T was sensitive to co-trimoxazole, levofloxacin, minocycline, and resistant to ceftazidime by MIC.
Bacteria display extensive diversity in fatty acid chains and fatty acids are major chemotaxonomic markers. The fatty acid analysis is particularly useful in the case of closely related species with uniform phenotypic characteristics [34]. As SM16975T is closely related to S. maltophilia and was originally miss-classified as S. maltophilia, we use fatty acid analysis to inspect the novel position of SM16975T. For fatty acid analysis, strains were grown on tryptic soy broth agar (TSBA) medium for 24 hours at 37 ˚C. Total fatty acids of cells were separated from a loopful of culture as methyl esters using the method as described (Buyer 2002). Like Smc type strains, the fatty acid profile of SM16975T has predominated with unsaturated fatty acids iso-C15:0 and anteiso-C15 (Table 1). Further, iso-C17: 0, iso-C13:0 3-OH, summed features 3 and 8 were present in significant amounts in SM16975T as compared to other strains of Smc. However, C16:1 w9c was found in low quantity in SM16975T as compared to other Smc strains.
Table 1.
Cellular fatty acid profile of strain SM16975T and its comparison to the other type strains of related species in the genus Stenotrophomonas
| Fatty acid | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| Straight chain fatty acid | ||||||
| C10:0 | 0.68 | 1.49 | 0.89 | 0.87 | 0.94 | 1.81 |
| C14:0 | 2.43 | 3.58 | 4.57 | 2.69 | 3.05 | 4.51 |
| C16:0 | 10.83 | 12.14 | 12.88 | 7.29 | 11.96 | 12.35 |
| C18:0 | 0.32 | 2.12 | 0.61 | 0.59 | 0.63 | 0.33 |
| Branched fatty acid | ||||||
| Iso C11:0 | 3.45 | 3.04 | 2.56 | 2.64 | 2.77 | 3.35 |
| Iso C13:0 | 0.36 | 0.21 | 0.34 | 0.2 | 0.35 | 0.41 |
| Iso C14:0 | 0.7 | 0.35 | 0.88 | 0.46 | 0.59 | 0.42 |
| Anteiso C11:0 | 0.12 | 0.13 | 0.1 | 0.14 | 0.20 | 0.21 |
| Anteiso C13:0 | 0.22 | - | 0.14 | - | 0.13 | 0.3 |
| Iso C15:0 | 36.92 | 23.47 | 26.19 | 26.85 | 26.48 | 21.98 |
| Iso C15:1 F | 0.63 | 0.82 | 0.91 | 0.62 | 0.67 | 0.64 |
| Anteiso-C15:0 | 15.42 | 10.16 | 11.62 | 16.60 | 15.67 | 14.45 |
| Iso C16:0 | 1.69 | 0.68 | 1.5 | 1.07 | 1.46 | 0.82 |
| Iso C17:0 | 4.27 | 2.91 | 2.38 | 3.53 | 3.71 | 2.30 |
| Anteiso-C17:0 | 0.54 | 0.39 | 0.3 | 0.77 | 0.76 | 0.53 |
| Iso C19:0 | 0.27 | 0.32 | 0.10 | 0.3 | 0.35 | |
| Unsaturated fatty acid | ||||||
| C14:1 w5c | 0.1 | 0.22 | 0.13 | 0.17 | 0.12 | 0.26 |
| C16:1 W9c | 1.49 | 5.66 | 5.15 | 4.59 | 4.03 | 6.59 |
| C17:1ω8c | 0.13 | 0.24 | 0.24 | 0.29 | ||
| C18:1ω9c | 1.06 | 3.21 | 1.88 | 2.37 | 2.77 | 1.91 |
| C18:3 W6c | - | - | - | 0.72 | - | - |
| Hydroxy fatty acid | ||||||
| C10:0 3-OH | 0.13 | 0.19 | 0.24 | - | 0.06 | 0.06 |
| C12:0 3-OH | 2.32 | 3.05 | 3.38 | 1.46 | 1.71 | 1.06 |
| C13:0 2-OH | 0.51 | 0.24 | 0.4 | 0.3 | 0.39 | |
| Iso C11:0 3-OH | 1.75 | 1.14 | 1.32 | 0.75 | 0.78 | 0.66 |
| iso-C13:0 3-OH | 2.75 | 1.69 | 1.94 | 1.01 | 1.88 | 0.38 |
| Summed feature | ||||||
| 3 | 6.18 | 17.68 | 14.04 | 17.42 | 11.79 | 18.32 |
| 8 | 0.56 | 1.57 | 1.01 | 1.7 | 1.42 | 0.98 |
| 9 | 3.47 | 3.55 | 3.83 | 4.03 | 3.98 | 2.97 |
1. SM16975T; 2. S. maltophilia ATCC 13637T; 3. P. geniculata JCM 13324T; 4. Pseudomonas hibiscicola JCM 13361T; 5. S. pavanii DSM 25135T; 6. P. beteli LMG 00978T. Summed feature 3 comprises 16:1 w7c/16:1 w6c, summed feature 8 comprises 18:1 w7c, and summed feature 9 comprises 17:1 iso w9c.
Biochemical characterization such as carbohydrate utilization, acid production, and various enzymatic activities was performed using BIOLOG GEN III MICROPLATETM on OMNILOG GEN III system (BIOLOG) according to manufacturer’s instructions (Table 2). Strains belonging to Smc along with SM16975T were investigated. Smc strains are able to utilize Dextrin, D-maltose, D-cellobiose, gentiobiose, N-acetyl-D-glucosamine, N-acetyl-D-galactosamine, α-D-glucose, D-mannose, D-fructose, 1% sodium lactate, D-serine, D-fructose-6-phosphate, gelatin, glycyl-L - proline, L-lactic acid, citric acid, L-malic acid, bromo-succinic acid, propionic acid, acetic acid. Strains were resistant to antibiotics like rifamycin SV, troleandomycin, lincomycin, vancomycin, aztreonam.
Table 2.
Comparison of biochemical characteristics of strain SM16975T and the type strains of related species in the genus Stenotrophomonas
| 1 | 2 | 3 | 4 | 5 | 6 | |
|---|---|---|---|---|---|---|
| Dextrin | + | + | + | + | + | + |
| D-Maltose | + | + | + | + | + | + |
| D-Trehalose | - | + | + | - | + | + |
| D-Cellobiose | + | + | + | + | + | + |
| Gentiobiose | + | + | + | + | + | + |
| Sucrose | + | + | + | - | + | + |
| D-Turanose | - | - | + | - | + | + |
| Stachyose | - | - | - | - | - | - |
| pH 6 | + | + | + | + | + | + |
| pH 5 | + | - | - | - | - | - |
| D-Raffinose | - | - | - | - | - | - |
| α-D-Lactose | - | - | + | - | - | + |
| D-Melibiose | + | + | + | + | - | + |
| β-Methyl-D-Glucoside | + | - | + | - | + | + |
| D-Salicin | + | - | + | + | - | + |
| N-Acetyl-D-Glucosamine | + | + | + | + | + | + |
| N-Acetyl-β-D-Mannosamine | - | + | - | + | - | - |
| N-Acetyl-D-Galactosamine | + | + | + | + | + | + |
| N-Acetyl Neuraminic Acid | - | - | - | - | - | - |
| 1% NaCl | + | + | + | + | + | + |
| 4% NaCl | + | + | + | + | + | + |
| α-D-Glucose | + | + | + | + | + | + |
| D-Mannose | + | + | + | + | + | + |
| D-Fructose | + | + | + | + | + | + |
| D-Galactose | - | + | - | + | - | - |
| 3-Methyl Glucose | - | - | - | - | - | - |
| D-Fucose | + | + | - | + | + | - |
| L-Fucose | + | + | - | + | - | - |
| L-Rhamnose | - | - | - | + | - | - |
| Inosine | - | - | - | - | - | - |
| 1% Sodium Lactate | + | + | + | + | + | + |
| Fusidic Acid | - | + | + | + | - | - |
| D-Serine | + | + | + | + | + | + |
| D-Sorbitol | - | - | - | - | - | - |
| D-Mannitol | - | - | - | - | - | - |
| D-Arabitol | - | - | - | - | - | - |
| myo-Inositol | - | - | - | - | - | - |
| Glycerol | - | - | - | - | - | - |
| D-Glucose6-PO4 | - | + | + | + | - | - |
| D-Fructose6-PO4 | + | + | + | + | + | + |
| D-Aspartic Acid | - | - | - | - | - | - |
| D-Serine | - | + | - | + | - | - |
| Troleandomycin | + | + | + | + | + | + |
| Rifamycin SV | + | + | + | + | + | + |
| Minocycline | - | - | - | - | - | - |
| Gelatin | + | + | + | + | + | + |
| Glycyl-L-Proline | + | + | + | + | + | + |
| L-Alanine | + | + | + | + | + | + |
| L-Arginine | - | + | - | + | - | - |
| L-Aspartic Acid | - | + | - | + | - | - |
| L-Glutamic Acid | - | + | - | + | - | - |
| L-Histidine | - | + | - | + | + | - |
| L-Pyroglutamic acid | - | - | - | - | - | - |
| L-Serine | + | + | + | + | + | + |
| Lincomycin | + | + | + | + | + | + |
| Guanidine HCl | + | + | + | + | + | + |
| Niaproof 4 | + | + | + | + | + | + |
| Pectin | - | - | - | + | + | - |
| D-Galacturonic acid | + | + | + | + | + | + |
| L-Galactonic Acid Lactone | - | - | - | + | - | - |
| D-Gluconic Acid | - | - | - | + | - | - |
| D-Glucuronic acid | + | + | + | - | + | + |
| Glucuronamide | + | + | + | + | + | + |
| Mucic Acid | - | + | - | + | - | - |
| Quinic Acid | - | - | - | + | - | - |
| D-saccharic Acid | - | - | - | + | - | - |
| Vancomycin | + | + | + | + | + | + |
| Tetrazolium Violet | + | + | + | + | + | + |
| Tetrazolium blue | + | + | + | + | + | + |
| p-hydroxy phenylacetic acid | - | - | - | - | - | - |
| Methyl Pyruvate | + | + | + | + | - | + |
| D-Lactic Acid Methyl Ester | - | + | - | + | - | - |
| L-lactic Acid | + | + | + | + | + | + |
| Citric Acid | + | + | + | + | + | + |
| α-Keto-Glutaric acid | + | + | + | + | + | + |
| D-Malic Acid | - | + | - | + | - | - |
| L-Malic Acid | + | + | + | + | + | + |
| Bromo-Succinic acid | + | + | + | + | + | + |
| Nalidixic Acid | - | - | + | + | + | + |
| Lithium Chloride | + | + | + | + | + | + |
| Potassium tellurite | - | - | - | - | - | - |
| Tween 40 | + | + | + | + | + | + |
| γ-Amino-Butryric acid | - | - | - | - | - | - |
| α-HydroxyButyric Acid | + | + | - | + | - | - |
| β-Hydroxy-D, LButyric Acid | - | - | - | + | - | - |
| α-Keto-Butyric acid | + | + | - | + | - | - |
| Acetoacetic Acid | + | + | - | + | - | - |
| Propionic Acid | + | + | + | + | + | + |
| Acetic Acid | + | + | + | + | + | + |
| Formic Acid | - | - | - | + | - | - |
| Aztreonam | + | + | + | + | + | + |
| Sodium Butyrate | + | + | + | + | + | + |
| Sodium Bromate | - | - | - | - | - | - |
1. SM16975T, 2. Pseudomonas hibiscicola JCM 13361T, 3. P. geniculata JCM 13324T, 4. S. maltophilia ATCC 13637T, 5. P. beteli LMG 00978T, 6. S. pavanii DSM 25135T Symbols represents; ‘+’: positive, ‘-’: negative.
Strain identification and phylogenomics
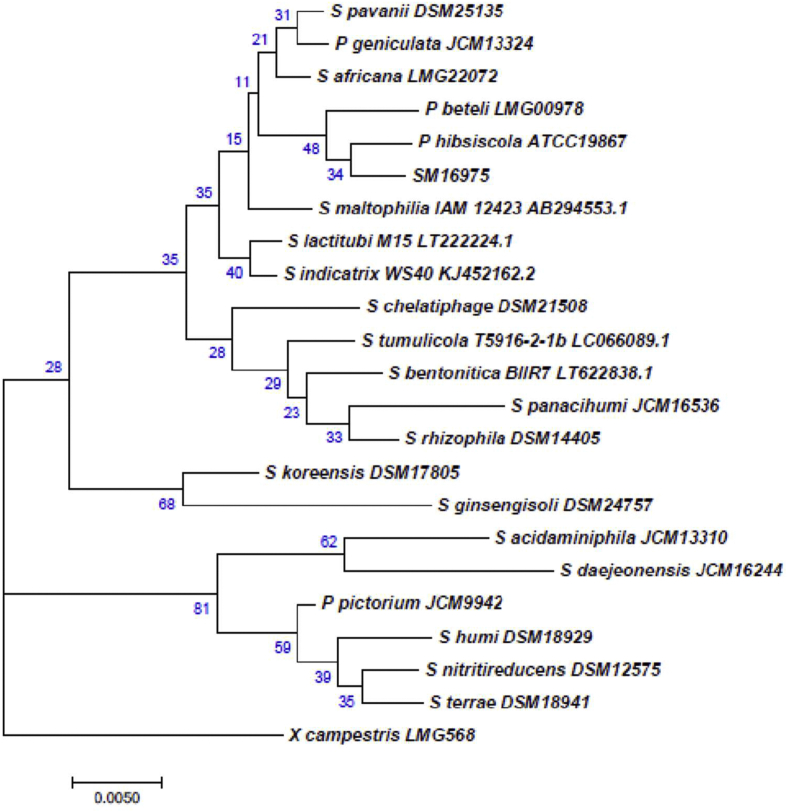
In an earlier study, we reported the genome of strain SM16975T with NCBI Accession number LXXZ00000000 [30]. The genome is 4,582,512 bp long with a 66.4 % G + C content. The complete 16S rRNA gene was extracted from the genome by using RNAmmer [35], and the phylogenetic tree was constructed with all the other validly defined species along with misclassified species of the genus Stenotrophomonas. Further, 16S rRNA of SM16975T had99.73%, 99.58%, and 99.52% identities with S. pavanii, P. beteli, and S. maltophilia, respectively. The 16S rRNA-based identity and phylogenetic analysis suggested that the strain SM16975T belongs to the Smc of genus Stenotrophomonas (Fig. 1).
Fig. 1.
Maximum-likelihood phylogenetic tree based on the alignment of complete 16s rRNA gene sequence of the Strain SM16975T with type strains of species of the genus Stenotrophomonas, as well as species misclassified as members of the genus Pseudomonas.
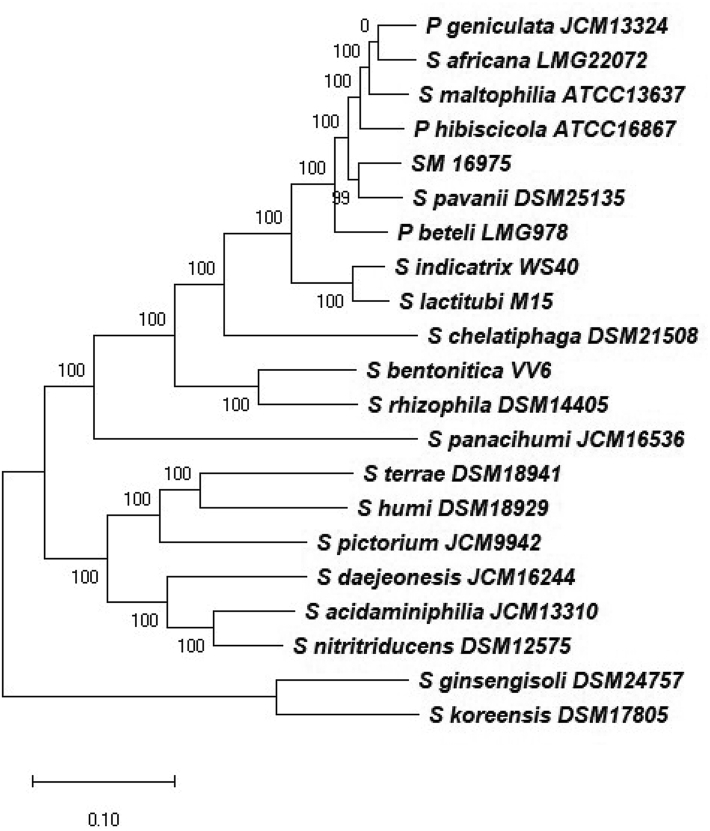
For more detailed phylogenetic analysis, we constructed core genome-based phylogeny using the PhyML v3.0 [36], which uses the core genome fetched using the Mauve v20150226 [37] to perform phylogenetic construction. The Phylogenetic tree obtained using PhyML [36] with the type strains of the genus Stenotrophomonas placed the strain SM16975T into a separate clade Fig. 2.
Fig. 2.
Maximum-likelihood phylogenetic tree based on the alignment of the core genome of the Strain SM16975T with the type strains of species of the genus Stenotrophomonas, as well as species misclassified as members of the genus Pseudomonas.
Taxonogenomics
The degree of genome similarity of strain SM16975T with the closely related species of genus Stenotrophomonas was estimated using average nucleotide identity (ANI) [38] using Jspecies [24], orthoANI using OAU [39], Average amino acid identity (AAI) using CompareM (https://github.com/dparks1134/CompareM) and digital DNA-DNA Hybridization [40] using GGDC online server (https://ggdc-test.dsmz.de/ggdc_background.php). The similarity values of ANI, orthoANI, AAI, and dDDH were 91.98%, 92.15%, 94.49%, and 46.4%, respectively (Table 3). It is pertinent to mention that values for all the taxonogenomic parameters were below the recommended cut-off values, depicting the novelty of the strain S. sepilia SM16975T.
Table 3.
Genome-based comparisons of SM16975T and other type strains of members of the genus Stenotrophomonas and Pseudomonas (misclassified)
| Reference strain | ANI (%) | OrthoANI (%) | AAI (%) | GDDC (%) |
|---|---|---|---|---|
| Pseudomonas hibiscicola ATCC 19867T | 91.98 | 92.15 | 94.49 | 46.4 |
| Stenotrophomonas pavanii DSM 25135T | 91.96 | 92.22 | 94.61 | 46.6 |
| Pseudomonas geniculata JCM 13324T | 91.13 | 91.48 | 93.91 | 44.2 |
| Stenotrophomonas maltophilia ATCC 13637T | 91.11 | 91.37 | 94.07 | 44.3 |
| Stenotrophomonas africana LMG22072T | 90.64 | 91.15 | 93.59 | 43.2 |
| Pseudomonas beteli LMG 978T | 90.57 | 90.78 | 93.39 | 41.8 |
| Stenotrophomonas lactitubi DSM 104152T | 86.49 | 87.1 | 88.29 | 32.1 |
| Stenotrophomonas indicatrix DSM28278T | 86.42 | 86.95 | 88.55 | 31.9 |
| Stenotrophomonas chelatiphaga DSM 21508T | 81.52 | 82.29 | 69.31 | 25.4 |
| Stenotrophomonas rhizophila DSM 14405T | 81.11 | 91.64 | 79.49 | 24.5 |
| Stenotrophomonas bentonitica LMG 29893T | 80.6 | 81.62 | 77.64 | 25 |
| Stenotrophomonas daejeonensis JCM 16244T | 79.79 | 80.35 | 76.05 | 23.6 |
| Stenotrophomonas acidaminiphila JCM 13310T | 79.49 | 80.28 | 75.81 | 23.8 |
| Stenotrophomonas pictorum JCM 9942T | 78.56 | 79.21 | 76.03 | 22.6 |
| Stenotrophomonas panacihumi JCM 16536T | 77.95 | 78.89 | 75.53 | 22.3 |
| Stenotrophomonas terrae DSM 18941T | 77.8 | 78.81 | 72.28 | 22.7 |
| Stenotrophomonas humi DSM 18929T | 77.09 | 78.72 | 75.11 | 22.5 |
| Stenotrophomonas nitritireducens DSM 12575T | 76.69 | 80.55 | 75.89 | 23.8 |
| Stenotrophomonas ginsengisoli DSM 24757T | 76.05 | 77.16 | 70.47 | 21.2 |
| Stenotrophomonas koreensis DSM 17805T | 76.01 | 76.71 | 70.07 | 21.3 |
Stenotrophomonas sepilia (sepilia referring to sepsis in the given patient known to be caused by this organism and isolated from blood specimen). Based on differential biochemical tests, fatty acid composition, along with genome-based taxonomic criteria, we propose to classify this strain as a member of a new species within the genus Stenotrophomonas, family Lysobacteraceae, phylum Proteobacteria.
Furthermore, S. sepilia is the third clinical species apart from S. maltophilia and S. africana to be formally reported from the genus. As revealed in earlier studies [30,31,41], it is pertinent to formally start referring S. maltophilia and closely related S. geniculata, S. africana, S. maltophilia, P. hibiscicola, S. sepilia, S. pavanii that form a monophyletic clade based on core genome phylogeny as Stenotrophomonas maltophilia complex (Smc).
Conclusion
Based on all the phenotypic, biochemical, and genomic tests performed on the bacterial strain, we conclude that strain SM16975T belongs to the genus Stenotrophomonas. The taxonogenomic evidence from this study, such as the sequence similarity obtained using ANI, orthoANI, AAI, and dDDH were less than the defined cut-offs for species delineation. Therefore, SM16975 T is proposed as the type strain of the new species Stenotrophomonas sepilia sp. nov.
Nucleotide sequence accession number
The genome sequences were deposited in Genbank under accession number LXXZ00000000.
Deposit in culture collections
Strain SM16975T was deposited in two different strain collections under the numbers (= JCM 32102; = KCTC 62052).
Transparency declaration
The authors declare that there are no conflicts of interest.
Funding sources
We acknowledge funding from the Council of Scientific and Industrial research (CSIR) to PBP as Niche Centric Project (MLP0016) titled ‘GEAR- Genomic, Evolutionary, and Big Data Analytic strategies to address antimicrobial resistance’.
Ethical statement
No experiments with humans or animals were carried out. The patient consent form is not required as the strain SM16975T has been isolated as a part of routine diagnostic services.
Acknowledgements
We acknowledge all staff in the clinical microbiology unit of PGIMER for their help with isolation of the strain SM16975T.
Contributor Information
V. Gautam, Email: r_vg@yahoo.co.uk.
P.B. Patil, Email: pbpatil@imtech.res.in.
References
- 1.Palleroni N.J., Bradbury J.F. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int J Syst Evol Microbiol. 1993;43(3):606–609. doi: 10.1099/00207713-43-3-606. [DOI] [PubMed] [Google Scholar]
- 2.Hugh R., Ryschenkow E. Pseudomonas maltophilia, an alcaligenes-like species. Microbiology. 1961;26(1):123–132. doi: 10.1099/00221287-26-1-123. [DOI] [PubMed] [Google Scholar]
- 3.Swings J., De Vos P., den Mooter M.V., De Ley J. Transfer of Pseudomonas maltophilia Hugh 1981 to the genus Xanthomonas as Xanthomonas maltophilia (Hugh 1981) comb. nov. Int J Syst Evol Microbiol. 1983;33(2):409–413. [Google Scholar]
- 4.Finkmann W., Altendorf K., Stackebrandt E., Lipski A. Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov. and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. Int J Syst Evol Microbiol. 2000;50(1):273–282. doi: 10.1099/00207713-50-1-273. [DOI] [PubMed] [Google Scholar]
- 5.Assih E.A., Ouattara A.S., Thierry S., Cayol J.-L., Labat M., Macarie H. Stenotrophomonas acidaminiphila sp. nov., a strictly aerobic bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int J Syst Evol Microbiol. 2002;52(2):559–568. doi: 10.1099/00207713-52-2-559. [DOI] [PubMed] [Google Scholar]
- 6.Wolf A., Fritze A., Hagemann M., Berg G. Stenotrophomonas rhizophila sp. nov., a novel plant-associated bacterium with antifungal properties. Int J Syst Evol Microbiol. 2002;52(6):1937–1944. doi: 10.1099/00207713-52-6-1937. [DOI] [PubMed] [Google Scholar]
- 7.Yang H.-C., Im W.-T., Kang M.S., Shin D.-Y., Lee S.-T. Microbiology e. Stenotrophomonas koreensis sp. nov., isolated from compost in South Korea. Int J Syst Evol Microbiol. 2006;56(1):81–84. doi: 10.1099/ijs.0.63826-0. [DOI] [PubMed] [Google Scholar]
- 8.Heylen K., Vanparys B., Peirsegaele F., Lebbe L., De Vos P. Stenotrophomonas terrae sp. nov. and Stenotrophomonas humi sp. nov., two nitrate-reducing bacteria isolated from soil. Int J Syst Evol Microbiol. 2007;57(9):2056–2061. doi: 10.1099/ijs.0.65044-0. [DOI] [PubMed] [Google Scholar]
- 9.Kaparullina E., Doronina N., Chistyakova T., Trotsenko Y.J.S. Microbiology a. Stenotrophomonas chelatiphaga sp. nov., a new aerobic EDTA-degrading bacterium. Int J Syst Evol Microbiol. 2009;32(3):157–162. doi: 10.1016/j.syapm.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Kim H.-B., Srinivasan S., Sathiyaraj G., Quan L.-H., Kim S.-H., Bui T.P.N. Stenotrophomonas ginsengisoli sp. nov., isolated from a Ginseng Field. Int J Syst Evol Microbiol. 2010;60(7):1522–1526. doi: 10.1099/ijs.0.014662-0. [DOI] [PubMed] [Google Scholar]
- 11.Yi H., Srinivasan S., Kim M.K.J.T. Stenotrophomonas panacihumi sp. nov., isolated from soil of a Ginseng Field. Int J Syst Evol Microbiol. 2010;48(1):30–35. doi: 10.1007/s12275-010-0006-0. [DOI] [PubMed] [Google Scholar]
- 12.Lee M., Woo S.-G., Chae M., Shin M.-C., Jung H.-M., Ten LNJIjos Stenotrophomonas daejeonensis sp. nov., isolated from Sewage. Int J Syst Evol Microbiol. 2011;61(3):598–604. doi: 10.1099/ijs.0.017780-0. [DOI] [PubMed] [Google Scholar]
- 13.Ramos P.L., Van Trappen S., Thompson F.L., Rocha R.C., Barbosa H.R., De Vos P. Screening for endophytic nitrogen-fixing bacteria in Brazilian sugar cane varieties used in organic farming and description of Stenotrophomonas pavanii sp. nov. Int J Syst Evol Microbiol. 2011;61(4):926–931. doi: 10.1099/ijs.0.019372-0. [DOI] [PubMed] [Google Scholar]
- 14.Handa Y., Tazato N., Nagatsuka Y., Koide T., Kigawa R., Sano C. Stenotrophomonas tumulicola sp. nov., a major contaminant of the stone chamber interior in the Takamatsuzuka Tumulus. Int J Syst Evol Microbiol. 2016;66(3):1119–1124. doi: 10.1099/ijsem.0.000843. [DOI] [PubMed] [Google Scholar]
- 15.Sánchez-Castro I., Ruiz-Fresneda M.A., Bakkali M., Kämpfer P., Glaeser S.P., Busse H.J. Stenotrophomonas bentonitica sp. nov., isolated from bentonite formations. Int J Syst Evol Microbiol. 2017;67(8):2779–2786. doi: 10.1099/ijsem.0.002016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ouattara A.S., Le Mer J., Joseph M., Macarie H. Transfer of Pseudomonas pictorum Gray and Thornton 1928 to genus Stenotrophomonas as Stenotrophomonas pictorum comb. nov., and emended description of the genus Stenotrophomonas. Int J Syst Evol Microbiol. 2017;67(6):1894–1900. doi: 10.1099/ijsem.0.001880. [DOI] [PubMed] [Google Scholar]
- 17.Weber M., Schünemann W., Fuß J., Kämpfer P., AJ Lipski. Stenotrophomonas lactitubi sp. nov. and Stenotrophomonas indicatrix sp. nov., isolated from surfaces with food contact. Int J Syst Evol Microbiol. 2018;68(6):1830–1838. doi: 10.1099/ijsem.0.002732. [DOI] [PubMed] [Google Scholar]
- 18.Ryan R.P., Monchy S., Cardinale M., Taghavi S., Crossman L., Avison M.B. The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nature Rev Microbiol. 2009;7(7):514. doi: 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- 19.Kumar S., Bansal K., Patil P.P., Kaur A., Kaur S., Jaswal V. Genomic insights into evolution of extensive drug resistance in Stenotrophomonas maltophilia complex. Genomics. 2020;112(6):4171–4178. doi: 10.1016/j.ygeno.2020.06.049. [DOI] [PubMed] [Google Scholar]
- 20.Anzai Y., Kim H., Park J.-Y., Wakabayashi H., Oyaizu H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol. 2000;50(4):1563–1589. doi: 10.1099/00207713-50-4-1563. [DOI] [PubMed] [Google Scholar]
- 21.Lin C.W., Chiou C.S., Chang Y.C., Yang T.C. Comparison of pulsed-field gel electrophoresis and three rep-PCR methods for evaluating the genetic relatedness of Stenotrophomonas maltophilia isolates. Lett Appl Microbiol. 2008;47(5):393–398. doi: 10.1111/j.1472-765X.2008.02443.x. [DOI] [PubMed] [Google Scholar]
- 22.Kaiser S., Biehler K., Jonas D. A Stenotrophomonas maltophilia multilocus sequence typing scheme for inferring population structure. J Bacteriol. 2009;191(9):2934–2943. doi: 10.1128/JB.00892-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vidigal P.G., Dittmer S., Steinmann E., Buer J., Rath P.-M., Steinmann J. Adaptation of Stenotrophomonas maltophilia in cystic fibrosis: molecular diversity, mutation frequency and antibiotic resistance. Int J Med Microbiol. 2014;304(5–6):613–619. doi: 10.1016/j.ijmm.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Richter M., Rosselló-Móra R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Nat Acad Sci. 2009;106(45):19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Auch A.F., von Jan M., Klenk H.-P., Göker M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand Genomic Sci. 2010;2(1):117–134. doi: 10.4056/sigs.531120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konstantinidis K.T., Tiedje J.M. Genomic insights that advance the species definition for prokaryotes. Proc Nat Acad Sci. 2005;102(7):2567–2572. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson C.C., Chimetto L., Edwards R.A., Swings J., Stackebrandt E., Thompson FL. Microbial genomic taxonomy. BMC Genomics. 2013;14: 913 doi: 10.1186/1471-2164-14-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar S., Bansal K., Patil P.P., Patil P.B. Phylogenomics insights into order and families of Lysobacterales. Access Microbiol. 2019;1(2) doi: 10.1099/acmi.0.000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bansal K., Kumar S., Kaur A., Sharma S., Patil P., Patil P.B. Deep phylo-taxono-genomics (DEEPT genomics) reveals misclassification of Xanthomonas species complexes into Xylella, Stenotrophomonas and Pseudoxanthomonas. bioRxiv. 2020 [Google Scholar]
- 30.Patil P.P., Kumar S., Midha S., Gautam V., Patil P.B. Taxonogenomics reveal multiple novel genomospecies associated with clinical isolates of Stenotrophomonas maltophilia. Microb Genomics. 2018;4(8) doi: 10.1099/mgen.0.000207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patil P.P., Midha S., Kumar S., Patil P.B. Genome sequence of type strains of genus Stenotrophomonas. Front Microbiol. 2016;7:309. doi: 10.3389/fmicb.2016.00309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gautam V., Kumar S., Kaur P., Deepak T., Singhal L., Tewari R. Antimicrobial susceptibility pattern of Burkholderia cepacia complex & Stenotrophomonas maltophilia over six years (2007-2012) Indian J Med Res. 2015;142(4):492. doi: 10.4103/0971-5916.169225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gautam V., Ray P., Vandamme P., Chatterjee S., Das A., Sharma K. Identification of lysine positive non-fermenting gram negative bacilli (Stenotrophomonas maltophilia and Burkholderia cepacia complex) Indian J Med Microbiol. 2009;27(2):128. doi: 10.4103/0255-0857.49425. [DOI] [PubMed] [Google Scholar]
- 34.Diogo A., Veríssimo A., Nobre M.F., da Costa M.S.J. Vol. 37. 1999. Usefulness of Fatty Acid Composition for Differentiation of Legionella Species; pp. 2248–2254. (7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lagesen K., Hallin P., Rødland E.A., Stærfeldt H.-H., Rognes T., DWJNar Ussery. Vol. 35. 2007. RNAmmer: Consistent and Rapid Annotation of Ribosomal RNA Genes; pp. 3100–3108. (9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 37.Darling A.C., Mau B., Blattner F.R., Perna N.T. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14(7):1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goris J., Konstantinidis K.T., Klappenbach J.A., Coenye T., Vandamme P., Tiedje J.M. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57(1):81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 39.Lee I., Kim Y.O., Park S.-C., Chun J. OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol. 2016;66(2):1100–1103. doi: 10.1099/ijsem.0.000760. [DOI] [PubMed] [Google Scholar]
- 40.Meier-Kolthoff J.P., Auch A.F., Klenk H.-P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 2013;14(1):1–14. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gröschel M.I., Meehan C.J., Barilar I., Diricks M., Gonzaga A., Steglich M. The phylogenetic landscape and nosocomial spread of the multidrug-resistant opportunist Stenotrophomonas maltophilia. Nature Commun. 2020;11(1):1–12. doi: 10.1038/s41467-020-15123-0. [DOI] [PMC free article] [PubMed] [Google Scholar]